Abstract
Liver regeneration is a hyperplastic phenomenon induced by partial hepatectomy (PH) or hepatic damage. A large number of genes have been indicated to be involved in the process of liver regeneration. It was recently reported that natural antisense transcripts are involved in the regulation of gene expression. However, no antisense transcript expressions in liver regeneration have been reported thus far. Therefore, the present study aimed to comprehensively identify up- or downregulated sense and antisense transcripts in liver regeneration using PH mice and a sense/antisense custom-microarray. The results showed that 97 genes were upregulated and 7 genes were downregulated for sense transcripts, whereas 15 genes were upregulated and 2 genes were downregulated for antisense transcripts in regenerating livers as compared to normal livers (P<0.05 and fold change >2.0). Sense and antisense transcripts of the genes, Apoa4, Hp, Fgb and Fgg, exhibited concordant upregulation during the course of liver regeneration. Apoa4, Hp and Fgb transcripts were further investigated by strand-specific reverse transcription-quantitative polymerase chain reaction (RT-qPCR), revealing results consistent with those of the microarray. In conclusion, the up- or downregulated sense and antisense transcripts identified in the present study are suggested to be involved in liver regeneration.
Keywords: 70%, partial hepatectomy, liver regeneration, microarray, sense/antisense transcripts, strand-specific reverse transcription-quantitative polymerase chain reaction
Introduction
Liver regeneration is a hyperplastic phenomenon of residual livers in response to partial hepatectomy (PH) or liver damage. Liver regeneration has been an advantage in liver cancer surgery and transplantation from a medical viewpoint (1). Therefore, it is important to elucidate the mechanisms underlying liver regeneration for the development of regenerative medicine.
Rodent PH models have been a useful tool in the investigation of signals that regulate this regenerative response (2,3). Growth factors, such as hepatocyte growth factor (HGF), epidermal growth factor (EGF) or insulin growth factor (IGF) and cytokines, such as IL-6 or TNF-α, have been shown to play important roles in the early phase of liver regeneration by using rodent PH models (4–6). HGF is synthesized by non-parenchymal (particularly stellate) cells in the liver and binds to the c-Met receptor of hepatocytes. HGF activates a variety of downstream pathways, including the mitogen-activated protein kinase (MAPK) signaling pathway which is involved in cell growth and enhances DNA synthesis of hepatocytes during liver regeneration (4–6). TNF-α, as an initiation factor of liver regeneration, stimulates Kupffer cells in the liver. Subsequently, IL-6 is produced in Kupffer cells by the activation of NF-κB and stimulates janus kinase (JAK)/signal transduction and activator of transcription (STAT) by binding to its receptor (IL-6R) in hepatocytes, resulting in hepatocyte proliferation (4–6). However, studies on TNF-α and IL-6 knockout mice have demonstrated that these factors are not key to liver regeneration (7,8). Thus, multiple pathways are currently considered to be involved in the initiation of liver regeneration.
Recently, microarray analyses were performed to identify novel genes involved in liver regeneration. Togo et al reported 23 genes, including Karyopherin α1 and interleukin-1 receptor associated kinase-1 (IRAK-1), as novel genes involved in the early phase of liver regeneration by microarray analysis (9). Furthermore, we previously demonstrated that Rab30 and S100a8/S100a9 genes were upregulated >20-fold at 2 and 6 h post 70% PH in mice (10).
Antisense transcripts, which were transcribed from the DNA strand opposite the sense strand, have been identified by cDNA sequencing projects in human and mouse (11,12). Using cDNA sequence database, Kiyosawa et al predicted that sense and antisense transcripts were produced from 15% of mouse gene loci (13) and demonstrated via microarray analysis that 1,947 sense and antisense transcripts were expressed in mice (14). Tsix, the antisense transcript of X-inactive specific transcript (Xist) gene, was demonstrated to be involved in the regulation of the Xist gene which mediates X chromosome inactivation (15). Furthermore, β-site APP cleaving enzyme 1 (BACE1) antisense transcript was shown to be involved in the stability of BACE1 sense transcript via RNA duplex formation with BACE1 sense transcripts (16). In a previous study, we identified antisense transcripts as up- and downregulated in human colorectal cancer tissues as compared to normal colonic tissues (17). These studies suggested that antisense transcripts were expressed in various tissues and may be involved in the regulation of several biological activities.
In the present study, in order to identify the sense and antisense transcripts possibly involved in liver regeneration, the up- or downregulated sense and antisense transcripts were investigated using 70% PH mice and custom-microarray containing mouse sense/antisense probe for ∼21,000 genes.
Materials and methods
Animals
Eight-week-old specific pathogen-free (SPF) male BALB/c mice were purchased from Clea Japan Inc. (Tokyo, Japan). Mice were maintained in a temperature-controlled room on a 12-h light-dark cycle, with free access to water and standard chow. Seventy percent PH was performed on anesthetized mice, according to the procedure described by Higgins and Anderson (2). Mice were sacrificed at five time points: 0, 2, 6, 12 and 24 h post PH (n=3 at each time point). All animal experiments in the present study were performed according to the Guidelines of the University of Tsukuba for the Care of Laboratory Animals and the Regulation for Animal Experiments.
Tissue samples
For microarray analysis, the hepatectomized 8-week-old BALB/c mice were sacrificed at the time points indicated above and the residual livers of mice were collected, immediately frozen in liquid nitrogen and stored at −80°C until use. For histological analysis, the residual livers at each time point were fixed with 4% (w/v in phosphate-buffered saline; PBS) ice-cold paraformaldehyde solution overnight. The fixed livers were washed in PBS, dehydrated in ethanol and embedded in paraffin blocks.
Hematoxylin and eosin staining
The livers were sliced into 4-μm sections. The sections were placed on glass slides and deparaffinized in xylene, followed by treatment with ethanol. Following immersion in water, the sections were treated with hematoxylin solution for 1 min, followed by washing with warm water for 15 min. The sections were then treated with eosin solution for 10 sec, followed by washing with water. The stained sections were dehydrated with ethanol followed by xylene. The resulting sections were photographed using the MIRAX Desk scanner (CarlZeiss, Oberkochen, Germany).
RNA extraction and quality check
Total RNAs from frozen livers were isolated using the ISOGEN kit (Nippon Gene, Tokyo, Japan), according to the manufacturer’s instructions. The quality and concentration of total RNAs were assessed with a NanoDrop Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA), according to the manufacturer’s instructions. Total RNA samples exhibited 260/280-nm absorbance ratios of 1.8–2.0. The integrity of the RNAs was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and the RNA 6000 Nano LabChip kit (Agilent Technologies), revealing RNA integrity numbers (RIN) of the samples to be >8.0; these RNA samples were considered suitable for microarray analysis.
Sense and antisense microarray
Cyanine 3 (Cy3)-labeled cDNA samples were synthesized from 10 μg total RNAs of regenerating or normal livers, using the LabelStar Array kit (Qiagen, Valencia, CA, USA), Cy3-dCTP (GE Healthcare, Buckinghamshire, UK) and random nonamer primers. The custom-designed mouse sense and antisense 60-nt sequences were arranged in an Agilent 44K × 4 system (Agilent eArray Design ID=021137; Tsukuba GeneTech Laboratories, Ibaraki, Japan). Agilent 44K × 4 mouse sense and antisense custom-microarray slides were hybridized with Cy3-labeled cDNAs (2 μg) in hybridization solution prepared with Gene Expression Hybridization kit (Agilent Technologies), following the manufacturer’s instructions. Cy3 fluorescence signal images on the slides were obtained by DNA microarray scanner (Agilent Technologies) and processed using Feature Extraction software (Agilent Technologies), according to the manufacturer’s instructions.
The expression data obtained were processed using GeneSpring GX12 software (Agilent Technologies) to perform a log transformation and normalization to the 75th percentile of all values on respective microarrays, followed by normalization of the median expression level of all samples. Additionally, the normalized data of gene expression were filtered on flags and genes classified as either flag-[Detected] or flag-[Not Detected] were allowed to pass the filter. The expression profiles at each time point, i.e., 2, 6, 12 and 24 h post PH against 0 h were compared using GeneSpring GX12 software. To identify genes with statistically significant differences at each time point, a Kruskal-Wallis test with a P-value cutoff of 0.05 was performed. The up- and downregulated sense/antisense transcripts were selected by fold changes (>2.0) at each time point against 0 h and by signal intensities (>100.0).
Strand-specific reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
To examine the expression of sense and antisense transcripts obtained by the microarray analysis, strand-specific RT-qPCR was performed in selected genes. The synthesis of strand-specific cDNAs derived from sense or antisense transcripts was performed using reverse or forward primers as previously described (17). In order to normalize the values of sense and antisense transcripts among samples, EGFP (enhanced green fluorescent protein) RNA was mixed with total RNAs prior to strand-specific RT-qPCR (17). These strand-specific cDNAs were used as a template for qPCR. The qPCRs were performed using Power SYBR-Green PCR Master Mix (Life Technologies, Carlsbad, CA, USA), primer pair sets and StepOnePlus Real-Time PCR system (Life Technologies) under the conditions of 10 min at 95°C followed by 40 cycles at 95°C for 15 sec and at 60°C for 60 sec. The values of sense or antisense transcripts were normalized based on the values of the control.
Results and Discussion
In order to confirm the histological characteristics of liver regeneration, hematoxylin and eosin staining was performed on the sections of regenerating livers as described above. As shown in Fig. 1, hepatocellular morphological changes were insignificant at 2 h post PH; however, fat droplets appeared in hepatocytes at 6 h post PH. Hepatocellular expansions were recognized at 12 h post PH and hepatocellular proliferation images were observed at 24 h post PH. These observations were consistent with those previously reported regarding liver regeneration (18).
Figure 1.
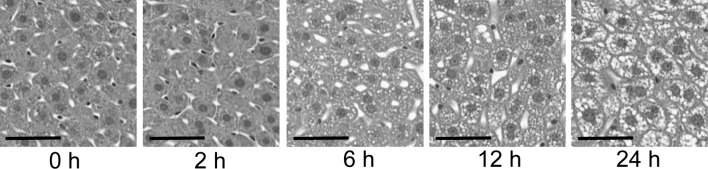
Histological appearance of regenerating livers at each time point post PH. Mice were sacrificed at five time points: 0, 2, 6, 12 and 24 h post PH. Regenerating livers were embedded in paraffin blocks. In order to visualize the fine structures of regenerating livers, hematoxylin and eosin staining was performed on the 4-μm sections. Scale bar, 50 mm.
Subsequently, using the liver samples as described above, up- or downregulation of sense and antisense transcripts in the liver regeneration process was investigated. The liver samples were collected from three animals at each time point, i.e., 0, 2, 6, 12 and 24 h post PH, and subjected to expression analysis using the custom-microarray. When the expression profiles of sense and antisense transcripts in the liver samples were assessed with the Kruskal-Wallis test using GeneSpring GX12 software, 104 sense and 17 antisense transcripts were observed to be differentially expressed with a magnitude of >2.0-fold (P<0.05) during the course of liver regeneration. Among the 104 sense transcripts, 97 were upregulated and 7 were down-regulated (Table I). The upregulation of Igfbp1, Btg2, Socs3, Gadd45g, S3-12 (Plin4), Apoa4 and Hp transcripts in the 104 sense transcripts has already been documented in liver regeneration (9,18–21); however, to the best of our knowledge, the upregulation of the remaining transcripts was first described in the present study. As regards the 17 antisense transcripts, 15 and 2 transcripts were up- and downregulated, respectively, as described for the first time in the present study (Table II).
Table I.
Upregulated 97 (20 of the 97 indicated) and downregulated 7 sense transcripts in the early phase of liver regeneration.
| Fold change (vs. 0 h)
|
||||||
|---|---|---|---|---|---|---|
| Accession no. | 2 h | 6 h | 12 h | 24 h | Gene name | Regulation |
| NM_008341 | 64.94 | 52.31 | 21.22 | 7.20 | Mus musculus insulin-like growth factor binding protein 1 (Igfbp1) | Up |
| NM_007570 | 26.45 | 28.70 | 9.16 | 4.47 | Mus musculus B-cell translocation gene 2, anti-proliferative (Btg2) | Up |
| NM_007707 | 18.36 | 8.78 | 5.21 | 2.61 | Mus musculus suppressor of cytokine signaling 3 (Socs3) | Up |
| NM_021897 | 15.99 | 4.10 | 2.73 | 2.31 | Mus musculus transformation-related protein 53-inducible nuclear protein 1 (Trp53inp1) | Up |
| NM_054098 | 14.80 | 24.71 | 11.68 | 2.74 | Mus musculus STEAP family member 4 (Steap4) | Up |
| NM_011817 | 14.50 | 14.67 | 5.65 | 4.54 | Mus musculus growth arrest and DNA-damage-inducible 45 γ (Gadd45g) | Up |
| NM_011803 | 14.05 | 25.39 | 11.39 | 10.75 | Mus musculus Kruppel-like factor 6 (Klf6) | Up |
| NM_025459 | 11.44 | 14.01 | 3.45 | 2.89 | Mus musculus family with sequence similarity 134, member B (Fam134b) | Up |
| NM_022424 | 10.50 | 7.67 | 4.41 | 2.18 | Mus musculus fibronectin type III domain-containing 4 (Fndc4) | Up |
| NM_144792 | 10.47 | 22.33 | 4.35 | 3.52 | Mus musculus sphingomyelin synthase 1 (Sgms1) | Up |
| NM_019653 | 10.11 | 5.73 | 2.64 | 2.90 | Mus musculus WD repeat and SOCS box-containing 1 (Wsb1) | Up |
| NM_011809 | 10.03 | 9.19 | 5.61 | 3.61 | Mus musculus E26 avian leukemia oncogene 2, 3′ domain (Ets2) | Up |
| NM_016896 | 9.88 | 12.72 | 6.28 | 3.14 | Mus musculus mitogen-activated protein kinase kinase kinase 14 (Map3k14) | Up |
| NM_029844 | 9.51 | 19.37 | 11.82 | 4.59 | Mus musculus melanocortin 2 receptor accessory protein (Mrap) | Up |
| NM_008362 | 8.36 | 15.05 | 7.75 | 2.98 | Mus musculus interleukin 1 receptor, type I (Il1r1) | Up |
| NM_007468 | 7.64 | 9.95 | 13.76 | 15.49 | Mus musculus apolipoprotein A-IV (Apoa4) | Up |
| NM_017370 | 3.20 | 4.85 | 6.42 | 3.82 | Mus musculus haptoglobin (Hp) | Up |
| NM_133862 | 2.59 | 3.92 | 4.77 | 2.43 | Mus musculus fibrinogen γ chain (Fgg) | Up |
| NM_181849 | 2.43 | 4.01 | 5.33 | 2.50 | Mus musculus fibrinogen β chain (Fgb) | Up |
| NM_020568 | 6.54 | 6.55 | 3.71 | 2.13 | Mus musculus perilipin 4, S3-12 (Plin4) | Up |
| NM_145434 | −7.07 | −10.06 | −21.76 | −5.68 | Mus musculus nuclear receptor subfamily 1, group D, member 1 (Nr1d1) | Down |
| NM_021704 | −6.92 | −8.73 | −3.66 | −3.45 | Mus musculus chemokine (C-X-C motif) ligand 12 (Cxcl12) | Down |
| NM_011535 | −2.83 | −5.04 | −3.08 | −2.64 | Mus musculus T-box 3 (Tbx3) | Down |
| NM_010570 | −2.60 | −9.03 | −3.71 | −2.02 | Mus musculus insulin receptor substrate 1 (Irs1) | Down |
| NM_016974 | −2.26 | −24.91 | −61.47 | −18.86 | Mus musculus D site albumin promoter-binding protein (Dbp) | Down |
| NM_145152 | −2.16 | −7.81 | −5.11 | −2.08 | Mus musculus leucine-rich repeat-containing 3 (Lrrc3) | Down |
| NM_153584 | −2.14 | −12.74 | −6.50 | −2.79 | Mus musculus cDNA sequence BC031353 (BC031353) | Down |
Up, upregulation; Down, downregulation.
Table II.
Fifteen upregulated and two downregulated antisense transcripts in the early phase of liver regeneration.
| Fold change (vs. 0 h)
|
||||||
|---|---|---|---|---|---|---|
| Accession no. | 2 h | 6 h | 12 h | 24 h | Gene name | Regulation |
| NM_026300 | 31.12 | 52.99 | 68.18 | 16.48 | Mus musculus RIKEN cDNA 4930549C01 gene (4930549C01Rik) | Up |
| NM_021478 | 10.82 | 13.23 | 6.03 | 2.95 | Mus musculus tubby-like protein 1 (Tulp1) | Up |
| NM_133862 | 6.44 | 27.91 | 24.97 | 11.12 | Mus musculus fibrinogen γ chain (Fgg) | Up |
| NM_017370 | 5.28 | 14.96 | 17.19 | 12.40 | Mus musculus haptoglobin (Hp) | Up |
| NM_175439 | 5.26 | 6.56 | 3.22 | 2.01 | Mus musculus methionine-tRNA synthetase 2 (mitochondrial) (Mars2) | Up |
| NM_181849 | 4.38 | 11.18 | 12.71 | 5.99 | Mus musculus fibrinogen β chain (Fgb) | Up |
| NM_009984 | 4.28 | 8.65 | 5.26 | 3.96 | Mus musculus cathepsin L (Ctsl) | Up |
| NM_007468 | 4.19 | 5.84 | 10.06 | 17.23 | Mus musculus apolipoprotein A-IV (Apoa4) | Up |
| NM_177867 | 3.76 | 7.30 | 9.35 | 2.69 | Mus musculus spermatogenesis associated 21 (Spata21) | Up |
| NM_133653 | 3.65 | 12.96 | 8.90 | 2.52 | Mus musculus methionine adenosyltransferase I, α (Mat1a) | Up |
| NM_010196 | 3.48 | 12.21 | 14.08 | 6.33 | Mus musculus fibrinogen α chain (Fga) | Up |
| NM_181325 | 2.60 | 4.92 | 3.28 | 3.63 | Mus musculus solute carrier family 25 (mitochondrial carrier ornithine transporter), member 15 (Slc25a15) | Up |
| NM_018746 | 2.45 | 10.18 | 10.67 | 3.77 | Mus musculus inter α-trypsin inhibitor, heavy chain 4 (Itih4) | Up |
| NM_008716 | 2.09 | 9.08 | 35.23 | 6.45 | Mus musculus Notch gene homolog 3 (Drosophila) (Notch3) | Up |
| NM_008407 | 2.04 | 4.04 | 5.36 | 2.99 | Mus musculus inter-α trypsin inhibitor, heavy chain 3 (Itih3) | Up |
| NM_010107 | −2.14 | −2.04 | −3.23 | −4.63 | Mus musculus ephrin A1 (Efna1) | Down |
| NM_001033537 | −2.07 | −2.08 | −4.53 | −5.25 | Mus musculus post-GPI attachment to proteins 3 (Pgap3) | Down |
Up, upregulation; Down, downregulation.
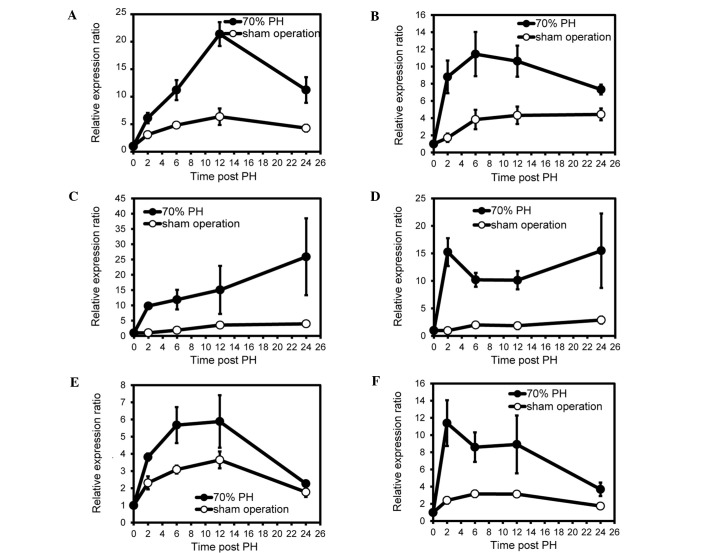
When we investigated whether the same genes were observed in the sense and antisense transcripts, four genes (Apoa4, Hp, Fgb and Fgg) exhibited upregulation in the sense and antisense transcripts during liver regeneration (Fig. 2, Tables I and II). Apolipoprotein A-IV (APOA4) is a component of chylomicrons and high-density lipoprotein (http://omim.org/entry/107690). Haptoglobin (HP), combines with free plasma hemoglobin, preventing loss of iron through the kidneys (http://omimorg/entry/140100). FGB and FGG are members of the fibrinogen family and function as factor I of blood coagulation for hemostasis (http://omim.org/entry/134830; http://omim.org/entry/134850). The expression of these genes was indicated to be under complex control in liver regeneration. To verify that these expression levels were the results of liver regeneration and were not due to the skin incision, we attempted to design strand-specific primers in the sequences used for the microarray probe and succeeded in synthesis of primer pairs of the Apoa4, Hp and Fgb transcripts (Table III). Subsequently, the amounts of the Apoa4, Hp and Fgb transcripts were measured in regenerating liver samples using strand-specific RT-qPCR and compared to liver samples of sham-operated mice. As shown in Fig. 3, the strand-specific RT-qPCR demonstrated that the expression patterns of the genes were essentially identical to those described above.
Figure 2.
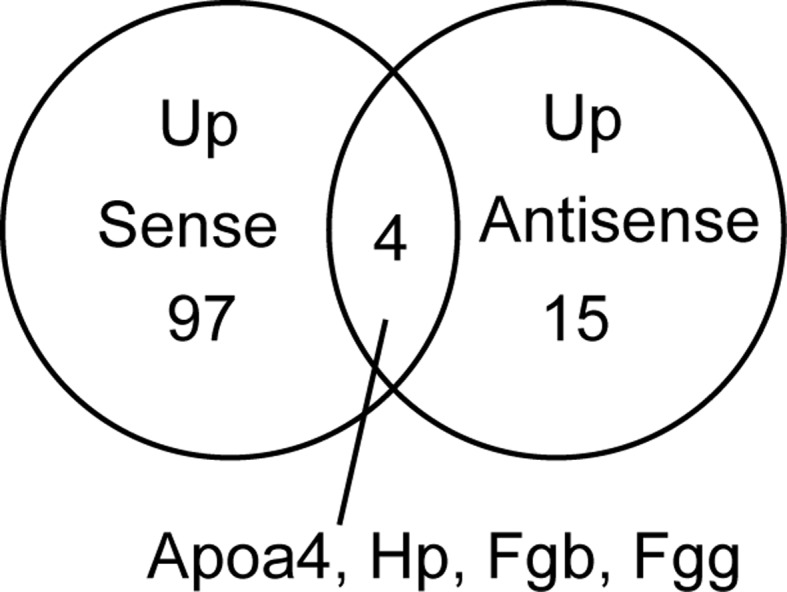
Venn diagram of upregulated (Up) sense and antisense transcripts in liver regeneration based on microarray analysis.
Table III.
Primer sequences for detection of sense/antisense transcripts.
| Primer name | Sequences | Size (mer) | PCR products (bp) |
|---|---|---|---|
| Apoa4 forward | 5′-ATGTGGACAACCTGCACAC-3′ | 19 | 60 |
| Apoa4 reverse | 5′-CCACCAACTTAAAGGACAAGTTTA-3′ | 24 | |
| Hp forward | 5′-GGGTGTACACCTTAAACGAC-3′ | 20 | 60 |
| Hp reverse | 5′-TCGCTGGAGAGAAACTCC-3′ | 18 | |
| Fgb forward | 5′-TTCTCATTGAAATGGAGGACTGGA-3′ | 24 | 60 |
| Fgb reverse | 5′-TGAAGCCTCCATAATGTGCCT-3′ | 21 |
PCR, polymerase chain reaction.
Figure 3.
Relative expression ratio of Apoa4, Hp and Fgb sense and antisense transcripts in the early phase of live regeneration. Total RNAs were isolated from regenerating and sham-operated livers (n=3 at each time point). Strand-specific cDNA derived from sense or antisense transcripts was synthesized using the reverse or forward primers described in Table III. qPCRs were performed using these primer pairs, Power SYBR-Green PCR Master Mix and StepOnePlus Real-Time PCR system. Means ± standard error of the mean (SEM) were obtained using the standard-curve method and calculated taking the values of strand-specific RT-qPCR at 0 h post PH (normal livers) as 1.0. (A) Apoa4 sense transcripts. (B) Apoa4 antisense transcripts. (C) Hp sense transcripts. (D) Hp antisense transcripts. (E) Fgb sense transcripts. (F) Fgb antisense transcripts.
In previous studies, two types of regulation were reported in sense and antisense transcripts of a single gene: one type was associated with a positive correlation between the regulation of sense and antisense transcripts and the other type with a negative correlation. An example of the former correlation was reported between breast tumor and normal cells (22) and an example of the latter correlation between leukemia and normal cells (23). These findings indicated that an antisense transcript has different control mechanisms in gene expression regulation. The Apoa4, Hp, Fgb and Fgg sense and antisense transcripts in the liver regeneration were positively correlated, as in the case of the breast tumor and normal cells. We were not able to determine the function of antisense transcripts, since it has not yet been elucidated. To elucidate the function of the transcripts in liver regeneration, protein synthesis from the sense transcripts should be measured. In the future, the up- and downregulation of transcripts other than Apoa4, Hp, Fgb and Fgg should be verified as being the results of liver regeneration and not those of the skin incision, using liver samples of sham-operated mice.
In conclusion, 104 sense and 17 antisense transcripts were indicated to be liver regeneration-responsive transcripts. Of these transcripts, the upregulation of Apoa4, Hp, Fgb and Fgg sense and antisense transcripts was confirmed during liver regeneration using sham-operated mice. Loss-of-function analyses of sense and antisense transcripts may provide additional information to further elucidate the function of antisense transcripts in liver regeneration.
References
- 1.Clavien PA, Petrowsky H, DeOliveira ML, Graf R. Strategies for safer liver surgery and partial liver transplantation. N Engl J Med. 2007;356:1545–1559. doi: 10.1056/NEJMra065156. [DOI] [PubMed] [Google Scholar]
- 2.Higgins GM, Anderson RM. Experimental pathology of the liver. Arch Pathol. 1931;12:186–202. [Google Scholar]
- 3.Martins PN, Theruvath TP, Neuhaus P. Rodent models of partial hepatectomies. Liver Int. 2008;28:3–11. doi: 10.1111/j.1478-3231.2007.01628.x. [DOI] [PubMed] [Google Scholar]
- 4.Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology. 2006;43:S45–S53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- 5.Michalopoulos GK. Liver regeneration. J Cell Physiol. 2007;213:286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taub R. Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Biol. 2004;5:836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- 7.Cressman DE, Greenbaum LE, DeAngelis RA, et al. Liver failure and defective hepatocyte regeneration in interleukin-6-deficient mice. Science. 1996;274:1379–1383. doi: 10.1126/science.274.5291.1379. [DOI] [PubMed] [Google Scholar]
- 8.Yamada Y, Kirillova I, Peschon JJ, Fausto N. Initiation of liver growth by tumor necrosis factor: deficient liver regeneration in mice lacking type I tumor necrosis factor receptor. Proc Natl Acad Sci USA. 1997;94:1441–1446. doi: 10.1073/pnas.94.4.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Togo S, Makino H, Kobayashi T, et al. Mechanism of liver regeneration after partial hepatectomy using mouse cDNA microarray. J Hepatol. 2004;40:464–471. doi: 10.1016/j.jhep.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Chiba M, Murata S, Myronovych A, et al. Elevation and characteristics of Rab30and S100a8/S100a9expression in an early phase of liver regeneration in the mouse. Int J Mol Med. 2011;27:567–574. doi: 10.3892/ijmm.2011.614. [DOI] [PubMed] [Google Scholar]
- 11.Katayama S, Tomaru Y, Kasukawa T, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 12.Røsok Ø, Sioud M. Systematic identification of sense-antisense transcripts in mammalian cells. Nat Biotechnol. 2004;22:104–108. doi: 10.1038/nbt925. [DOI] [PubMed] [Google Scholar]
- 13.Kiyosawa H, Yamanaka I, Osato N, Kondo S, Hayashizaki Y. Antisense transcripts with FANTOM2 clone set and their implications for gene regulation. Genome Res. 2003;13:1324–1334. doi: 10.1101/gr.982903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiyosawa H, Mise N, Iwase S, Hayashizaki Y, Abe K. Disclosing hidden transcripts: mouse natural sense-antisense transcripts tend to be poly(A) negative and nuclear localized. Genome Res. 2005;15:463–474. doi: 10.1101/gr.3155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 16.Faghihi MA, Modarresi F, Khalil AM, et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohno K, Chiba M, Murata S, et al. Identification of natural antisense transcripts involved in human colorectal cancer development. Int J Oncol. 2010;37:1425–1432. doi: 10.3892/ijo_00000794. [DOI] [PubMed] [Google Scholar]
- 18.Shteyer E, Liao Y, Muglia LJ, Hruz PW, Rudnick DA. Disruption of hepatic adipogenesis is associated with impaired liver regeneration in mice. Hepatology. 2004;40:1322–1332. doi: 10.1002/hep.20462. [DOI] [PubMed] [Google Scholar]
- 19.White P, Brestelli JE, Kaestner KH, Greenbaum LE. Identification of transcriptional networks during liver regeneration. J Biol Chem. 2005;280:3715–3722. doi: 10.1074/jbc.M410844200. [DOI] [PubMed] [Google Scholar]
- 20.Jensen SA. Liver gene regulation in rats following both 70 or 90% hepatectomy and endotoxin treatment. J Gastroenterol Hepatol. 2001;16:525–530. doi: 10.1046/j.1440-1746.2001.02475.x. [DOI] [PubMed] [Google Scholar]
- 21.Riehle KJ, Campbell JS, McMahan RS, et al. Regulation of liver regeneration and hepatocarcinogenesis by suppressor of cytokine signaling 3. J Exp Med. 2008;205:91–103. doi: 10.1084/jem.20070820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grigoriadis A, Oliver GR, Tanney A, et al. Identification of differentially expressed sense and antisense transcript pairs in breast epithelial tissues. BMC Genomics. 2009;10:324. doi: 10.1186/1471-2164-10-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu W, Gius D, Onyango P, et al. Epigenetic silencing of tumor suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]