Abstract
Streptococcus mutans, the primary aetiological agent of dental caries, possesses an YjeE-like protein that is encoded by locus SMU.409, herein designated brpB. In this study, a BrpB-deficient mutant, JB409, and a double mutant deficient of BrpB and BrpA (a paralogue of the LytR–CpsA–Psr family of cell wall-associated proteins), JB819, were constructed and characterized using function assays and microscopy analysis. Both JB409 and JB819 displayed extended lag phases and drastically slowed growth rates during growth in brain heart infusion medium as compared to the wild-type, UA159. Relative to UA159, JB409 and JB819 were more than 60- and 10-fold more susceptible to acid killing at pH 2.8, and more than 1 and 2 logs more susceptible to hydrogen peroxide, respectively. Complementation of the deficient mutants with a wild-type copy of the respective gene(s) partly restored the acid and oxidative stress responses to a level similar to the wild-type. As compared to UA159, biofilm formation by JB409 and JB819 was drastically reduced (P<0.001), especially during growth in medium containing sucrose. Under a scanning electron microscope, JB409 had significantly more giant cells with an elongated, rod-like morphology, and JB819 formed marble-like super cells with apparent defects in cell division. As revealed by transmission electron microscopy analysis, BrpB deficiency in both JB409 and JB819 resulted in the development of low electron density patches and formation of a loose nucleoid structure. Taken together, these results suggest that BrpB likely functions together with BrpA in regulating cell envelope biogenesis/homeostasis in Strep. mutans. Further studies are under way to elucidate the mechanism that underlies the BrpA- and BrpB-mediated regulation.
Introduction
Streptococcus mutans, a major causative agent of dental caries (Loesche, 1986), lives primarily on the tooth surface in tenacious biofilms commonly known as dental plaque. The oral cavity is a microenvironment that is known for its unpredictable fluctuations in conditions such as carbohydrate source and availability, acid and alcohol concentration, oxygen tension, sheering forces, and the presence of antimicrobial agents (Burne, 1998; Lemos & Burne, 2008). This bacterium has evolved strategies to endure and cope with drastic changes in the environmental conditions (Lemos & Burne, 2008). The cell envelope plays a vital role during these processes as it protects the cell from the environment, acts as a molecular sieve, and provides a platform for components of the cell involved in sensing and transmission of environmental signals. In addition, maintenance and turnover of the cell envelope govern the fundamental properties of cell growth, shape and division. Ensuring envelope integrity is therefore crucial for bacterial cells to survive.
Our recent studies have shown that BrpA, for biofilm regulatory protein A, a paralogue of the LytR–CpsA–Psr (LCP) family of cell wall-associated proteins highly conserved in Gram-positive bacteria (Hübscher et al., 2008), plays a major role in cell envelope biogenesis/homeostasis in Strep. mutans (Bitoun et al., 2012a, 2013). Relative to the wild-type, Strep. mutans strains deficient of BrpA displayed significant increases in susceptibility to cell envelope antimicrobials, and drastic reductions in acid and oxidative stress tolerance responses (Bitoun et al., 2012a; Chatfield et al., 2005; Wen et al., 2006). In addition, the deficient mutants also had an elevated autolysis rate and major defects in cell division, cell morphology and biofilm formation (Bitoun et al., 2012a, 2013; Wen et al., 2006). Similar results were also observed with Psr, the only other LCP paralogue Strep. mutans possesses (Bitoun et al., 2013). A Psr-deficient, BrpA-down mutant exists primarily in long chains of dividing, giant cells with multiple septa, and deficiency of both BrpA and Psr appears to be lethal in Strep. mutans under the conditions studied.
The YjeE-like proteins are a unique, new family of P-loop ATP/GTPase proteins widespread in virtually all eubacteria, but not in archaea and eukaryotes (Teplyakov et al., 2002), although their exact function in cellular physiology remains unclear (Allali-Hassani et al., 2004; Karst et al., 2009). In Escherichia coli, YjeE is required for growth, and strains with decreased expression of YjeE had an elongated lag phase and a decreased growth rate (Allali-Hassani et al., 2004). Repression of YjeE expression also caused alterations of cell morphology, including massive cells with curving and branching, and an unusual distribution of the cell nucleoid (Handford et al., 2009). Unlike E. coli, however, Bacillus subtilis deficient of YdiB orthologue was shown to be viable, but had a much reduced growth rate, when compared to the wild-type (Karst et al., 2009). Recombinant proteins of YjeE from E. coli and Haemophilus influenzae, and YdiB from B. subtilis, all possessed low ATPase activity (Allali-Hassani et al., 2004; Karst et al., 2009; Teplyakov et al., 2002), and in B. subtilis ATPase activity was shown to be essential for the function of YdiB protein in vivo (Karst et al., 2009). More recently, an in vitro study has also provided evidence that in E. coli YjeE is part of a three essential protein complex required for biosynthesis of threonylcarbamoyl adenosine (t6A), a modified tRNA nucleoside involved in codon decoding (Deutsch et al., 2012). However, certain component(s) of the complex were shown to be species specific, which again suggests diversity in function of the YjeE-like proteins in different species.
Strep. mutans possesses a gene in locus SMU.409, here designated brpB, which encodes a protein with homology to YjeE of E. coli and YdiB of B. subtilis (Allali-Hassani et al., 2004; Karst et al., 2009). Previous studies showed that brpB can be co-transcribed with brpA, suggesting likely involvement of its product, BrpB, in BrpA-regulated cell envelope biogenesis/homeostasis (Bitoun et al., 2012a, 2013). The objective of this study was to determine the role of BrpB in the cellular physiology and fitness of Strep. mutans. Here a BrpB-deficient mutant, JB409, and a BrpB/BrpA-deficient double mutant, JB819, were constructed by allelic replacement. Characterization of these mutants revealed that BrpB deficiency weakened the ability of Strep. mutans to cope with both acid and oxidative stress relative to the parent strain; biofilm formation by the deficient mutants was also greatly compromised. In addition, electron microscopy analyses also showed BrpB deficiency in Strep. mutans caused alteration in cell size and morphology of the deficient mutants.
Methods
Plasmids, bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Strep. mutans strains were maintained in brain heart infusion (BHI) medium (Difco Laboratories). Solid media were prepared similarly, but Bacto agar (Difco Laboratories) was added at a concentration of 1.5 % (w/v). When needed, kanamycin (Kan, 1 mg ml−1) and/or spectinomycin (Spe) (1 mg ml−1) were added to the growth medium. Unless stated otherwise, cultures were grown aerobically in a 37 °C chamber containing 5 % CO2 under static conditions. For growth studies, overnight cultures were diluted 1 : 100 into fresh BHI and allowed to continue growing until mid-exponential phase (OD600 0.4–0.5), when they were further diluted 1 : 100 and the optical density of the cultures at 600 nm was continuously monitored using a Bioscreen C (Oy Growth Curves) at 37 °C with and without sterile mineral oil overlay (Bitoun et al., 2012b). For biofilm assays, Strep. mutans were grown in modified biofilm medium (BM) with glucose (20 mM, BMG), sucrose (10 mM, BMS) or glucose (18 mM) plus sucrose (2 mM) (BMGS) as the supplementary carbohydrate sources (Li et al., 2002; Loo et al., 2000).
Table 1. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Relevant characteristics | Source or reference |
| Strains | ||
| Strep. mutans UA159 | Wild-type, serotype c | Ajdić et al. (2002) |
| Strep. mutans JB409 | Derivative of UA159, ΔbrpB (SMU.409), Kanr | This study |
| Strep. mutans JB409C | JB409 carrying pDL278:brpB, Sper, Kanr | This study |
| Strep. mutans JB819 | Derivative of UA159, ΔbrpBA (SMU.409-410), Kanr | This study |
| Strep. mutans JB819C | JB819 carrying pDL278:brpBA, Sper, Kanr | This study |
| Plasmids | ||
| pDL278 | Shuttle vector, Sper | LeBlanc & Lee (1991) |
| pDL278:brpB | Shuttle carrying brpB, Sper | This study |
| pDL278:brpBA | Shuttle carrying brpBA, Sper | This study |
Kanr and Sper are kanamycin and spectinomycin resistance, respectively.
Construction of mutants and complemented strains.
To construct the BrpB-deficient mutant, a PCR-ligation-mutagenesis strategy was used as described elsewhere (Lau et al., 2002; Wen & Burne, 2004). Briefly, the 5′ and 3′ regions flanking brpB were amplified by PCR using Phusion high-fidelity DNA polymerase (New England Biolabs) with the gene-specific primers shown in Table 2. Following proper restriction enzyme digestions, the flanking regions were ligated to a non-polar Kan-resistance element (Zeng et al., 2006) that was digested similarly. The resulting ligation mixtures were used to directly transform Strep. mutans UA159 in the presence of competence stimulating peptide (CSP) (Li et al., 2001). An allelic replacement mutant, JB409, with brpB deficiency was isolated on BHI-Kan plates, and further confirmed using PCR and sequencing. For complementation of mutant JB409, the brpB coding sequence plus its cognate promoter region were amplified by PCR, digested and cloned directly into shuttle vector pDL278. After sequence confirmation, the resulting construct, pDL:brpB, was then used to transform JB409, generating complemented strain, JB409C. To construct the brpB and brpA double mutant, a similar PCR-ligation-mutagenesis strategy was used with the 5′ region flanking upstream of brpB and the 3′ region flanking downstream of brpA. The brpBA operon deletional mutant resulted in the removal of nucleotide 47 within the ORF of brpB through nucleotide 1208 of brpA. Allelic replacement mutant, JB819, with brpB and brpA deficiencies, was isolated on BHI-Kan plates, and further confirmed using PCR and sequencing. JB819C, a complemented strain that carried the brpBA coding sequence plus its cognate promoter region (Bitoun et al., 2012a) in shuttle vector pDL278, was constructed in a similar manner to that described above.
Table 2. Primers used in this study.
Underlined nucleotides are restriction sites engineered for cloning. qPCR, Quantitative PCR.
| Name | Nucleotide sequence (5′-3′) | Application |
| 5′-5′ 409 up | 5′-TACAGCTAACTCTTCTGCAACACCATC-3′ | 5′ fragment for brpB and brpBA mutation |
| 5′-3′ 409 RI | 5′-TGGCTCTTGGACAGAGAATTCGTCA-3′ | 5′ fragment for brpB and brpBA mutation |
| 3′-5′ 409 RI | 5′-TGCGGAATTCGAGCATGACTGA-3′ | 3′ fragment for brpB mutation |
| 3′-3′ 409 | 5′-AGCTTTAGACAGTGTCAGCAAGTATCGT-3′ | 3′ fragment for brpB mutation |
| 3′-5′ 409-410 RI | 5′-TGCTGCAGGAACGGGAATTCGTA-3′ | 3′ fragment for brpBA mutation |
| 3′-3′ 409-410 | 5′-TCATAGGCCTCAATGGTGCAGGA-3′ | 3′ fragment for brpBA mutation |
| 5′ 409comp RI | 5′-GCATATATAGGAATTCAGATAAGGCTGAGCT-3′ | brpB and brpBA complementation |
| 3′ 409comp Hd | 5′-CAGTTATATGAGGGCAAGCTTCATCT-3′ | brpB complementation |
| 3′ 409-410comp Bm | 5′-GTATAACTGAAAAACCGGGATCCTTCCA-3′ | brpBA complementation |
| brpB Fw | 5′- TAAGAGTCCAACCTATACGATT-3′ | 138 bp, qPCR of brpB |
| brpB Rev | 5′-ATCACTGTTACGCCATCT-3′ | 138 bp, qPCR of brpB |
| brpA Fw | 5′-CGTGAGGTCATCAGCAAGGTC-3′ | 148 bp, qPCR of brpA |
| brpA Rev | 5′-CGCTGTACCCCAAAAGTTTAGG-3′ | 148 bp, qPCR of brpA |
| 16S rRNA Fw | 5′-CACACCGCCCGTCACACC-3′ | 160 bp, qPCR of SMU_r01 (16S rRNA) |
| 16S rRNA Rev | 5′-CAGCCGCACCTTCCGATACG-3′ | 160 bp, qPCR of SMU_r01 (16S rRNA) |
Induced autolysis assay.
For the autolysis assay, Strep. mutans strains were grown in BHI overnight. Bacterial cells were harvested by centrifugation at 3250 g, at 4 °C for 10 min, washed once with PBS (100 mM potassium phosphate buffer, pH 7.2, sodium chloride, 0.9 %, w/v), and then resuspended in PBS containing 0.2 % (v/v) Triton X-100 (Sigma) (Bitoun et al., 2013; Wen & Burne, 2002). The optical density of the cell suspension was then monitored automatically using a Bioscreen C at 37 °C every 30 min with moderate shaking (10 s) before measurement.
Acid and hydrogen peroxide killing assays.
The impact of brpB and brpBA deficiencies on the ability of Strep. mutans to withstand acid and hydrogen peroxide stress was determined by using acid killing and hydrogen peroxide challenge assays as described elsewhere (Wen & Burne, 2004). In brief, planktonic cultures were grown in BHI until early exponential phase (OD600 0.3) and biofilms cells were grown in BMGS on sterile glass slides in 50 ml Falcon tubes for 24 h (Wen et al., 2010). The cells were then collected by centrifugation at 3250 g at 4 °C for 5 min, and washed twice with 0.1 M glycine buffer, pH 7.0. For the acid killing assay, the washed cells were resuspended in 0.1 M glycine buffer, pH 2.8, and incubated at room temperature for periods of 15, 30, 45 and 60 min. For preparation of cells that had undergone an adaptive acid tolerance response, cultures with an OD600 0.3 were washed once with 0.1 M glycine buffer, pH 7.0, as described above, and the cell pellets were resuspended in fresh BHI that was adjusted to pH 5.0 with HCl. Following an additional hour of incubation, cells were harvested, washed and subjected to acid killing as described above. For the hydrogen peroxide killing assays, washed cells were resuspended in 0.1 M glycine buffer, pH 7.0, and hydrogen peroxide was added at a final concentration of 0.2 % (58 mM, final concentration). Surviving cells were appropriately diluted, plated on BHI plates in triplicate and incubated in 5 % CO2 at 37 °C for 24 to 48 h.
Biofilm analysis.
Strep. mutans biofilms were grown in semi-defined BM (BMG, BMS and BMGS) as described elsewhere (Li et al., 2002; Loo et al., 2000). Overnight cultures were inoculated 1 : 100 into fresh BHI and allowed to grow until OD600 0.5 when the biofilm media was inoculated by 1 : 100 dilutions. For quantitative and structural analysis, biofilms were allowed to grow on hydroxylapatite (HA) discs that were placed vertically in fabricated orthodontic wire holders made to fit 24-well plates in a 5 % CO2 incubator at 37 °C under static conditions (Lemos et al., 2010). Biofilm samples were collected after 24 h and were stained with either a BacLight Live/Dead bacterial viability kit (Invitrogen), SYTO 62 (Invitrogen) or SYTO 59/Alexa488 Concanavalin A (Invitrogen). Optical dissections of the biofilms were carried out using a laser scanning confocal microscope (Olympus Fluoview BX61) with pixel size on x- and y-axis representing 0.414 µm, and the z-axis was set at 2.0 microns. At least three independent image stacks were acquired at ×600 magnification on the HA surface using a ×60 water-immersion objective lens for each sample at random positions on the HA surface. The resulting image stacks were analysed using Slidebook 5.0 (Olympus). comstat 2.0 was used to quantify the biovolume, surface area, mean thickness and maximum thickness of the biofilms as described elsewhere (Heydorn et al., 2000; Wen et al., 2011). Biovolume is defined as the biofilm volume (µm3) per unit area (µm2) (Heydorn et al., 2000).
Electron microscopy analysis.
For field emission-scanning electron microscopy (FE-SEM) analysis, Strep. mutans overnight cultures were grown in BMG, BMGS or BMS on HA discs that were placed horizontally in 24-well plates as described elsewhere (Bitoun et al., 2011; Wen & Burne, 2004). After 24 h of growth, the HA discs were removed and washed twice with PBS, pH 7.4, before overnight fixation with 2.5 % glutaraldehyde (Polysciences) at 4 °C. The samples were dehydrated using a graded ethanol series, dried using a critical point dryer and then coated with carbon using a sputter coater. Microscopy analysis was performed with a Hitachi S-4800 high-resolution microscope (at Tulane University, New Orleans, LA, USA).
For transmission electron microscopy (TEM) analysis, Strep. mutans strains were grown in BHI until mid-exponential phase, harvested by centrifugation at 3250 g at 4 °C for 10 min, washed twice with PBS and then fixed in 2.5 % glutaraldehyde/2 % paraformaldehyde (Polysciences) in PBS for 1 h at room temperature (Bitoun et al., 2013). Cells were washed in PBS and post-fixed in 1 % osmium tetroxide (Polysciences) for 1 h. Samples were then rinsed extensively in dH20 prior to en bloc staining with 1 % aqueous uranylacetate (Ted Pella Inc.) for 1 h. The cells were then washed with dH20 and dehydrated in a graded series of ethanol solutions and embedded in Eponate 12 resin (Ted Pella Inc.). Sections of 90–100 nm were prepared, stained with uranyl acetate and lead citrate, and viewed under a JEOL 1200 EX transmission electron microscope (JEOL USA) equipped with an AMT 8 megapixel digital camera (Advanced Microscopy Techniques).
Statistical analysis.
Quantitative data were analysed using the paired Student’s t-test.
Results
BrpB is a member of YjeE family of P-loop ATPase and the brpA/brpB cluster is highly conserved among streptococci
As annotated (Ajdić et al., 2002), the brpB gene (locus SMU.409) encodes a polypeptide with high similarity to YjeE of E. coli, YdiB of B. subtilis and other members of the highly conserved UPF0079 family of uncharacterized P-loop hydrolases (www.expasy.org) (Allali-Hassani et al., 2004; Handford et al., 2009; Karst et al., 2009; Teplyakov et al., 2002). As shown by pairwise sequence alignment analysis, BrpB displays the highest similarity in amino acid composition to the streptococcal homologues, and the least homology was observed between BrpB and YjeE of E. coli (data not shown). However, like YjeE of E. coli, BrpB and its streptococcal homologues all show conservation of a Walker A motif (GX4GKTT) that is typical of nucleotide-binding proteins, including ATP/GTPase (data not shown) (Hanson & Whiteheart, 2005).
Our previous studies showed that locus SMU.409 could be co-transcribed with brpA (SMU.410) despite the presence of a large (323 bp) intergenic region (Bitoun et al., 2012a). Based upon this genetic linkage and the function relationship as suggested by data presented here, the gene encoded in SMU.409 was designated brpB. Indeed, analysis of the genetic structure also revealed that the regions flanking the loci for the YjeE homologues are highly conserved among streptococci, although differences exist between Strep. mutans and the other streptococci (Fig. S1, available in Microbiology Online). Like Strep. mutans, the gene encoding the YjeE homologue in locus SP_1944 of Streptococcus pneumoniae is located upstream of lytR (locus SP_1942) (Johnsborg & Håvarstein, 2009). Unlike Strep. mutans, however, the yjeE locus in Strep. pneumoniae represents the leading gene of an apparent three gene operon that includes in the middle locus SP_1943 encoding a putative acyltransferase, followed by lytR. Similar features were also found in group A Streptococcus pyogenes, group B Streptococcus agalactiae and all other streptococcal species, including other oral streptococci whose genome sequences are available and accessible (Fig. S1). Interestingly, when compared to other streptococci, Strep. mutans species are the only streptococci lacking the acyltransferase locus, a likely result of genetic deletions during the evolutionary processes and an attributing factor to the presence of a large intergenic region between brpA and brpB (Bitoun et al., 2012a). Unlike streptococci, however, no such linkage could be identified between the loci for YjeE (or YdiB for B. subtilis) homologues and the genes for any LCP proteins in Staphylococcus aureus, B. subtilis and other Gram-positive bacteria (Eberhardt et al., 2012; Johnsborg & Håvarstein, 2009; Kawai et al., 2011; Over et al., 2011).
Deficiency of BrpB causes major growth defects in Strep. mutans
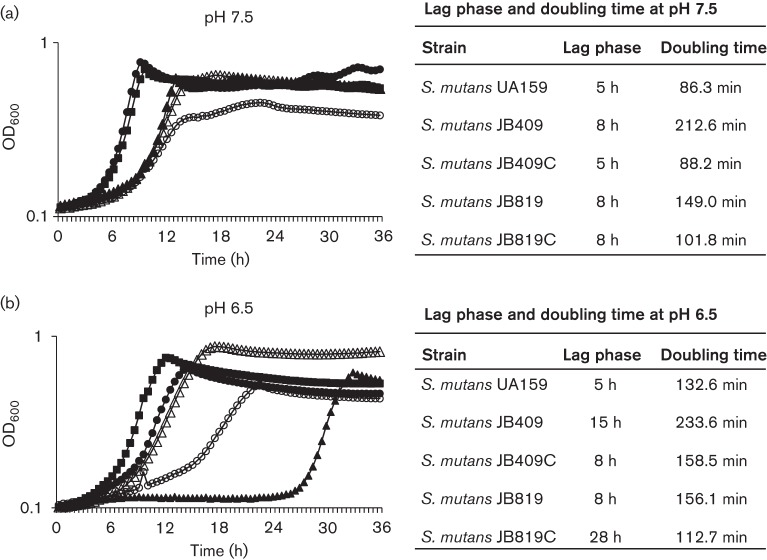
A BrpB-deficient Strep. mutans strain, JB409, was constructed with a region of 380 bp from nucleotides 47 to 427 of its coding sequence (435 bp total) relative to the start codon deleted and replaced with a nonpolar Kan-resistance cassette. As compared to the parent strain, UA159, the BrpB-deficient mutant, JB409, took an additional day to form discernible colonies on solid medium (data not shown), suggesting growth defects as a result of BrpB deficiency. During planktonic growth in BHI with pH adjusted to 7.5 (Fig. 1), the maximal culture density (OD600) of JB409 was found to be reduced by more than 40 %, when compared to UA159. Also, relative to UA159, JB409 had an extended lag phase and increased its doubling time by more than 2 h (Fig. 1a). Such defects were further magnified during growth in BHI adjusted to pH 6.5 (Fig. 1b). As expected, complementation in trans with a wild-type copy of brpB plus its cognate promoter in multiple copy shuttle vector pDL278 was able to restore the slow growth phenotype to a level similar to UA159 (Fig. 1a). A BrpB/BrpA-deficient double mutant, JB819, was constructed with the coding sequences of both brpB and brpA deleted (from nucleotide 47 of brpB relative to the start codon through nucleotide 1208 of brpA) and replaced with a nonpolar Kan-resistance cassette (Tables 1 and 2). JB819 also showed major defects in growth with both its lag phase and doubling time significantly increased, when compared to the wild-type. Interestingly, however, the doubling time of the BrpA/BrpB-deficient double mutant was significantly faster than that of the BrpB single mutant (Fig. 1). Previous studies showed that BrpA deficiency did not result in any major effect on cell growth when cells were grown in regular BHI (Bitoun et al., 2013; Wen & Burne, 2002; Wen et al., 2006). When complemented with a wild-type copy of the brpA/brpB operon plus their cognate promoter(s) in multiple copy shuttle vector pDL:brpB/A, the complemented strain, JB819C, showed a further decreased doubling time with a level closer to the wild-type (Fig. 1). Interestingly, during growth in BHI adjusted to pH 6.5, however, the complemented strain, JB819C, demonstrated a severely delayed lag phase although the doubling time was shorter than that of UA159 (Fig. 1b). Paraquat (methyl viologen; Sigma) is commonly used to test for the ability of cultures to adapt to superoxide stress. The addition of paraquat did not further exacerbate the growth deficiencies; albeit, all strains grew at slower, but proportional rates in the presence of paraquat (data not shown).
Fig. 1.
Growth study of Strep. mutans. Strep. mutans wild-type, UA159 (▪), BrpB-deficient mutant, JB409 (○), BrpB-complemented strain, JB409C (•), BrpB- and BrpA-deficient double mutant, JB819 (Δ), and BrpB- and BrpA-complemented strain, JB819C (▴), were grown in BHI adjusted to pH 7.5 (a) and pH 6.5 (b) in a Bioscreen C at 37 °C with a sterile mineral oil overlay (50 µl), and the optical density of the cultures was continuously monitored at 600 nm. Results showed that BrpB deficiency in JB409 greatly affects bacterial cell growth. Interestingly, the BrpB and BrpA double deficiency in JB819 partially alleviated the slow growth phenotype of the BrpB-deficient mutant, JB409. During growth in BHI adjusted to pH 6.5, the complemented strain, JB819C, showed an extremely delayed lag phase, when compared to the wild-type and the parent strain, JB819. The data presented here are representative of three independent experiments. The lag phase duration and the doubling time of each strain under the conditions tested are expressed as means and presented in the tables to the right of the growth curves.
BrpB deficiency results in exacerbated sensitivity to acid and oxidative stress
It was apparent that during growth in BHI adjusted to pH 6.5, the deficient mutants, JB409 and JB819, were challenged to adapt to the acidic conditions (Fig. 1b). When analysed by acid killing with mid-exponential phase cultures grown in BHI adjusted to pH 7.4, the survival rate of JB409 and JB819 was decreased by 60- and 11-fold, respectively, after 30 min of incubation in 0.1 M glycine buffer of pH 2.8, as compared to UA159 (Fig. 2a). Similar results were obtained using planktonic cells grown in regular BHI and cells derived from 48 h biofilms grown in BMGS on glass slides (data not shown). As expected, complementation with the wild-type brpB and brpA/brpB in a multiple copy plasmid in strains JB409C and JB819C, respectively, partly restored the acid-sensitive phenotypes to a level similar to that of UA159 (Fig. 2a).
Fig. 2.
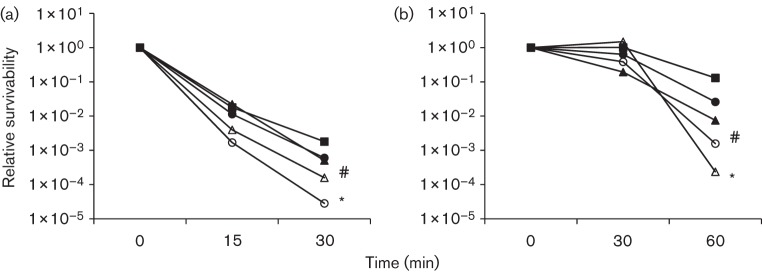
Acid killing assay of non-adapted (a) and adapted (b) Strep. mutans strains. (a) Strep. mutans wild-type, UA159 (▪), BrpB-deficient mutant, JB409 (○), BrpB-complemented strain, JB409C (•), BrpB- and BrpA-deficient double mutant, JB819 (Δ), and BrpB- and BrpA-complemented strain, JB819C (▴), were grown in BHI until mid-exponential phase (OD600 0.3). Bacterial cells were then harvested by centrifugation and washed twice in 0.1 M glycine, pH 7.0. Acid killing was performed by incubating the bacterial cells in 0.1 M glycine buffer, pH 2.8, for periods of 15, 30 and 45 min. Data presented here are representative of three independent experiments; significant difference is indicated by *, P<0.001, and #, P<0.01, as compared to the wild-type under similar conditions. (b) For adaptive conditions, bacterial cells were incubated in BHI, pH 5.0, for 1 h prior to acid killing. Other details are the same as for (a).
For adaptive acid tolerance response, Strep. mutans strains were allowed to adapt and grow in BHI adjusted to pH 5.0 for 1 h before being subjected to acid killing (Wen & Burne, 2004). The results showed that like wild-type UA159, the BrpB-deficient mutant, JB409, also significantly increased its survival rate after 1 h incubation in BHI adjusted to pH 5.0 (Fig. 2b). Relative to UA159, however, the survival rate of JB409 was still 2 logs less. Unlike UA159 and JB409, the BrpB/BrpA-deficient double mutant, JB819, did not show any adaptive response after initial exposure to low pH. Consequently, the survival rate of JB819 was further reduced by more than 500-fold after 60 min of acid killing (Fig. 2b).
When challenged with hydrogen peroxide (0.2 %, w/v, or 58 mM), the BrpB-deficient strain, JB409, and the BrpB/BrpA-deficient double mutant, JB819, were at least 1 and 2 logs more sensitive than the wild-type, UA159 (Fig. 3). Complementation with a wild-type copy of brpB plus its cognate promoter in strain JB409C fully restored the phenotype to the wild-type UA159 (Fig. 3). Interestingly, complementation in strain JB819C, which carries a wild-type copy of both brpB and brpA plus their promoter(s), showed little effect in reducing the increased sensitivity to hydrogen peroxide observed in JB819.
Fig. 3.
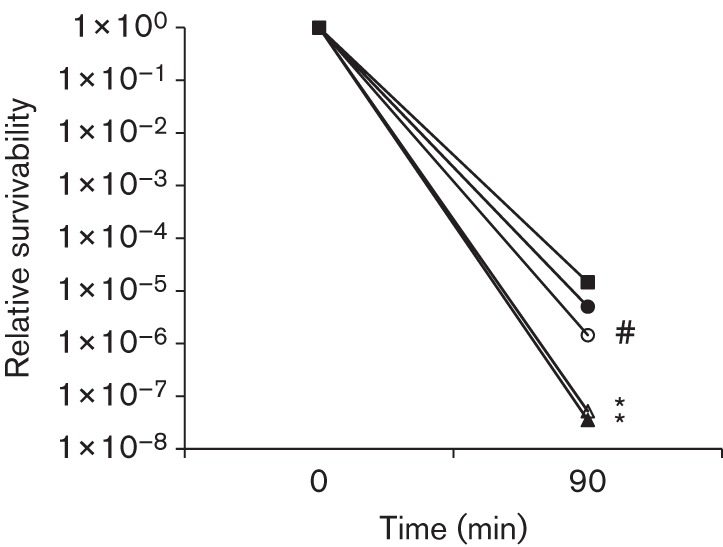
Hydrogen peroxide killing assay. Strep. mutans wild-type, UA159 (▪), BrpB-deficient mutant, JB409 (○), BrpB-complemented strain, JB409C (•), BrpB- and BrpA-deficient double mutant, JB819 (Δ), and BrpB- and BrpA-complemented strain, JB819C (▴), were grown in regular BHI broth, harvested during mid-exponential phase (OD600 0.3), and then washed twice with 0.1 M glycine buffer before being subjected to hydrogen peroxide killing by incubating the cells in 0.1 M glycine buffer with 0.2 % (w/v) hydrogen peroxide for a period of 90 and 110 min. Results showed that BrpB deficiency weakened the ability of the mutants, JB409 and JB819, to withstand hydrogen peroxide stress. Data presented here are representative of three independent experiments with significant difference being indicated by *, P<0.001, and #, P<0.01, as compared to the wild-type under similar conditions.
BrpB deficiency causes drastic reductions in biofilm formation by the deficient mutants
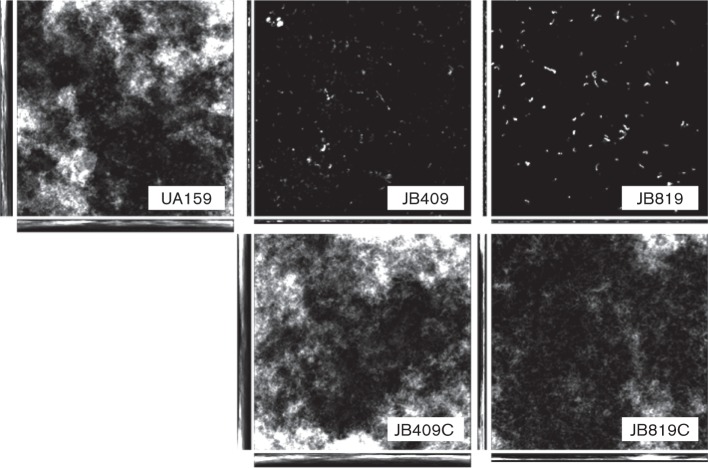
When grown on vertically positioned HA discs, a commonly used in vitro tooth model, and analysed using laser scanning confocal microscopy following fluorescent staining using SYTO 62 (Invitrogen), UA159 accumulated robust biofilms after 24 h, with a biovolume of 14.48 (±5.26) µm3 µm−2 (Fig. 4). In comparison, biofilm formation by the BrpB-deficient mutant, JB409, and BrpB- and BrpA-deficient double mutant, JB819, was drastically reduced, with a biovolume of 0.38±0.24 µm3 µm−2 (P<0.001) for JB409 and 0.02±0.01 µm3 µm−2 (P<0.001) for JB819, respectively. Further analyses using comstat 2.0 also revealed that JB409 and JB819 had reduced the surface area of the biofilms by 1 log and 2 logs, respectively, when compared to UA159. The mean and maximum thicknesses of the biofilms formed by JB409 and JB819 were also significantly reduced, as compared to UA159 (Fig. S2). As expected, complementation in strains JB409C and JB819C restored biofilm formation to a level similar to that of UA159 (Figs 4 and S2). In addition, fluorescence staining using Concanavalin A-conjugated antibody also revealed that the BrpB-deficient mutant, JB409, possessed reduced glucans as compared to the wild-type UA159 (data not shown).
Fig. 4.
Confocal laser scanning microscopy analysis of biofilms. Strep. mutans wild-type, UA159, BrpB-deficient mutant, JB409, BrpB-complemented strain, JB409C, BrpB- and BrpA-deficient double mutant, JB819, and BrpB- and BrpA-complemented strain, JB819C, were cultivated in BMGS for 24 h with biofilms growing on HA discs deposited vertically in 24-well plates. Following staining using SYTO62, biofilms were dissected using an Olympus laser scanning confocal microscope, and post-acquisition analyses were performed with slidebook 5.0 and comstat 2.0. As shown, biofilm formation on vertical HA discs for the BrpB-deficient mutant, and the BrpB- and BrpA-deficient double mutant, was significantly reduced, as compared to the wild-type. Images shown are representative stacks composed of xyz, xz and yz dimensions (512×512). At least five independent fields were dissected at ×600 magnification, and more than three independent experiments were carried out for quantitative analysis using comstat, which is shown in Fig. S2.
BrpB deficiency causes defects in cell division and alters cell morphology
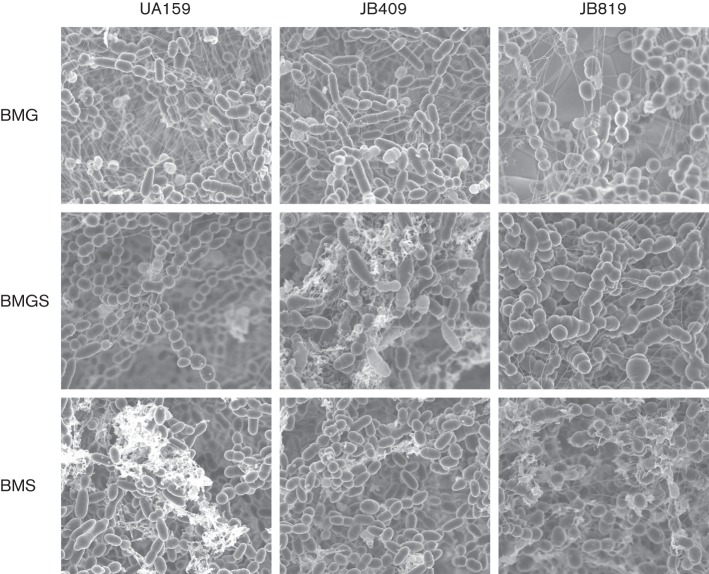
Unlike the parent strain, UA159, the BrpB-deficient mutant, JB409, formed aggregates and precipitated to the bottom of the culture tubes during growth in BHI broth. FE-SEM analysis revealed that when grown in BM with glucose, sucrose, or glucose plus sucrose, as the supplementary carbohydrate sources, the BrpB-deficient mutant, JB409, displayed elongated, rod-like cell morphology and slightly longer chaining properties (Fig. 5), and such characteristics appeared to be most evident during growth in BMGS. The BrpB- and BrpA-deficient double mutant, JB819, grown in BMG, displayed irregular cell septa as compared to the wild-type UA159 (Fig. 5). The inclusion of 2 mM sucrose in BMGS exacerbated the predominance of irregular cell septa and the overall biofilm architecture of both JB409 and JB819, resulting in the formation of swollen marble-like cell clusters, which is likely a reflection of defects in cell division, and is similar to recent findings with mutants deficient of BrpA (Bitoun et al., 2013). When grown in BM with sucrose only, differences between JB409 and UA159 became less obvious, while the irregular cell septa of JB819 and marble-like cell phenotypic characteristics remained evident, and more cell debris was seen, possibly resulting from increased cell lysis.
Fig. 5.
FE-SEM analysis of biofilms. Strep. mutans wild-type, UA159, BrpB-deficient mutant, JB409, and BrpB- and BrpA-deficient double mutant, JB819, were grown in BMG, BMS or BMGS. After 24 h, the biofilms grown on HA discs deposited horizontally in 24-well plates were fixed with 2.5 % glutaraldehyde overnight followed by a series of ethanol dehydrations, and dried at the critical point of CO2, and then carbon coated and viewed with a Hitachi S-4800 high-resolution microscope. Unlike UA159, JB409 and JB819 formed long chains with multiple sites of cell division when grown in BMG. When grown in BMGS, both JB409 and JB819 showed swollen, giant cells possibly due to incomplete and/or aberrant cell division. However, when grown in BMS, the chaining phenotype was less severe, but relative to UA159, JB819 exhibited more cell debris, which is consistent with the findings that JB409 and JB819 had a higher autolysis rate (Fig. 7). Images shown are representative of three independent experiments at ×10 000 magnification.
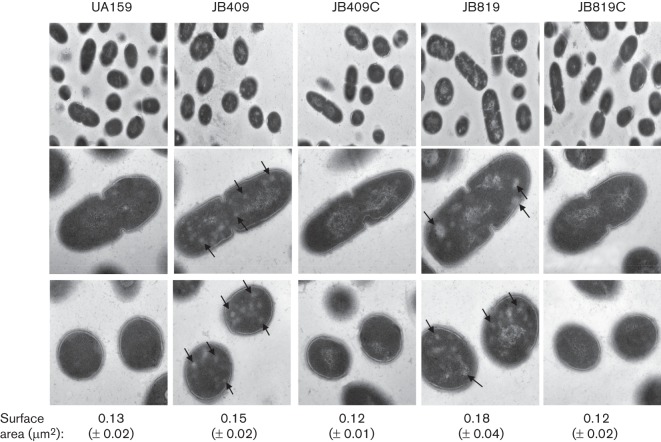
Under TEM with cultures grown in regular BHI broth, the BrpB mutant, JB409, and the BrpB/BrpA-deficient double mutant, JB819, displayed striking defects in cell morphology, when compared to the wild-type, UA159. As shown in Fig. 6, both JB409 and JB819 were littered with regions of low electron density in both non-dividing and dividing cells. In addition, the cross-sectional surface areas of the deficient mutants were also increased from 0.13 (±0.02) µm2 for UA159 to 0.15 (±0.03) µm2 for JB409 (P<0.01) and to 0.18 (±0.04) µm2 for JB819 (P<0.01), consistent with the results of FE-SEM analysis (Fig. 5).The nucleoid also appears very grainy and disorganized in JB409 and JB819, when compared to UA159. As expected, the complementation strains, JB409C and JB819C, all had morphological characteristics that were similar to the wild-type.
Fig. 6.
TEM analysis. Strep. mutans wild-type, UA159, BrpB-deficient mutant, JB409, BrpB-complemented strain, JB409C, BrpB- and BrpA-deficient double mutant, JB819, and BrpB- and BrpA-complemented strain, JB819C, were grown in BHI, pH 7.4, until mid-exponential phase. When compared to the parent strain, UA159, both JB409 and JB819 displayed patches of low-density areas (shown by arrows in the middle and bottom panels), as a result of BrpB deficiency. In addition, the nucleoid of JB409 and JB819 also appeared rougher when compared to UA159. Complementation with a copy of the respective wild-type gene plus its cognate promoter(s) in JB409C and JB819C was able to restore the wild-type phenotype. Images in the top panel were taken at a magnification of ×10 000 and those in the middle and bottom panels were taken at ×50 000. Numbers under the image panels are surface areas as means (±sd) of the cell sections.
BrpB deficiency causes elevation of autolysis rates
As compared to the parent strain, UA159, both the BrpB-deficient strain, JB409, and the BrpB/BrpA-deficient double mutant, JB819, showed increased autolysis rates, especially upon the inclusion of 0.2 % Triton X-100 (Fig. 7).
Fig. 7.
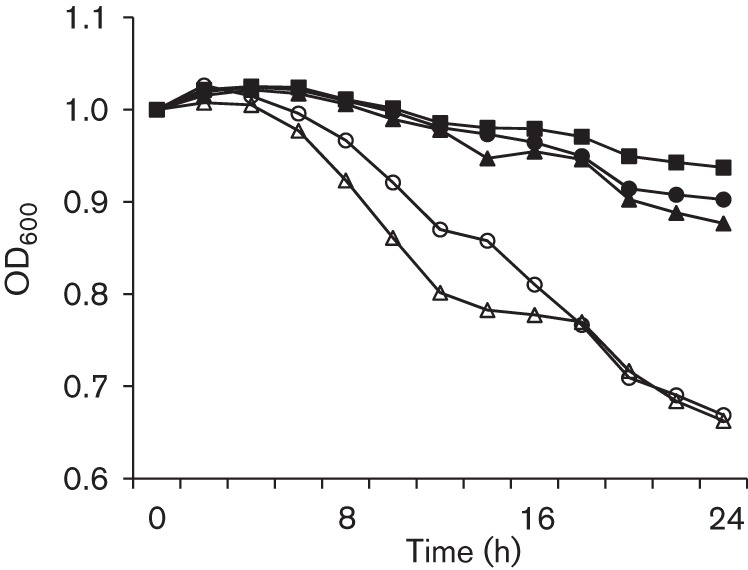
Autolysis assay. Overnight cultures of Strep. mutans wild-type, UA159 (▪), BrpB-deficient mutant, JB409 (○), BrpB-complemented strain, JB409C (•), BrpB- and BrpA-deficient double mutant, JB819 (Δ), and BrpB- and BrpA-complemented strain, JB819C (▴), were washed and resuspended in 100 mM PBS, pH 7.2, containing 0.2 % (v/v) Triton X-100, and the optical density at 600 nm was then monitored continuously using a Bioscreen C for 24 h. The autolysis rates of JB409 and JB819 were significantly increased as compared to the wild-type UA159. Complementation strains JB409C and JB819C showed autolysis rates restored to a level similar to the wild-type. Data presented here are representative of three independent experiments.
Discussion
As the outermost barrier between the cell and the environment, the cell envelope is known to play a crucial role in a number of biological processes vital for bacterial growth and survival, including environmental sensing and signal transduction, maintenance of osmotic pressure, cell shape and size, and cell division. As part of a two-gene operon, brpB is genetically linked to brpA, which encodes a paralogue of a highly conserved LCP family of cell wall-associated proteins. Data presented here have shown that like BrpA, BrpB plays an important role in Strep. mutans cellular physiology. BrpB deficiency increases autolysis rates, drastically weakens acid and oxidative stress tolerance responses, and causes major defects in biofilm formation in the deficient mutants. These results further suggest that BrpB and BrpA are linked in genetic structure as well as functionality in the regulation of cell division, stress tolerance response and cell envelope biogenesis/homeostasis.
Like YdiB in B. subtilis, but different from YjeE in E. coli (Allali-Hassani et al., 2004; Handford et al., 2009; Karst et al., 2009), BrpB deficiency in Strep. mutans is viable, but causes an extended lag phase and increased doubling time of the deficient mutants, which could be attributed to defects in stress tolerance response and cell division. Similar to YjeE in E. coli, whose depletion was shown to result in unusual cell morphology, including alterations in nucleoid distribution (Handford et al., 2009), Strep. mutans strains deficient of BrpB also displayed higher numbers of elongated, giant cells, and seemingly, disorganization of the nucleoid under FE-SEM and TEM, which is also indicative of defects in cell division and cell envelope biogenesis/homeostasis. The increased bacterial cell size seen in JB409 and JB819 also suggests alterations in the osmotic pressure on the cell; therefore, this could in part explain the increased autolysis rates, whereby swelling causes the cells to lyse. Characteristically, all BrpB-deficient mutants possessed low-density patches under TEM, which appears to be unique to BrpB in Strep. mutans. It is worth pointing out that the low-density patches seem to have a structure/texture different from the nucleoid, although their exact nature awaits further investigation. Nevertheless, defects in cell envelope biogenesis/homeostasis and nucleoid organization would likely affect the ability of the deficient mutants to survive and adapt to harsh environmental conditions, influence cell division and growth, cause alterations in morphology, and induce elevations in autolysis rates as observed with the BrpB-deficient mutants, JB409 and JB819.
Strep. mutans is known to possess an adaptive acid tolerance response, featuring an elevated acid tolerance after initial exposure to low pH conditions. The ability of the BrpB-deficient mutant, JB409, to adapt to low pH and mount an elevated survival rate after 1 h incubation in BHI of pH 5.0 suggests that BrpB deficiency primarily affects the constitutive acid tolerance response, such as changes related to cell envelope biogenesis and integrity, but not the adaptive acid tolerance response. Like several members of the LCP proteins (Hanson et al., 2011, 2012; Lazarevic et al., 1992), BrpA in Strep. mutans can also function as a transcriptional regulator (Bitoun et al., 2012a, 2013; Wen et al., 2006). Previous studies showed that deficiency of BrpA caused substantial alterations in the transcriptional profile of the deficient mutant, including changes in transcripts involved in peptidoglycan biosynthesis, DNA repair and stress tolerance response, and biofilm formation (Bitoun et al., 2012a, 2013; Wen et al., 2006). Consistent with our previous findings that BrpA is a key regulator of the acid tolerance response in Strep. mutans (Wen et al., 2006), deficiency of both BrpA and BrpB further reduced the survival rate of the deficient mutant after acid killing at pH 2.8. Different from the respective BrpB and BrpA single mutants, however, the BrpB/BrpA-deficient double mutant showed limited capacity to mount an adaptive acid tolerance response after initial incubation in low pH medium. Considering both the BrpA (Bitoun et al., 2013; Wen et al., 2006) and BrpB single mutants were still capable of launching an adaptive acid tolerance response, a logical explanation is that BrpB and BrpA double deficiency imposed an additive effect on the deficient mutant that severely damaged or destroyed the constitutive acid tolerance responses, such as those on cell envelope integrity and nucleoid structure. Even if not directly affected, the adaptive acid tolerance response in JB819 failed to sufficiently compensate the resulting defects in acid tolerance response. When introduced via a multiple copy shuttle vector, pDL278, the higher level of BrpA and BrpB in the complemented strain, JB819C, would likely impose a dominant negative effect, which could be in part attributed to the extremely extended lag phase of the complemented strain during growth in pH 6.5 (Fig. 1b) and the failure to restore the increased sensitivity to hydrogen peroxide of the double mutant (Fig. 3).
Strep. mutans has adapted biofilms as its primary life style. It produces at least three glucosyltransferases that utilize sucrose to produce adhesive glucans, which function as scaffolding matrix, playing a crucial role in biofilm formation. Accumulating data also suggests that formation of mature biofilms requires coordination of an array of physiological and biochemical reactions in response to environmental conditions. This study showed that BrpB deficiency in Strep. mutans affects biofilm formation by the deficient mutants, especially during growth in BM supplemented with glucose and sucrose. In comparison with the wild-type, BrpB-deficient mutants had limited ability to bind to and accumulate on a surface in biofilms, as evidenced by drastic reductions in surface area, thickness and biovolume of the mutant during growth on HA discs. The weakened tolerance responses to acid and oxidative stress, the elevated autolysis rate, and especially the growth defects demonstrated by the extended lag phase and delayed doubling time of the deficient mutants are likely the major contributing factors. Defects in cell envelope as a result of BrpB deficiency will also affect cell-surface and cell–cell interactions, modulating biofilm formation. In addition, fluorescence staining using Concanavalin A-conjugated antibody also revealed that the BrpB-deficient mutants possess reduced glucans. Among others, the growth defects of the deficient mutants are likely the major contributor to the reduced glucan production. Nevertheless, modulation of glucan production in response to BrpB deficiency also explains why the most drastic impact on biofilm formation was seen with cultures grown in medium with sucrose.
While the YjeE-like proteins are highly conserved in eubacteria, many intracellular and symbiotic bacteria do not possess any of these homologues. As shown by data presented here, differences also exist between the YjeE homologues in primary sequence and protein function, as well as the genetic structure of the coding regions. Phylogenetic analysis revealed that in E. coli and most γ-proteobacteria, the yjeE locus is closely linked to genes encoding proteins with potential functions in DNA repair and peptidoglycan remodelling, which is consistent with a role for YjeE in stress tolerance and cell envelope biogenesis (Handford et al., 2009; Teplyakov et al., 2002). All streptococci whose genomes have been sequenced possess a gene encoding a YjeE-like protein. Unlike E. coli and the proteobacteria, the loci encoding the YjeE-like proteins in all streptococci are closely linked to genes encoding a member of the LCP family of proteins. This genetic structure appears to be unique to streptococci, as no such linkage could be identified in other Gram-positive bacteria such as Staph. aureus and B. subtilis that have been shown to possess LCP paralogues. Highly conserved in Gram-positive bacteria, LCP proteins are widely considered as cell wall-associated transcriptional regulators (Hübscher et al., 2008), although there is recent evidence suggesting that LCP proteins are responsible for covalent attachment of anionic cell wall polymers, such as teichoic acid and capsular polysaccharides, to peptidoglycan (Eberhardt et al., 2012; Kawai et al., 2011). BrpA and several other LCP paralogues have been shown to play a major role in cell envelope biogenesis (Bitoun et al., 2012a, 2013; Eberhardt et al., 2012; Johnsborg & Håvarstein, 2009; Kawai et al., 2011; Over et al., 2011; Wen et al., 2006). This highly conserved genetic linkage, along with the characteristic phenotypes of the BrpB-deficient mutants, all support a role for BrpB and its streptococcal homologues in cell wall biogenesis/homeostasis. To assess whether brpB is regulated by BrpA, real-time PCR was also used to analyse the transcription of brpB in response to BrpA deficiency. The results showed no significant differences between the parent strain and the deficient mutant, TW14D (Bitoun et al., 2012a; Wen et al., 2006), during growth in early exponential phase (OD600 0.3), suggesting that brpB is not directly under the control of BrpA under the conditions studied. In E. coli and Salmonella typhimurium, YjeE was shown to interact with YeaZ (a protease) and YeaZ–YgiD (an atypical DNA-binding protein) complex (Handford et al., 2009; Nichols et al., 2013). Unlike YjeE and YeaZ, Strep. mutans and some other streptococci do not seem to possess the YgiD orthologue. The absence of YgiD again suggests differences in the function of BrpB in Strep. mutans, and probably the other YjeE-like proteins in other streptococci, from their counterparts in E. coli and Salm. typhimurium. Considering the drastic differences in the growth and stress responses of the BrpB/BrpA-deficient double mutant, JB819, from the respective single mutants, JB409 and TW14D (Bitoun et al., 2013; Wen et al., 2006), and especially its complemented strain, JB819C, which possesses a high copy number of both brpA and brpB, it is highly possible that BrpB and BrpA interact in the regulation of cellular physiology in Strep. mutans. Like the two-component system LiaRS of B. subtilis (Schrecke et al., 2013), proper stoichiometry of BrpB and BrpA in Strep. mutans may be crucial to a functional Brp system. Overexpression of BrpB and BrpA in the complemented strain may cause dysregulation of the Brp system, resulting in an imbalance in cell envelope composition and compromises in cell envelope integrity, which could also be in part attributed to the severely delayed lag phase and the failure to restore the increased susceptibility to hydrogen peroxide.
In summary, the results presented here suggest that while YjeE-like proteins are highly conserved in eubacteria, differences exist in amino acid composition, function and genetic structure between these proteins in different species. In Strep. mutans, BrpB is genetically linked to and appears to function in concert with BrpA in the regulation of cell envelope biogenesis/homeostasis. Current effort is being directed to elucidation of the mechanism by which these important regulatory proteins interact in related functions.
Acknowledgements
This study was supported in part by NIH/NIDCR grant DE19452 to Z. T. W and by the Southern Louisiana Institute for Infectious Disease Research.
Abbreviations:
- FE-SEM
field emission-scanning electron microscopy
- HA
hydroxylapatite
- Kan
kanamycin
- TEM
transmission electron microscopy
Footnotes
Two supplementary figures are available with the online version of this paper.
References
- Ajdić D., McShan W. M., McLaughlin R. E., Savić G., Chang J., Carson M. B., Primeaux C., Tian R., Kenton S. & other authors (2002). Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci U S A 99, 14434–14439. 10.1073/pnas.172501299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allali-Hassani A., Campbell T. L., Ho A., Schertzer J. W., Brown E. D. (2004). Probing the active site of YjeE: a vital Escherichia coli protein of unknown function. Biochem J 384, 577–584. 10.1042/BJ20041082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitoun J. P., Nguyen A. H., Fan Y., Burne R. A., Wen Z. T. (2011). Transcriptional repressor Rex is involved in regulation of oxidative stress response and biofilm formation by Streptococcus mutans. FEMS Microbiol Lett 320, 110–117. 10.1111/j.1574-6968.2011.02293.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitoun J. P., Liao S., Yao X., Ahn S. J., Isoda R., Nguyen A. H., Brady L. J., Burne R. A., Abranches J., Wen Z. T. (2012a). BrpA is involved in regulation of cell envelope stress responses in Streptococcus mutans. Appl Environ Microbiol 78, 2914–2922. 10.1128/AEM.07823-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitoun J. P., Liao S., Yao X., Xie G. G., Wen Z. T. (2012b). The redox-sensing regulator Rex modulates central carbon metabolism, stress tolerance response and biofilm formation by Streptococcus mutans. PLoS ONE 7, e44766. 10.1371/journal.pone.0044766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitoun J. P., Liao S., McKey B. A., Yao X., Fan Y., Abranches J., Beatty W. L., Wen Z. T. (2013). Psr is involved in regulation of glucan production, and double deficiency of BrpA and Psr is lethal in Streptococcus mutans. Microbiology 159, 493–506. 10.1099/mic.0.063032-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burne R. A. (1998). Oral streptococci... products of their environment. J Dent Res 77, 445–452. 10.1177/00220345980770030301 [DOI] [PubMed] [Google Scholar]
- Chatfield C. H., Koo H., Quivey R. G., Jr (2005). The putative autolysin regulator LytR in Streptococcus mutans plays a role in cell division and is growth-phase regulated. Microbiology 151, 625–631. 10.1099/mic.0.27604-0 [DOI] [PubMed] [Google Scholar]
- Deutsch C., El Yacoubi B., de Crécy-Lagard V., Iwata-Reuyl D. (2012). Biosynthesis of threonylcarbamoyl adenosine (t6A), a universal tRNA nucleoside. J Biol Chem 287, 13666–13673. 10.1074/jbc.M112.344028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardt A., Hoyland C. N., Vollmer D., Bisle S., Cleverley R. M., Johnsborg O., Håvarstein L. S., Lewis R. J., Vollmer W. (2012). Attachment of capsular polysaccharide to the cell wall in Streptococcus pneumoniae. Microb Drug Resist 18, 240–255. 10.1089/mdr.2011.0232 [DOI] [PubMed] [Google Scholar]
- Handford J. I., Ize B., Buchanan G., Butland G. P., Greenblatt J., Emili A., Palmer T. (2009). Conserved network of proteins essential for bacterial viability. J Bacteriol 191, 4732–4749. 10.1128/JB.00136-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson P. I., Whiteheart S. W. (2005). AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol 6, 519–529. 10.1038/nrm1684 [DOI] [PubMed] [Google Scholar]
- Hanson B. R., Lowe B. A., Neely M. N. (2011). Membrane topology and DNA-binding ability of the streptococcal CpsA protein. J Bacteriol 193, 411–420. 10.1128/JB.01098-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson B. R., Runft D. L., Streeter C., Kumar A., Carion T. W., Neely M. N. (2012). Functional analysis of the CpsA protein of Streptococcus agalactiae. J Bacteriol 194, 1668–1678. 10.1128/JB.06373-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydorn A., Nielsen A. T., Hentzer M., Sternberg C., Givskov M., Ersbøll B. K., Molin S. (2000). Quantification of biofilm structures by the novel computer program comstat. Microbiology 146, 2395–2407. [DOI] [PubMed] [Google Scholar]
- Hübscher J., Lüthy L., Berger-Bächi B., Stutzmann Meier P. (2008). Phylogenetic distribution and membrane topology of the LytR-CpsA-Psr protein family. BMC Genomics 9, 617. 10.1186/1471-2164-9-617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsborg O., Håvarstein L. S. (2009). Pneumococcal LytR, a protein from the LytR-CpsA-Psr family, is essential for normal septum formation in Streptococcus pneumoniae. J Bacteriol 191, 5859–5864. 10.1128/JB.00724-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst J. C., Foucher A.-E., Campbell T. L., Di Guilmi A.-M., Stroebel D., Mangat C. S., Brown E. D., Jault J.-M. (2009). The ATPase activity of an ‘essential’ Bacillus subtilis enzyme, YdiB, is required for its cellular function and is modulated by oligomerization. Microbiology 155, 944–956. 10.1099/mic.0.021543-0 [DOI] [PubMed] [Google Scholar]
- Kawai Y., Marles-Wright J., Cleverley R. M., Emmins R., Ishikawa S., Kuwano M., Heinz N., Bui N. K., Hoyland C. N. & other authors (2011). A widespread family of bacterial cell wall assembly proteins. EMBO J 30, 4931–4941. 10.1038/emboj.2011.358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P. C. Y., Sung C. K., Lee J. H., Morrison D. A., Cvitkovitch D. G. (2002). PCR ligation mutagenesis in transformable streptococci: application and efficiency. J Microbiol Methods 49, 193–205. 10.1016/S0167-7012(01)00369-4 [DOI] [PubMed] [Google Scholar]
- Lazarevic V., Margot P., Soldo B., Karamata D. (1992). Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-l-alanine amidase and its modifier. J Gen Microbiol 138, 1949–1961. 10.1099/00221287-138-9-1949 [DOI] [PubMed] [Google Scholar]
- LeBlanc D., Lee L. (1991). Replication function of pVA380-1. In Genetics and Molecular Biology of Streptococci, Lactococci, and Enterococci, pp. 235–239. Edited by Dunny G., Cleary P. P., Mckay L. L.. Washington, DC: ASM Press. 10.1099/mic.0.2008/023770-0 [DOI] [Google Scholar]
- Lemos J. A., Burne R. A. (2008). A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology 154, 3247–3255. 10.1099/mic.0.2008/023770-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos J. A., Abranches J., Koo H., Marquis R. E., Burne R. A. (2010). Protocols to study the physiology of oral biofilms. Methods Mol Biol 666, 87–102. 10.1007/978-1-60761-820-1_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. H., Lau P. C., Lee J. H., Ellen R. P., Cvitkovitch D. G. (2001). Natural genetic transformation of Streptococcus mutans growing in biofilms. J Bacteriol 183, 897–908. 10.1128/JB.183.3.897-908.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y. H., Lau P. C., Tang N., Svensäter G., Ellen R. P., Cvitkovitch D. G. (2002). Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J Bacteriol 184, 6333–6342. 10.1128/JB.184.22.6333-6342.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loesche W. J. (1986). Role of Streptococcus mutans in human dental decay. Microbiol Rev 50, 353–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo C. Y., Corliss D. A., Ganeshkumar N. (2000). Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J Bacteriol 182, 1374–1382. 10.1128/JB.182.5.1374-1382.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols C. E., Lamb H. K., Thompson P., El Omari K., Lockyer M., Charles I., Hawkins A. R., Stammers D. K. (2013). Crystal structure of the dimer of two essential Salmonella typhimurium proteins, YgjD & YeaZ and calorimetric evidence for the formation of a ternary YgjD–YeaZ–YjeE complex. Protein Sci 22, 628–640. 10.1002/pro.2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Over B., Heusser R., McCallum N., Schulthess B., Kupferschmied P., Gaiani J. M., Sifri C. D., Berger-Bächi B., Stutzmann Meier P. (2011). LytR-CpsA-Psr proteins in Staphylococcus aureus display partial functional redundancy and the deletion of all three severely impairs septum placement and cell separation. FEMS Microbiol Lett 320, 142–151. 10.1111/j.1574-6968.2011.02303.x [DOI] [PubMed] [Google Scholar]
- Schrecke K., Jordan S., Mascher T. (2013). Stoichiometry and perturbation studies of the LiaFSR system of Bacillus subtilis. Mol Microbiol 87, 769–788. 10.1111/mmi.12130 [DOI] [PubMed] [Google Scholar]
- Teplyakov A., Obmolova G., Tordova M., Thanki N., Bonander N., Eisenstein E., Howard A. J., Gilliland G. L. (2002). Crystal structure of the YjeE protein from Haemophilus influenzae: a putative ATPase involved in cell wall synthesis. Proteins 48, 220–226. 10.1002/prot.10114 [DOI] [PubMed] [Google Scholar]
- Wen Z. T., Burne R. A. (2002). Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl Environ Microbiol 68, 1196–1203. 10.1128/AEM.68.3.1196-1203.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z. T., Burne R. A. (2004). LuxS-mediated signaling in Streptococcus mutans is involved in regulation of acid and oxidative stress tolerance and biofilm formation. J Bacteriol 186, 2682–2691. 10.1128/JB.186.9.2682-2691.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z. T., Baker H. V., Burne R. A. (2006). Influence of BrpA on critical virulence attributes of Streptococcus mutans. J Bacteriol 188, 2983–2992. 10.1128/JB.188.8.2983-2992.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z. T., Yates D., Ahn S. J., Burne R. A. (2010). Biofilm formation and virulence expression by Streptococcus mutans are altered when grown in dual-species model. BMC Microbiol 10, 111. 10.1186/1471-2180-10-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z. T., Nguyen A. H., Bitoun J. P., Abranches J., Baker H. V., Burne R. A. (2011). Transcriptome analysis of LuxS-deficient Streptococcus mutans grown in biofilms. Mol Oral Microbiol 26, 2–18. 10.1111/j.2041-1014.2010.00581.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L., Wen Z. T., Burne R. A. (2006). A novel signal transduction system and feedback loop regulate fructan hydrolase gene expression in Streptococcus mutans. Mol Microbiol 62, 187–200. 10.1111/j.1365-2958.2006.05359.x [DOI] [PubMed] [Google Scholar]