Abstract
Fluorescent reporter proteins are widely used for the non-invasive monitoring of gene expression patterns, but dynamic measurements are hampered by the extremely high stability of GFP and homologue proteins. In this study, we used SsrA-mediated peptide tagging for the construction of unstable variants of the GFP derivative eYFP (enhanced yellow fluorescent protein) and applied those for transient gene expression analysis in the industrial platform organism Corynebacterium glutamicum.
The Gram-positive soil bacterium Corynebacterium glutamicum was isolated in 1957 in Japan due to its ability to excrete large amounts of the amino acid l-glutamate (Kinoshita, 1957). Within the last decades C. glutamicum was proven to be an excellent production platform not only for amino acids, but also for a variety of other metabolites, including organic acids, vitamins and polymer precursors (Leuchtenberger, 1996; Eggeling and Bott, 2005; Burkovski, 2008). Efficient metabolic engineering, however, depends on a detailed understanding of gene expression patterns and adaptive responses of the respective organism. Reporter proteins, such as β-galactosidase (lacZ), bacterial luciferase (luxCDABE), and autofluorescent proteins (gfp, yfp, etc.) represent convenient tools for the detection and quantification of molecular and genetic events (Ghim et al., 2010). Among these, fluorescent proteins offer the advantage of broad-host applicability, no need for substrate addition, and a non-destructive measurement at the single cell level (Chalfie et al., 1994; Tsien, 1998). A major drawback of several reporter proteins is, however, their extremely long half-life (GFP > 24 h), which leads to accumulation of the protein within the cell and hampers the study of transient changes in gene expression. To address this issue, Andersen and co-workers used C-terminal SsrA peptide tagging for destabilization of GFP in Escherichia coli and Pseudomonas putida (Andersen et al., 1998). This approach was in the following successfully applied to generate unstable GFP variants in, e.g. Mycobacterium species or for destabilization of the Photorhabdus luminescens luciferase (Triccas et al., 2002; Blokpoel et al., 2003; Allen et al., 2007).
The SsrA tag is encoded by the tmRNA (tmRNA, ssrA or 10Sa RNA), which functions as both transfer and messenger RNA and acts as a rescue system of ribosomes stalled on broken or damaged mRNA (Keiler, 2008). By translation of the messenger part of tmRNA a peptide tag of 11 amino acids is added to the C-terminus of the premature protein (E. coli SsrA tag: AANDENYALAA), thereby rendering it susceptible for tail-specific proteases, such as ClpXP, ClpAP or FtsH (Gottesman et al., 1998; Herman et al., 2009). Previous studies showed that variation of the terminal three amino acids of the tag can be used to generate variants of different protein stability (Andersen et al., 1998).
The ssrA gene is highly conserved in bacterial genomes and a homologue was also annotated in the C. glutamicum genome (Kalinowski et al., 2003). In this study we used SsrA peptide tagging to construct a set of destabilized fluorescent protein variants (eYFP and GFPuv) with significantly reduced half-lives, compared with the native proteins. These proteins represent valuable tools for the monitoring of dynamic gene expression patterns in the biotechnological organism C. glutamicum.
For construction of unstable fluorescent protein variants the native E. coli (AANDENYALAA) and C. glutamicum SsrA tag (AAEKSQRDYALAA) or tags varying in their terminal three amino acids of the corynebacterial tag (in the following designated as ASV, AAV and LVA) were fused to the C-terminus of eYFP and GFPuv, which are both commonly used as reporter proteins in C. glutamicum. The fusion variants were cloned into the vector pEKEx2, under control of Ptac, and transferred into C. glutamicum ATCC 13032 by electroporation (Table 1). For fluorescence measurements cells were grown in 48-well microtiter Flowerplates containing CGXII minimal medium with 4% glucose in a BioLector microbioreactor system (m2p labs, Germany) (Kensy et al., 2009); gene expression was induced by addition of 1 mM IPTG. In case of the eYFP variants we observed significant fluorescent signals for fusion constructs with ASV tag (46% of native eYFP) and AAV tag (20% of native eYFP) (Table 2). No signal was detected for the eYFP variants fused either to the native SsrA tag (E. coli or C. glutamicum) or to the LVA modified version (data not shown). To assess the stability of the ASV and AAV variants, transcription and translation of the respective strains were inhibited by the addition of tetracycline and rifampicin (100 μg ml−1 and 250 μg ml−1, respectively) to the culture medium. Both antibiotics were added 1.5 h after induction with IPTG. The decrease in fluorescence was measured in 10 min intervals in the BioLector system (Fig. 1A and B). In the course of these measurements, a stable signal was observed for native eYFP for > 24 h whereas a rapid decrease in signal was observed for eYFP-ASV and eYFP-AAV. The half-life of both variants was determined via Western blot analysis using anti-GFP antibodies cross-reacting towards eYFP. A rapid decrease in eYFP protein level was detected for both, ASV and AAV, tagged variants and half-lives of 22 ± 4 min (ASV) and 8 ± 3 min (AAV) were calculated (Fig. 1C and Table 2). The reference protein, isocitrate dehydrogenase (Icd), exhibited a stable signal in Western blot analysis within the period of measurement (Fig. 1C). These data are in agreement with studies of SsrA-tagged variants in E. coli or Mycobacterium species. In almost all cases ASV and AAV variants resulted in a moderate destabilization of reporter proteins, whereas the native tag and the LVA variant are very unstable and hardly useful for the study of expression kinetics (Andersen et al., 1998; Triccas et al., 2002; Blokpoel et al., 2003; Allen et al., 2007).
Table 1.
Plasmids and oligonucleotides used in this study.
| Plasmid | Properties | Reference |
|---|---|---|
| pEKEx2 | KanR; expression vector with lacIq, Ptac and pUC18 multiple cloning site | Eikmanns et al. (1991) |
| pEKEx2-eyfp | KanR, pEKEx2 containing eyfp with artificial RBS, under control of Ptac | This study |
| pEKEx2-eyfp-asv | KanR, pEKEx2 containing eyfp, with artificial RBS, under control of Ptac, modified to include the gene sequence for the C-terminal C. glutamicum SsrA tag variation AAEKSQRDYAASV | This study |
| pEKEx2-eyfp-aav | Similar to pEKEx2-eyfp-ASV using the alternative tag AAEKSQRDYAAAV | This study |
| pEKEx2-eyfp-lva | Similar to pEKEx2-eyfp-ASV using the alternative tag AAEKSQRDYALVA | This study |
| pEKEx2-eyfp-laa | Similar to pEKEx2-eyfp-ASV using the native C. glutamicum tag AAEKSQRDYALAA | This study |
| pEKEx2-eyfp-Ec_laa | Similar to pEKEx2-eyfp-ASV using the native E. coli tag AANDENYALAA | This study |
| pJC1 | KanR, AmpR; C. glutamicum shuttle vector | Cremer et al. (1991) |
| pJC1-PgntK-eyfp | KanR, pJC1 containing eyfp under control of the promoter of gntK (153 bp) | This study |
| pJC1-PgntK-eyfp-asv | Similar to pJC1-PgntK-eyfp using eyfp modified to include the gene sequence for the C-terminal C. glutamicum SsrA tag AAEKSQRDYAASV | This study |
| pEKEx2-eyfp-tetR | KanR, pEKEx2 derivative containing yfp-tetR, encoding a eYFP–TetR fusion protein under the control of the Ptac | Frunzke et al. (2008a) |
| pEKEx2-gfp | KanR pEKEx2 containing gfpuv with artificial RBS, under control of Ptac | This study |
| pEKEx2-gfp-asv | KanR, pEKEx2 containing gfpuv, with artificial RBS, under control of Ptac, modified to include the gene sequence for the C-terminal C. glutamicum SsrA tag variation AAEKSQRDYAASV | This study |
| pEKEx2-gfp-aav | Similar to pEKEx2-gfpuv-ASV using the alternative tag AAEKSQRDYAAAV | This study |
| pEPR1-gfp | gfpuv, KanR, rep, per, T1 (T-trpE) (T-trpA), T2 (T-rrnB) (T-leuB); promoter probe vector | Knoppova et al. (2007) |
| Oligonucleotide | Sequence 5′ -> 3′a | Applicationb |
| eYFP-ASV-C.g.-EcoRI-rv | CGCGAATTCTTAAACTGATGCAGCGTAATCACGTTGGCTCTTTTCTGCTGCTCTAGACTTGTACAGCTCGTC (EcoRI) | Rv for eyfp-Cg-ASV |
| eYFP-AAV-C.g.-EcoRI-rv | CGCGAATTCTTAAACAGCTGCTGCGTAATCACGTTGGCTCTTTTCTGCTGCTCTAGACTTGTACAGCTCGTC (EcoRI) | Rv for eyfp-Cg-AAV |
| eYFP-LVA-C.g.-EcoRI-rv | CGCGAATTCTTAAGCTACTAAAGCGTAATCACGTTGGCTCTTTTCTGCTGCTCTAGACTTGTACAGCTCGTC (EcoRI) | Rv for eyfp-Cg-LVA |
| eYFP-LAA-C.g.-EcoRI-rv | CGCGAATTCTTAAGCTGCTAAAGCGTAATCACGTTGGCTCTTTTCTGCTGCTCTAGACTTGTACAGCTCGTC (EcoRI) | Rv for eyfp-Cg-LAA |
| eYFP-LAA-E.c.-EcoRI-rv | CGCGAATTCTTAAGCTGCTAAAGCGTAGTTTTCGTCGTTTGCTGCTCTAGACTTGTACAGCTCGTC (EcoRI) | Rv for eyfp-Ec-LAA |
| eYFP-EcoRI-rv | CGCGAATTCTTATCTAGACTTGTACAGCTCGTC (EcoRI) | Rv for eyfp |
| eYFP-RBS-BamHI-fw | CGCGGATCCAAGGAGATATGATATGGTGAGCAAGGGCGAGGAG (BamHI) | Fw for eyfp and destabilized variants |
| eYFP-ASV-C.g.-SalI-rv | CGCGTCGACTTAAACTGATGCAGCGTAATCACG (SalI) | Rv for eyfp-Cg-ASV |
| eYFP-AAV-C.g.-SalI-rv | CGCGTCGACTTAAACAGCTGCTGCGTAATCACG (SalI) | Rv for eyfp-Cg-AAV |
| eYFP-NdeI-fw | CGCCATATGGTGAGCAAGGGCGAGGAG (NdeI) | Fw for eyfp and destabilized variants |
| PgntK-BamHI-fw | CGCGGATCCACATACAGTCCCCGTGATGTGAC (BamHI) | Fw for promoter region of gntK |
| PgntK-NdeI-rv | CGC CATATGGTCTTATCCTTTCTTTGGTGGCG (NdeI) | Rv for promotor region of gntK |
| GFPuv-AAV-C.g.-EcoRI-rv | GCGCGATATCGAATTCTCATTAAACAGCTGCTGCGTAATCACGTTGGCTCTTTTCTGCTGCTTTGTAGAGCTCATCCATGCCATG (EcoRI) | Rv for gfpuv-Cg-AAV |
| GFPuv-ASV-C.g.-EcoRI-rv | GCGCGATATCGAATTCTCATTAAACTGATGCAGCGTAATCACGTTGGCTCTTTTCTGCTGCTTTGTAGAGCTCATCCATGCCATG (EcoRI) | Rv for gfpuv-Cg-ASV |
| GFPuv-Acc65I-fw | GCGCGGTACCGGTAGAAAAAATGAG (Acc65I) | Fw for gfpuv and destabilized variants |
| GFPuv-EcoRI-rv | GCGCGCGAATTCGAGAGTTATCTCGGCCAGCCAC (EcoRI) | Rv for gfpuv |
Some oligonucleotides were designed with restriction sites (underlined), ribosome binding sites (bold) and include SsrA-tag sequences (italic) as indicated.
Rv, reverse primer; Fw, forward primer for amplification.
Table 2.
Overview of SsrA tag variants.
| SsrA-tag | Amino acid sequence | Half-life (min)a | Signal intensity (%)b |
|---|---|---|---|
| E. c. LAA-tag native | AANDENYALAA | n.d. | n.d. |
| C. g. LAA-tag native | AAEKSQRDYALAA | n.d. | n.d. |
| C. g. LVA-tag variation | AAEKSQRDYALVA | n.d. | n.d. |
| C. g. ASV-tag variation | AAEKSQRDYAASV | 22 ± 4 | 46 |
| C. g. AAV-tag variation | AAEKSQRDYAAAV | 8 ± 3 | 20 |
Protein half-lives were calculated from Western blot analysis (Fig. 1C) corresponding to the decay law 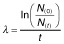
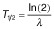 . Given values represent the average values with standard deviation of three independent experiments. For indicated constructs rapid degradation occurred and half-lives could not be determined (n.d.).
. Given values represent the average values with standard deviation of three independent experiments. For indicated constructs rapid degradation occurred and half-lives could not be determined (n.d.).
Signal intensity relative to native eYFP at the time of measurement.
Figure 1.
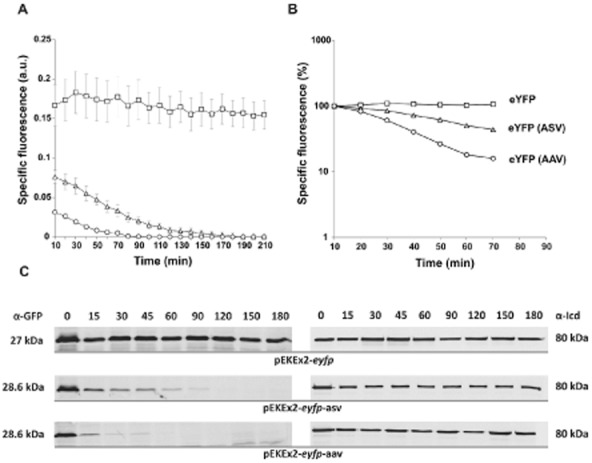
Stability of eYFP variants in C. glutamicum.(A) Fluorescence of recombinant C. glutamicum ATCC 13032 strains expressing eyfp variants: ATCC 13032/pEKEx2-eyfp (squares), ATCC 13032/pEKEx2-eyfp-asv (triangles), ATCC 13032/pEKEx2-eyfp-aav (circles). Prior induction, cells were inoculated to an OD600 of 1 in 750 μl of CGXII minimal medium containing 4% glucose and cultivated in 48-well microtiter plates in the BioLector system (m2p labs, Germany). Gene expression was induced by addition of 1 mM IPTG. To estimate the stability of the eYFP variants 250 μg ml−1 rifampicin (Rif) and 100 μg ml−1 tetracycline (Tet) were added 1.5 h after induction to stop the transcription and translation. In the BioLector system the growth (backscatter signal of 620 nm light) and eYFP fluorescence (excitation 510 nm/emission 532 nm) were monitored in 10 min intervals. The specific fluorescence was calculated as fluorescence signal per backscatter signal (given in arbitrary units, a.u.). Results represent average values with standard deviation of three independent experiments.(B) Logarithmic scale blotting of data shown in 1A. Specific fluorescence of each strain to the time of antibiotic addition was set to 100%.(C) Determination of half-lives via Western blot analysis of eYFP (27.0 kDa), eYFP-ASV (28.6 kDa) and eYFP-AAV (28.6 kDa). Cells were cultivated in 70 ml of BHI medium with 2% glucose to an OD600 of 3–4. Prior and after addition of Tet and Rif (addition of antibiotics after 1.5 h) 5 ml of cells were harvested by centrifugation and subsequently frozen in liquid nitrogen. For isolation of crude extract cells were ruptured with glass beads in TE buffer (10 mM Tris, 1 mM EDTA, pH 8) with complete protease inhibitor (Roche, Germany). Samples (25 μg) were loaded on two identical SDS gels and proteins were separated by SDS-PAGE and analysed via Western blot analysis using anti-GFP (cross-reacting to eYFP) and anti-Icd for referencing (80 kDa). The intensity of bands was analysed with the AIDA software version 4.15 (Raytest GmbH, Germany). The images are representative ones out of three independent biological replicates.
We also investigated the suitability of SsrA-tagged GFPuv variants as reporters for transient gene expression, since GFPuv has been used as an appropriate reporter for C. glutamicum promoters in previous studies (Knoppová et al., 2007; Hänßler et al., 2009). As observed for the eYFP variants, no signal was detectable for proteins fused to the native SsrA tag or the LVA version whereas for GFPuv tagged with either ASV or AAV, fluorescence could be monitored (Fig. 2). However, in contrast to eYFP, tagging of GFPuv led to an immense loss in fluorescent signal with a very low residual fluorescence of 10% (AAV) and 12% (ASV) compared with native GFPuv. Since these fluorescence levels were only slightly above background level, determination of reporter half-lives was hardly feasible (Fig. 2).
Figure 2.
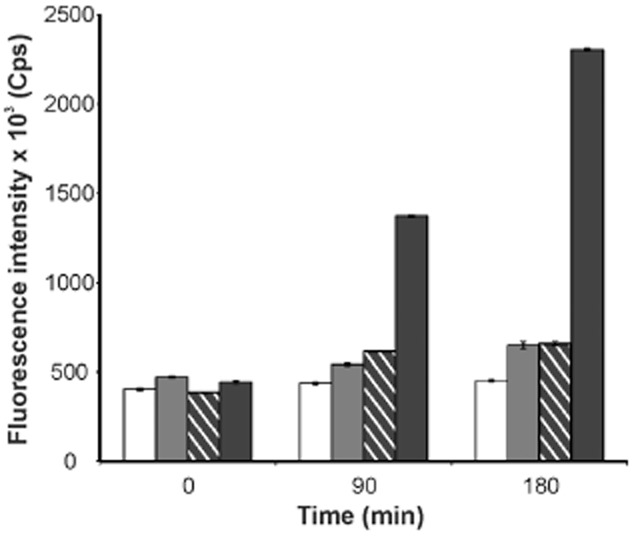
Fluorescence of recombinant C. glutamicum strains expressing gfp variants. Shown is ATCC 13032/pEKEx2-gfpuv (black), ATCC 13032/pEKEx2-gfpuv-aav (patterned), ATCC 13032/pEKEx2-gfpuv-asv (grey) and ATCC 13032/pEKEx2 (white) as control. Cells were cultivated in CGXII minimal medium containing 1 mM IPTG to an OD600 of 3–5; 2 ml of cells were harvested by centrifugation and subsequently frozen in liquid nitrogen. Fluorescence was determined using Fluorolog 3 Double Spectrometer (Spex, USA). For this purpose, cells were thawed on ice, resuspended in 10 ml of cold TE buffer (10 mM Tris, 1 mM EDTA, pH 7.5) and the OD600 was determined. GFPuv fluorescence was carried out in triplicates using an excitation wavelength of 395 nm and recording emission at 509 nm. The graph shows the mean maximum fluorescence referred to the cell dry weight (1 ml of cell suspension of an OD600 of 1 corresponds to 0.36 mg of dry weight).
The results observed for eYFP and GFPuv show that the destabilizing effect conferred by a specific degradation tag depends very much on the protein it is fused to. In fact, when fusing the same SsrA tag variants (ASV and AAV) to the far-red dsRed derivative E2-Crimson (Strack et al., 2009), only a slightly decreased protein stability was obtained, whereas a major fraction of the proteins seemed stable after blocking translation and transcription with antibiotics (data not shown). This effect might be due to the tetrameric structure of E2-Crimson masking the SsrA tag towards recognition by tail-specific proteases. A similar assumption was made by Allen and co-workers who constructed an SsrA-tagged version of the luciferase subunits LuxB and LuxA, respectively (Allen et al., 2007). In their study, tagging of LuxB did not result in significant protein degradation; modification of both subunits, LuxA and LuxB, resulted in a rapid decay of bioluminescence. Therefore, a masking of the SsrA tag within a protein complex is an aspect which has to be considered for the destabilization of multimeric proteins.
As proof of principle we applied the eYFP-ASV variant to study the dynamic expression of the gntK gene in C. glutamicum. Expression of gntK, encoding gluconate kinase, is stringently regulated by the transcriptional regulators GntR1 and GntR2 in response to carbon source availability (Frunzke et al., 2008b). When gluconate, the substrate of GntK, is present, repression of gntK by GntR1/2 is relieved and expression of gntK is strongly induced in the exponential growth phase. To assess the suitability of the unstable eYFP-ASV variant, fusions of the PgntK promoter with eyfp or eyfp-asv, respectively, were constructed and cloned into the vector pJC1. As expected, both promoter fusions gave rise to a significant fluorescent signal when the cells were cultivated in minimal medium containing gluconate as carbon source (Fig. 3A: 100 mM gluconate; Fig. 3B: 50 mM of glucose and gluconate). No signal was observed in minimal medium with 100 mM glucose (data not shown). In contrast to the reporter with native eYFP, the signal of the eYFP-ASV variant dropped back to zero when entering the stationary phase which is in agreement with enzyme activity measurements of GntK (Frunzke et al., 2008b). The signal of native eYFP, however, did not reach background level within 24 h of measurement. This illustrates the suitability of unstable reporter variants to mirror the dynamic expression pattern of a gene of interest. Notably, the kink in specific fluorescence residing at the entrance into the stationary phase is due to delayed chromophore maturation in the log phase caused by oxygen limitation (Tsien, 1998; Shaner et al., 2005; Drepper et al., 2010). This effect is most likely less distinct for the unstable variant due to a lower amount of protein requiring oxygen for chromophore maturation. The kink in fluorescence was not observed with carbon source concentrations lower than 50 mM (data not shown). Interestingly, C. glutamicum expressing native eyfp showed a slight delay in growth in comparison with cells expressing the eyfp-asv variant. This indicates that expression of unstable reporter protein variants might even diminish the burden for the cell due to a rapid protein turnover and the avoidance of protein accumulation. A drawback of unstable variants is, however, the lowered reporter output (about twofold lower for eYFP-ASV) compared with the native reporter protein, which might lead to problems when monitoring genes with a low expression level. Consequently, the choice of the fluorescent protein variant, regarding spectral properties and protein half-life, clearly depends on the strength and dynamics of the promoter to be measured. Enlarging the tool box of reporter protein variants is required for optimal experimental design and output.
Figure 3.
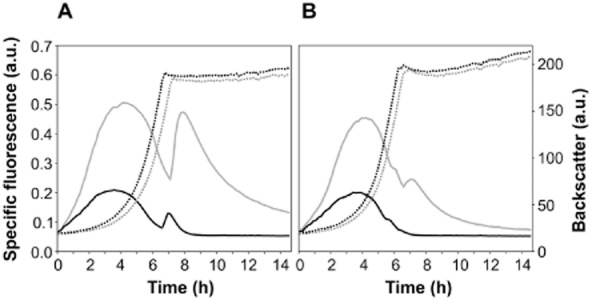
Application of eYFP (grey) and destabilized eYFP-ASV (black) for dynamic gene expression analysis of gntK in C. glutamicum. Cells were inoculated to an OD600 of 1 in 750 μl of CGXII minimal medium containing either 100 mM gluconate (A) or 50 mM glucose and gluconate (B) in 48-well microtiter plates in the BioLector cultivation system (m2p labs, Germany). The final backscatter corresponds to a maximum OD600 of 35 and 38 for growth on 100 mM gluconate and 50 mM glucose plus gluconate, respectively. For pre-cultures, cells were cultivated in CGXII with 100 mM glucose. Growth (dashed line) and fluorescence (solid line) were recorded in 15 min intervals (for details see Fig. 1).
In a recent study, introduction of an SsrA-tagged variant of the enzyme TyrA into a phenylalanine producing E. coli strain was an elegant approach to improve the accumulation of phenylalanine (Doroshenko et al., 2010). Our data reveal the applicability of corynebacterial SsrA tags and variants thereof for the efficient destabilization of eYFP and GFPuv in C. glutamicum. The use of SsrA peptide tagging is, yet, not limited to reporter proteins, but can be a valuable tool for the engineering of synthetic gene circuits (Elowitz and Leibler, 2000) or fine-tuning of protein levels in metabolic engineering of this important platform organism.
Funding Information
This work was supported by the Helmholtz Association (Helmholtz Grant VH-NG-716 to J.F.) and by the BMBF in frame of the SysMo2 programme.
Conflict of interest
None declared.
References
- Allen SA, Wilgus JR, Chewning CS, Sayler GS, Simpson ML. A destabilized bacterial luciferase for dynamic gene expression studies. Syst Synth Biol. 2007;1:3–9. doi: 10.1007/s11693-006-9001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JB, Sternberg C, Poulsen LK, Bjorn SP, Givskov M, Molin S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol. 1998;64:2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokpoel MCJ, O'Toole R, Smeulders MJ, Williams HD. Development and application of unstable GFP variants to kinetic studies of mycobacterial gene expression. J Microbiol Methods. 2003;54:203–211. doi: 10.1016/s0167-7012(03)00044-7. [DOI] [PubMed] [Google Scholar]
- Burkovski A. Corynebacteria: Genomics and Molecular Biology. Norfolk, UK: Caister Academic Press; 2008. [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Cremer J, Eggeling L, Sahm H. Control of the lysine biosynthesis in Corynebacterium glutamicum as analyzed by overexpression of the individual corresponding genes. Appl Environ Microbiol. 1991;57:1746–1752. doi: 10.1128/aem.57.6.1746-1752.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroshenko VG, Shakulov RS, Kazakova SM, Kivero AD, Yampolskaya TA, Mashko SV. Construction of an l-phenylalanine-producing tyrosine-prototrophic Escherichia coli strain using tyrA ssrA-like tagged alleles. Biotechnol Lett. 2010;32:1117–1121. doi: 10.1007/s10529-010-0265-1. [DOI] [PubMed] [Google Scholar]
- Drepper T, Huber R, Heck A, Circolone F, Hillmer AK, Büchs J, Jaeger KE. Flavin mononucleotide-based fluorescent reporter proteins outperform green fluorescent protein-like proteins as quantitative in vivo real-time reporters. Appl Environ Microbiol. 2010;76:5990–5994. doi: 10.1128/AEM.00701-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggeling L, Bott M. Handbook of Corynebacterium glutamicum. Boca Raton, FL, USA: Academic Press; 2005. [Google Scholar]
- Eikmanns BJ, Kleinertz E, Liebl W, Sahm H. A family of Corynebacterium glutamicumEscherichia coli shuttle vectors for cloning, controlled gene expression, and promoter probing. Gene. 1991;102:93–98. doi: 10.1016/0378-1119(91)90545-m. [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Leibler S. A synthetic oscillatory network of transcriptional regulators. Nature. 2000;403:335–338. doi: 10.1038/35002125. [DOI] [PubMed] [Google Scholar]
- Frunzke J, Bramkamp M, Schweitzer JE, Bott M. Population heterogeneity in Corynebacterium glutamicum ATCC 13032 caused by prophage CGP3. J Bacteriol. 2008a;190:5111–5119. doi: 10.1128/JB.00310-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frunzke J, Engels V, Hasenbein S, Gätgens C, Bott M. Co-ordinated regulation of gluconate catabolism and glucose uptake in Corynebacterium glutamicum by two functionally equivalent transcriptional regulators, GntR1 and GntR2. Mol Microbiol. 2008b;67:305–322. doi: 10.1111/j.1365-2958.2007.06020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghim CM, Lee SK, Takayama S, Mitchell RJ. The art of reporter proteins in science: past, present and future applications. BMB Rep. 2010;43:451–460. doi: 10.5483/bmbrep.2010.43.7.451. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Roche E, Zhou YN, Sauer RT. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 1998;12:1338–1347. doi: 10.1101/gad.12.9.1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänßler E, Müller T, Palumbo K, Patek M, Brocker M, Krämer R, Burkovski A. A game with many players: control of gdh transcription in Corynebacterium glutamicum. J Biotechnol. 142:114–122. doi: 10.1016/j.jbiotec.2009.04.007. [DOI] [PubMed] [Google Scholar]
- Herman C, Thevenet D, Bouloc P, Walker GC, D'Ari R. Degradation of carboxy-terminal-tagged cytoplasmic proteins by the Escherichia coli protease HflB (FtsH) Genes Dev. 2009;12:1348–1355. doi: 10.1101/gad.12.9.1348. 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski J, Bathe B, Bartels D, Bischoff N, Bott M, Burkovski A, et al. The complete Corynebacterium glutamicum ATCC 13032 genome sequence and its impact on the production of L-aspartate-derived amino acids and vitamins. J Biotechnol. 2003;104:5–25. doi: 10.1016/s0168-1656(03)00154-8. [DOI] [PubMed] [Google Scholar]
- Keiler KC. Biology of trans-translation. Annu Rev Microbiol. 2008;62:133–151. doi: 10.1146/annurev.micro.62.081307.162948. [DOI] [PubMed] [Google Scholar]
- Kensy F, Zang E, Faulhammer C, Tan RK, Buchs J. Validation of a high-throughput fermentation system based on online monitoring of biomass and fluorescence in continuously shaken microtiter plates. Microb Cell Fact. 2009;8:31. doi: 10.1186/1475-2859-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita JH. The stimulation of the phosphogluconate oxidation pathway by pyruvate in bovine corneal epithelium. J Biol Chem. 1957;228:247–253. [PubMed] [Google Scholar]
- Knoppová M, Phensaijai M, Veselý M, Zemanova M, Neŝvera J, Paték M. Plasmid vectors for testing in vivo promoter activities in Corynebacterium glutamicum and Rhodococcus erythropolis. Curr Microbiol. 2007;55:234–239. doi: 10.1007/s00284-007-0106-1. [DOI] [PubMed] [Google Scholar]
- Leuchtenberger W. Amino acids – technical production and use. In: Rehm H, Reed G, Pühler A, Stadler P, editors. Biotechnology. Weinheim: VCH; 1996. pp. 465–502. [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Strack RL, Hein B, Bhattacharyya D, Hell SW, Keenan RJ, Glick BS. A rapidly maturing far-red derivative of dsRed-Express2 for whole-cell labeling. Biochemistry. 2009;48:8279–8281. doi: 10.1021/bi900870u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triccas JA, Pinto R, Britton WJ. Destabilized green fluorescent protein for monitoring transient changes in mycobacterial gene expression. Res Microbiol. 2002;153:379–383. doi: 10.1016/s0923-2508(02)01327-x. [DOI] [PubMed] [Google Scholar]
- Tsien RY. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]


