Abstract
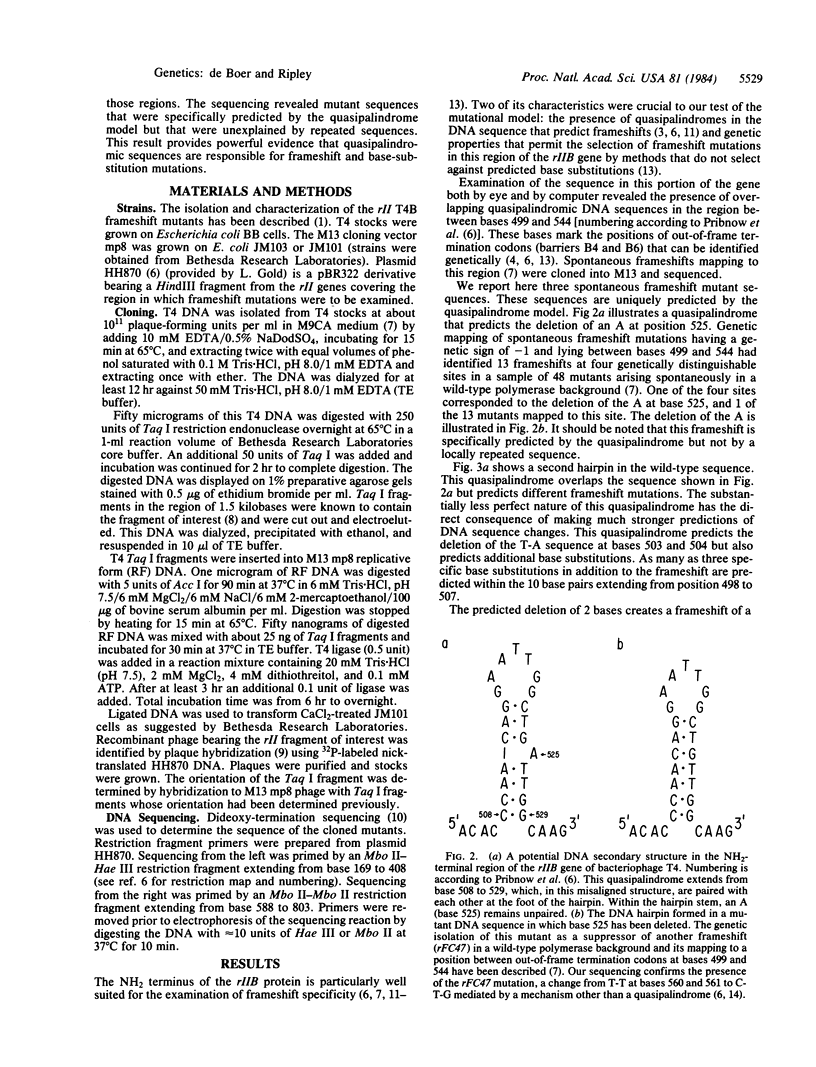
The in vivo production of frameshift and base-substitution mutations predicted as a consequence of the metabolic processing of misaligned quasipalindromic DNA sequences has been confirmed. Spontaneous frameshift mutations of the T4 rII gene that had been genetically mapped to quasipalindromic DNA sequences were sequenced. Some of the mutant sequences are exactly those predicted by a mutational model based on misaligned quasipalindromes. Furthermore, these sequences are distinct from those predicted by the classical frameshift model based on misaligned repeated sequences. The rII frameshift mutant sequences reported here result from the deletion of a specific base or bases that would remain looped out should the quasipalindromes assume a hairpin secondary structure. One hairpin predicted not only the deletion of two bases (a frameshift) but the concomitant production of nearby but noncontiguous base substitutions. The substitution of as many as three bases as well as the frameshift were predicted to arise as a consequence of a single mutational event in the palindrome. Two independent examples of the predicted deletion frameshift were found among the small sample of sequenced spontaneous frameshifts examined and both were associated with the predicted transversion and transition base substitutions.
Full text
PDF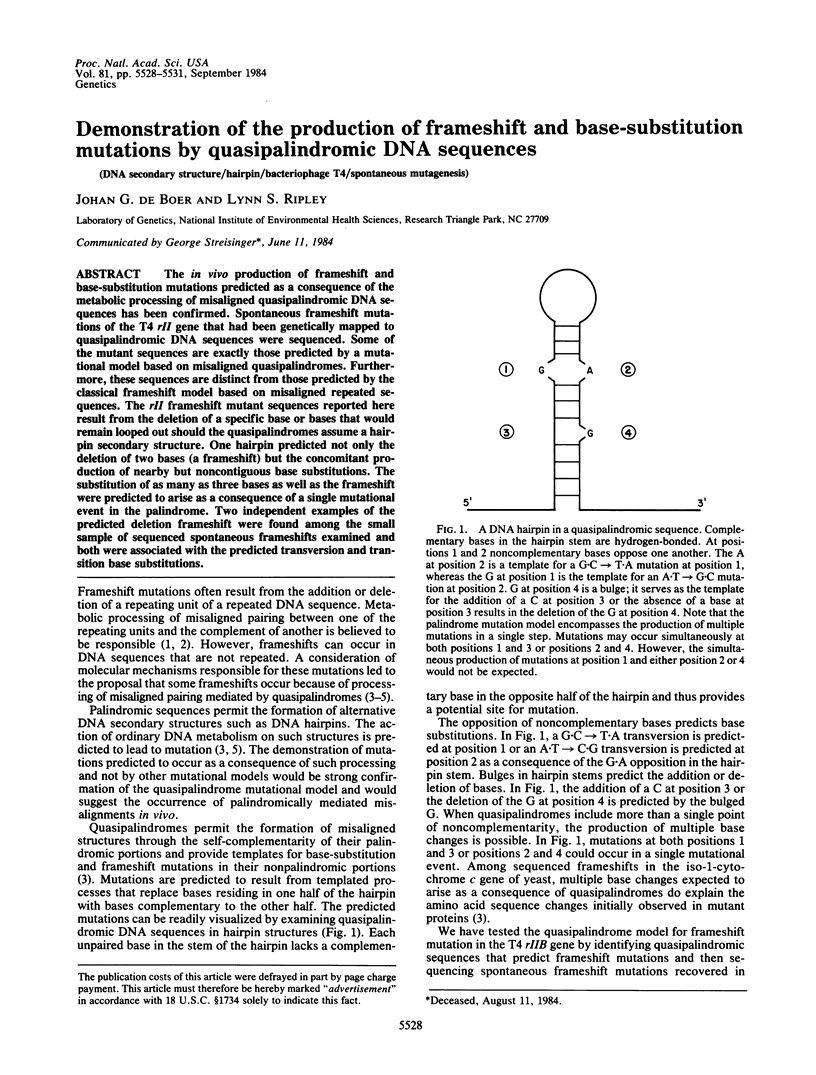


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- CRICK F. H., BARNETT L., BRENNER S., WATTS-TOBIN R. J. General nature of the genetic code for proteins. Nature. 1961 Dec 30;192:1227–1232. doi: 10.1038/1921227a0. [DOI] [PubMed] [Google Scholar]
- Okada Y., Streisinger G., Owen J. E., Newton J., Tsugita A., Inouye M. Molecular basis of a mutational hot spot in the lysozyme gene of bacteriophage T4. Nature. 1972 Apr 14;236(5346):338–341. doi: 10.1038/236338a0. [DOI] [PubMed] [Google Scholar]
- Pribnow D., Sigurdson D. C., Gold L., Singer B. S., Napoli C., Brosius J., Dull T. J., Noller H. F. rII cistrons of bacteriophage T4. DNA sequence around the intercistronic divide and positions of genetic landmarks. J Mol Biol. 1981 Jul 5;149(3):337–376. doi: 10.1016/0022-2836(81)90477-0. [DOI] [PubMed] [Google Scholar]
- Ripley L. S., Glickman B. W., Shoemaker N. B. Mutator versus antimutator activity of a T4 DNA polymerase mutant distinguishes two different frameshifting mechanisms. Mol Gen Genet. 1983;189(1):113–117. doi: 10.1007/BF00326062. [DOI] [PubMed] [Google Scholar]
- Ripley L. S., Glickman B. W. Unique self-complementarity of palindromic sequences provides DNA structural intermediates for mutation. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):851–861. doi: 10.1101/sqb.1983.047.01.097. [DOI] [PubMed] [Google Scholar]
- Ripley L. S. Model for the participation of quasi-palindromic DNA sequences in frameshift mutation. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4128–4132. doi: 10.1073/pnas.79.13.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripley L. S., Shoemaker N. B. A major role for bacteriophage T4 DNA polymerase in frameshift mutagenesis. Genetics. 1983 Mar;103(3):353–366. doi: 10.1093/genetics/103.3.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Sugino A., Drake J. W. Modulation of mutation rates in bacteriophage T4 by a base-pair change a dozen nucleotides removed. J Mol Biol. 1984 Jun 25;176(2):239–249. doi: 10.1016/0022-2836(84)90422-4. [DOI] [PubMed] [Google Scholar]


