Abstract
Background
GATA transcription factors are essential for self-renewal of the small intestinal epithelium. Gata4 is expressed in the proximal 85% of small intestine while Gata6 is expressed throughout the length of small intestine. Deletion of intestinal Gata4 and Gata6 results in an altered proliferation/differentiation phenotype, and an up-regulation of SAM pointed domain containing ETS transcription factor (Spdef), a transcription factor recently shown to act as a tumor suppressor. The goal of this study is to determine to what extent SPDEF mediates the downstream functions of GATA4/GATA6 in the small intestine. The hypothesis to be tested is that intestinal GATA4/GATA6 functions through SPDEF by repressing Spdef gene expression. To test this hypothesis, we defined the functions most likely regulated by the overlapping GATA6/SPDEF target gene set in mouse intestine, delineated the relationship between GATA6 chromatin occupancy and Spdef gene regulation in Caco-2 cells, and determined the extent to which prevention of Spdef up-regulation by Spdef knockout rescues the GATA6 phenotype in conditional Gata6 knockout mouse ileum.
Results
Using publicly available profiling data, we found that 83% of GATA6-regulated genes are also regulated by SPDEF, and that proliferation/cancer is the function most likely to be modulated by this overlapping gene set. In human Caco-2 cells, GATA6 knockdown results in an up-regulation of Spdef gene expression, modeling our mouse Gata6 knockout data. GATA6 occupies a genetic locus located 40 kb upstream of the Spdef transcription start site, consistent with direct regulation of Spdef gene expression by GATA6. Prevention of Spdef up-regulation in conditional Gata6 knockout mouse ileum by the additional deletion of Spdef rescued the crypt cell proliferation defect, but had little effect on altered lineage differentiation or absorptive enterocytes gene expression.
Conclusion
SPDEF is a key, immediate downstream effecter of the crypt cell proliferation function of GATA4/GATA6 in the small intestine.
Keywords: GATA6, SPDEF, Crypt cell proliferation, Intestinal differentiation
Background
The mature mammalian small intestine is lined by a highly specialized epithelium that regenerates itself in a tightly controlled manner resulting in a lineage distribution and gene expression patterning that is perfectly suited for the absorption of nutrients. The epithelium is organized into crypt-villus structures in which the Crypts of Lieberkühn contain stem cells that produce proliferating, transit-amplifying (TA) cells that differentiate into five principal post-mitotic cell types comprised of one type of absorptive cell (absorptive enterocytes) and four types of secretory cells (enteroendocrine, goblet, Paneth, and tuft cells [1]). Absorptive enterocytes express digestive enzymes and transporters necessary for the terminal digestion and absorption of nutrients. Mucus-secreting goblet cells and defensin-secreting Paneth cells maintain a dynamic mucosal defensive barrier. Enteroendocrine cells secrete hormones that regulate gastrointestinal processes. Tuft cells, recently shown to be an independent secretory lineage [2], secrete opioids and produce enzymes that synthesize prostaglandins, suggesting a role in inflammation. Absorptive enterocytes, goblet cells, enteroendocrine cells, and tuft cells migrate up the crypt to populate the villi, whereas Paneth cells migrate to the base of crypts. The differentiated cells eventually undergo apoptosis and are shed into the lumen. Cells of the villus epithelium turn over in 3–4 days, whereas Paneth cells at the base of crypts turn over at a slower rate of 3–6 weeks.
Current models of intestinal epithelial renewal suggest that long-lived, multipotent stem cells produce progenitors that undergo a series of transitions that ultimately give rise to the individual cell lineages [1,3]. The Wnt, hedgehog, and bone morphogenetic protein signaling pathways regulate intestinal proliferation and differentiation, while the Notch signaling pathway plays a central role in determining epithelial cell fate. The first decision selects absorptive vs. secretory progenitors. Activated Notch signaling results in the transcriptional activation of its principal intestinal target, hairy and enhancer of split 1 (Hes1), which encodes a transcription factor that selects the absorptive enterocyte lineage. Progenitor cells that escape Notch signaling and activation of Hes1 gene transcription express atonal homolog 1 (Atoh1, formerly called Math1), which encodes a transcription factor that selects the secretory cells. Additional regulators that function in secretory cell differentiation include: growth factor independent 1 (Gfi1) that distinguishes enteroendocrine from goblet/Paneth progenitors; neurogenin 3 (Neurog3) that specifies the enteroendocrine lineage; SAM pointed domain-containing Ets transcription factor (Spdef), a GFI1 target, that promotes goblet differentiation; and SRY-box containing gene 9 (Sox9) and ephrin type B receptor 3 (Ephb3), Wnt targets that are necessary for the differentiation of Paneth cells and their localization to the crypt base, respectively.
Recently, we showed that members of the GATA family, an ancient family of transcription factors that bind WGATAR motifs in DNA, play essential roles in crypt cell proliferation, secretory cell differentiation, and absorptive enterocyte gene expression. Gata4 and Gata6 are expressed in the intestinal epithelium, but whereas Gata6 is expressed throughout the length of the small intestine, Gata4 is expressed in the proximal 85% of small intestine and is sharply down-regulated in the distal ileum [4-6]. Using conditional knockout technology, we [5,7] and others [8] have shown that GATA4 functions to promote a 'jejunal’ pattern of absorptive enterocyte gene expression and function while repressing an 'ileal’ pattern. Using single and double conditional knockout approaches for Gata4 and Gata6, we found that in the ileum of single Gata6 conditional knockout mice, where Gata4 is not normally expressed, or throughout the small intestine of double Gata4/Gata6 conditional knockout mice, crypt cell proliferation and enteroendocrine cell specification are decreased, Paneth cells are replaced by a goblet-like cell type, and the expression of specific absorptive enterocyte genes is altered [4]. We also noted that Spdef, a transcription factor expressed in secretory progenitors, goblet cells and Paneth cells that functions in goblet and Paneth cell differentiation [9,10], was up-regulated [4]. Using a conditional Spdef over-expression model, Noah et al. [10] described an intestinal phenotype that, with the exception of the changes in absorptive enterocyte gene expression, essentially phenocopies that of our Gata4/Gata6 conditional knockout mice: crypt cell proliferation is decreased, enteroendocrine and Paneth cells are decreased, and goblet-like cells are increased. GATA6 is co-expressed with SPDEF in the same lineages in the small intestine [4]. Based on these findings [4], we hypothesized that GATA4/GATA6 regulates crypt cell proliferation and secretory cell differentiation in the small intestine by repressing Spdef gene expression. To test this hypothesis, we defined the functions most likely regulated by the overlapping GATA6/SPDEF target gene set in mouse intestine, delineated the relationship between GATA6 chromatin occupancy and Spdef gene regulation in Caco-2 cells, and determined the extent to which prevention of Spdef up-regulation by Spdef knockout rescues the GATA6 phenotype in conditional Gata6 knockout mouse ileum.
Results and discussion
GATA6 and SPDEF regulate similar subsets of genes
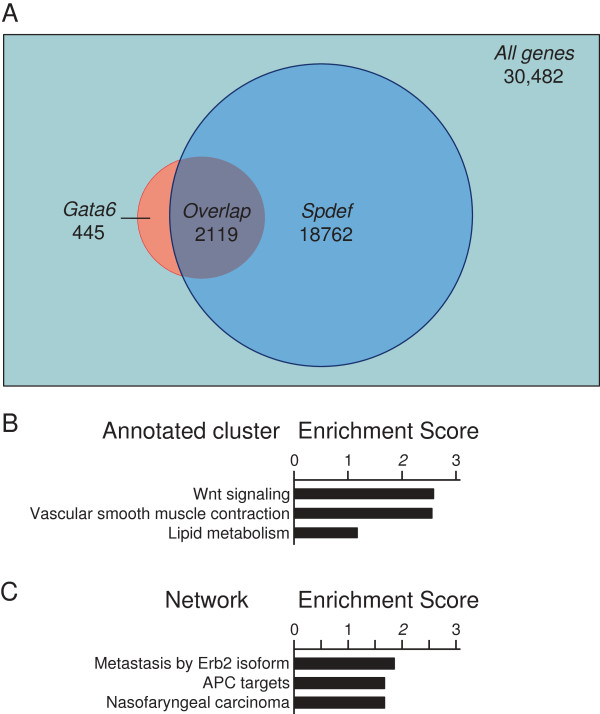
To gain insight on the relationship between GATA6 and SPDEF in the small intestine, we scanned the overlap of gene targets using publicly available gene profiling data from conditional Gata6 and Spdef knockout mouse intestine [4,9]. Previously, we identified 2564 genes whose expression is altered in ileum by conditional Gata6 deletion [4]. Network analysis of this gene set indicated an up-regulation of p53 targets and a down-regulation of c-MYC targets (Additional file 1: Figure S1), consistent with a decrease in cellular proliferation. Of the 2564 genes altered by conditional Gata6 knockout, 83% (2119) were also altered by Spdef knockout (Figure 1A), a far greater overlap than would be expected from a similar-sized, randomized allocation of genes (P < 10-60, Fisher’s Exact Test). The changes in expression of this subset when Gata6 is deleted were analyzed by gene set array analysis. Using Database for Annotation, Visualization and Integrated Discovery (DAVID), we conducted functional annotation clustering of all the major pathways (listed in the KEGG, Biocarta and BBID databases). We found that Wnt signaling was the function most likely to be effected (Figure 1B), consistent with regulation of crypt cell proliferation. Using Gene Set Enrichment Analysis (GSEA), a publically available bioinformatics tool that delineates gene expression data for enrichment of pre-defined gene-sets, APC target network was one of the top three networks affected by Gata6 deletion with a Normalized Enrichment Score (NES) of -1.57 (Figure 1C). This analysis reveals a very strong overlap in gene targets between GATA6 and SPDEF, and suggests that the principal function of this overlap involves cellular proliferation.
Figure 1.
Overlap of genes in small intestine modulated by conditional Gata6 or Spdef knockout. (A) Publicly available gene profiling data from conditional Gata6 (GSE22416) and Spdef (GSE14892) knockout mouse intestine show a significant overlap. (B) Functional annotation clustering of pathways shows that the top-enriched cluster, in overlapping Gata6 and Spdef genes, contains the Wnt pathway as its main function (ES score 2.58). (C) GSEA analysis shows, in the overlapping Gata6 and Spdef gene segment, APC targets as the second most highly enriched cluster (ES score 1.57).
GATA6 regulates Spdef gene expression and occupies a locus in the Spdef 5′-flanking region in human Caco-2 cells
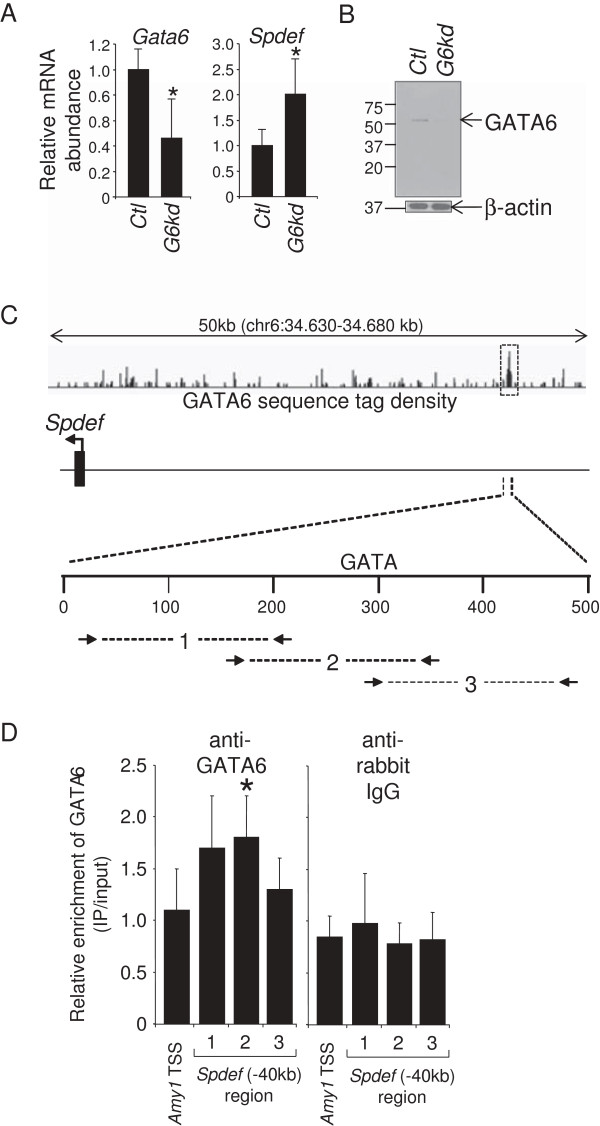
Conditional Gata6 deletion in mice produces an up-regulation of Spdef gene expression in ileum while knockout or over-expression of Spdef has no effect on Gata6 expression [4], suggesting that Spdef is regulated downstream by GATA6. To determine the extent to which this process is conserved, and to further explore the role of GATA6 in regulating Spdef gene expression, we characterized the effect of Gata6 knock-down on Spdef gene expression in Caco-2 cells. The Caco-2 cell line is a human colorectal adenocarcinoma-derived cell line that expresses GATA6, but very little GATA4 [11], similar to the ileum. We screened five GATA6 short-hairpin RNA (shRNA) knockdown lentiviral constructs, and found that only one resulted in a statistically significant knockdown of Gata6 mRNA (55%, P < 0.05, Figure 2A) and a concomitant decrease in GATA6 protein (Figure 2B). Spdef mRNA was up-regulated 2-fold in Gata6 knock-down (G6kd) cells (P < 0.05, Figure 2A), demonstrating that the Caco-2 cells model the up-regulation of Spdef gene expression observed in conditional Gata6 knockout mouse ileum [4].
Figure 2.
GATA6 regulates Spdef expression and occupies an enhancer in the Spdef 5′-flanking region in Caco-2 cells. (A) Quantitative RT-PCR analysis showing reduced expression of Gata6 mRNA, and enhanced expression of Spdef mRNA in Caco-2 cells infected with a lentivirus vector expressing an shRNA for human Gata6 mRNA (G6kd) (mean ± SD, n = 5, *P < 0.05). An shRNA vector for GFP was used as a control (Ctl). (B) Western analysis showing reduced abundance of GATA6 in G6kd Caco-2 cells. Β-actin was used as an internal loading control. (C) Schematic representation of the human Spdef 5′-flanking region showing GATA6 occupancy at a locus ~40 kb upstream of the transcription start site (TSS). GATA6 sequence tag density is shown as a 'wiggle file’, and a statistically significant GATA6-occupied locus was defined by MACS peak analysis [29] (dotted box). (D) ChIP assays on chromatin obtained from Caco-2 cells using a GATA6 antibody and three sets of overlapping primers centered on the GATA motif showing increased GATA6 occupancy (mean ± SD, n = 4, *P < 0.05). The salivary amylase-α1a (Amy-1) TSS was used as a negative control. ChIP assays using anti-rabbit IgG was used as a control for non-specific immunoprecipitation and primer efficiency.
To gain insight on the relationship between GATA6 and Spdef gene expression, we examined GATA6 chromatin occupancy at the Spdef gene locus. Because we were unable to immunoprecipitate bound chromatin from crosslinked mouse intestinal epithelial cells using existing antisera, we utilized the human Caco-2 intestinal cell culture model in which GATA6 ChIP assays have been performed previously [12]. Using a publicly available GATA6 chromatin immunoprecipitation-high throughput sequencing (ChIP-seq) database in Caco-2 cells [12], we identified a GATA6-occupied cis-regulatory region approximately 40 kb upstream of the Spdef transcription start site (TSS) that mapped to the Spdef gene based on closest distance to any TSS (Figure 2C). To confirm and localize GATA6 occupancy at this locus, we performed GATA6 ChIP assays using a multiple, overlapping primer strategy with amplicons of ~200 bp per primer pair (Figure 2C) centered at the only GATA motif within this region. We found increases of all three amplicons with amplicon from primer set 2 being significantly greater (~70%, P < 0.05, n = 4) than the Amy1 TSS negative control (Figure 2D). Although our ChIP data reveal a modest increase in enrichment, it is nonetheless statistically significant and thus confirmatory of ChIP-seq data conducted by others [12] (see MACS peak data, Figure 2C). We also conducted an IgG control ChIP assay to show that enrichment with primer set 2 was not due to differences in primer efficiency from that of the Amy1 TSS, or to non-specific antibody binding (Figure 2D). These data confirm previous ChIP-seq analysis that GATA6 occupies a cis-regulatory region that maps to the Spdef gene [12], and, together with Spdef up-regulation in G6kd cells, is consistent with the notion that GATA6 directly represses Spdef gene transcription. While mapping of occupancy sites to the nearest TSS is a well recognized method for defining transcription factor targets on a global basis [12,13], direct regulation of Spdef gene expression by GATA6 at this site will need to be confirmed using chromosome conformation capture or other techniques [14].
Transcriptional repression is highly complex, especially in eukaryotes, generally involving the recruitment of specific co-repressors and the local modification of histone tails and chromatin structure [15]. GATA factors are well known to mediate gene repression, and the mechanisms are beginning to be understood. GATA4 has been shown to directly repress cardiac genes [16] while GATA1 has been shown to directly repress hematopoietic genes [17,18]. Both GATA4 and GATA1 interact directly with corepressor complexes including the nucleosome remodeling and histone deacetylase (NuRD) complex and the polycomb repressive complex 2 (PRC2) [16], and are necessary for the multiple modifications of histone tails, but the extent to which GATA factors recruit co-repressors and/or modulate histone tails in the small intestine remains to be determined.
Spdef knockout rescues the proliferation defect in conditional Gata6 knockout mice
We next asked to what extent Spdef deletion rescues the conditional Gata6 knockout phenotype by analyzing single and double Gata6/Spdef knockout mice. In previous studies, conditional deletion of Gata6[4] or deletion of Spdef[9] resulted in greatly diminished levels of Gata6 or Spdef mRNA, respectively, in the intestine that correlated with reduced protein levels and altered intestinal phenotypes. Analysis of mRNA in the present study showed that Gata6 and Spdef mRNAs were both expressed in Ctl mice, Gata6 mRNA was nearly undetectable while Spdef mRNA was expressed in Gata6ΔIE mice, Gata6 mRNA was expressed while Spdef mRNA was nearly undetectable in SpdefKO mice, and both Gata6 and Spdef mRNAs were nearly undetectable in the double knockout (DKO) mice (Additional file 1: Figure S2), verifying our models.
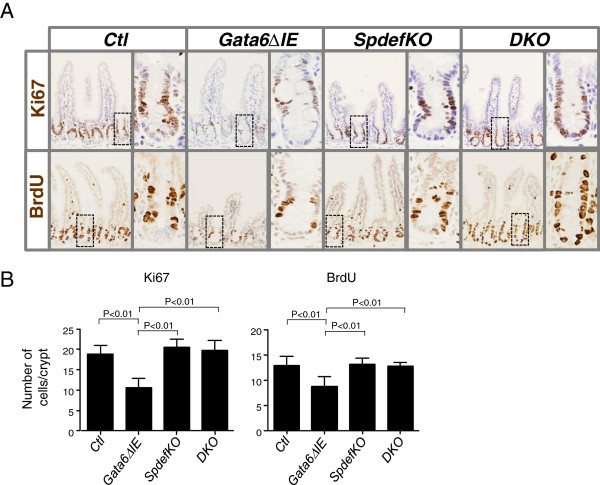
Crypt cell proliferation is essential for the continuation of intestinal epithelial renewal. One of the principal phenotypic outcomes of conditional Gata4/Gata6 deletion is a ~30% reduction in the number of Ki67- and BrdU-positive cells, and a concomitant decrease in villus height and villus epithelial cell number [4] resulting in a reduction in absorptive surface area. To define the role of SPDEF in mediating the decrease in crypt cell proliferation when Gata6 is conditionally deleted, we stained ileal segments for Ki67 and BrdU (Figure 3A), markers of the non-G0 and S-phases of the cell cycle, respectively. The number of Ki67-positive and BrdU-positive crypt cells was significantly reduced ~40% in Gata6ΔIE mice (Figure 3B), as previously reported [4], but unchanged in SpdefKO mice, as compared to controls. The number of Ki67- and BrdU-positive crypt cells in DKO mice was also similar to controls, indicating that the additional deletion of Spdef rescues the decrease in crypt cell proliferation observed in Gata6ΔIE mice. These data support the notion that GATA6 maintains crypt cell proliferation by down-regulating Spdef gene expression.
Figure 3.
Crypt cell proliferation is decreased in Gata6ΔIE mice. (A) Immunostaining in ileum for the proliferation markers Ki67 (top row) and BrdU (bottom row). (B) Quantification of Ki67- and BrdU-positive cells/crypt in each group. Cells were counted as described in Methods. Gata6ΔIE mice had significantly fewer Ki67- or BrdU-positive cells than each of the other three groups (P < 0.01 in each case), consistent with a decrease in crypt cell proliferation.
SPDEF does not regulate the secretory cell differentiation function of GATA6
Previously, conditional deletion of Gata6 in the ileum resulted in a 25-40% reduction in the number of chromogranin A (CHGA)-positive cells, and in the mRNA abundances of Chga and Neurog3[4], consistent with a decrease in enteroendocrine lineage commitment. Though not statistically significant, we found a 15-50% reduction in the number of CHGA-positive cells (Figure 4A), and in the mRNA abundances of Chga and Neurog3 (Figure 4B) in the Gata6ΔIE mice, in general agreement with our previous study [4]. We further found that although the pattern for these three measurements in the SpdefKO mice was similar to that in Ctl mice, consistent with previous data for distal intestine [9], the pattern in the DKO mice was similar to that in the Gata6ΔIE mice (Figure 4A and B). Together, these data show that enteroendocrine cell commitment is reduced in both Gata6ΔIE and DKO mice, indicating that GATA6 promotes enteroendocrine cell commitment independently of SPDEF.
Figure 4.
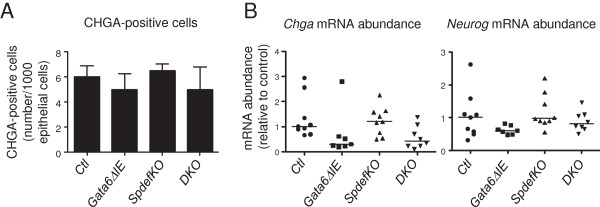
Enteroendocrine cell allocation is decreased in Gata6ΔIE and DKO mice. (A) Quantification of CHGA-positive cells in each group. Sections of ileum were stained for CHGA, and the number of positive cells/1000 epithelial cells was determined as described in Methods. (B)Chga and Neurog3 mRNA abundance in each group. Gata6ΔIE and DKO mice had generally lower numbers of CHGA-positive cells and Chga and Neurog3 mRNA abundances than Ctl mice, consistent with a decrease in enteroendocrine lineage allocation.
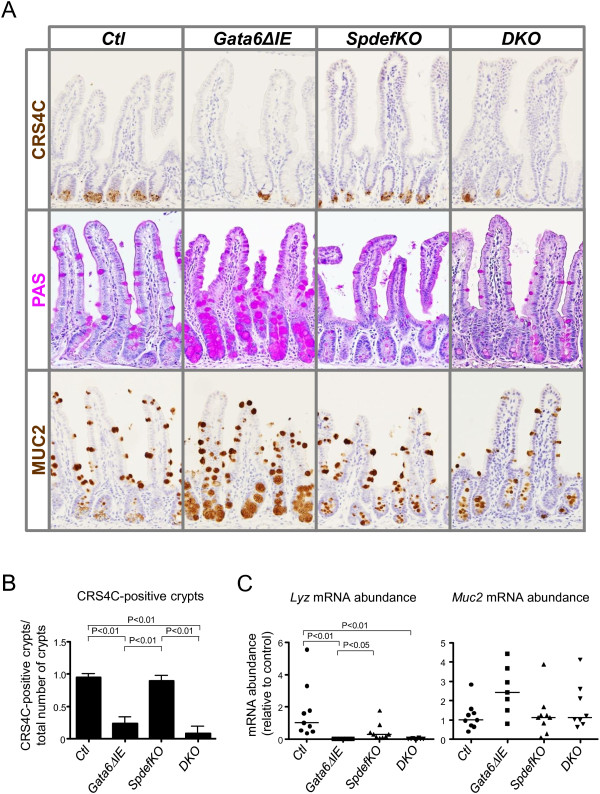
Previously, we also showed that conditional Gata6 deletion resulted in a transformation of Paneth cells into Mucin-2 (MUC2)-enriched goblet-like cells at the base of crypts [4]. In the present study, immunostaining for CRS4C, a Paneth-specific marker, was slightly reduced in SpdefKO mice as compared to controls, consistent with previous data [9], but was greatly reduced in both Gata6ΔIE and DKO mice as compared to controls (Figure 5A). The number of crypt cross-sections with at least one CRS4C-positive cell was nearly 100% in Ctl and SpdefKO mice, but was less than 30% in both Gata6ΔIE and DKO mice (P < 0.01 for each, Figure 5B). The mRNA abundance for lysozyme (Lyz), another Paneth marker, was not significantly different between Ctl and SpdefKO mice, but was significantly reduced in Gata6ΔIE and DKO mice as compared to controls (P < 0.01 for each, Figure 5C). PAS staining was greatly increased in crypts of Gata6ΔIE mice, as previously shown [4], noticeably reduced in SpdefKO mice, especially in villus goblet cells, as previously shown [9], and similar to controls in DKO mice (Figure 5A). MUC2 immunostaining was greatly enriched in crypts of Gata6ΔIE mice, consistent with our previous report [4], but similar to controls in SpdefKO and DKO mice (Figure 5A). Generally consistent with this observation, Muc2 mRNA abundance in Gata6ΔIE mice was 2.5-fold higher as compared to controls, though not statistically significant, whereas Muc2 mRNA abundances in SpdefKO and DKO mice were similar to controls (Figure 5C). These data indicate that the additional deletion of Spdef did not rescue the decrease in the terminal differentiation of Paneth cells observed in Gata6ΔIE mice, but did at least partially prevent their conversion to MUC2-enriched goblet-like cells. Thus, SPDEF is not necessary for the GATA-mediated differentiation of Paneth cells, but does function in their default differentiation into goblet-like cells in the absence of Gata6. This is consistent with the general function of SPDEF to promote the differentiation of goblet cells [9,10].
Figure 5.
Paneth cells are decreased in Gata6ΔIE and DKO mice. (A) Immunostaining in ileum for CRS4C (first row) and MUC2 (third row), and chemical staining using the PAS reaction (second row). (B) Quantification of CRS4C-positive cells in each group. Sections of ileum were stained for CRS4C, and the number of positive cells/crypt cross section was determined as described in Methods. (C)Lyz and Muc2 mRNA abundance in each group. Gata6ΔIE and DKO mice had significantly lower numbers of CRS4C-positive cells and Lyz mRNA abundance than Ctl mice, consistent with a decrease in mature Paneth cells.
In spite of the common secretory cell phenotypes in Gata6 deletion and Spdef over-expression models [4,10], our data show that Spdef deletion did not rescue the Gata6 knockout defects in enteroendocrine lineage commitment or in Paneth cell differentiation. Closer scrutiny suggests that the secretory cell phenotypes in the Gata6 deletion and Spdef over-expression models are not identical. While the decline in enteroendocrine and Paneth cells, and accumulation of goblet cells, appear similar, the underlying alteration in Paneth and goblet cell differentiation is different. Conditional Gata6 deletion has no effect on the normal commitment and differentiation of goblet cells on villi but reveals a conversion of Paneth cells at the base of crypts into a goblet-like cell type. These cells do not express defensins, but express abundant Muc2; they also express abundant Sox9 and Ephb3[4], indicating that they are committed and targeted Paneth cells. On the other hand, conditional Spdef over-expression results in a generalized increase in goblet cells at the expense of all other epithelial cell types [10]. Hence, these mice show a decline in Paneth cell specification rather than a defect in the terminal differentiation of committed Paneth cells, as observed in our conditional Gata6 knockout model.
SPDEF does not regulate the absorptive enterocyte gene expression function of GATA6
We next examined whether Spdef loss affected GATA6-dependent absorptive enterocyte gene expression. Conditional Gata6 deletion resulted in a down-regulation of specific absorptive enterocyte genes in ileum that include lipid transporters and apolipoproteins, and an up-regulation of genes in absorptive enterocytes normally not expressed or expressed at low levels in small intestine, but expressed at high levels in colon [4]. Using marker genes for these two patterns, apolipoprotein A1 (Apoa1) and carbonic anyhdrase 1 (Car1), respectively, we found that their mRNA abundances were down-regulated and up-regulated, respectively, in Gata6ΔIE mice (Figure 6), consistent with our previous findings [4]. Apoa1 and Car1 mRNA abundance in SpdefKO mice was similar to controls, whereas that in DKO mice was similar to Gata6ΔIE, indicating that SPDEF does not play a role in mediating the conditional Gata6 knockout alteration in absorptive enterocyte gene expression.
Figure 6.
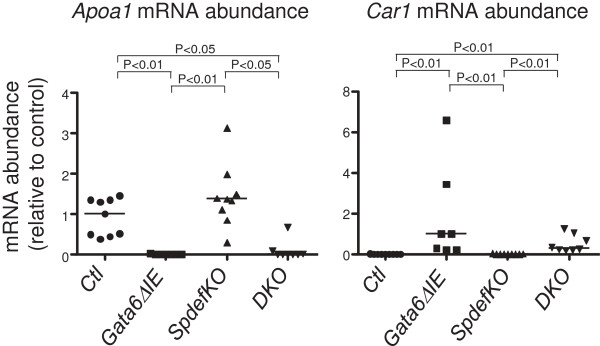
Absorptive enterocyte gene expression is altered in Gata6ΔIE and DKO mice. Gata6ΔIE and DKO mice had significantly lower Apoa1, and significantly higher Car1 mRNA abundances than Ctl mice.
GATA6 is expressed in crypts and in mature absorptive enterocytes [4,5], whereas SPDEF is expressed in secretory progenitors, and in goblet and Paneth cells, but is not expressed in mature absorptive enterocytes [9,10]. While it is possible that SPDEF could instruct gene expression in absorptive enterocytes through a process that originates in progenitors early in the differentiation process, we found that SPDEF did not regulate the GATA6 targets studied here. As in the GATA4-specific pathway, in which GATA4 regulates its targets in mature absorptive enterocytes on villi rather than in crypt progenitors [19], we believe that GATA6 also regulates absorptive enterocyte gene expression within mature absorptive enterocytes on villi.
Conclusion
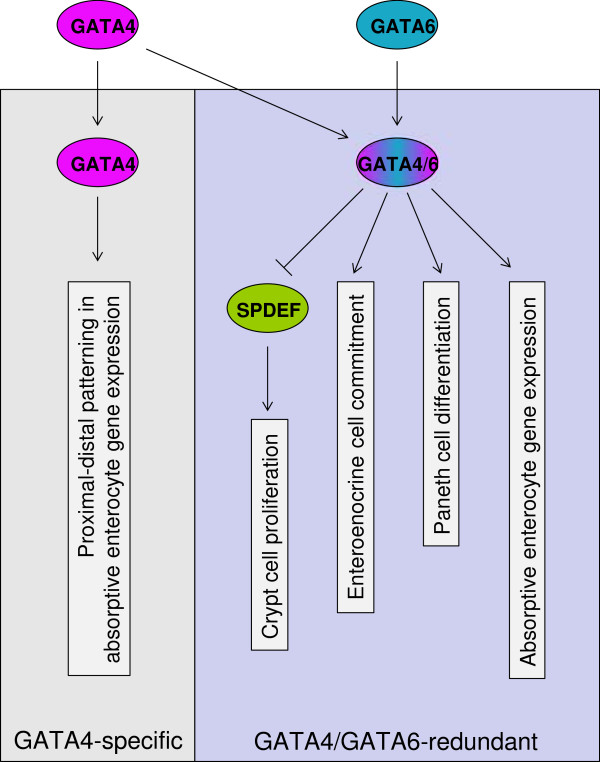
Previously, we defined two fundamental pathways of GATA regulation in the small intestine, one mediated exclusively by GATA4 (GATA4-specific pathway), and one regulated by GATA4 or GATA6 (GATA4/GATA6-redundant pathway) (Figure 7). In the GATA4-specific pathway, GATA4, but not GATA6, activates and represses a subset of absorptive enterocyte genes, and by virtue of its expression in the proximal 85% of small intestine and lack of expression in distal ileum [7], distinguishes proximal intestinal from distal ileal gene expression and function [5,7,8]. For the GATA4/GATA6-redundant pathway, either GATA4 or GATA6 (which is expressed throughout the small intestine, including distal ileum) promote intestinal epithelial renewal by supporting crypt cell proliferation, enteroendocrine lineage commitment, Paneth cell differentiation, and absorptive enterocyte gene expression [4]. Here, we show that the maintenance of crypt cell proliferation function of the intestinal GATA4/GATA6-redundant pathway is dependent on SPDEF (Figure 7). Specifically, our data support the notion that GATA4/GATA6 promotes crypt cell proliferation by directly repressing Spdef gene expression.
Figure 7.
Model for the known GATA pathways in mature small intestine, and the placement of SPDEF within those pathways. GATA4 (purple), but not GATA6 (blue), activates and represses a subset of absorptive enterocyte genes (GATA4-specific), whereas either GATA4 or GATA6 (purple/blue) promote crypt cell proliferation, enteroendocrine lineage commitment, Paneth cell differentiation, and absorptive enterocyte gene expression (GATA4/GATA6-redundant). GATA4/GATA6 promotes the crypt cell proliferation function by repressing the gene encoding SPDEF (green).
GATA6 has been suggested to be oncogenic in multiple cancers and pre-cancerous lesions, including Barrett’s esophagus, gastric cancer, pancreatic cancer, and colon cancer [20]. Gata6 expression is up-regulated in colon cancer epithelial cells [21-23], as well as in non-malignant cells along the stromal margins in human colorectal cancer [22], but it should be noted that it has not been determined whether this is a correlative, causative or protective event. Recently, Noah et al. [24] showed that SPDEF functions as a colorectal tumor suppressor. In colorectal tumors from patients, loss of SPDEF was observed in approximately 85% of tumors and correlated with progression from normal tissue, to adenoma, to adenocarcinoma. Further, SPDEF inhibited the expression of β-catenin-target genes in mouse colon tumors, and interacted with β-catenin to block its transcriptional activity in colorectal cancer cell lines, resulting in lower levels of cyclin D1 and c-MYC. Pathway analysis showed that Gata6 deletion also modulated c-MYC targets (Additional file 1: Figure S1) and that the GATA6/SPDEF overlapping gene set likely functions in Wnt signaling (Figure 1). Thus, our data here suggest that the possible tumorigenic effects of GATA6 could be mediated by its repression of Spdef gene expression.
Methods
Analysis of publicly available gene profiling data
From the National Center for Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/), we downloaded gene profiling data from the ileum of conditional Gata6 knockout mice (GSE22416) [4], and from the small intestine of Spdef-/- mice (GSE14892) [9]. Data analysis was performed using Cistrome (http://www.cistrome.org), a flexible bioinformatics workbench with an analysis platform for ChIP-seq and gene expression microarray analysis [25]. Significant over-representation of overlapping target genes was determined by the Fisher’s Exact Test. Gene ontology analysis and pathway analysis was conducted using Database for Annotation, Visualization and Integrated Discovery (DAVID) [26]. Gene set enrichment analysis was performed by using publicly available Gene Set Enrichment Analysis (GSEA) tools v2.3 from the Broad Institute [27].
Cell culture and lentiviral infection
Lentiviral infection was conducted as previously described [28]. 293 T and Caco-2 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Mediatech, Inc., Manassas, VA) containing glucose (4.5 g/liter), l-glutamine, and sodium pyruvate. The media was supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) and penicillin (100 U/ml)-streptomycin (1 mg/ml) (Sigma Chemical Company, St. Louis, MO). Both cell lines were maintained in 5% CO2 at 95% relative humidity and 37°C. Media were replaced every 2 to 3 days. To produce lentivirus for the knockdown of Gata6, 293 T cells were plated at 105 cells/ml on 6-cm plates and transfected after 24 h with 1 μg of vesicular stomatitis virus glycoprotein G (VSV-G) envelope-expressing plasmid pMD.G, 1 μg of pCMV-dR8.91 (Delta 8.9) plasmid containing gag, pol, and rev genes, and 1 μg of shRNA-pLKO.1 plasmids expressing a knockdown short hairpin RNA (shRNA) (Sigma) for green fluorescent protein (GFP) (used as a control [Ctl]) (SHC005) or human Gata6 (G6kd) using 6 μl of FuGENE 6 reagent (Roche Diagnostic Corporation, Indianapolis, IN). A total of five human Gata6 knockdown constructs were screened, and only one (TRCN0000005392) was subsequently found to reduce Gata6 mRNA significantly. Media from the transfected 293 T cells were changed 16 h after transfection. Culture media containing lentiviral particles were collected and filtered (0.45 μm pore size) 48 h after transfection and transferred immediately to Caco-2 cells plated at 30% confluence to infect for 2 h using 0.5 μl of polybrene/ml. Infection was repeated the next day followed by selection of infected cells using 4 μg/ml puromycin. Infected cells were kept under conditions of selection until the day of harvesting. Trypsinized cells were homogenized using a QIA shredder (Qiagen, Inc., Valencia, CA), and RNA was isolated as described above for mouse tissue. Nuclear extracts were isolated as described previously [11]. Gata6 knockdown was determined by quantitative reverse transcriptase- (qRT-) PCR for human Gata6 (see Additional file 1: Figure S3A for primers) and by Western blot analysis as previously described [5] using rabbit anti-GATA6 (Cell Signaling Technology, Inc, Danvers, MA; Cat. No. 4253S).
Chromosomal immunoprecipitation (ChIP) assays
Using a publicly available GATA6 chromatin immunoprecipitation-high throughput sequencing (ChIP-seq) database in Caco-2 cells [12], sequences were mapped to reference genome Homo Sapiens build 18 (HG18) using ELAND tools, allowing 0 to 2 mismatches (Illumina), and binding peaks were identified by model-based analysis of ChIP-seq (MACS) [29] using default parameters and P value cutoffs of 10-10.
Chromosomal immunoprecipitation (ChIP) assays were conducted as previously described [28]. Caco-2 cells were incubated in DMEM containing 1% formaldehyde (Fisher Scientific, Pittsburgh, PA) for 10 min at 37°C. The cells were washed 2 times with PBS, scraped, and resuspended in lysis buffer (50 mM Tris-Cl [pH 8.1], 10 mM EDTA, 1% sodium dodecyl sulfate [SDS]) containing protease inhibitor cocktail and PMSF (10 μl/ml). The samples were sonicated to obtain chromatin fragments of between 400 and 1,000 bp. Sonicated samples were resuspended in ChIP dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris–HCl, pH 8.1) and incubated overnight at 4°C with Dynabeads beads (Life Technologies, Grand Island, NY) conjugated with rabbit anti-GATA6 (Cell Signaling; Cat. No. 4253) or rabbit IgG (negative control) (Millipore, Temecula, CA; Cat. No. PP64B). The IP samples were washed 6 times with radioimmunoprecipitation (RIPA) buffer (50 mM HEPES [pH 7.6], 0.5 M LiCl, 1 mM EDTA, 1% Nonidet P-40, 0.7% sodium deoxycholate), and the DNA was recovered by reverse cross-linking in 1% SDS–0.1 M NaHCO3 for 7 h at 65°C. DNA was purified using a QIAquick PCR purification kit (Qiagen) and quantified by Picogreen (Life Technologies). One nanogram of DNA was used per qPCR reaction using primers shown in Additional file 1: Figure S3B.
Mice
Previously established and confirmed Gata6 loxP/loxP [30], Spdef-/-[9], and transgenic VillinCreERT2[31] mice were used in this study to produce four groups of mice, including controls (Ctl), conditional Gata6 deletion in the intestinal epithelium (Gata6ΔIE), germline Spdef knockout (SpdefKO), and double Gata6/Spdef knockout (DKO). The genotypes of each group are as follows:
Ctl: Gata6 loxP/loxP , Spdef +/-, VillinCreERT2-negative
Gata6ΔIE: Gata6 loxP/loxP , Spdef +/-, VillinCreERT2-positive
SpdefKO: Gata6 loxP/loxP , Spdef -/-, VillinCreERT2-negative
DKO: Gata6 loxP/loxP , Spdef -/-, VillinCreERT2-positive
DNA was extracted from tail biopsies, and genotypes were determined by semiquantitive polymerase chain reaction (PCR) using previously validated primers (Additional file 1: Figure S3C). Male and female mice four weeks of age were treated with a single dose of tamoxifen (Sigma) (0.1 ml, 10 mg/ml; dissolved in ethanol/sunflower oil = 1:9 [vol/vol]) daily for five consecutive days as described [5], followed by a single dose two weeks later (see timeline, Additional file 1: Figure S4). Mice were killed and tissue was collected 28 days after the start of tamoxifen treatment. Bromodeoxyuridine (BrdU, 0.1 ml of 10 mg/ml) was injected two hours before dissection. Approval was obtained from the Institutional Animal Care and Use Committee.
Tissue isolation and processing
Mice were anesthetized for dissection as previously described [28]. The most distal 1.0 cm segment of small intestine adjacent to the ileocecal valve was snap frozen for RNA isolation, and the 6 cm segment proximal to that was removed and transferred to a glass plate on wet ice and prepared for sectioning. This segment was flushed with ice cold 4% paraformaldehyde (PFA) in PBS, cut longitudinally, pinned open onto paraffin wax in a petri dish filled with 4% PFA in PBS with the epithelium facing upward. After a 5 min incubation period, the pins were removed and the proximal end was grasped with forceps, rolled with the epithelium facing outwards, and a pin was inserted transversely to secure the roll in place. The roll was placed in 10 ml of 4% PFA in PBS and mixed gently on a tube rotator for 16–18 hr at 4°C. The PFA was decanted and the tissue was washed in PBS 3 times for 20 min each at 4°C, and then dehydrated in 70% ethanol. Tissue was processed at the Rodent Histopathology Core at the Dana Farber/Harvard Cancer Center (Boston, MA) for paraffin embedding and sectioning. Selected slides were stained using the periodic acid Schiff (PAS) reaction.
RNA isolation and gene expression analysis
RNA was isolated using the RNeasy kit (Qiagen), and mRNA abundances were determined by qRT-PCR as previously described [4], using validated primer pairs (Additional file 1: Figure S3A). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA abundance was measured for each sample and used to normalize the data. Data were expressed relative to the median value of control ileum. A minimum of five mice in each group was analyzed.
Immunohistochemistry
Tissue sections were immunostained as previously described [28]. Primary antibodies included rabbit anti-Ki67 (Thermo Fisher Scientific, Inc., Fremont, CA; Cat. No. RM-9106-S1) (1:200), mouse anti-BrdU (Thermo; Cat. No. MS-1058-PO) (1:250), goat anti-cryptdin related sequence 4C (CRS4C) (gift from Dr. A. J. Ouellette, University of Southern California, Los Angeles, CA) [32] (1:2000), and rabbit anti-MUC2 (Santa Cruz Biotechnology, Inc, Santa Cruz, CA; Cat. No. sc15334) (1:100). Secondary antibodies included biotinylated donkey anti-rabbit IgG, donkey anti-goat IgG, and donkey anti-mouse IgG (all from Vector Labs, Burlingame, CA). Biotinylated antibodies were linked to avidin-horseradish peroxidase conjugates (Vector Labs), visualized using 3,3′-diamino benzidine (Sigma) for 2 to 5 min, and lightly counterstained with hematoxylin.
Cell counting
The total number of Ki67-, BrdU-, or CRS4C-positive cells in crypts was determined as the total number per crypt. Only well oriented crypts with the epithelial layer on at least one side continuous with the villus epithelial layer were counted, and a minimum of 6 crypts per slide were analyzed. The average number of CHGA–positive cells was expressed as a fraction of total epithelial cells (villi and crypts) from a minimum of 5000 epithelial cells per slide, with equal representation of crypts and villi. All determinations were blinded and conducted on a minimum of 5 animals per group.
Statistical analyses
In Caco-2 cells, qRT-PCR data was compared by the student’s t-test and ChIP data were compared by the analysis of variance (ANOVA) followed by the Tukey-Kramer multiple comparison test. In mice, mRNA measurements had unequal variances across groups requiring nonparametric statistics, and were thus compared by the Kruskal-Wallis test followed by the Dunn multiple comparison test, and presented as individual data points and medians. Cell count determinations had equal variances across groups allowing parametric statistics, and were thus compared by ANOVA followed by the Tukey-Kramer multiple comparison test, and presented as mean ± SD. Differences were considered statistically significant at a P-value of less than 0.05.
Competing interests
The authors declare no competing interest.
Authors’ contributions
BEA, LATMV, ES and HB performed and interpreted the Caco-2 cell culture experiments. KAS performed and interpreted the in vivo mouse experiments. BEA, KAS and SDK contributed to the study design and interpretation of results, and the writing of the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Intestinal Gata6 deletion alters gene networks controlling cell proliferation in the mature ileum. Network analyses on microarray data of Ctl and Gata6ΔIE ileum (n=3 in each group) revealed (A) an increase in targets of the tumor surpressor gene p53, and (B) a decrease in targets of the proto-oncogene c-MYC. (C) Legend defining symbols used in the networks. Arrows indicate the direction of the interaction. Red circles = up-regulated transcripts; Blue circles = down-regulated transcripts. Differentially expressed transcripts were determined at the 5% FDR level using Significance Analysis of Microarrays (SAM) and interaction networks were developed from the differentially expressed transcripts using Metacore. Figure S2. Gata6 and Spdef mRNA abundance in ileum in each group of mice. Gata6ΔIE and DKO mice had significantly lower Gata6 mRNA abundances than Ctl and SpdefKO mice. SpdefKO and DKO mice had significantly lower Spdef mRNA abundances than Ctl and Gata6ΔIE mice. Figure S3. Primers used for: (A) qRT-PCR, (B) ChIP assays, and (C) genotyping. Figure S4. Timeline for study. Mice 4 wks of age were given Tamoxifen as indicated (black circle) beginning on Day 0, and an injection of BrdU 2 hr before tissue collection at Day 28.
Contributor Information
Boaz E Aronson, Email: boazaronson@gmail.com.
Kelly A Stapleton, Email: kstapleton@gmail.com.
Laurens ATM Vissers, Email: sandervissers61@hotmail.com.
Eva Stokhuijzen, Email: e.stokhuijzen@gmail.com.
Hanneke Bruijnzeel, Email: h.bruijnzeel@gmail.com.
Stephen D Krasinski, Email: stephen.krasinski@childrens.harvard.edu.
Acknowledgements
We would like to thank Drs. E. Beuling and M. P. Verzi for helpful suggestions; Dr. A. J. Ouellette for the CRS4C antibody; and Dr. H. Clevers for the Spdef-/- mice.
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases grant RO1-DK-061382 (S.D.K.), the Harvard Digestive Disease Center (5P30-DK-34854), the Nutricia Research Foundation (B.E.A.), KWF Kankerbestrijding (B.E.A.), Prins Bernhard Cultuurfonds (B.E.A.) in The Netherlands, and the European Society for Pediatric Research (B.E.A.) in Switzerland.
References
- Noah TK, Donahue B, Shroyer NF. Intestinal development and differentiation. Exp Cell Res. 2011;317(19):2702–2710. doi: 10.1016/j.yexcr.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbe F, van Es JH, Makrini L, Brulin B, Mellitzer G, Robine S, Romagnolo B, Shroyer NF, Bourgaux JF, Pignodel C. et al. Distinct ATOH1 and Neurog3 requirements define tuft cells as a new secretory cell type in the intestinal epithelium. J Cell Biol. 2011;192(5):767–780. doi: 10.1083/jcb.201010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung TM, Chia LA, Kosinski CM, Kuo CJ. Regulation of self-renewal and differentiation by the intestinal stem cell niche. Cell Mol Life Sci. 2011;68(15):2513–2523. doi: 10.1007/s00018-011-0687-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuling E, Baffour-Awuah NY, Stapleton KA, Aronson BE, Noah TK, Shroyer NF, Duncan SA, Fleet JC, Krasinski SD. GATA factors regulate proliferation, differentiation, and gene expression in small intestine of mature mice. Gastroenterology. 2011;140(4):1219–1229. doi: 10.1053/j.gastro.2011.01.033. e1211-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosse T, Piaseckyj CM, Burghard E, Fialkovich JJ, Rajagopal S, Pu WT, Krasinski SD. Gata4 is essential for the maintenance of jejunal-ileal identities in the adult mouse small intestine. Mol Cell Biol. 2006;26(23):9060–9070. doi: 10.1128/MCB.00124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wering HM, Bosse T, Musters A, de Jong E, de Jong N, Hogen Esch CE, Boudreau F, Swain GP, Dowling LN, Montgomery RK. et al. Complex regulation of the lactase-phlorizin hydrolase promoter by GATA-4. Am J Physiol Gastrointest Liver Physiol. 2004;287(4):G899–G909. doi: 10.1152/ajpgi.00150.2004. [DOI] [PubMed] [Google Scholar]
- Beuling E, Kerkhof IM, Nicksa GA, Giuffrida MJ, Haywood J, aan de Kerk DJ, Piaseckyj CM, Pu WT, Buchmiller TL, Dawson PA. et al. Conditional Gata4 deletion in mice induces bile acid absorption in the proximal small intestine. Gut. 2010;59(7):888–895. doi: 10.1136/gut.2009.204990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle MA, Bondow BJ, Iverson MA, Adams SJ, Jandacek RJ, Tso P, Duncan SA. GATA4 is essential for jejunal function in mice. Gastroenterology. 2008;135(5):1676–1686. doi: 10.1053/j.gastro.2008.07.074. e1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorieff A, Stange DE, Kujala P, Begthel H, van den Born M, Korving J, Peters PJ, Clevers H. The ets-domain transcription factor Spdef promotes maturation of goblet and paneth cells in the intestinal epithelium. Gastroenterology. 2009;137(4):1333–1345. doi: 10.1053/j.gastro.2009.06.044. e1331-1333. [DOI] [PubMed] [Google Scholar]
- Noah TK, Kazanjian A, Whitsett J, Shroyer NF. SAM pointed domain ETS factor (SPDEF) regulates terminal differentiation and maturation of intestinal goblet cells. Exp Cell Res. 2010;316(3):452–465. doi: 10.1016/j.yexcr.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasinski SD, Van Wering HM, Tannemaat MR, Grand RJ. Differential activation of intestinal gene promoters: functional interactions between GATA-5 and HNF-1 alpha. Am J Physiol Gastrointest Liver Physiol. 2001;281(1):G69–G84. doi: 10.1152/ajpgi.2001.281.1.G69. [DOI] [PubMed] [Google Scholar]
- Verzi MP, Shin H, He HH, Sulahian R, Meyer CA, Montgomery RK, Fleet JC, Brown M, Liu XS, Shivdasani RA. Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev Cell. 2010;19(5):713–726. doi: 10.1016/j.devcel.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Mitra A, Dojer N, Fu S, Rowicka M, Brasier AR. A probabilistic approach to learn chromatin architecture and accurate inference of the NF-kappaB/RelA regulatory network using ChIP-Seq. Nucleic Acids Res. 2013;41(15):7240–7259. doi: 10.1093/nar/gkt493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev. 2012;26(1):11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payankaulam S, Li LM, Arnosti DN. Transcriptional repression: conserved and evolved features. Curr Biol. 2010;20(17):R764–R771. doi: 10.1016/j.cub.2010.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, He A, Pu WT. Regulation of GATA4 transcriptional activity in cardiovascular development and disease. Curr Top Dev Biol. 2012;100:143–169. doi: 10.1016/B978-0-12-387786-4.00005-1. [DOI] [PubMed] [Google Scholar]
- Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, Yu D, Hardison R, Weiss MJ, Orkin SH. et al. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol Cell. 2009;36(4):682–695. doi: 10.1016/j.molcel.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, O’Geen H, Keles S, Blahnik K, Linnemann AK, Kang YA, Choi K, Farnham PJ, Bresnick EH. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol Cell. 2009;36(4):667–681. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuling E, Bosse T, aan de Kerk DJ, Piaseckyj CM, Fujiwara Y, Katz SG, Orkin SH, Grand RJ, Krasinski SD. GATA4 mediates gene repression in the mature mouse small intestine through interactions with friend of GATA (FOG) cofactors. Dev Biol. 2008;322(1):179–189. doi: 10.1016/j.ydbio.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayanbule F, Belaguli NS, Berger DH. GATA factors in gastrointestinal malignancy. World J Surg. 2011;35(8):1757–1765. doi: 10.1007/s00268-010-0950-1. [DOI] [PubMed] [Google Scholar]
- Belaguli NS, Aftab M, Rigi M, Zhang M, Albo D, Berger DH. GATA6 promotes colon cancer cell invasion by regulating urokinase plasminogen activator gene expression. Neoplasia. 2010;12(11):856–865. doi: 10.1593/neo.10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haveri H, Westerholm-Ormio M, Lindfors K, Maki M, Savilahti E, Andersson LC, Heikinheimo M. Transcription factors GATA-4 and GATA-6 in normal and neoplastic human gastrointestinal mucosa. BMC Gastroenterol. 2008;8:9. doi: 10.1186/1471-230X-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shureiqi I, Zuo X, Broaddus R, Wu Y, Guan B, Morris JS, Lippman SM. The transcription factor GATA-6 is overexpressed in vivo and contributes to silencing 15-LOX-1 in vitro in human colon cancer. FASEB J. 2007;21(3):743–753. doi: 10.1096/fj.06-6830com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noah TK, Lo YH, Price A, Chen G, King E, Washington MK, Aronow BJ, Shroyer NF. SPDEF Functions as a Colorectal Tumor Suppressor by Inhibiting beta-Catenin Activity. Gastroenterology. 2013;144(5):1012–1023. doi: 10.1053/j.gastro.2013.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y, Shin H, Wong SS, Ma J, Lei Y. et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011;12(8):R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Zheng X, Yang J, Imamichi T, Stephens R, Lempicki RA, Baxevanis AD. et al. Extracting biological meaning from large gene lists with DAVID. Current Protoc Bioinformatics / editoral board. 2009;Chapter 13:Unit 13 11. doi: 10.1002/0471250953.bi1311s27. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuling E, Aronson BE, Tran LM, Stapleton KA, ter Horst EN, Vissers LA, Verzi MP, Krasinski SD. GATA6 is required for proliferation, migration, secretory cell maturation, and gene expression in the mature mouse colon. Mol Cell Biol. 2012;32(17):3392–3402. doi: 10.1128/MCB.00070-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W. et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodhi CP, Li J, Duncan SA. Generation of mice harbouring a conditional loss-of-function allele of Gata6. BMC Dev Biol. 2006;6:19. doi: 10.1186/1471-213X-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el Marjou F, Janssen KP, Chang BH, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, Robine S. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39(3):186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- Ouellette AJ, Lualdi JC. A novel mouse gene family coding for cationic, cysteine-rich peptides. Regulation in small intestine and cells of myeloid origin. J Biol Chem. 1990;265(17):9831–9837. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Intestinal Gata6 deletion alters gene networks controlling cell proliferation in the mature ileum. Network analyses on microarray data of Ctl and Gata6ΔIE ileum (n=3 in each group) revealed (A) an increase in targets of the tumor surpressor gene p53, and (B) a decrease in targets of the proto-oncogene c-MYC. (C) Legend defining symbols used in the networks. Arrows indicate the direction of the interaction. Red circles = up-regulated transcripts; Blue circles = down-regulated transcripts. Differentially expressed transcripts were determined at the 5% FDR level using Significance Analysis of Microarrays (SAM) and interaction networks were developed from the differentially expressed transcripts using Metacore. Figure S2. Gata6 and Spdef mRNA abundance in ileum in each group of mice. Gata6ΔIE and DKO mice had significantly lower Gata6 mRNA abundances than Ctl and SpdefKO mice. SpdefKO and DKO mice had significantly lower Spdef mRNA abundances than Ctl and Gata6ΔIE mice. Figure S3. Primers used for: (A) qRT-PCR, (B) ChIP assays, and (C) genotyping. Figure S4. Timeline for study. Mice 4 wks of age were given Tamoxifen as indicated (black circle) beginning on Day 0, and an injection of BrdU 2 hr before tissue collection at Day 28.