Abstract
Scanning of the mRNA transcript by the preinitiation complex (PIC) requires a panel of eukaryotic initiation factors including eIF1 and eIF1A, the main transducers of stringent AUG selection. eIF1A plays an important role in start codon recognition; however, its molecular contacts with eIF5 are unknown. Using NMR, we unveil eIF1A’s binding surface on the carboxyl-terminal domain of eIF5 (eIF5-CTD). We validated this interaction by observing that eIF1A does not bind to an eIF5-CTD mutant, altering the revealed eIF1A-interaction site. We also found that the interaction between eIF1A:eIF5-CTD is conserved between human and yeast. Using GST pull down assays of purified proteins, we showed that the N-terminal tail (NTT) of eIF1A mediates the interaction with eIF5-CTD and eIF1. Genetic evidence indicates that overexpressing eIF1 or eIF5 suppresses the slow growth phenotype of eIF1A-NTT mutants. These results suggest that the eIF1A:eIF5-CTD interaction during scanning PICs contributes to the maintenance of eIF1 within the open PIC.
INTRODUCTION
Accumulating evidence indicates that a sophisticated scanning system has evolved to efficiently locate the proper start codon on the mRNA in eukaryotes. This scanning process involves the dynamic interplay of translation initiation factors, ultimately regulating the conformational change of the ribosomal pre-initiation complex (PIC) (Aitken and Lorsch, 2012; Asano and Sachs, 2007; Hinnebusch, 2011; Pestova et al., 1998; Pestova and Kolupaeva, 2002). To begin translation, the 40S small ribosomal subunit is pre-loaded with initiation factors eIF1A, eIF1, eIF2, eIF3, eIF5 and Met-tRNAiMet in the 43S PIC (Asano et al., 2000; Pestova et al., 1998; Sokabe et al., 2012). The 43S PIC binds the 5′ end of the mRNA that had been primed by eIF4F and eIF4B and scans downstream until reaching a start codon (Sonenberg and Hinnebusch, 2009). The scanning PIC thus formed (43S PIC, which becomes 48S after it finds the start codon) is thought to exist in equilibrium between two conformations: open (scanning competent) and closed (scanning incompetent) (Hinnebusch, 2011; Pestova and Kolupaeva, 2002). Upon binding of eIF1 and eIF1A to the 40S subunit, these two initiation factors induce a conformational rearrangement of the 40S subunit from a closed to an open state (Passmore et al., 2007). During scanning, eIF1, eIF1A, and perhaps other assembled factors in vivo (Singh et al., 2012) facilitate the scanning of the PIC and prevent it from shifting to the closed state. Once the correct start codon is reached (with AUG in a proper sequence context), eIF1 is physically excluded from the decoding site, shifting the PIC into the closed conformation and arresting it at the start codon. Compared to bacterial initiation allowing the commencement of translation from UUG or GUG codons (Asano et al., 1999a), eukaryotic initiation strictly discriminates against these non-AUG codons.
Multiple eukaryotic initiation factors regulate the fidelity of start codon recognition by strictly coupling AUG recognition to the ribosomal conformational change (Lorsch and Dever, 2010). It has been shown that overexpression of eIF1 increases the stringency of start codon recognition at its own AUG, which itself is in poor context (Ivanov et al., 2010; Martin-Marcos et al., 2011), whereas eIF5 overexpression reduces the stringency of start codon recognition at upstream ORFs on its own mRNA (Loughran et al., 2012). These studies highlight the importance of understanding the mechanism by which eIF1, eIF1A and eIF5 regulate the PIC conformations strictly in response to AUG base-pairing to tRNAiMet anticodon.
The structures of two domains of eIF5 have been solved by NMR-spectroscopy and X-ray crystallography. The first structural domain of eIF5 is the GTPase activating region located at the amino-terminal end (eIF5-NTD; residues 1-170) (Conte et al., 2006). The second structural domain is located at the carboxyl terminal end (eIF5-CTD; residues 225-409) or eIF5-HEAT (Bieniossek et al., 2006). The HEAT domains were so named because of the structural resemblance of four proteins, all containing a series of α-helices [Huntingtin, elongation factor 3 (EF3), the regulatory A subunit of protein phosphatase 2A and TOR1 (a target of rapamycin)] (Andrade and Bork, 1995; Bieniossek et al., 2006; Wei et al., 2006). In yeast Saccharomyces cerevisiae, key eukaryotic initiation factors assemble off (or away) from the ribosome by forming the multifactor complex (MFC), consisting of eIF3, eIF5, eIF1, eIF2-ternary complex (TC) (Asano et al., 2000; Asano et al., 2001). Studies using yeast have shown that eIF5, in particular its CTD, serves a critical role in the assembly of the MFC via interactions with eIF1, eIF2β-NTD and eIF3 (Asano et al., 2000; Yamamoto et al., 2005). Mammalian eIF5-CTD has also been shown to directly bind to each of these partners (Bieniossek et al., 2006; Das et al., 1997; Das and Maitra, 2000; Luna et al., 2012). In humans, a MFC similar to yeast complex has also been observed (Sokabe et al., 2012).
Previously, we showed that the CTD of eIF5 promotes start codon recognition by its dynamic interplay with eIF1 and subsequently eIF2β (Luna et al., 2012). We provided evidence that the eIF2β interaction with eIF5-CTD drives the ribosomal PICs into the closed conformation by promoting the release of eIF1 (Luna et al., 2012). In the present study, we propose that eIF1A plays a contributing role in supporting eIF1 in the open PIC through its interaction with eIF5-CTD. The amino terminal tail (NTT) of eIF1A was previously shown to increase initiation accuracy by promoting the closed conformation (Fekete et al., 2007; Saini et al., 2010).
Based on a previous study, the position of eIF1A-NTT indicates that it binds to the 40S and could also interact directly with Met-tRNAi, consistent with stabilizing the closed conformation (Yu et al., 2009). However, it has been unclear how the NTT of eIF1A mediates its function within the open PICs prior to its closure on start codons. Interestingly, alanine substitution mutation tif1117-21 altering amino acids 17-21 of the NTT of eIF1A displayed a slow growth phenotype as well as a strong PIC assembly defect, both of which were suppressed by overexpression of eIF1 (Fekete et al., 2007). Thus, at least a part of the NTT of eIF1A is responsible for retention of eIF1 within the scanning PICs (open state). Consistent with the additional role played by the NTT of eIF1A, this segment of eIF1A had been known to mediate the interaction with eIF2, eIF3 or eIF5 (Olsen et al., 2003). In this study, our NMR spectroscopic data reveal that eIF1A interacts directly with the CTD of eIF5. Combining our biophysical and yeast genetics results, we propose that the interaction between eIF1A and the CTD of eIF5 contributes to the retention of eIF1 in the open scanning compatible PIC in vivo.
MATERIALS AND METHODS
NMR Resonance Assignments and Chemical Shift Perturbation Assay
NMR chemical shift mapping experiments were performed as described previously (Luna et al., 2012; Marintchev et al., 2007). NMR spectra were recorded at 298 K on a Varian Inova 600 MHz spectrometer, equipped with a cryoprobe. Protein samples for NMR measurements contained 200 μM protein in buffer containing 200 mM NaCl, 20 mM Tris-HCl, 2 mM DTT, 1 mM EDTA, and 10% D2O (pH 7.2). We utilized the backbone resonance assignments of human eIF5-CTD (Luna et al., 2012).
Small-angle X-ray Scattering (SAXS) Reconstitution Assay
SAXS experiments were performed as previously described (Akabayov et al., 2013; Luna et al., 2012). Briefly, SAXS is a biophysical method that uses the elastic scattering of X-rays to probe sample features in the nanometer scale. SAXS allows the characterization of structure and interactions of macromolecules and their complexes in solution. Protein samples were measured in the following buffer conditions: 20 mM Tris.HCl, pH 7.4, 300 mM NaCl, 0.5 mM TCEP. SAXS experiments were performed for the following protein samples: 1.) eIF5-CTD, 2.) eIF1A, and 3.) eIF1A:eIF5-CTD. eIF5-CTD at a final concentration of 90 μM, while eIF1A titration concentrations were in the range from 90 μM, 180 μM and 360 μM. As a control, the concentration of eIF1A alone was 180 μM. The Rg values for eIF5-CTD, in the free state and in the eIF1A-bound state, were derived from SAXS intensities and determined using Guinier analysis.
Plasmid constructions
All of the plasmids used for NMR and SAXS experiments are bacterial expression vectors encoding human initiation factors that contain either N-terminal or C-terminal hexahistidine tags. The purification of proteins were purified as previously described (Luna et al., 2012). Briefly, the initiation factors were purified through standard Ni-NTA columns and subsequent gel filtration. The peak fractions were collected and tested for the presence of the target protein using SDS-PAGE followed by staining with Coomassie blue. The human eIF5 plasmid clone encodes the eIF5-CTD amino acid residues 225-409 with a C-terminal His6-tag. His6-tagged human eIF1A constructs were kindly provided by Assen Marintchev. The expression constructs for yeast eIF5-CTD clone (TIF5-B6241-405) was described previously (Reibarkh et al., 2008). pET-TIF11, the expression plasmid for yeast eIF1A, was constructed by cloning 0.4-kb NdeI-BglII fragment of pGAD-TIF11 (pKA129; Katsura Asano, personal stock) into the NdeI-BamHI sites of pET15b. For eIF5 overexpression, we subcloned the 2.2-kb EcoRI-HindIII TIF5 fragment of YEpL-TIF5, YEpL-TIF5-7A (Asano et al, 1999b) or YEpL-TIF5-Quad (Luna et al., 2012) into YEplac112 to generate YEpW-TIF5, YEpW-TIF5-7A or YEpW-TIF5-Quad, respectively. These plasmids overproduce wild-type or mutant versions of C-terminally FLAG-tagged yeIF5.
Yeast genetics experiments
Yeast genetics experiments and reporter assays were performed as described previously (Lee et al., 2007). Strains KAY955 (TIF11), KAY956 (tif117-11) and KAY957 (tif1112-16) were constructed by transforming H3582 (tif11Δ p[URA3 TIF11]) with the LEU2 FL-TIF11 plasmid pDSO157, its tif11 derivatives, pCF84, and pCF85 (Fekete et al., 2007), respectively, and evicting the URA3 TIF11 plasmid in H3582 by 5 fluoroorotic acid (plasmid shuffling). To overexpress eIF1, we used pCF82 (2μ TRP1 SUI1) (Alan G Hinnebusch personal collection) or YEpU-SUI1 (2μ URA3 SUI1) (He et al., 2003). Assays of β-galactosidase activity in whole cell extracts (WCEs) were performed as described previously (Lee et al., 2007).
RESULTS
eIF1A interacts with eIF5-CTD at a site that is targeted by eIF2β-NTT and overlaps the eIF1-binding surface
eIF5-CTD interacts with eIF1 and eIF2β-NTT at overlapping but distinct surfaces (Luna et al., 2012). In this study, we employed an NMR chemical shift perturbation assay to study the interaction of heIF1A with heIF5-CTD. The domain organization of these factors and the constructs used in this work are shown in Figure 1A. The NMR chemical shift perturbation (CSP) assay exploits the sensitivity of the chemical shift of a nucleus to its environment (reviewed in (Marintchev et al., 2007)). In order to determine whether the heIF5 interaction with heIF1A occurs through the amino-terminal domain (NTD) of heIF5, we used the CSP assay. However, we found no interaction between these two proteins (Fig. S1). We were also not able to detect an interaction between heIF1A and heIF1 (Fig. S2).
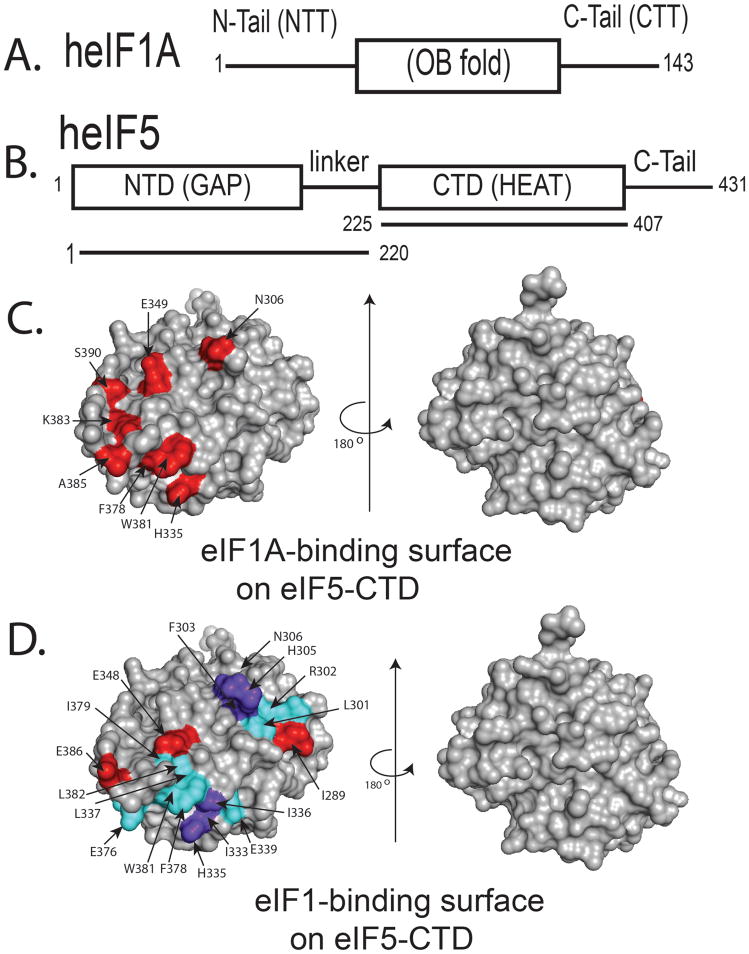
Figure 1. eIF1A and eIF1 bind to overlapping but distinct binding surfaces on eIF5-CTD.
(A) Domain organization of eIF1A, Abbreviations: NTT, amino (N)-terminal tail; CTT, carboxyl(C)-terminal tail; OB (oligonucleotide/oligosaccharide) fold domain structured as an elliptically shaped β-barrel. (B) Domain organization of eIF5, Abbreviations: NTD, amino(N)-terminal domain; GAP, GTPase-activating protein; CTD, carboxyl(C)-terminal domain. eIF5-CTD is a member of the HEAT domain family consisting of a series of α-helices. (C) NMR mapping of the heIF1A binding surface on heIF5-CTD (1IU1). Contacts are only observed on one face of the domain. heIF5-CTD residues wherein heIF1A causes chemical shift perturbations (CSPs) are painted red. Two orientations of the heIF5-CTD molecule are shown as surface representations: (left), interaction interface, and (right), a rotation of 180° along the Y-axis shows no interaction on the other side of the molecule. The left molecule of heIF5-CTD is in the similar orientation as the molecule in D. (D) NMR mapping of the heIF1 binding surface on heIF5-CTD was adapted from our previously published study, wherein the red colored residues experience CSPs and the residues painted cyan are broadened due to paramagnetic relaxation enhancement (PRE) experiments, while purple colored residues experience both PRE-induced broadening and CSPs (Luna et al., 2012): (left), interaction interface, and (right), a rotation of 180° along the Y-axis shows no interaction on the other side of the molecule.
As shown in Fig. 1C, we discovered a novel interaction between heIF1A and heIF5-CTD. In the NMR time scale, this interaction is in the intermediate exchange regime. At physiological salt concentrations of 150mM NaCl, addition of heIF1A to 15N-labeled heIF5-CTD completely broadens the spectrum with the exception of the few unstructured residues. However, increasing the salt concentration weakens the interaction between heIF5-CTD and heIF1A, thereby pushing it towards a fast exchange regime. This is reflected in the appearance of resonances in the structured region of the heIF5-CTD spectrum at concentrations of 200mM and 300mM NaCl. (Figure S3). At a salt concentration of 300 mM NaCl, we were able to monitor chemical shift perturbations and map the binding of heIF1A (Figure S4).
We found that the eIF1A-binding site involves residues (F378, W381, K383, A385 and S390) closely overlapping with the eIF2β-binding site. The addition of unlabeled eIF5-CTD significantly broadened the resonances of 15N-eIF1A (Fig. 2B, left panel), which may be due to a larger size resulting from complex formation, from multiple bound conformations, or from reduced solubility of the formed complex. Utilizing our previously characterized quadruple mutation (H305D/N306D/E347K/E348K), wherein one face of eIF5-CTD is altered to disrupt its interaction with eIF1 and eIF2β, we discovered that the CTD-Quad mutant significantly reduces eIF5’s interaction with eIF1A (Fig. 2B, right panel). Previously, we identified the heIF1 and heIF2β-NTT binding surfaces on the surface of heIF5-CTD (Fig. 1D), wherein the binding affinity for heIF1 was undetectable and the affinity for heIF2β-NTT was detected ~17μM (Luna et al., 2012). Given the proximity of eIF1 and eIF1A within the PIC (Lomakin et al., 2003; Lomakin and Steitz, 2013; Rabl et al., 2011; Weisser et al., 2013; Yu et al., 2009), it is reasonable to postulate that the CTD of eIF5 bridges the interaction between eIF1 and eIF1A.
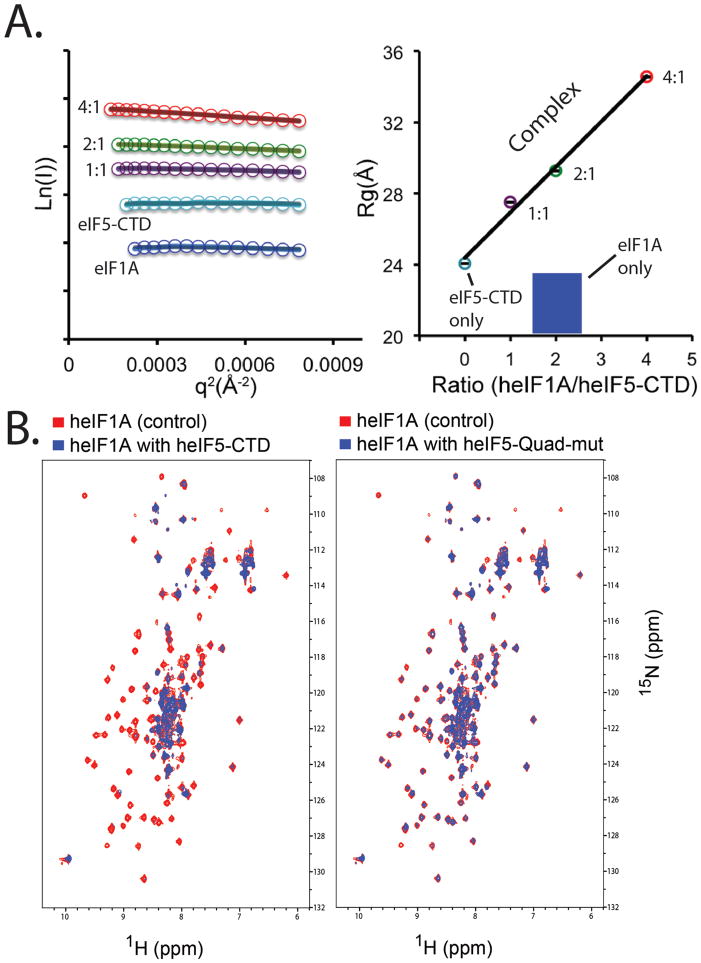
Figure 2. SAXS and NMR experiments indicate that eIF1A binds eIF5-CTD, while the eIF5-CTD-Quad mutant significantly weakens the interaction with eIF1A.
(A) Left panel: SAXS Guinier plots shown for different heIF1A:heIF5-CTD molar ratios with color codes as used in the right panel. Right panel: SAXS results plotting the radius of gyration (Rg) (Y-axis) versus heIF1A:heIF5-CTD protein ratio (X-axis). The Rg was derived from the Guiner plots (left panel). Rg serves as an indicator for the formation of higher-order protein complexes. To exclude the possibility that the Rg is increased due solely to higher concentration of heIF1A, we used a higher concentration of heIF1A alone (180uM; Blue) as a control. Cyan corresponds to heIF5-CTD alone (90 μM). Data were collected for heIF5-CTD (90 μM; cyan circle) titrated with increasing amounts of heIF1A (90 μM-purple, 180 μM-green, 360 μM-red). (B) Left panel: Overlay of 1H-15N HSQC spectra of 0.2 mM 15N-labeled heIF1A alone (red) and in the presence of 0.4 mM (blue) unlabeled wild-type heIF5-CTD domain. Right panel: Overlay of 1H-15N HSQC spectra of 0.2 mM 15N-labeled heIF1A alone (red) and in the presence of 0.4 mM (blue) unlabeled heIF5-CTD-Quad mutant.
SAXS reconstitution assay shows that eIF1A and eIF5-CTD interact
We employed an alternative approach to show that heIF1A binds heIF5-CTD. As an orthogonal binding assay to NMR, we used small-angle X-ray scattering (SAXS) to monitor the weak binding interaction of the heIF1A: heIF5-CTD complex in solution, which was also used previously to characterize the binding between heIF1 and heIF5-CTD (Luna et al., 2012). In this SAXS reconstitution assay, increasing amounts of heIF1A were titrated into a fixed concentration of heIF5-CTD, and the mixture at each point was subjected to SAXS analysis. In this assay, the radius of gyration (Rg) reflects the conformational/binding state of the proteins in solution, either free or in complex with each other (Akabayov et al., 2013; Luna et al., 2012). Titrating heIF1A into a solution of fixed concentration of eIF5-CTD results in a steady increase in the Rg, consistent with complex formation between heIF1A and heIF5-CTD (Figure 2A, colored lines, left panel and colored spheres, right panel). Free heIF1A (high concentration) has an Rg value of 23.1 Å. At the same concentration of heIF5-CTD in the presence of twice the amount of heIF1A (2:1 molar ratio), the Rg value is dramatically increased to 29.3 Å indicating complex formation. High amount of eIF1A alone (180μM) did not increase the Rg indicating that this elevation is not due to interparticle interference. The results support the notion that heIF1A binds weakly to heIF5-CTD, because complex formation did not reach saturation even though we added a large excess of the heIF1A titrant. A similar observation was made in the SAXS study of the eIF1: eIF5-CTD complex (Luna et al., 2012).
The interaction between eIF1A and eIF5-CTD is evolutionarily conserved
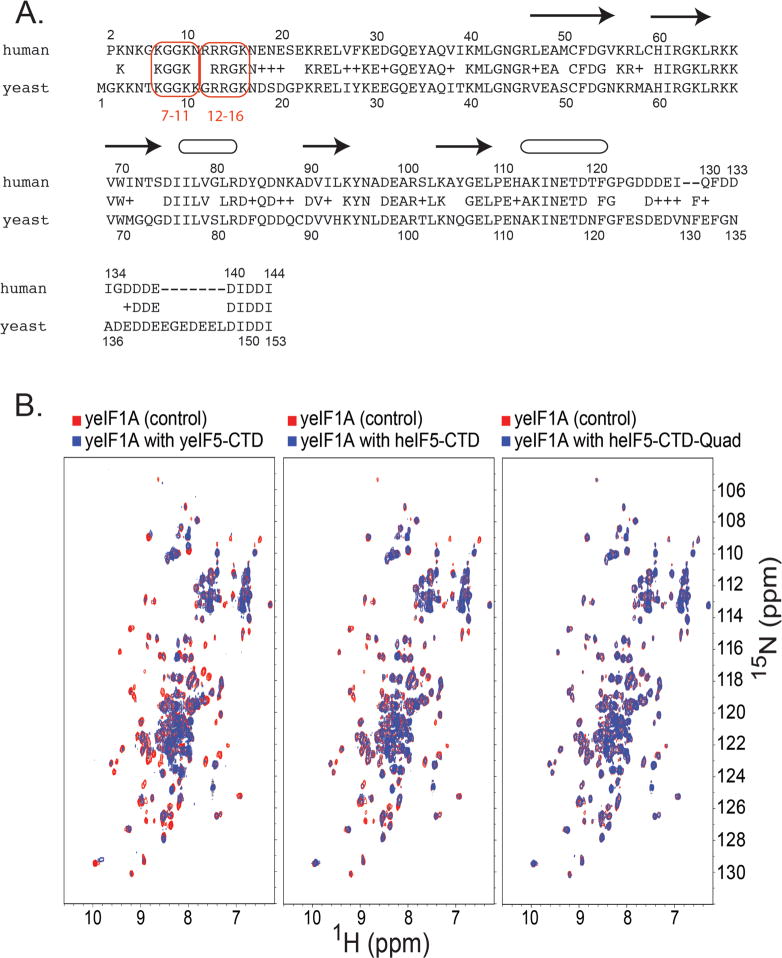
Our data clearly show that eIF1A interacts with eIF5-CTD. The eIF1A amino acid sequence alignment shows a striking similarity of basic residues (lysines and arginines) within the NTT regions of human and yeast eIF1A (Fig. 3A). We assessed whether yeast eIF5-CTD (aa 241-405; Tif5p-B6) also interacted with yeast eIF1A (Tif11p or yeIF1A), which has not been previously assessed. In the left panel of Figure 3B, we show that yeast eIF5-CTD (Tif5p-B6) does bind to yeIF1A, hence the eIF1A: eIF5-CTD interaction is conserved between human and yeast proteins. We proceeded to evaluate whether the heterologous proteins could bind each other. In the middle panel of Figure 3B, we noticed that 15N-labeled yeIF1A (Tif11p) binds to unlabeled human eIF5-CTD, as evidenced by chemical shift perturbations and peak broadening. We evaluated whether 15N-labeled yeIF1A (Tif11p) would bind to the human eIF5-CTD-Quad mutant protein. In the right panel of Figure 3B, we clearly see diminished binding between 15N-labeled yeIF1A (Tif11p) and the human eIF5-CTD-Quad protein, as evidenced by the return of the previously broadened signals in the spectra and significantly less chemical shift perturbations, when compared to Figure 3B middle panel. Hence yeast eIF1A binds to the face on the human eIF5-CTD molecule that binds to human eIF1A. Since yeast eIF1A binds to yeast eIF5-CTD protein and heterologously with human eIF5-CTD, we suggest that the eIF1A: eIF5-CTD complex serves a conserved regulatory role during the scanning process of open PICs.
Figure 3. Interactions between eIF1A and eIF5-CTD are conserved in yeast and humans and disrupted by the Quad mutation.
(A) Amino acid sequence comparison between human and yeast eIF1A. Previously identified mutations on the NTT of yeIF1A are circumscribed in light orange (tif117-11 and tif1112-16). Arrows and ellipses indicate β-sheet and helical secondary structure. (B) Left panel: Overlay of 1H-15N HSQC spectra of 0.2 mM 15N-labeled yeIF1A alone (red) and in the presence of 0.4 mM (blue) unlabeled wild-type yeIF5-CTD (Tif5-B6) domain. Middle panel: Overlay of 1H-15N HSQC spectra of 0.2 mM 15N-labeled yeIF1A alone (red) and in the presence of 0.4 mM (blue) unlabeled wild-type heIF5-CTD domain. Right panel: Overlay of 1H-15N HSQC spectra of 0.2 mM 15N-labeled yeIF1A alone (red) and in the presence of 0.4 mM (blue) unlabeled heIF5-CTD-Quad mutant domain (H305D, N306D, E347K, E348K mutant).
eIF1A forms a higher-order complex with eIF1 and eIF5-CTD
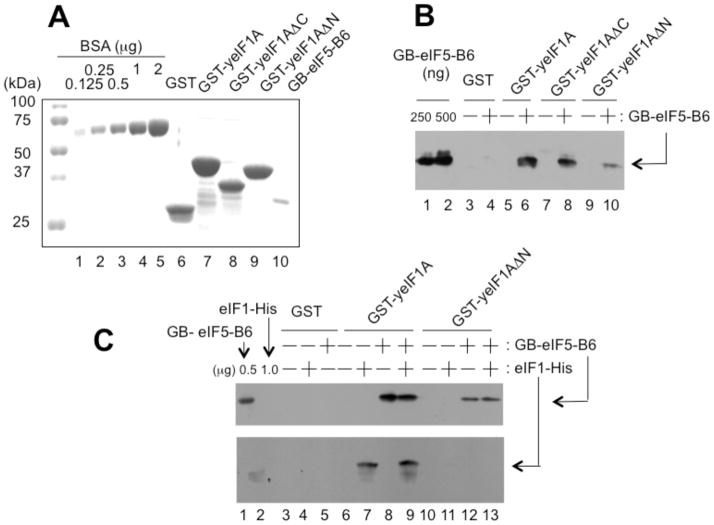
To examine whether eIF1A forms a higher-order complex with eIF1 and eIF5-CTD and if so, whether the complex formation is mediated by eIF1A-NTT, we performed the GST pull down assay with yeast proteins (see Fig. 4A for GST fusion proteins used). As shown in Fig. 4B, the full-length GST-yeIF1A or its derivative lacking the CTT (ΔC lacking aa. 108-153) bound yeIF5-B6 (lanes 6 & 8), but GST-yeIF1A lacking the NTT (ΔN lacking aa. 1-25) bound it only weakly (lane 10), indicating that the interaction between yeIF1A and yeIF5-CTD depends on yeIF1A-NTT. As shown in Fig. 4C, GST-yeIF1A bound yeIF1 and yeIF5-B6 equally well, regardless of whether yeIF1 and yeIF5-B6 were added separately (lane 7–8) or simultaneously (lane 9). Thus, yeIF1A binds simultaneously to yeIF1 and yeIF5-B6, likely forming a higher-order complex. This complex was disrupted by the ΔNTT mutation introduced to GST-yeIF1A (Fig. 4C, lane 13). Thus, yeIF1A-NTT mediates these interactions. Interestingly, yeIF1 interacts with yeIF1A (Fig. 4C, lane 7), in contrast to humans (Fig. S2). This fact may explain our failure to observe a strong enough eIF1A/eIF5-CTD/eIF1 complex with human proteins.
Figure 4. eIF1A-NTT mediates interaction with eIF5-CTD and eIF1 in yeast.
(A) Coomassie staining of GST fusion proteins used in this study (lanes 6–9). Lanes 1–5, BSA standards. Lane 10, GB-yeIF5-B6. (B) GST-yeIF1A binds GB-yeIF5-B6. Equal quantities (~5 μg) of GST or indicated GST fusion proteins were mixed with (+) or without (-) 10 μg of GB-yeIF5-B6. After pulled down by glutathione resin and washed, the bound proteins were visualized by immunoblotting with anti-His antibodies. (C) Higher-order complex of yeIF1A, yeIF1 and yeIF5-B6. Equal quantities (~5 μg) of GST or indicated GST fusion proteins were mixed with (+) or without (-) GB-yeIF5-B6 or with (+) or without (-) eIF1-His (10 μg each), and the bound proteins were analyzed by immunoblotting, as in (B).
Genetic evidence that eIF1A-NTT retains eIF1 and eIF5 within the open scanning PIC in vivo
Previous evidence that the NTT of eIF1A contributes to retaining eIF1 within the PIC prior to the closure on the start codon was presented as follows: tif1117-21, altering amino acids NDSDG17-21 of the NTT of eIF1A, reduced the amount of eIF1A, eIF1, eIF2, eIF5 and eIF3 in the 43/48S complexes isolated by sucrose gradient velocity sedimentation, in a manner restored by eIF1 overexpression (Fekete et al., 2007). In keeping with the PIC assembly defect, tif1117-21 and two other five-alanine substitution mutations tif117-11 and tif1112-16 (altering KGGKK7-11 and GRRGK12-16) respectively, of eIF1A-NTT, showed a slow growth phenotype in a manner suppressed by eIF1 overexpression (for tif117-11 and tif1112-16, see Fig. 5A). These basic residues in the NTT of eIF1A (KGGKK7-11 and GRRGK12-16) are similar to the three K-box regions of the NTD of eIF2β, which facilitate binding to the CTD of eIF5.
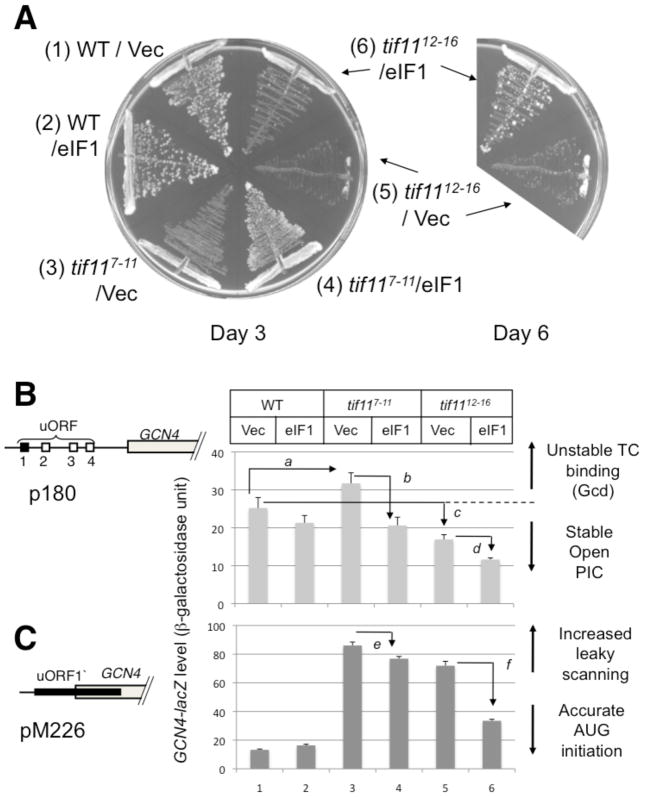
Figure 5. Genetic evidence that the lysine- and arginine-rich eIF1A-NTT contributes to stable formation of the open PIC requiring high eIF1 occupancy for its function.
(A) In the left panel, tif11 (yeast eIF1A) mutants (carrying p180) were streaked on plates to verify slow growth phenotypes prior to the assays shown in (B). The plates were incubated for 3 (left) and 6 (right) days. The six quadrants in the left plate express WT eIF1A and mutants (tif117-11 and tif1112-16) paired with either vector alone or high copy eIF1. (B) and (C) GCN4-lacZ expression in eIF1A WT and eIF1A-NTT mutations (tif117-11 and tif1112-16). Yeast strains used in (A) were doubly transformed with p180 (B) or pM226 (C) carrying GCN4-lacZ and with pCF82 (high copy eIF1) or vector control and assayed for β-galactosidase. Schematics to the left depict the arrangement of uORFs in the GCN4 leader region of the GCN4-lacZ fusion plasmid employed. P values for differences observed are; a, 0.001; b, 0.0005; c, 0.02; d, 0.005; e and f, <0.000001 (n=8~10).
In this study, we focused on the effect of two NTT mutants of yeast eIF1A on GCN4 expression, since GCN4 is a sensitive reporter to detect changes in the stability of the open, scanning PIC. We chose tif117-11 and tif1112-16, since three of the five substituted amino acids in these mutants are Arginines or Lysines (red boxes in Fig. 3A). This resembles the NTD of eIF2β, which targets a similar binding surface on eIF5-CTD. We first examined the effect on wild-type GCN4-lacZ expression (encoded by plasmid p180). The GCN4 leader region contains four regulatory upstream ORFs (uORFs). This leader region normally functions to inhibit GCN4 translation; however in response to amino acid starvation, the GCN4 leader region induces GCN4 translation. Under normal growth conditions, the ribosome that has translated uORF1 stays associated with the mRNA leader, resumes scanning, re-initiates at uORF 2, 3 or 4 and dissociate after its translation. Under starvation conditions, eIF2 is phosphorylated, which reduces the level of active eIF2-GTP-Met-tRNAiMet ternary complexes (TC). This results in the delay of TC binding to the ribosome, wherein the resumed scanning preinitiation complex (PIC) binds to the TC after uORF1 thus bypassing the inhibitory uORFs (2–4), allowing initiation at GCN4 start codon by the ribosome that has translated uORF1. Mutations that delay TC binding or increase TC dissociation from the open PIC allow the bypass of uORFs 2–4 by 40S subunits scanning downstream from uORF1 even under normal conditions, increasing the expression of GCN4 (general control derepressed or Gcd− phenotype). As expected, tif117-11 increased GCN4-lacZ expression by 20%. (Fig. 5B, columns 1 and 3), as observed previously with tif1117-21 (Fekete et al., 2007). Importantly, the increased GCN4-lacZ level in tif117-11 was significantly diminished by overexpression of eIF1 (Fig. 5B, columns 3 and 4). Thus, the weak Gcd− phenotype of tif117-11, suggestive of destabilized TC retention in the open PIC, is due to the weakened PIC retention of eIF1. In the case of tif1112-16, the most severe slow growth mutant (Fig. 5A), GCN4-lacZ level was decreased (Fig. 5B, columns 1 and 5) possibly due to the strong bypass of uORF1 and GCN4 start codons, a condition known to dampen GCN4 expression (Hinnebusch, 2005). The further decrease in GCN4-lacZ level by eIF1 overexpression (Fig. 5B, columns 5 and 6) is consistent with the idea that TC retention is also destabilized in this stronger mutant, in a manner restored by increasing eIF1 occupancy of the PIC.
Next, we examined the effect on GCN4 expression from a modified construct, pM226. In this plasmid, uORF1 is elongated and overlaps with GCN4 (Fig. 5C). Therefore, GCN4-lacZ is translated only when the ribosome has bypassed the uORF1 start codon. As observed with tif1117-21 (Fekete et al., 2007), both eIF1A-NTT mutations dramatically increased the expression from this plasmid (Fig. 5C, columns 1, 3 and 5), indicative of a strong bypass of uORF1 start codon. Overexpression of eIF1 significantly decreased the strong bypass (Fig. 5C, columns 4–6), reinforcing that the leaky scanning arises at least in part owing to the weakened retention of eIF1 and attendant dissociation of Met-tRNAi. Together, we provided further evidence that eIF1A-NTT rich in basic residues plays a crucial role in maintaining eIF1 in the open, scanning-competent PIC, presumably in part by eIF1A’s interaction with eIF5-CTD.
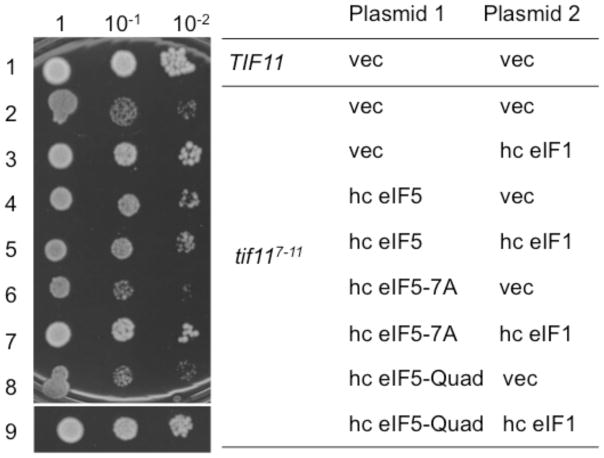
Having obtained strong evidence that the slow growth phenotypes caused by tif117-11 and tif1112-16 are due to unstable eIF1 anchoring to the open PIC, we next examined whether eIF5 contributes to stabilizing the open PIC in vivo. For this purpose, we overproduced eIF5 in yeast carrying tif117-11. As shown in Fig. 6, rows 2 and 4, the overexpression of wild-type eIF5 partially suppressed the slow growth caused by tif117-11. Importantly, this partial suppression was eliminated when we overexpressed eIF5 mutants carrying tif5-7A disrupting its CTD (Asano et al., 1999) (Fig. 6, row 6) or tif5-Quad weakening the interaction with eIF2β, eIF1 (Luna et al 2012) and/or eIF1A (Fig. 3) (Fig. 6, row 8). These results provide in vivo evidence for the mutual interaction between eIF5 and eIF1A-NTT via the Quad residues in the CTD surface (Fig. S5).
Figure 6. Suppression of tif117-11 phenotypes by eIF1 and eIF5 overexpression.
5 μl of 0.15 A600 culture and its 10-fold dilutions of KAY955 (TIF11) or KAY956 (tif117-11) transformants carrying indicated combinations of the plasmids were spotted onto SC-ura-trp medium plate and incubated for 4 days at 30 °C. Plasmids used were, for Plasmid 1, YEplac112 (TRP1) (vec), YEpW-TIF5 (hc eIF5), YEpW-TIF5-7A (hc eIF5-7A), YEpW-TIF5-Quad (hc eIF5-Quad), and, for Plasmid 2, YEplac195 (URA3) (vec), and YEpU-SUI1 (hc eIF1).
Interestingly, we also observed that eIF5 co-overexpression attenuates the suppression of tif117-11 by hc eIF1, again dependent on the intact CTD (disrupted by tif5-7A) or the eIF1/eIFA/eIF2β binding surface (altered by tif5-Quad) (Fig. 6, rows 3, 5, 7, and 9). This suggests that, in the absence of the intact eIF1A-NTT, eIF1 binds eIF5 on the open PIC in a partially competing manner. This is agreement with the antagonism of eIF5 against the ability of eIF1 to keep tRNAiMet anticodon out of the P-site (Pout state) within the scanning-competent open PIC (Nanda et al. 2009). Together, the results shown in Fig. 6 support the hypothesis that the higher-order complex interaction between eIF1A-NTT, eIF5-CTD and eIF1 plays a crucial role in maintaining the open conformation of the PIC.
DISCUSSION
A body of biochemical and genetic experiments from yeast studies provide evidence that the NTT of eIF1A, containing the proposed scanning-inhibitor (SI) element, is involved in the closure of the 40S ribosome conformation in response to AUG recognition by Met-tRNAiMet (Fekete et al., 2007; Saini et al., 2010). In agreement with this model, eIF1A has been mapped by hydroxyl radical cleavage to the vicinity of the A site on the 40S subunit, and its NTT lines the bed of the mRNA channel in the direction towards the P-site (Yu et al., 2009). The current molecular architecture of the eukaryotic PIC supports the notion that the closure in response to AUG recognition involves the direct interaction of eIF1A-NTT with the 40S subunit P-site, which would stabilize the positioning of the Met-tRNAiMet in the P-site (Pin state). In the present study, we identified the CTD of eIF5 as an additional partner of eIF1A, which could occur within the 48S PIC in the open conformation, prior to AUG recognition by Met-tRNAiMet.
Our biophysical and genetic analyses provides the first evidence that eIF1A directly binds eIF5. In our previous study with human proteins, we were not able to obtain binding affinities for the eIF1:eIF5-CTD complex, leading us to conclude that other initiation factors assist the CTD of eIF5 to maintain eIF1 in position within the scanning PIC (Luna et al., 2012). Based on the results in our present study, we suggest that one of these other factors include eIF1A. Because the concentrations used in the interaction assays are much higher than at physiological conditions, the proposed eIF1A:eIF5-CTD:eIF1 complex is weak both in humans and yeasts and therefore likely to occur only on the ribosome. However, taking advantage of the tif117-11 mutation, which apparently makes the open PIC formation rate-limiting for yeast growth, we provided evidence that the interaction occurs in vivo in the open PIC (Fig. 5 and 6).
Our finding that the eIF5-Quad mutation (H305D, N306D, E347K and E348K) dramatically weakened the interaction with eIF1A (this study), along with both eIF1 and eIF2β (Luna et al., 2012), strengthens the idea that the altered eIF5-CTD surface, which is in part made of conserved AA-boxes (Asano et al., 1999b), is the “business end” of this factor crucial for PIC assembly. Since eIF1A-NTT and eIF2β-NTD contain lysine-rich segments along with the evidence that eIF1A and eIF2β bind overlapping acidic surfaces on eIF5-CTD (Fig. S5), we propose that the putative nexus of eIF1A/eIF5-CTD/eIF1 interactions protects eIF5-CTD from interacting with eIF2β prematurely, which would otherwise promote the release of eIF1 and the subsequent closure of the PIC on the start codon (Fig. 7). In agreement with this model, the eIF1-binding surface of eIF5-CTD is more extended towards the area including R298-N306 (towards the right in Fig. 1D), compared to eIF1A- or eIF2β-binding sites (Fig. 1C and S5). This would provide sufficient surface area on eIF5 for simultaneous interactions with eIF1 and eIF1A.
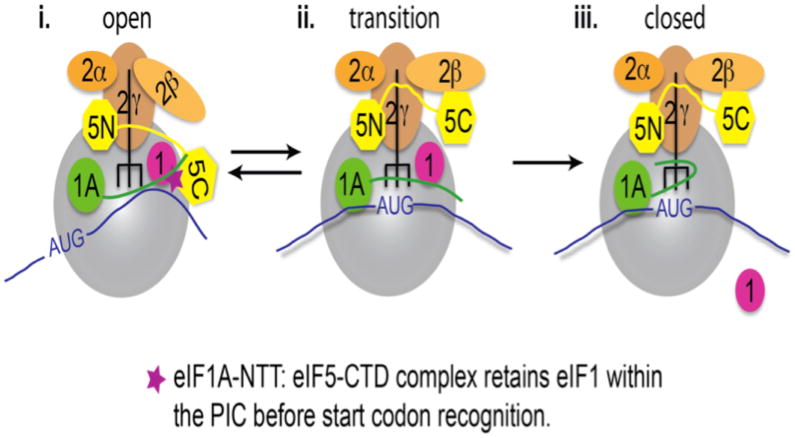
Figure 7. Model of events occurring within PICs, wherein the eIF1A-NTT: eIF5-CTD interaction retains eIF1 in position before start codon recognition.
(i.) During the assembly stage of the open scanning-compatible open PIC, the eIF5-CTD interacts with eIF1. It is at this period during scanning that we propose that the NTT of eIF1A reaches and binds eIF5-CTD, hence the eIF1A-NTT: eIF5-CTD interaction effectively retains eIF1 in position during the scanning process. (ii.) The 43S PIC continues to scan the mRNA in an open conformation until start codon recognition. eIF2β binds to eIF5-CTD on an overlapping binding surface with eIF1A. The eIF2β disruption of the eIF5-CTD interaction with the NTT of eIF1A allows for dislodging of eIF1 from the 43S PIC. Upon release of eIF1, the free phosphate is subsequently released. (iii.) The eIF5-CTD:eIF2β interaction stabilizes the closed ribosomal conformation of PICs upon start codon selection.
Studies using a yeast reconstitution system (components: eIF1, eIF1A, eIF2, eIF5, Met-tRNAiMet, mRNA and the 40S subunit) provided clues that led us to suggest that eIF1A, along with eIF5-CTD, stabilize eIF1 within the PIC during the scanning process (Maag et al., 2006; Nanda et al., 2013). 1.) eIF1 and eIF1A alone can bind to 40S subunits and induce the open conformation, which clears the mRNA channel (Passmore et al., 2007). A stably bound eIF2-TC (tRNA anticodon is base-paired to the start codon) and eIF1 are mutually exclusive, although eIF1A can clearly promote initial TC binding to the 40S (Passmore et al., 2007). 2.) The NTT of eIF1A was mapped to the mRNA channel in the vicinity of the P-site (Yu et al., 2009). These findings, when combined, suggest the following: (i) the ability of eIF1A-NTT to stabilize TC binding to the P-site is mediated through its direct interaction with Met-tRNAiMet and the ribosomal P-site, and (ii) eIF1 prevents tRNA accommodation upon start codon recognition. 3.) eIF5 was shown to bind to PICs in an antagonistic fashion to eIF1 (Nanda et al., 2009) and more recently it was shown that this function was mediated through the CTD of eIF5 (Nanda et al., 2013). Since the antagonism is mediated via eIF5-CTD binding to eIF2β to release eIF1 and end the scanning event on the proper start codon (Luna et al., 2012), it is reasonable to assume that the eIF2β-binding site on eIF5-CTD is masked by a PIC component until the closure on the start codon. eIF1A-NTT appears to be bound to the 40S subunit near the P-site in the presence or absence of eIF1 but differently in each condition (Lomakin and Steitz, 2013; Weisser et al., 2013; Yu et al., 2009). Therefore, eIF1A-NTT binding by eIF5-CTD (missing in these previous structural studies) in the scanning PIC is plausible and could help stabilize the positioning of eIF1 before start codon recognition.
This model is supported by complementary genetic findings in yeast. eIF1A-NTT mutations altering three consecutive 5 amino acid (aa)-long segments exhibited slow-growth phenotypes, in a manner suppressed by overexpressing eIF1 in vivo (Fekete et al., 2007) (also see Fig. 5A). Since the open PIC is characterized as eIF1-loaded PIC, the slow growth phenotype here most likely results from defective eIF1 loading to the open PIC caused by eIF1A-NTT mutations. Any phenotype suppressed by mass action effects would result from failure to retain the overproduced component. The first biological evidence suggesting a possible interaction between the NTT of eIF1A and eIF5 was presented in a study showing that a deletion of the first 25 amino acids (NTT) of yeast eIF1A (Δ1-25) inhibited GST-eIF1A binding to eIF5 in whole cell extracts (Olsen et al., 2003). Here we verified this interaction using purified proteins (Fig. 4). GST-eIF1A was also shown to interact with purified eIF3 (Olsen et al., 2003), which often copurifies with eIF5 (Asano et al., 1998; Phan et al., 1998). Since the low affinity between eIF1A and eIF5 does not explain the bridging interaction observed between GST-eIF1A and eIF3 (Olsen et al., 2003), it is likely that eIF3 is involved in indirectly anchoring eIF1 in the open PIC through unidentified interaction with eIF1A, in addition to direct eIF1 anchoring through eIF3c-NTD (Singh et al., 2012; Valasek et al., 2003). There is also good evidence that eIF1A promotes eIF5 and eIF3 recruitment to the PIC independent of TC binding (Fekete et al., 2005). While the present study links eIF1A to eIF5-CTD and eIF1, other studies link eIF1A-CTT more intimately to eIF2 TC. In strong support of the idea that eIF1A-CTT is the direct binding partner of eIF2 in TC recruitment, eIF1A-CTT deletion mutation (Δ108-153 or ΔC) displays a strong Gcd− phenotype that is suppressible by overexpression of eIF2 and tRNAiMet, without disrupting eIF1A-ΔC binding to the PIC (Fekete et al., 2005). It was also presumed that the scanning enhancer (SE) elements in eIF1A-CTT directly bind tRNAiMet, preventing it from positioning tightly into the P-site (Saini et al., 2010).
Our biological studies provides additional evidence for the model that the eIF1A-NTT plays an important role in retaining eIF1 within PICs before start codon recognition (Fig. 5): We showed that 1.) the weak Gcd− phenotype of eIF1A-NTT mutants tif117-11 is suppressed by overexpressing eIF1 2.) the leaky scanning phenotype of eIF1A-NTT mutant tif1112-16 is suppressed by overexpressing eIF1. These results indicate that the skipping of uORFs or GCN4 start codons (which would cause the above-mentioned phenotypes) is at least in part due to the weakened interaction between eIF1 and the (open) PIC during the process of scanning. As observed in Fig. 4B with the eIF1A ΔN mutation, the substitution mutations in the NTT of eIF1A would weaken the interaction with eIF1 and eIF5-CTD, ultimately reducing the ability to anchor eIF1 within the open PIC. We further propose that the eIF1A: eIF5-CTD interaction normally functions to position eIF1 closer to the P-site via the eIF1/eIF5/eIF1A linkage, such that eIF1 is poised to leave PIC effectively upon anticodon binding of the initiator tRNA to the P-site. If the mutant NTT of eIF1A cannot position eIF1 properly via the CTD of eIF5, then eIF1 cannot be ejected efficiently, allowing the scanning PIC to bypass the AUG start codon.
In conclusion, our results suggest that eIF1A-NTT does not strongly contribute to TC recruitment, but contributes to maintaining eIF1 through eIF5-CTD in the open PIC. A breadth of studies on eIF5-CTD suggests that it binds eIF2β twice during the pathway of translation initiation: 1.) during the formation of the MFC and 2.) when the PIC closes on the start codon. During the interim scanning period, our results suggest that eIF1A along with eIF1 masks the acidic eIF2β-binding site on eIF5-CTD while eIF1 is positioned close to the P-site. Since eIF1A binds weakly to the CTD of eIF5 at a site also targeted by the stronger binder eIF2β-NTT (Luna et al., 2012), it appears that eIF1A binds to eIF5-CTD prior the closure of the PIC as eIF2β binding terminates the initiation process.
Supplementary Material
Acknowledgments
This work was supported by NIH grants CA68262 and GM47467 to GW, GM64781, Kansas COBRE-PSF Pilot grant, and Innovative Award from KSU Terry Johnson Cancer Center to KA, and NIDDK-K01-DK085198 to H.A. Use of the National Synchrotron Light Source, Brookhaven National Laboratory, was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-98CH10886.
We thank Alan Hinnebusch and Assen Marintchev for their useful discussion. We would like to also thank L. Yang & M. Allier (Beamline X-9 NSLS, BNL) at Brookhaven National Laboratory for assistance with the SAXS experiments.
Footnotes
Supporting Information Available
Additional NMR data and the summary diagram are reported in Fig. S1–S5. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nature structural & molecular biology. 2012;19:568–576. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- Akabayov B, Akabayov SR, Lee SJ, Wagner G, Richardson CC. Impact of macromolecular crowding on DNA replication. Nature communications. 2013;4:1615. doi: 10.1038/ncomms2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade MA, Bork P. HEAT repeats in the Huntington’s disease protein. Nat Genet. 1995;11:115–116. doi: 10.1038/ng1095-115. [DOI] [PubMed] [Google Scholar]
- Asano K, Clayton J, Shalev A, Hinnebusch AG. A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNA(Met) is an important translation initiation intermediate in vivo. Genes Dev. 2000;14:2534–2546. doi: 10.1101/gad.831800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Hama C, Inoue S, Moriwaki H, Mizobuchi K. The plasmid ColIb-P9 antisense Inc RNA controls expression of the RepZ replication protein and its positive regulator repY with different mechanisms. The Journal of biological chemistry. 1999a;274:17924–17933. doi: 10.1074/jbc.274.25.17924. [DOI] [PubMed] [Google Scholar]
- Asano K, Krishnamoorthy T, Phan L, Pavitt GD, Hinnebusch AG. Conserved bipartite motifs in yeast eIF5 and eIF2Bepsilon, GTPase-activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. Embo J. 1999b;18:1673–1688. doi: 10.1093/emboj/18.6.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asano K, Phan L, Anderson J, Hinnebusch AG. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J Biol Chem. 1998;273:18573–18585. doi: 10.1074/jbc.273.29.18573. [DOI] [PubMed] [Google Scholar]
- Asano K, Sachs MS. Translation factor control of ribosome conformation during start codon selection. Genes & development. 2007;21:1280–1287. doi: 10.1101/gad.1562707. [DOI] [PubMed] [Google Scholar]
- Asano K, Shalev A, Phan L, Nielsen K, Clayton J, Valasek L, Donahue TF, Hinnebusch AG. Multiple roles for the C-terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. Embo J. 2001;20:2326–2337. doi: 10.1093/emboj/20.9.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniossek C, Schutz P, Bumann M, Limacher A, Uson I, Baumann U. The crystal structure of the carboxy-terminal domain of human translation initiation factor eIF5. J Mol Biol. 2006;360:457–465. doi: 10.1016/j.jmb.2006.05.021. [DOI] [PubMed] [Google Scholar]
- Conte MR, Kelly G, Babon J, Sanfelice D, Youell J, Smerdon SJ, Proud CG. Structure of the eukaryotic initiation factor (eIF) 5 reveals a fold common to several translation factors. Biochemistry. 2006;45:4550–4558. doi: 10.1021/bi052387u. [DOI] [PubMed] [Google Scholar]
- Das S, Maiti T, Das K, Maitra U. Specific interaction of eukaryotic translation initiation factor 5 (eIF5) with the beta-subunit of eIF2. J Biol Chem. 1997;272:31712–31718. doi: 10.1074/jbc.272.50.31712. [DOI] [PubMed] [Google Scholar]
- Das S, Maitra U. Mutational analysis of mammalian translation initiation factor 5 (eIF5): role of interaction between the beta subunit of eIF2 and eIF5 in eIF5 function in vitro and in vivo. Mol Cell Biol. 2000;20:3942–3950. doi: 10.1128/mcb.20.11.3942-3950.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete CA, Applefield DJ, Blakely SA, Shirokikh N, Pestova T, Lorsch JR, Hinnebusch AG. The eIF1A C-terminal domain promotes initiation complex assembly, scanning and AUG selection in vivo. Embo J. 2005;24:3588–3601. doi: 10.1038/sj.emboj.7600821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete CA, Mitchell SF, Cherkasova VA, Applefield D, Algire MA, Maag D, Saini AK, Lorsch JR, Hinnebusch AG. N- and C-terminal residues of eIF1A have opposing effects on the fidelity of start codon selection. Embo J. 2007;26:1602–1614. doi: 10.1038/sj.emboj.7601613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, von der Haar T, Singh CR, Ii M, Li B, Hinnebusch AG, McCarthy JE, Asano K. The yeast eukaryotic initiation factor 4G (eIF4G) HEAT domain interacts with eIF1 and eIF5 and is involved in stringent AUG selection. Mol Cell Biol. 2003;23:5431–5445. doi: 10.1128/MCB.23.15.5431-5445.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiology and molecular biology reviews: MMBR. 2011;75:434–467. doi: 10.1128/MMBR.00008-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Loughran G, Sachs MS, Atkins JF. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1) Proc Natl Acad Sci U S A. 2010;107:18056–18060. doi: 10.1073/pnas.1009269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B, Udagawa T, Singh CR, Asano K. Yeast phenotypic assays on translational control. Methods in enzymology. 2007;429:105–137. doi: 10.1016/S0076-6879(07)29006-8. [DOI] [PubMed] [Google Scholar]
- Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev. 2003;17:2786–2797. doi: 10.1101/gad.1141803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Steitz TA. The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature. 2013;500:307–311. doi: 10.1038/nature12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch JR, Dever TE. Molecular view of 43 S complex formation and start site selection in eukaryotic translation initiation. J Biol Chem. 2010;285:21203–21207. doi: 10.1074/jbc.R110.119743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughran G, Sachs MS, Atkins JF, Ivanov IP. Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic acids research. 2012;40:2898–2906. doi: 10.1093/nar/gkr1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna RE, Arthanari H, Hiraishi H, Nanda J, Martin-Marcos P, Markus MA, Akabayov B, Milbradt AG, Luna LE, Seo HC, et al. The C-Terminal Domain of Eukaryotic Initiation Factor 5 Promotes Start Codon Recognition by Its Dynamic Interplay with eIF1 and eIF2beta. Cell reports. 2012;1:689–702. doi: 10.1016/j.celrep.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag D, Algire MA, Lorsch JR. Communication between eukaryotic translation initiation factors 5 and 1A within the ribosomal pre-initiation complex plays a role in start site selection. J Mol Biol. 2006;356:724–737. doi: 10.1016/j.jmb.2005.11.083. [DOI] [PubMed] [Google Scholar]
- Marintchev A, Frueh D, Wagner G. NMR methods for studying protein-protein interactions involved in translation initiation. Methods Enzymol. 2007;430:283–331. doi: 10.1016/S0076-6879(07)30012-8. [DOI] [PubMed] [Google Scholar]
- Martin-Marcos P, Cheung YN, Hinnebusch AG. Functional elements in initiation factors 1, 1A and 2β discriminate against poor AUG context and non-AUG start codons. Mol Cell Biol. 2011;31:4814–4831. doi: 10.1128/MCB.05819-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda JS, Cheung YN, Takacs JE, Martin-Marcos P, Saini AK, Hinnebusch AG, Lorsch JR. eIF1 controls multiple steps in start codon recognition during eukaryotic translation initiation. J Mol Biol. 2009;394:268–285. doi: 10.1016/j.jmb.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda JS, Saini AK, Munoz AM, Hinnebusch AG, Lorsch JR. Coordinated Movements of Eukaryotic Translation Initiation Factors eIF1, eIF1A and eIF5 Trigger Phosphate Release from eIF2 in response to Start Codon Recognition by the Ribosomal Pre-initiation Complex. J Biol Chem. 2013;88:5316–5329. doi: 10.1074/jbc.M112.440693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen DS, Savner EM, Mathew A, Zhang F, Krishnamoorthy T, Phan L, Hinnebusch AG. Domains of eIF1A that mediate binding to eIF2, eIF3 and eIF5B and promote ternary complex recruitment in vivo. Embo J. 2003;22:193–204. doi: 10.1093/emboj/cdg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Borukhov SI, Hellen CU. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan L, Zhang X, Asano K, Anderson J, Vornlocher HP, Greenberg JR, Qin J, Hinnebusch AG. Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol Cell Biol. 1998;18:4935–4946. doi: 10.1128/mcb.18.8.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- Reibarkh M, Yamamoto Y, Singh CR, del Rio F, Fahmy A, Lee B, Luna RE, Ii M, Wagner G, Asano K. Eukaryotic initiation factor (eIF) 1 carries two distinct eIF5-binding faces important for multifactor assembly and AUG selection. J Biol Chem. 2008;283:1094–1103. doi: 10.1074/jbc.M708155200. [DOI] [PubMed] [Google Scholar]
- Saini AK, Nanda JS, Lorsch JR, Hinnebusch AG. Regulatory elements in eIF1A control the fidelity of start codon selection by modulating tRNA(i)(Met) binding to the ribosome. Genes Dev. 2010;24:97–110. doi: 10.1101/gad.1871910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh CR, Watanabe R, Chowdhury W, Hiraishi H, Murai MJ, Yamamoto Y, Miles D, Ikeda Y, Asano M, Asano K. Sequential Eukaryotic Translation Initiation Factor 5 (eIF5) Binding to the Charged Disordered Segments of eIF4G and eIF2beta Stabilizes the 48S Preinitiation Complex and Promotes Its Shift to the Initiation Mode. Mol Cell Biol. 2012;32:3978–3989. doi: 10.1128/MCB.00376-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokabe M, Fraser CS, Hershey JW. The human translation initiation multi-factor complex promotes methionyl-tRNAi binding to the 40S ribosomal subunit. Nucleic Acids Res. 2012;40:905–913. doi: 10.1093/nar/gkr772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L, Mathew AA, Shin BS, Nielsen KH, Szamecz B, Hinnebusch AG. The yeast eIF3 subunits TIF32/a, NIP1/c, and eIF5 make critical connections with the 40S ribosome in vivo. Genes Dev. 2003;17:786–799. doi: 10.1101/gad.1065403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Z, Xue Y, Xu H, Gong W. Crystal structure of the C-terminal domain of S.cerevisiae eIF5. J Mol Biol. 2006;359:1–9. doi: 10.1016/j.jmb.2006.03.037. [DOI] [PubMed] [Google Scholar]
- Weisser M, Voigts-Hoffmann F, Rabl J, Leibundgut M, Ban N. The crystal structure of the eukaryotic 40S ribosomal subunit in complex with eIF1 and eIF1A. Nature structural & molecular biology. 2013;20:1015–1017. doi: 10.1038/nsmb.2622. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Singh CR, Marintchev A, Hall NS, Hannig EM, Wagner G, Asano K. The eukaryotic initiation factor (eIF) 5 HEAT domain mediates multifactor assembly and scanning with distinct interfaces to eIF1, eIF2, eIF3, and eIF4G. Proc Natl Acad Sci U S A. 2005;102:16164–16169. doi: 10.1073/pnas.0507960102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Marintchev A, Kolupaeva VG, Unbehaun A, Veryasova T, Lai SC, Hong P, Wagner G, Hellen CU, Pestova TV. Position of eukaryotic translation initiation factor eIF1A on the 40S ribosomal subunit mapped by directed hydroxyl radical probing. Nucleic Acids Res. 2009;37:5167–5182. doi: 10.1093/nar/gkp519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.