Abstract
A unique mRNA produced in leukemic cells from a t(15;17) acute promyelocytic leukemia (APL) patient encodes a fusion protein between the retinoic acid receptor α (RARα) and a myeloid gene product called PML. Studies have reported that neutrophil elastase (NE) cleaves bcr-1-derived PML-RARα in early myeloid cells, leaving only the nuclear localization signal (NLS) of PML attached to RARα. The resultant NLS-RARα fusion protein mainly localizes to, and functions within, the cell nucleus. It is speculated that NLS-RARα may act in different ways from the wild-type RARα, but its biological characteristics have not been reported. This study takes two approaches. Firstly, the NLS-RARα was silenced with pNLS-RARα-shRNA. The mRNA and protein expression of NLS-RARα were detected by RT-PCR and Western blot respectively. Cell proliferation in vitro was assessed by MTT assay. Flow cytometry (FCM) was used to detect the differentiation of cells. Secondly, the NLS-RARα was over-expressed by preparation of recombinant adenovirus HL-60/pAd-NLS-RARα. The assays of mRNA and protein expression of NLS-RARα, and cell proliferation, were as above. By contrast, cell differentiation was stimulated by all trans retinoic acid (ATRA) (2.5µmol/L) at 24h after virus infection of pAd-NLS-RARα, and then detected by CD11b labeling two days later. The transcription and translation of C-MYC was detected in HL-60/pAd-NLS-RARα cells which treated by ATRA. Our results showed that compared to the control groups, the expression of NLS-RARα was significantly reduced in the HL-60/pNLS-RARα-shRNA cells, and increased dramatically in the HL-60/pAd-NLS-RARα cells. The proliferation was remarkably inhibited in the HL-60/pNLS-RARα-shRNA cells in a time-dependent manner, but markedly promoted in the HL-60/pAd-NLS-RARα cells. FCM outcome revealed the differentiation increased in HL-60/pNLS-RARα-shRNA cells, and decreased in the HL-60/pAd-NLS-RARα cells treated with 2.5µmol/L ATRA. The expression of C-MYC increased strikingly in HL-60/pAd-NLS-RARα cells treated with 2.5µmol/L ATRA. Down-regulation of NLS-RARα expression inhibited the proliferation and induced the differentiation of HL-60 cells. On the contrary, over-expression of NLS-RARα promoted proliferation and reduced the ATRA-induced differentiation of HL-60 cells.
Keywords: NLS-RARα, shRNA, over-expression, HL-60 cell, proliferation, differentiation
Introduction
The retinoic acid receptor-alpha (RARA) gene is located on chromosome 17q21, in proximity to the breakpoint of APL1, and is thought to play a critical role in the pathogenesis of APL. The product of the RARA gene, the RARα protein, is a member of the nuclear steroid/thyroid hormone receptor superfamily 2, 3, 4. It modulates the expression of target genes by acting as a ligand-dependent transcription factor, which is stimulated by ATRA 5, 6, 7. The amino acid sequence of RARα has been divided into six regions (A-F) based on different degrees of conservation, with regions C and E being highly conserved, binding to target DNA and the ligand separately 8. Promyelocytic leukemia (PML) which contains nuclear localization signal (NLS), B-Boxes and α-helical coiled-coil region, is encoded by PML gene mapped on chromosome 15q22 in humans. 9 In 1991, it was discovered that the consistent chromosomal translocation of APL, t(15:17), fused the retinoic acid receptor a (RARa) gene to the promyelocytic leukemia (PML) gene on chromosome 1510, yielding the fusion protein PML-RARα 11, which represents the etiologic agent of APL. This translocation is detected in as many as 90% of APL patients and has become the definitive marker for the disease 12. The aberrant PML-RARα fusion product plays a vital role in APL 13. Some studies have demonstrated that the transgenic and knock-in animals expressing PML-RARα in early myeloid cells 14, 15, 16 developed acute promyelocytic leukemia (APL), but APL was absent when PML-RARα was expressed in late myeloid cells 17. However, the mechanisms by which PML-RARα predisposes early myeloid cells to eventual leukemic transformation are not yet completely clear. Recently, some studies declared that neutrophil elastase (NE), an early myeloid-specific serine protease, is important for the development of APL in mice. Furthermore, NE can cleave bcr-1 derived PML-RARα protein in early myeloid cells 18, resulting in removal of NLS from PML, and attaching to RARα]. The resultant fusion of RARα with NLS was named NLS-RARα. The NLS-RARα gene is about 1506 bp in length and encodes a protein of 61 kD. These data suggested that the disruption of RARα protein may be the major cause of APL. So far, few studies have focused on the biological functions of NLS-RARα which probably has remarkable value for the genesis and development of APL. At present, gene therapy is developing rapidly, and every gene which plays a significant role and has significant values in APL can be therapeutic target gene. So, we contribute ourselves to study the gene function of NLS-RARα gene, in order to conform that it has potential therapeutic value. This work is mainly aimed to the biological function of NLS-RARα gene.
In order to explore the biological characteristics of NLS-RARα gene, the NLS-RARα was alternately silenced with shRNA, or over-expressed by from an adenovirus vector. Differentiation of the acute premyeloid leukemia (APL) cell line HL-60 can be induced by all trans-retinoic acid (ATRA) 19. Thus, HL-60 cells were infected with adenovirus vector and treated with 2.5 μmol/L ATRA, then examined for cell differentiation.
Materials and methods
Vector maps
Cell line and culture
Human HL-60 cells were purchased from the Shanghai Institute for Biological Science and maintained in IMDM (Gibco, MD, USA) supplemented with 20% fetal bovine serum (FBS; Gibco, MD, USA) in an environment with 5% CO2 at 37 °C.
Human K562 cells were saved by our own laboratory and maintained in IMDM (Gibco, MD, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, MD, USA) in an environment with 5% CO2 at 37 °C.
AD293 cells were saved by our own laboratory and cultured in DMEM (Gibco, MD, USA) containing 10% fetal bovine serum (FBS; Gibco, MD, USA) in an environment with 5% CO2 at 37 °C.
Transient Transfection
HL-60 cells (2.5×105/ml) in logarithmic growth phase were seeded in 6-well plate. For transfection, 2.5 μg of plasmid pPML(NLS-)-shRNA was diluted in 500 μl Opti-MEM I, which was added to each well. Then, 2.5 μl of PLUS™ Reagent was directly added to dilute DNA. After gentle mixing and incubation for 15 min at room temperature, 13.75 μl of Lipofectamine™ LTX was added into above DNA solution, followed by incubation for 25 min at room temperature. The resultant solution was added to each well. After incubation at 37°C for 6 h, 1 ml of fresh complete medium was added. These transfected cells were named as HL-60/pNLS-RARα-shRNA. In the negative control group, HL-60/pshRNA-NC cells were processed similarly. At 48 h after transfection, the successfully transfected cells were verified by fluorescent microscopy. The transfection efficiency was expressed as the percentage of GFP-positive cells. There were three groups in this experiment: HL-60 cells group, HL-60/pshRNA-NC group and HL-60/pNLS-RARα-shRNA group.
Cell infection by adenovirus
HL-60 cells in logarithmic growth phase (5×105/well) were seeded in 6-well plate. These cells were infected with adenovirus vectors, pAd-KZ or the NLS-RARα-expressing recombinant adenovirus, pAd-NLS-RARα, and both of them express RFP (Red Fluorescent Protein). Meanwhile, polybrene was also added, as per manufacturer's instructions. After culture for 8-12 h, the medium was refreshed. The fluorescence was then detected 48h later. There were three groups in this experiment: HL-60 cells group, HL-60/pAd-KZ groups, and HL-60/pAd-NLS-RARα group.
RNA isolation and RT-PCR
Total RNA was extracted from cells in each group using Trizol reagent, as per manufacturer's instructions (Invitrogen, Carlsbad, California). The first-strand cDNA was synthesized from 1μg of total RNA using a Prime Script Kit (TAKARA, Dalian, China). NLS-RARα gene expression was quantified by reverse transcriptase polymerase chain reaction (RT-PCR) with cDNA Synthesis Kit (TAKARA, Dalian, China). β-actin was used as an endogenous control. All samples were run in duplicate for each experiment. Do 1% agarose gel lelctrophoresis as soon as we acquire the PCR product. And 5µl PCR product was used in each lane. Gene expression analysis was performed with the Quantity One Software (BIO-RAD, USA). The PCR conditions were: pre-denaturation at 95°C for 5 min, 35 cycles of denaturation at 95 °C for 30 s, annealing at 64 °C for 30 s, and extension at 72 °C for 100s, and a final extension at 72 °C for 5 min. The amplification of β-actin gene was the as for NLS-RARα. The mRNA expression levels of the target gene were normalized to those of β-actin. The specific primers for NLS-RARα were 5'- ATGTACGCCTTCTCCATCAAA -3' (forward) and 5'- TCACGGGGAGTGGGTG -3' (reverse). Those for β-actin were 5′-CACCACACCTTCTACAATGAGC-3′ (forward) and 5′-GTGATCTCCTTCTGCATCCTGT-3′ (reverse).
Western blot analysis
Cells in each group were washed with ice-cold phosphate-buffered saline and lysed in RIPA solution containing protease inhibitor cocktail. Protein concentration was determined with BCA method. A total of 100 μg of protein was added in 12% sodium dodecyl sulfate-polyacrylamide gel, and then transferred to polyvinylidene difluoride membrane. The membrane was blocked with 5% non-fat milk for 4 h, then incubated with primary antibody overnight at 4 °C (rabbit polyclonal antibody against RARα; 1:1000; Santa Cruz, SC-551, USA) and then with secondary antibody (goat anti-rabbit antibody, 1:1000, Zhongshan Goldenbridge Biotechnology Co. Ltd., Beijing, China [ conjugate]) for 1 h at 37 °C. After washing with Tris-Buffered Saline Tween-20 (TBST), the autoradiograms were scanned and subjected to densitometry. β-actin (mouse monoclonal antibody against β-actin, 1:500; Zhongshan Goldenbridge Biotechnology Co., Ltd., Beijing, China) was used as an internal control. Protein bands were visualized using the Quantity One Software (BIO-RAD, USA).
MTT proliferation assay
To assess cell viability, cells in each group were seeded in 96-well plates at a density of 1.0×104 cells/well. Cell proliferation was quantified at 4 days post-treatment by MTT assay. In brief, 20 μl of MTT (5 mg/ml; Sigma, MO, USA) was added to each well followed by incubation for 4 h at 37°C. The medium was then replaced with 150 μl of dimethylsulphoxide (DMSO, Sigma, MO, USA). The cell viability was assessed by detection of absorbance at 492 nm using a spectrophotometer. Cells growth curves were plotted. The experiment was repeated at least three times.
Flow cytometry
For detection of the cell differentiation antigen CD11b, 1 × 106 cells in each group were washed twice with PBS, incubated with PE-conjugated CD11b antibody or PE-conjugated IgG1isotype control antibody at 4°C for 30 min and analyzed by flow cytometry using a FACScan flow cytometer and Cell Quest software (Becton Dickinson, Mountain View, CA). The expression rate of CD11b positive cells was determined from 1 × 104 cells for each group.
To preferably evaluate the differentiation level of infection group, besides the treatment of polybrene and a variety of adenoviruses, 2.5 μmol/L ATRA was added into each well in each group to induce the differentiation of HL-60 cell of each group. After incubation for 48h, cells of each group were processed as described above.
Detection of expression of differentiation-related genes by RT-PCR and Western blot assay
The extraction of total RNA and reverse transcription into cDNA were the same as described above. The specific primers for the fusion gene were 5'-CCTACCCTCTCAACGACAGC-3' (forward) and 5'-CTCTGACCTTTTGCAAGGAG-3' (reverse). Those for β-actin were the same as the above. PCR condition was as follow: pre-denaturation at 95°C for 5 min; 35 cycles of denaturation at 95°C for 30 s, annealing at 56°C for 30 s and extension at 72°C for 45s; a final extension at 72°C for 5 min. The products were electrophoresed on 1.5% agarose gel and analyzed with Quantity One Software (BIO-RAD, USA).
Protein concentrations in the cell lysates were determined by BCA protein assay kit (Bi Yun Tian, Shang Hai). Equal amounts of proteins (50 μg) were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and blotted onto nitrocellulose sheets. The proteins were then probed with the mouse monoclonal antibody C-MYC (Zhongshan Goldenbridge Biotechnology Co., Ltd., Beijing, China) and goat anti-mouse secondary antibodies (Zhongshan Goldenbridge Biotechnology Co., Ltd., Beijing, China). Protein bands were visualized using the Quantity One Software (BIO-RAD, USA).
Statistical analysis
Data was expressed as mean ± standard deviation (SD). Statistical analysis was performed with SPSS version 16.0. Independent sample t-test was employed for comparing the means between two groups. A value of P<0.05 was considered statistically significant.
Results
Fluorescent protein expression in HL-60 cells
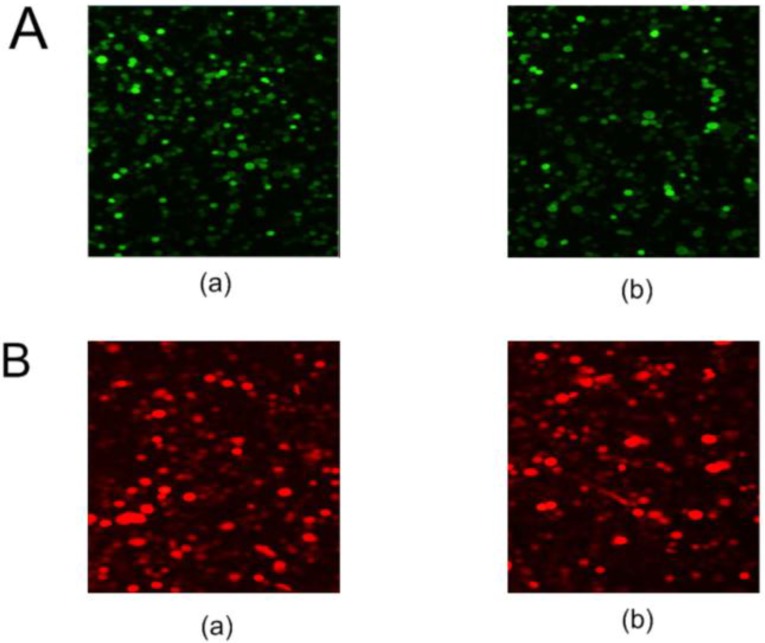
The pNLS-RARα-shRNA was introduced in HL-60 cells to silence NLS-RARα expression. The GFP-positive cells were 84% at 48 h after transfection (Figure 1A).
Figure 1.
Fluorescent protein expression in HL-60 cells of each group. A, GFP protein expression in transfected cells was detected by fluorescence microscopy (×100). (a) HL-60/pshRNA-NC cells; (b) HL-60/pNLS-RARα-shRNA cells. B, RFP protein expression in infected cells was detected by fluorescence microscopy (×100). (a) HL-60/pAd-KZ cells; (b) HL-60/pAd-NLS-RARα cells.
Meanwhile, recombinant adenovirus pAd-NLS-RARα was used to infect HL-60 cells to induce NLS-RARα over-expression. The percentage of RFP-positive cells was about 70% at 48 h after infection (Figure 1B).
Down-regulation of NLS-RARα in HL-60/pNLS-RARα-shRNA cells
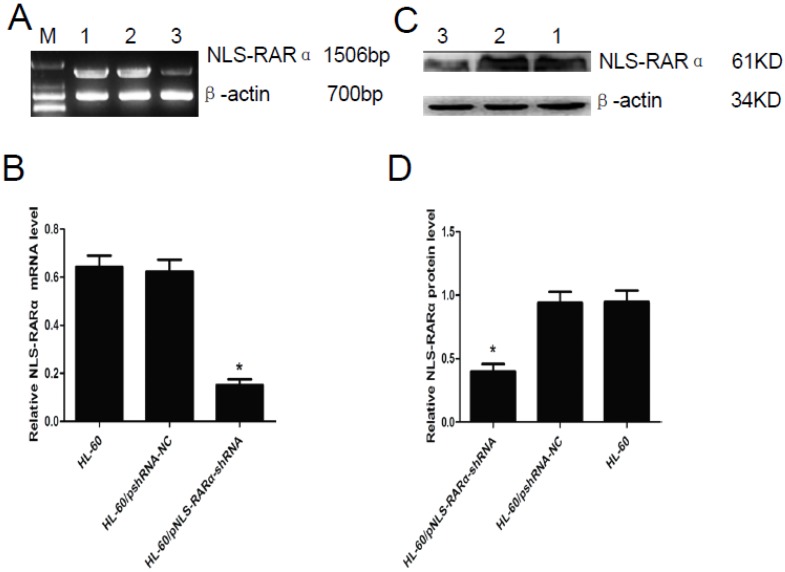
RT-PCR and Western blot assay showed that mRNA and protein expression of NLS-RARα were significantly lowered in the HL-60/pNLS-RARα-shRNA cells when compared with control group (P<0.05, Figure 2). No significant difference in the mRNA and protein expressions of NLS-RARα was found between HL-60 cells and HL-60/pshRNA-NC cells (P>0.05, Figure 2).
Figure 2.
Expression of NLS-RARα in transfected cells. A and B, mRNA expression of NLS-RARα was detected by RT-PCR. The mRNA expression of NLS-RARα in the HL-60/pNLS-RARα-shRNA cells was significantly decreased when compared with control group. Data was expressed as means ±SD. *P<0.05. C and D, Protein expression of NLS-RARα was assessed by Western blot assay. The protein expression of NLS-RARα in the HL-60/pNLS-RARα-shRNA cells was significantly decreased when compared with the controls. Data was expressed as means±SD. * P<0.05 (lane 1: HL-60 cells; lane 2: HL-60/pshRNA-NC cells; lane 3: HL-60/pNLS-RARα-shRNA cells).
Up-regulation of NLS-RARα in HL-60/pAd-NLS-RARα cells
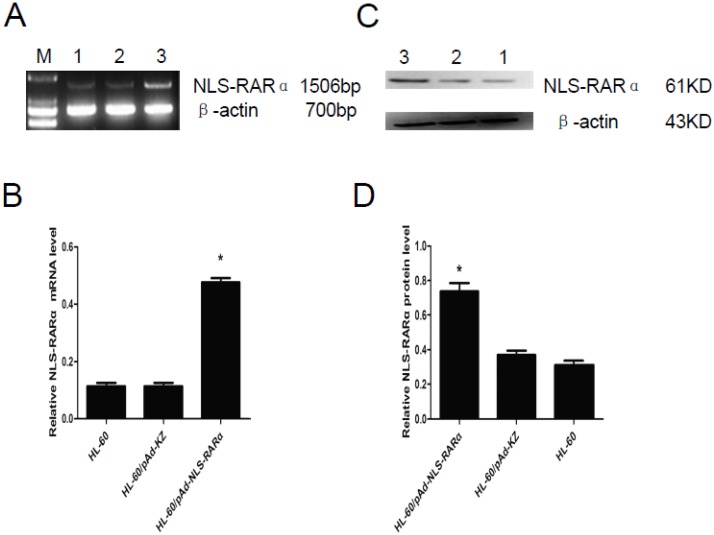
The mRNA and protein expressions of NLS-RARα were also detected in cells with NLS-RARα over-expression by RT-PCR and western blot assay. Results revealed that NLS-RARα expression in the HL-60/pAd-NLS-RARα cells was significantly higher than those in the control groups (P<0.05, Figure 3).
Figure 3.
Expression of NLS-RARα in infected cells. A and B, mRNA expression of NLS-RARα was detected by RT-PCR. The relative mRNA expression of NLS-RARα in the HL-60/pAd-NLS-RARα cells was significantly increased when compared with the control group. Data was expressed as means ± SD. *P<0.05. C and D, The protein expression of PNLS-RARα was assessed by Western blot assay. The protein expression of NLS-RARα in the HL-60/pAd-NLS-RARα cells was significantly higher than control groups. Data was expressed as means ± SD. *P<0.05 (lane 1: HL-60 cells; lane 2: HL-60/pAd-KZ cells; lane 3: HL-60/pAd-NLS-RARα cells).
Effect of NLS-RARα on proliferation of HL-60 cells
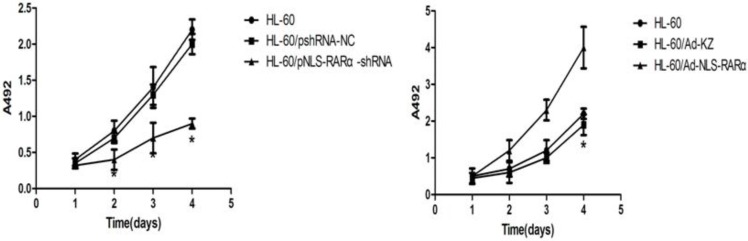
MTT assay showed that, when compared with the HL-60/pshRNA-NC cells, the proliferation of HL-60/pshRNA-NLS-RARα cells was significantly inhibited in a time-dependent manner (P<0.05, Figure 4A). In addition, up-regulation of NLS-RARα could promote the proliferation of HL-60 cells (Figure 4B).
Figure 4.
Proliferation of HL-60 cells in different groups. MTT assays were used to measure[/record/assay] cell proliferation on days 1, 2, 3 and 4 post-transfection/infection. Absorbance was measured at 492 nm. A, The proliferation of pNLS-RARα-shRNA cells was significantly inhibited in a time-dependent manner. Data was expressed as means ± SD. *P<0.05. B, The proliferation of HL-60/pAd-NLS-RARα infected cells increased significantly when compared with controls. Data was expressed as mean ± SD. * P<0.05.
Cell differentiation
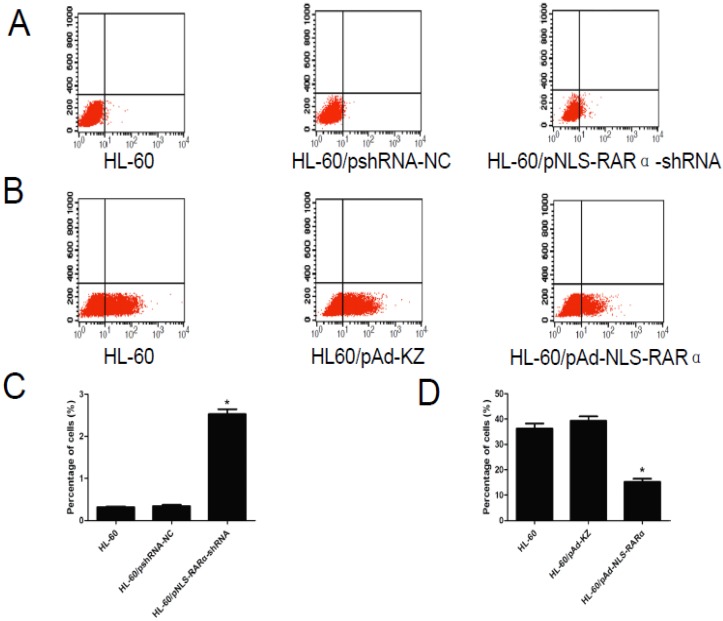
As shown in Figure 5, the differentiation rate of HL-60/pNLS-RARα-shRNA group (2.58±0.21%) was significantly higher than that of the control groups (0.32±0.03% and 0.34±0.06%) 48 h after transfection (P < 0.05). The differentiation rate of the HL-60/pAd-NLS-RARα transfected group (15.99±2.08%) was markedly lower than that of the control groups (36.37±3.20% and 39.33±3.00%) after treatment with ATRA for 48 h (P < 0.05).
Figure 5.
Differentiation rate in different cells. A and C, The differentiation rate of transfected HL-60 cells was detected by FCM. The differentiation rate of HL-60/pNLS-RARα-shRNA cells increased significantly (*P< 0.05). B and D, The differentiation rate of infected HL-60 cells treated with ATRA was detected by FCM. The differentiation rate of HL-60/pAd-NLS-RARα cells was reduced dramatically (*P< 0.05).
Expressions of C-MYC gene in HL-60/ pAd-NLS-RARα cells
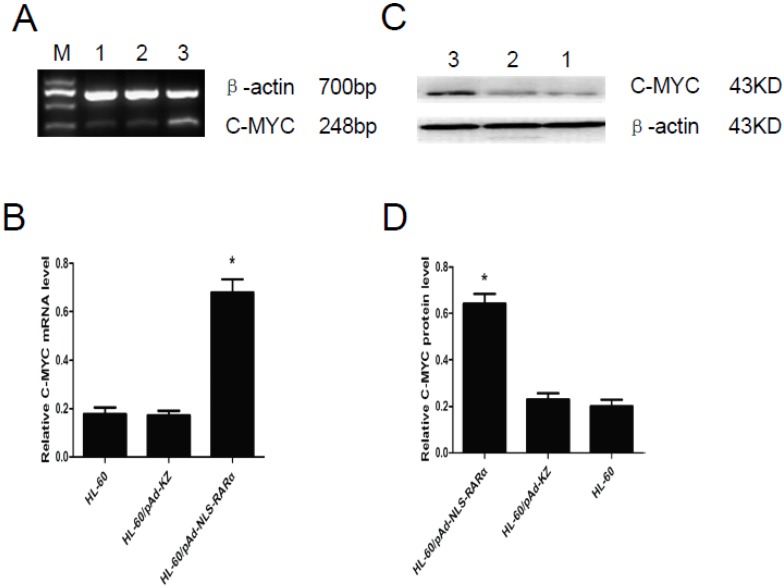
The expression of C-MYC gene was detected by RT-PCR and Western blot assay. Results showed that, when compared with controls, the mRNA (Figure 6, A and B) and protein (Figure 6, C and D) expressions of C-MYC in HL-60/pAd-NLS-RARα cells increased markedly (P<0.05). These findings suggested that NLS-RARα could inhibit cell differentiation through promoting the expression of C-MYC.
Figure 6.
Expression of C-MYC gene in HL-60/ pAd-NLS-RARα cells. A and B, Detection of mRNA expression of C-MYC by RT-PCR. When compared with control groups, the expression of C-MYC in HL-60/pAD-NLS-RARα cells increased significantly (P<0.05). C and D, Detection of protein expression of C-MYC gene by Western blot assay. The change in the protein expression of C-MYC gene was consistent with that in mRNA expression (lane 1: HL-60 cells; lane 2: HL-60/pAd-KZ cells; lane 3: HL-60/pAd-NLS-RARα cells).
Discussion
The fusion gene PML-RARα is a marker of APL and plays an important role in the occurrence and development of this disease. Its importance has been confirmed in transgenic mice 20. However, there is evidence that the entire PML-RARα fusion protein is not necessary for the development of APL 21, 22. Studies have shown that in early myeloid cells, the PML-RARα protein can be cleaved by NE within the PML component, leaving the NLS domain fused to RARα, resulting in a new mutant of RARα, named NLS-RARα. We speculate that the NLS-RARα fusion functions differently to the wild-type RARα, and may play a vital role in APL. In addition, the effect of NLS-RARα on leukemia cells has not been completely elucidated.
Human wild-type RARα acts as a ligand-dependent transcription factor involved in multiple functions 23, including cellular proliferation and differentiation. Our present findings demonstrate that NLS-RARα promotes the proliferation of leukemic cells and interrupts the pro-differentiation signal of ATRA.
The NLS-RARα was silenced with shRNA. RNA interference is a gene silencing process mediated by double-stranded RNA, and an effective method for gene silencing.
Recombinant adenovirus technology represents a powerful method to genetically modify leukemia cells. Therefore the recombinant adenovirus pAd-NLS-RARα was constructed and used to express the NLS-RARα gene, achieving high levels of stable expression in leukemia cell lines (HL-60 cells).
As shown in Figure 2 and Figure 3, the NLS-RARα was successfully silenced and over-expressed, respectively. The proliferation and differentiation of leukemic cells in vitro were investigated in cells with down-regulation or up-regulation of NLS-RARα expression. MTT assays showed that the proliferation of HL-60/pNLS-RARα-shRNA transfected cells was inhibited in a time-dependent manner, but that of HL-60/pAd-NLS-RARα infected cells was markedly promoted when compared with controls. These findings indicated that NLS-RARα could promote the proliferation of HL-60 cells. Furthermore, when compared with controls, the differentiation rate of the HL-60/pNLS-RARα-shRNA transfected group increased significantly. However, the differentiation rate of the HL-60/pAd-NLS-RARα infected group decreased significantly. These findings demonstrated that over-expression of NLS-RARα could promote the proliferation of APL cells and inhibit their differentiation induced by 2.5µmol/L ATRA, in marked contrast to the biological effect of RARα. Thus, we confirmed that NLS-RARα played important role in the differentiation of HL-60 cell, and it is likely to be fairly important to the genesis and development of APL. The further study of NLS-RARα gene will be very necessary and valuable.
It has been noted that MYC was one of the most prevalent oncogenes in human cancer, and particularly in hematopoietic neoplasias 24, 25. It is also involved in a number of fundamental functions of cell biology, such as control of proliferation a differentiation, energy production, cell size 26, 27, 28. Moreover, the inhibition of myeloid cell differentiation was one of the first biological effects described for MYC 28. It has been shown that PML-RARα induced MYC expression 29, 30. In this paper, C-MYC was detected by RT-PCR and Western blot assays, showing that over-expression of NLS-RARα could increase the C-MYC expression. This suggests that C-MYC is a downstream target of NLS-RARα gene.
In summary, wild-type RARα inhibits cell growth and promotes differentiation in response to ATRA, while NLS-RARα has the opposite effect. We speculated that this difference could be associated with abnormal cellular localization of the RARα domain, which is likely to result in abnormal protein-protein interactions. This would alter the original signal pathway and biological function, thus promoting the occurrence and development of APL.
Figure A.
Vector maps
Acknowledgments
This study was supported by the National Natural Science Foundation of China (No. 81171658) and Natural Science Foundation Project of CQ CSTC (No. 2011BA5037).
References
- 1.Mattei MG, Petkovich M, Mattei JF. et al. Mapping of the human retinoic acid receptor to the Q21 band of chromosome 17.Hum. Genet. 1988;80:186–188. doi: 10.1007/BF00702866. [DOI] [PubMed] [Google Scholar]
- 2.Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Escriva H, Bertrand S, Laudet V. The evolution of the nuclear receptor superfamily. Essays Biochem. 2004;40:11–26. doi: 10.1042/bse0400011. Review. [DOI] [PubMed] [Google Scholar]
- 4.McEwan IJ. Nuclear receptors: one big family. Methods Mol Biol. 2009;505:3–18. doi: 10.1007/978-1-60327-575-0_1. [DOI] [PubMed] [Google Scholar]
- 5.Glass CK, DiRenzo J, Kurokawa R, Han ZH. Regulation of gene expression by retinoic acid receptors. DNA Cell Biol. 1991;10:623–638. doi: 10.1089/dna.1991.10.623. Review. [DOI] [PubMed] [Google Scholar]
- 6.Rochette-Egly C, Germain P. Dynamic and combinatorial control of gene expression by nuclear retinoic acid receptors. Nucl Recepet Signal. 2009;7:e005. doi: 10.1621/nrs.07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gronemeyer H, Gustafsson JA, Laudet V. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov. 2004;3:950–964. doi: 10.1038/nrd1551. [DOI] [PubMed] [Google Scholar]
- 8.Aranda A, Pascual A. Nuclear hormone receptors and gene expression. Physiol Rev. 2001;81:1269–1304. doi: 10.1152/physrev.2001.81.3.1269. [DOI] [PubMed] [Google Scholar]
- 9.Saurin AJ, Borden KL, Boddy MN. et al. Does this have a familiar RING? Trends Biochem Sci. 1996;21:208–14. [PubMed] [Google Scholar]
- 10.Kakizuka A, Miller Jr WH, Umesono K. et al. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 11.Soo JY, Seob EJ, Jae HL. A complex, four-way variant t(15;17) in acute promyelocytic leukemia. Cancer Genetics and Cytogenetics. 2006;167:168–71. doi: 10.1016/j.cancergencyto.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 12.Pollock JL, Westervelt P, Walter MJ. et al. Mouse models of acute. Promyelocytic leukemia. Curr Opin Hematol. 2001;8(4):206–211. doi: 10.1097/00062752-200107000-00005. [DOI] [PubMed] [Google Scholar]
- 13.Laurenzana A, Pettersson F, Miller WH. Role of PML/RARα in the pathogenesis of APL. Drug Discovery Today: Disease Mechanisms. 2006;3:499–505. [Google Scholar]
- 14.Grisolano JL, Wesselschmidt RL, Pelicci PG. et al. Altered myeloid development and acute leukemia in transgenic mice expressing PML-RARα under control of cathepsin G regulatory sequences. Blood. 1997;89:376–87. [PubMed] [Google Scholar]
- 15.He LZ, Tribioli C, Riv R. et al. Acute leukemia with promyelocytic features in PML/RARα transgenic mice. Proc Natl Acad Sci USA. 1997;94:5302–7. doi: 10.1073/pnas.94.10.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Westervelt P, Lane AA, Pollock JL. et al. High-penetrance mouse model of acute promyelocytic leukemia with very low levels of PML-RARα expression. Blood. 2003;102:1857–65. doi: 10.1182/blood-2002-12-3779. [DOI] [PubMed] [Google Scholar]
- 17.Early E, Moore MA, Kakizuka A. et al. Transgenic expression of PML/RARα impairs myelopoiesis. Proc Natl Acad Sci USA. 1996;93:7900–4. doi: 10.1073/pnas.93.15.7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lane AA, Ley TJ. Neutrophil elastase cleaves PML-RARα and is important for the development of acute promyelocytic leukemia in mice. Cell. 2003;115:305–18. doi: 10.1016/s0092-8674(03)00852-3. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi N. Induction of cell differentiation and development of new anticancer drugs. Yakugaku Zasshi. 2002;122:547–563. doi: 10.1248/yakushi.122.547. [DOI] [PubMed] [Google Scholar]
- 20.Wartman LD, Welch LJ, Uy GL. et al. Expression and function of PML-RARA in the hemotopoietic progenitor cells of Ctsg-PML-RARA mice. PLoS One. 2012;7:e46529. doi: 10.1371/journal.pone.0046529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayakawa F, Privalsky ML. Phosphorylation of PML by mitogen-activated protein kinases plays a key role in arsenic trioxide-mediated apoptosis. Cancer Cell. 2004;5:389–401. doi: 10.1016/s1535-6108(04)00082-0. [DOI] [PubMed] [Google Scholar]
- 22.Shima Y, Shima T, et a1. PML activates transcription by protecting HIPK2 and p300 from SCFFbx3-mediated degradation. Mol Cell Biol. 2008;28:7126–38. doi: 10.1128/MCB.00897-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Melnick A, Licht JD. Deconstructing a disease: RARα, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood. 1999;93:3167–3215. [PubMed] [Google Scholar]
- 24.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 25.Vita M, Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol. 2006;16:318–330. doi: 10.1016/j.semcancer.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 26.Oster SK, Ho CS, Soucie EL, Penn LZ. The myc oncogene: marvelouslY complex. Adv Cancer Res. 2002;84:81–154. doi: 10.1016/s0065-230x(02)84004-0. [DOI] [PubMed] [Google Scholar]
- 27.Eilers M, Eisenman RN. Myc's broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leon J, Ferrandiz N, Acosta JC, Delgado MD. Inhibition of cell differentiation: a critical mechanism for MYC-mediated carcinogenesis? Cell Cycle. 2009;8:1148–1157. doi: 10.4161/cc.8.8.8126. [DOI] [PubMed] [Google Scholar]
- 29.Muller-Tidow C, Steffen B, Cauvet T, Tickenbrock L, Ji P. et al. Translocation products in acute myeloid leukemia activate the Wnt signaling pathway in hematopoietic cells. Mol Cell Biol. 2004;24:2890–2904. doi: 10.1128/MCB.24.7.2890-2904.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rice KL, Hormaeche I, Doulatov S. et al. Comprehensive genomic screens identify a role for PLZF-RARalpha as a positive regulator of cell proliferation via direct regulation of c-MYC. Blood. 2009;114:5499–5511. doi: 10.1182/blood-2009-03-206524. [DOI] [PMC free article] [PubMed] [Google Scholar]