Abstract
IQGAP1 stimulates branched actin filament nucleation by activating N-WASP, which then activates the Arp2/3 complex. N-WASP can be activated by other factors, including GTP-bound Cdc42 or Rac1, which also bind IQGAP1. Here we report the use of purified proteins for in vitro binding and actin polymerization assays, and Förster (or fluorescence) resonance energy transfer (FRET) microscopy of cultured cells to illuminate functional interactions among IQGAP1, N-WASP, actin, and either Cdc42 or Rac1. In pyrene-actin assembly assays containing N-WASP and Arp2/3 complex, IQGAP1 plus either small G protein cooperatively stimulated actin filament nucleation by reducing the lag time before 50% maximum actin polymerization was reached. Similarly, Cdc42 and Rac1 modulated the binding of IQGAP1 to N-WASP in a dose-dependent manner, with Cdc42 enhancing the interaction and Rac1 reducing the interaction. These in vitro reconstitution results suggested that IQGAP1 interacts by similar, yet distinct mechanisms with Cdc42 versus Rac1 to regulate actin filament assembly through N-WASP in vivo. The physiological relevance of these multi-protein interactions was substantiated by 3-color FRET microscopy of live MDCK cells expressing various combinations of fluorescent N-WASP, IQGAP1, Cdc42, Rac1 and actin. This study also establishes 3-color FRET microscopy as a powerful tool for studying dynamic intermolecular interactions in live cells.
Keywords: N-WASP, Arp2/3 complex, Cdc42, Rac1, actin, Förster resonance energy transfer
Introduction
The IQGAP proteins are broadly represented among eukaryotic species ranging from unicellular organisms to humans, and are defined by a common, minimal set of structural features. All IQGAPs contain a RasGAP-related C-terminal region that is not associated with GAP activity, and function in cellular processes that require actin filaments (Briggs and Sacks 2003; Mateer and Bloom 2003). Vertebrates express three such proteins, IQGAP1 (Weissbach et al. 1994), IQGAP2 (Brill et al. 1996) and IQGAP3 (Wang et al. 2007), which share a uniform domain organization and are ~60–70% identical to each other in primary sequence. In addition to the signature domains of all IQGAPs, the vertebrate members of this family contain a variety of domains that have been functionally characterized, primarily for IQGAP1. These include, but are not limited to an N-terminal calponin homology domain (CHD); a series of tandem, α-helical coiled coil repeats for homodimerization; multiple IQ motifs that bind calmodulin or related calcium binding EF hand proteins; binding sites for MAP kinases, cell adhesion molecules, microtubule plus end tracking proteins, N-WASP, the Dia1 formin, growth factor receptors and the exocyst-septin complex; and a GAP-related domain, or GRD, which is necessary, but not sufficient for binding activated forms of the Rho family GTPases, Cdc42 and Rac1 (Bashour et al. 1997; Benseñor et al. 2007; Brandt et al. 2007; Fukata et al. 2002; Hart et al. 1996; Joyal et al. 1997; Kuroda et al. 1996; Kuroda et al. 1998; Le Clainche et al. 2007; Mateer et al. 2002; Mateer et al. 2004; McCallum et al. 1996; Rittmeyer et al. 2008; Roy et al. 2004; Roy et al. 2005; Sakurai-Yageta et al. 2008; Watanabe et al. 2004; Weissbach et al. 1998; Yamaoka-Tojo et al. 2004). This structural richness of IQGAP1 underlies its roles as a scaffolding protein that couples extracellular signals to regulation of cellular motility, morphogenesis and adhesion via control of actin assembly and secretion.
IQGAP1 regulates actin assembly through alternate pathways that respectively require either the Dia1 formin (Brandt et al. 2007) or the Arp2/3 complex (Benseñor et al. 2007; Le Clainche et al. 2007). In the latter case, IQGAP1 binds directly to N-WASP and thereby enables it to activate the Arp2/3 complex, which then nucleates new actin filaments from the sides of pre-existing filaments. This process is initiated upstream by direct binding of IQGAP1 to ligand activated growth factor receptors, such as VEGFR2 and FGFR1, which causes branched actin filament networks to be assembled in the immediate vicinity of the activated receptors (Benseñor et al. 2007; Yamaoka-Tojo et al. 2004). Growth of these filament networks provides the force for plasma membrane protrusion during cell motility and morphogenesis, which also requires IQGAP1-dependent secretion of new membrane into the expanding region of the cell surface (Rittmeyer et al. 2008; Sakurai-Yageta et al. 2008). Superimposed on the ability of IQGAP1 to stimulate branched actin filament nucleation is its ability to bind actin filaments directly via its CHD (Bashour et al. 1997; Fukata et al. 1997; Mateer et al. 2004). This property of IQGAP1 may further stimulate actin filament nucleation by helping to recruit activated Cdc42 or Rac1, and N-WASP to pre-existing actin filaments.
Further complicating actin assembly regulation by IQGAP1 is the fact that activated Cdc42 and Rac1 directly bind both IQGAP1 and N-WASP (Hart et al. 1996; Kuroda et al. 1996; McCallum et al. 1996; Miki et al. 1998); also see Figs. 1A and S1, Supplementary Information), and activate N-WASP independently of IQGAP1 (Tomasevic et al. 2007). In the absence of N-WASP, activated Cdc42 has been reported to enhance the ability of IQGAP1 to cross-link actin filaments, which implies that Cdc42 binding increases the affinity of IQGAP1 for F-actin (Fukata et al. 1997). In addition, we reported preliminary evidence that full length IQGAP1 and activated Cdc42 co-stimulate actin filament nucleation in vitro by N-WASP and the Arp2/3 complex (Benseñor et al. 2007).
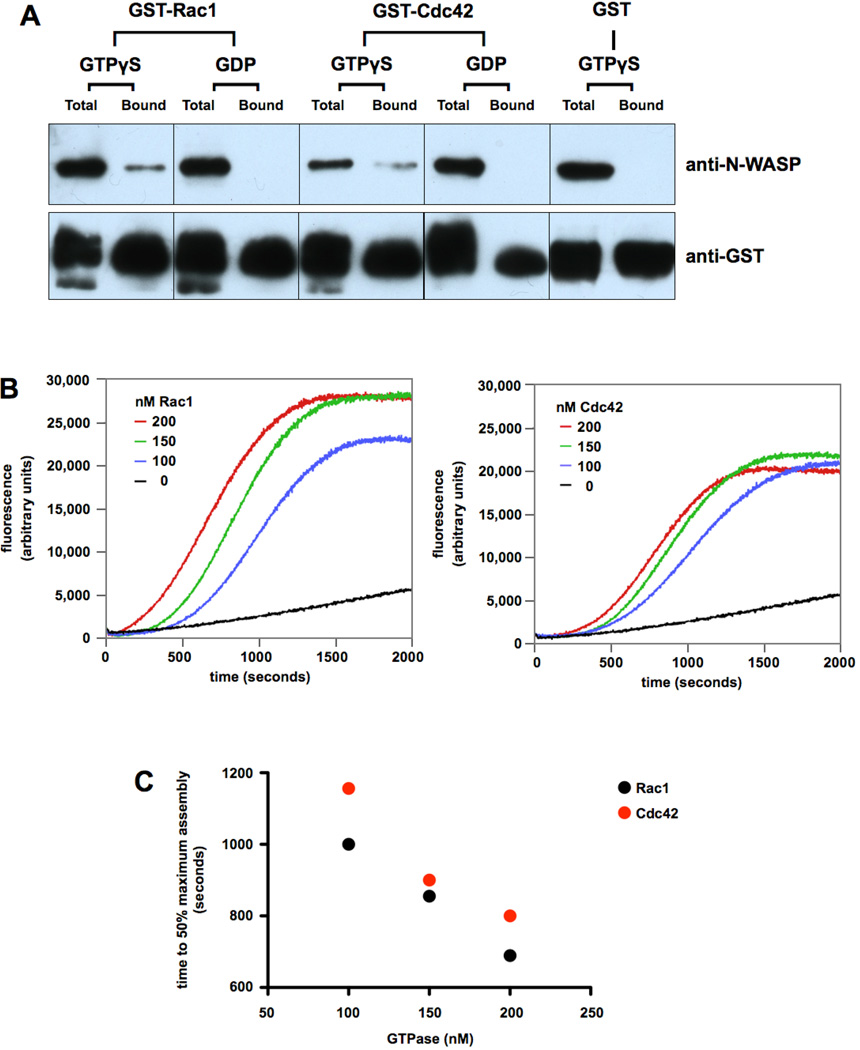
Fig. 1. Rac1, like Cdc42, binds N-WASP and stimulates actin assembly through the Arp2/3 complex.
A) 200 nM GST-Rac1 or GST-Cdc42 were loaded with GTPγS or GDP, and mixed with 200 nM N-WASP. Protein complexes were then adsorbed to glutathione-Sepharose beads, and western blotting was used to compare Total versus Bound protein. B) Effects of GTPγS-loaded Rac1 and Cdc42 on actin assembly were monitored by spectrofluorometry. Samples contained 2 µM G-actin (5% labeled with pyrene), 50 nM N-WASP, 50 nM Arp2/3 complex and the indicated concentrations of GST-Rac1 or GST-Cdc42. Note: small G protein concentrations refer to Rac1 and Cdc42 monomers in this and all subsequent figures. C) The number of seconds required for each sample shown in panel B to reach 50% maximum assembly was plotted versus GTPase concentration.
Förster (or fluorescence) resonance energy transfer (FRET), particularly when expressed as energy transfer efficiency (E%), represents a powerful tool to investigate and quantify protein-protein interactions and molecular co-localization. FRET is the non-radiative transfer of excited-state energy from a donor fluorophore to a nearby acceptor fluorophore via a long-range dipole-dipole coupling mechanism (Jares-Erijman and Jovin 2003; Sun et al. 2011; Wallrabe and Periasamy 2005). Evidence for FRET is sought by exciting donors that can transfer energy to nearby acceptors, resulting in fluorescence emitted by the acceptor. FRET requires four conditions: 1) significant overlap between the donor emission spectrum and the acceptor absorption spectrum; 2) the average distance between donor and acceptor should be ~1–10 nm; 3) optimal dipole-dipole orientation of donor and acceptor; and 4) sufficient donor quantum yield and acceptor extinction coefficient. Since energy transfer is a dipole-dipole interaction, no photons are transferred.
Quantitative FRET microscopy requires rigorous attention to spectral bleedthrough, which contaminates the raw FRET signal. There are two principal sources of spectral bleedthrough: direct acceptor excitation by the donor excitation beam and donor emission into the acceptor emission channel. Several algorithms that correct raw FRET signals for spectral bleedthrough have been developed and were reviewed recently (Berney and Danuser 2003). Most notable for the present study is our own algorithm that uses singly labeled reference specimens to correct contaminated FRET signals on a pixel-by-pixel basis (Chen et al. 2005; Elangovan et al. 2003; Sun and Periasamy 2010).
Quantitative 2-color FRET microscopy is now widely used, but is poorly suited for analyzing processes, like actin filament nucleation, that require interactions among more than two proteins. Until recently, FRET analysis of such complex processes required numerous double-label experiments, which still provided limited spatial and temporal information about multi-protein complexes. To overcome this constraint of FRET microscopy, we recently developed an algorithm for 3-color FRET, in which multiple 2-color FRET interactions involving three different fluorochromes are analyzed, and provided proof-of-principle evidence that it can be used to examine interactions among 3 fluorophore-tagged proteins in live cells (Sun et al. 2010).
The present study describes the combined application of in vitro reconstitution of purified proteins and live cell, 3-color FRET imaging to determine how actin filament assembly is affected by the simultaneous action of Rho family GTPases, IQGAP1 and N-WASP, and to establish whether Cdc42 and Rac1 are functionally equivalent or distinct in this context. Using purified proteins in assays of multiprotein complex formation and pyrene-labeled actin polymerizaton, we found only subtle functional differences between Cdc42 and Rac1. To evaluate the cell physiological significance of these findings, we co-expressed various combinations of fluorescent fusion proteins of Cdc42, Rac1, N-WASP, IQGAP1 and actin in MDCK epithelial cells, which were then analyzed by 3-color FRET microscopy. These live cell experiments confirmed that the protein-protein interactions we had studied in vitro also occur similarly in live cells. This study also emphasizes the capabilities of this novel 3-color FRET microscopy for analysis of dynamic, multi-molecular interactions that naturally occur in living cells, and thus illustrates the capabilities of this approach.
Results
To characterize in vitro interactions among IQGAP1, N-WASP, Cdc42 and Rac1, a series of binding experiments were performed using purified recombinant versions of these proteins. The same proteins, along with purified native versions of actin and the Arp2/3 complex, were also used to examine the effects of IQGAP and each small GTPase on actin nucleation and polymerization by Arp2/3 complex and N-WASP (Fig. S1, Supporting Information). Note that although the Cdc42 and Rac1 used throughout this study were often in the form of dimeric GST fusion proteins (Guthenberg and Mannervik 1981), and full length IQGAP1 is a natural homodimer (Bashour et al. 1997), all protein molarities specified in this report refer to Cdc42 or Rac1 monomers, or IQGAP1 dimers. Moreover, GST-tagged and untagged small G proteins yielded indistinguishable results in both binding and actin assembly assays at comparable concentrations of Cdc42 or Rac1.
In Vitro Bimolecular Interactions and Effects on Actin Assembly
Full length IQGAP1 bound in solution to Cdc42 or Rac1 activated with GTPγS (Fig. S2, Supporting Information), consistent with reports from other groups (Hart et al. 1996; Kuroda et al. 1996; McCallum et al. 1996). Prior studies of N-WASP (Miki et al. 1998) and the closely related protein, WASP (Symons et al. 1996) indicated that both proteins bind activated Cdc42, but not activated Rac1. Although we did not investigate WASP in the present study, we did find that N-WASP bound to both Cdc42 and Rac1 when these small G proteins were pre-loaded with GTPγS, but not with GDP (Fig. 1A).
To study effects on actin assembly of N-WASP binding to activated Cdc42 and Rac1, we used a spectrofluorometric assay to quantify rates of actin polymerization from the increase in pyrene-actin fluorescence (Benseñor et al. 2007; Bryan and Coluccio 1985). As shown in Fig. 1B, GTPγS-loaded Rac1 or Cdc42 stimulated actin assembly in the presence of N-WASP and the Arp2/3 complex in a dose-dependent manner as deduced from the decreased lag time before half maximal polymerization (50%Pmax) was achieved (Fig 1C). In light of the original claim that N-WASP does not bind Rac1 (Miki et al. 1998), the binding and actin assembly results presented here explain, confirm and extend a recent report that activated Rac1, like activated Cdc42, stimulates actin filament nucleation by N-WASP and the Arp2/3 complex (Tomasevic et al. 2007).
In Vitro Trimolecular Interactions and Effects on Actin Assembly
To determine how Cdc42 and Rac1 influence binding of IQGAP1 to N-WASP, GTPγS-loaded versions of each small G protein were mixed at various concentrations between 5 nM and 1.28 μM with fixed concentrations of IQGAP1 (50 nM) and N-WASP (150 nM), after which the multi-protein complexes that formed were immunoprecipitated (IP'd) with anti-IQGAP1. Finally, western blotting was used to reveal the content of IQGAP1, N-WASP, and Cdc42 or Rac1 in each IP.
As shown in Fig. 2A, Cdc42 increased the amount of N-WASP in anti-IQGAP1 IPs. Between 0 and 20 nM Cdc42, approximately 2/3 of the available N-WASP pelleted with IQGAP1, but nearly all of the N-WASP was present in the pellet fraction at Cdc42 concentrations >80 nM. Maximal levels of Cdc42 in the pellet fraction were achieved at a total Cdc42 concentration of ~40 nM, when ~2/3 of the Cdc42 was complexed with IQGAP1 and N-WASP. Taken together, these data indicate that Cdc42 promotes formation of complexes of N-WASP and IQGAP1.
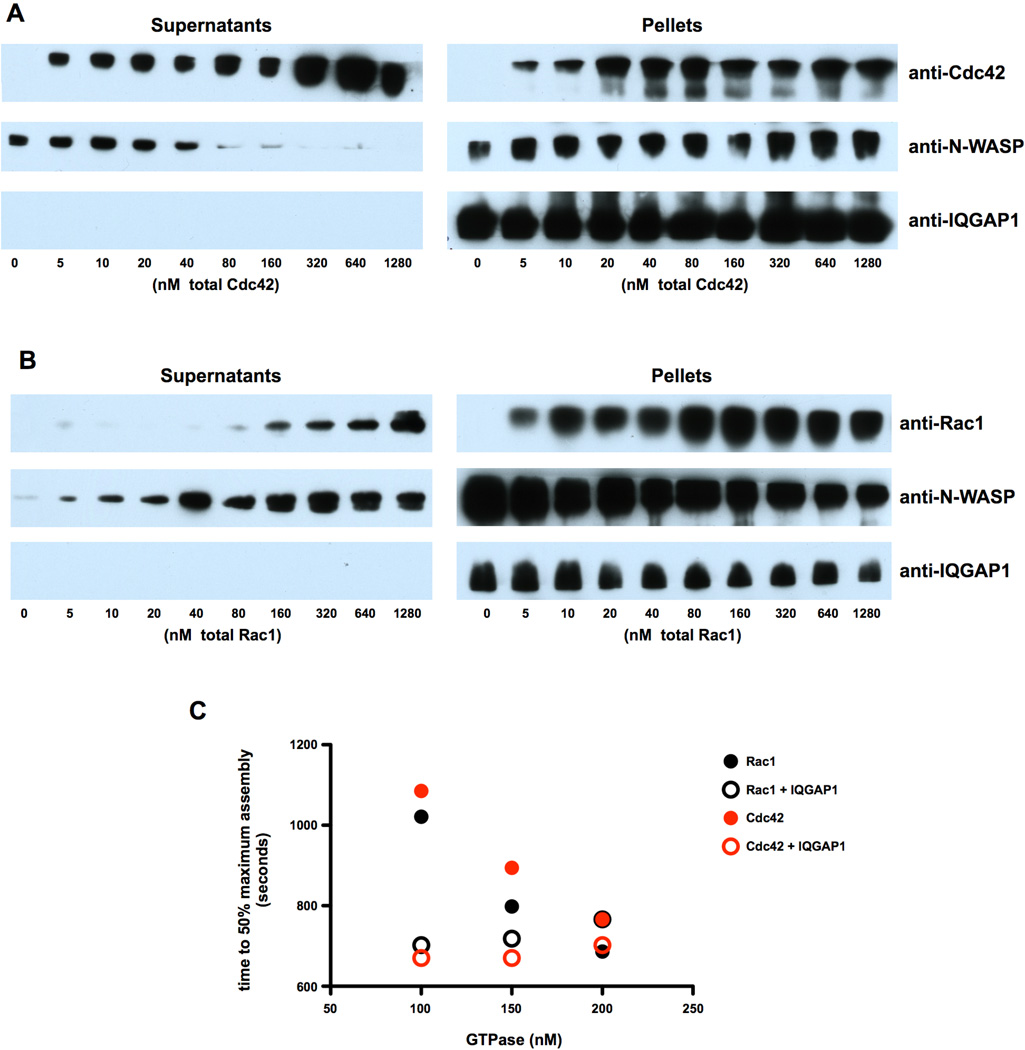
Fig. 2. Cdc42 and Rac1 differentially affect association of N-WASP with IQGAP1.
Solutions of 100 nM IQGAP1 plus 150 nM N-WASP were adsorbed to Protein G-Sepharose beads that were pre-saturated with anti-IQGAP1, and then were mixed with the indicated concentrations of GST-Cdc42 (A) or GST-Rac1 (B) that had been pre-loaded with GTPγS. The beads were then collected by centrifugation, and the Pellets and Supernatants were analyzed by western blotting. C) Actin assembly was monitored by spectrofluorometry. Samples contained 2 µM total G-actin (5% labeled with pyrene), 50 nM N-WASP, 50 nM Arp2/3 complex, 45 nM IQGAP1 (homodimers) and the indicated concentrations of GTPγS-loaded GST-Cdc42 or GST-Rac1. The number of seconds required for each sample to reach 50% maximum assembly was plotted versus GTPase concentration; the original spectrofluorometer tracings are shown in Fig. S2, Supplementary Information.
When binding experiments were performed using mixtures of Rac1, IQGAP1 and N-WASP, a different result was obtained. In contrast to Cdc42, Rac1 negatively influenced binding of N-WASP to IQGAP1 (Fig. 2B). Nearly all of the available N-WASP pelleted with IQGAP1 in the absence of Rac1, but addition of Rac1 caused a dose-dependent shift of N-WASP from the pellet to the supernatant fraction. Although binding of N-WASP to IQGAP1 in these examples was more complete in the absence of Rac1 (Fig 2B) than in the absence of Cdc42 (Fig. 2A), this difference may simply reflect experimental variation. At the upper range of total Rac1, nearly half of the available N-WASP was excluded from the pellet fraction. Conversely, the amount of Rac1 in the pellet with IQGAP and N-WASP increased in a dose-dependent manner until saturation binding was achieved at a Rac1 concentration of ~80 nM. Rac1 and Cdc42 thus have opposite effects of the association of IQGAP1 with N-WASP, with Rac1 inhibiting and Cdc42 promoting association. This effect is far from dramatic, however, as a substantial amount of N-WASP co-pelleted with IQGAP1 even at saturating levels of Rac1.
To determine if the different effects of Rho GTPases on N-WASP-IQGAP1 interactions influence the actin nucleation-promoting activity of N-WASP, pyrene-actin assembly assays were performed using fixed concentrations of actin (2 μM), N-WASP (50 nM), Arp2/3 complex (50 nM) and IQGAP1 (45 nM), and in the presence or absence of either 100–200 nM Cdc42 or Rac1 that had been activated with GTPγS (Fig. 2C; fluorescence time course plots from the identical experiments are shown in Fig. S3, Supporting Information). The maximal rate of actin assembly by Arp2/3 complex and N-WASP was much faster in the presence of either Cdc42 or Rac1 at all concentrations tested than with IQGAP1 alone. However, in combination with IQGAP1, low concentrations of either Cdc42 or Rac1 supported robust actin assembly comparable to that achieved with the highest concentrations of either Rho GTPase alone, as indicated by a reduction in the lag before 50%Pmax was achieved (Fig. 2C). Thus, IQGAP1 cooperates with Cdc42 or Rac1 to stimulate actin filament nucleation under conditions in which either Rho GTPase is limiting.
Live Cell Imaging Strategy
To assess the physiological significance of the in vitro binding and actin assembly assays, live MDCK cells were analyzed by 3-color FRET microscopy while transiently co-expressing fluorescent fusion proteins of low (F1), intermediate (F2) and high (F3) wavelength relative to each other (Fig. S4, Supporting Information). The following fluorophore-tagged protein combinations were used, with each set listed by ascending wavelength: 1) CFP-Cdc42 + Venus-IQGAP1 + mCherry-actin; 2) CFP-Rac1 + Venus-IQGAP1 + mCherry-actin; 3) Teal-N-WASP + Venus-IQGAP1 + mRFP1-Cdc42; or 4) Teal-N-WASP + Venus-IQGAP1 + mRFP1-Rac1. As controls, we expressed corresponding combinations of the unfused fluorescent proteins (CFP + Venus + mCherry, and Teal + Venus + mRFP1).
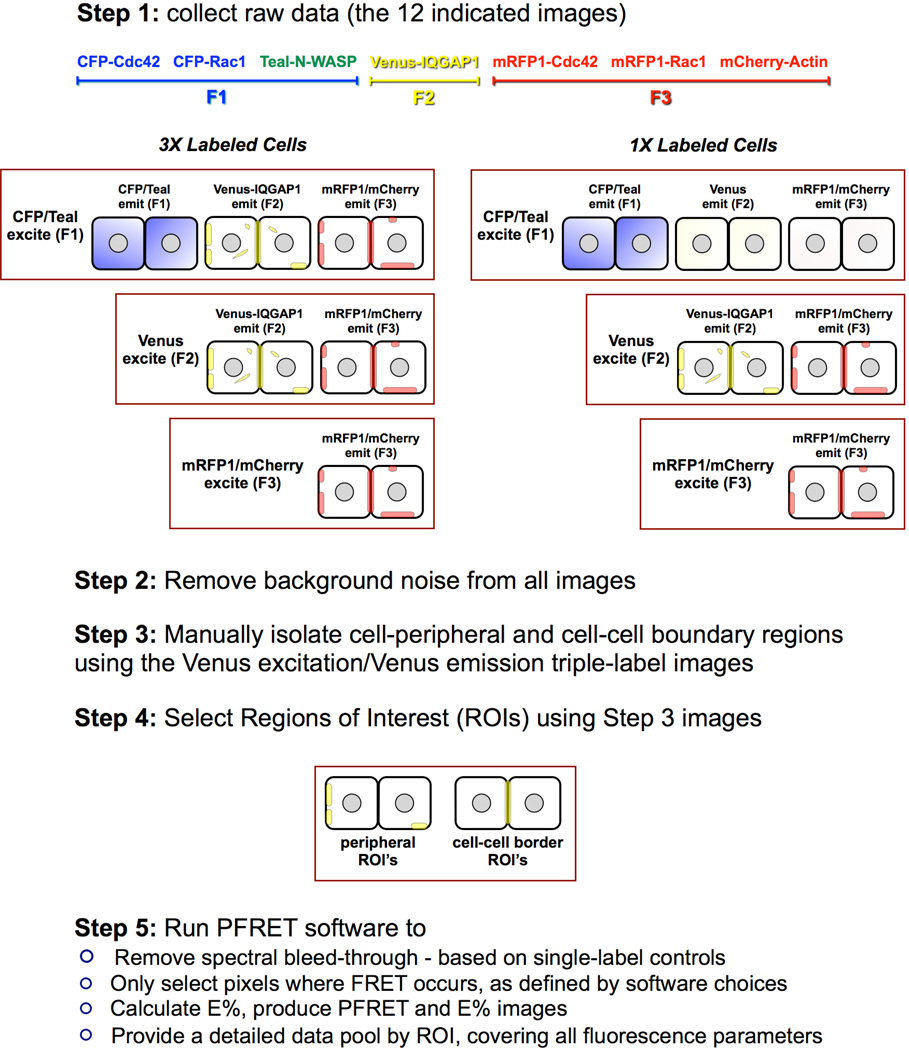
As illustrated in Fig. 3, and described in greater detail in the Materials and Methods section, a 5 step process was followed to produce 3-color FRET data. All such data were derived from either of two subcellular regions where IQGAP1 is known to function: at peripheral regions in lamellipodia, where IQGAP1 modulates plasma membrane protrusion during cellular motility and morphogenesis (Mataraza et al. 2003; Yamaoka-Tojo et al. 2004) through control of actin assembly (Benseñor et al. 2007; Le Clainche et al. 2007); and at sites of cell-cell contact, where IQGAP1 regulates intercellular adhesion through E-cadherin (Kuroda et al. 1998). Figs. 4, 5, 7 and 8 include micrographs highlighting 3x3 pixel regions of interest (ROIs) where 3-color FRET was detected in at least one pixel, as well as the conventional fluorescence images from which the FRET images were derived.
Fig. 3. 3-color FRET image acquisition, processing and analysis procedure.
Step 1. Triple-label and single-label control specimens were imaged using identical conditions. The algorithm requires the acquisition of single-label control images with comparable fluorescence ranges to the triple-label specimens for reliable spectral bleedthrough correction. Step 2. Based on non-transfected cell specimens, imaged at identical conditions, the background noise level was established and deducted from all labeled images. Step 3. Using the Venus-IQGAP1 images (Venus excitation/Venus emission) of the triple-labels as a reference, cell-peripheral and cell-cell boundary regions were manually isolated in ImageJ. Step 4. 3x3 pixel ROI's where 3-color FRET occured were selected within the regions isolated in the prior step. Step 5. The 3-color FRET software processed all selected ROI's to remove spectral bleed-through in each pixel, and calculated a complete set of data by ROI. These data included fluorescence gray levels, corrected FRET (PFRET), intensity ratios, E% and distances between fluorochromes exhibiting PFRET.
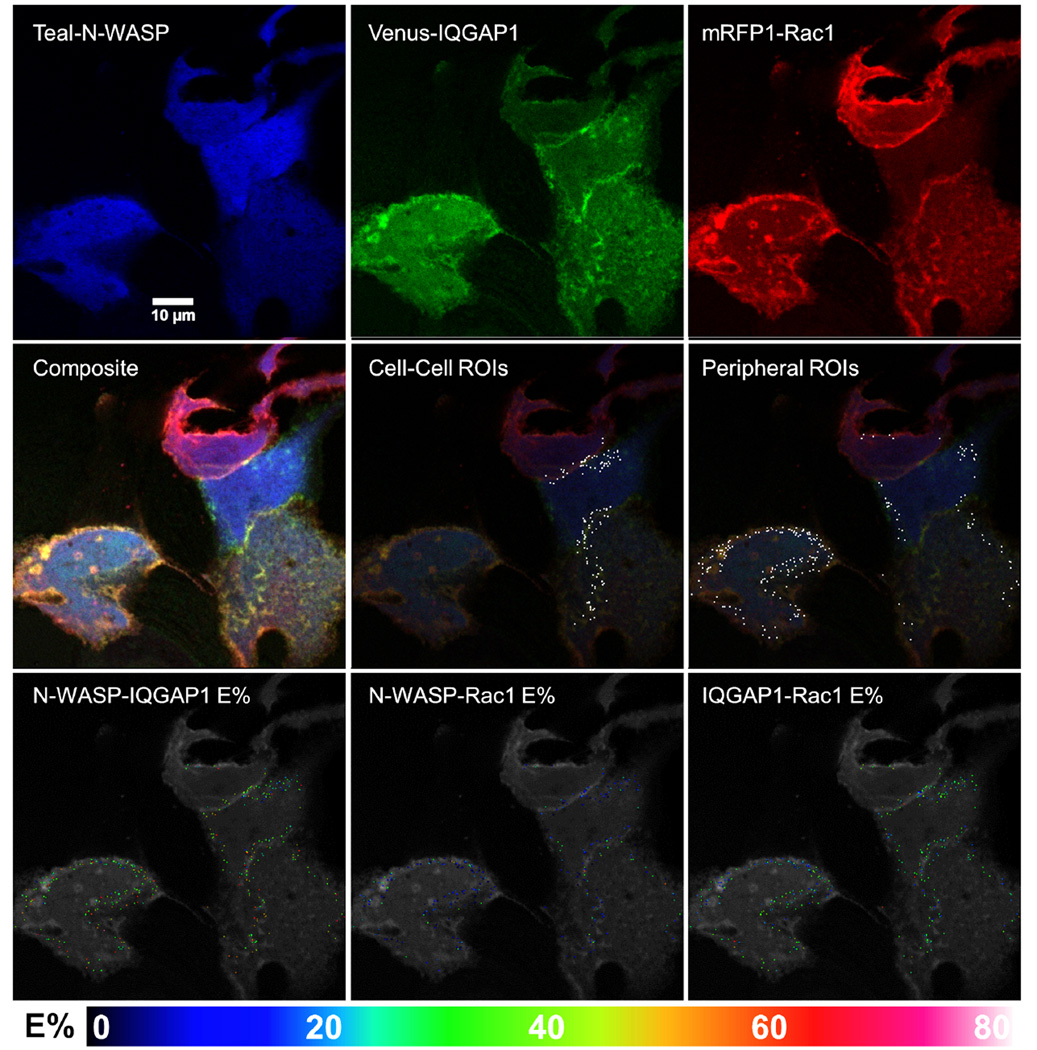
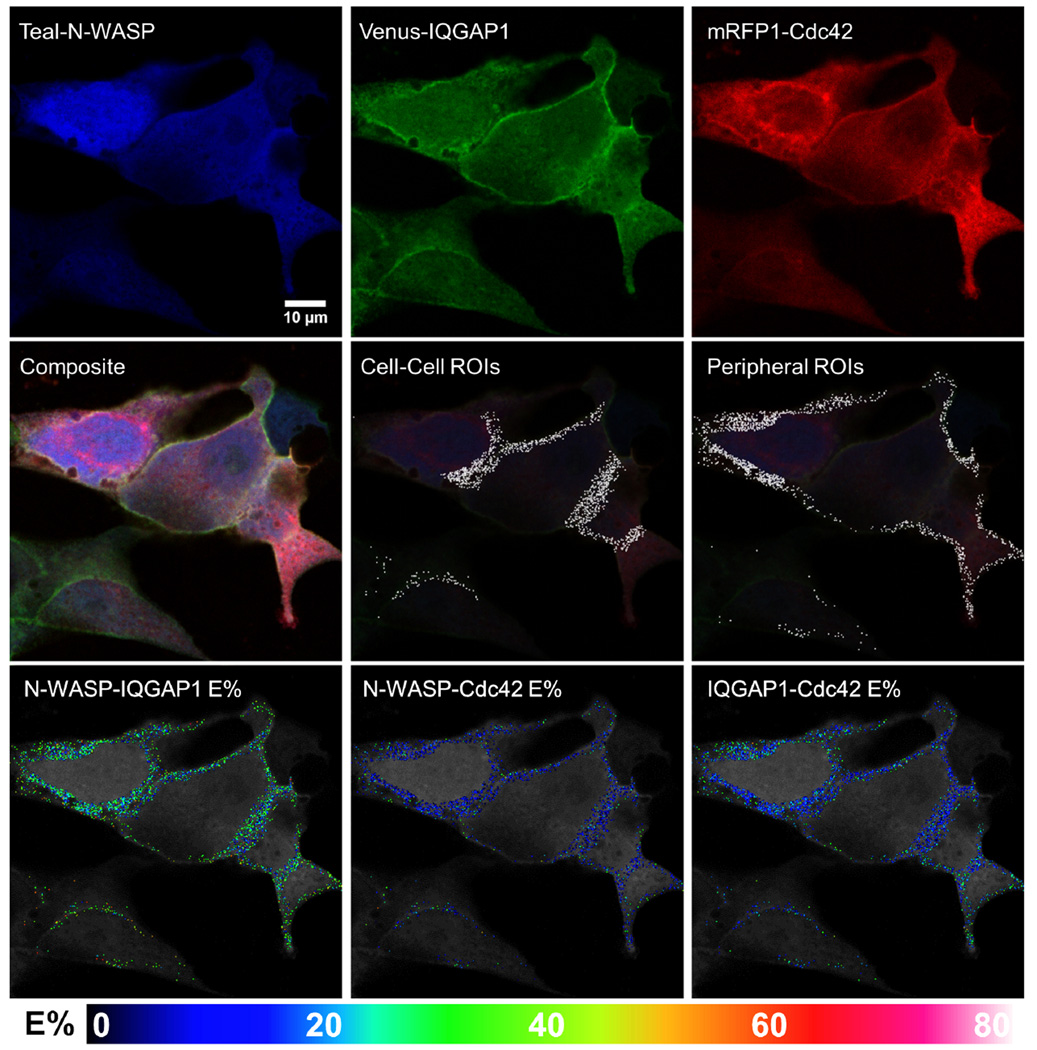
Fig. 4. Representative images of cells co-expressing Teal-N-WASP, Venus-IQGAP1 and mRFP1-Rac1, and exhibiting 3-color FRET.
Single fluorescence and composite images are shown, along with ROI's and E%'s for the indicated protein pairs that exhibited FRET within each ROI.
Fig. 5. Representative images of cells co-expressing Teal-N-WASP, Venus-IQGAP1 and mRFP1-Cdc42, and exhibiting 3-color FRET.
Single fluorescence and composite images are shown, along with ROI's and E%'s for the indicated protein pairs that exhibited FRET within each ROI.
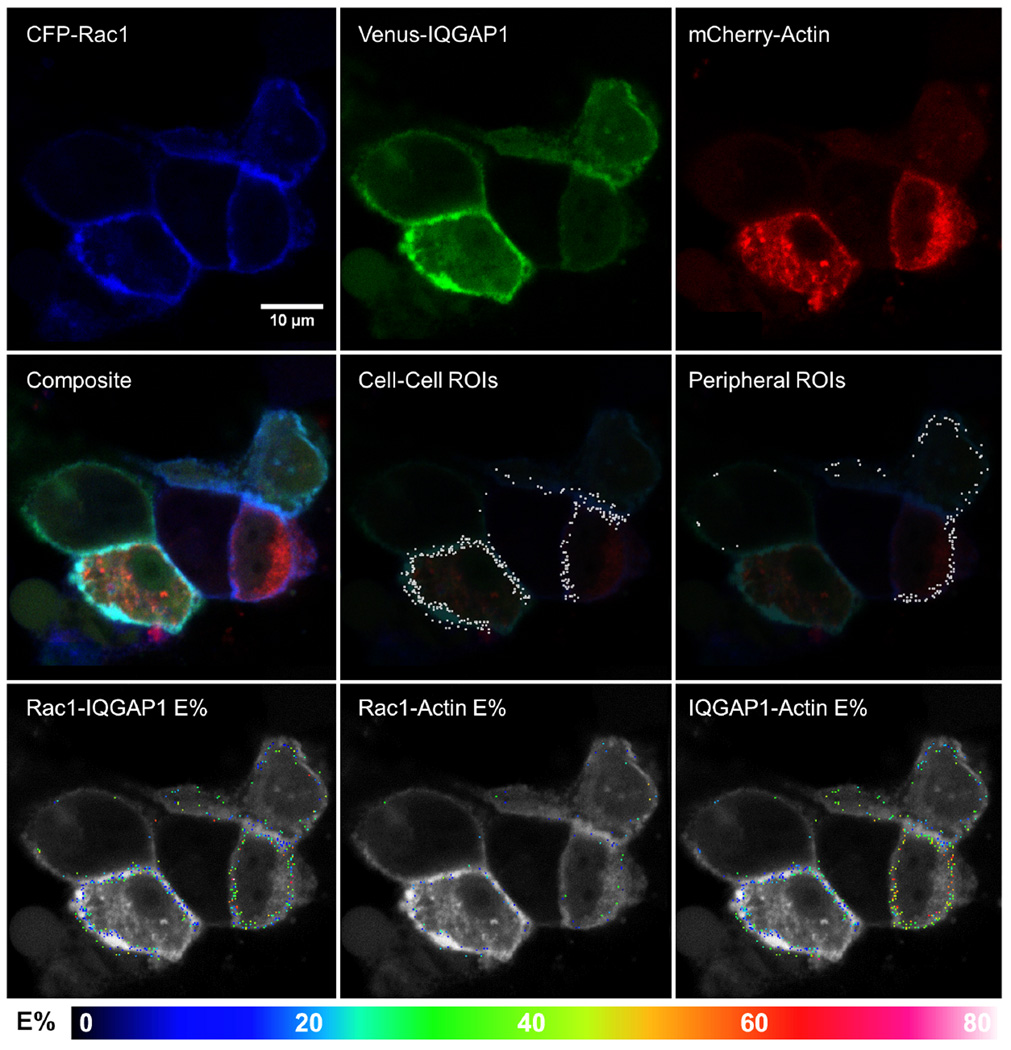
Fig. 7. Representative images of cells co-expressing CFP-Rac1, Venus-IQGAP1 and mCherry-actin, and exhibiting 3-color FRET.
Single fluorescence and composite images are shown, along with ROI's and E%'s for the indicated protein pairs that exhibited FRET within each ROI.
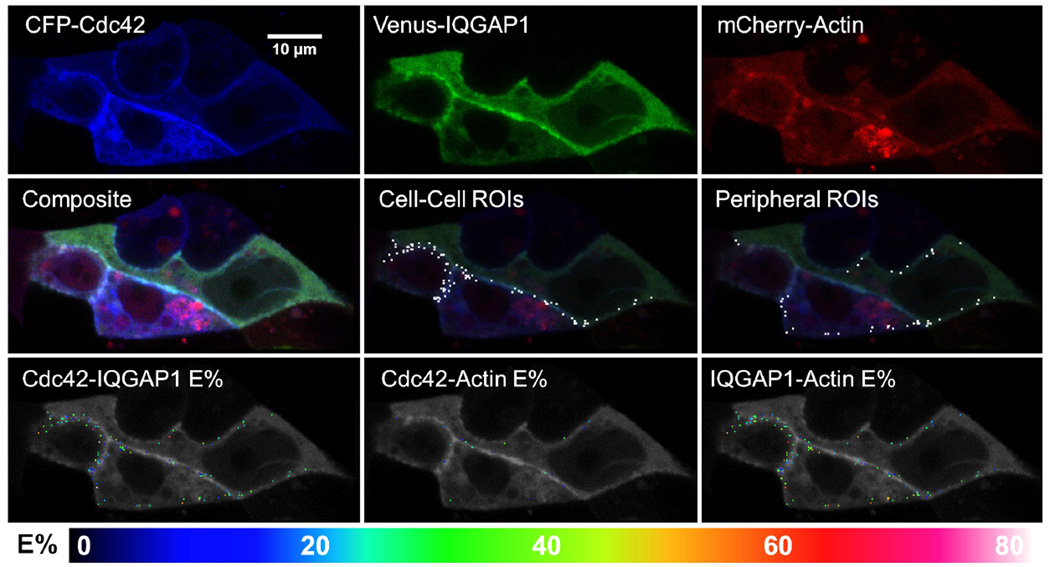
Fig. 8. Representative images of cells co-expressing CFP-Cdc42, Venus-IQGAP1 and mCherry-actin, and exhibiting 3-color FRET.
Single fluorescence and composite images are shown, along with ROI's and E%'s for the indicated protein pairs that exhibited FRET within each ROI.
To be classified as a 3-color FRET site, a pixel must simultaneously exhibit 2-color FRET between each pair of fluorescent fusion proteins that are known to bind directly to each other. For example, in cells expressing empty vector combinations or Teal-N-WASP (F1) + Venus-IQGAP1 (F2) + mRFP1-Rac1 or mRFP1-Cdc42 (F3), a 3-color FRET pixel must comprise all of the following 2-color FRET interactions: F1➛F2, F1➛F3 and F2➛F3. In contrast, co-expression of CFP-Rac1 or CFP-Cdc42 (F1) + Venus-IQGAP1 (F2) + mCherry-actin (F3) would require only F1➛F2 and F2➛F3 FRET to yield 3-color FRET because neither Rac1 nor Cdc42 binds directly to actin (Ma et al. 1998). Based on these criteria, 9–14% of the IQGAP1-positive ROI's that were located at cell peripheries or sites of cell-contact exhibited 3-color FRET.
To discriminate FRET dominated by specific versus random interactions, we adapted an assay developed earlier for 2-color FRET analysis of clustered polymeric IgA and transferrin receptors after ligand binding in crowded membranes (Wallrabe et al. 2007; Wallrabe et al. 2003; Wallrabe and Periasamy 2005). The assay is based on the principle that FRET can result from either specific binding of a donor (D) to an acceptor (A) or by chance D-A encounters, and that scatter plots of E% (FRET efficiency) versus D:A ratios, as in Figures 6 and 9, yield least squares regression lines that indicate the degree of randomness. All donor molecules, including those not engaged in FRET, are included in the E% calculation. Because E% for specifically clustered fluorophores is sensitive not only to the distance between them, but also to the proportion of donors engaged in FRET, E% decreases as the D:A ratio approaches or exceeds a level that yields maximum, specific D:A binding. For randomly clustered fluorophores, however, as D:A ratios rise, E% also tends to rise because of a concentration-dependent increase in random D-A interactions. When absolute concentrations of randomly interacting donor and acceptor pairs are sufficiently low, however, E% can be insensitive to the D:A ratio. Thus, least squares regression lines for E% versus D:A ratio plots have negative slopes when specific interactions dominate, and positive or neutral slopes when random interactions prevail (Wallrabe et al. 2007; Wallrabe et al. 2003; Wallrabe and Periasamy 2005).
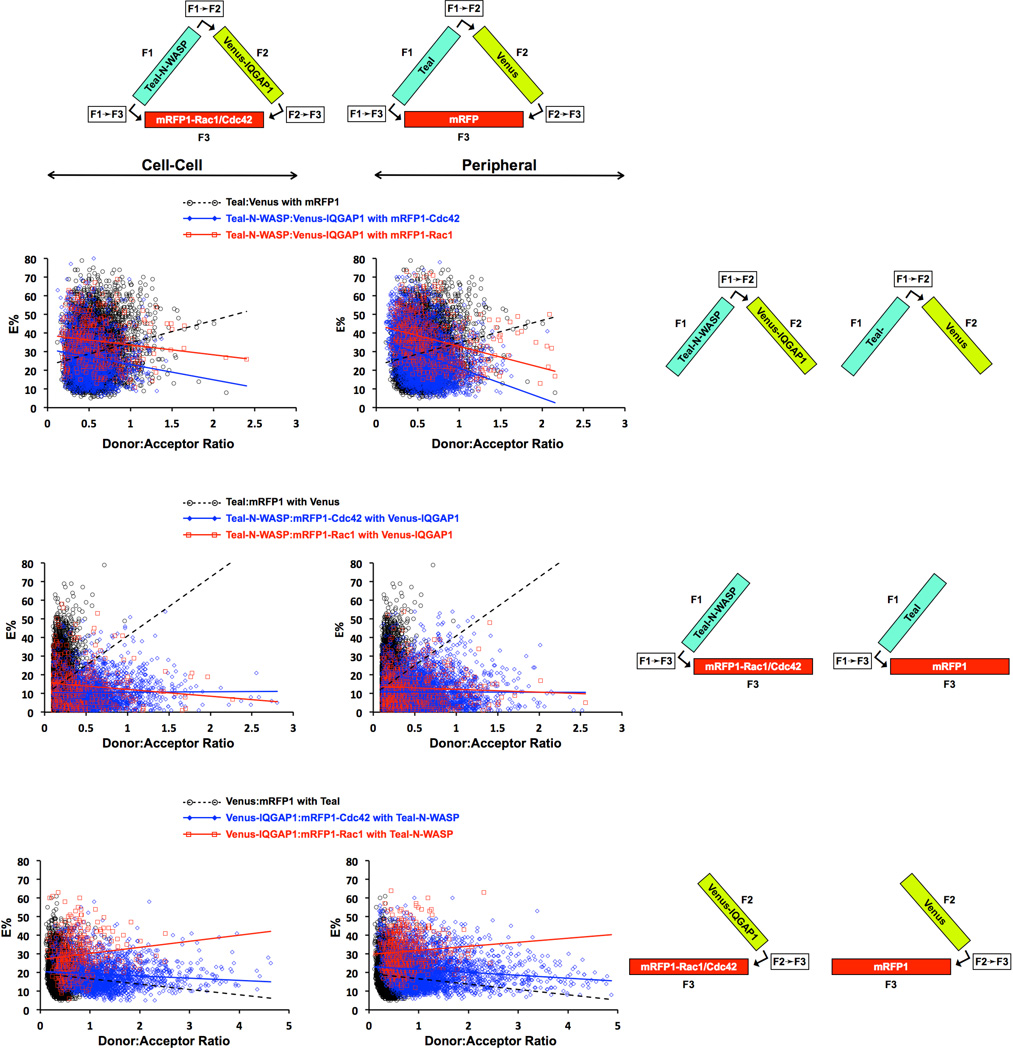
Fig. 6. FRET Efficiencies (E%'s) plotted as a function of donor-to-acceptor (D:A) ratio for cells co-expressing Teal-N-WASP, Venus-IQGAP1, and mRFP-Rac1 or mRFP1-Cdc42, and exhibiting 3-color FRET.
Each data point represents a single ROI at either the cell periphery or cell-cell boundary. Data for cell expressing unfused Teal + Venus + mRFP1 are also shown. Least squares linear regression was used to generate the straight line plot for each data set, and p-values derived from T-tests for each pair of data sets are shown in Table S1, Supplementary Information..
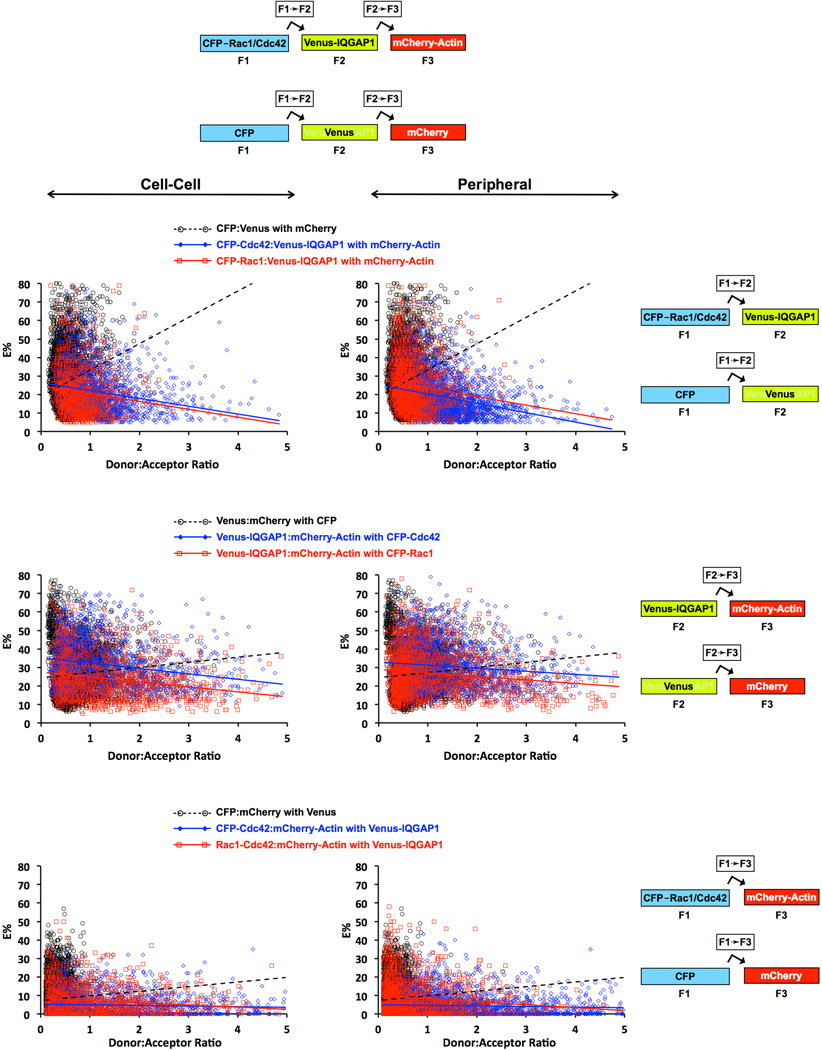
Fig. 9. FRET Efficiencies (E%'s) plotted as a function of donor-to-acceptor (D:A) ratio for cells co-expressing CFP-Rac1 or CFP-Cdc42 plus Teal-N-WASP, Venus-IQGAP1, and mRFP-Rac1 or mRFP1-Cdc42Cherry-actin, and exhibiting 3-color FRET.
Each data point represents a single ROI at either the cell periphery or cell-cell boundary. Data for cell expressing unfused CFP + Venus + mCherry are also shown. Least squares linear regression was used to generate the straight line plot for each data set, and p-values derived from T-tests for each pair of data sets are shown in Table S1, Supplementary Information. Least squares linear regression was used to generate the straight line plot for each data set.
Intracellular Interactions Among Rac1/Cdc42, IQGAP1 and N-WASP
To determine if Cdc42 and Rac1 form complexes with IQGAP1 and N-WASP, as we observed in vitro (Fig. 2), cells were transfected to co-express Teal-N-WASP, Venus-IQGAP1, and mRFP1-Rac1 (Fig. 4) or mRFP1-Cdc42 (Fig. 5). Because the range of arbitrary gray-level fluorescence values for N-WASP in the Cdc42 combination was approximately twice (10–100) that of the Rac1 combination (10–45), the two data sets were made statistically comparable by ignoring within the Cdc42 group all N-WASP data points whose arbitrary fluorescence level exceeded 45. As shown in Figs. 4 and 5, the most robust FRET was between Teal-N-WASP and Venus-IQGAP1, with average E% for this pair at ~36% for the Rac1 combination and ~27% for the Cdc42 group. Based on the correspondence between E% and theoretical intermolecular distance for any pair of fluorochromes that exhibit FRET (Sun et al. 2009), these E% values equate to an ~6 nm separation between the Teal and Venus tags when co-expressed with mRFP1-Rac1 (Fig. 4) and an ~6.6 nm separation when co-expressed with mRFP1-Cdc42 (Fig. 5). This pattern was repeated for E% and intermolecular distances for each small G protein versus N-WASP and IQGAP1, with average distances to Rac1 always being closer than to Cdc42. Average E% values for FRET from Venus-IQGAP1 to mRFP1-Rac1 and mRFP1-Cdc42 were ~30% and ~20%, respectively, corresponding to average distances of 6.1 nm and 6.6 nm between the fluorescent moieties. Average E% between Teal-N-WASP and mRFP1-Rac1 or mRFP1-Cdc42 was lower, at ~14% or 11% respectively, signifying intermolecular distances of 6.9 nm or 7.4 nm. It should be noted that Teal-N-WASP could interact with mRFP1-Rac1 or mRFP1-Cdc42 either directly, or indirectly through Venus-IQGAP1 (Fig. S4C, Supporting Information), but prior evidence from an analogous study suggests that the indirect route contributed negligibly to the measured overall FRET (Sun et al. 2010).
Peripheral Versus Cell-Cell Border Complexes of Rac1/Cdc42, IQGAP1 and N-WASP
Endogenous IQGAP1 is concentrated in lamellipodia, where it helps to drive leading edge protrusion through stimulation of branched actin filament assembly (Benseñor et al. 2007; Le Clainche et al. 2007), and is also enriched at sites of cell-cell interaction, where it regulates intercellular adhesion through interactions with E-cadherin and β-catenin (Kuroda et al. 1998). Venus-IQGAP1 and the other fluorescent fusion proteins studied here also localized to lamellipodia and cell-cell interaction sites (Figs. 4, 5, 7 and 8), prompting us to investigate the spatial distributions of complexes formed by exogenously encoded, fluorescently-tagged IQGAP1, N-WASP, Rac1 and Cdc42 using 3-color FRET microscopy. Such comparisons were made by plotting E% versus D:A fluorescence ratio for each D:A pair at both peripheral and cell-cell contact sites, and as described earlier in the "Live Cell Imaging Strategy" section, using least squares regression analysis of the raw data points to evaluate the specificity of FRET interactions (Fig. 6). Cells expressing unmodified vectors for Teal, Venus and mRFP1 were used as controls.
We found predominantly specific interactions between Teal-N-WASP and Venus-IQGAP1, with the linear regression lines having negative slopes, particularly at the periphery. In contrast, linear regression line slopes were positive for the unfused fluorescent proteins, a sign of largely random interactions. The fusion proteins demonstrated higher E% values (~30–40%) at the lowest D:A ratios, with substantial E% values (~10–25%) even at the highest D:A ratios. The E% values were slightly more when Teal-N-WASP and Venus-IQGAP1 were co-expressed with mRFP1-Rac1 than with mRFP-Cdc42, and were also slightly higher at sites of cell-cell adhesion than at lamellipodia (Fig. 6, upper panels). Paired T-tests comparing E% values between Teal-N-WASP plus Venus-IQGAP1 in the presence of either mRFP1-Cdc42 or mRFP1-Rac1 indicated significant differences (p=1.26E-32 at cell-cell boundaries and p=4.03E-38 at cell peripheries; Table S1, Supplementary Information). These results imply that Rac1 and Cdc42 differentially modulate intracellular complexes comprising N-WASP and IQGAP1, as we observed in vitro using purified proteins (Figs. 1 and 2). The majority of the E% values for the unfused fluorescent proteins (Teal + Venus with mRFP1) ranged from 5–90%, a considerably different pattern than observed for the fluorescent fusion proteins. Paired T-tests also demonstrated robust differences between the unfused flourescent protein set and each of the two fluorescent fusion protein combinations (p-values ranging from 4.6E-05 to 1.35E-31; Table S1, Supplementary Information).
The linear regression lines for E% versus the D:A ratio of Teal-N-WASP paired with mRFP1-Rac1 or mRFP-Cdc42 in the presence of Venus-IQGAP1 had slightly negative or neutral slopes, indicating a greater degree of random D:A interactions than we observed for the Teal-N-WASP: Venus-IQGAP1 pair (Fig. 6, middle panels). In addition, the E% values were lower, in the range of ~10–15%, and were virtually indistinguishable for mRFP1-Rac1 versus mRFP1-Cdc42. By T-test analysis, though, the E% populations for Teal-N-WASP:mRFP1-Rac1 were significantly different than for Teal-N-WASP:mRFP1-Cdc42 (p=1.70E-09 at cell-cell borders and p=3.33E-06 at the cell peripheries). There was no striking difference in the average E% level between cell-cell boundary and peripheral sites, and the unfused fluorescent proteins (Teal + mRFP1 with Venus) were dramatically different than either corresponding set of fluorescent fusion proteins (p-values ranging from 1.47E-01 to 3.28E-101; Table S1, Supplementary Information). Taken together, these data indicate that at both cell-cell boundaries and cell peripheries N-WASP forms specific complexes differentially with Rac1 and Cdc42.
Finally, we also noted a difference between Rac1 and Cdc42 in cells that co-expressed Venus-IQGAP1 with mRFP1-Rac1 or mRFP1-Cdc42 in the presence of N-WASP (Fig. 6, bottom panels). At low D:A ratios the linear regression lines indicated higher E% values for mRFP-Rac1 (~30%) than for mRFP1-Cdc42 (~20%) at cell-cell interaction and peripheral sites, but the slopes of the E% versus D:A ratios were positive for mRFP1-Rac1 and negative for mRFP1-Cdc42 at both intercellular locations. Compared to mRFP1-Cdc42, mRFP-1Rac1 therefore seems to have more frequent random interactions with Venus-IQGAP1. Curiously, the linear regression lines for the unfused fluorescent proteins (Venus + mRFP1 with Teal) had negative slopes for the E% versus D:A ratio plots, raising the unexpected possibility that naked Venus and mRFP1 can interact specifically. This result, along with the surprising finding that Venus-IQGAP1 interacts in a predominantly random manner with mRFP1-Rac1 (Fig. 6, bottom panels), but as expected, in a predominantly specific manner with CFP-Rac1 (Figure 9, upper panels), signifies anomalous FRET behavior of Venus and mRFP1 as a D:A pair.
Intracellular Interactions Among Rac1/Cdc42, IQGAP1 and Actin
To extend the 3-color FRET approach to actin we co-expressed CFP-Rac1 or CFP-Cdc42 with Venus-IQGAP1 and mCherry-actin (Figs. 7–8). Significant FRET occurred between CFP-Rac1 or CFP-Cdc42 and Venus-IQGAP1, and between Venus-IQGAP1 and mCherry-actin at both cell peripheries and sites of intercellular contact. FRET was similar between CFP-Rac1 or CFP-Cdc42 and Venus-IQGAP1 with average E% values of ~23% and ~20%, corresponding to average separation distances of ~6 nm and ~6.2 nm for the Rac1 and Cdc42 pairs, respectively. The corresponding average E% values for Venus-IQGAP1 to mCherry-actin were ~26% (~6.7 nm) in the presence of CFP-Rac1 and ~31% (~6.4 nm) in cells expressing CFP-Cdc42. In contrast, FRET from CFP-Rac1 or CFP-Cdc42 to mCherry-actin was considerably lower with E% averages ranging from 4.5–8.7% and a large proportion being less than 5%, corresponding to distances at the limit of FRET detection. These data, along with prior evidence that neither Rac1 nor Cdc42 binds directly to actin (Ma et al. 1998), strongly indicate that the infrequent, low level FRET between CFP-tagged small G Proteins and mCherry-actin represented random interactions.
Peripheral Versus Cell-Cell Border Complexes of Rac1/Cdc42, IQGAP1 and Actin
Scatter plots and linear regression analyses derived from cells co-expressing CFP-Rac1 or CFP-Cdc42 with Venus-IQGAP1 and mCherry-actin are shown in Figure 9, along with analogous data for cells expressing the corresponding unfused fluorescent proteins (CFP, Venus and mCherry). Venus-IQGAP1 served as an effective and specific FRET acceptor for CFP-Rac1 and CFP-Cdc42 (Fig. 9, upper panels), with negatively sloped linear regression lines demonstrating E% values of ~25% at low D:A ratios for both small G proteins at both cell-cell boundaries and peripheral sites. In all cases for these FRET pairs, E% values dropped steeply as D:A ratios rose, indicating that specific, clustered interactions dominated FRET from either CFP-Rac1 or CFP-Cdc42 to Venus-IQGAP1, irrespective of subcellular location. In contrast, when the unfused fluorescent proteins were expressed, the plots of E% versus D:A ratios had large positive slopes, signifying predominantly random interactions. These results firmly imply that the binding of Rac1 and Cdc42 to IQGAP1 observed in vitro (Fig. S2, Supplementary Information) does, indeed, occur in the cytoplasm of live cells. T-test p-values comparing relevant data set pairs for these experiments and the others presented in Fig. 9 are shown in Table S1, Supplementary Information.
Similar results were obtained for FRET from Venus-IQGAP1 to mCherry-actin (Fig. 9, middle panels). At low D:A ratios, negatively sloped linear regression lines indicated E% values of ~25–35%, with the higher values appearing in cells that co-expressed CFP-Cdc42, as opposed to CFP-Rac1. Again, the negative slopes of the linear regression lines signified that clustered, rather than random interactions dominated the FRET signals from Venus-IQGAP1 to mCherry-actin, and no obvious differences between peripheral versus cell-cell interaction sites were observed. In contrast, cells that expressed unfused Venus and mCherry in the presence of CFP the linear regression slopes were strongly positive, as expected for mainly random interactions. These results imply direct binding of IQGAP1 to actin at both locations, modulated similarly by Rac1 and Cdc42.
Lastly, FRET from CFP-Rac1 or CFP-Cdc42 to mCherry-actin was barely detectable, with ~80% of ROI's having E% values of 10% or less and ~60% having values no more than 5%, independently of D:A ratios, and at both peripheral and cell-cell interaction sites (Figure 9, bottom panels). These values were nearly identical to the E% values we found when the corresponding unfused fluorescent proteins (CFP + mCherry) were expressed along with unfused Venus. Taken together, these data constitute strong evidence that neither Rac1 nor Cdc42 binds directly to F-actin in cells, as was previously established by in vitro biochemical assays (Ma et al. 1998).
Discussion
IQGAP1 is one of several proteins that stimulates actin filament assembly by direct activation of N-WASP, which then activates the Arp2/3 complex, thereby promoting nucleation of new "daughter" filaments from the sides of pre-existing "mother" filaments (Benseñor et al. 2007; Le Clainche et al. 2007). N-WASP can also be activated by GTP-bound forms of Cdc42 (Rohatgi et al. 2000; Rohatgi et al. 1999) or Rac1 (Tomasevic et al. 2007) (see also Fig, 1A), both of which are tight binding partners for IQGAP1 as well (Hart et al. 1996; Kuroda et al. 1996; McCallum et al. 1996) (see also Fig. S1, Supplementary Information). We therefore sought to determine how actin assembly mediated by N-WASP and the Arp2/3 complex is affected by the simultaneous presence of IQGAP1 and either Cdc42 or Rac1, and whether these closely related Rho GTPases are functionally equivalent in this context.
Using purified proteins for in vitro binding and actin assembly assays, we found that Cdc42 and Rac1 have opposing effects in modulating interactions between N-WASP and IQGAP1, but similarly support actin nucleation in the presence of IQGAP1 (Fig, 2). Whereas Cdc42 promoted association of N-WASP with IQGAP1, Rac1 antagonized that association. A substantial amount of N-WASP remained associated with IQGAP1 at supramolar levels of Rac1, however, and in the context of actin filament nucleation in vitro, low concentrations of either Rac1 or Cdc42 worked cooperatively with IQGAP1 to activate the nucleation promoting function of N-WASP. Rac1 and Cdc42 therefore appear to work similarly with IQGAP1 to activate N-WASP.
To assess the cell physiological relevance of these results, we adapted a recently developed method for live cell, 3-color FRET microscopy (Sun et al. 2010) to analyze MDCK epithelial cells that simultaneously expressed fluorescently tagged versions of IQGAP1, Cdc42 or Rac1, and N-WASP or actin (Figs. 3–9 and Fig. S4, Supplementary Information). These live cell experiments supported the conclusions that all of the direct binding interactions we observed using purified proteins in vitro (Cdc42-IQGAP1, Rac1-IQGAP1, Cdc42-N-WASP, Rac1-N-WASP and IQGAP1-N-WASP) occur in living cells, as well, and that protein pairs that bind only indirectly in vitro (Cdc42-actin and Rac1-actin) also do not interact directly in live cells.
3-Color FRET
It is important to emphasize that the 3-color FRET approach employed here identifies individual 2-color FRET events (F1➛F2, F1➛F3 and F2➛F3; see Figs. 3–9 and S4, Supplementary Information) that occur simultaneously in space and time within an individual cell. This is a major advance beyond conventional 2-color FRET, for which the closest equivalent data would have to be generated in three separate experiments with two components each, with each experiment requiring its own unique cell culture. This multiple 2-color FRET approach would still be limited to analysis of just one type of intermolecular interaction per culture, however, whereas three different intermolecular interactions can be monitored in each culture by the 3-color FRET method.
Since the actin nucleation process is fast, with transient associations among the proteins of interest, we frequently detected pixels in which just two of the three fluorescent fusion proteins were within FRET distance when images were captured. We ignored these pixels for the present study, and with one caveat, focused solely on pixels with confirmed FRET between all fusion protein pairs. The caveat concerned FRET between CFP-Rac1 or CFP-Cdc42 (F1), and mCherry-actin (F3) in cells that also expressed Venus-IQGAP1 (F2). Because neither small G protein binds directly to actin (Ma et al. 1998), we neither expected nor detected non-random F1➛F3 FRET in these experiments.
Having selected only pixels in which processed FRET (PFRET: corrected for background and spectral bleed-through) occurred, it became possible to interpret fluorescence data by a variety of quantitative strategies. Because it has the advantage of being insensitive to donor or acceptor fluorescence levels, we also explored fluorescence lifetime imaging microscopy (FLIM) as a readout for FRET (Periasamy and Clegg 2010) in cells expressing three FRET-compatible fluorescent fusion proteins. In practice, however, the long image acquisition times (>1minute) required to collect adequate FLIM signals with our available equipment made this approach impractical for monitoring the dynamic interactions that are the subject of this study and occur on a much shorter time scale.
An obvious unique value of 3-color FRET is its capacity to monitor interactions among three proteins in live cells in real time with spatial resolution that vastly exceeds the resolution afforded by conventional live cell fluorescence microscopy or even by super-resolution microscopy (1–10 nm for FRET versus ~200 nm or ~20 nm for conventional or super-resolution fluorescence, respectively). Immunoprecipitation out of cell or tissue extracts is widely used to isolate and characterize multimolecular complexes, but cannot typically discriminate between direct and indirect protein-protein interactions, will not provide detailed information about where proteins that interact in extracts were actually localized within cells or tissues, and requires destruction of the biological specimen, thereby precluding its subsequent evaluation.
Live cell 3-color FRET largely overcomes these limitations, but there are restrictions to what can be accomplished by the 3-color FRET approach described here. For example, we initially hoped that 3-color FRET would allow us to determine if Cdc42 and Rac1 respectively promote and inhibit binding of IQGAP1 to N-WASP in live cells, as observed in vitro (Fig. 2), but at least a few confounding factors prevented any such conclusions to be made. Most notably, the in vitro binding and actin assembly experiments allowed us to use known concentrations of all of the proteins being studied and to vary their concentrations systematically, but as explained in the following paragraph, this was not feasible for the 3-color FRET experiments.
We could determine relative concentrations of exogenously encoded fluorescent fusion proteins based on their fluorescence intensities, and it would even be possible to equate fluorescence intensities with absolute concentrations of the respective proteins. Because each fusion protein was expressed on the background of unlabeled, endogenously encoded counterparts, whose concentrations may have varied from cell to cell and within distinct intracellular regions, however, we were reluctant to use the fluorescence signals as reliable indicators of the total amount (labeled plus endogenous) of Rac1, Cdc42, N-WASP, IQGAP1 or actin present within each ROI where 3-color FRET was detected. This restriction could be overcome in principle by using RNA interference to reduce levels of endogenous counterparts of exogenously encoded fluorescent fusion proteins, or by expressing fluorescent versions of proteins that are not found naturally in a particular cell type. To cite one hypothetical example, it should be possible to express fluorescent versions of each of the three neurofilament subunit proteins in fibroblasts, whose endogenous neurofilament genes are silent, and use 3-color FRET microscopy to study neurofilament assembly under conditions in which the concentrations of all three subunit proteins can be accurately measured.
The study documented here revealed peculiar FRET behavior between Venus and mRFP1 as a D:A pair for two reasons. First, the linear regression line slopes of E% versus D:A ratio for unfused Venus and mRFP1 were negative, implying that these fluorescent proteins can interact specifically when they are not fused to other proteins. Secondly, and in contrast, the linear regression line slopes for Venus-IQGAP1 and mRFP1-Rac1 were positive (Fig. 6, bottom panels), indicating mostly random interactions, even though purified, untagged IQGAP1 and Rac1 bind directly to each other (Fig. S2, Supplementary Information) and linear regression slopes for the CFP-Rac1 and Venus-IQGAP1 D:A pair were strongly negative, signifying predominantly specific interactions (Fig. 9, upper panels). These results suggest that Venus and mRFP1 are not reliable FRET partners, despite the apparently favorable overlap of the emission spectrum of Venus with the excitation spectrum of mRFP1. Perhaps this anomalous behavior reflects the fact that mRFP1 has two excitation peaks, one of which nearly coincides with the single excitation peak of Venus (Fig. S4, Supplementary Information). None of the other FRET pairs used in this study exhibited any such irregular behavior.
Notwithstanding the expressed caveats of 3-color FRET, the data reported here confirmed that numerous protein-protein interactions detected in vitro using purified proteins, do indeed occur in live cells at physiologically relevant intracellular locations. Furthermore, the results indicate that the association of IQGAP1 with other proteins that regulate actin dynamics occurs similarly at lamellipodia and cell-cell interaction sites, and that interactions of IQGAP1 with Rac1 or Cdc42 lead to very similar functional consequences.
Materials and Methods
Protein Purification
Previously published methods were used to purify rabbit muscle actin (Bashour et al. 1997), recombinant, his-tagged IQGAP1 (Mateer et al. 2004), recombinant untagged rat N-WASP (Miki et al. 1998) and native bovine calf thymus Arp2/3 complex (Higgs et al. 1999), and for covalently coupling pyrene to actin (Bryan and Coluccio 1985). GST-Cdc42, GST-Rac1 and unmodified GST were expressed in transformed E. coli (strain BL21), and were lysed into a GDP containing buffer (50mM Tris pH 7.5, 100nM GDP, 20 mM NaF, 1 mM PMSF, and 2 μg/ml each of chymostatin, leupeptin and pepstatin A). The proteins were then purified using glutathione-Sepharose 4B beads (Pharmacia) and stored at −80° C in GDP containing buffer. Upon thawing, they were incubated for 10 minutes at 30° C in loading buffer (5mM Tris pH 7.5, 20mM KCl, 6.25mM EDTA, 1mM fresh DTT, 1mM GTPγS or GDP). 25 mM MgCl2 was then added to stabilize the loaded conformation for Cdc42 and Rac1, and the proteins were then kept on ice and used within two hours of preparation.
Affinity Pull Down and Pyrene-Actin Assembly Assays
All assays were performed using purified proteins that were dialyzed overnight in buffer A (50 mM Hepes pH 7.4, 50 mM NaCl, 20 mM NaF, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 2 μg/ml each of chymostatin, leupeptin and pepstatin A). To monitor bimolecular interactions, glutathione-Sepaharose 4B beads saturated with GTPγS-loaded GST-Cdc42 or GST-Rac1, or with GST were mixed with 150 nM N-WASP, and were subsequently incubated for 1 hour at 4° C (Fig. 1A). Alternatively, EZview red protein-G affinity beads (Sigma) saturated with polyclonal anti-IQGAP1 (Mateer et al. 2002) were incubated with 100 nM his-IQGAP1. Then 0.5 μM GST-Cdc42, GST-Rac1 or unmodified GST that had been loaded with GTPγS or GDP were added and incubated for an additional 2 hours at 4° C (Fig. S1, Supplementary Information). To analyze trimolecular interactions (Figs. 2A/B), IQGAP1-N-WASP complexes were pre-formed by incubating 100 nM his-IQGAP1 and 150 nM N-WASP at 4° C for one hour, and then immobilizing the complexes to protein G-Sepharose beads (Sigma) saturated with polyclonal anti-IQGAP1 (Mateer et al. 2002). GST-Cdc42 or GST-Rac1 loaded with GTPγS were then added to final concentrations of 0, 5, 10, 20, 40, 80, 160, 320, 640 and 1280 nM, after which the bead suspensions were incubated for an additional hour. Both bimolecular and trimolecular complexes were collected by brief centrifugation, washed in buffer B (50 mM Tris pH 7.4, 150 mM NaCl, 0.5% Triton X-100, 1 mM PMSF), and analyzed by immunoblotting using rabbit polyclonal antibodies to IQGAP1 (Mateer et al. 2002), N-WASP (Santa Cruz), GST (Invitrogen), Rac1 (Santa Cruz) or Cdc42 (Santa Cruz), and SuperSignal chemiluminescent reagents (Pierce).
Pyrene-actin assembly assays were performed using a Photon Technology Incorporated model QM-4/5000 spetrofluorometer with 365 nm excitation and 386 nm emission exactly as described previously (Benseñor et al. 2007).
Plasmid Construction
The Venus-IQGAP1 was derived from a pmGFP-C1-IQGAP1 vector generously provided by Dr. Geri Kreitzer of Weill-Cornell Medical College. Venus cDNA was amplified by PCR using pVenus-C1 as template and following primers: 5' primer: 5'-TTT ACC GGT CGC CAC CAT GGT GAG CAA GGG C-3'; 3' primer: 5'-CGT CGA CTG CAG AAT TCG AAG CTT GAG CTC GAG-3'. The GFP coding sequence in the pmGFP-C1-hIQGAP1 vector was replaced by PCR-amplified Venus cDNA using AgeI and XhoI restriction digest sites, and the resulting modified vector was ligated using T4 DNA ligase (NEB) to create Venus-IQGAP1.
The Teal-N-WASP vector was constructed by a 3 piece ligation process, as follows. The pYFP-C1 vector (digested by NdeI and EcoRI), Teal cDNA (digested from pmTFPC1rC-EBPa by NdeI, BsrGI; kindly provided by Dr. Richard Day, Indiana University School of Medicine) and hNWASp (PCR products digested by BsrGI and EcoRI) were ligated using T4 DNA ligase. hNWASp cDNA was amplified by PCR from pBluescriptR-hWASL clone (OpenBiosystems; Clone ID #5264663) and the following primers were used: 5' primer: 5'-AAA AAA AAA TGT ACA AGT CCG GAA TGA GCT CCG TCC AGC AGC A-3'; 3' primer: 5'-CCG GAA TTC TCA GTC TTC CCA CTC ATC-3'.
Plasmids encoding constitutively active human mRFP-Rac1V12 and mRFP-Cdc42V12 were kindly provided by Dr. Maria Carla Parrini of l'Institut Curie, and were converted back to wild type sequences by replacement of each valine at position 12 with a glycine using QuickChange Site-Directed Mutagenesis Kit (Stratagene). To generate mRFP-Rac1WT, the following primers were used: 5' primer: 5'-GGT GGT GGG AGA CGG AGC TGT AGG TAA AAC-3'; 3'-primer: 5'-GTT TTA CCT ACA GCT CCG TCT CCC ACC ACC-3'). To generate mRFP-Cdc42WT, the following primers were used: 5' primer: 5'-GTG TGT TGT TGT GGG CGA TGG TGC TGT TGG TAA AAC-3'; 3' primer: 5'-GTT TTA CCA ACA GCA CCA TCG CCC ACA ACA ACA CAC-3'.
The mCherry-actin plasmid was a kind gift of Dr. Derek Applewhite of the University of North Carolina at Chapel Hill. CFP-Rac1 and CFP-Cdc42 were made by inserting the human coding sequences for each small G protein into an pECFP-C1 vector. The coding sequence for WT human Rac1 was derived from the mRFP1-Rac1WT vector described in the prior paragraph. WT human Cdc42 cDNA was amplified by PCR from pGEX2T-Cdc42-wt (Adgene) using the following primers: 5' primer: 5'-CCG CTC GAG CT ATG CAG ACA ATT AAG TG-3'; 3' primer: 5'-CG GGA TCC TCA TAG CAG CAC ACA CC-3'. The pECFP-C1 vector and Cdc42 PCR product were digested by XhoI and BamHI (NEB), and ligated using T4 DNA ligase.
All of the newly created expression vectors for fluorescent fusion proteins described here were analyzed by restriction digestion, verified by sequencing to be correct, amplified in DH5α E. coli cells, and extracted using HiSpeed Plasmid Maxi Kits (Qiagen).
Cell Culture and Transfection
MDCK cells growing in 10 cm dishes in Dulbecco's MEM (GIBCO) supplemented with HyClone Cosmic Calf Serum (Thermo Scientific) to 10% by volume and gentamycin (Sigma) to 50 mg/l were detached using TrypLE (Invitrogen) and plated at 1 × 106 cells per well on 25mm cover slips in 6-well dishes. Immediately after plating onto the coverslips, the cells were transfected for simultaneous expression of one, two or three fluorescent proteins using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Transfected cells were grown for 24 hours before imaging.
3-Color Confocal FRET Microscopy
The 3-color confocal FRET microscopy method was described in detail recently and requires proprietary analysis software (Sun et al. 2010). Imaging required a total of three excitation wavelengths (Ex1, Ex2, Ex3) and three emission channels (Em1, Em2, Em3) for detecting fluorophore 1 (F1: Teal or CFP), fluorophore 2 (F2: Venus) and fluorophore 3 (F3: mCherry or mRFP1). As shown in Figure 3, a 5-step procedure was followed to yield 3-color FRET data. Step 1 involved collecting 6 images from cells that co-expressed 3 fluorescent proteins and 6 additional images from singly labeled cells. Images from triply labeled cells included the following: F1 excite-F1 emit; F1 excite-F2 emit (raw/uncorrected F1-F2 FRET); F1 excite-F3 emit (raw/uncorrected F1-F3 + F2-F3 FRET); F2 excite-F2 emit; F2 excite-F3 emit (raw/uncorrected F2-F3 FRET); and F3 excite-F3 emit. In addition to the raw, uncorrected FRET signals, these images also contained spectral bleed-through, which could be measured after collection of 6 analogous images from singly labeled cells. Step 2 involved removal of background noise from the raw images by the 3-color FRET software. Next, Venus-IQGAP1 (F2 excite-F2 emit) images were modified manually in step 3 to highlight two distinct cellular regions where IQGAP1 functions: peripheral, lamellipodia-like areas and sites of cell-cell contact. In non-contact regions IQGAP1 modulates plasma membrane protrusion during cellular motility and morphogenesis (Mataraza et al. 2003; Yamaoka-Tojo et al. 2004), whereas at cell-cell borders it helps to regulate intercellular adhesion (Kuroda et al. 1998). Using the 12 background-corrected images, at step 4 the software automatically selected, within the previously chosen peripheral and cell-cell border sites, 3x3 pixel regions of interest (ROIs) defined by having at least one pixel that displayed 2-color FRET interactions between all fluorescently tagged proteins known to bind directly to each other. Then (step 5), the software subtracted spectral bleedthrough, calculated E% and theoretical intermolecular distances, and produced the fully corrected images (Sun et al. 2010) of the ROIs shown in Figs. 4, 5, 7 and 8 and the graphical analyses shown in Figs. 6 and 9. Each E% value displayed in those figures corresponds either to the E% for the sole pixel within an ROI that displayed 3-color FRET, or to the average E% for all 3-color FRET pixels within an ROI that contained more than one such pixel.
A Leica TCS SP5 X confocal microscope was used to collect all light microscopic data, as described earlier (Sun et al. 2009). Specimens were placed in a temperature-controlled stage maintained at 37° in a chamber that maintained an atmosphere of 5% CO2 in air. All micrographs were acquired as 8-bit images at 512 × 512 or 1024 × 1024 pixels using a 60X, 1.2 NA water-immersion objective. Fluorophore excitation relied on a 458 nm argon laser line (Ex1) for Teal or CFP, a 514 nm argon laser line (Ex2) for Venus, and a 581 nm line produced by the tunable white laser for mCherry or mRFP1. Laser power was controlled through acousto-optical tunable filters (AOTF), was optimized for each excitation wavelength, and was kept constant within each individual 3-color FRET experiment. Three emission channels were defined using an acousto-optical beamsplitter (AOBS): 468–515 nm (Em1) for Teal or CFP, 525–585 nm (Em2) for Venus, and 595–750 nm (Em3) for mCherry or mRFP1. Fluorescence was detected by 3 identical photomultiplier tubes (1 per channel), using the same confocal pinhole, and gain and offset settings.
Micrographs were exported from the Leica SP5X as TIFF files and were processed by the aforementioned, proprietary 3-color FRET software (Sun et al. 2010), which yielded fully corrected, processed FRET (PFRET) images and data, E% images and inter-fluorochrome distances, and a spreadsheet file of all ROIs in each image. Separating ROIs into sub-populations, charts and statistics were performed using Excel spreadsheet software (Microsoft).
Supplementary Material
Acknowledgements
This work was supported by NIH grants NS051746 (GSB) and HL101871 (AP).
References
- Bashour A-M, Fullerton AT, Hart MJ, Bloom GS. IQGAP1, a Rac- and Cdc42-binding protein, directly binds and cross-inks microfilaments. J. Cell Biol. 1997;137:1555–1566. doi: 10.1083/jcb.137.7.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benseñor LB, Kan HM, Wang N, Wallrabe H, Davidson LA, Cai Y, Schafer DA, Bloom GS. IQGAP1 regulates cell motility by linking growth factor signaling to actin assembly. J Cell Sci. 2007;120(Pt 4):658–669. doi: 10.1242/jcs.03376. [DOI] [PubMed] [Google Scholar]
- Berney C, Danuser G. FRET or no FRET: a quantitative comparison. Biophysical Journal. 2003;84(6):3992–4010. doi: 10.1016/S0006-3495(03)75126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia S, Edidin M, Almo SC, Nathenson SG. Different cell surface oligomeric states of B7-1 and B7-2: implications for signaling. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15569–15574. doi: 10.1073/pnas.0507257102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt DT, Marion S, Griffiths G, Watanabe T, Kaibuchi K, Grosse R. Dia1 and IQGAP1 interact in cell migration and phagocytic cup formation. J Cell Biol. 2007;178(2):193–200. doi: 10.1083/jcb.200612071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs MW, Sacks DB. IQGAP proteins are integral components of cytoskeletal regulation. EMBO Rep. 2003;4(6):571–574. doi: 10.1038/sj.embor.embor867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill S, Li S, Lyman CW, Church DM, Wasmuth JJ, Weissbach L, Bernards A, Snijders A. The Ras GTPase-activating-protein-related human protein IQGAP2 harbors a potential actin binding domain and interacts with calmodulin and Rho family GTPases. Mol. Cell. Biol. 1996;16:4869–4878. doi: 10.1128/mcb.16.9.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan J, Coluccio LM. Kinetic analysis of F-actin depolymerization in the presence of platelet gelsolin and gelsolin-actin complexes. J Cell Biol. 1985;101(4):1236–1244. doi: 10.1083/jcb.101.4.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Elangovan M, Periasamy A. FRET data analysis: the algorithm. Chapter 7. In: Periasamy A, Day R, editors. Molecular Imaging: FRET Microscopy and Spectroscopy. New York: Oxford University Press; 2005. pp. 126–145. [Google Scholar]
- Elangovan M, Wallrabe H, Chen Y, Day RN, Barroso M, Periasamy A. Characterization of one- and two-photon excitation fluorescence resonance energy transfer microscopy. Methods. 2003;29(1):58–73. doi: 10.1016/s1046-2023(02)00283-9. [DOI] [PubMed] [Google Scholar]
- Fukata M, Kuroda S, Fujii K, Nakamura T, Shoji I, Matsuura Y, Okawa K, Iwamatsu A, Kikuchi A, Kaibuchi K. Regulation of cross-linking activity of actin filament by IQGAP1, a target for Cdc42. J. Biol. Chem. 1997;272:29579–29583. doi: 10.1074/jbc.272.47.29579. [DOI] [PubMed] [Google Scholar]
- Fukata M, Watanabe T, Noritake J, Nakagawa M, Yamaga M, Kuroda S, Matsuura Y, Iwamatsu A, Perez F, Kaibuchi K. Rac1 and Cdc42 Capture Microtubules through IQGAP1 and CLIP-170. Cell. 2002;109:873–885. doi: 10.1016/s0092-8674(02)00800-0. [DOI] [PubMed] [Google Scholar]
- Guthenberg C, Mannervik B. Glutathione S-transferase (transferase pi) from human placenta is identical or closely related to glutathione S-transferase (transferase rho) from erythrocytes. Biochim. Biophys. Acta. 1981;661(2):255–260. doi: 10.1016/0005-2744(81)90012-7. [DOI] [PubMed] [Google Scholar]
- Hart MJ, Callow MG, Souza B, Polakis P. IQGAP1, a calmodulin-binding protein with a RasGAP-related domain, is a potential effector for Cdc42Hs. EMBO J. 1996;15:2997–3005. [PMC free article] [PubMed] [Google Scholar]
- Higgs HN, Blanchoin L, Pollard TD. Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochem. 1999;38(46):15212–15222. doi: 10.1021/bi991843+. [DOI] [PubMed] [Google Scholar]
- Jares-Erijman EA, Jovin TM. FRET imaging. Nature biotechnology. 2003;21(11):1387–1395. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- Joyal JL, Annan RS, Ho Y-D, Huddleston ME, Carr SA, Hart MJ, Sacks DB. Calmodulin modulates the interaction between IQGAP1 and Cdc42. J. Biol. Chem. 1997;272:15419–15425. doi: 10.1074/jbc.272.24.15419. [DOI] [PubMed] [Google Scholar]
- Kuroda S, Fukata M, Kobayashi K, Nakafuku M, Nomura N, Iwamatsu A, Kaibuchi K. Identification of IQGAP as a putative target for the small GTPases, Cdc42 and Rac1. J. Biol. Chem. 1996;271:23363–23367. doi: 10.1074/jbc.271.38.23363. [DOI] [PubMed] [Google Scholar]
- Kuroda S, Fukata M, Nakagawa M, Fujii K, Nakamura T, Ookubo T, Izawa I, Nagase T, Nomura N, Tani H, et al. Role of IQGAP1, a target of the small GTPases Cdc42 and Rac1, in regulation of E-cadherin- mediated cell-cell adhesion. Science. 1998;281(5378):832–835. doi: 10.1126/science.281.5378.832. [DOI] [PubMed] [Google Scholar]
- Le Clainche C, Schlaepfer D, Ferrari A, Klingauf M, Grohmanova K, Veligodskiy A, Didry D, Le D, Egile C, Carlier MF, et al. IQGAP1 stimulates actin assembly through the N-WASP-Arp2/3 pathway. J Biol Chem. 2007;282(1):426–435. doi: 10.1074/jbc.M607711200. [DOI] [PubMed] [Google Scholar]
- Ma L, Rohatgi R, Kirschner MW. The Arp2/3 complex mediates actin polymerization induced by the small GTP-binding protein Cdc42. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(26):15362–15367. doi: 10.1073/pnas.95.26.15362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mataraza JM, Briggs MW, Li Z, Entwistle A, Ridley AJ, Sacks DB. IQGAP1 promotes cell motility and invasion. J. Biol. Chem. 2003;278(42):41237–41245. doi: 10.1074/jbc.M304838200. [DOI] [PubMed] [Google Scholar]
- Mateer SC, Bloom GS. IQGAPs: integrators of the cytoskeleton, cell adhesion machinery and signaling networks. Cell Motil. Cytoskel. 2003;55:147–155. doi: 10.1002/cm.10118. [DOI] [PubMed] [Google Scholar]
- Mateer SC, McDaniel AE, Nicolas V, Habermacher GM, Lin M-JS, Cromer DA, King ME, Bloom GS. The Mechanism for Regulation of the F-actin Binding Activity of IQGAP1 by Calcium/Calmodulin. J. Biol. Chem. 2002;277:12324–12333. doi: 10.1074/jbc.M109535200. [DOI] [PubMed] [Google Scholar]
- Mateer SC, Morris LE, Cromer DA, Benseñor LB, Bloom GS. Actin filament binding by a monomeric IQGAP1 fragment with a single calponin homology domain. Cell Motil. Cytoskel. 2004;58:231–241. doi: 10.1002/cm.20013. [DOI] [PubMed] [Google Scholar]
- McCallum SJ, Wu WJ, Cerione RA. Identification of a putative effector of Cdc42Hs with high sequence similarity to the RasGAP-related protein IQGAP1 and a Cdc42Hs binding partner with similarity to IQGAP2. J. Biol. Chem. 1996;271:21732–21737. doi: 10.1074/jbc.271.36.21732. [DOI] [PubMed] [Google Scholar]
- Miki H, Sasaki T, Takai Y, Takenawa T. Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature. 1998;391:93–96. doi: 10.1038/34208. [DOI] [PubMed] [Google Scholar]
- Periasamy A, Clegg RM. FLIM microscopy in biology and medicine. Boca Raton: Taylor & Francis; 2010. [Google Scholar]
- Rittmeyer EN, Daniel S, Hsu S-C, Osman MA. A dual role for IQGAP1 in regulating exocytosis. J Cell Sci. 2008;121(3):391–403. doi: 10.1242/jcs.016881. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Ho HY, Kirschner MW. Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4, 5-bisphosphate. J. Cell Biol. 2000;150(6):1299–1310. doi: 10.1083/jcb.150.6.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner MW. The interaction between N-WASP and the Arp2/3 complex links Cdc42-dependent signals to actin assembly. Cell. 1999;97(2):221–231. doi: 10.1016/s0092-8674(00)80732-1. [DOI] [PubMed] [Google Scholar]
- Roy M, Li Z, Sacks DB. IQGAP1 binds ERK2 and modulates its activity. J. Biol. Chem. 2004;279(17):17329–17337. doi: 10.1074/jbc.M308405200. [DOI] [PubMed] [Google Scholar]
- Roy M, Li Z, Sacks DB. IQGAP1 is a scaffold for mitogen-activated protein kinase signaling. Mol. Cell Biol. 2005;25(18):7940–7952. doi: 10.1128/MCB.25.18.7940-7952.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai-Yageta M, Recchi C, Le Dez G, Sibarita JB, Daviet L, Camonis J, D'Souza-Schorey C, Chavrier P. The interaction of IQGAP1 with the exocyst complex is required for tumor cell invasion downstream of Cdc42 and RhoA. J Cell Biol. 2008;181(6):985–998. doi: 10.1083/jcb.200709076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Booker CF, Kumari S, Day RN, Davidson M, Periasamy A. Characterization of an orange acceptor fluorescent protein for sensitized spectral fluorescence resonance energy transfer microscopy using a white-light laser. Journal of Biomedical Optics. 2009;14(5):054009. doi: 10.1117/1.3227036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Periasamy A. Additional correction for energy transfer efficiency calculation in filter-based Forster resonance energy transfer microscopy for more accurate results. Journal of Biomedical Optics. 2010;15(2):020513. doi: 10.1117/1.3407655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Wallrabe H, Booker CF, Day RN, Periasamy A. Three-color spectral FRET microscopy localizes three interacting proteins in living cells. Biophysical journal. 2010;99(4):1274–1283. doi: 10.1016/j.bpj.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Wallrabe H, Seo SA, Periasamy A. FRET microscopy in 2010: the legacy of Theodor Forster on the 100th anniversary of his birth. Chemphyschem. 2011;12(3):462–474. doi: 10.1002/cphc.201000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons M, Derry JM, Karlak B, Jiang S, Lemahieu V, McCormick F, Francke U, Abo A. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell. 1996;84(5):723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- Tomasevic N, Jia Z, Russell A, Fujii T, Hartman JJ, Clancy S, Wang M, Beraud C, Wood KW, Sakowicz R. Differential regulation of WASP and N-WASP by Cdc42, Rac1, Nck, PI(4,5)P2. Biochemistry. 2007;46(11):3494–3502. doi: 10.1021/bi062152y. [DOI] [PubMed] [Google Scholar]
- Wallrabe H, Bonamy G, Periasamy A, Barroso M. Receptor complexes cotransported via polarized endocytic pathways form clusters with distinct organizations. Molecular biology of the cell. 2007;18(6):2226–2243. doi: 10.1091/mbc.E06-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrabe H, Elangovan M, Burchard A, Periasamy A, Barroso M. Confocal FRET microscopy to measure clustering of ligand-receptor complexes in endocytic membranes. Biophysical journal. 2003;85(1):559–571. doi: 10.1016/S0006-3495(03)74500-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrabe H, Periasamy A. Imaging protein molecules using FRET and FLIM microscopy. Current opinion in biotechnology. 2005;16(1):19–27. doi: 10.1016/j.copbio.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Wang S, Watanabe T, Noritake J, Fukata M, Yoshimura T, Itoh N, Harada T, Nakagawa M, Matsuura Y, Arimura N, et al. IQGAP3, a novel effector of Rac1 and Cdc42, regulates neurite outgrowth. J Cell Sci. 2007;120(Pt 4):567–577. doi: 10.1242/jcs.03356. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Wang S, Noritake J, Sato K, Fukata M, Takefuji M, Nakagawa M, Izumi N, Akiyama T, Kaibuchi K. Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev. Cell. 2004;7(6):871–883. doi: 10.1016/j.devcel.2004.10.017. [DOI] [PubMed] [Google Scholar]
- Weissbach L, Bernards A, Herion DW. Binding of myosin essential light chain to the cytoskeleton-associated protein IQGAP1. Biochem. Biophys. Res. Commun. 1998;251(1):269–276. doi: 10.1006/bbrc.1998.9371. [DOI] [PubMed] [Google Scholar]
- Weissbach L, Settleman J, Kalady MF, Snijders AJ, Murthy AE, Yan Y-X, Bernards A. Identification of a human rasGAP-related protein containing calmodulin-binding domains. J. Biol. Chem. 1994;269:20517–20521. [PubMed] [Google Scholar]
- Yamaoka-Tojo M, Ushio-Fukai M, Hilenski L, Dikalov SI, Chen YE, Tojo T, Fukai T, Fujimoto M, Patrushev NA, Wang N, et al. IQGAP1, a Novel VEGF Receptor Binding Protein Involved in Reactive Oxygen Species-dependent Endothelial Migration and Proliferation. Circ. Res. 2004;95:276–283. doi: 10.1161/01.RES.0000136522.58649.60. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.