Abstract
The shortage of oil resources, the steadily rising oil prices and the impact of its use on the environment evokes an increasing political, industrial and technical interest for development of safe and efficient processes for the production of chemicals from renewable biomass. Thus, microbial fermentation of renewable feedstocks found its way in white biotechnology, complementing more and more traditional crude oil-based chemical processes. Rational strain design of appropriate microorganisms has become possible due to steadily increasing knowledge on metabolism and pathway regulation of industrially relevant organisms and, aside from process engineering and optimization, has an outstanding impact on improving the performance of such hosts. Corynebacterium glutamicum is well known as workhorse for the industrial production of numerous amino acids. However, recent studies also explored the usefulness of this organism for the production of several organic acids and great efforts have been made for improvement of the performance. This review summarizes the current knowledge and recent achievements on metabolic engineering approaches to tailor C. glutamicum for the bio-based production of organic acids. We focus here on the fermentative production of pyruvate, l-and d-lactate, 2-ketoisovalerate, 2-ketoglutarate, and succinate. These organic acids represent a class of compounds with manifold application ranges, e.g. in pharmaceutical and cosmetics industry, as food additives, and economically very interesting, as precursors for a variety of bulk chemicals and commercially important polymers.
Funding Information Work in the laboratories of the authors was supported by the Fachagentur Nachwachsende Rohstoffe (FNR) of the Bundesministerium für Ernährung, Landwirtschaft und Verbraucherschutz (BMELV; FNR Grants 220-095-08A and 220-095-08D; Bio-ProChemBB project, ERA-IB programme), by the Deutsche Bundesstiftung Umwelt (DBU Grant AZ13040/05) and the Evonik Degussa AG.
Introduction
The depletion of earth's fossil energy resources, accompanied by the strong impact of their use on the environment, particularly in form of higher CO2 emissions, raises the demand and the consumer pull for sustainable, safe and efficient substitution of hitherto crude oil derived chemicals and chemical building blocks from renewable resources. Besides chemical manufacturing of renewable feedstocks to valuable compounds, biotechnological processes afford more and more opportunities to produce fuels, building blocks, and solvents in a cost-effective way from biomass (Bozell and Petersen, 2010). Chemical buildings blocks, such as some organic acids, serve as precursors for a variety of bulk chemicals and commercially important polymers (Werpy and Petersen, 2004). The cost-effective bio-based production of these chemicals is a most relevant goal for the future and has to meet economic and environmental requirements. Therefore, the microbial production systems have to perform excellent with regard to yield, productivity, product purity and flexibility to substrate consumption.
Corynebacterium glutamicum is a Gram-positive facultative anaerobic organism that grows on a variety of sugars, organic acids, and alcohols as single or combined carbon and energy sources (Eggeling and Bott, 2005; Liebl, 2006; Nishimura et al., 2007; Takeno et al., 2007). The organism is generally regarded as safe (GRAS status) and is traditionally employed for large scale production of amino acids, such as l-glutamate (> 2 million t/a) and l-lysine (> 1.4 million t/a) (Eggeling and Bott, 2005; Takors et al., 2007; Ajinomoto, 2010; 2011). In order to improve the production performance by metabolic engineering approaches, the central carbon metabolism, the physiology and the regulation of main and specific pathways of C. glutamicum were analysed in detail and genetic tools as well as systems biology approaches on the ‘omics’ level have been developed and employed (overviews in Kirchner and Tauch, 2003; Eggeling and Bott, 2005; Sauer and Eikmanns, 2005; Wendisch et al., 2006a; Bott, 2007; Takors et al., 2007; Burkowski, 2008; Brinkrolf et al., 2010; Becker and Wittmann, 2011; Teramoto et al., 2011; Vertes et al., 2012). Since C. glutamicum is regarded as a robust and easily manageable production host, recent studies also focused on the suitability of this organism for the production of other commodity chemicals, such as the biofuels isobutanol and ethanol (Inui et al., 2004b; Smith et al., 2010; Blombach and Eikmanns, 2011; Blombach et al., 2011), the diamines cadaverine and putrescine (Mimitsuka et al., 2007; Schneider and Wendisch, 2010; 2011; Kind et al., 2010a,2010b; Kind and Wittmann, 2011), the sugar alcohol xylitol (Sasaki et al., 2010), gamma-amino butyric acid (Takahashi et al., 2012), polyhodroxybutyrate (Song et al., 2012), and also several organic acids (reviewed in this article).
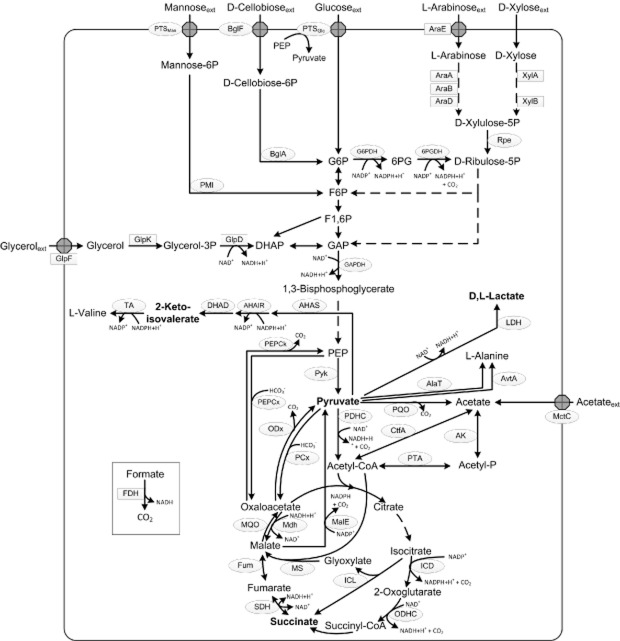
Six years ago, Wendisch and colleagues (2006b) reviewed the metabolic engineering of C. glutamicum and Escherichia coli for the biotechnological production of organic acids and amino acids. At that time, E. coli was the superior platform organism for the production of organic acids and it was hardly known that C. glutamicum forms lactate and succinate under oxygen-deprivation conditions (Dominguez et al., 1993; Inui et al., 2004a; Okino et al., 2005). However, it was foreseeable that genetically modified C. glutamicum strains will become promising biocatalysts for the production of at least some organic acids. As outlined in the following, great efforts have been made in the last 7 years to successfully implement and/or to improve the production of several organic acids with C. glutamicum. This review focuses on the metabolic/genetic engineering approaches to tailor C. glutamicum for the fermentative production of pyruvate, d-and l-lactate, 2-ketoisovalerate, 2-ketoglutarate, and succinate from renewable carbon sources. Figure 1 gives an overview on pathways and enzymes of the central metabolism of C. glutamicum, including the pathways for the degradation of selected substrates and those for the synthesis of organic acids produced with this organism.
Figure 1.
Schematic presentation of the central carbon metabolism of C. glutamicum including pathways for the degradation of carbon sources (glucose, glycerol, d-cellobiose, l-arabinose, d-xylose, mannose, formate, acetate) used for the production of pyruvate, d,l-lactate, 2-ketoisovalerate, 2-ketoglutarate and succinate. Ellipses represent enzymes and transport systems present in C. glutamicum. Rectangles represent heterologous enzymes. Abbreviations: Coding genes are given in brackets. 6PG, 6P-gluconate; 6PGDH (gnd), 6PG dehydrogenase; AHAIR (ilvC), acetohydroxyacid isomeroreductase; AHAS (ilvBN), acetohydroxyacid synthase; AK (ack), acetate kinase; AlaT (alaT), alanine aminotransferase; AraA (araA from E. coli), arabinose isomerase; AraB (araB from E. coli), ribulokinase; AraD (araD from E. coli), l-ribulose-5-phosphate 4-epimerase; AraE (araE from E. coli), l-arabinose transporter; AvtA (avtA), valine-pyruvate aminotransferase; BglA (bglA1, bglA2), phospho-β-glucosidases; BglF (bglFV317A), mutated PTS permease enabling d-cellobiose import; CtfA (cat), CoA transferase A; DHAD (ilvD), dihydroxyacid dehydratase; DHAP, dihydroxyacetone-P; F1,6P, fructose-1,6P; F6P, fructose-6P; FDH (fdh from Mycobacterium vaccae), formate dehydrogenase; Fum (fum), fumarase; GAP, glyceraldehyde-3P; GAPDH (gapA), GAP dehydrogenase; GlpD (glpD from E. coli), glycerol-3P dehydrogenase; GlpF (glpF from E. coli), glycerol facilitator; GlpK (glpK from E. coli), glycerol kinase; G6P, glucose-6P; G6PDH (zwf, opcA), G6P dehydrogenase; ICD (icd), isocitrate dehydrogenase; ICL (aceA), isocitrate lyase; LDH (native ldhA or ldhA from L. delbrueckii), l-and d-lactate dehydrogenase respectively; MalE (malE), malic enzyme; MctC (mctC) monocarboxylic acid transporter; Mdh (mdh), malate dehydrogenase; MQO (mqo), malate:quinone oxidoreductase; MS (aceB), malate synthase; ODHC (odhA, aceF, lpd), 2-oxoglutarate dehydrogenase complex; ODx (odx), oxaloacetate decarboxylase; PCx (pyc), pyruvate carboxylase; P, phosphate; PDHC (aceE, aceF, lpd), pyruvate dehydrogenase complex; PEP phosphoenolpyruvate; PEPCk (pck), PEP carboxykinase; PEPCx (ppc), PEP carboxylase; Pyk (pyk), pyruvate kinase; PMI (manA), phosphomannose isomerase; PQO (pqo), pyruvate: quinone oxidoreductase; PTA (pta), phosphotransacetylase; PTS (ptsG, hpr; ptsI), phosphotransferase system; Rpe (rpe), ribulose-5-phosphate epimerase; SDH (sdhABC), succinate dehydrogenase; TA (ilvE), transaminase B; XylA (xylA from E. coli), xylose isomerase; XylB (xylB from E. coli), xylulokinase.
Production of pyruvate
Pyruvate is broadly used as ingredient or additive in food, cosmetics and pharmaceuticals, but also for the synthesis of various chemicals and polymers (Li et al., 2001; Zhu et al., 2008). Chemical production of pyruvate is realized by dehydration and decarboxylation of tartaric acid, but in a cost-ineffective way (Howard and Fraser, 1932; Li et al., 2001). Different approaches were made for pyruvate production with eukaryotic microorganisms like multi-auxotrophic yeasts (reviewed in Li et al., 2001); however, prokaryotic microorganisms, such as E. coli and C. glutamicum, also were successfully engineered to produce pyruvate.
Pyruvate is a central intermediate in the carbon and energy metabolism (see Fig. 1) in all organisms and thus, for construction of an efficient pyruvate-producing C. glutamicum strain, the major pyruvate-drawing reactions had to be downregulated or even eliminated. In the course of the molecular analysis of the pyruvate dehydrogenase complex (PDHC), Schreiner and colleagues (2005) inactivated this complex in C. glutamicum by deletion of the aceE gene, encoding the E1p subunit of the PDHC. The resulting strain C. glutamicum ΔaceE required acetate or ethanol as an additional carbon source for growth on glucose (Schreiner et al., 2005; Blombach et al., 2009). In an approach to engineer C. glutamicum for l-valine production, Blombach and colleagues (2007) observed that C. glutamicum ΔaceE showed a relatively high intracellular concentration of pyruvate and, when acetate was exhausted from the medium and growth stopped, secreted significant amounts of l-alanine (30 mM), l-valine (30 mM), and pyruvate (30 mM) from glucose. In subsequent studies, the PDHC-deficient strain turned out to be an excellent starting point to engineer C. glutamicum for the efficient production of l-valine (Blombach et al., 2007; 2008; 2009; Krause et al., 2009), isobutanol (Blombach et al., 2011), and also of 2-ketoisovalerate (Krause et al., 2010; see below), succinate (see below) and pyruvate. The additional inactivation of the pyruvate:quinone oxidoreductase (PQO) and NADH-dependent l-lactate dehydrogenase (l-LDH) significantly improved pyruvate formation (Wieschalka et al., 2012). In shake-flask experiments, C. glutamicum ΔaceE Δpqo ΔldhA accumulated in a growth-decoupled manner about 50 mM pyruvate with a substrate-specific product yield (YP/S) of 0.48 mol per mol of glucose, aside from l-alanine (29 mM) and l-valine (21 mM) as by-products (Wieschalka et al., 2012). To abolish overflow metabolism towards l-valine, the native acetohydroxyacid synthase (AHAS) was substituted by a leaky variant (ΔC-T IlvN) leading to an almost threefold increased YP/S of 1.36 mol pyruvate per mol of glucose, and a strong increase of pyruvate production (up to 193 mM), while l-valine and l-alanine formation were reduced to 1 mM and 9 mM respectively (Wieschalka et al., 2012). Additional deletion of the genes encoding alanine aminotransferase (AlaT) and valine-pyruvate aminotransferase (AvtA) resulted in cumulative reduction of l-alanine as undesired by-product by 50% (Wieschalka et al., 2012). With the final strain C. glutamicum ΔaceE Δpqo ΔldhA ΔC-T ilvN ΔalaT ΔavtA (designated as C. glutamicum ELB-P; see Fig. 2) up to 200 mM pyruvate were formed in shake-flask experiments, with a YP/S of 1.49 mol per mol of glucose. The yields of the by-products l-alanine and l-valine were evanescent low with 0.03 and 0.01 mol per mol of glucose respectively (Wieschalka et al., 2012). To study the relevance for industrial applications, fed-batch fermentations were performed with C. glutamicum ELB-P. When C. glutamicum ELB-P was cultivated with a constant pO2 of about 30% a twofold lower glucose consumption rate (0.28 mmol g cell dry weight (CDW)−1 h−1) and a significantly lower YP/S (0.8 mol pyruvate per mol of glucose) were observed when compared with shake-flask experiments (0.58 mmol g(CDW)−1 h−1 and 1.49 mol pyruvate per mol of glucose respectively). Implementation of low oxygen tension from the middle until the end of growth phase restored the production performance and led to the formation of more than 500 mM (45 g l−1) pyruvate with a YP/S of 0.97 mol pyruvate per mol of glucose in the production phase (Wieschalka et al., 2012). In comparison, the best pyruvate-producing E. coli strains (E. coli YYC202 and ALS1059) produced under optimized process conditions about 720 mM (63 g l−1) and 1 M (90 g l−1) pyruvate, with YP/S of 1.74 and 1.39 mol pyruvate per mol of glucose respectively (Zelic et al., 2003; Zhu et al., 2008). Since the yield of C. glutamicum ELB-P in shake-flask experiments is in the same range as in these E. coli strains, further process optimization might disclose the whole potential of C. glutamicum ELB-P for a further improved pyruvate production process.
Figure 2.
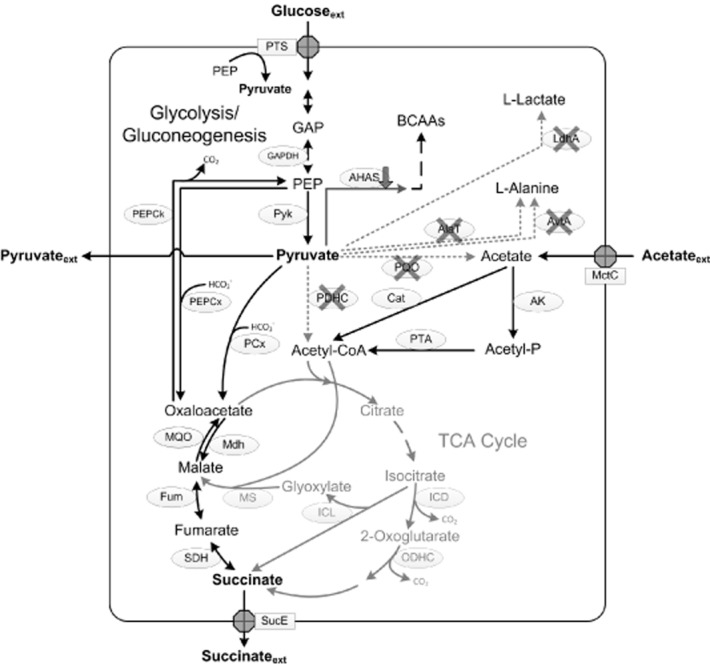
Schematic presentation of the central carbon metabolism of C. glutamicum ELB-P with the corresponding enzymes and modifications, leading to pyruvate production under aerobic conditions and reductive succinate production under anaerobic conditions. For most abbreviations see legend to Fig. 1. Further abbreviations: BCAAs, branched-chain amino acids; LdhA, NAD+-dependent l-lactate dehydrogenase; SucE, succinate exporter; TCA, tricarboxylic acid. Down arrow at AHAS indicates decreased activity of the truncated AHAS derivative, crosses indicate inactivation of the enzyme by deletion of the respective gene. Dotted arrows indicate pathways not present due to gene inactivation, grey arrows indicate pathways not used in C. glutamicum ELB-P.
Production of lactate
Lactate is widely used as both d-and l-isomers for pharmaceutical, cosmetic, leather and textile, chemical, biomedical and food industries, as well as for green solvent and biodegradable fibre and polymer production (Hofvendahl and Hahn-Hägerdal, 2000; Bozell and Petersen, 2010; Okano et al., 2010). Especially the latter, in form of the stereocomplex of d-and l-polylactic acid is a fully biodegradable substitute for polyethylene terephthalates with high melting temperatures and mechanical strength, and therefore, of great economical interest (Lorenz and Zinke, 2005; Dodds and Gross, 2007; Fukushima et al., 2007; Uehara et al., 2010). In the past and still today, wild-type and recombinant lactic acid bacteria have been mainly employed for the production of both d-and l-lactate (reviewed in Okano et al., 2010). However, these bacteria have a demand for complex media, which makes the cultivation of the organisms and the purification of the product relatively cost-intensive. Therefore, other less fastidious organisms, such as metabolically engineered E. coli, Saccharomyces cerevisiae and C. glutamicum have also been developed for efficient l-and d-lactic acid production (Okano et al., 2010).
Corynebacterium glutamicum is facultatively anaerobic and grows aerobically and anaerobically in the presence of oxygen and nitrate respectively (Nishimura et al., 2007; Takeno et al., 2007). The lack of oxygen or nitrate as external electron acceptors results in growth arrested cells, which still have the capability to ferment C6 sugars to l-lactate and succinate as major products. Dominguez and colleagues (1993) firstly reported that C. glutamicum forms lactate, succinate and acetate at small amounts when oxygen is limited during aerobic growth. Inui and colleagues (2004a) further studied this phenomenon in an attempt to utilize corynebacterial properties for the industrial production of lactate and succinate. These authors reported of organic acid production with C. glutamicum strain R and described that the bacteria showed no growth under oxygen-deprivation conditions, but produced significant amounts of l-lactate (∼ 220 mM) and succinate (∼ 20 mM) from about 130 mM glucose. Addition of bicarbonate to the medium led to an increase of the NAD+/NADH ratio and, probably as consequence of a derepression of the glyceraldehyde-3-phosphate dehydrogenase gene gapA, to an increased glucose consumption (Inui et al., 2004a). Furthermore, the addition of bicarbonate led to an altered product spectrum, i.e. the formation of succinate and lactate increased by a factor of two to four and significant concentrations (about 10 mM) of acetate were formed (Inui et al., 2004a; Okino et al., 2005). In a high cell density [30 g dry cell weight (DCW) l−1] fed-batch system, C. glutamicum R already produced 574 mM l-lactate (i.e. 53 g l−1), with only small amounts (< 10 mM) of succinate and acetate as side-products (Okino et al., 2005). Addition of 400 mM bicarbonate raised the l-lactate concentration to more than 1 M (97.5 g l−1), but also the concentrations of the by-products succinate (192 mM) and acetate (50 mM) (Okino et al., 2005). Even without genetic modification of C. glutamicum, the resulting l-lactate titre from glucose and the YP/S of 1.79 mol l-lactate per mol of glucose (i.e. 0.90 g g−1) are highly competitive as e.g. the best known metabolically engineered l-lactate-producing E. coli strain SZ85 (pflB, frdBC, adhE, ackA, ldhA::ldhL, overexpressed ldhL gene from Pediococcus acidilactici) accumulated 505 mM l-lactate (i.e. 46 g l−1) with an optical purity of > 99% and a YP/S of 1.9 mol per mol of glucose (i.e. 0.95 g g−1; Zhou et al., 2003b) (see Table 1).
Table 1.
Maximal titres, substrate-specific yields (YP/S), productivities, by-products and the respective references of the so far most efficient processes for organic acid production with C. glutamicum and E. coli strains
| Strain | Medium | Maximal titre (mM) (g l−1) | YP/S (molproduct per molsubstrate) (g g−1) | Productivitya (mM h−1) (g l−1 h−1) | By-productsb | Reference |
|---|---|---|---|---|---|---|
| Pyruvate | ||||||
| C. glutamicum ELB-P | minimal medium, glucose | 512 (44.5) | 1.49 (0.72) | 5.6 (0.49) | –c | Wieschalka et al. (2012) |
| E. coli ALS1059 | minimal medium, glucose, l-isoleucine, betaine | 1022 (88.9) | 1.39 (0.67) | 23.9 (2.08) | – | Zhu et al. (2008) |
| E. coli YYC202 | minimal medium, glucose | 720 (62.6) | 1.74 (0.84) | 37.0 (3.22) | – | Zelic et al. (2003) |
| l-Lactate | ||||||
| C. glutamicum R | minimal medium, glucose | 574 (51.1) | 1.42 (0.70) | 71.8 (6.39) | – | Okino et al. (2005) |
| C. glutamicum R | minimal medium, glucose, bicarbonate | 1061 (94.4) | 1.79 (0.89) | 176.8 (15.74) | acetate, succinate | Okino et al. (2005) |
| E. coli SZ85 | minimal medium, glucose | 505 (44.9) | 1.90 (0.94) | 7.2 (0.64) | – | Zhou et al. (2003b) |
| d-Lactate | ||||||
| C. glutamicum R ΔldhA pCRB204 | minimal medium, glucose | 1340 (119.3) | 1.73 (0.86) | 44.5 (3.96) | acetate, succinate | Okino et al. (2008b) |
| E. coli JP203 | complex medium, glucose | 691 (61.5) | 1.80 (0.89) | 11.6 (1.03) | – | Chang et al. (1999) |
| E. coli SZ63 | minimal medium, glucose | 528 (47.0) | 1.92 (0.95) | 9.8 (0.87) | – | Zhou et al. (2003a) |
| 2-Ketoisovalerate | ||||||
| C. glutamicum ΔaceE Δpqo ΔilvE (pJC4ilvBNCD) | minimal medium, glucose, yeast extract | 188 (21.8) | 0.56 (0.36) | 4.6 (0.53) | l-valine | Krause et al. (2010) |
| 2-Ketoglutarate | ||||||
| C. glutamicum R Δgdh ΔgltB ΔaceA | complex medium, glucose, molasses, soybean hydrolysate | 325 (47.5) | n.s.d | 2.7 (0.39) | – | Jo et al. (2012) |
| Succinic acid (anaerobic) | ||||||
| C. glutamicum R ΔldhA pCRA717 | minimal medium, glucose, bicarbonate | 1240 (146.3) | 1.40 (0.92) | 27 (3.19) | acetate | Okino et al. (2008a) |
| C. glutamicum ELB-P | minimal medium, glucose | 330 (38.9) | 1.02 (0.67) | 5.6 (0.66) | pyruvate | S. Wieschalka and B.J. Eikmanns, own unpubl. data |
| C. glutamicum BOL-3/pAN6-gap | saline, glucose, formate, bicarbonate | 1134 (133.8) | 1.67 (1.09) | 21 (2.48) | 2-oxoglutarate, acetate, fumarate, malate | Litsanov et al. (2012b) |
| E. coli SBS550MG/pHL413 | complex medium, glucose | 330 (38.9) | 1.61 (1.06) | 10 (1.18) | acetate, formate | Sánchez et al. (2005) |
| E. coli KJ134 | minimal medium, glucose | 606 (71.5) | 1.53 (1.00) | 6.4 (0.76) | acetate, pyruvate, malate | Jantama et al. (2008) |
| Succinic acid (aerobic) | ||||||
| C. glutamicum BL-1/pAN6-pycP458Sppc | minimal medium, glucose | 90 (10.6) | 0.45 (0.30) | 0.8 (0.09) | 2-oxoglutarate, acetate, pyruvate | Litsanov et al. (2012a) |
| C. glutamicum BL-1 pVWEx1-glpFKD | minimal medium, glycerol | 79 (9.3) | 0.21 (0.27) | 3.6 (0.42) | acetate | Litsanov et al. (2012c) |
| E. coli HL51276k(pKK313) | complex medium, glucose, bicarbonate | 70 (8.3) | 1.09 (0.71) | 1.2 (0.14) | acetate, pyruvate | Lin et al. (2005) |
| E. coli HL27659k(pKK313) | complex medium, glucose, bicarbonate | 60 (7.1) | 0.95 (0.62) | 2.3 (0.27) | acetate | Lin et al. (2005) |
During production phase.
Significant concentrations above 10 mM.
– = byproducts below 10 mM.
n.s. = not specified.
For d-lactate production with C. glutamicum, a l-LDH-deficient mutant was constructed, expressing the d-lactate dehydrogenase (d-LDH) from Lactobacillus delbrueckii (Okino et al., 2008b). Under oxygen-deprivation conditions, this mutant (C. glutamicum R ΔldhA/pCRB204) produced in a high cell density system (60 g(DCW) l−1) about 1.34 M d-lactate (i.e. 120 g l−1) within 30 h with an optical purity of > 99.9% and a YP/S of 1.73 mol per mol of glucose. But also significant amounts of succinate (146 mM) and actetate (52 mM) were formed, underlining product purity as major problem (Okino et al., 2008b). However, C. glutamicum R ΔldhA/pCRB204 produced more d-lactate than E. coli JP203 (pta, ppc) (Chang et al., 1999) and SZ63 (W3110; pflB, frdBC, adhE, ackA) (Zhou et al., 2003a) (Table 1), the best known genetically defined d-lactate producing E. coli strains, harbouring the native d-LDH of E. coli. With about 690 mM (62 g l−1) and 530 mM (48 g l−1), these strains formed approximately half of the titre obtained with C. glutamicum R ΔldhA/pCRB204, however, with comparable YP/S of between 1.76 and 1.92 mol d-lactate per mol of glucose (0.90–0.99 g d-lactate per g of glucose; Chang et al., 1999; Zhou et al., 2003a).
It has to be noted that all described C. glutamicum and E. coli processes have to compete with those using recombinant yeast strains (Saccharomyces and Kluyveromyces) that produce l-lactic acid with titres of up to 1.3 M, optical purity of > 99.9%, and YP/S of up to 1.6 mol l-lactate per mol of glucose (Saitoh et al., 2005; Okano et al., 2010).
Production of 2-ketoisovalerate and 2-ketoglutarate
In nature, 2-ketoisovalerate (3-methyl-2-oxobutanoic acid) is a precursor for l-valine, l-leucine, and pantothenate synthesis in bacteria and plants. In these organisms, it is formed from two molecules of pyruvate via the reactions catalysed by AHAS, acetohydroxyacid isomeroreductase (AHAIR), and dihydroxyacid dehydratase (DHAD) (see Fig. 1). 2-Ketoisovalerate is used as substitute for l-valine or l-leucine in chronic kidney disease patients (Teschan et al., 1998; Feiten et al., 2005; Aparicio et al., 2009; 2012) and also has been used in therapy for uremic hyperphosphatemia (Schaefer et al., 1994). To our knowledge, 2-ketoisovalerate for these purposes is mainly synthesized chemically by different methods (Cooper et al., 1983) and only very recently, directed fermentative production of 2-ketoisovalerate with microorganisms has been reported for the first time (Krause et al., 2010; see below).
Since 2-ketoisovalerate stems from two molecules of pyruvate (see Fig. 1) and a PDHC-deficient C. glutamicum secreted significant amounts of pyruvate and l-valine (see above), C. glutamicum ΔaceE was an excellent basis to engineer C. glutamicum for the production of this 2-ketoacid. To avoid transamination of 2-ketoisovalerate to l-valine, the ilvE gene encoding transaminase B was deleted, leading to an auxotrophy for branched chain amino acids. Aerobically, C. glutamicum ΔaceE ΔilvE formed about 76 mM pyruvate, 25 mM l-alanine, and 40 mM 2-ketoisovalerate in a growth-decoupled manner from glucose (Krause et al., 2010). Overexpression of the AHAS, AHAIR and DHAD genes shifted the product spectrum towards 2-ketoisovalerate and the resulting strain C. glutamicum ΔaceE ΔilvE (pJC4ilvBNCD) produced in fed-batch fermentations about 85 mM 2-ketoisovalerate with a volumetric productivity of 1.9 mM h−1 and a YP/S of about 0.38 mol per mol of glucose. Although the PQO has been found to be dispensable for growth and a deletion was only slightly beneficial on l-valine production (Schreiner et al., 2006; Blombach et al., 2008), PQO inactivation turned out to be highly beneficial for 2-ketoisovalerate production. Compared with the parental strain, C. glutamicum ΔaceE ΔilvE Δpqo (pJC4ilvBNCD) showed in fed-batch fermentations more than two times higher final titres (up to 220 mM = 25.5 g l−1) and volumetric productivities of 4.6 mM h−1 (Krause et al., 2010; Table 1).
It is noteworthy to mention that the 2-ketoisovalerate-producer C. glutamicum ΔaceE ΔilvE Δpqo (pJC4ilvBNCD) was used as a basis for the generation of a series of C. glutamicum strains producing isobutanol via the so-called ‘Ehrlich pathway’ (Blombach and Eikmanns, 2011; Blombach et al., 2011). The most promising strain of this series, C. glutamicum Iso7, carries additional deletions of the l-LDH and malate dehydrogenase genes (ΔldhA and Δmdh respectively) and overexpresses additionally the E. coli transhydrogenase genes pntAB, the Lactococcus lactis ketoacid decarboxylase gene kivD, and the homologous alcohol dehydrogenase gene adhA (Blombach et al., 2011).
2-Ketoglutarate is an intermediate of the tricarboxylic acid (TCA) cycle and the precursor for the synthesis of glutamate and the glutamate family of amino acids. 2-Ketoglutarate is used in dairy industry (Banks et al., 2001; Gutiérrez-Mendéz et al., 2008) and also is suitable to treat chronic renal insufficiency in hemodialysis patients (Riedel et al., 1996). An enzymatic process to synthesize 2-ketoglutarate from glutamate via the coupled reactions of glutamate dehydrogenase and NADH oxidase has been established (Ödmann et al., 2004), however, this bioconversion seems not very efficient. Therefore, Jo and colleagues (2012) very recently used a glutamate-overproducing mutant of C. glutamicum for the construction of a 2-ketoglutarate-producer. Inactivation of the genes encoding glutamate dehydrogenase, glutamate synthase and isocitrate lyase (gdh, gltB, and aceA respectively) led to a drastic reduction of glutamate formation (< 10 mM) and concomitantly to 2-ketoglutarate accumulation to concentrations of up to 325 mM (47.5 g l−1) after 120 h of cultivation in medium containing glucose, molasses, glutamate, and soybean hydrolysate (Jo et al., 2012). To our knowledge, there were no other approaches to produce 2-ketoglutarate by fermentation with any other bacterium. However, Zhou and colleagues (2012) recently reported efficient 2-ketoglutarate production (up to about 380 mM) with a recombinant (‘non-conventional’) yeast strain of Yarrowia lipolytica with enhanced acetyl-CoA availability.
Production of succinate
The C4 dicarboxylate succinate has been denoted as ‘a LEGO® of chemical industry’ (Sauer et al., 2008) and as such, can be used as precursor for known petrochemical bulk products, such as 1,4-butanediol, tetrahydrofuran, γ-butyrolactone, adipic acid, maleic anhydride, various n-pyrrolidinones, and linear aliphatic esters (Zeikus et al., 1999; Sauer et al., 2008; Bozell and Petersen, 2010). Moreover, succinate (or succinic acid) is directly used as surfactant, ion chelator, and as an additive in pharmaceutical, and food industry (McKinlay et al., 2007). The market potential for succinic acid and its direct derivatives has been estimated to be 245 000 tons per year, that for succinic acid-derived polymers about 25 000 000 tons per year, and with the transition to cost-efficient bio-based production of succinate or succinic acid, the market is predicted to steadily increase (Werpy and Petersen, 2004; Bozell and Petersen, 2010).
Aside from l-lactate and acetate, succinate is a natural fermentative end-product of the wild type of C. glutamicum, when incubated with glucose under oxygen deprivation (Dominguez et al., 1993; Inui et al., 2004a). Under these conditions, succinate is formed via glycolysis, carboxylation of phosphoenolpyruvate (PEP) or pyruvate to oxaloacetate (OAA) by PEP carboxylase (PEPCx) and/or pyruvate carboxylase (PCx), and subsequent conversion of OAA by malate dehydrogenase (Mdh), fumarase (Fum), and succinate dehydrogenase (SDH) (Inui et al., 2004a; see Fig. 1).
A two-stage succinate production process with C. glutamicum strain R was developed by Okino and colleagues (2008a), using a derivative devoid of LDH activity and overexpressing the native PCx gene (pyc), C. glutamicum R ΔldhA pCRA717. In a first step, cells of this strain were grown under fully aerobic conditions. Then, the cells were harvested, washed and transferred to closed bottles, to give a high cell density of about 50 g(DCW) l−1. With repeated intermittent addition of glucose and sodium bicarbonate, a final titre of 1.24 M succinate (146 g l−1) was obtained within 46 h, with a YP/S of 1.4 mol per mol of glucose (Okino et al., 2008a). The cells did not form any lactate; however, they produced significant amounts of acetate (0.3 M = 16 g l−1) as by-product.
Recently, also Litsanov and colleagues (2012b) engineered C. glutamicum ATCC 13032 for high yield succinate production by further extending the experimental approach by Okino et al. (see above). Deletion of the LDH gene, chromosomal integration of an allele for a deregulated PCx (pycP458S) and deletion of the genes encoding enzymes responsible for acetate synthesis (Δcat, Δpqo, Δpta-ack) resulted in C. glutamicum BOL-2, that produces up to 116 mM succinate with a YP/S of 1.03 mol per mol of glucose, and pyruvate (23 mM) as well as 2-ketoglutarate (12 mM) as major by-products (Litsanov et al., 2012b). To increase NADH and CO2 availability and to increase the glycolytic flux, the authors then integrated the formate dehydrogenase gene fdh from Mycobacterium vaccae into the genome of C. glutamicum BOL-2 and additionally overexpressed the homologous glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene (gapA) from plasmid. In a fed-batch fermentation with glucose, formate and bicarbonate as substrates, the ultimate strain C. glutamicum BOL-3/pAN6-gap (see Fig. 3) produced 1.13 M succinate (134 g l−1) with a YP/S of 1.67 mol per mol of glucose (Litsanov et al., 2012b). Aside from succinate, 2-ketoglutarate (35 mM), malate (33 mM), acetate (20 mM), fumarate (13 mM), and pyruvate (6 mM) were formed as by-products.
Figure 3.
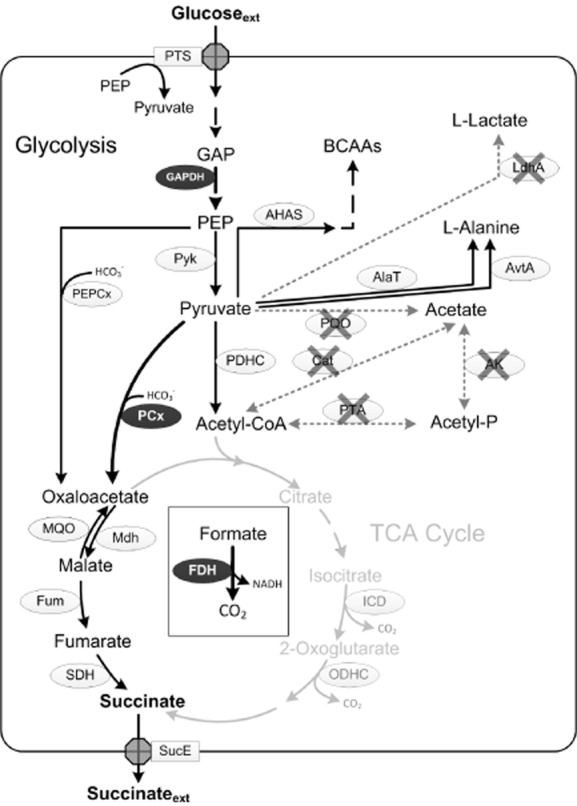
Schematic presentation of the central carbon metabolism of C. glutamicum BOL-3/pAN6-gap during anaerobic succinate production. For most abbreviations see legend to Fig. 1. Further abbreviations: BCAAs, branched-chain amino acids; LdhA, NAD+-dependent l-lactate dehydrogenase; SucE, succinate exporter; TCA, tricarboxylic acid. Dark ellipses indicate homologous/heterologous enzymes, crosses indicate inactivation of the enzyme by deletion of the respective gene. Dotted arrows indicate pathways not present due to gene inactivation, grey arrows indicate pathways not used in C. glutamicum BOL-3/pAN6-gap.
In a further approach, the pyruvate-producing strain C. glutamicum ELB-P (see above and Fig. 2) was employed for succinate production (S. Wieschalka and B. J. Eikmanns, unpublished). Due to the inactivation of the PDHC, PQO, and LDH, this strain does not form significant amounts of acetate or lactate as by-products under any aerobic and anaerobic condition tested (Wieschalka et al., 2012). In contrast to the two-stage processes described above (i.e. aerobic growth in complex or minimal media and, after harvest of the cells and resuspension in new medium, transfer to sealed bottles or fermenters respectively; Okino et al., 2008a; Litsanov et al., 2012b), a one-stage fed-batch fermentation process with C. glutamicum ELB-P was established, combining biomass formation and succinate production in a single bioreactor. This process includes three phases: (i) an aerobic growth phase on glucose plus acetate, (ii) a self-induced microaerobic phase at the end of the exponential growth by minimal aeration, and (iii) an anaerobic production phase, realized by gassing the fermenter with CO2 (Fig. 4). This optimized process led to growth-decoupled succinate production of more than 330 mM (i.e. 39 g l−1) with a YP/S of 1.02 mol succinate per mol of glucose. The final YP/S obtained, together with the formation of pyruvate (about 30 mM) as by-product, however, still indicates a limitation, which might be overcome by increasing the carbon flux from PEP/pyruvate to OAA or by integration of the M. vaccae fdh gene and the use of formate as an additional substrate for reduction equivalents, as described above by Litsanov and colleagues (2012b).
Figure 4.
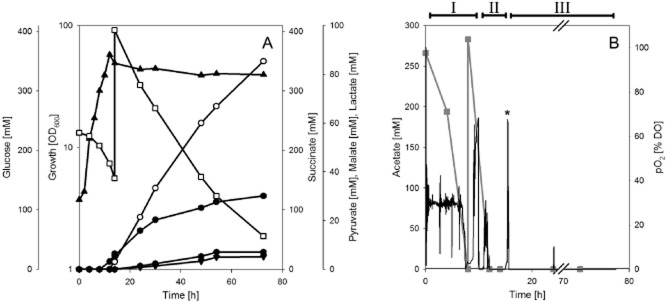
Growth, glucose consumption, and product formation (A) and acetate consumption and the course of the pO2 (B) during a representative pH-controlled tri-phasic fed-batch cultivation of C. glutamicum ELB-P in a 400 ml bioreactor with minimal medium, initially containing 4% (w/v) glucose, 1% (w/v) acetate and 6 mM l-alanine. (A) ▴, growth; □, glucose; •, pyruvate; ○, succinate; ▾, malate;  , lactate. (B) Black line, pO2; grey line, acetate. Roman numerals indicate (I) aerobic growth phase, (II) self-induced microaerobic phase, and (III) oxygen deprivation by CO2 gassing. The pO2 peak at the end of the microaerobic phase (marked with the asterisk in 4B) indicated the end of aerobic growth, as O2 consumption stopped, leading to increasing DO in the medium. Immediately, aeration was replaced by CO2 sparging and the production phase started. A batch of glucose at the beginning of phase III should ensure carbon availability for succinate production. At least five independent fermentations were performed, showing comparable results.
, lactate. (B) Black line, pO2; grey line, acetate. Roman numerals indicate (I) aerobic growth phase, (II) self-induced microaerobic phase, and (III) oxygen deprivation by CO2 gassing. The pO2 peak at the end of the microaerobic phase (marked with the asterisk in 4B) indicated the end of aerobic growth, as O2 consumption stopped, leading to increasing DO in the medium. Immediately, aeration was replaced by CO2 sparging and the production phase started. A batch of glucose at the beginning of phase III should ensure carbon availability for succinate production. At least five independent fermentations were performed, showing comparable results.
The experimental setup of a one-stage process (consecutive aerobic growth and anaerobic production in a single bioreactor) as done with C. glutamicum ELB-P, see above) represents an industrially feasible process. However, a recent study on isobutanol production with C. glutamicum disclosed the differences in the production performance between two-stage fermentations (aerobic growth in complex or minimal media and anaerobic production in different containments, see above) and one-stage fermentations in a single bioreactor: The isobutanol YP/S in the one-stage fermentation was significantly lower (0.48 mol vs 0.77 mol of isobutanol per mol of glucose), indicating that the transition from the aerobic environment (growth phase) to the anaerobic environment (production phase) has a strong impact on the overall production behaviour (Blombach and Eikmanns, 2011; Blombach et al., 2011). Similarly, Martínez and colleagues (2010) recently observed that introducing a microaerobic phase at the end of the aerobic growth phase of an E. coli succinate-producer led to an adjustment of the enzymatic machinery and to improved succinate production under anaerobic conditions. To our knowledge, the physiological changes of C. glutamicum during a (slow or fast) shift from aerobic to anaerobic conditions have so far not been investigated. However, it can be foreseen that the insight into the metabolic adaptation of the cells to such alternating culture conditions will help to further optimize organic acid production by novel metabolic engineering approaches and also by applying optimally adapted process conditions.
The YP/S of the most efficient E. coli strains producing succinate under anaerobic conditions, E. coli SBS550MG/pHL413 and E. coli KJ134, were 1.60 mol and 1.53 mol succinate per mol of glucose respectively (Sánchez et al., 2005; Jantama et al., 2008; Table 1). Thus, both recombinant E. coli strains and in particular, C. glutamicum BOL-3/pAN6-gap (Table 1) showed higher YP/S than all known natural succinate-producing bacteria, such as Anaerospirillium succiniproducens (1.37 mol mol−1 of glucose; Glassner and Datta, 1992) or Mannheimia succiniproducens (1.16 mol mol−1 glucose; Lee et al., 2006). A further advantage of employing the recombinant C. glutamicum or E. coli strains is the potential use of mineral media, keeping production and purification costs lower than with Mannheimia or Anaerospirillum, which both require complex media. C. glutamicum BOL-3/pAN6-gap and C. glutamicum R ΔldhA pCRA717 produced about threefold higher succinate titres than E. coli SBS550MG/pHL413 (Table 1) and thus, Corynebacterium seems to be the superior organism for succinate production.
Very recently, Litsanov and colleagues (2012a) reported also on aerobic succinate production with C. glutamicum for the first time. Deletion of the SDH genes initiated aerobic succinate production in C. glutamicum via glycolysis, PEP and/or pyruvate carboxylation, the oxidative branch of the TCA cycle, and the glyoxylate shunt. Acetate formation was mostly prohibited by shutdown of the known pathways for acetate synthesis, resulting in C. glutamicum BL-1 (genotype: ΔsdhCAB, Δcat, Δpqo, Δpta-ack; Litsanov et al., 2012a). To reduce carbon-loss into cell mass, nitrogen-limited growth conditions were established, forcing the cells into a resting state after a certain period. With additional, plasmid-bound overproduction of both PEPCx and the PCxP458S-variant, final succinate titres and YP/S of up to 90 mM and 0.45 mol succinate per mol of glucose, respectively, were observed (Litsanov et al., 2012a; Table 1). Concerning the specific productivity of 1.6 mmol g(CDW)−1 h−1, C. glutamicum BL-1/pAN6-pycP458Sppc showed the highest value described so far for aerobic succinate production from glucose with bacteria.
In comparison to other bacterial succinate producers, C. glutamicum BL-1/pAN6-pycP458Sppc is exceedingly competitive in aerobic succinate production (Table 1). Lin and colleagues (2005) described various E. coli strains approaching the maximal theoretical YP/S of about 1 mol succinate per mol of glucose under aerobic conditions. Corynebacterium glutamicum BL-1/pAN6-pycP458Sppc did not reach this high YP/S, but the recombinant C. glutamicum strains produced significantly higher final succinate titres in minimal instead of complex media (Table 1).
Production of organic acids with C. glutamicum from alternative substrates
Economical relevant and sustainable production of organic acids with microorganisms in an industrial scale is dependent on the use of low-cost carbon sources, in particular from renewable resources. So far, we focused in this review on the fermentative organic acid production from pretreated and purified carbon sources, such as glucose and glucose plus formate, since the most promising attempts to produce organic acids with C. glutamicum were made with these substrates. To simplify feedstock purchase and to improve the economic efficiency, utilization of alternative, crude materials is of great interest. However, C. glutamicum naturally cannot utilize certain industrially relevant substrates, such as glycerol, starch (from corn, wheat, rice, or potato), whey, straw, or hemi-and lignocellulose. Especially lignocellulose, consisting largely of cellulose, hemicellulose, and lignin, is a widely abundant and potentially attractive source of renewable feedstock. Hemicellulose, consisting mainly of glucose but also to a significant portion of C5 sugars (xylose and arabinose) (Wiselogel et al., 1996; Aristidou and Penttilä, 2000), can be depolymerized by chemical or enzymatic processes, and the resulting sugar mixtures are also of interest as alternative feedstock for C. glutamicum. Whereas some organisms (e.g. E. coli) are naturally able to consume the majority of sugars in the mixtures resulting from saccharification from hemicellulose, C. glutamicum needs metabolic engineering to expand the spectrum of sugars that can be utilized. Thus, the extension of the substrate spectrum of C. glutamicum to cheap, easily accessible and renewable monomeric and polymeric carbon sources is desired and therefore, an ongoing field of intensive research (Wendisch et al., 2006b; Blombach and Seibold, 2010; Rumbold et al., 2010; Okano et al., 2010; Becker and Wittmann, 2011).
Several attempts have been made to broaden the natural substrate spectrum of C. glutamicum towards starch (Seibold et al., 2006; Tateno et al., 2007), whey (Barret et al., 2004), rice straw and wheat bran hydrolysates (Gopinath et al., 2011), grass and corn silages (Neuner et al., 2012), glucosides and d-cellobiose (Kotrba et al., 2003), glycerol (Rittmann et al., 2008), amino sugars (Gruteser et al., 2012; Uhde et al., 2012) or pentose sugars for growth and for the production of amino acids or other value-added products (Blombach and Seibold, 2010; Jojima et al., 2010; Buschke et al., 2011; Schneider et al., 2011; Gopinath et al., 2012). The first approaches to extend the substrate spectrum especially for organic acid production were performed by Kawaguchi and colleagues (2006; 2008) and Sasaki and colleagues (2008; 2009). Plasmid-bound introduction of the xylose isomerase and xylulokinase genes (xylA and xylB respectively) from E. coli into C. glutamicum R enabled both aerobic growth on xylose as sole carbon source and production of l-lactate and succinate with resting cells under oxygen deprivation conditions (Kawaguchi et al., 2006). Although the sugar consumption rate and the specific productivity of the recombinant C. glutamicum CRX2 was lower with xylose than with glucose, the YP/S for succinate was even higher on xylose (0.42 mol mol−1) than on glucose (0.23 mol mol−1). In contrast, the YP/S for l-lactate was lower with xylose as substrate (1.06 and 1.36 mol mol−1 respectively; Kawaguchi et al., 2006). A similar behaviour was shown for succinate and l-lactate production from arabinose with C. glutamicum CRA1, which expresses the E. coli genes araA, araB and araD (encoding l-arabinose isomerase, l-ribulokinase, and l-ribulose-5-phosphate 4-epimerase respectively) and therefore is able to metabolize this C5 sugar (Kawaguchi et al., 2008). In this case, with 200 mM arabinose as substrate, the YP/S for succinate and l-lactate were 0.67 mol mol−1 and 0.75 mol mol−1 respectively (Kawaguchi et al., 2008).
Co-utilization of different C5 sugars with C6 sugars was investigated to study catabolite repression effects in C. glutamicum and to expand sugar utilization on conditioned hemi-and lignocellulosic biomass hydrolysates (Sasaki et al., 2008). These efforts resulted in a C. glutamicum strain harbouring xylA and xylB as well as bglFV317A and bglA (encoding PTS β-glucoside-specific enzyme IIBCA component and phospho-β-glucosidase respectively). This strain produced from a mixture of d-cellobiose (10 g l−1), glucose (40 g l−1), and d-xylose (20 g l−1) about 460 mM l-lactate, 110 mM succinate, and 30 mM acetate under anaerobic conditions, with a combined yield of 0.85 g acids per g of sugar (Sasaki et al., 2008). A combined strain, containing all named modifications for d-xylose, l-arabinose and d-cellobiose consumption, and additionally overexpressing the arabinose transporter gene araE from C. glutamicum ATCC31831, was even able to consume glucose (35 g l−1), d-xylose (17.5 g l−1), l-arabinose (7 g l−1), and cellobiose (7 g l−1) simultaneously and completely under oxygen-deprived conditions within 14 h (Sasaki et al., 2009).
Recently, Sasaki and colleagues (2011) developed a C. glutamicum strain overexpressing the mannose 6-phosphate isomerase and fructose permease genes manA and ptsF respectively. This strain consumed mannose and glucose simultaneously and produced about 400 mM l-lactate, 100 mM succinate and 30 mM acetate from a sugar mixture of 200 mM glucose and 100 mM mannose under oxygen deprivation conditions (Sasaki et al., 2011).
Litsanov and colleagues (2012c) very recently showed aerobic succinate production with glycerol as sole carbon source, by plasmid-bound transfer of the glycerol utilizing genes glpFKD from E. coli into C. glutamicum BL-1. Glycerol is a main by-product of biodiesel and bioethanol production (Yazdani and Gonzalez, 2007) and using this carbon source for the production of value-added chemicals (such as succinate), the economic efficiency of these biofuel production processes can be increased (Wendisch et al., 2011). Plasmid pVWEx1-glpFKD has previously been shown to enable growth and amino acid production of C. glutamicum on glycerol as sole carbon source (Rittmann et al., 2008). Consequently, using the conditions established for C. glutamicum BL-1/pAN6-pycP458Sppc (see above), C. glutamicum BL-1 (pVWEx1-glpFKD) aerobically produced up to 79 mM succinate (9.3 g l−1) with a YP/S of 0.21 mol per mol of glycerol (Litsanov et al., 2012c). The specific succinate productivity of C. glutamicum BL-1 pVWEx1-glpFKD on glycerol was as high as for C. glutamicum BL-1/pAN6-pycP458Sppc on glucose with 1.6 mmol g(CDW)−1 h−1. However, the volumetric productivity of 3.59 mM h−1 is the highest productivity so far described for aerobic succinate production (Litsanov et al., 2012c; Table 1).
In summary, the above mentioned studies showed the feasibility to expand the substrate spectrum of C. glutamicum to the main C5 and C6 sugars found in agricultural residues, in hydrolysed hemicellulose and lignocellulosic biomass, and to glycerol. For directed production of organic acids from hemicellulose feedstock, the modifications made for broadening the substrate spectrum and those made for optimal carbon flux to a desired organic acid must be combined. The successful aerobic production of succinate from glycerol instead of glucose by introduction of the glycerol utilizing genes from E. coli to C. glutamicum (Litsanov et al., 2012c; see above), is one such example and promises the feasibility of such approaches.
Summary and outlook
Driven by old and new knowledge and genome-based metabolic and genetic engineering strategies, C. glutamicum has become a major candidate as platform organism for bio-based, industrial production of a variety of organic acids from renewable biomass. As outlined above and highlighted in Table 1, titres, YP/S, and productivities of recently developed C. glutamicum producer strains are highly competitive, in several cases already superior in comparison to other bacterial, well-established production systems. From current studies on transcriptome, proteome, metabolome, and intracellular fluxes (Vertes et al., 2012), from recent advances in evolutionary engineering tools (Becker and Wittmann, 2011), and from recent development of plasmid addiction systems (Schneider et al., 2012) and of single cell approaches (Binder et al., 2012; Mustafi et al., 2012), it can be expected that a variety of further metabolic (or genetic) targets for strain development/improvement will be identified in C. glutamicum. Future approaches to optimize organic acid production certainly will not only aim at substrate flexibility (low cost and eco-efficient feedstocks), product extension (e.g. fumarate, malate, or itaconate), and/or the paths from a substrate or substrate mixtures to the desired products (i.e. substrate uptake, central metabolism, precursor supply, synthetic pathways, and export of the respective organic acid). They will also focus on maintenance of a well-balanced redox state within the cells, on optimal adaptation of the cells to alternating culture conditions (e.g. shift from aerobic to anaerobic conditions), on strain robustness, and on an increased acid-resistance of the producer strains. Tolerance to organic acid stress represents a highly relevant factor for process design and downstream processing of large-scale production processes, since organic acid recovery from low pH fermentation broth in general is more cost-efficient than from neutral broth. However, the achievements obtained in the last 6 years and the wealth of new knowledge about the physiology, the metabolism and its regulation, and the proven production capabilities of C. glutamicum bode well for the implementation of this organism as a platform for new and even more (cost-)efficient processes for the production of a variety of organic acids and also of other specialty, fine and bulk chemicals.
Conflict of interest
None declared.
References
- Ajinomoto. 2010. Food products business [WWW document]. URL http://www.ajinomoto.com/ir/pdf/Food-Oct2010.pdf.
- Ajinomoto. 2011. Feed-use amino acid business [WWW document]. URL http://www.ajinomoto.com/ir/pdf/Feed-useAA-Oct2011.pdf.
- Aparicio M, Cano NJM, Cupisti A, Ecder T, Fouque D, Garneata L, et al. Keto-acid therapy in predialysis chronic kidney disease patients: consensus statements. J Ren Nutr. 2009;19:33–35. doi: 10.1053/j.jrn.2009.06.013. [DOI] [PubMed] [Google Scholar]
- Aparicio M, Bellizzi V, Chauveau P, Cupisti A, Ecder T, Fouque D, et al. Keto acid therapy in predialysis chronic kidney disease patients: final consensus. J Ren Nutr. 2012;22:22–24. doi: 10.1053/j.jrn.2011.09.006. [DOI] [PubMed] [Google Scholar]
- Aristidou A, Penttilä M. Metabolic engineering applications to renewable resource utilization. Curr Opin Biotechnol. 2000;11:187–198. doi: 10.1016/s0958-1669(00)00085-9. [DOI] [PubMed] [Google Scholar]
- Banks JM, Yvon M, Gripon JC, ‘la Fuente MA, Brechany EY, Williams AG, Muir DD. Enhancement of amino acid catabolism in cheddar cheese using α-ketoglutarate: amino acid gegradation in relation to volatile compounds and aroma character. Int Dairy J. 2001;11:235–243. [Google Scholar]
- Barret E, Stanton C, Zelder O, Fitzgerald G, Ross RP. Heterologous expression of lactose-and galactose-utilizing pathways from lactic acid bacteria in Corynebacterium glutamicum for production of lysine in whey. Appl Environ Microbiol. 2004;70:2861–2866. doi: 10.1128/AEM.70.5.2861-2866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J, Wittmann C. Bio-based production of chemicals, materials and fuels – Corynebacterium glutamicum as versatile cell factory. Curr Opinion Biotechnol. 2011;23:1–10. doi: 10.1016/j.copbio.2011.11.012. [DOI] [PubMed] [Google Scholar]
- Binder S, Schendzielorz G, Stäbler N, Ktumbach K, Hoffmann K, Bott M, Eggeling L. A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level. Genome Biol. 2012;13:R40. doi: 10.1186/gb-2012-13-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blombach B, Eikmanns BJ. Current knowledge on isobutanol production with Escherichia coli Bacillus subtilis and Corynebacterium glutamicum. Bioeng Bugs. 2011;2:346–350. doi: 10.4161/bbug.2.6.17845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blombach B, Seibold G. Carbohydrate metabolism in Corynebacterium glutamicum and applications for the metabolic engineering of L-lysine production strains. Appl Microbiol Biotechnol. 2010;86:1313–1322. doi: 10.1007/s00253-010-2537-z. [DOI] [PubMed] [Google Scholar]
- Blombach B, Schreiner ME, Holátko J, Bartek T, Oldiges M, Eikmanns BJ. L-valine production with pyruvate dehydrogenase complex-deficient Corynebacterium glutamicum. Appl Environ Microbiol. 2007;73:2079–2084. doi: 10.1128/AEM.02826-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blombach B, Schreiner ME, Bartek T, Oldiges M, Eikmanns BJ. Corynebacterium glutamicum tailored for high-yield L-valine production. Appl Microbiol Biotechnol. 2008;79:471–479. doi: 10.1007/s00253-008-1444-z. [DOI] [PubMed] [Google Scholar]
- Blombach B, Arndt A, Auchter M, Eikmanns BJ. L-valine production during growth of pyruvate dehydrogenase complex-deficient Corynebacterium glutamicum in the presence of ethanol or by inactivation of the transcriptional regulator SugR. Appl Environ Microbiol. 2009;75:1197–1200. doi: 10.1128/AEM.02351-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blombach B, Riester T, Wieschalka S, Ziert C, Youn JW, Wendisch VF, Eikmanns BJ. Corynebacterium glutamicum tailored for efficient isobutanol production. Appl Environ Microbiol. 2011;77:3300–3310. doi: 10.1128/AEM.02972-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bott M. Offering surprises: TCA cycle regulation in Corynebacterium glutamicum. Trends Microbiol. 2007;15:417–425. doi: 10.1016/j.tim.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Bozell JJ, Petersen GR. Technology development for the production of biobased products from biorefinery carbohydrates – the US Department of Energy's ‘Top 10’ revisited. Green Chem. 2010;12:539–554. [Google Scholar]
- Brinkrolf K, Schröder J, Pühler A, Tauch A. The transcriptional regulatory repertoire of Corynebacterium glutamicum: reconstruction of the network controlling pathways involved in lysine and glutamate production. J Biotechnol. 2010;149:173–182. doi: 10.1016/j.jbiotec.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Burkowski A. Corynebacteria – Genomics and Molecular Biology. Norfolk, UK: Caister Academic Press; 2008. [Google Scholar]
- Buschke N, Schröder H, Wittmann C. Metabolic engineering of Corynebacterium glutamicum for production of 1,5-diaminopentane from hemicellulose. Biotechnol J. 2011;6:306–317. doi: 10.1002/biot.201000304. [DOI] [PubMed] [Google Scholar]
- Chang DE, Jung HC, Rhee JS, Pan JG. Homofermentative production of D-or L-lactate in metabolically engineered Escherichia coli RR1. Appl Environ Microbiol. 1999;65:1384–1389. doi: 10.1128/aem.65.4.1384-1389.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AJI, Ginos JZ, Meister A. Synthesis and properties of the alpha-keto acids. Chem Rev. 1983;83:321–358. [Google Scholar]
- Dodds DR, Gross RA. Chemicals from biomass. Science. 2007;318:1250–1251. doi: 10.1126/science.1146356. [DOI] [PubMed] [Google Scholar]
- Dominguez H, Nezondet C, Lindley ND, Cocaign M. Modified carbon flux during oxygen-limited growth of Corynebacterium glutamicum and the consequences for amino acid overproduction. Biotechnol Lett. 1993;15:449–454. [Google Scholar]
- Eggeling L, Bott M, editors. Handbook of Corynebacterium glutamicum. Boca Raton, FL, USA: CRC Press LLC; 2005. [Google Scholar]
- Feiten SF, Draibe SA, Watanabe R, Duenhas MR, Baxmann AC, Nerbass FB, Cuppari L. Short-term effects of a very-low-protein diet supplemented with ketoacids in nondialyzed chronic kidney disease patients. Eur J Clin Nutr. 2005;59:129–136. doi: 10.1038/sj.ejcn.1602050. [DOI] [PubMed] [Google Scholar]
- Fukushima K, Chang YH, Kimura Y. Enhanced stereocomplex formation of poly(L-lactic acid) and poly(d-lactic acid) in the presence of stereoblock poly(lactic acid) Macromol Biosci. 2007;7:829–835. doi: 10.1002/mabi.200700028. [DOI] [PubMed] [Google Scholar]
- Glassner DA, Datta R. 1992. Process for the production and purification of succinic acid US Patent, patent number 5,143,834.
- Gopinath V, Meiswinkel TM, Wendisch VF, Nampoothiri KM. Amino acid production from rice straw and wheat bran hydrolysates by recombinant pentose-utilizing Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2011;92:985–996. doi: 10.1007/s00253-011-3478-x. [DOI] [PubMed] [Google Scholar]
- Gopinath V, Murali A, Dhar KS, Nampoothiri KM. Corynebacterium glutamicum as a potent biocatalyst for the bioconversion of pentose sugars to value-added products. Appl Microbiol Biotechnol. 2012;93:95–106. doi: 10.1007/s00253-011-3686-4. [DOI] [PubMed] [Google Scholar]
- Gruteser N, Marin K, Krämer R, Thomas GH. Sialic acid utilization by the soil bacterium Corynebacterium glutamicum. FEMS Microbiol Lett. 2012;336:131–138. doi: 10.1111/j.1574-6968.2012.02663.x. [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Mendéz N, Vallejo-Cordoba B, González-Córdova AF, Nevárez-Moorillón GV, Rivera-Chavira B. Evaluation of aroma generation of Lactococcus lactis with an electronic nose and sensory analysis. J Dairy Sci. 2008;91:49–57. doi: 10.3168/jds.2007-0193. [DOI] [PubMed] [Google Scholar]
- Hofvendahl K, Hahn-Hägerdal B. Factors affecting the fermentative lactic acid production from renewable resources. Enzyme Microb Technol. 2000;26:87–107. doi: 10.1016/s0141-0229(99)00155-6. [DOI] [PubMed] [Google Scholar]
- Howard JW, Fraser WA. Preparation of pyruvic acid. Org Synth Coll. 1932;1:475–480. [Google Scholar]
- Inui M, Murakami S, Okino S, Kawaguchi H, Vertès AA, Yukawa H. Metabolic analysis of Corynebacterium glutamicum during lactate and succinate productions under oxygen deprivation conditions. J Mol Microbiol Biotechnol. 2004a;7:182–196. doi: 10.1159/000079827. [DOI] [PubMed] [Google Scholar]
- Inui M, Kawaguchi H, Murakami S, Vertès AA, Yukawa H. Metabolic engineering of Corynebacterium glutamicum for fuel ethanol production under oxygen deprivation-conditions. J Mol Microbiol Biotechnol. 2004b;8:243–225. doi: 10.1159/000086705. [DOI] [PubMed] [Google Scholar]
- Jantama K, Zhang X, Moore JC, Shanmugam KT, Svoronos SA, Ingram LO. Eliminating side products and increasing succinate yields in engineered strains of Escherichia coli C. Biotechnol Bioeng. 2008;101:881–893. doi: 10.1002/bit.22005. [DOI] [PubMed] [Google Scholar]
- Jo J-H, Seol H-Y, Lee Y-B, Kim M-H, Hyun H-H, Lee H-H. Disruption of genes for enhanced biosynthesis of α-ketoglutarate in Corynebacterium glutamicum. Can J Microbiol. 2012;58:278–286. doi: 10.1139/w11-132. [DOI] [PubMed] [Google Scholar]
- Jojima T, Omumasaba CA, Inui M, Yukawa H. Sugar transporters in efficient utilization of mixed sugar substrates: current knowledge and outlook. Appl Microbiol Biotechnol. 2010;85:471–480. doi: 10.1007/s00253-009-2292-1. [DOI] [PubMed] [Google Scholar]
- Kawaguchi H, Vertès AA, Okino S, Inui M, Yukawa H. Engineering of a xylose metabolic pathway in Corynebacterium glutamicum. Appl Environ Microbiol. 2006;72:3418–3428. doi: 10.1128/AEM.72.5.3418-3428.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi H, Sasaki M, Vertès AA, Inui M, Yukawa H. Engineering of an L-arabinose metabolic pathway in Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2008;77:1053–1062. doi: 10.1007/s00253-007-1244-x. [DOI] [PubMed] [Google Scholar]
- Kind S, Wittmann C. Bio-based production of the platform chemical 1,5-diaminopentane. Appl Microbiol Biotechnol. 2011;5:1287–1296. doi: 10.1007/s00253-011-3457-2. [DOI] [PubMed] [Google Scholar]
- Kind S, Jeong WK, Schröder H, Wittmann C. Systems-wide metabolic pathway engineering in Corynebacterium glutamicum for bio-based production of diaminopentane. Metab Eng. 2010a;12:341–351. doi: 10.1016/j.ymben.2010.03.005. [DOI] [PubMed] [Google Scholar]
- Kind S, Jeong WK, Schröder H, Zelder O, Wittmann C. Identification and elimination of the competing N-acetyldiaminopentane pathway for improved production of diaminopentane by Corynebacterium glutamicum. Appl Environ Microbiol. 2010b;76:5175–5180. doi: 10.1128/AEM.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner O, Tauch A. Tools for genetic engineering in the amino acid-producing bacterium Corynebacterium glutamicum. J Biotechnol. 2003;104:287–299. doi: 10.1016/s0168-1656(03)00148-2. [DOI] [PubMed] [Google Scholar]
- Kotrba P, Inui M, Yukawa H. A single V317A or V317M substitution in enzyme II of a newly identified beta-glucoside phosphotransferase and utilization system of Corynebacterium glutamicum R extends its specificity towards cellobiose. Microbiology. 2003;149:1569–1580. doi: 10.1099/mic.0.26053-0. [DOI] [PubMed] [Google Scholar]
- Krause FS, Henrich A, Blombach B, Krämer R, Eikmanns BJ, Seibold GM. Increased glucose utilization in Corynebacterium glutamicum by use of maltose, and its application for the improvement of L-valine productivity. Appl Environ Microbiol. 2009;76:370–374. doi: 10.1128/AEM.01553-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause FS, Blombach B, Eikmanns BJ. Metabolic engineering of Corynebacterium glutamicum for 2-ketoisovalerate production. Appl Environ Microbiol. 2010;76:8053–8061. doi: 10.1128/AEM.01710-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Song H, Lee SY. Genome-based metabolic engineering of Mannheimia succiniciproducens for succinic acid production. Appl Environ Microbiol. 2006;72:1939–1948. doi: 10.1128/AEM.72.3.1939-1948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Chen J, Lun SY. Biotechnological production of pyruvic acid. Appl Microbiol Biotechnol. 2001;57:451–459. doi: 10.1007/s002530100804. [DOI] [PubMed] [Google Scholar]
- Liebl W, Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. The Prokaryotes. 3rd edn. Vol. 3. New York, USA: Springer; 2006. The genus Corynebacterium – nonmedical; pp. 796–818. Vol. [Google Scholar]
- Lin H, Bennett GN, San KY. Metabolic engineering of aerobic succinate production systems in Escherichia coli to improve process productivity and achieve the maximum theoretical succinate yield. Metab Eng. 2005;7:116–127. doi: 10.1016/j.ymben.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Litsanov B, Kabus A, Brocker M, Bott M. Efficient aerobic succinate production from glucose in minimal medium with Corynebacterium glutamicum. Microb Biotechnol. 2012a;5:116–128. doi: 10.1111/j.1751-7915.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litsanov B, Brocker M, Bott M. Towards homosuccinate fermentation: metabolic engineering of Corynebacterium glutamicum for anaerobic succinate production from glucose and formate. Appl Environ Microbiol. 2012b;78:3325–3337. doi: 10.1128/AEM.07790-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litsanov B, Brocker M, Bott M. Glycerol as a substrate for aerobic succinate production in minimal medium with Corynebacterium glutamicum. Microb Biotechnol. 2012c doi: 10.1111/j.1751-7915.2012.00347.x. doi: 10.1111/j.1751-7915.2012.00347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz P, Zinke H. White biotechnology: differences in US and EU approaches? Trends Biotechnol. 2005;23:570–574. doi: 10.1016/j.tibtech.2005.10.003. [DOI] [PubMed] [Google Scholar]
- McKinlay JB, Vieille C, Zeikus JG. Prospects for a bio-based succinate industry. Appl Microbiol Biotechnol. 2007;76:727–740. doi: 10.1007/s00253-007-1057-y. [DOI] [PubMed] [Google Scholar]
- Martínez I, Bennett GN, San KY. Metabolic impact of the level of aeration during cell growth on anaerobic succinate production by an engineered Escherichia coli strain. Metab Eng. 2010;12:499–509. doi: 10.1016/j.ymben.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Mimitsuka T, Sawai H, Hatsu M, Yamada K. Metabolic engineering of Corynebacterium glutamicum for cadaverine fermentation. Biosci Biotechnol Biochem. 2007;71:2130–2135. doi: 10.1271/bbb.60699. [DOI] [PubMed] [Google Scholar]
- Mustafi N, Grünberger A, Kohlheyer D, Bott M, Frunzke J. The development and application of a single-cell biosensor for the detection of L-methionine and branched-chain amino acids. Metab Eng. 2012;14:449–457. doi: 10.1016/j.ymben.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Neuner A, Wagner I, Sieker T, Ulber R, Schneider K, Peifer S, Heinzle E. Production of L-lysine on different silage juices using genetically engineered Corynebacterium glutamicum. J Biotechnol. 2012 doi: 10.1016/j.jbiotec.2012.07.190. Aug 9. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Nishimura T, Vertès AA, Shinoda Y, Inui M, Yukawa H. Anaerobic growth of Corynebacterium glutamicum using nitrate as a terminal electron acceptor. Appl Microbiol Biotechnol. 2007;75:889–897. doi: 10.1007/s00253-007-0879-y. [DOI] [PubMed] [Google Scholar]
- Ödmann P, Welborn W, Bommarius AS. An enzymatic process to α-ketoglutarate from L-glutamate: the coupled system L-glutamate dehydrogenase/NADH oxidase. Tetrahedron: Asymmetry. 2004;15:2933–2937. [Google Scholar]
- Okano K, Tanaka T, Ogino C, Fukuda H, Kondo A. Biotechnological production of enantiomeric pure lactic acid from renewable resources: recent achievements, perspectives, and limits. Appl Microbiol Biotechnol. 2010;85:413–423. doi: 10.1007/s00253-009-2280-5. [DOI] [PubMed] [Google Scholar]
- Okino S, Inui M, Yukawa H. Production of organic acids by Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol. 2005;68:475–480. doi: 10.1007/s00253-005-1900-y. [DOI] [PubMed] [Google Scholar]
- Okino S, Noburyu R, Suda M, Jojima T, Inui M, Yukawa H. An efficient succinic acid production process in a metabolically engineered Corynebacterium glutamicum strain. Appl Microbiol Biotechnol. 2008a;81:459–464. doi: 10.1007/s00253-008-1668-y. [DOI] [PubMed] [Google Scholar]
- Okino S, Suda M, Fujikura K, Inui M, Yukawa H. Production of d-lactic acid by Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol. 2008b;78:449–454. doi: 10.1007/s00253-007-1336-7. [DOI] [PubMed] [Google Scholar]
- Riedel E, Nundel M, Hampl H. α-ketoglutarate application in hemodialysis patients improves amino acid metabolism. Nephron. 1996;74:261–265. doi: 10.1159/000189319. [DOI] [PubMed] [Google Scholar]
- Rittmann D, Lindner SN, Wendisch VF. Engineering of a glycerol utilization pathway for amino acid production by Corynebacterium glutamicum. Appl Environ Microbiol. 2008;74:6216–6222. doi: 10.1128/AEM.00963-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbold K, van Buijsen HJJ, Gray VM, van Groenestijn JW, Overkamp KM, Slomp RS, et al. Microbial renewable feedstock utilization – a substrate-oriented approach. Bioeng Bugs. 2010;1:359–366. doi: 10.4161/bbug.1.5.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh S, Ishida N, Onishi T, Tokuhiro K, Nagamori E, Kitamoto K, Takahashi H. Genetically engineered wine yeast produces a high concentration of L-lactic acid of extremely high optical purity. Appl Environ Microbiol. 2005;71:2789–2792. doi: 10.1128/AEM.71.5.2789-2792.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez AM, Bennett GN, San KY. Novel pathway engineering design of the anaerobic central metabolic pathway in Escherichia coli to increase succinate yield and productivity. Metab Eng. 2005;7:229–239. doi: 10.1016/j.ymben.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Sasaki M, Jojima T, Inui M, Yukawa H. Simultaneous utilization of d-cellobiose, d-glucose, and d-xylose by recombinant Corynebacterium glutamicum under oxygen-deprived conditions. Appl Microbiol Biotechnol. 2008;81:691–699. doi: 10.1007/s00253-008-1703-z. [DOI] [PubMed] [Google Scholar]
- Sasaki M, Jojima T, Kawaguchi H, Inui M, Yukawa H. Engineering of pentose transport in Corynebacterium glutamicum to improve simultaneous utilization of mixed sugars. Appl Microbiol Biotechnol. 2009;85:105–115. doi: 10.1007/s00253-009-2065-x. [DOI] [PubMed] [Google Scholar]
- Sasaki M, Jojima T, Inui M, Yukawa H. Xylitol production by recombinant Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol. 2010;86:1057–1066. doi: 10.1007/s00253-009-2372-2. [DOI] [PubMed] [Google Scholar]
- Sasaki M, Teramoto H, Inui M, Yukawa H. Identification of mannose uptake and catabolism genes in Corynebacterium glutamicum and genetic engineering for simultaneous utilization of mannose and glucose. Appl Microbiol Biotechnol. 2011;89:1905–1916. doi: 10.1007/s00253-010-3002-8. [DOI] [PubMed] [Google Scholar]
- Sauer M, Porro D, Mattanovich D, Branduardi P. Microbial production of organic acids: expanding the markets. Trends Biotechnol. 2008;26:100–108. doi: 10.1016/j.tibtech.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Sauer U, Eikmanns BJ. The PEP–pyruvate–oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol Rev. 2005;29:765–794. doi: 10.1016/j.femsre.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Schaefer K, von Herrath D, Erley CM, Asmus G. Calcium ketovaline as new therapy for uremic hyperphosphatemia. Miner Electrolyte Metab. 1994;16:362–364. [PubMed] [Google Scholar]
- Schneider J, Wendisch VF. Putrescine production by engineered Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2010;88:859–868. doi: 10.1007/s00253-010-2778-x. [DOI] [PubMed] [Google Scholar]
- Schneider J, Wendisch VF. Biotechnological production of polyamines by bacteria: recent achievements and future perspectives. Appl Microbiol Biotechnol. 2011;91:17–30. doi: 10.1007/s00253-011-3252-0. [DOI] [PubMed] [Google Scholar]
- Schneider J, Niermann K, Wendisch VF. Production of amino acids L-glutamate, L-lysine, L-ornithine and L-arginine from arabinose by recombinant Corynebacterium glutamicum. J Biotechnol. 2011;154:191–198. doi: 10.1016/j.jbiotec.2010.07.009. [DOI] [PubMed] [Google Scholar]
- Schneider J, Eberhard D, Wendisch VF. Improving putrescine production by Corynebacterium glutamicum by fine-tuning ornithine transcarbamoylase activity using a plasmid addiction system. Appl Microbiol Biotechnol. 2012;95:169–178. doi: 10.1007/s00253-012-3956-9. [DOI] [PubMed] [Google Scholar]
- Schreiner ME, Fiur D, Holátko J, Pátek M, Eikmanns BJ. E1 enzyme of the pyruvate dehydrogenase complex in Corynebacterium glutamicum: molecular analysis of the gene and phylogenetic aspects. J Bacteriol. 2005;187:6005–6018. doi: 10.1128/JB.187.17.6005-6018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner ME, Riedel C, Holátko J, Pátek M, Eikmanns BJ. Pyruvate:quinone oxidoreductase in Corynebacterium glutamicum: molecular analysis of the pqo gene, significance of the enzyme, and phylogenetic aspect. J Bacteriol. 2006;188:1341–1350. doi: 10.1128/JB.188.4.1341-1350.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibold G, Auchter M, Berens S, Kalinowski J, Eikmanns BJ. Utilization of soluble starch by a recombinant Corynebacterium glutamicum strain: growth and lysine production. J Biotechnol. 2006;124:381–391. doi: 10.1016/j.jbiotec.2005.12.027. [DOI] [PubMed] [Google Scholar]
- Smith KM, Cho KM, Liao JC. Engineering Corynebacterium glutamicum for isobutanol production. Appl Microbiol Biotechnol. 2010;87:1045–1055. doi: 10.1007/s00253-010-2522-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Matsumoto K, Tanaka T, Kondo A, Taguchi S. Single-step production of polyhydroxybutyrate from starch by using α-amylase cell-surface displaying system of Corynebacterium glutamicum. J Biosci Bioeng. 2012 doi: 10.1016/j.jbiosc.2012.08.004. Sep 4. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Takahashi C, Shirakawa J, Tsuchidate T, Okai N, Hatada K, Nakayama H, et al. Robust production of gamma-amino butyric acid using recombinant Corynebacterium glutamicum expressing glutamate decarboxyölase from Escherichia coli. Enzyme Microb Technol. 2012;51:171–176. doi: 10.1016/j.enzmictec.2012.05.010. [DOI] [PubMed] [Google Scholar]
- Takeno S, Ohnishi J, Komatsu T, Masaki T, Sen K, Ikeda M. Anaerobic growth and potential for amino acid production by nitrate respiration in Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2007;75:1173–1182. doi: 10.1007/s00253-007-0926-8. [DOI] [PubMed] [Google Scholar]
- Takors R, Bathe B, Rieping M, Hans S, Kelle R, Huthmacher K. Systems biology for industrial strains and fermentation processes – example: amino acids. J Biotechnol. 2007;129:181–190. doi: 10.1016/j.jbiotec.2007.01.031. [DOI] [PubMed] [Google Scholar]
- Tateno T, Fukuda H, Kondo A. Direct production of L-lysine from raw corn starch by Corynebacterium glutamicum secreting Streptococcus bovis alpha-amylase using cspB promoter and signal seuence. Appl Microbiol Biotechnol. 2007;77:533–541. doi: 10.1007/s00253-007-1191-6. [DOI] [PubMed] [Google Scholar]
- Teramoto H, Inui M, Yukawa H. transcriptional regulators of multiple genes involved in carbon metabolism in Corynebacterium glutamicum. J Biotechnol. 2011;154:114–125. doi: 10.1016/j.jbiotec.2011.01.016. [DOI] [PubMed] [Google Scholar]
- Teschan PE, Beck GJ, Dwyer JT, Greene T, Klahr S, Levy AS, et al. Effect of a ketoacid-aminoacid-supplemented very low protein diet on the progression of advanced renal disease: a reanalysis of the MDRD feasibility study. Clin Nephrol. 1998;50:273–283. [PubMed] [Google Scholar]
- Uehara H, Karaki Y, Wada S, Yamanobe T. Stereo-complex crystallization of poly(lactic acid)s in block-copolymer phase separation. ACS Appl Mater Interfaces. 2010;2:2707–2710. doi: 10.1021/am1005755. [DOI] [PubMed] [Google Scholar]
- Uhde A, Youn JW, Maeda T, Clermont L, Matano C, Krämer R, et al. Glucosamine as carbon source for amino acid-producing Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2012 doi: 10.1007/s00253-012-4313-8. Aug 2. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Vertes AA, Inui M, Yukawa H. Postgenomic approaches to using corynebacteria as biocatalysts. Annu Rev Microbiol. 2012;66:521–550. doi: 10.1146/annurev-micro-010312-105506. [DOI] [PubMed] [Google Scholar]
- Wendisch VF, Bott M, Kalinowski J, Oldiges M, Wiechert W. Emerging Corynebacterium glutamicum systems biology. J Biotechnol. 2006a;124:74–92. doi: 10.1016/j.jbiotec.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Wendisch VF, Bott M, Eikmanns BJ. Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for biotechnological production of organic acids and amino acids. Curr Opin Microbiol. 2006b;9:268–274. doi: 10.1016/j.mib.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Wendisch VF, Lindner SN, Meiswinkel TM, Montero G, Stoytcheva M. Biodiesel – Quality, Emissions and By-Products. Rijeka, Croatia: InTech; 2011. Use of glycerol in biotechnological applications; pp. 305–341. [Google Scholar]
- Werpy T, Petersen GR. Top Value Added Chemicals from Biomass. Volume I: Results of Screening for Potential Candidates from Sugars and Synthesis Gas. Oak Ridge, TN, USA: U.S. Department of Energy; 2004. [Google Scholar]
- Wieschalka S, Blombach B, Eikmanns BJ. Engineering Corynebacterium glutamicum for the production of pyruvate. Appl Microbiol Biotechnol. 2012;94:449–459. doi: 10.1007/s00253-011-3843-9. [DOI] [PubMed] [Google Scholar]
- Wiselogel A, Tyson S, Johnson D. Biomass Feedstock Resources and Composition. Washington, DC, USA: Taylor & Francis; 1996. [Google Scholar]
- Yazdani SS, Gonzalez R. Anaerobic fermentation of glycerol: a path to economic viability for the biofuels industry. Curr Opin Biotechnol. 2007;18:213–219. doi: 10.1016/j.copbio.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Zeikus JG, Jain MK, Elankovan P. Biotechnology of succinic acid production and markets for derived industrial products. Appl Microbiol Biotechnol. 1999;51:545–552. [Google Scholar]
- Zelic B, Gerharz T, Bott M, Vasic-Racki D, Wandrey C, Takors R. Fed-batch process for pyruvate production by recombinant Escherichia coli YYC202 strain. Eng Life Sci. 2003;3:299–305. [Google Scholar]
- Zhou J, Yin X, Madzak C, Du G, Chen J. Enhanced α-ketoglutarate production in Yarrowia lipolytica WSH-Z06 by alteration of the acetyl-CoA metabolism. J Biotechnol. 2012;161:257–264. doi: 10.1016/j.jbiotec.2012.05.025. [DOI] [PubMed] [Google Scholar]
- Zhou S, Causey TB, Hasona A, Shanmugam KT, Ingram LO. Production of optically pure d-lactic acid in mineral salts medium by metabolically engineered Escherichia coli W3110. Appl Environ Microbiol. 2003a;69:399–407. doi: 10.1128/AEM.69.1.399-407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Shanmugam KT, Ingram LO. Functional replacement of the Escherichia coli D-(-)-lactate dehydrogenase gene (ldhA) with the L-(+)-lactate dehydrogenase gene (ldhL) from Pediococcus acidilactici. Appl Environ Microbiol. 2003b;69:2237–2244. doi: 10.1128/AEM.69.4.2237-2244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Eiteman MA, Altman R, Altman E. High glycolytic flux improves pyruvate production by a metabolically engineered Escherichia coli strain. Appl Environ Microbiol. 2008;74:6649–6655. doi: 10.1128/AEM.01610-08. [DOI] [PMC free article] [PubMed] [Google Scholar]