Summary
Because of their abundance in hemicellulosic wastes arabinose and xylose are an interesting source of carbon for biotechnological production processes. Previous studies have engineered several Corynebacterium glutamicum strains for the utilization of arabinose and xylose, however, with inefficient xylose utilization capabilities. To improve xylose utilization, different xylose isomerase genes were tested in C. glutamicum. The gene originating from Xanthomonas campestris was shown to have the highest effect, resulting in growth rates of 0.14 h−1, followed by genes from Bacillus subtilis, Mycobacterium smegmatis and Escherichia coli. To further increase xylose utilization different xylulokinase genes were expressed combined with X. campestris xylose isomerase gene. All combinations further increased growth rates of the recombinant strains up to 0.20 h−1 and moreover increased biomass yields. The gene combination of X. campestris xylose isomerase and C. glutamicum xylulokinase was the fastest growing on xylose and compared with the previously described strain solely expressing E. coli xylose isomerase gene delivered a doubled growth rate. Productivity of the amino acids glutamate, lysine and ornithine, as well as the diamine putrescine was increased as well as final titres except for lysine where titres remained unchanged. Also productivity in medium containing rice straw hydrolysate as carbon source was increased.
Funding Information No funding information provided.
Introduction
Lignocellulosic hydrolysates contain glucose as well as significant amounts of the pentoses xylose (5–20%) and arabinose (1–5%) (Aristidou and Penttila, 2000). Lignocellulosic hydrolysates may be obtained from agricultural wastes such as rice straw and are therefore cheap carbon sources. However, lignocellulosic hydrolysates are not fully capitalized on since several industrially relevant microorganisms are not able to utilize pentose sugars as substrates (Jeffries and Jin, 2000). Metabolic engineering of pentose utilization has been successful in some cases, e.g. of Saccharomyces cerevisiae, while in other cases absent or inefficient pentose utilization is still a major bottleneck to be overcome for industrial processes based on lignocellulosic biomass (Aristidou and Penttila, 2000; Becker and Boles, 2003; Karhumaa et al., 2006; Hahn-Hagerdal et al., 2007a,2007b).
Corynebacterium glutamicum as a workhorse of industrial microbiology is well known for fermentative production of amino acids and has been engineered for the production of diamines like 1,4-diaminobutane (Schneider and Wendisch, 2010) and 1,5-diaminopentane (Mimitsuka et al., 2007; Kind et al., 2010; 2011), of ketoacids such as pyruvate (Wieschalka et al., 2012) and 2-ketoisovalerate (Krause et al., 2010), diacids such as succinate (Okino et al., 2008; Litsanov et al., 2012a,2012b,2012c) and the alcohols ethanol (Inui et al., 2004) and isobutanol (Blombach et al., 2011). Traditionally, technical substrates like starch hydrolysates and molasses are used in industrial processes. The respective sugars glucose (starch hydrolysate), fructose and sucrose (molasses) are taken up and are phosphorylated by the phosphoenolpyruvate-dependent carbohydrate phosphotransferase (PTS) system or, in the case of glucose, alternatively also by myo-inositol permeases with subsequent phosphorylation by ATP-and/or polyphosphate-dependent glucokinases (Lindner et al., 2010; 2011). The natural substrate spectrum of C. glutamicum further includes sugars like ribose or maltose, alcohols like ethanol or myo-inositol and organic acids like acetate, citrate, lactate, propionate and pyruvate and amino acids like l-glutamate (Kramer et al., 1990; Dominguez et al., 1998; Kiefer et al., 2002; Gerstmeir et al., 2003; Eikmanns, 2005; Moon et al., 2005; Polen et al., 2005; Stansen et al., 2005; Krings et al., 2006; Frunzke et al., 2008; Kato et al., 2010; Neuner and Heinzle, 2011). Within the flexible feedstock concept, the substrate spectrum of C. glutamicum has been extended by metabolic engineering to allow access to starch, cellobiose, lactose, galactose and glycerol as well as succinate, fumarate and malate as carbon sources (Brabetz et al., 1991; Cadenas et al., 1992; Kotrba et al., 2003; Barrett et al., 2004; Seibold et al., 2006; Tateno et al., 2007; Rittmann et al., 2008; Youn et al., 2008; 2009).
Similarly, C. glutamicum has been engineered for growth with the pentoses arabinose and xylose and for the production of ethanol, organic acids, amino acids and diamines from arabinose and/or xylose (Kawaguchi et al., 2006; 2008; Sasaki et al., 2009, 2010; Gopinath et al., 2011; Kind and Wittmann, 2011; Schneider et al., 2011). Metabolic engineering relied on bacterial pathway genes. In Escherichia coli and other bacteria able to utilize arabinose and/or xylose, arabinose is catabolized via arabinose isomerase (encoded by araA), ribulokinase (araB) and ribulose-5-phosphate-4-epimerase (araD) while xylose catabolism requires xylose isomerase (xylA) and xylulokinase (xylB) (Lin, 1996; Hahn-Hagerdal et al., 2007a,2007b). Heterologous expression of araA, araB and araD from E. coli resulted in C. glutamicum recombinants able to grow with arabinose as sole source of carbon (Kawaguchi et al., 2008). When the arabinose importer gene araE from C. glutamicum ATCC31831 was expressed in addition, faster growth with arabinose entailed (Sasaki et al., 2009). In the case of xylose, heterologous expression of a single E. coli gene, xylA, was sufficient to allow growth with xylose as sole carbon source (Kawaguchi et al., 2006) since the C. glutamicum genome encodes xylulokinase (Kalinowski et al., 2003). Corynebacterium glutamicum has proven a good choice for utilizing complex mixtures of carbon sources such as hemicellulosic hydrolysates because, unlike E. coli and S. cerevisiae, C. glutamicum efficiently co-utilizes different carbon sources when present in blends (Wendisch, 2006; Arndt and Eikmanns, 2008; Blombach and Seibold, 2010; Gopinath et al., 2011). Consequently, besides proof-of-concept using pure chemicals, growth and production with hemicellulosic hydrolysates obtained, e.g. from rice straw could be achieved (Gopinath et al., 2011). In the present study, we address xylose catabolism as a possible rate-limiting step of xylose-based production by C. glutamicum.
Results
Comparative analysis of recombinant C. glutamicum strains with different plasmid encoded xylose isomerases
Corynebacterium glutamicum possesses a xylulokinase and heterologous production of E. coli xylose isomerase allowed C. glutamicum to grow with xylose, however, the observed growth rates were low (0.09 h−1) as compared to growth rates, e.g. with glucose (0.32 h−1), ribose (0.23 h−1) or acetate (0.28 h−1) (Wendisch et al., 2000; Wendisch, 2003; Netzer et al., 2004). In order to test whether xylose isomerase activity is limiting growth with xylose of C. glutamicum recombinants expressing xylA from E. coli, several recombinants were constructed expressing xylose isomerase genes from different sources. The xylose isomerase genes of well-understood model organisms, plant pathogens and strains closely to C. glutamicum related E. coli, Bacillus subtilis, Xanthomonas campestris and Mycobacterium smegmatis, were cloned into the IPTG-inducible expression vector pEKEx3 and transformed into C. glutamicum WT (Table 1C). Xylose isomerase (XI) activity measured as described in Experimental procedures was not detectable in empty vector control strains (< 0.005 U mg−1) (Table 2). High and comparable XI activities were observed in crude extracts of WT(pEKEx3-xylAEc) (0.095 ± 0.010 U mg−1) and WT(pEKEx3-xylAXc) (0.090 ± 0.008 U mg−1), while about three times less activity was found for WT(pEKEx3-xylABs) (0.023 ± 0.003 U mg−1) and WT(pEKEx3-xylAMs) (0.033 ± 0.007 U mg−1). While XI activity increase due to overexpression of xylA could be detected in each case, the values are difficult to compare as a single enzyme assay was used without optimization for the enzymes of various origin.
Table 1.
List of sequences of oligonucleotide primers, plasmids and strains used
| Name | Sequence (5′–3′) or function | Relevant characteristics or reference |
|---|---|---|
| A. Oligonucleotides | ||
| xylB_fw_Bsu | GAGAAAGGAGGCCCTTCAGATGAAGTATGTCATTGGAATAGATCTTGG | HE of Bsu xylB; RBS; start |
| xylB_rv_Bsu | GATCTAGATTAGTTTTTTCGAAAGCTCTTCAAAGC | HE of Bsu xylB; XbaI; stop |
| xylB_fw_Cgl | GAGAAAGGAGGCCCTTCAGATGGCTTTGGTTCTTGGAATCG | OE of Cgl xylB; RBS; start |
| xylB_rv_Cgl | GATCTAGACTAGTACCAACCCTGCGTTG | OE of Cgl xylB; XbaI; stop |
| xylB_fw_Eco | GAGAAAGGAGGCCCTTCAGATGTATATCGGGATAGATCTTGGCAC | HE of Eco xylB; RBS; start |
| xylB_rv_Eco | GATCTAGATTACGCCATTAATGGCAGAAGTTG | HE of Eco xylB; XbaI; stop |
| xylA_fw_Bsu | GATCTAGAGAAAGGAGGCCCTTCAGATGGCTCAATCTCATTCCAGTTCA | HE of Bsu xylA; XbaI; RBS; start |
| xylA_rv_Bsu | GAGAGCTCTTATACTTCTAAAATGTATTGGTTCAATATCGCT | HE of Bsu xylA; Ecl136II; stop |
| xylA_fw_Eco | GATCTAGAGAAAGGAGGCCCTTCAGATGCAAGCCTATTTTGACCAGC | HE of Eco xylA; XbaI; RBS; start |
| xylA_rv_Eco | GAGAGCTCTTATTTGTCGAACAGATAATGGTTTACCAG | HE of Eco xylA; Ecl136II; stop |
| xylA_fw_Msm | GATCTAGAGAAAGGAGGCCCTTCAGATGACCGTGTTGGAGTCGAA | HE of Msm xylA; XbaI; RBS; start |
| xylA_rv_Msm | GAGAGCTCTCATCGCGCGCCCATCAG | HE of Msm xylA; Ecl136II; stop |
| xylA_fw_Xcc | GATCTAGAGAAAGGAGGCCCTTCAGATGAGCAACACCGTTTTCATCG | HE of Xcc xylA; XbaI; RBS; start |
| xylA_rv_Xcc | GAGAGCTCTCAACGCGTCAGGTACTGATT | HE of Xcc xylA; Ecl136II; stop |
| B. Plasmids | ||
| pEKEx3 | SpecR; C. glutamicum/E. coli shuttle vector (Ptac, lacIq; pBL1, OriVCg, OriVEc) | Stansen et al. (2005) |
| pEKEx3-xylAEc | Derived from pEKEx3, for regulated expression of xylAEc (b3565) of E. coli | Gopinath et al. (2011) |
| pEKEx3-xylABs | Derived from pEKEx3, for regulated expression of xylABs (BSU17600) of B. subtilis | This work |
| pEKEx3-xylAMs | Derived from pEKEx3, for regulated expression of xylAMs (MSMEG_6021) of M. smegmatis | This work |
| pEKEx3-xylAXc | Derived from pEKEx3, for regulated expression of xylAXc (XCC1758) of X. campestris | This work |
| pEKEx3-xylAXc-xylBEc | Derived from pEKEx3, for regulated expression of xylAXc (XCC1758) of X. campestris and xylBEc (b3580) of E. coli | This work |
| pEKEx3-xylAXc-xylBBs | Derived from pEKEx3, for regulated expression of xylAXc (XCC1758) of X. campestris and xylBBs (BSU17610) of B. subtilis | This work |
| pEKEx3-xylAXc-xylBCg | Derived from pEKEx3, for regulated expression of xylAXc (XCC1758) of X. campestris and xylBCg (cg0147) of C. glutamicum | This work |
| pVWEx1 | KanR; C. glutamicum/E. coli shuttle vector (Ptac, lacIq; pHM1519, OriVCg, OriVEc) | Peters-Wendisch et al. (1998) |
| pVWEx1-araBAD | Derived from pVWEx1, for regulated expression of araB (b0063) and araA (b0062) and araD (b0061) of E. coli | Schneider et al. (2011) |
| C. Strains | ||
| E. coli | ||
| DH5α | Fthi-1 endA1 hsdr17(r−, m−) supE44 _lacU169 (ф80lacZ_M15) recA1 gyrA96 relA1 | Hanahan (1983) |
| C. glutamicum | ||
| ATCC13032 | Wild type (WT) | Kinoshita et al. (1957) |
| DM1729 | lysC P458S, homV59A, pyc T311I | Georgi et al. (2005) |
| ORN1 | l-ornithine overproducing strain derived from ATCC13032, auxotrophic for l-arginine due to argF deletion | Schneider et al. (2011) |
| PUT21 | ORN1 carrying pVWEx1-speC-5′21-argF | Schneider et al. (2011) |
Restriction sites are underlined, ribosomal binding sites are shown in bold, stop and start codons are in italics.
OE, overexpression; HE, heterologous expression; RBS, ribosomal binding site; Cgl, C. glutamicum; Eco, E. coli; Bsu, B. subtilis; Msm, M. smegmatis; Xcc, X. campestris.
Table 2.
Specific activities of different xylose isomerase and xylulokinase
| Specific activity (U mg−1 total protein) | ||||||||
|---|---|---|---|---|---|---|---|---|
| WT(pEKEx3-x) |
||||||||
| – | xylAEc | xylABs | xylAMs | xylAXc | xylAXc-xylBEc | xylAXc-xylBBs | xylAXc-xylBCg | |
| Xylose isomerase | < 0.005 | 0.095 ± 0.010 | 0.023 ± 0.003 | 0.033 ± 0.007 | 0.090 ± 0.008 | 0.062 ± 0.004 | 0.026 ± 0.003 | 0.077 ± 0.010 |
| Xylulokinase | 0.013 ± 0.005 | 0.020 ± 0.000 | 0.024 ± 0.000 | 0.019 ± 0.000 | 0.021 ± 0.000 | 0.541 ± 0.063 | 0.020 ± 0.004 | 0.468 ± 0.018 |
All tests were carried out with crude extracts at 30°C.
To check the performance of the C. glutamicum strains harbouring the different XI genes growth experiments in CgXII minimal medium with 100 mM xylose as sole carbon source were performed (Fig. 1A). All recombinant strains expressing xylose isomerase genes were able to grow with xylose as sole carbon source (Fig. 1A). Corynebacterium glutamicum WT(pEKEx3-xylAXc) showed the fastest growth (0.144 ± 0.001 h−1) and reached the highest biomass concentration (3.37 ± 0.12 gCDW l−1), followed by WT(pEKEx3-xylABs) (0.118 ± 0.007 h−1; 1.66 ± 0.15 gCDW l−1), WT(pEKEx3-xylAMs) (0.093 ± 0.003 h−1; 1.29 ± 0.05 gCDW l−1) and WT(pEKEx3-xylAEc) (0.090 ± 0.005 h−1; 2.79 ± 0.05 gCDW l−1). Thus, heterologous expression of the xylose isomerase gene from X. campestris improved xylose-utilization by recombinant C. glutamicum significantly reducing generation times from 7.7 h to 4.8 h.
Figure 1.
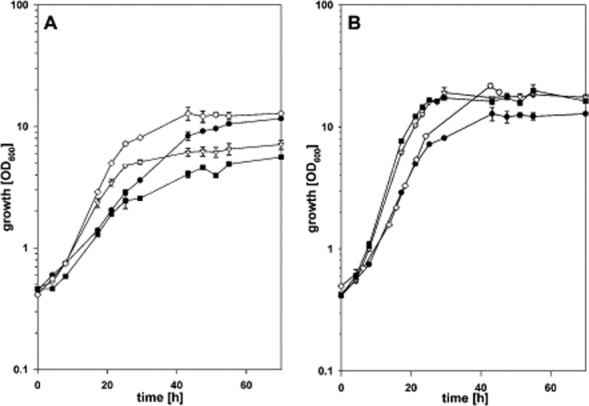
Growth of C. glutamicum strains in CgXII medium containing 100 mM xylose.A. Corynebacterium glutamicum strains WT(pEKEx3-xylAXc) (open diamonds), WT(pEKEx3-xylABs) (open triangles), WT(pEKEx3-xylAEc) (closed circles) and WT(pEKEx3-xylAMs) (closed squares) were analysed.B. Corynebacterium glutamicum strains WT(pEKEx3-xylAXc-xylBBs) (open diamonds), WT(pEKEx3-xylAXc-xylBEc) (open triangles), WT(pEKEx3-xylAXc) (closed circles) and WT(pEKEx3-xylAXc-xylBCg) (closed squares) were analysed. Data represents mean values and standard deviations of three independent cultivations.
Comparative analysis of recombinant C. glutamicum strains overexpressing endogenous or heterologous xylulokinase genes
Corynebacterium glutamicum WT contains xylulokinase, however, xylulokinase activities determined as described in Experimental procedures were low in crude extracts of C. glutamicum WT, the empty vector control strain and of all recombinants expressing only a heterologous xylose isomerase gene (between 0.013 and 0.024 U mg−1) (Table 2). Ectopic expression xylA from X. campestris was combined either with overexpression of endogenous xylB or with overexpression of xylulokinase genes from E. coli or B. subtilis (Table 1C). Xylulokinase (XK) activity was not increased significantly in strain WT(pEKEx3-xylAXc-xylBBs) (Table 2). In contrast, ectopic expression of xylB from E. coli and overexpression of endogenous xylB increased XK activity in crude extracts about 25-fold. To test the effect of xylB overexpression in addition to xylA overexpression, growth of C. glutamicum strains overproducing the different XK's along with XI from X. campestris in CgXII minimal medium with 100 mM xylose as sole carbon source was compared (Fig. 1B). The control strain WT(pEKEx3-xylAXc) reached a lower biomass concentration (3.37 ± 0.12 gCDW l−1) and grew with a slower growth rate (0.144 ± 0.001 h−1) than the strains overproducing XK gene in addition. Corynebacterium glutamicum WT(pEKEx3-xylAXc-xylBCg) grew fastest (0.199 ± 0.009 h−1) and reached the highest biomass concentration (4.87 ± 0.53 gCDW l−1) followed by WT(pEKEx3-xylAXc-xylBEc) (0.189 ± 0.001 h−1; 4.82 ± 0.33 gCDW l−1) and WT(pEKEx3-xylAXc-xylBBs) (0.162 ± 0.001 h−1; 5.30 ± 0.22 gCDW l−1). Thus, heterologous expression of the endogenous xylulose kinase gene from C. glutamicum in addition to the xylose isomerase gene from X. campestris further improved xylose utilization significantly reducing generation times from 4.8 h to 3.5 h.
Amino acid and diamine production from xylose by the improved strain
Previously, we showed production of amino acids like l-glutamate and l-lysine as well as the diamine putrescine from xylose minimal medium by strains harbouring the basic xylose utilization plasmid pEKEx3-xylAEc (Gopinath et al., 2011). The improved plasmid pEKEx3-xylAXc-xylBCg was transformed into the model lysine producer DM1729, the model ornithine producer ORN1 and the model 1,4-diaminobutane producer PUT21. l-glutamate production in CgXII minimal medium with 100 mM xylose as sole carbon source by C. glutamicum WT(pEKEx3-xylAEc) and by WT(pEKEx3-xylAXc-xylBCg) was triggered by ethambutol addition and the improved strain reached higher titres (14.5 ± 0.1 mM as compared with 0.8 ± 0.1 mM) and exhibited an increased productivity (29.7 ± 0.2 as compared with 1.6 ± 0.3 mg l−1 h−1, Fig. 2). Lysine production by DM1729(pEKEx3-xylAXc-xylBCg) was characterized by a volumetric productivity improved from 25.5 ± 0.8 to 35.4 ± 1.4 mg l−1 h−1. The volumetric ornithine productivity by ORN1(pEKEx3-xylAXc-xylBCg) was higher than that of the control (43.2 ± 4.3 as compared with 14.8 ± 2.2 mg l−1 h−1) and higher ornithine concentrations were achieved (19.6 ± 1.9 mM as compared with 9.4 ± 1.4 mM). Also putrescine production was faster (27.8 ± 2.0 as compared with 15.7 ± 1.2 mg l−1 h−1) and titres rose from 12.9 ± 1.0 mM to 15.1 ± 1.1 mM. Taken together, all recombinants carrying the improved plasmid pEKEx3-xylAXc-xylBCg showed significantly increased volumetric productivities in medium with pure xylose.
Figure 2.
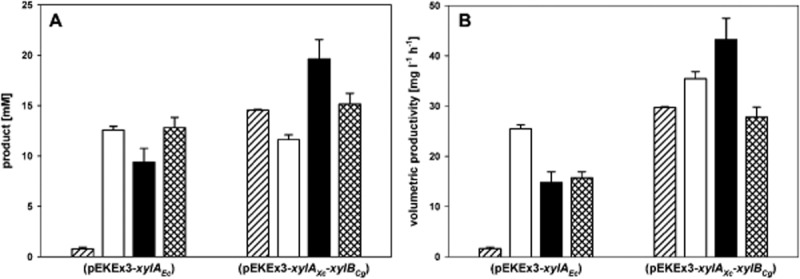
Product concentrations (A) and volumetric productivities (B) for l-glutamate, l-lysine, l-ornithine and putrescine production in CgXII medium containing 100 mM xylose. Corynebacterium glutamicum strains with pEKEx3-xylAEc or pEKEx3-xylAXc-xylBCg were analysed. l-glutamate was produced with WT (hatched bars), l-lysine with DM1729 (open bars), l-ornithine with ORN1 (closed bars) and putrescine with PUT21 (checked bars). Data represent mean values and experimental imprecision of two independent cultivations.
Amino acid production on rice straw hydrolysate
To characterize l-glutamate and l-lysine production from hemicellulosic hydrolysates in particular rice straw hydrolysate (52 mM glucose, 203 mM xylose, 55 mM arabinose) derivatives of C. glutamicum WT or l-lysine model producer DM1729 harbouring either empty vectors, pVWEx1-araBAD and pEKEx3-xylAEc or pVWEx1-araBAD and pEKEx3-xylAXc-xylBCg were used. The empty vector control utilized glucose for biomass formation and amino acid production, while the pentose-utilizing recombinants grew to higher biomass concentrations and produced more l-glutamate and l-lysine as they utilized arabinose and xylose in addition to glucose (Fig. 3). In case of l-glutamate the empty vector control reached 16 ± 5.4 mM and a volumetric productivity of 98.1 ± 33.1 mg l−1 h−1 in contrast to the pentose-utilizing strain WT(pVWEx1-araBAD)(pEKEx3-xylAEc) with 39 ± 1.9 mM l-glutamate and a productivity of 79.7 ± 3.9 mg l−1 h−1. The strain improved for xylose utilization reached a comparable level of l-glutamate at 37 ± 5 mM and the highest productivity at 113.4 ± 15.3 mg l−1 h−1. As expected for growth-coupled l-glutamate production the specific productivities were similar [around 2.6, 3.0 and 3.1 mg gCDW−1 h−1 for the empty vector control, WT(pVWEx1-araBAD)(pEKEx3-xylAEc) and WT(pVWEx1-araBAD)(pEKEx3-xylAXc-xylBCg) respectively].
Figure 3.
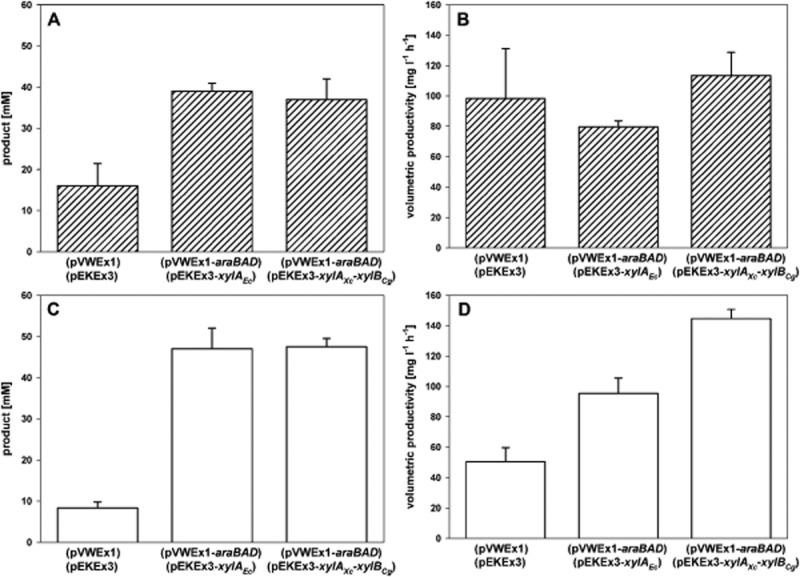
Product concentrations (A, C) and volumetric productivities (B, D) for l-glutamate (A, B) and l-lysine (C, D) production in CgXII medium containing rice straw hydrolysate. Corynebacterium glutamicum strains with empty vectors, pVWEx1-araBAD and pEKEx3-xylAEc or pVWEx1-araBAD and pEKEx3-xylAXc-xylBCg were analysed. l-glutamate was produced with WT (hatched bars) and l-lysine with DM1729 (open bars). Data represent mean values and experimental imprecision of two independent cultivations.
As observed in the l-glutamate production experiment, product formation and productivity strongly depends on the ability to utilize the pentose fraction of the rice straw hydrolysate due to the expression of araBAD and xylA and/or xylA along with xylB. Therefore empty vector control reached 8.3 ± 1.5 mM l-lysine and a volumetric productivity of 50.6 ± 9.1 mg l−1 h−1 in contrast to the pentose-utilizing strain DM1729(pVWEx1-araBAD)(pEKEx3-xylAEc) with clearly increased 47 ± 5 mM l-lysine and a productivity of 95.4 ± 10.2 mg l−1 h−1. The strain improved for xylose utilization reached a similar level of l-lysine at 47.5 ± 2 mM together with the highest productivity at 144.7 ± 6.1 mg l−1 h−1. As l-lysine production was growth-coupled the specific productivities were similar [around 2.6, 3.0 and 3.1 mg gCDW−1 h−1 for the empty vector control, DM1729 (pVWEx1-araBAD)(pEKEx3-xylAEc) and DM1729 (pVWEx1-araBAD)(pEKEx3-xylAXc-xylBCg) respectively].
Discussion
The newly engineered strain WT(pEKEx3-xylAXc-xylBCg) was shown to grow significantly faster (0.199 ± 0.009 h−1) on minimal medium containing xylose as sole carbon source compared with the previously described strain WT(pEKEx3-xylAEc) (0.090 ± 0.005 h−1) (Gopinath et al., 2011) expressing xylA from E. coli only. A first improvement was already achieved by expressing different xylA genes, where xylA from X. campestris performed best (0.144 ± 0.001 h−1). By additional production of xylulokinase from different organisms further growth acceleration was observed with the fastest growing strain mentioned above. These findings let to the construction of production strains for lysine, glutamate, ornithine and putrescine for optimized utilization of xylose. The newly engineered xylose utilization strains showed a significantly higher volumetric productivity (l-glutamate: 29.7 ± 0.2 mg l−1 h−1; l-lysine: 35.4 ± 1.4 mg l−1 h−1; l-ornithine: 43.2 ± 4.3 mg l−1 h−1; putrescine: 27.8 ± 2.0 mg l−1 h−1) compared with production strains using the previously reported (Gopinath et al., 2011) xylose utilization plasmid (l-glutamate: 1.6 ± 0.3 mg l−1 h−1; l-lysine: 25.5 ± 0.8 mg l−1 h−1; l-ornithine: 14.8 ± 2.2 mg l−1 h−1; putrescine: 15.7 ± 1.2 mg l−1 h−1). Also during growth and production on rice straw hydrolysate a clear increase in volumetric productivity was observed for strains carrying pEKEx3-xylAXc-xylBCg (l-glutamate: 113.4 ± 15.3 mg l−1 h−1; l-lysine: 144.7 ± 6.1 mg l−1 h−1) compared with strains with pEKEx3-xylAEc (l-glutamate: 79.7 ± 3.9 mg l−1 h−1; l-lysine: 95.4 ± 10.2 mg l−1 h−1) and in case of lysine production as well compared with the empty vector control strain, which is only capable of utilizing the glucose part of rice straw hydrolysate. As expected for growth-coupled amino acid production the specific productivities normalized to the biomass concentrations were similar.
Engineering for a better use of second-generation feedstock like rice straw hydrolysate, one possible bottleneck regarding catabolism of those carbon sources was successfully dealt with in this work. Further possible bottlenecks are transport of the carbon sources into the cell and the process of breaking the poly-and oligomeric sugars into their monomeric compounds to make them accessible for the producing microorganisms. Concerning the later point in this study a mild sulfuric acid treatment was used to hydrolysate the rice straw (Gopinath et al., 2011). A potential formation of typical fermentation inhibitors, e.g. 5-HMF or weak acids, may result in slower growth and lower production (Palmqvist et al., 1999; Zaldivar and Ingram, 1999; Zaldivar et al., 1999; 2000; Klinke et al., 2004; Heer and Sauer, 2008; Gopinath et al., 2011) and could be an aim to analyse in more detail in future studies for further optimization. However it was already described that in case of ethanol production by growth-arrested cells, typical inhibitors like organic acids, phenolic inhibitors or furans did not substantially disturb C. glutamicum (Sakai et al., 2007). In principal overcoming inhibition can be achieved by different ways, e.g. by simple resistance to the inhibitory substances due to efflux pump or prevention of uptake, by degradation of the relevant substances (Koopman et al., 2010) or by simply not creating inhibitors during processing of the substrates.
Dealing with the potential bottleneck of transport, the xylose and/or arabinose transporting system in the used C. glutamicum wild-type strain ATCC13032 is still unknown and therefore the heterologous expression of xylose and/or arabinose transport systems, e.g. araE (Sasaki et al., 2009), might result in faster substrate uptake and also higher productivity. AraE from C. glutamicum ATCC31831 might be a promising target for transport optimization because this uptake system accepts both arabinose and xylose and allows growth on even very low arabinose or xylose concentrations (Sasaki et al., 2009) and the donor strain is closely related to the used C. glutamicum ATCC13032.
With the potential for further improvements this study has clearly shown the ability of the already industrially intensively used C. glutamicum (Eggeling and Bott, 2005; Wendisch, 2006) to play a key role in utilization of second-generation feedstocks with respect to a wide product spectra reaching from products like amino acids to products like fine chemicals as diamines, e.g. putrescine.
Experimental procedures
Microorganisms and cultivation conditions
E. coli strain DH5α (Hanahan, 1985) was used for cloning and was cultivated in lysogeny broth medium (LB) (Sambrook et al., 1989). Corynebacterium glutamicum strains used in this work are wild-type strain ATCC13032 (WT) (Abe et al., 1967), l-lysine producing model strain DM1729 (Georgi et al., 2005), l-ornithine producing strain ORN1 and putrescine producing strain PUT21 (Schneider et al., 2011). Pre-cultures of C. glutamicum strains were inoculated from brain heart infusion (BHI) plates into BHI medium. For growth experiments with C. glutamicum 50 ml of BHI overnight cultures were harvested by centrifugation (10 min; 3220 g), washed in CgXII (Eggeling and Bott, 2005), centrifuged again and inoculated in CgXII medium to a final optical density (λ = 600 nm) (OD600) of 0.5. All growth and production experiments were carried out with CgXII medium in baffled shake flasks at 30°C and 120 r.p.m. When appropriate 100 μg ml−1 spectinomycin, 25 μg ml−1 kanamycin and 1 mM isopropyl-β-d-thiogalactopyranosid (IPTG) were added to the medium. l-glutamate excretion was triggered by addition of 500 μg ml−1 ethambutol (Radmacher et al., 2005). Ethambutol triggering of glutamate production was preferred over triggering by biotin limitation to avoid effects of residual biotin in hydrolysates. ORN1 was supplemented by addition of 500 μM l-arginine to minimal medium (Schneider et al., 2011). Growth was followed by OD600 determination until the stationary phase. OD600 was measured using a UV-1650 PC photometer (Shimadzu, Duisburg, Germany) in dilutions resulting in an OD600 between 0.05 and 0.25. The plasmids used in this study are listed in Table 1B.
Heterologous expression of xylA and xylB genes from B. subtilis, E. coli, M. smegmatis and X. campestris and overexpression of xylB from C. glutamicum
For heterologous expression of genes encoding xylose isomerase (xylA) and xylulokinase (xylB) from B. subtilis, E. coli, M. smegmatis and X. campestris the genes were amplified via PCR from genomic DNA of E. coli MG1655, B. subtilis strain 168, M. smegmatis MC2 155 and X. campestris pv. campestris ATCC33913.
DNA from of E. coli, B. subtilis, M. smegmatis and X. campestris was prepared by using DNA isolation Kit (Roche, Mannheim, Germany). For overexpression of xylB, the gene was amplified via PCR from genomic DNA of C. glutamicum WT, which was prepared as described previously (Eikmanns et al., 1995).
Genes were amplified by PCR using the oligonucleotide primer pairs xylA_fw_Eco and xylA_rv_Eco, xylA_fw_Bsu and xylA_rv_Bsu, xylA_fw_Msm and xylA_rv_Msm, xylA_fw_Xcc and xylA_rv_Xcc, xylB_fw_Eco and xylB_rv_Eco, xylB_fw_Bsu and xylB_rv_Bsu and xylB_fw_Cgl and xylB_rv_Cgl (oligonucleotide sequences are listed in Table 1A). PCR products encoding XylA were cloned blunt into SmaI-restricted vector pEKEx3 (Stansen et al., 2005) and xylB genes were cloned into Ecl136II-restricted vector pEKEx3-xylAXc resulting in the pEKEx3 derivatives listed in Table 1B, pEKEx3 allows IPTG-inducible gene expression. All resulting vectors were sequenced to confirm their sequence integrity.
Enzyme activity measurements
Enzyme activity measurements were analysed in crude extracts of C. glutamicum (Guyer et al., 1981). Cells were inoculated from LB overnight cultures to an OD600 of 0.5 in 50 ml of LB medium containing 1 mM IPTG. Cells were harvested by centrifugation at a final OD600 of 4 and stored at −20°C until use.
Xylose isomerase and xylulokinase activity was measured by the determination of NADH using sorbitol dehydrogenase in the case of xylose isomerase and pyruvate kinase as well as lactate dehydrogenase in case of xylulokinase. Xylose isomerase assays were carried out at 30°C in a total volume of 1 ml containing 100 mM TRIS/HCl, pH 7.5, 10 mM MgCl2, 0.23 mM NADH and sorbitol dehydrogenase (1 U) (Brat et al., 2009), in the case of xylulokinase the assay contained pyruvate kinase (6.8 U), lactate dehydrogenase (9.9 U), 2 mM phosphoenolpyruvate, 0.2 mM NADH, 1 mM ATP, 2 mM MgCl2 and 50 mM TRIS/HCl, pH 7.5 (Eliasson et al., 2000). Tests were started by addition of d-xylose (2 M) or d-xylulose (167 mM) respectively. Enzymatic activities are displayed in μmol min−1 mg−1, defined as one unit (U). Continuous measurements were carried out using a Shimadzu UV-1650 PC photometer (Shimadzu, Duisburg, Germany).
Protein concentrations in crude extracts were determined using Bradford reagents (Sigma, Taufkirchen, Germany) and concentrations were calculated against bovine serum albumin standards.
Acid hydrolysis of agricultural residues
Hydrolysis of rice straw has been carried out as described before (Gopinath et al., 2011).
Determination of amino acid and diamine concentrations
Amino acids l-lysine, l-glutamate and l-ornithine were quantified via HPLC as described previously (Georgi et al., 2005). Putrescine was quantified via HPLC as described before (Schneider et al., 2011).
Conflict of interest
None declared.
References
- Abe S, Takayarna K, Kinoshita S. Taxonomical studies on glutamic acid producing bacteria. J Gen Appl Microbiol. 1967;13:279–301. [Google Scholar]
- Aristidou A, Penttila M. Metabolic engineering applications to renewable resource utilization. Curr Opin Biotechnol. 2000;11:187–198. doi: 10.1016/s0958-1669(00)00085-9. [DOI] [PubMed] [Google Scholar]
- Arndt A, Eikmanns BJ, Burkovski A. Corynebacteria: Genomics and Molecular Biology. Wymondham, UK: Caister Academic Press; 2008. Regulation of carbon metabolism in Corynebacterium glutamicum; pp. 155–182. [Google Scholar]
- Barrett E, Stanton C, Zelder O, Fitzgerald G, Ross RP. Heterologous expression of lactose-and galactose-utilizing pathways from lactic acid bacteria in Corynebacterium glutamicum for production of lysine in whey. Appl Environ Microbiol. 2004;70:2861–2866. doi: 10.1128/AEM.70.5.2861-2866.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J, Boles E. A modified Saccharomyces cerevisiae strain that consumes l-Arabinose and produces ethanol. Appl Environ Microbiol. 2003;69:4144–4150. doi: 10.1128/AEM.69.7.4144-4150.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blombach B, Seibold GM. Carbohydrate metabolism in Corynebacterium glutamicum and applications for the metabolic engineering of l-lysine production strains. Appl Microbiol Biotechnol. 2010;86:1313–1322. doi: 10.1007/s00253-010-2537-z. [DOI] [PubMed] [Google Scholar]
- Blombach B, Riester T, Wieschalka S, Ziert C, Youn JW, Wendisch VF, Eikmanns BJ. Corynebacterium glutamicum tailored for efficient isobutanol production. Appl Environ Microbiol. 2011;77:3300–3310. doi: 10.1128/AEM.02972-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabetz W, Liebl W, Schleifer KH. Studies on the utilization of lactose by Corynebacterium glutamicum, bearing the lactose operon of Escherichia coli. Arch Microbiol. 1991;155:607–612. doi: 10.1007/BF00245357. [DOI] [PubMed] [Google Scholar]
- Brat D, Boles E, Wiedemann B. Functional expression of a bacterial xylose isomerase in Saccharomyces cerevisiae. Appl Environ Microbiol. 2009;75:2304–2311. doi: 10.1128/AEM.02522-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadenas RF, Gil JA, Martin JF. Expression of Streptomyces genes encoding extracellular enzymes in Brevibacterium lactofermentum: secretion proceeds by removal of the same leader peptide as in Streptomyces lividans. Appl Microbiol Biotechnol. 1992;38:362–369. doi: 10.1007/BF00170087. [DOI] [PubMed] [Google Scholar]
- Dominguez H, Rollin C, Guyonvarch A, Guerquin-Kern JL, Cocaign-Bousquet M, Lindley ND. Carbon-flux distribution in the central metabolic pathways of Corynebacterium glutamicum during growth on fructose. Eur J Biochem. 1998;254:96–102. doi: 10.1046/j.1432-1327.1998.2540096.x. [DOI] [PubMed] [Google Scholar]
- Eggeling L, Bott M. Handbook of Corynebacterium glutamicum. Boca Raton, USA: CRC Press; 2005. [Google Scholar]
- Eikmanns BJ, Eggeling L, Bott M. Handbook on Corynebacterium glutamicum. Boca Raton, USA: CRC Press; 2005. Central metabolism: tricarboxylic acid cycle and anaplerotic reactions; pp. 241–276. [Google Scholar]
- Eikmanns BJ, Rittmann D, Sahm H. Cloning, sequence analysis, expression, and inactivation of the Corynebacterium glutamicum icd gene encoding isocitrate dehydrogenase and biochemical characterization of the enzyme. J Bacteriol. 1995;177:774–782. doi: 10.1128/jb.177.3.774-782.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliasson A, Christensson C, Wahlbom CF, Hahn-Hagerdal B. Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl Environ Microbiol. 2000;66:3381–3386. doi: 10.1128/aem.66.8.3381-3386.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frunzke J, Engels V, Hasenbein S, Gatgens C, Bott M. Co-ordinated regulation of gluconate catabolism and glucose uptake in Corynebacterium glutamicum by two functionally equivalent transcriptional regulators, GntR1 and GntR2. Mol Microbiol. 2008;67:305–322. doi: 10.1111/j.1365-2958.2007.06020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgi T, Rittmann D, Wendisch VF. Lysine and glutamate production by Corynebacterium glutamicum on glucose, fructose and sucrose: roles of malic enzyme and fructose-1,6-bisphosphatase. Metab Eng. 2005;7:291–301. doi: 10.1016/j.ymben.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Gerstmeir R, Wendisch VF, Schnicke S, Ruan H, Farwick M, Reinscheid D, Eikmanns BJ. Acetate metabolism and its regulation in Corynebacterium glutamicum. J Biotechnol. 2003;104:99–122. doi: 10.1016/s0168-1656(03)00167-6. [DOI] [PubMed] [Google Scholar]
- Gopinath V, Meiswinkel TM, Wendisch VF, Nampoothiri KM. Amino acid production from rice straw and wheat bran hydrolysates by recombinant pentose-utilizing Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2011;92:985–996. doi: 10.1007/s00253-011-3478-x. [DOI] [PubMed] [Google Scholar]
- Guyer MS, Reed RR, Steitz JA, Low KB. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb Symp Quant Biol. 1981;45(Part 1):135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- Hahn-Hagerdal B, Karhumaa K, Fonseca C, Spencer-Martins I, Gorwa-Grauslund MF. Towards industrial pentose-fermenting yeast strains. Appl Microbiol Biotechnol. 2007a;74:937–953. doi: 10.1007/s00253-006-0827-2. [DOI] [PubMed] [Google Scholar]
- Hahn-Hagerdal B, Karhumaa K, Jeppsson M, Gorwa-Grauslund MF. Metabolic engineering for pentose utilization in Saccharomyces cerevisiae. Adv Biochem Eng Biotechnol. 2007b;108:147–177. doi: 10.1007/10_2007_062. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Glover DM. DNA Cloning: A Practical Approach. Vol. 1. Oxford, UK: IRL Press; 1985. Techniques for transformation of E. coli; pp. 109–135. [Google Scholar]
- Heer D, Sauer U. Identification of furfural as a key toxin in lignocellulosic hydrolysates and evolution of a tolerant yeast strain. Microb Biotechnol. 2008;1:497–506. doi: 10.1111/j.1751-7915.2008.00050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inui M, Kawaguchi H, Murakami S, Vertes AA, Yukawa H. Metabolic engineering of Corynebacterium glutamicum for fuel ethanol production under oxygen-deprivation conditions. J Mol Microbiol Biotechnol. 2004;8:243–254. doi: 10.1159/000086705. [DOI] [PubMed] [Google Scholar]
- Jeffries TW, Jin YS. Ethanol and thermotolerance in the bioconversion of xylose by yeasts. Adv Appl Microbiol. 2000;47:221–268. doi: 10.1016/s0065-2164(00)47006-1. [DOI] [PubMed] [Google Scholar]
- Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, et al. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of l-aspartate-derived amino acids and vitamins. J Biotechnol. 2003;104:5–25. doi: 10.1016/s0168-1656(03)00154-8. [DOI] [PubMed] [Google Scholar]
- Karhumaa K, Wiedemann B, Hahn-Hagerdal B, Boles E, Gorwa-Grauslund MF. Co-utilization of l-arabinose and d-xylose by laboratory and industrial Saccharomyces cerevisiae strains. Microb Cell Fact. 2006;5:18. doi: 10.1186/1475-2859-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato O, Youn JW, Stansen KC, Matsui D, Oikawa T, Wendisch VF. Quinone-dependent d-lactate dehydrogenase Dld (Cg1027) is essential for growth of Corynebacterium glutamicum on d-lactate. BMC Microbiol. 2010;10:321. doi: 10.1186/1471-2180-10-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi H, Vertes AA, Okino S, Inui M, Yukawa H. Engineering of a xylose metabolic pathway in Corynebacterium glutamicum. Appl Environ Microbiol. 2006;72:3418–3428. doi: 10.1128/AEM.72.5.3418-3428.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi H, Sasaki M, Vertes AA, Inui M, Yukawa H. Engineering of an l-arabinose metabolic pathway in Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2008;77:1053–1062. doi: 10.1007/s00253-007-1244-x. [DOI] [PubMed] [Google Scholar]
- Kiefer P, Heinzle E, Wittmann C. Influence of glucose, fructose and sucrose as carbon sources on kinetics and stoichiometry of lysine production by Corynebacterium glutamicum. J Ind Microbiol Biotechnol. 2002;28:338–343. doi: 10.1038/sj/jim/7000252. [DOI] [PubMed] [Google Scholar]
- Kind S, Wittmann C. Bio-based production of the platform chemical 1,5-diaminopentane. Appl Microbiol Biotechnol. 2011;91:1287–1296. doi: 10.1007/s00253-011-3457-2. [DOI] [PubMed] [Google Scholar]
- Kind S, Jeong WK, Schroder H, Wittmann C. Systems-wide metabolic pathway engineering in Corynebacterium glutamicum for bio-based production of diaminopentane. Metab Eng. 2010;12:341–351. doi: 10.1016/j.ymben.2010.03.005. [DOI] [PubMed] [Google Scholar]
- Kind S, Kreye S, Wittmann C. Metabolic engineering of cellular transport for overproduction of the platform chemical 1,5-diaminopentane in Corynebacterium glutamicum. Metab Eng. 2011;13:617–627. doi: 10.1016/j.ymben.2011.07.006. [DOI] [PubMed] [Google Scholar]
- Kinoshita S, Udaka S, Shimono M. Studies on the amino acid fermentation. Production of l-glutamic acid by various microorganisms. J Gen Appl Microbiol. 1957;3:193–205. [PubMed] [Google Scholar]
- Klinke HB, Thomsen AB, Ahring BK. Inhibition of ethanol-producing yeast and bacteria by degradation products produced during pre-treatment of biomass. Appl Microbiol Biotechnol. 2004;66:10–26. doi: 10.1007/s00253-004-1642-2. [DOI] [PubMed] [Google Scholar]
- Koopman F, Wierckx N, de Winde JH, Ruijssenaars HJ. Identification and characterization of the furfural and 5-(hydroxymethyl)furfural degradation pathways of Cupriavidus basilensis HMF14. Proc Natl Acad Sci USA. 2010;107:4919–4924. doi: 10.1073/pnas.0913039107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotrba P, Inui M, Yukawa H. A single V317A or V317M substitution in Enzyme II of a newly identified beta-glucoside phosphotransferase and utilization system of Corynebacterium glutamicum R extends its specificity towards cellobiose. Microbiology. 2003;149:1569–1580. doi: 10.1099/mic.0.26053-0. [DOI] [PubMed] [Google Scholar]
- Kramer R, Lambert C, Hoischen C, Ebbighausen H. Uptake of glutamate in Corynebacterium glutamicum. 1. Kinetic properties and regulation by internal pH and potassium. Eur J Biochem. 1990;194:929–935. doi: 10.1111/j.1432-1033.1990.tb19488.x. [DOI] [PubMed] [Google Scholar]
- Krause FS, Blombach B, Eikmanns BJ. Metabolic engineering of Corynebacterium glutamicum for 2-ketoisovalerate production. Appl Environ Microbiol. 2010;76:8053–8061. doi: 10.1128/AEM.01710-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krings E, Krumbach K, Bathe B, Kelle R, Wendisch VF, Sahm H, Eggeling L. Characterization of myo-inositol utilization by Corynebacterium glutamicum: the stimulon, identification of transporters, and influence on l-lysine formation. J Bacteriol. 2006;188:8054–8061. doi: 10.1128/JB.00935-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin ECC, Neidhardt FC, Curtiss R, III, Ingraham JL, Lin ECC, Low KB, Magasanik B, et al. Escherichia coliSalmonella. 2nd edn. Washington, DC, USA: ASM Press; 1996. Dissimilatory pathways for sugars, polyols and carboxylates; pp. 307–342. and Cellular and Molecular Biology. [Google Scholar]
- Lindner SN, Knebel S, Pallerla SR, Schoberth SM, Wendisch VF. Cg2091 encodes a polyphosphate/ATP-dependent glucokinase of Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2010;87:703–713. doi: 10.1007/s00253-010-2568-5. [DOI] [PubMed] [Google Scholar]
- Lindner SN, Seibold GM, Henrich A, Kramer R, Wendisch VF. Phosphotransferase system-independent glucose utilization in Corynebacterium glutamicum by inositol permeases and glucokinases. Appl Environ Microbiol. 2011;77:3571–3581. doi: 10.1128/AEM.02713-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litsanov B, Brocker M, Bott M. Glycerol as a substrate for aerobic succinate production in minimal medium with Corynebacterium glutamicum. Microb Biotechnol. 2012a doi: 10.1111/j.1751-7915.2012.00347.x. doi: 10.1111/j.1751-7915.2012.00347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litsanov B, Brocker M, Bott M. Toward homosuccinate fermentation: metabolic engineering of Corynebacterium glutamicum for anaerobic production of succinate from glucose and formate. Appl Environ Microbiol. 2012b;78:3325–3337. doi: 10.1128/AEM.07790-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litsanov B, Kabus A, Brocker M, Bott M. Efficient aerobic succinate production from glucose in minimal medium with Corynebacterium glutamicum. Microb Biotechnol. 2012c;5:116–128. doi: 10.1111/j.1751-7915.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitsuka T, Sawai H, Hatsu M, Yamada K. Metabolic engineering of Corynebacterium glutamicum for cadaverine fermentation. Biosci Biotechnol Biochem. 2007;71:2130–2135. doi: 10.1271/bbb.60699. [DOI] [PubMed] [Google Scholar]
- Moon MW, Kim HJ, Oh TK, Shin CS, Lee JS, Kim SJ, Lee JK. Analyses of enzyme II gene mutants for sugar transport and heterologous expression of fructokinase gene in Corynebacterium glutamicum ATCC 13032. FEMS Microbiol Lett. 2005;244:259–266. doi: 10.1016/j.femsle.2005.01.053. [DOI] [PubMed] [Google Scholar]
- Netzer R, Krause M, Rittmann D, Peters-Wendisch PG, Eggeling L, Wendisch VF, Sahm H. Roles of pyruvate kinase and malic enzyme in Corynebacterium glutamicum for growth on carbon sources requiring gluconeogenesis. Arch Microbiol. 2004;182:354–363. doi: 10.1007/s00203-004-0710-4. [DOI] [PubMed] [Google Scholar]
- Neuner A, Heinzle E. Mixed glucose and lactate uptake by Corynebacterium glutamicum through metabolic engineering. Biotechnol J. 2011;6:318–329. doi: 10.1002/biot.201000307. [DOI] [PubMed] [Google Scholar]
- Okino S, Noburyu R, Suda M, Jojima T, Inui M, Yukawa H. An efficient succinic acid production process in a metabolically engineered Corynebacterium glutamicum strain. Appl Microbiol Biotechnol. 2008;81:459–464. doi: 10.1007/s00253-008-1668-y. [DOI] [PubMed] [Google Scholar]
- Palmqvist E, Grage H, Meinander NQ, Hahn-Hagerdal B. Main and interaction effects of acetic acid, furfural, and p-hydroxybenzoic acid on growth and ethanol productivity of yeasts. Biotechnol Bioeng. 1999;63:46–55. doi: 10.1002/(sici)1097-0290(19990405)63:1<46::aid-bit5>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Peters-Wendisch PG, Kreutzer C, Kalinowski J, Patek M, Sahm H, Eikmanns BJ. Pyruvate carboxylase from Corynebacterium glutamicum: characterization, expression and inactivation of the pyc gene. Microbiology. 1998;144:915–927. doi: 10.1099/00221287-144-4-915. [DOI] [PubMed] [Google Scholar]
- Polen T, Kramer M, Bongaerts J, Wubbolts M, Wendisch VF. The global gene expression response of Escherichia coli to l-phenylalanine. J Biotechnol. 2005;115:221–237. doi: 10.1016/j.jbiotec.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Radmacher E, Stansen KC, Besra GS, Alderwick LJ, Maughan WN, Hollweg G, et al. Ethambutol, a cell wall inhibitor of Mycobacterium tuberculosis, elicits l-glutamate efflux of Corynebacterium glutamicum. Microbiology. 2005;151:1359–1368. doi: 10.1099/mic.0.27804-0. [DOI] [PubMed] [Google Scholar]
- Rittmann D, Lindner SN, Wendisch VF. Engineering of a glycerol utilization pathway for amino acid production by Corynebacterium glutamicum. Appl Environ Microbiol. 2008;74:6216–6222. doi: 10.1128/AEM.00963-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai S, Tsuchida Y, Okino S, Ichihashi O, Kawaguchi H, Watanabe T, et al. Effect of lignocellulose-derived inhibitors on growth of and ethanol production by growth-arrested Corynebacterium glutamicum R. Appl Environ Microbiol. 2007;73:2349–2353. doi: 10.1128/AEM.02880-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sasaki M, Jojima T, Kawaguchi H, Inui M, Yukawa H. Engineering of pentose transport in Corynebacterium glutamicum to improve simultaneous utilization of mixed sugars. Appl Microbiol Biotechnol. 2009;85:105–115. doi: 10.1007/s00253-009-2065-x. [DOI] [PubMed] [Google Scholar]
- Sasaki M, Jojima T, Inui M, Yukawa H. Xylitol production by recombinant Corynebacterium glutamicum under oxygen deprivation. Appl Microbiol Biotechnol. 2010;86:1057–1066. doi: 10.1007/s00253-009-2372-2. [DOI] [PubMed] [Google Scholar]
- Schneider J, Wendisch VF. Putrescine production by engineered Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2010;88:859–868. doi: 10.1007/s00253-010-2778-x. [DOI] [PubMed] [Google Scholar]
- Schneider J, Niermann K, Wendisch VF. Production of the amino acids l-glutamate, l-lysine, l-ornithine and l-arginine from arabinose by recombinant Corynebacterium glutamicum. J Biotechnol. 2011;154:191–198. doi: 10.1016/j.jbiotec.2010.07.009. [DOI] [PubMed] [Google Scholar]
- Schneider J, Eberhardt D, Wendisch VF. Improving putrescine production by Corynebacterium glutamicum by fine-tuning ornithine transcarbamoylase activity using a plasmid addiction system. Appl Microbiol Biotechnol. 2012;95:169–178. doi: 10.1007/s00253-012-3956-9. [DOI] [PubMed] [Google Scholar]
- Seibold G, Auchter M, Berens S, Kalinowski J, Eikmanns BJ. Utilization of soluble starch by a recombinant Corynebacterium glutamicum strain: growth and lysine production. J Biotechnol. 2006;124:381–391. doi: 10.1016/j.jbiotec.2005.12.027. [DOI] [PubMed] [Google Scholar]
- Stansen C, Uy D, Delaunay S, Eggeling L, Goergen JL, Wendisch VF. Characterization of a Corynebacterium glutamicum lactate utilization operon induced during temperature-triggered glutamate production. Appl Environ Microbiol. 2005;71:5920–5928. doi: 10.1128/AEM.71.10.5920-5928.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateno T, Fukuda H, Kondo A. Production of l-Lysine from starch by Corynebacterium glutamicum displaying alpha-amylase on its cell surface. Appl Microbiol Biotechnol. 2007;74:1213–1220. doi: 10.1007/s00253-006-0766-y. [DOI] [PubMed] [Google Scholar]
- Wendisch VF. Genome-wide expression analysis in Corynebacterium glutamicum using DNA microarrays. J Biotechnol. 2003;104:273–285. doi: 10.1016/s0168-1656(03)00147-0. [DOI] [PubMed] [Google Scholar]
- Wendisch VF. Genetic regulation of Corynebacterium glutamicum metabolism. J Microbiol Biotechnol. 2006;16:999. [Google Scholar]
- Wendisch VF, de Graaf AA, Sahm H, Eikmanns BJ. Quantitative determination of metabolic fluxes during coutilization of two carbon sources: comparative analyses with Corynebacterium glutamicum during growth on acetate and/or glucose. J Bacteriol. 2000;182:3088–3096. doi: 10.1128/jb.182.11.3088-3096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieschalka S, Blombach B, Eikmanns BJ. Engineering Corynebacterium glutamicum for the production of pyruvate. Appl Microbiol Biotechnol. 2012;94:449–459. doi: 10.1007/s00253-011-3843-9. [DOI] [PubMed] [Google Scholar]
- Youn JW, Jolkver E, Kramer R, Marin K, Wendisch VF. Identification and characterization of the dicarboxylate uptake system DccT in Corynebacterium glutamicum. J Bacteriol. 2008;190:6458–6466. doi: 10.1128/JB.00780-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn JW, Jolkver E, Kramer R, Marin K, Wendisch VF. Characterization of the dicarboxylate transporter DctA in Corynebacterium glutamicum. J Bacteriol. 2009;191:5480–5488. doi: 10.1128/JB.00640-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaldivar J, Ingram LO. Effect of organic acids on the growth and fermentation of ethanologenic Escherichia coli LY01. Biotechnol Bioeng. 1999;66:203–210. doi: 10.1002/(sici)1097-0290(1999)66:4<203::aid-bit1>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Zaldivar J, Martinez A, Ingram LO. Effect of selected aldehydes on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol Bioeng. 1999;65:24–33. doi: 10.1002/(sici)1097-0290(19991005)65:1<24::aid-bit4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Zaldivar J, Martinez A, Ingram LO. Effect of alcohol compounds found in hemicellulose hydrolysate on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol Bioeng. 2000;68:524–530. doi: 10.1002/(sici)1097-0290(20000605)68:5<524::aid-bit6>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]


