Summary
Radionuclides in the environment are a major human and environmental health concern. Like the Chernobyl disaster of 1986, the Fukushima Daiichi nuclear disaster in 2011 is once again causing damage to the environment: a large quantity of radioactive waste is being generated and dumped into the environment, and if the general population is exposed to it, may cause serious life-threatening disorders. Bioremediation has been viewed as the ecologically responsible alternative to environmentally destructive physical remediation. Microorganisms carry endogenous genetic, biochemical and physiological properties that make them ideal agents for pollutant remediation in soil and groundwater. Attempts have been made to develop native or genetically engineered (GE) microbes for the remediation of environmental contaminants including radionuclides. Microorganism-mediated bioremediation can affect the solubility, bioavailability and mobility of radionuclides. Therefore, we aim to unveil the microbial-mediated mechanisms for biotransformation of radionuclides under various environmental conditions as developing strategies for waste management of radionuclides. A discussion follows of ‘-omics’-integrated genomics and proteomics technologies, which can be used to trace the genes and proteins of interest in a given microorganism towards a cell-free bioremediation strategy.
Introduction
In addition to the occasional disastrous accidents at nuclear facilities such as the Chernobyl disaster of 1986 and the Fukushima Daiichi nuclear disaster in 2011, the extensive use of radioactive materials at research and development, biomedical, and industrial sites has created a great accumulation of radioactive waste. Fredrickson and colleagues (2004) reported that during World War II, ∼90 million gallons of high-level radioactive waste accumulated across the USA. Most radioactive wastes are generated by nuclear power plants, which contribute ∼95% of the radioactivity generated from all sources (Ahier and Tracy, 1995; Tamponnet and Declerck, 2008). In the environment, even a small concentration of radionuclides can have an impact for a prolonged period of time due to their long half-life. As a result, the impact of radionuclide pollutants is growing with time. The commonly encountered radionuclides include cobalt-60 (60Co), plutonium-239 (239Pu), radium-226 (226Ra), radon-222 (222Rn), technetium-99 (99Tc), thorium-232 (232Th) and uranium-238 (238U). However, the typical radionuclides produced through nuclear reactors via the splitting of elemental atoms are thallium-201 (201Tl), iridium-192 (192Ir), caesium-137 (137Cs) and strontium-90 (90Sr), which take a significantly long time to decay (Kurnaz et al., 2007). Additionally, 238U decays to form 226Ra, which has a half-life of 1600 years.
Exposure to radionuclides or radiation causes acute health effects that begin with nausea, vomiting and headaches. With increased exposure a person may also experience fatigue, weakness, fever, hair loss, dizziness disorientation, diarrhoea, blood in stool, low blood pressure and ultimately death. Foetuses are particularly vulnerable to the effects of radiation at the cellular level, which can result in smaller head or brain size, poorly formed eyes, abnormal growth and mental retardation (Nussbaum, 2007; Al-Zoughool and Krewski, 2009; Bogutskaya et al., 2011). Studies have revealed that long-term exposure to radionuclides leads to an elevated risk of leukaemia, leucopenia, kidney damage and genetic damage, which can result in lethal health problems that can pass into the next generation (Mohner et al., 2006).
Currently, excavation and shipping to a distant waste disposal location is the most common means of eradicating soil contaminated with radionuclides. The costs of cleaning up these sites are estimated to be in excess of a trillion dollars in the USA and 50 billion pounds sterling in the UK (Lloyd and Renshaw, 2005). Given the high costs of physiochemical approaches, there has been an unprecedented interest in microbes and plants with radionuclides for decontamination of sediments and waters impacted by nuclear waste (Lloyd et al., 2003; Kumar et al., 2007). Figure 1 summarizes the various biotechnological approaches for bioremediation of radionuclides. Bioremediation via microorganisms can be an attractive alternative to excavating contaminated soil. Microorganisms such as Rhodanobacter sp. and Desulfuromus aferrireducens were observed to be able to interact with these contaminants (Amachi et al., 2010; Green et al., 2012). The interaction of site-specific microorganisms initiates solubility of transformed radionuclides by addition or removal of electrons, thus increasing the mobility of the contaminant and allowing it to be easily flushed from the environment (Amachi et al., 2010). This microbial-mediated biotransformation presents opportunities for bioremediation of radionuclides in the environment, either to immobilize them in place or to accelerate their removal. This article aims to interpret the procedure by which microorganisms assist in eliminating radionuclides from the environment and how they influence the toxicity and transport of radionuclides.
Figure 1.
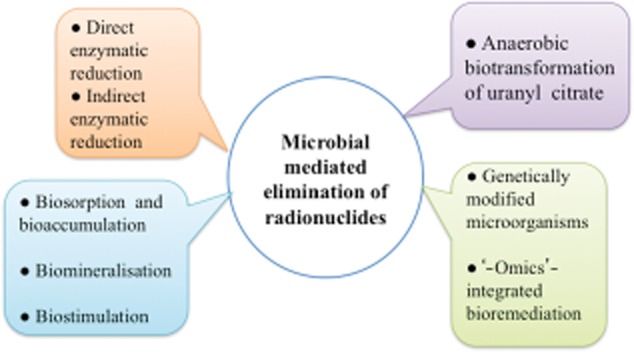
Summarization of various biotechnological approaches for bioremediation of radionuclides.
Microorganisms: an asset in elimination of radionuclides
Bioremediation of environmental niches such as soil, sediments and water contaminated with radionuclides can be achieved through biologically encoded changes in the oxidation state. Changes in speciation such as detoxification of mercury by methylation [Hg(CH3)2] can alter the solubility, transport properties and toxicity of radionuclides (Wang et al., 2012). The bioremedial strategies for radionuclides depend on the active metabolizing capabilities of microorganisms. Radionuclides can be solubilized by direct and indirect enzymatic reduction through oxidation–reduction, change in pH and Eh (activity of electrons), biosorption by mass, biodegradation of radionuclide–organic complexes or biosorption by biomass (Holker et al., 2002; Law et al., 2010; Hegazy and Emam, 2011). Microbial activity during the biotransformation of radionuclides is greatly influenced by electron donors and acceptors, nutrients, and environmental factors. Figure 2 shows the possible mechanistic linkages of metals with microorganism: key interaction for bioremediation.
Figure 2.
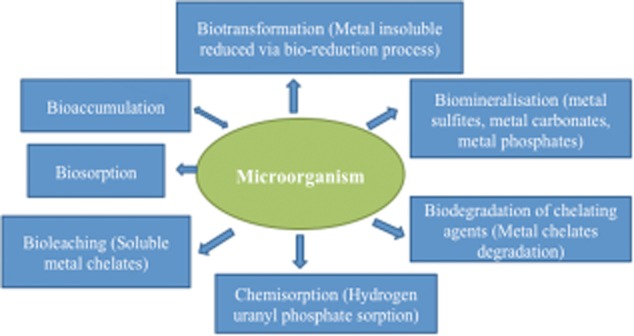
Linkage of metals with microorganism: key interaction for bioremediation.
Direct enzymatic reduction of radionuclides
The oxidized forms of radionuclides are highly soluble in aqueous medium, which makes them mobile in groundwater, whereas the reduced species are highly insoluble and often precipitate from the solution. A direct enzymatic reduction of soluble U(VI) to insoluble species is presented in Fig. 3. Wildung and colleagues (2000) reported enzymatic reduction of U(VI) on the surface of the microorganism Shewanella putrefaciens. A c-type cytochrome of mass 9.6 kDa was observed in the periplasm of S. putrefaciens required for U(VI) reduction. The studies showed that the mutant was unable to synthesize c-type cytochrome, so it did an in vitro reduction of U(VI). The purified tetrahaem cytochrome c3 protein from Desulfovibrio vulgaris was observed to reduce U(VI) in vitro using U(VI) reductase in combination with hydrogenase as its physiological electron donor (Lovley, 2003; Lovley and Phillips, 1994). In vivo studies showed that a homologue of Desulfovibrio desulfuricans (G20) confirmed the role of cytochrome c3 in hydrogen-dependent U(VI) reduction. Similarly, Lloyd and colleagues (2003) identified a homologous cytochrome (PpcA), a trihaem periplasmic cytochrome c7 of the Fe(III)-reducing bacterium Geobacter sulfurreducens that may also play a role in U(VI) reduction in vitro.
Figure 3.
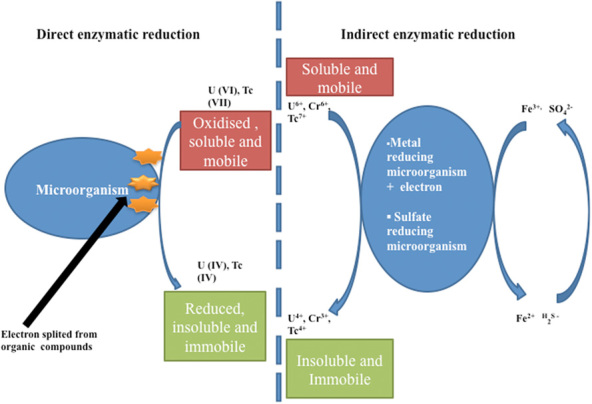
Depiction of direct enzymatic reduction and indirect mobilization of radionuclides by metal-reducing microorganisms via capturing of electrons derived by organic compounds (lactate or acetate).
99Tc, another long-lived radionuclide (half-life 2.13 × 105 years) that mostly occurs in nuclear wastes, has attracted considerable interest (Tamponnet and Declerck, 2008; Alliot et al., 2009). Tc(VII) has feeble ligand-complexing capabilities and is difficult to remove from solution using conventional chemical methods. Metal-reducing microorganisms can reduce Tc(VII) and precipitate the radionuclide into low-valency oxide TC(IV). The studies demonstrated reduction of Tc solubility (Henrot, 1989; Pignolet et al., 1989). However, the direct microbial enzymatic reduction of Tc(VII) was first observed by Lloyd and Macaskie (1996) using S. putrefaciens and Geobacter metallireducens. The reduction of Tc(VII) was quantified using a phosphorimager technique. In another study, X-ray absorption spectroscopy showed Tc(IV) as the final insoluble oxidation state after Tc(VII) was reduced by G. sulfurreducens and S. putrefaciens (Wildung et al., 2000). It is well reported that Tc(VII) reduction has produced fortuitous biochemical side reactions in the organisms studied to date.
Further, the biochemical basis of Tc(VII) reduction has been studied in Escherichia coli. Initial studies demonstrated that anaerobic cultures of E. coli reduced Tc(VII) with the reduced radionuclide precipitated within the cell (Lloyd et al., 1997). Results obtained with wild-types and mutants defective in the synthesis of regulatory or electron transfer proteins were used to construct a model for Tc(VII) reduction by E. coli. The model demonstrates that three major components of formate hydrogenlyase catalyse the transfer of electrons from dihydrogen to Tc(VII). However, according to the model, the formate dehydrogenase component is required only if formate is supplied as an electron donor for Tc(VII) reduction in place of hydrogen. The model was authenticated by a mutant strain unable to synthesize all three components of hydrogenase to reduce Tc(VII) when either hydrogen or formate was supplied as an electron donor (Lloyd et al., 1997). The identification of three components of formate hydrogenlyase and Tc(VII) reductase of E. coli opened up a new avenue to screen for organisms with naturally enhanced ability to act against Tc(VII).
Several other organisms are known to have naturally high formate hydrogenlyase activity. In addition, the uptake of hydrogenase was established to couple the oxidation of formate or hydrogen to Tc(VII) reduction (Lloyd et al., 2001). Desulfovibrio desulfuricans and related organisms were observed to utilize formate as an efficient electron donor for Tc(VII) reduction (Lloyd et al., 1999). This observation is consistent with the existence of a rudimentary formate hydrogenlyase (FHL) complex located in the periplasm of microorganisms (Peck, 1993). These studies have also been confirmed by the role of periplasmic Ni-Fe hydrogenase in Tc(VII) reduction by a relative in the N subclass of Desulfovibrio fructosovorans (De Luca et al., 2001). Subsequent studies on the development of a bioprocess to decontaminate Tc(VII)-contaminated water have focused on the use of immobilized cells of sulfate-reducing bacteria such as D. desulfuricans, which are capable of treating low concentrations of Tc(VII) against a high concentration of nitrate ions, which is frequently found in nuclear waste (Lloyd et al., 1999). Fujimoto and Morita (2006) reported a novel strain of Halomonas (Tc-202), isolated from a marine environment, that was capable of removing Tc(VII) from solid- and aqueous-phase material aerobically.
Although U and Tc remain the highest-priority radionuclide pollutants in most medium- and low-level radioactive wastes, other actinides including Th, Np, Pu and Am also occur at the contaminated sites (Lloyd and Macaskie, 2000; Tamponnet and Declerck, 2008). Iron-reducing bacteria such as Geobacter sp. and Rhodoferax ferrireducens have the metabolic potential to reduce these radionuclides enzymatically (Kim et al., 2012). These findings are significant, as most tetravalent actinides are acquiescent to bioremediation due to their high ligand-complexing abilities. In addition, these actinides could be immobilized in sediments containing active biomass (Peretrukhin et al., 1996). Although it is possible for Fe(III)-reducing bacteria to reduce and precipitate actinides in one step [e.g. soluble U(VI) to insoluble U(IV)], few studies support the direct formation of an insoluble mineral stage; rather they indicate the formation of a cation prone to bioprecipitation (Lloyd et al., 2000). This phenomenon is illustrated when considering highly soluble Np(V), which was reduced to insoluble Np(IV) by S. putrefaciens, then removed as an insoluble phosphate biomineral by a phosphate-liberating Citrobacter sp. Lloyd and colleagues (2000) studies have also suggested that the reduction of Pu(IV) to Pu(III) can be achieved by Fe(III)-reducing bacteria and Pu(III) was reported to reoxidize spontaneously (Rusin et al., 1994).
Indirect enzymatic reduction of radionuclides
The enzymatic bioreduction of radionuclides can be triggered through indirect reduction of soluble contaminants in sedimentary and subsurface environments by metal-reducing or sulfate-reducing microorganisms. One approach is to couple the oxidation of organic compounds or hydrogen to the reduction of iron Fe(III) or sulfur S(IV) in the form of sulfate. The iron Fe(III) can be bioreduced into Fe(II) and S(VI) into S(II) (hydrogen sulfide, H2S). The product can be further reduced chemically to yield separate or multi-component insoluble species (van Hullebusch et al., 2005). The reduced forms of these metals are insoluble and can precipitate as either reduced oxide or hydroxide minerals. Similarly, sulfate-reducing bacteria may be stimulated to produce hydrogen sulfide. Several microorganisms such as Microbacterium flavescens grown in the presence of other radionuclides (i.e. U, Th, Am and Pu) produced unidentified compounds such as organic acids, siderophores and extracellular metabolites capable of dissolving and mobilizing radionuclides into the soil. These compounds may also help to transport radionuclides within the cells (John et al., 2001). Figure 3 depicts the direct enzymatic reduction and indirect mobilization of radionuclides by metal-reducing microorganisms via capturing of electrons derived by organic compounds (lactate or acetate).
Pu(IV), Th(IV), U(VI) and Fe(III) have chemical and biochemical similarities because of the ubiquity of siderophore-producing microbes. The iron-sequestering agents produced by such microorganisms are crucial to increase the solubility and bioavailability of radionuclides. Premuzic and colleagues (1985) reported the production of extracellular chelating agents in Pseudomonas aeruginosa that can bioaccumulate uranium. This report prompted several chelating agents for thorium and uranium when grown with these metals. Brainard and colleagues (1992) solubilized hydrous PuO2(s) using the siderophores enterobactin, desferrioxamin, carboxylate amino polycarboxylate and catecholate ligands. The report conclusively showed that enterobactin siderophores are extremely effective in solubilizing actinide oxides of plutonium, among the tested other chelators.
Several microorganisms produced extracellular complexes in the presence of Pu and Th that increased the concentration of Pu and Th in soil-column elutes compared with controls. The increased mobility of Pu and Th in soil resulted from the formation of neutral and negatively charged Pu and Th complexes. In the presence of known microbial metabolites such as synthetic ligand [ethylenediamine tetraacetic acid (EDTA)] and citric acid, Pu(VI) and Th(IV) were reduced to Pu(IV) and Th(IV) respectively before complex formation, suggesting that the latter valence state would be the dominant one associated with the organic complexes in soil (Panak and Nitsche, 2001).
Biosorption and bioaccumulation
Biosorption is the sequestration of positively charged metal ions to the negatively charged cell membranes and polysaccharides secreted on the outer surfaces of bacteria through slime and capsule formation. Sorption of metals to intact cells is directed by a multiplicity of mechanisms and interactions that are not yet fully understood. Langley and Beveridge (1999) described the role of carboxyls in the binding of metal cations to O-side-chains of lipopolysaccharide (LPS) and concluded that metal was most likely to bind to phosphoryl groups in the core-lipid ‘A’ of LPS, and the negatively charged side-chains affected cell hydrophobicity in Gram-negative bacteria. Similarly, Khani and colleagues (2005) described the effective adsorption of radionuclide U(VI) by the brown marine alga Cystoseira indica and observed that the pre-treatment of the alga with calcium could enhance the adsorption efficiency of several radionuclides. Several microorganisms including Citrobacter freudii and Firmicutes have been reported as radionuclide biosorbents (Haferburg et al., 2007; N'Guessan et al., 2008; Xie et al., 2008).
Sorption of metals plays a key role in microbe-metal interactions. The precipitation of uranyl phosphate by Citrobacter is initiated by interaction (electrostatic) with phosphate groups in the LPS. This interaction provides nucleation sites for mineral formation and protects an outer membrane phosphatase (Macaskie et al., 2000). Panak and colleagues (2002) examined the sorption of Pu(VI) with bacterial resting cells and concluded that the interaction with bacteria caused changes in the oxidation state of Pu(V) due to endogenous respiration. Extended X-ray Absorption Fine Structure (EXAFS) analysis of Pu associated with the cells showed that Pu(VI) was primarily bound to the phosphate groups on the cell surface; the biosorption efficiency looked to be positively related to temperature and could occur within hours. This process is substantially faster than direct bioreduction. However, radionuclide-contaminated ground is normally poor in biomass concentration due to the higher toxicity of radionuclides. Therefore, biosorption alone may not be sufficient to bioremediate radionuclides unless the ground biomass content is increased. To improve the efficiency of biosorption, elegant strategies such as recombinant DNA technology and stimulated growth of microbes in the contaminated ground could enhance radionuclide remediation.
Biostimulation
Biostimulation using specific communities of microorganisms is another mechanism to enhance the bioremediation of radionuclides. An in situ remediation method was developed to reduce the bioavailability and prevent the further spread of uranium in groundwater by promoting the activity of dissimilatory sulfate- and iron-reducing organisms (Vrionis et al., 2005). In biostimulation, nitrate serves as an energetically favourable electron acceptor for metal-reducing bacteria in nitric acid co-contaminated sediments (DiChristina, 1992). A lack of microbial reduction in U(VI) has been reported if the sediment is co-contaminated with nitrate (Finneran et al., 2002). Sakadevan and colleagues (1999) reported that nitrate reduction was inhibited due to the presence of several heavy metals. Therefore, an attempt was made to resolve this issue by the ex situ treatment and removal of heavy metals and nitrate prior to in situ biostimulation to reduce the U(VI) (Wu et al., 2006).
At sites co-contaminated with metallic ions beyond toxic levels, the microbial resistance of endogenous microbial populations is critical for in situ biostimulation. A number of microbes has been shown to carry out reductive precipitation of radionuclides (e.g. Desulfovibrio sp., Geobacter sp. and Shewanella sp.); however, the resistance of these organisms to non-reducible heavy metals could possibly limit their in situ activity. Martinez and colleagues (2006) reported the presence of heavy-metal-resistance genes within the endogenous microbial community at the Oak Ridge Field Research Center (FRC) site. In the presence of heavy metals, the potential for biostimulation was improved by using ethanol for bioreduction of nitrate, followed by the successful reduction and in situ immobilization of U(VI) (Istok et al., 2004; Nyman et al., 2006). Therefore, the addition of a suitable carbon source was recommended to stimulate radionuclide bioreduction at co-contaminated sites.
Another novel method was developed to bioremediate areas contaminated with high concentrations of uranium (> 1000 μm) at low pH and high nitric acid concentrations (Wu et al., 2006). The method was based on raising pH with the addition of carbon (ethanol) to stimulate the growth of denitrifying and radionuclide-reducing bacteria, using a hydraulic recirculation system composed of an outer and inner loop. An ex situ system was also developed to prevent clogging of the hydraulic recirculation system with nitrogen gas, aluminium, calcium precipitation and biomass due to denitrification. After 1 year at optimized conditions, the concentrations of U(VI) and nitrate had been reduced to 5 μm and 0.5 μm respectively. After 2 years of preconditioning, the level of U(VI) was reduced to 0.126 μm – below the EPA's recommended level. These studies suggested that alternation of geochemical and hydrological parameters can effectively biostimulate radionuclide-reducing bacteria in situ. Remediation of high U(VI) concentrations with ethanol-biostimulated biofilm is an alternative approach that can effectively reduce 87% of U(VI) (Marsili et al., 2007).
Biomineralization of radionuclides
Microorganisms can interact with metal ions and immobilize them to transformation. Some microorganisms generate biofilms to bind significant quantities of metallic ions, which can serve as a platform for the precipitation of insoluble minerals. Microorganism Citrobacter sp. was reported to be able to produce deposits of metal phosphate enzymatically. Polycrystalline NaUO2Po4 accumulates in and around the cell wall of Citrobacter by sorption to LPS and the activity of an outer membrane acid-phosphatase (Keasling et al., 2000). The mineral formation is driven by two gradients in the outer membranes: an incoming UO4 and an outgoing PO4, resulting in total removal of U from the solution and binding of 1 mg NaUO2Po4 per mg of the cell. Keasling and colleagues (2000) prepared a recombinant version of this mechanism by cloning the gene encoding polyphosphate kinase in P. aeruginosa, resulting in the precipitation of a complex containing both phosphorous and uranium on the cell surface.
Biomineralization via microbial-generated ligands
Chelating agents are present in wastes because they are widely used for the decontamination of nuclear reactors and equipment, cleanup operations, and the separation of radionuclides. Several organic agents, such as citric acid, hydroxyl-acetic acid, oxalic acid, tartaric acid, EDTA, diethylenetriamine pentaacetic acid (DTPA), nitrilotriacetic acid (NTA) and N-hydroxyethylenediamine triacetic acid (HEDTA) have been used to complex with radionuclides. These metal chelates undergo aerobic or anaerobic biodegradation and cause the precipitation of released ions as water-insoluble hydroxides or oxides, thus retarding their migration into groundwater. Citrate has been found useful as a chelating agent in decontamination because it forms highly soluble metal–citrate complexes that can be degraded by microorganisms, resulting in subsequent re-precipitation of metals. Stable complexes such as bidentate, tridentate and polynuclear complexes can be formed with citrate and radionuclides. Francis (1998) reported that bioidentate uranium complexed with citric acid was readily biodegraded, whereas tridentate was recalcitrant.
Anaerobic and aerobic biotransformation of uranyl–citrate
Sulfate-reducing D. desulfuricans and facultative iron-reducing Schwanella alga reduced U(VI) complexes with citrate to U(IV) anaerobically, but little uranium was precipitated (Ganesh et al., 1997). Similarly, Clostridium sp. (ATCC 53464) metabolized glucose but not citrate and reduced U(VI)–citrate into U(IV)–citrate only in the presence of glucose. Another Clostridium sphenoides (ATCC 19403) metabolized bidentate Fe(III)–citrate complex. This bacterium reduced Fe(III) to Fe(II) and concomitantly metabolized citric acid. In contrast, U(VI)–citrate was reduced to U(IV)–citrate by the bacterium in the presence of electron donor glucose or uncomplex citric acid. Therefore, these results indicate that the complex of uranium with citric acid is readily available for organisms as an electron acceptor, despite their incapability to metabolize organic ligands of radionuclides.
Some organisms metabolize citric acid aerobically using the enzymes aconitase and citrate lyase. The enzyme aconitase isomerizes citrate into isocitrate in the TCA cycle. However, citrate lyase drives the anaerobic metabolism of citric acid. Francis (1998) tested the ability of Acinetobacter, Citrobacter and Pseudomonas from low-level radioactive waste sites to metabolize citric acid into a uranyl–citrate complex. These cultures failed to metabolize a stable binuclear complex of uranyl–citrate, possibly because the uranyl–citrate complex did not transport inside the cells, as revealed by 14C labelling analysis. Furthermore, a cell-free extract of the tested organism showed that the binuclear U–citrate complex was completely degraded. Francis and colleagues (2002) tested the speciation of uranium and citric acid as a function of pH and concluded that the amount of U–citrate complex decreased rapidly above pH 6.0, whereas free citric acid was not biodegraded and transported.
Similarly, citric acid can form ternary mixed-metal complexes with a number of metals, and the complex formed affects the biodegradation and mobility of metal–citrate complexes (Dodge et al., 2002). For example, the biotransformation of Fe–U–citrate complex was recalcitrant. When onefold excess citric acid was added to the 1:1:2 Fe–U–citrate complex, the excess citric acid was completely degraded. However, with twofold excess citric acid, 1:1:1 Fe–U-citric acid remained in solution after the excess citric acid was biodegraded. Therefore, this study suggested that Fe–U–mixed-metal citric acid complexes resist biodegradation and may persist in the environment, which is a limitation of current technology and a challenge to the microbial-mediated bioremediation of radionuclides (Dodge et al., 2002).
Genetically modified microorganisms
Genetic engineering (GE) and recombinant DNA technology have been employed to generate character-specific microorganisms for efficient removal of metal by sorption. Different protein constructs have been generated in which the bacterial cell surface is equipped with metal-binding polypeptides by fusion-binding domains to outer-membrane-anchored proteins that include metallothioneins (Valls et al., 2000a), randomly generated polypeptides (Schembri et al., 1999), polyhistidines and synthetic phytochelatines (Schembri et al., 1999; Bae et al., 2000). These protein constructs proved an increase in metal binding. The metallothioneins were also tested in microcosm field study (Valls et al., 2000b). Another approach attempted to enhance the metal accumulation by combining the specific metal transporter with MTs in the cytoplasm (Wolfram and Bauerfeind, 2009). A recombinant strain of E. coli was generated with five times the sorption ability for U(VI) radionuclides by altering the transporter genes nixA (Helicobacter pylori) and merTP (Serratia marcescens) respectively (Beckwith et al., 2001). Therefore, the expression of both metal transporter proteins and metal-binding peptides may enhance a strain's ability to accumulate metal ions.
The microorganism Deinococcus radiodurans has been studied to detoxify Cr(VI), U(VI) and Tc(VII) from soil (Fredrickson et al., 2000). A genetically engineered D. radiodurans strain was created by cloning the E. coli gene (merA) that provides the ability to utilize carbon and energy from catabolism of toluene and mercury (radioactive contaminants) (Brim et al., 2000). Progress has been made in constructing strains of D. radiodurans for radionuclide remediation; however, an in situ bioremediation strategy has yet to be discovered. The microorganisms Deinococcus murrayi and Deinococcus geothermalis were characterized as growing at a higher temperature range (55°C), and showing remarkable resistance against chronic irradiation (50 Gy h−1) (Brim et al., 2003).
The microbial family Geobacteriaceae has shown potential for radioactive metal reduction (Lloyd et al., 2003). The gene dcuB from G. sulfurreducens, which encodes a fumarate transporter, was engineered in G. metallireducens to grow with fumarate as a terminal electron acceptor, revealing the ruling approach of GE with expanded respiratory capabilities (Butler et al., 2006). Undoubtedly, GE microbes show promise, but their implementation for in situ bioremediation will require additional steps to develop safe routes of environmental cleanup.
‘-Omics’-implemented radionuclide bioremediation
The genome of an organism comprises its entire set of hereditary information that is converted to mRNA (the transcriptome) for protein translation. The proteome of an organism is the entire set of proteins, including enzymes, that are expressed in the organism under specific environmental conditions. In order to identify genes, proteins and enzymes involved in the bioremediation of radionuclides, it is important to study the structural and functional interactions between proteins and other metabolites. Potential genes and proteins involved in the metabolism of radionuclides can be identified and studied via advanced genomics and proteomics techniques (Nagaraj and Singh, 2010; Singh et al., 2011). Recent advances in next-generation sequencing, genomics and proteomics allow the expression of required proteins and enzymes of interest into radionuclide-resistant organisms for bioremediation. Further, genome-wide transcriptome analysis can provide us with a better understanding of the metabolic pathways and the physiology of the microorganisms.
The genome sequences of many microorganisms are now available and can be used for genome organization, including comparisons through microarrays (Ishii et al., 2007). Among several genome-wide studies of genes and proteins involved in radionuclide reduction pathways, Methe and colleagues (2003) reported that G. sulfurreducens had more than 100 c-type cytochrome gene coding regions in its genome and many of its translated proteins were involved with radionuclide reduction pathways. DNA-microarray-mediated analysis revealed 121 genes found to be upregulated in Shewanella oneidensis during U(VI) reduction, compared with Cr(VI) reduction, where only 83 genes were overexpressed (Bencheikh-Latmani et al., 2005). In a comparative genomics study, the organism Thermococcus gammatolerans was observed to be radioresistant among Archaea, expressing thioredoxin reductase (tgo180), a glutaredoxin-like protein (tg1302) and two peroxiredoxins (tg1253 and tg1220), which allowed the organism to cope with the stress of radionuclides (Zivanovic et al., 2009). It was found that the expression of the NiCoT gene in Rhodopseudomonas palustris CGA009 and Novosphingobium aromaticivorans F-199 was highly increased when the organisms were grown in the presence of radioactive cobalt (Raghu et al., 2008).
In addition to genomics technology, proteomics technology has been proven effective in studying proteins involved in radionuclide bioremediation. Tian and colleagues (2010) found a total of 552 differentially regulated proteins, including a cytochrome bd ubiquinol oxidase, on the membrane of D. geothermalis that were involved in the radioresistance of the organism. Many of these proteins comprised function groups including nutrient transport and metabolism, energy production and conversion, and cell wall/membrane biogenesis (Tian et al., 2010). In another study, cascades of enzymatic proteins in the outer membrane of S. oneidensis were found to effectively reduce the levels of radionuclides such as uranium and chromium (Marshall et al., 2006). In a well-known radioresistant bacterium, D. radiodurans, 2DE and MALDI-TOF MS revealed that 31 radiation-responsive proteins were significantly upregulated, including RecA and PprA, which are well known for DNA replication and repair (Lu et al., 2009). Other proteins included those involved in the stress response, energy metabolism, transcriptional regulation, protein turnover and chaperoning (Lu et al., 2009).
Understanding specific genes and their protein products is essential to understand the pathways involved in the bioremediation of radionuclides. Combining ‘-omics’-based approaches may assist in the identification of specific microbes effective for in situ bioremediation of radionuclides. Table 1 summarizes the major transcripts and proteins investigated in various radionuclides remediation studies.
Table 1.
Major transcripts and their sources for radionuclides remediation
| Radionuclides | Organisms | Gene/protein | Function | Reference |
|---|---|---|---|---|
| Uranium | Deinococcus radiodurans | DrPhoN | Surface-associated precipitation of uranium (5.7 g uranium g−1 biomass) | Misra et al. (2012) |
| Cobalt | Rhodopseudomonas palustris CGA0009 | NiCoT | 85% removal of Cobalt was achieved in a two-cycle treatment with recombinant E. coli | Raghu et al. (2008) |
| Cobalt | Novosphingobium aromaticivorans F-199 | NiCoT | Raghu et al. (2008) | |
| Uranium | Sphingomonas sp. BSAR-1 | phoK | Recombinant E. coli precipitated > 90% of 0.5–5 mM of uranyl carbonate in less than 2 h | Nilgiriwala et al. (2008) |
| Chromate, uranyl | Escherichia coli | ChrR6 | Expressed chromate reductase activity in addition to convert soluble U(VI) to insoluble with U(IV) with Vmax of 8,812 ± 611 | Barak et al. (2006) |
| Uranium | Salmonella enterica serovar Typhi | phoN | Recombinant strain precipitated over 90% of the uranium from a 0.8 mM uranyl nitrate solution in 6 h | Appukuttan et al. (2006) |
| Mercury | Escherichia coli BL308 | merA | Biotransformation of Hg (II) to the less toxic volatile Hg (I) | Brim et al. (2003) |
| Cadminum, zinc, cobalt | Ralstonia eutrophus CH34 | czc | Diels et al. (1995) | |
| Fumarate, nitrate, dimethyl sulfoxide, trimethylamine N-oxide (TMAO), nitrite and insoluble iron and manganese oxides | Shewanella oneidensis | fccA | Periplasmic flavocytochrome c fumarate reductase | Saffarini et al. (2003) |
| Uranium | Desulfovibrio vulgaris | ctyc3 | Reduce uranium(VI) to uranium(IV) with hydrogen as the electron donor | Payne et al. (2002) |
| Fe(III) | Geobacter sulfurreducens | PpcA | Restoration of Fe(III) reduction with acetate | Lloyd et al. (2003) |
Challenges, limitations and 5-year view
Despite the progress that has been made in the field of radionuclide bioremediation using microorganisms, many challenges lie ahead. A basic concern is to optimize the conditions and procedures for sustained and effective bioremediation in the presence of competing anions, toxic metals, organic compounds and chelating agents. Another aspect that is of interest is the reoxidation and remobilization of reduced radionuclides by microbial metabolism and abiotic mechanisms. Careful consideration needs to be given to in situ biostimulation or bioaugmentation where various additives (microbes or chemical ingredients) are employed to enhance microbial activity; such additives might be disruptive to natural microbiota. It is still an issue to enhance the metabolic activity by maintaining the required growth conditions, such as pH, temperature, and levels of contaminants and nutrients, including diverse chemical parameters of selective microbial populations for in situ bioremediation. In addition, it is necessary to recognize the heterogeneous nature of contaminants on sites that can lead to an uneven flow of the liquid or gas containing the microbes. Due to the slow speed of achieving acceptable levels of decontamination at diverse sites, it is difficult to predict the performance of bench-scale bioremediation in field operations.
Genetically modified microorganisms have limitations in terms of liability and ethical issues from regulatory agencies such as the United States Environmental Protection Agency (USEPA). The release of GE organisms is a tedious task that requires site dependence compatibility and comparability with other organisms on site. After remediation, removal of GE microorganisms is also not an easy task. The direct immobilization of enzymes on support material could be useful for bioremediation, but it requires abundant optimization of site-specific substrates.
Recent developments in ‘-omics’-based technologies, i.e. genomics, transcriptomics and proteomics, are set to revolutionize many aspects of the biological sciences. A whole-genome transcription profile of E. coli has already uncovered potential strategies for metal detoxification (Brocklehurst and Morby, 2000). Because of the current availability of complete genome sequences of many radioresistant microorganisms, this approach will prove useful for determining the precise mechanisms of biologically relevant radionuclide–microbe interactions. However, these discoveries in environmental science are still in their infancy stage and have yet to prove their usefulness in environmental cleanup. In the future, coordinated multidisciplinary efforts need to be made to understand the overall process by which microorganisms degrade radionuclides as well as to implement the conditions that most effectively degrade the radionuclides. Focused research exploring the necessary genes and proteins of the microbial metabolic pathway towards cell-free bioremediation will pave the way to refine these bioremediation technologies before successful field practice.
Conclusion
Microbial transformations of radionuclides, heavy metals and minerals are a vital part of natural biosphere processes and can have beneficial consequences for the human community. As we know, the interactions between microorganisms and radionuclides are far from simple, and it is not easy to understand the wide range of environments these organisms inhabit. Study of the molecular mechanisms behind the microbial transformation of radionuclides using ‘-omics’-based approaches and exploiting them in applications such as bioremediation would assist in tracking the responsible microbial metabolic products towards cell-free bioremediation and further assist in efficient removal of radionuclides from the environment.
Conflict of interest
None declared.
Funding Information
No funding information provided
References
- Ahier BA, Tracy BL. Radionuclides in the Great Lakes basin. Environ Health Perspect. 1995;103(Suppl. 9):89–101. doi: 10.1289/ehp.95103s989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alliot I, Alliot C, Vitorge P, Fattahi M. Speciation of technetium(IV) in bicarbonate media. Environ Sci Technol. 2009;43:9174–9182. doi: 10.1021/es9021443. [DOI] [PubMed] [Google Scholar]
- Al-Zoughool M, Krewski D. Health effects of radon: a review of the literature. Int J Radiat Biol. 2009;85:57–69. doi: 10.1080/09553000802635054. [DOI] [PubMed] [Google Scholar]
- Amachi S, Minami K, Miyasaka I, Fukunaga S. Ability of anaerobic microorganisms to associate with iodine: 125I tracer experiments using laboratory strains and enriched microbial communities from subsurface formation water. Chemosphere. 2010;79:349–354. doi: 10.1016/j.chemosphere.2010.02.028. [DOI] [PubMed] [Google Scholar]
- Appukuttan D, Rao AS, Apte SK. Engineering of Deinococcus radiodurans R1 for bioprecipitation of uranium from dilute nuclear waste. Appl Environ Microbiol. 2006;72:7873–7878. doi: 10.1128/AEM.01362-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae W, Chen W, Mulchandani A, Mehra RK. Enhanced bioaccumulation of heavy metals by bacterial cells displaying synthetic phytochelatins. Biotechnol Bioeng. 2000;70:518–524. doi: 10.1002/1097-0290(20001205)70:5<518::aid-bit6>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Barak Y, Ackerley DF, Dodge CJ, Banwari L, Alex C, Francis AJ, Matin A. Analysis of novel soluble chromate and uranyl reductases and generation of an improved enzyme by directed evolution. Appl Environ Microbiol. 2006;72:7074–7082. doi: 10.1128/AEM.01334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckwith CS, McGee DJ, Mobley HL, Riley LK. Cloning, expression, and catalytic activity of Helicobacter hepaticus urease. Infect Immun. 2001;69:5914–5920. doi: 10.1128/IAI.69.9.5914-5920.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bencheikh-Latmani R, Williams SM, Haucke L, Criddle CS, Wu L, Zhou J, Tebo BM. Global transcriptional profiling of Shewanella oneidensis MR-1 during Cr(VI) and U(VI) reduction. Appl Environ Microbiol. 2005;71:7453–7460. doi: 10.1128/AEM.71.11.7453-7460.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogutskaya NG, Zuykov MA, Naseka AM, Anderson EB. Normal axial skeleton structure in common roach Rutilus rutilus (Actinopterygii: Cyprinidae) and malformations due to radiation contamination in the area of the Mayak (Chelyabinsk Province, Russia) nuclear plant. J Fish Biol. 2011;79:991–1016. doi: 10.1111/j.1095-8649.2011.03078.x. [DOI] [PubMed] [Google Scholar]
- Brainard JR, Strietelmeier BA, Smith PH, Langston-Unkefer PJ. Actinide binding and solubilization by microbial siderophores. Radiochim Acta. 1992;58–59:357–363. [Google Scholar]
- Brim H, McFarlan SC, Fredrickson JK, Minton KW, Zhai M, Wackett LP, Daly MJ. Engineering Deinococcus radiodurans for metal remediation in radioactive mixed waste environments. Nat Biotechnol. 2000;18:85–90. doi: 10.1038/71986. [DOI] [PubMed] [Google Scholar]
- Brim H, Venkateswaran A, Kostandarithes HM, Fredrickson JK, Daly MJ. Engineering Deinococcus geothermalis for bioremediation of high-temperature radioactive waste environments. Appl Environ Microbiol. 2003;69:4575–4582. doi: 10.1128/AEM.69.8.4575-4582.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocklehurst KR, Morby AP. Metal-ion tolerance in Escherichia coli: analysis of transcriptional profiles by gene-array technology. Microbiology. 2000;146(Part 9):2277–2282. doi: 10.1099/00221287-146-9-2277. [DOI] [PubMed] [Google Scholar]
- Butler JE, Glaven RH, Esteve-Nunez A, Nunez C, Shelobolina ES, Bond DR, Lovley DR. Genetic characterization of a single bifunctional enzyme for fumarate reduction and succinate oxidation in Geobacter sulfurreducens and engineering of fumarate reduction in Geobacter metallireducens. J Bacteriol. 2006;188:450–455. doi: 10.1128/JB.188.2.450-455.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca G, de Philip P, Dermoun Z, Rousset M, Vermeglio A. Reduction of technetium(VII) by Desulfovibrio fructosovorans is mediated by the nickel-iron hydrogenase. Appl Environ Microbiol. 2001;67:4583–4587. doi: 10.1128/AEM.67.10.4583-4587.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiChristina TJ. Effects of nitrate and nitrite on dissimilatory iron reduction by Shewanella putrefaciens 200. J Bacteriol. 1992;174:1891–1896. doi: 10.1128/jb.174.6.1891-1896.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diels L, Dong Q, van der Lelie D, Baeyens W, Mergeay M. The czc operon of Alcaligenes eutrophus CH34: from resistance mechanism to the removal of heavy metals. J Ind Microbiol. 1995;14:142–153. doi: 10.1007/BF01569896. [DOI] [PubMed] [Google Scholar]
- Dodge CJ, Francis AJ, Gillow JB, Halada GP, Eng C, Clayton CR. Association of uranium with iron oxides typically formed on corroding steel surfaces. Environ Sci Technol. 2002;36:3504–3511. doi: 10.1021/es011450+. [DOI] [PubMed] [Google Scholar]
- Finneran KT, Housewright ME, Lovley DR. Multiple influences of nitrate on uranium solubility during bioremediation of uranium-contaminated subsurface sediments. Environ Microbiol. 2002;4:510–516. doi: 10.1046/j.1462-2920.2002.00317.x. [DOI] [PubMed] [Google Scholar]
- Francis AJ. Biotransformation of uranium and other actinides in radioactive wastes. J Alloys Comp. 1998;271–273:78–84. [Google Scholar]
- Francis AJ, Joshi-Tope GA, Dodge CJ, Gillow JB. Biotransformation of uranium and transition metal citrate complexes by Clostridia. J Nucl Sci Technol. 2002;3:935–938. [Google Scholar]
- Fredrickson JK, Kostandarithes HM, Li SW, Plymale AE, Daly MJ. Reduction of Fe(III), Cr(VI), U(VI), and Tc(VII) by Deinococcus radiodurans R1. Appl Environ Microbiol. 2000;66:2006–2011. doi: 10.1128/aem.66.5.2006-2011.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrickson JK, Zachara JM, Balkwill DL, Kennedy D, Li SM, Kostandarithes HM, et al. Geomicrobiology of high-level nuclear waste-contaminated vadose sediments at the Hanford site, Washington state. Appl Environ Microbiol. 2004;70:4230–4241. doi: 10.1128/AEM.70.7.4230-4241.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto K, Morita T. Aerobic removal of technetium by a marine Halomonas strain. Appl Environ Microbiol. 2006;72:7922–7924. doi: 10.1128/AEM.00819-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesh R, Robinson KG, Reed GD, Sayler GS. Reduction of hexavalent uranium from organic complexes by sulfate- and iron-reducing bacteria. Appl Environ Microbiol. 1997;63:4385–4391. doi: 10.1128/aem.63.11.4385-4391.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green SJ, Prakash O, Jasrotia P, Overholt WA, Cardenas E, Hubbard D, et al. Denitrifying bacteria from the genus Rhodanobacter dominate bacterial communities in the highly contaminated subsurface of a nuclear legacy waste site. Appl Environ Microbiol. 2012;78:1039–1047. doi: 10.1128/AEM.06435-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haferburg G, Merten D, Buchel G, Kothe E. Biosorption of metal and salt tolerant microbial isolates from a former uranium mining area. Their impact on changes in rare earth element patterns in acid mine drainage. J Basic Microbiol. 2007;47:474–484. doi: 10.1002/jobm.200700256. [DOI] [PubMed] [Google Scholar]
- Hegazy AK, Emam MH. Accumulation and soil-to-plant transfer of radionuclides in the Nile Delta coastal black sand habitats. Int J Phytoremediation. 2011;13:140–155. doi: 10.1080/15226511003753961. [DOI] [PubMed] [Google Scholar]
- Henrot J. Bioaccumulation and chemical modification of Tc by soil bacteria. Health Phys. 1989;57:239–245. doi: 10.1097/00004032-198908000-00001. [DOI] [PubMed] [Google Scholar]
- Holker U, Schmiers H, Grosse S, Winkelhofer M, Polsakiewicz M, Ludwig S, et al. Solubilization of low-rank coal by Trichoderma atroviride: evidence for the involvement of hydrolytic and oxidative enzymes by using 14C-labelled lignite. J Ind Microbiol Biotechnol. 2002;28:207–212. doi: 10.1038/sj/jim/7000232. [DOI] [PubMed] [Google Scholar]
- van Hullebusch ED, Peerbolte A, Zandvoort MH, Lens PN. Sorption of cobalt and nickel on anaerobic granular sludges: isotherms and sequential extraction. Chemosphere. 2005;58:493–505. doi: 10.1016/j.chemosphere.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Ishii N, Nakahigashi K, Baba T, Robert M, Soga T, Kanai A, et al. Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science. 2007;316:593–597. doi: 10.1126/science.1132067. [DOI] [PubMed] [Google Scholar]
- Istok JD, Senko JM, Krumholz LR, Watson D, Bogle MA, Peacock A, et al. In situ bioreduction of technetium and uranium in a nitrate-contaminated aquifer. Environ Sci Technol. 2004;38:468–475. doi: 10.1021/es034639p. [DOI] [PubMed] [Google Scholar]
- John SG, Ruggiero CE, Hersman LE, Tung CS, Neu MP. Siderophore mediated plutonium accumulation by Microbacterium flavescens (JG-9) Environ Sci Technol. 2001;35:2942–2948. doi: 10.1021/es010590g. [DOI] [PubMed] [Google Scholar]
- Keasling JD, Van Dien SJ, Trelstad P, Renninger N, McMahon K. Application of polyphosphate metabolism to environmental and biotechnological problems. Biochemistry (Mosc) 2000;65:324–331. [PubMed] [Google Scholar]
- Khani M, Keshtkar A, Meysami B, Zarea M, Jalali R. Biosorption of uranium from aqueous solutions by nonliving biomass of marine algae Cystoseiraical heterogeneity in an in situ uranium bioremediation field site. Appl Environ Microbiol. 2005;71:6308–6318. doi: 10.1128/AEM.71.10.6308-6318.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Koh DC, Park SJ, Cha IT, Park JW, Na JH, et al. Molecular analysis of spatial variation of iron-reducing bacteria in riverine alluvial aquifers of the Mankyeong River. J Microbiol. 2012;50:207–217. doi: 10.1007/s12275-012-1342-z. [DOI] [PubMed] [Google Scholar]
- Kumar R, Singh S, Singh OV. Bioremediation of radionuclides: emerging technologies. OMICS. 2007;11:295–304. doi: 10.1089/omi.2007.0013. [DOI] [PubMed] [Google Scholar]
- Kurnaz A, Kucukomeroglu B, Keser R, Okumusoglu NT, Korkmaz F, Karahan G, Cevik U. Determination of radioactivity levels and hazards of soil and sediment samples in Firtina Valley (Rize, Turkey) Appl Radiat Isot. 2007;65:1281–1289. doi: 10.1016/j.apradiso.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Langley S, Beveridge TJ. Effect of O-side-chain-lipopolysaccharide chemistry on metal binding. Appl Environ Microbiol. 1999;65:489–498. doi: 10.1128/aem.65.2.489-498.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law GT, Geissler A, Lloyd JR, Livens FR, Boothman C, Begg JD, et al. Geomicrobiological redox cycling of the transuranic element neptunium. Environ Sci Technol. 2010;44:8924–8929. doi: 10.1021/es101911v. [DOI] [PubMed] [Google Scholar]
- Lloyd JR, Macaskie LE. A novel PhosphorImager-based technique for monitoring the microbial reduction of technetium. Appl Environ Microbiol. 1996;62:578–582. doi: 10.1128/aem.62.2.578-582.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd JR, Macaskie LE. Bioremediation of radionuclide-containing waste waters. In: Lovley DR, editor. Environmental Microbe–Metal Interactions. Washington, DC, USA: ASM Press; 2000. pp. 277–327. [Google Scholar]
- Lloyd JR, Renshaw JC. Microbial transformations of radionuclides: fundamental mechanisms and biogeochemical implications. Met Ions Biol Syst. 2005;44:205–240. [PubMed] [Google Scholar]
- Lloyd JR, Cole JA, Macaskie LE. Reduction and removal of heptavalent technetium from solution by Escherichia coli. J Bacteriol. 1997;179:2014–2021. doi: 10.1128/jb.179.6.2014-2021.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd JR, Ridley J, Khizniak T, Lyalikova NN, Macaskie LE. Reduction of technetium by Desulfovibrio desulfuricans: biocatalyst characterization and use in a flowthrough bioreactor. Appl Environ Microbiol. 1999;65:2691–2696. doi: 10.1128/aem.65.6.2691-2696.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd JR, Sole VA, Van Praagh CV, Lovley DR. Direct and Fe(II)-mediated reduction of technetium by Fe(III)-reducing bacteria. Appl Environ Microbiol. 2000;66:3743–3749. doi: 10.1128/aem.66.9.3743-3749.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd JR, Mabbett A, Williams DR, Macaskie LE. Metal reduction by sulfate-reducing bacteria: physiological diversity and metal specificity. Hydrometallurgy. 2001;59:327–337. [Google Scholar]
- Lloyd JR, Leang C, Hodges Myerson AL, Coppi MV, Cuifo S, Methe B, et al. Biochemical and genetic characterization of PpcA, a periplasmic c-type cytochrome in Geobacter sulfurreducens. Biochem J. 369:153–161. doi: 10.1042/BJ20020597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley DR. Dissimilatory Fe(III) and Mn(IV) reduction 1991. Microbiol Rev. 2003;55:259–287. doi: 10.1128/mr.55.2.259-287.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley DR, Phillips EJ. Reduction of chromate by Desulfovibrio vulgaris and Its c(3) cytochrome. Appl Environ Microbiol. 1994;60:726–728. doi: 10.1128/aem.60.2.726-728.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Gao G, Xu G, Fan L, Yin L, Shen B, Hua Y. Deinococcus radiodurans PprI switches on DNA damage response and cellular survival networks after radiation damage. Mol Cell Proteomics. 2009;8:481–494. doi: 10.1074/mcp.M800123-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaskie LE, Bonthrone KM, Yong P, Goddard DT. Enzymically mediated bioprecipitation of uranium by a Citrobacter sp.: a concerted role for exocellular lipopolysaccharide and associated phosphatase in biomineral formation. Microbiology. 2000;146(Part 8):1855–1867. doi: 10.1099/00221287-146-8-1855. [DOI] [PubMed] [Google Scholar]
- Marshall MJ, Beliaev AS, Dohnalkova AC, Kennedy DW, Shi L, Wang Z, et al. c-Type cytochrome-dependent formation of U(IV) nanoparticles by Shewanella oneidensis. PLoS Biol. 2006;4:e268. doi: 10.1371/journal.pbio.0040268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsili E, Beyenal H, Di Palma L, Merli C, Dohnalkova A, Amonette JE, Lewandowski Z. Uranium immobilization by sulfate-reducing biofilms grown on hematite, dolomite, and calcite. Environ Sci Technol. 2007;41:8349–8354. doi: 10.1021/es071335k. [DOI] [PubMed] [Google Scholar]
- Martinez RJ, Wang Y, Raimondo MA, Coombs JM, Barkay T, Sobecky PA. Horizontal gene transfer of PIB-type ATPases among bacteria isolated from radionuclide- and metal-contaminated subsurface soils. Appl Environ Microbiol. 2006;72:3111–3118. doi: 10.1128/AEM.72.5.3111-3118.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methe BA, Nelson KE, Eisen JA, Paulsen IT, Nelson W, Heidelberg JF, et al. Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science. 2003;302:1967–1969. doi: 10.1126/science.1088727. [DOI] [PubMed] [Google Scholar]
- Misra CS, Appukuttan D, Kantamreddi VS, Rao AS, Apte SK. Recombinant D. radiodurans cells for bioremediation of heavy metals from acidic/neutral aqueous wastes. Bioeng Bugs. 2012;3:44–48. doi: 10.4161/bbug.3.1.18878. [DOI] [PubMed] [Google Scholar]
- Mohner M, Lindtner M, Otten H, Gille HG. Leukemia and exposure to ionizing radiation among German uranium miners. Am J Ind Med. 2006;49:238–248. doi: 10.1002/ajim.20289. [DOI] [PubMed] [Google Scholar]
- N'Guessan AL, Vrionis HA, Resch CT, Long PE, Lovley DR. Sustained removal of uranium from contaminated groundwater following stimulation of dissimilatory metal reduction. Environ Sci Technol. 2008;42:2999–3004. doi: 10.1021/es071960p. [DOI] [PubMed] [Google Scholar]
- Nagaraj NS, Singh OV. Using genomics to develop novel antibacterial therapeutics. Crit Rev Microbiol. 2010;36:340–348. doi: 10.3109/1040841X.2010.495941. [DOI] [PubMed] [Google Scholar]
- Nilgiriwala KS, Alahari A, Rao AS, Apte SK. Cloning and overexpression of alkaline phosphatase PhoK from Sphingomonas sp. strain BSAR-1 for bioprecipitation of uranium from alkaline solutions. Appl Environ Microbiol. 2008;74:5516–5523. doi: 10.1128/AEM.00107-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaum RH. The Chernobyl nuclear catastrophe: unacknowledged health detriment. Environ Health Perspect. 2007;115:A238–A239. doi: 10.1289/ehp.115-a238. ; author reply A239–A240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyman JL, Marsh TL, Ginder-Vogel MA, Gentile M, Fendorf S, Criddle C. Heterogeneous response to biostimulation for U(VI) reduction in replicated sediment microcosms. Biodegradation. 2006;17:303–316. doi: 10.1007/s10532-005-9000-3. [DOI] [PubMed] [Google Scholar]
- Panak P, Nitsche H. Interaction of aerobic soil bacteria and plutonium (VI) Radiochim Acta. 2001;89:499–504. [Google Scholar]
- Panak PJ, Booth CH, Caulder DL, Bucher JJ, Shuh DK, Nitsche H. X-ray absorption fine structure spectroscopy of plutonium complexes with Bacillus sphaericus. Radiochimica Acta. 2002;90:315–321. [Google Scholar]
- Payne RB, Gentry DM, Rapp-Giles BJ, Casalot L, Wall JD. Uranium reduction by Desulfovibrio desulfuricans strain G20 and a cytochrome c3 mutant. Appl Environ Microbiol. 2002;68:3129–3132. doi: 10.1128/AEM.68.6.3129-3132.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck HD., Jr . Bioenergetic strategies of the sulfate-reducing bacteria. In: Odom JM, Singleton R, editors. Sulfate-Reducing Bacteria: Contemporary Perspectives. New York, USA: Springer-Verlag; 1993. pp. 41–76. [Google Scholar]
- Peretrukhin VF, Khizhniak NN, Lyalikova NN, German KE. Biosorption of technetium-99 and some actinides by bottom sediments of Lake Belsso Kosino of the Moscow region. Radiochem. 1996;38:440–443. [Google Scholar]
- Pignolet L, Auvray F, Fonsny K, Capot F, Moureau Z. Role of various microorganisms on Tc behavior in sediments. Health Phys. 1989;57:791–800. doi: 10.1097/00004032-198911000-00013. [DOI] [PubMed] [Google Scholar]
- Premuzic ET, Francis AJ, Lin M, Schubert J. Induced formation of chelating agents by Pseudomonas aeruginosa grown in presence of thorium and uranium. Arch Environ Contam Toxicol. 1985;14:759–768. doi: 10.1007/BF01055783. [DOI] [PubMed] [Google Scholar]
- Raghu G, Balaji V, Venkateswaran G, Rodrigue A, Maruthi Mohan P. Bioremediation of trace cobalt from simulated spent decontamination solutions of nuclear power reactors using E. coli expressing NiCoT genes. Appl Microbiol Biotechnol. 2008;81:571–578. doi: 10.1007/s00253-008-1741-6. [DOI] [PubMed] [Google Scholar]
- Rusin PA, Quintana L, Brainard JR, Strietelmeier BA, Tait CD, Ekberg SA, et al. Solubilization of plutonium hydrous oxide by iron-reducing bacteria. Environ Sci Technol. 1994;28:1686–1690. doi: 10.1021/es00058a021. [DOI] [PubMed] [Google Scholar]
- Saffarini DA, Schultz R, Beliaev A. Involvement of cyclic AMP (cAMP) and cAMP receptor protein in anaerobic respiration of Shewanella oneidensis. J Bacteriol. 2003;185:3668–3671. doi: 10.1128/JB.185.12.3668-3671.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakadevan K, Zheng H, Bavor HJ. Impact of heavy metals on denitrification in surface wetland sediments receiving wastewater. Water Sci Technol. 1999;40:349–355. [Google Scholar]
- Schembri MA, Kjaergaard K, Klemm P. Bioaccumulation of heavy metals by fimbrial designer adhesins. FEMS Microbiol Lett. 1999;170:363–371. doi: 10.1111/j.1574-6968.1999.tb13396.x. [DOI] [PubMed] [Google Scholar]
- Singh OV, Nagaraj NS, Gabani P. Systems biology: integrating ‘-omics’ oriented approaches to determine foodborne microbial toxins. In: Sahu SC, Casciano DA, editors. Handbook of Systems Toxicology: From Omcis Technology to Nanotechnology. New York, USA: John Wiley & Sons; 2011. pp. 469–488. [Google Scholar]
- Tamponnet C, Declerck S. Radionuclide (RN) pollution is a worldwide problem that arises from human activities. J Environ Radioact. 2008;99:773–774. doi: 10.1016/j.jenvrad.2007.10.006. [DOI] [PubMed] [Google Scholar]
- Tian B, Wang H, Ma X, Hu Y, Sun Z, Shen S, et al. Proteomic analysis of membrane proteins from a radioresistant and moderate thermophilic bacterium Deinococcus geothermalis. Mol Biosyst. 2010;6:2068–2077. doi: 10.1039/c004875e. [DOI] [PubMed] [Google Scholar]
- Valls M, Atrian S, de Lorenzo V, Fernandez LA. Engineering a mouse metallothionein on the cell surface of Ralstonia eutropha CH34 for immobilization of heavy metals in soil. Nat Biotechnol. 2000a;18:661–665. doi: 10.1038/76516. [DOI] [PubMed] [Google Scholar]
- Valls M, de Lorenzo V, Gonzalez-Duarte R, Atrian S. Engineering outer-membrane proteins in Pseudomonas putida for enhanced heavy-metal bioadsorption. J Inorg Biochem. 2000b;79:219–223. doi: 10.1016/s0162-0134(99)00170-1. [DOI] [PubMed] [Google Scholar]
- Vrionis HA, Anderson RT, Ortiz-Bernad I, O'Neill KR, Resch CT, Peacock AD, et al. Microbiological and geochemical heterogeneity in an in situ uranium bioremediation field site. Appl Environ Microbiol. 2005;71:6308–6318. doi: 10.1128/AEM.71.10.6308-6318.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Feng X, Anderson CW, Xing Y, Shang L. Remediation of mercury contaminated sites – a review. J Hazard Mater. 2012;221–222:1–18. doi: 10.1016/j.jhazmat.2012.04.035. [DOI] [PubMed] [Google Scholar]
- Wildung RE, Gorby YA, Krupka KM, Hess NJ, Li SW, Plymale AE, et al. Effect of electron donor and solution chemistry on products of dissimilatory reduction of technetium by Shewanella putrefaciens. Appl Environ Microbiol. 2000;66:2451–2460. doi: 10.1128/aem.66.6.2451-2460.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfram L, Bauerfeind P. Activities of urease and nickel uptake of Helicobacter pylori proteins are media- and host-dependent. Helicobacter. 2009;14:264–270. doi: 10.1111/j.1523-5378.2009.00685.x. [DOI] [PubMed] [Google Scholar]
- Wu WM, Carley J, Fienen M, Mehlhorn T, Lowe K, Nyman J, et al. Pilot-scale in situ bioremediation of uranium in a highly contaminated aquifer. 1. Conditioning of a treatment zone. Environ Sci Technol. 2006;40:3978–3985. doi: 10.1021/es051954y. [DOI] [PubMed] [Google Scholar]
- Xie S, Yang J, Chen C, Zhang X, Wang Q, Zhang C. Study on biosorption kinetics and thermodynamics of uranium by Citrobacter freudii. J Environ Radioact. 2008;99:126–133. doi: 10.1016/j.jenvrad.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Zivanovic Y, Armengaud J, Lagorce A, Leplat C, Guerin P, Dutertre M, et al. Genome analysis and genome-wide proteomics of Thermococcus gammatolerans, the most radioresistant organism known amongst the Archaea. Genome Biol. 2009;10:R70. doi: 10.1186/gb-2009-10-6-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]


