Abstract
Monoclonal antibodies (mAbs) are powerful therapeutics, and their characterization has drawn considerable attention and urgency. Unlike small-molecular drugs (150-600 Da) that have rigid structures, mAbs (~150 kDa) are engineered proteins that undergo complicated folding and can exist in a number of low-energy structures, posing a challenge for traditional methods in structural biology. Mass spectrometry (MS)-based biophysical characterization approaches can provide structural information, bringing high sensitivity, fast turnaround, and small sample consumption. This review outlines various MS-based strategies for protein biophysical characterization and then reviews how these strategies provide structural information of mAbs at the protein level (intact or top-down approaches), peptide, and residue level (bottom-up approaches), affording information on higher order structure, aggregation, and the nature of antibody complexes.
Keywords: Monoclonal Antibody, Mass Spectrometry, Native ESI, Top-down and Bottom-up, Protein Footprinting, hydrogen/deuterium exchange, FPOP, ion mobility
1. Therapeutic monoclonal antibodies (mAbs)
Therapeutic mAbs may have become the most popular drug candidates following their introduction into the clinic in the late 1980s [1]. Their high specificity and low side effects make mAbs powerful human therapeutics for oncology, autoimmunity/inflammation, infectious diseases, and metabolic disorders [2]. At present, approximately 30 therapeutic mAbs are being marketed. The sales contributed approximately $18.5 billion to the US economy in 2010 [1]. The high demands for new therapeutic mAbs have trigged a burst of mAb-based drug development. For example, 16 human mAbs entered the clinic during 1985-1996, whereas during 1997-2008, 131 human mAbs became available [3]. In 2011, more than 300 mAb-based thereapeutics were in clinical trials [2]. As older mAbs come off patent and go into production as generic drugs, the need for characterizing their higher order structure in quality control becomes even more important, motivating this review.
1.1 Introduction to mAbs
Therapeutic mAbs are glycoproteins that belong to the immunoglobulin (Ig) family. Ig’s are used by the immune system to identify and neutralize foreign organisms or antigens [4,5]. Ig’s are classified in five groups, IgA, IgD, IgE, IgG and IgM (as α, δ, ε, and μ), based on the structure of their constant regions [6]. At present, most approved mAbs are from IgG’s (γ-immunoglobulin). IgG’s have the typical “Y”-shaped structure comprised of two identical heavy and light chains (H and L chains) (Figure 1). All heavy and light chains are covalently linked by disulfide bonds. IgGs can be further classified into four groups, IgG1, 2, 3, and IgG4 (as γ-1, γ-2, γ-3 and γ-4) on the basis of different patterns of inter-chain disulfide bonds and heavy-chain sequences. IgG1, 2 and 4 are widely used in therapeutics, whereas IgG3, which has a shorter serum half-life, is rarely used.
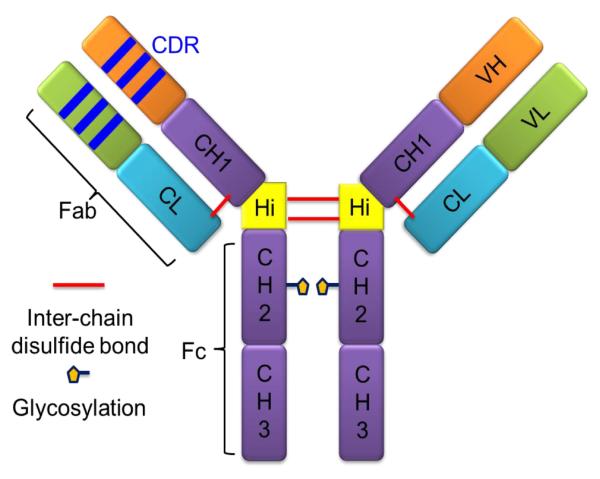
Figure 1.
IgG structure (IgG1). The global structure of IgG1 has two identical heavy chains and light chains. Four chains are attached covalently with inter-chain disulfide bonds and also by non-covalent interactions. The two constant regions from heavy chains (CH2 and CH3, Fc regions) respond to the binding to Fc gamma and FcRn receptors. The variable region from both light and heavy chains contains antigen-binding regions (CDRs). Variable regions with the close constant region together are called the Fab region. The Fab and Fc region are linked by the hinge region in the heavy chain. In the IgG1, there is a glycosylation site on the second constant region (CH2).
Each heavy chain contains one variable (VH) and three constant domains (CH1, CH2 and CH3), whereas each light chain contains one variable (VL) and one constant domain (CL). In the heavy chain, CH1 and CH2 are linked by a hinge region that contains inter-heavy chain disulfide bonds (IgG1 and IgG4 have two disulfide bounds in hinge region, whereas IgG2 has four). Antigen binding is mediated by the variable region, mainly by three loops connecting individual β-strands, which are called the complementarity determining regions (CDRs), from both heavy and light chains. Upstream of the heavy chain (VH and CH1) is the disulfide-bond-linked light chain (VL and CL), known as the fragment antigen-binding (Fab) region. The downstream constant regions (CH2 and CH3) of the heavy chain are called the fragment crystallizable (Fc) region, which is responsible for effector function during recycling [7].
A milestone in the development of therapeutic engineered mAbs was the introduction of murine mAbs from hybridomas [8]. Clinical applications of murine mAbs (suffix: -omab) began in the late 1980s [9]. Dissimilarities between murine and human immune systems led to clinical failure of those antibodies. Murine antibodies are engineered to generate chimeric mouse-human mAbs (~65% human in molecules, suffix: -ximab) by fusing the murine-variable regions onto human-constant regions [10]. The humanized mAbs (~95% human in molecules, suffix: -zumab) are produced by grafting murine hyper-variable regions on amino acid domains of human antibodies [11]. Both chimeric mouse-human and humanized mAbs have reduced immunogenicity and increased serum half-life [12]. With the development of phage-display technology and various transgenic mouse strains expressing human variable domains [13,14], fully human mAbs (suffx: -umab) with significantly reduced immunogenic potential and high similarity to human endogenous IgGs, have become rich sources of new therapeutics [1,3,15].
1.2 The challenge of verifying higher order structure of therapeutic mAbs
Unlike traditional small molecular drugs (150-600 Da), mAbs are large macromolecules (~150 KDa) with four polypeptide chains held in place by tens of inter- or intra-disulfide bonds as well as by non-covalent interactions [6]. For example, one approved therapeutic mAb, trastuzumab, has 6560 carbon atoms, 10132 hydrogens, 2090 oxygens, 1728 nitrogens and 44 sulfur atoms [16]. The functional form of the protein depends on its higher order structure (HOS), referring to the tertiary 3-D architecture determined by the secondary alpha-helices and beta-sheets, building upon the primary structure, and the quaternary complex formed by interacting/binding with other entities. Sources affecting the HOS of mAbs are not limited to primary structures. Variations in PTMs (post-translational modifications), mutations and modifications can trigger changes in HOS to affect binding to an antigen or to Fc-gamma and Fc-Rn receptors. Production and storage of therapeutic mAbs can introduce significant changes of HOS. From the view of patient health, HOS variations of these proteins can pose serious safety issues [17,18]. HOS can be fleeting for proteins; HOS is certainly more dynamic than primary structure. Although strategies to determine the primary structure of mAbs, including mutations, PTMs, and other modifications, have been available for decades, approaches to verify HOS are still needed. Although circular dichroism (CD), fluorescence and related optical spectroscopic methods are used for rapid HOS characterization [19], many regional but important structural changes are missed [20]. In the recently published draft guidelines for quality control of biosimilars (copies of therapeutic mAbs that are coming off patent), the US Food and Drug Administration (FDA) acknowledged that “a protein’s three-dimensional conformation can often be difficult to define precisely using current physicochemical analytical technology”. New approaches are under development to meet this challenge [16,20]. In this review, we focus on the new and promising MS-based biophysical approaches as means for characterization of mAb HOS.
2. Mass spectrometry based protein biophysics
2.1 Overview
The advantages of MS in biology are attracting the attention of structural biologists who address the biophysical properties of proteins [21]. Modern MS instrumentation and proteomics methods offer two major approaches to interrogate protein biophysics. One is an intact or top-down approach that employs native electrospray ionization (ESI), ion-mobility measurements, and fragmentation, usually by interaction with electrons, to provide a global view of the protein of interest [22]. The other is a bottom-up approach combining either protein footprinting [23] or cross-linking [24] that provide detailed peptide and even amino-acid-residue information. These terms “top down” and “bottom-up” first appeared in the MS-based proteomics literature [25]. Compared to bottom-up, top-down is less mature, requiring the invention of electron capture dissociation (ECD) [26] in 1998. Both top-down and bottom-up MS approaches have the advantages of small sample consumption, nearly no limit to protein size, and ability to determine in the gas phase the native or near-native protein properties. Furthermore, MS can be combined with protein footprinting to give an approach that is tolerant to solution media containing MS-unfriendly small molecules. Native ESI and top-down sequencing offer high throughput and unique specificity for oligomeric states and stoichiometry of native protein samples. By combining these complementary MS methods, important structural information can be discovered with intermediate structural resolution. We will review the principles of both approaches in the following subsections.
2.2 Intact and top-down based approach
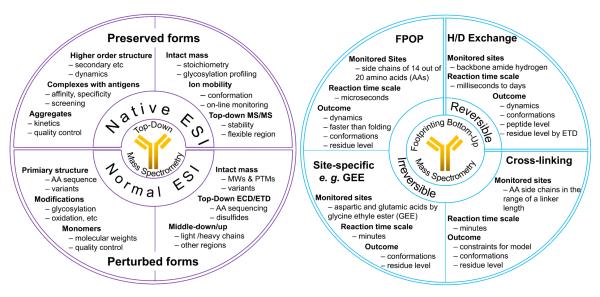
At present, proteins and protein complexes with MW even at the mega-Dalton range can be directly analyzed by MS [27]. Top-down protocols provide information without requiring proteolytic digestion of protein samples prior to MS analysis [28]. Removing the digestion step should significantly reduce analysis time. Intact proteins and protein complexes are then interrogated close to their functional forms, even as protein assemblies [29]. Species existing in different oligomeric states or with PTMs can be analyzed separately. Although targeted analysis (e.g., oligomer specific analysis) can also be accomplished by bottom-up approaches by adding a pre-separation step, top-down approaches are more efficient. Using an approach targeting intact proteins, we can directly monitor the charge-state distribution and obtain stoichiometry or measure ion mobility of intact proteins or of protein complexes and capture some information about shape and changes in shape by keeping the protein in near-native states in the gas phase. Tandem MS capabilities available on most commercial MS instruments can also be employed to elucidate the conformations by fragmenting portions of a protein or protein complex and interpreting the decomposition reactions. An overview of top-down MS strategies is summarized in Figure 2A where native ESI in the upper half preserves the protein structure; while normal ESI in the lower half deals with proteins that are denatured, and such proteins can be studied further by limited or specific proteolysis to generate large peptide fragments (middle-out) to improve the sequence coverage by MSMS in a way of middle-down/up that is similar to bottom-up approach but for large peptides.
Figure 2.
Overview of top-down and bottom-up MS based protein biophysical studies. The left circle is the summary of top-down approaches. The right circle is the summary of bottom-up approaches.
2.2.1 Native MS (or Native ESI)
Native MS has proved to be an alternative strategy to investigate structure in the near-native state of intact proteins and their complexes by using MS platforms that work for gas-phase species [22]. Prior to conducting an MS experiment, the original buffer solution that maintains the protein native structure is exchanged with a volatile ammonium acetate solution that mimics the native buffer but favors solvent (and salt) evaporation during ESI to release protein ions that bear a memory of the structure in solution [30]. In this way, the ionized proteins carry less charge (are less protonated on the surface) than those generated by normal ESI, which utilizes denaturing solutions comprised of water and organic solvents at low pH. As a result, the ions seen in the mass spectrum produced by a native ESI experiment appear at higher m/z.
The non-covalent interactions within a protein and between subunits of protein assemblies can also be preserved in the gas phase [29]. It is sometimes relatively straightforward to determine the stoichiometry of a complex by using native MS. More importantly, variations within proteins or protein complexes can be monitored based on their charge-state distribution in native MS [31]. Highly ordered or compact proteins and protein complexes have smaller surface areas than disordered or unfolded ones. This difference can be directly read out from the charge-state distribution [32]. Following the introduction of protein ions into the gas phase, they can be interrogated by ion mobility or tandem MS to obtain structural information, all in a top-down manner [33,34].
2.2.2 Ion mobility measurements
Gas-phase ion mobility provides a collisional cross-section (CCS) of a particular ion drifting through in inert buffer gas in a low electrical field [35]. In the drift region, an ion experiences many collisions with inert buffer gas molecules. The upshot of this ion drift is a two-dimension projection (or CCS) of the three-dimension shape of the ion in free rotation. The readout is the time for the ion to pass through the drift region (called an arrival time distribution (ATD) or drift time). Ions with a large cross section drift more slowly, affording an outcome that is similar to that of a native-gel experiment. Thus, proteins and protein complexes can be separated based on the difference of their native states. An important application of ion mobility is to provide experimental evidence on native protein conformation, which appears to be preserved, at least in part, in the gas phase when the protein is introduced by native MS [36]. Mobility can be applied to differentiate the structure of two ions of the same m/z, thus providing information on shape and size in the gas phase [37]. Robinson and coworkers [38,39] have used native ESI and ion mobility to establish a calibration curve for the CCS of proteins and protein complexes. Furthermore, these approaches have also facilitated the investigation of large membrane-embedded protein complexes [40].
2.2.3 Top-down fragmentation
MS/MS-based fragmentation of protein ions affords information on primary structure. A number of approaches can be used to activate ions: collisionally activated dissociation (CAD), sometimes called collision-induced dissociation (CID) [41], electron-capture dissociation (ECD) [26], electron-transfer dissociation (ETD) [42], surface-induced [43] dissociation, and photon-induced dissociation [44] . Top-down fragmentation in this context provides structural connectivity and compositions of the protein or protein assembly [34]. As exemplified by its applications in proteomics [45], top-down sequencing overcomes a disadvantage of bottom-up in which the digestion of the starting proteins leads to a loss of information about protein isoforms, especially for PTMs.
Thus far, CAD/CID in a quadruple/time-of-flight (TOF) instrument has played the dominant role for in characterization of native protein complexes; it reveals the stoichiometry and topology of a protein assembly [46]. Electron-capture dissociation (ECD) in a Fourier transform ion cyclotron resonance (FTICR) mass spectrometer can identify flexible regions of a protein [47] to reveal isomerization of small proteins from solution to the gas phase [48]. Surface-induced dissociation (SID) tends to release and to distribute charges symmetrically among the fragmented subcomplexes [49].
2.3 Bottom-up approaches
In bottom-up approaches, proteins are digested into peptides before MS analysis. Conformational information of a protein must be encoded into peptides beforehand. This encoding can be achieved by labeling solvent-accessible amino acid side chains or backbones. The labeling reagent can involve one reactive group that attaches the reagent to various amino acids (protein footprinting), or have two reactive groups that form a linkage between two amino acids of a protein (protein crosslinking).
2.3.1 Protein footprinting
Footprinting examines ligand binding and conformational changes by determining the solvent accessibility of macromolecules through their sensitive responses to chemical or enzymatic modification and cleavage reactions [50]. Protein footprinting as a complementary approach to probing protein conformation has rapidly developed during the last decade [51]. Footprinting strategies are labeling approaches that include amide hydrogen deuterium exchange (HDX) and hydroxyl-radical based labeling; they are becoming sufficiently characterized that they can have wide usage. MS-based protein footprinting approaches can be classified into two groups: reversible and irreversible, as is summarized in Figure 2B. Protein footprinting can also be combined with top-down MS, as was shown recently [52,53].
In this review, we focus on the original bottom-up approach for protein footprinting.
A. Reversible HDX footprints therapeutic proteins
The hydrogens of solvent-accessible amino-acid backbones and side chains can exchange with deuterium when a protein in normal water is diluted into a buffer containing D2O, initiating deuterium uptake by the amides and other active sites of the protein. The mass shift induced by deuterium uptake can be monitored by MS to reveal some structural features of a protein [54]. The resolution of the HDX MS platform can be further improved by adding proteolytic digestion accompanied by LC separation [55]. At present, even super protein complexes, like viral capsids and E coli ribosome, have been analyzed by HDX MS [56,57], well beyond the capabilities of NMR, which was the dominant tool in the early development of HDX.
There are three types of hydrogens in proteins: those on side-chain carbons, on heteroatoms, and on the backbone amide linkages. There is no measureable exchange for the carbon hydrogens, whereas the exchange rate for most hydrogens on heteroatoms is too fast to be followed during normal HDX LCMS experiments (except for those on histidine [58]). Those on the backbone amides of all amino acids (except proline) exchange at measurable rates. The amide hydrogen exchange can be significantly slowed (effectively quenched) such that the rate constant decreases by 10000 times when the conditions (pH 7 at 25 °C) are changed (pH 2.5 at 0 °C) prior to MS analysis [59]. The quenching condition (pH 2.5) is compatible with denaturing positive-ion electrospray ionization (ESI), making MS a good detector for the outcome of HDX. More importantly, mass shifts (induced by deuterium uptake) instead of peak intensities are measured in MS based HDX, which avoids the problems of changes in ionization efficiency for the various constituent peptides. Nevertheless, sample preparation and analysis of the peptides to give “regional information” still must be done quickly (e.g., < 10 min), limiting chromatographic resolution and restricting the use of complex matrices or samples for study.
B. Irreversible footprinting approaches
For irreversible approaches, the labeled protein sample can survive extensive separation and purification after the labeling experiment, whereas reversible HDX suffers back exchange. The labeling approaches can be relatively general or site-nonspecific [60] (e.g., hydroxyl radical labeling), or site-specific (such as carboxyl group labeling).
Hydroxyl radicals footprint therapeutic proteins
Radicals are usually highly reactive and have a short life time, making them good labeling reagents in protein footprinting. The generation and control of radicals, however, are not easy. Although development of other radical besides •OH as protein footprinting reagents have been reported [53,61], the most popular radical in protein footprinting remains the hydroxyl radical, which is similar in size as water molecules and is highly reactive toward approximately two thirds of the amino-acid side chains.
Hydroxyl radicals can be generated by electron-pulse radiolysis, synchrotron radiolysis of water, laser photolysis of hydrogen peroxide, Fenton and Fenton-like reactions, and high-voltage electrical discharges [62]. Although Fenton and Fenton-like reactions were used early on for protein footprinting [63], the speed of Fenton and Fenton-like reactions is relatively slow (minutes). To speed up the process and avoid label-induced unfolding, synchrotron radiolysis of water [64] and the laser photolysis of hydrogen peroxide to make radicals [65,66] are the most promising. The photolysis of water in the kilovolt X-ray range causes water to ionize and lose a proton to give hydroxyl radicals [67]. No reagent besides the solvent water is required in this experiment. The reaction time can be controlled by irradiation time. Chance’s group developed a systematic approach that uses synchrotron light source, found in national labs, for protein footprinting studies [68]. Recently, that group investigated the water distribution in the membrane-embedded channel complexes [69]. The access to synchrotron light sources, however, limits the general application of this method. Laser photolysis of hydrogen peroxide is an alternative that can be set up in most chemistry laboratories.
We developed a laser photolysis approach, which we call Fast Photochemical Oxidation of Protein (FPOP), to form •OH, in a few-nanosecond, 248 nm laser pulse that photolyzes low concentrations of hydrogen peroxide (0.04%, 15 mM) in a flowing solution containing the protein of interest. FPOP limits the radical lifetime by using a scavenger (free amino acids, like glutamine or histidine) to ensure the labeling reaction takes place within approximately 1 μs [70-72]. We took advantage of the fast labeling of FPOP to study fast protein folding by a “pump-probe” method whereby we use two lasers, one to supply a temperature jump, and the second to generate hydroxyl radicals that footprint the protein as a function of its folding time (hundreds of microsec) [73].
Glycl ethyl ester and other reagents label proteins in a site-specific manner
A variety of chemical reagents can be used to modify specifically certain amino acids [23]; an example is the carbodiimide-mediated coupling reaction between glycine ethyl ester and the carboxyl groups of a protein [74]. Another example of a reactive amino acid group is the thiol of cysteine, which can be modified by several reagents (e.g., iodoacetamide, NEM [75]). Major drawbacks of site-specific labeling are that the labeling is slow relative to FPOP, and less conformational information can be obtained because the target residues on the surface are limited in number (i.e., the method has limited structural resolution). Site-specific labeling is recommended for studies of very complicated systems that have hundreds of target residues. An advantage is the data analysis, identifying and quantifying labeled products, can be simplified because the target list can be narrowed. We reported several examples that utilized site-specific labeling for the study of complicated systems like membrane-embedded protein complexes implicated in photosynthesis and cancer [76,77].
2.3.2 Protein cross-linking
The chemical-labeling attribute of protein footprinting can be extended to chemical cross linking by using bi-functional labeling reagents (e.g., N-hydroxysuccinimide esters). Chemical cross-linkers can modify two amino-acid side chains within a designed distance and form a covalent linker between the two residues, provided the residues are with the distance constraint [24,78,79]. The cross-linked peptides can be identified by LC-MS/MS after proteolytic digestion. Information about the distance between the two residues, defined by the length of cross-linker reagent, can be used to determine adjoining proteins in a protein-protein interaction or to locate within a protein two interacting domains. With the development of new isotope-encoded linkers, cleavable linker, and tagged linkers [80], protein crosslinking has become a more effective tool to determine sites of protein-protein interaction in complicated biological systems.
3. MS-based characterization of antibodies
The higher order structural variations of mAbs and their dynamics must be addressed during discovery and development as therapeutics. Many approaches taken from structural biology can be applied [16,19,20]. Traditional biophysical techniques, like X-ray crystallography (X-ray), nuclear magnetic resonance (NMR) and cryo-electron microscopy, are hampered by the size of the protein, the amount available, and the need to determine dynamics of mAbs. MS has become an essential analytical tool for the therapeutic mAb development owing to its superior resolution and speed, allowing it to monitor primary structure and locate post-translational modifications (PTMs) [6,81]. Variations of primary structure, including those of disulfide-linkage location, amino-acid sequence, PTM location, and other in-storage modifications can be determined by MS in all phases of mAb production. Although MS-based structural approaches are still limited by the lower resolution compared to NMR and X-ray, they are more efficient because they have high sensitivity, can be applied to proteins in various environments, and have high throughput.
Applications by MS are rapidly growing [21,82]. Currently, many MS-based biophysical approaches focus on differentiation of mAb isomers [83,84]. Those isomers could arise as a consequence of primary structural variations or variables in production and storage. In this section, we review MS strategies that can address three important and challenging issues: quality control of high order structure (HOS), assessment of aggregation, and mapping of antibody-antigen interfaces. To provide future perspective, we include several new developments in the MS-based biophysical studies to demonstrate their potential for mAb characterization.
3.1 Higher order structure (HOS)
3.1.1 Intact and top-down approaches
The observation of intact mAbs in native MS is the most direct measurement of an antibody. We now can introduce mAbs into the gas phase with minimal perturbation of their native conformation. As a simple first approach, we can follow the lead of Kaltashov et al. [85]. Who reported that protein conformational variations can be directly observed by observing their charge-state variations when introduced to the mass spectrometer by native MS. This approach is recommended as an early one to apply for quality control of mAb HOS [86].
Combining native MS and ion mobility provides a simple and direct shape/size measurement of mAbs. Structural information obtained from ion mobility refers to the global conformations of species even with the same MW. The first demonstration of ion mobility for antibodies was by Bagal et al [87] to differentiate, using native ESI, the conformations of IgG2 isomers caused by disulfide linkage variations. They used IgG1 as a control because it does not have S-S isoforms, and found that IgG2 has a longer drift time than IgG1 and shows two distinct peaks in the ATD of each charge state. They concluded that these two peaks correspond to IgG2-A (shorter time) and IgG2-B forms, respectively. The double peaks were not caused by glycosylation, which was confirmed by redox-enrichment of A and B after refolding in the presence and absence of GuHCl. Heck and coworkers [88] used native MS to monitor the CH3 domain swapping between human IgG4 molecules, a process related to spontaneous Fab-arm exchange to form bi-specific antibodies [89]. Beck and coworkers [90] monitored the dynamics of this process by using native MS combined with time-resolved ion mobility; their results demonstrate the high potential of IM-MS for characterizing biopharmaceutical protein products.
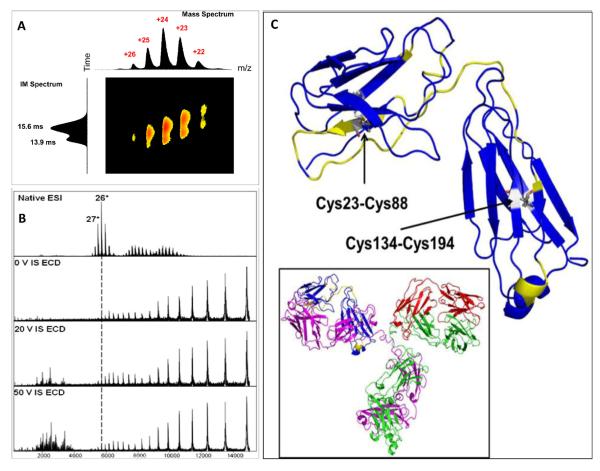
We in collaboration with colleagues at Pfizer [91] used a strategy combining native ESI, ion mobility, ECD top-down, and hydroxyl-radical footprinting (FPOP) to characterize five IgG2 disulfide isomers including the wild type. Ion mobility showed two major drift time peaks for the WT isoform, consistent with Bagal’s observation [87]. The shorter drift time peak of the WT is approximately the same as those of the other four mutants. ECD in a top-down mode sequenced a flexible portion of structure, providing a result that is consistent with the FPOP results (Figure 3).
Figure 3.
Native ESI, IM and ECD mass spectra of WT IgG2, (a) ion mobility separation, (b) ECD top-down with in-source activation, and (c) highlighted region in yellow of light chain in CDR showing ECD cleavage sites. (Copied with permission from JASMS)
3.1.2 Bottom-up approaches
At present, the most used bottom-up approach for characterizing mAb HOS is HDX. Most HDX studies differentiate mAb structural variations and provide evidence that the HOS of an unknown protein is that of a reference (i.e., both give nearly identical HDX kinetics and extents (footprints) at the peptide level). Differences in HDX pinpoint regions that have changed their HOS. Summaries of protocols for HDX MS applications can be found in recent publications [92,93]. Here, we describe several examples that demonstrate how HDX MS provides HOS information of mAbs.
Glycosylation changes antibody behavior, increases solubility, imparts longer shelf-life, increases resistance to unfolding and proteolysis, and lowers aggregation rates [94]. Little is known about the relevant structural changes caused by glycosylation. Houde et al. [95] utilized HDX MS to probe the conformational changes for IgG1 with and without glycans. HDX MS can also be used to probe structural variations and receptor binding induced by other PTMs (e.g., methionine oxidation and fucosylation) [96,97]. One major mAb isoform is a charge variant (or charge heterogeneity) [98] designed to maintain optimized electrostatic interactions of the favored structure and have the desired reactivity. Modification, degradation, and covalent adduction can result in net changes of the positive or negative charge (pI value) of a mAb, and ultimately change its structure and stability. Tang et al. [99] demonstrated how HDX and an extended approach to give affinity (i.e., SUPREX) can characterize conformation and stability of charge variants of human IgG1.
Variations of HOS may also result from storage of mAbs. Furthermore, effects of excipients used in the therapeutic preparation also need to be evaluated as to their effect on the stability of mAbs. Manikwar et al. [100] reported the use of HDX MS to measure local dynamics of mAbs stored in different environments. This approach can be extended to examine the effects of salts on stability, aggregation propensity, and local flexibility of mAbs [101].
Hydroxyl-radical-based protein footprinting methods are a more recent development than HDX, but they can also be applied to protein therapeutics to afford information similar to that of HDX. Watson et al. [102] demonstrated the use of FPOP in structural studies of the protein therapeutic, granulocyte colony-stimulating factor (GCSF). Recently, we reported the application of FPOP for the structural characterization of conformational differences of mAb IgG2 isomers. We found that the FPOP results are consistent with top-down ECD data and point to flexible regions of the protein. Furthermore, FPOP identifies local conformational changes and reveals solvent accessibility in the CDR [91]. Thus, FPOP complements HDX but may be more reliable and versatile because it imparts an irreversible change to the protein on the microsecond timescale. Moreover, analysis of the outcome of free-radical footprinting can take advantage of advances in analytical proteomics. Radical-labeling outcomes are a measure of solvent accessibility, and the speed of FPOP can be used to locate dynamic or flexible regions of a protein. Other slower labeling approaches including HDX give a time-averaged view.
Site-specific labeling can also measure solvent accessibility of certain residues. Zhang et al. [103] applied the specific labeling reagents, sulfosuccinimidyl acetate, for lysine and p-hydroxylphenylglyoxal for arginine, to reveal a positive-charge patch on an antibody. Free mAbs and a resin-bound antibody were labeled and analyzed by LC-MS/MS. The charge distributions as well as the antibody-resin binding interface were successfully located.
3.2 Protein Aggregation
Antibody aggregation is a common problem occurring in protein manufacturing and storage [104]. The importance arises because the functional form of a protein is often the monomer, whereas higher oligomers not only reduce the dose efficiency but also pose toxicological problems. This is a serious issue facing the biotechnology and pharmaceutical industries [105]. Understanding the mechanism of aggregation is important, therefore, not only for manufacturing and storage of proteins but also for shedding light on protein aggregation in general. It is not surprising that protein aggregation is attracting a considerable attention given that serious problems in human health can be caused by protein aggregation. The analytical tools used for analysis of protein aggregations in neurodegenerative disease were recently reviewed [106], and they may also be applicable to the problem of antibody aggregation.
3.2.1 Intact and top-down approaches
Native ESI can be used to analyze monomeric antibodies and their associated glycosylated forms. Given the narrower charge-state distribution produced by native ESI, ion signals are more concentrated than in normal ESI. Thus, the sensitivity for the low-abundant modified forms becomes higher. The Heck group in the Netherlands has observed IgG4 oligomers up to tetramers by using this approach and even monitored the dynamics of IgG4. When size-exclusion chromatography (SEC) is integrated with native ESI, and combined with fractionation, the antibody aggregates can be separated and analyzed in more detail [107].
3.2.2 Bottom-up approaches for aggregation
Understanding the mechanism of aggregation and locating the aggregation interface are vital to develop preventive measures. We [108] developed a pulsed HD exchange labeling strategy that can monitor the conformational changes during aggregation processes of Abeta-42 peptide, adding a new approach to the “toolbox” for protein aggregation studies. The approach should have applicability for study of aggregation of other proteins, possibly including antibodies. The advantages of this new platform are that no modification (e.g., addition of a fluorophore) of the protein is needed, information can be achieved at the peptide level, and factors that affect aggregation can be readily evaluated for following the HDX patterns.
Cross-linking can also be applied to proteins in different oligomeric states collected after SEC separation. Identification of resulting linked peptides by bottom-up MS pinpoint the interface and interaction regions.
Freeze-thaw stresses induce protein aggregation. Zhang et al. [109] reported the use of MS-based HDX to assess the impact of freeze-thaw cycling on protein structure, demonstrating that the aggregation mechanism of mAbs under thermal and freeze-thaw stresses can be revealed by HDX MS [110]. Engen’s group [111] used H/DX and other biophysical measurements to compare the monomeric and dimeric forms of two mAbs, respectively, to connect aggregation with the structural changes in antibodies, They found that the dimerization has no effect on the deuterium uptake between monomer and dimer forms of one of the mAbs. However, the other mAb monomer showed subtle changes in HDX in the CH 2 domain and the hinge region between CH 1 and CH 2 domains, as compared with its dimer form.
Cold storage of proteins, particularly with traces of hydrogen peroxide carried over from FPOP, for example, can lead to protein oxidation [112]. Usually removal by various solid-phase desalting methods, catalase treatment, or freeze-drying after protein footprinting is critical to insure no uncontrolled oxidation, which does occur even at −80 °C. Taking a positive view of this phenomenon, we see possibilities to use cold chemical oxidation to bring insights in a manner similar to FPOP or Fenton chemistry to protein aggregation occurring in cold storage.
The combination of size-exclusion chromatography and hydroxyl radical labeling should be considered for the characterization of mAb dimers [113]. The small amount of dimer in the mAb product can be separated by SEC and labeled by hydroxyl radicals generated via synchrotron irradiation (ionization of water). The dimer interface as well as the dimer orientation can be elucidated by this footprinting and site-specific digestion[113].
Gu and coworkers [114] used cross-linking to probe the interaction of the Fc moiety of a mAb expressed in Chinese hamster Ovary cells. They formed aggregates by incubation at 40 °C for 6 h, followed by reaction with the bifunctional crosslinker BS3. Their results suggest the interaction occurs at three Fc molecules in the CH2 and CH3 domains, indicating not only the Fab region but also the Fc region can aggregate.
3.3 mAb complexes
3.3.1 Intact and top-down approaches
Antibody aggregates can be treated as homogeneous complexes whereas antibody/antigen complexes are heterogeneous. Because of the fast turnaround of native MS, it can be used to screen small–molecules for drug candidates. For mixtures of small molecules, the stoichiometry, relative intensity, and affinity of mab/Ag complexes can be obtained in one experiment. The relative intensity should reflect the relative abundance of a complex, which can be used for relative quantification, assuming there is no discrimination in the native spray. Klassen and coworkers [115] have extended their program on protein/carbohydrate complexes to assess antigen-binding fragments in an assay of carbohydrates. Combining this approach with ECD top-down sequencing should locate the binding pocket in such systems.
3.3.2 Bottom-up approaches
Antibody-antigen interactions are the core function of mAb therapeutics. Understanding this interaction and locating the interface (epitopes) are crucial in mAb design. Epitope mapping is also involved in the patentability and protection of intellectual property [93]. Footprinting by HDX and MS and comparing the outcomes of free antigen and antibody-antigen complex can locate the epitope binding region. A number of examples of this approach were reported [109,116-119]. Similar approaches should be possible with FPOP, and an example is illustrative, underscoring the advantage of irreversible labeling for epitope mapping [120]. One should be cautious in the interpretation of epitope mapping data by FPOP or HDX. The changes induced by antigen binding may be away from the epitope binding, resulting from remote or allosteric effects [121], and these changes will also be picked up. Furthermore, the binding imparts thermodynamic stability to the protein that may cause overall protection. Complementary information from other techniques, like modeling, site-directed mutagenesis, and x-ray [119], should help resolve these issues.
4. Conclusions
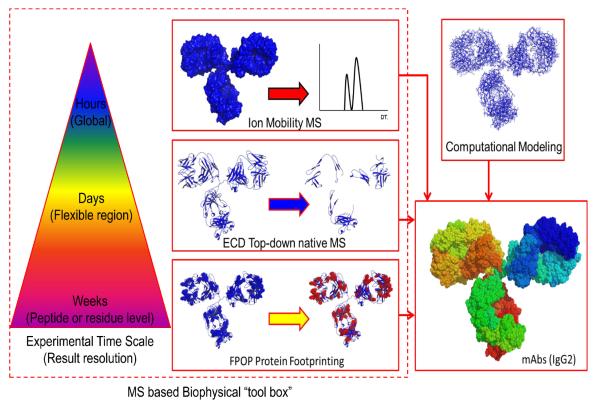
Recently, mass spectrometry has seen dramatic evolution in instrumentation and method. In the meantime, the biotechnology and pharmaceutical industries are in a stage of expansion that requires new analytical means to characterize their protein products at each step from preparation to storage and shipping. Mass spectrometry is poised to address higher order structure, aggregation, and binding with antigens, providing a powerful physicochemical approach that spins off its development for protein biophysics and structural biology. MS information can serve as guideline for production of the next generation of engineered therapeutic proteins. No single method, however, can provide complete information of a complex protein system. Rather, utilization of combined methods (Figure 4) including native MS, top-down and bottom-up sequencing, footprinting, and modeling should be effective in generating a full picture of a protein.
Figure 4.
The MS based biophysical “tool box” in characterizations of IgG2 with different S-S bond networks. This is an example of combining multiple approaches, not limited to MS, to provide complementary structural information of mAbs. The experimental time scale and structural resolution are labeled.
For the future, the technological advances of mass spectrometry will continue, driven in part by the complex problems in biological sciences. For example, the recently modified Orbitrap Q-Exactive instrument has demonstrated high sensitivity and resolving power for large proteins and protein complexes [122] as demonstrated by Heck and coworkers [123], who employed this instrument to analyze intact antibodies. The implementation ETD, SID, or UV photodissociation [124] for MSMS may provide a new dimension of information for mAbs. UV photodissociation, owing to its fast pulsing and high fragmentation efficiency, may drive top-down sequencing to a new level. These combinations coupled with SEC in the front end would increase the power of native ESI and top-down MS/MS. Trapped ion mobility spectrometry (TIMS) [125] has become a new member of the ion mobility family, showing high resolution in just several centimeters drift length and suggesting that improvement in IM are also forthcoming. All these new developments would add to the collection of tools for the characterization of protein therapeutics.
Acknowledgement
Preparation of this review was supported by funds from the National Institute of General Medical Sciences of the US National Institutes of Health (Grant P41 GM103422-35). H.Z was also supported by the Photosynthetic Antenna Research Center, an Energy Frontier Research Center funded by the U.S. DOE, Office of Basic Energy Sciences (Grant No. DE-SC 0001035 to Robert.E.Blankenship). H.Z. was funded equally by the DOE and NIH grants.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Buss NA, Henderson SJ, McFarlane M, Shenton JM, de Haan L. Monoclonal antibody therapeutics: history and future. Curr Opin Pharmacol. 2012;12:615–22. doi: 10.1016/j.coph.2012.08.001. [DOI] [PubMed] [Google Scholar]
- [2].Larrick JW. Conference scene: progress with promising human antibodies. Immunotherapy. 2012;4:257–61. doi: 10.2217/imt.12.5. [DOI] [PubMed] [Google Scholar]
- [3].Nelson AL, Dhimolea E, Reichert JM. Development trends for human monoclonal antibody therapeutics. Nat Rev Drug Discov. 2010;9:767–74. doi: 10.1038/nrd3229. [DOI] [PubMed] [Google Scholar]
- [4].Metzger H, Kinet JP. How antibodies work: focus on Fc receptors. FASEB J. 1988;2:3–11. doi: 10.1096/fasebj.2.1.3275562. [DOI] [PubMed] [Google Scholar]
- [5].Raghavan M, Bjorkman PJ. Fc receptors and their interactions with immunoglobulins. Annu Rev Cell Dev Biol. 1996;12:181–220. doi: 10.1146/annurev.cellbio.12.1.181. [DOI] [PubMed] [Google Scholar]
- [6].Zhang Z, Pan H, Chen X. Mass spectrometry for structural characterization of therapeutic antibodies. Mass Spectrom Rev. 2009;28:147–76. doi: 10.1002/mas.20190. [DOI] [PubMed] [Google Scholar]
- [7].Carter PJ. Potent antibody therapeutics by design. Nat Rev Immunol. 2006;6:343–357. doi: 10.1038/nri1837. [DOI] [PubMed] [Google Scholar]
- [8].Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–7. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- [9].Stern M, Herrmann R. Overview of monoclonal antibodies in cancer therapy: present and promise. Crit Rev Oncol Hematol. 2005;54:11–29. doi: 10.1016/j.critrevonc.2004.10.011. [DOI] [PubMed] [Google Scholar]
- [10].Boulianne GL, Hozumi N, Shulman MJ. Production of functional chimaeric mouse/human antibody. Nature. 1984;312:643–6. doi: 10.1038/312643a0. [DOI] [PubMed] [Google Scholar]
- [11].Jones PT, Dear PH, Foote J, Neuberger MS, Winter G. Replacing the complementarity-determining regions in a human antibody with those from a mouse. Nature. 1986;321:522–5. doi: 10.1038/321522a0. [DOI] [PubMed] [Google Scholar]
- [12].Presta LG. Engineering of therapeutic antibodies to minimize immunogenicity and optimize function. Adv Drug Deliv Rev. 2006;58:640–56. doi: 10.1016/j.addr.2006.01.026. [DOI] [PubMed] [Google Scholar]
- [13].Winter G, Griffiths AD, Hawkins RE, Hoogenboom HR. Making antibodies by phage display technology. Annu Rev Immunol. 1994;12:433–55. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]
- [14].Green LL, et al. Antigen-specific human monoclonal antibodies from mice engineered with human Ig heavy and light chain YACs. Nat Genet. 1994;7:13–21. doi: 10.1038/ng0594-13. [DOI] [PubMed] [Google Scholar]
- [15].Zwick MB, Gach JS, Burton DR. A welcome burst of human antibodies. Nat Biotechnol. 2008;26:886–7. doi: 10.1038/nbt0808-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Beck A, Wagner-Rousset E, Ayoub D, Van Dorsselaer A, Sanglier-Cianferani S. Characterization of Therapeutic Antibodies and Related Products. Analytical Chemistry. 2013;85:715–736. doi: 10.1021/ac3032355. [DOI] [PubMed] [Google Scholar]
- [17].Jiskoot W, van Schie RM, Carstens MG, Schellekens H. Immunological risk of injectable drug delivery systems. Pharm Res. 2009;26:1303–14. doi: 10.1007/s11095-009-9855-9. [DOI] [PubMed] [Google Scholar]
- [18].Hermeling S, Crommelin DJ, Schellekens H, Jiskoot W. Structure-immunogenicity relationships of therapeutic proteins. Pharm Res. 2004;21:897–903. doi: 10.1023/b:pham.0000029275.41323.a6. [DOI] [PubMed] [Google Scholar]
- [19].Lundblad RL. Approaches to the Conformational Analysis of Biopharmaceuticals. Chapman and Hall/CRC; 2009. Introduction to Biopharmaceutical Conformational Analysis; pp. 1–18. [Google Scholar]
- [20].Berkowitz SA, Engen JR, Mazzeo JR, Jones GB. Analytical tools for characterizing biopharmaceuticals and the implications for biosimilars. Nat Rev Drug Discov. 2012;11:527–40. doi: 10.1038/nrd3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kaltashov IA, Eyles SJ. Studies of biomolecular conformations and conformational dynamics by mass spectrometry. Mass Spectrom Rev. 2002;21:37–71. doi: 10.1002/mas.10017. [DOI] [PubMed] [Google Scholar]
- [22].Ronholm J, van Faassen H, MacKenzie R, Zhang ZY, Cao XD, Lin M. Monoclonal Antibodies Recognizing the Surface Autolysin IspC of Listeria monocytogenes Serotype 4b: Epitope Localization, Kinetic Characterization, and Cross-Reaction Studies. Plos One. 2013;8:12. doi: 10.1371/journal.pone.0055098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Mendoza VL, Vachet RW. Mass Spectrom. Rev. 2009;28:785–815. doi: 10.1002/mas.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sinz A. Chemical cross-linking and mass spectrometry to map three-dimensional protein structures and protein-protein interactions. Mass Spectrom Rev. 2006;25:663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- [25].Kelleher NL, Lin HY, Valaskovic GA, Aaserud DJ, Fridriksson EK, McLafferty FW. Top Down versus Bottom Up Protein Characterization by Tandem High-Resolution Mass Spectrometry. J. Am. Chem. Soc. 1999;121:806–812. [Google Scholar]
- [26].Zubarev RA, Kelleher NL, McLafferty FW. Electron Capture Dissociation of Multiply Charged Protein Cations. A Nonergodic Process. J. Am. Chem. Soc. 1998;120:3265–3266. [Google Scholar]
- [27].Uetrecht C, Versluis C, Watts NR, Roos WH, Wuite GJ, Wingfield PT, Steven AC, Heck AJ. High-resolution mass spectrometry of viral assemblies: molecular composition and stability of dimorphic hepatitis B virus capsids. Proc Natl Acad Sci U S A. 2008;105:9216–20. doi: 10.1073/pnas.0800406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Cui W, Rohrs HW, Gross ML. Top-down mass spectrometry: Recent developments, applications and perspectives. Analyst. 2011;136:3854–64. doi: 10.1039/c1an15286f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ruotolo BT, Robinson CV. Aspects of native proteins are retained in vacuum. Curr Opin Chem Biol. 2006;10:402–8. doi: 10.1016/j.cbpa.2006.08.020. [DOI] [PubMed] [Google Scholar]
- [30].Benesch JL, Ruotolo BT, Simmons DA, Robinson CV. Protein complexes in the gas phase: technology for structural genomics and proteomics. Chem Rev. 2007;107:3544–67. doi: 10.1021/cr068289b. [DOI] [PubMed] [Google Scholar]
- [31].Kaltashov IA, Mohimen A. Estimates of protein surface areas in solution by electrospray ionization mass spectrometry. Anal Chem. 2005;77:5370–9. doi: 10.1021/ac050511+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Abzalimov RR, Frimpong AK, Kaltashov IA. Detection and characterization of large-scale protein conformational transitions in solution using charge-state distribution analysis in ESI-MS. Methods Mol Biol. 2012;896:365–73. doi: 10.1007/978-1-4614-3704-8_24. [DOI] [PubMed] [Google Scholar]
- [33].Zhong Y, Hyung SJ, Ruotolo BT. Ion mobility-mass spectrometry for structural proteomics. Expert Rev Proteomics. 2012;9:47–58. doi: 10.1586/epr.11.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Benesch JL. Collisional activation of protein complexes: picking up the pieces. J Am Soc Mass Spectrom. 2009;20:341–8. doi: 10.1016/j.jasms.2008.11.014. [DOI] [PubMed] [Google Scholar]
- [35].Bowers MT, Kemper PR, von Helden G, van Koppen PA. Gas-phase ion chromatography: transition metal state selection and carbon cluster formation. Science. 1993;260:1446–1451. doi: 10.1126/science.260.5113.1446. [DOI] [PubMed] [Google Scholar]
- [36].Ruotolo BT, Giles K, Campuzano I, Sandercock AM, Bateman RH, Robinson CV. Evidence for macromolecular protein rings in the absence of bulk water. Science. 2005;310:1658–61. doi: 10.1126/science.1120177. [DOI] [PubMed] [Google Scholar]
- [37].Uetrecht C, Rose RJ, van Duijn E, Lorenzen K, Heck AJR. Ion mobility mass spectrometry of proteins and protein assemblies. Chem Soc Rev. 2010;39:1633–1655. doi: 10.1039/b914002f. [DOI] [PubMed] [Google Scholar]
- [38].Bush MF, Campuzano ID, Robinson CV. Ion mobility mass spectrometry of peptide ions: effects of drift gas and calibration strategies. Anal Chem. 2012;84:7124–30. doi: 10.1021/ac3014498. [DOI] [PubMed] [Google Scholar]
- [39].Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, Ruotolo BT. Collision cross sections of proteins and their complexes: a calibration framework and database for gas-phase structural biology. Anal Chem. 2010;82:9557–65. doi: 10.1021/ac1022953. [DOI] [PubMed] [Google Scholar]
- [40].Barrera NP, Di Bartolo N, Booth PJ, Robinson CV. Micelles protect membrane complexes from solution to vacuum. Science. 2008;321:243–6. doi: 10.1126/science.1159292. [DOI] [PubMed] [Google Scholar]
- [41].McLafferty FW, Bente PF, Kornfeld R, Tsai SC, Howe I. Collisional Activation Spectra of Organic Ions. J. Am. Chem. Soc. 1973;95:2120–2129. [Google Scholar]
- [42].Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and Protein Sequence Analysis by Electron Transfer Dissociation Mass Spectrometry. Proc. Natl. Acad. Sci. USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhou M, Jones CM, Wysocki VH. Dissecting the Large Noncovalent Protein Complex GroEL with Surface-Induced Dissociation and Ion Mobility-Mass Spectrometry. Analytical chemistry. 2013;85:8262–8267. doi: 10.1021/ac401497c. [DOI] [PubMed] [Google Scholar]
- [44].Brodbelt JS. Shedding Light on the Frontier of Phodissociation. J. Am. Soc. Mass Spectrom. 2011;22:197–206. doi: 10.1007/s13361-010-0023-6. [DOI] [PubMed] [Google Scholar]
- [45].Chait BT. Mass Spectrometry: Bottom-Up or Top-Down. Science. 2006;314:65–66. doi: 10.1126/science.1133987. [DOI] [PubMed] [Google Scholar]
- [46].Benesch JLP, Ruotolo BT, Simmons DA, Robinson CV. Protein Complexes in the Gas Phase: Technology for Structural Genomics and Proteomics. Chem. Rev. 2007;107:3544–3567. doi: 10.1021/cr068289b. [DOI] [PubMed] [Google Scholar]
- [47].Zhang H, Cui W, Wen J, Blankenship RE, Gross ML. Native electrospray and electron-capture dissociation FTICR mass spectrometry for top-down studies of protein assemblies. Anal Chem. 2011;83:5598–606. doi: 10.1021/ac200695d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zhang H, Cui W, Gross ML. Native Electrospray Ionization and Electron-Capture Dissociation for Comparison of Protein Structure in Solution and the Gas Phase. Int. J. Mass Spectrom. 2013 doi: 10.1016/j.ijms.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Zhou M, Dagan S, Wysocki VH. Impact of charge state on gas-phase behaviors of noncovalent protein complexes in collision induced dissociation and surface induced dissociation. Analyst. 2013;138:1353–1362. doi: 10.1039/c2an36525a. [DOI] [PubMed] [Google Scholar]
- [50].Xu G, Chance MR. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem Rev. 2007;107:3514–43. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- [51].Kaltashov IA, Bobst CE, Abzalimov RR. Mass spectrometry-based methods to study protein architecture and dynamics. Protein Sci. 2013;22:530–44. doi: 10.1002/pro.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kaltashov IA, Bobst CE, Abzalimov RR. H/D exchange and mass spectrometry in the studies of protein conformation and dynamics: is there a need for a top-down approach? Anal Chem. 2009;81:7892–9. doi: 10.1021/ac901366n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Chen J, Cui W, Giblin D, Gross ML. New protein footprinting: fast photochemical iodination combined with top-down and bottom-up mass spectrometry. J Am Soc Mass Spectrom. 2012;23:1306–18. doi: 10.1007/s13361-012-0403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Englander SW. Hydrogen exchange mass spectrometry: a historical perspective. J Am Soc Mass Spectrom. 2006;17:1481–1489. doi: 10.1016/j.jasms.2006.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- [56].Tuma R, Coward LU, Kirk MC, Barnes S, Prevelige PE., Jr Hydrogen-deuterium exchange as a probe of folding and assembly in viral capsids. J Mol Biol. 2001;306:389–96. doi: 10.1006/jmbi.2000.4383. [DOI] [PubMed] [Google Scholar]
- [57].Benjamin DR, Robinson CV, Hendrick JP, Hartl FU, Dobson CM. Mass spectrometry of ribosomes and ribosomal subunits. Proc Natl Acad Sci U S A. 1998;95:7391–5. doi: 10.1073/pnas.95.13.7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Miyagi M, Nakazawa T. Determination of pKa values of individual histidine residues in proteins using mass spectrometry. Anal Chem. 2008;80:6481–7. doi: 10.1021/ac8009643. [DOI] [PubMed] [Google Scholar]
- [59].Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25:158–70. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- [60].Wang L, Chance MR. Structural mass spectrometry of proteins using hydroxyl radical based protein footprinting. Anal Chem. 2011;83:7234–41. doi: 10.1021/ac200567u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Gau BC, Chen H, Zhang Y, Gross ML. Sulfate radical anion as a new reagent for fast photochemical oxidation of proteins. Anal Chem. 2010;82:7821–7. doi: 10.1021/ac101760y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Xu G, Chance MR. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chemical Reviews. 2007;107:3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- [63].Zhu Y, et al. Elucidating in vivo structural dynamics in integral membrane protein by hydroxyl radical footprinting. Mol Cell Proteomics. 2009;8:1999–2010. doi: 10.1074/mcp.M900081-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Takamoto K, Chance MR. Radiolytic protein footprinting with mass spectrometry to probe the structure of macromolecular complexes. Annu Rev Biophys Biomol Struct. 2006:251–276. doi: 10.1146/annurev.biophys.35.040405.102050. [DOI] [PubMed] [Google Scholar]
- [65].Hambly DE, Gross ML. Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale. J Am Soc Mass Spectrom. 2005;16:2057–2063. doi: 10.1016/j.jasms.2005.09.008. [DOI] [PubMed] [Google Scholar]
- [66].Aye TT, Low TY, Sze SK. Nanosecond laser-induced photochemical oxidation method for protein surface mapping with mass spectrometry. Analytical chemistry. 2005;77:5814–5822. doi: 10.1021/ac050353m. [DOI] [PubMed] [Google Scholar]
- [67].Maleknia SD, Brenowitz M, Chance MR. Millisecond radiolytic modification of peptides by synchrotron X-rays identified by mass spectrometry. Anal Chem. 1999;71:3965–73. doi: 10.1021/ac990500e. [DOI] [PubMed] [Google Scholar]
- [68].Takamoto K, Chance MR. Radiolytic protein footprinting with mass spectrometry to probe the structure of macromolecular complexes. Annu Rev Biophys Biomol Struct. 2006;35:251–76. doi: 10.1146/annurev.biophys.35.040405.102050. [DOI] [PubMed] [Google Scholar]
- [69].Gupta S, D’Mello R, Chance MR. Structure and dynamics of protein waters revealed by radiolysis and mass spectrometry. Proc Natl Acad Sci U S A. 2012;109:14882–7. doi: 10.1073/pnas.1209060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Zhang H, Gau BC, Jones LM, Vidavsky I, Gross ML. Fast photochemical oxidation of proteins for comparing structures of protein-ligand complexes: the calmodulin-peptide model system. Anal Chem. 2011;83:311–8. doi: 10.1021/ac102426d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Gau BC, Sharp JS, Rempel DL, Gross ML. Fast photochemical oxidation of protein footprints faster than protein unfolding. Anal Chem. 2009;81:6563–71. doi: 10.1021/ac901054w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Hambly DM, Gross ML. Laser flash photolysis of hydrogen peroxide to oxidize protein solvent-accessible residues on the microsecond timescale. J Am Soc Mass Spectrom. 2005;16:2057–63. doi: 10.1016/j.jasms.2005.09.008. [DOI] [PubMed] [Google Scholar]
- [73].Chen J, Rempel DL, Gross ML. Temperature jump and fast photochemical oxidation probe submillisecond protein folding. J Am Chem Soc. 2010;132:15502–4. doi: 10.1021/ja106518d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Wen J, Zhang H, Gross ML, Blankenship RE. Membrane orientation of the FMO antenna protein from Chlorobaculum tepidum as determined by mass spectrometry-based footprinting. Proceedings of the National Academy of Sciences of the United States of America, Early Edition. 2009:1–6. 6. doi: 10.1073/pnas.0901691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Su D, Delaplane S, Luo M, Rempel DL, Vu B, Kelley MR, Gross ML, Georgiadis MM. Interactions of apurinic/apyrimidinic endonuclease with a redox inhibitor: evidence for an alternate conformation of the enzyme. Biochemistry. 2011;50:82–92. doi: 10.1021/bi101248s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Zhang H, Shen W, Rempel D, Monsey J, Vidavsky I, Gross ML, Bose R. Carboxyl-group footprinting maps the dimerization interface and phosphorylation-induced conformational changes of a membrane-associated tyrosine kinase. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.005678. M110 005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Wen J, Zhang H, Gross ML, Blankenship RE. Membrane orientation of the FMO antenna protein from Chlorobaculum tepidum as determined by mass spectrometry-based footprinting. Proc Natl Acad Sci U S A. 2009;106:6134–9. doi: 10.1073/pnas.0901691106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Petrotchenko EV, Borchers CH. Crosslinking combined with mass spectrometry for structural proteomics. Mass Spectrom Rev. 2010;29:862–76. doi: 10.1002/mas.20293. [DOI] [PubMed] [Google Scholar]
- [79].Chavez JD, Weisbrod CR, Zheng C, Eng JK, Bruce JE. Protein Interactions, Post-translational Modifications and Topologies in Human Cells. Mol Cell Proteomics. 2013;12:1451–67. doi: 10.1074/mcp.M112.024497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Weisbrod CR, Chavez JD, Eng JK, Yang L, Zheng C, Bruce JE. In Vivo Protein Interaction Network Identified with a Novel Real-Time Cross-Linked Peptide Identification Strategy. J Proteome Res. 2013 doi: 10.1021/pr3011638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Wei H, Tymiak AA, Chen GD. High-resolution MS for structural characterization of protein therapeutics: advances and future directions. Bioanalysis. 2013;5:1299–1313. doi: 10.4155/bio.13.80. [DOI] [PubMed] [Google Scholar]
- [82].Bobst CE, Abzalimov RR, Houde D, Kloczewiak M, Mhatre R, Berkowitz SA, Kaltashov IA. Detection and characterization of altered conformations of protein pharmaceuticals using complementary mass spectrometry-based approaches. Anal Chem. 2008;80:7473–81. doi: 10.1021/ac801214x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Kaltashov IA, Bobst CE, Abzalimov RR, Wang G, Baykal B, Wang S. Advances and challenges in analytical characterization of biotechnology products: mass spectrometry-based approaches to study properties and behavior of protein therapeutics. Biotechnol Adv. 2012;30:210–22. doi: 10.1016/j.biotechadv.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Beck A, Sanglier-Cianferani S, Van Dorsselaer A. Biosimilar, Biobetter, and Next Generation Antibody Characterization by Mass Spectrometry. Analytical Chemistry. 2012;84:4637–4646. doi: 10.1021/ac3002885. [DOI] [PubMed] [Google Scholar]
- [85].Kaltashov IA, Abzalimov RR. Do ionic charges in ESI MS provide useful information on macromolecular structure? J Am Soc Mass Spectrom. 2008;19:1239–46. doi: 10.1016/j.jasms.2008.05.018. [DOI] [PubMed] [Google Scholar]
- [86].Zamani L, Lindholm J, Ilag LL, Jacobsson SP. Discrimination among IgG1-kappa monoclonal antibodies produced by two cell lines using charge state distributions in nanoESITOF mass spectra. J Am Soc Mass Spectrom. 2009;20:1030–6. doi: 10.1016/j.jasms.2009.01.008. [DOI] [PubMed] [Google Scholar]
- [87].Bagal D, Valliere-Douglass JF, Balland A, Schnier PD. Resolving Disulfide Structural Isoforms of IgG2 Monoclonal Antibodies by Ion Mobility Mass Spectrometry. Analytical chemistry. 2010;82:6751–6755. doi: 10.1021/ac1013139. [DOI] [PubMed] [Google Scholar]
- [88].Rose RJ, et al. Structure. 2011;19:1274–1282. doi: 10.1016/j.str.2011.06.016. [DOI] [PubMed] [Google Scholar]
- [89].Beck A, Reichert JM. Mabs. 2011;3:221–222. doi: 10.4161/mabs.3.3.15625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Debaene F, Wagner-Rousset E, Coales O, Ayoub D, Corvaia N, van Dorsselaer A, Beck A, Cianferani S. TIme Resolved Native Ion-Mobility Mass Spectrometry to Monitor Dynamics of IgG4 Fab Arm Exchange and “Bispecific” Monoclonal Antobody Formation. Analytical chemistry. 2013 doi: 10.1021/ac402237v. [DOI] [PubMed] [Google Scholar]
- [91].Jones LM, Zhang H, Cui WD, Kumar S, Sperry JB, Carroll JA, Gross ML. Complementary MS Methods Assist Conformational Characterization of Antibodies with Altered S-S Bonding Networks. J Am Soc Mass Spectrom. 2013;24:835–845. doi: 10.1007/s13361-013-0582-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Houde D, Engen JR. Conformational analysis of recombinant monoclonal antibodies with hydrogen/deuterium exchange mass spectrometry. Methods Mol Biol. 2013;988:269–89. doi: 10.1007/978-1-62703-327-5_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Wei H, Mo J, Tao L, Russell RJ, Tymiak AA, Chen G, Iacob RE, Engen JR. Hydrogen/deuterium exchange mass spectrometry for probing higher order structure of protein therapeutics: methodology and applications. Drug Discov Today. 2013 doi: 10.1016/j.drudis.2013.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Zheng K, Bantog C, Bayer R. The impact of glycosylation on monoclonal antibody conformation and stability. Mabs. 2011;3:568–76. doi: 10.4161/mabs.3.6.17922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Houde D, Arndt J, Domeier W, Berkowitz S, Engen JR. Characterization of IgG1 conformation and conformational dynamics by hydrogen/deuterium exchange mass spectrometry. Anal Chem. 2009;81:2644–51. doi: 10.1021/ac802575y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Houde D, Peng Y, Berkowitz SA, Engen JR. Post-translational modifications differentially affect IgG1 conformation and receptor binding. Mol Cell Proteomics. 2010;9:1716–28. doi: 10.1074/mcp.M900540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Burkitt W, Domann P, O’Connor G. Conformational changes in oxidatively stressed monoclonal antibodies studied by hydrogen exchange mass spectrometry. Protein Sci. 2010;19:826–35. doi: 10.1002/pro.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Khawli LA, Goswami S, Hutchinson R. Charge variants in IgG1. MAbs. 2010;2:613–624. doi: 10.4161/mabs.2.6.13333. al., e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Tang L, Sundaram S, Zhang J, Carlson P, Matathia A, Parekh B, Zhou Q, Hsieh MC. Conformational characterization of the charge variants of a human IgG1 monoclonal antibody using H/D exchange mass spectrometry. Mabs. 2013;5:114–25. doi: 10.4161/mabs.22695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Manikwar P, et al. Correlating excipient effects on conformational and storage stability of an IgG1 monoclonal antibody with local dynamics as measured by hydrogen/deuterium-exchange mass spectrometry. J Pharm Sci. 2013;102:2136–51. doi: 10.1002/jps.23543. [DOI] [PubMed] [Google Scholar]
- [101].Majumdar R, et al. Effects of Salts from the Hofmeister Series on the Conformational Stability, Aggregation Propensity, and Local Flexibility of an IgG1 Monoclonal Antibody. Biochemistry. 2013;52:3376–3389. doi: 10.1021/bi400232p. [DOI] [PubMed] [Google Scholar]
- [102].Watson C, Sharp JS. Conformational analysis of therapeutic proteins by hydroxyl radical protein footprinting. AAPS J. 2012;14:206–17. doi: 10.1208/s12248-012-9336-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Zhang L, Lilyestrom W, Li C, Scherer T, van Reis R, Zhang B. Revealing a positive charge patch on a recombinant monoclonal antibody by chemical labeling and mass spectrometry. Anal Chem. 2011;83:8501–8. doi: 10.1021/ac2016129. [DOI] [PubMed] [Google Scholar]
- [104].Philo JS. A Critical Review of Methods for Size Characterization of Non-Particulate Protein Aggregates. CUrr Pharm Biotechnol. 2009;10:359–372. doi: 10.2174/138920109788488815. [DOI] [PubMed] [Google Scholar]
- [105].Wang W, Roberts CJ, editors. Aggregation of Therapeutic Proteins. John Wiley and Sons; 2010. [Google Scholar]
- [106].Pedersen JT, Heegaard NHH. Analysis of Protein Aggregation in Neurodegenerative Disease. Analytical chemistry. 2013;85:4215–4227. doi: 10.1021/ac400023c. [DOI] [PubMed] [Google Scholar]
- [107].Kukrer B, Filipe V, van Duijn E, Kasper PT, Vreeken RJ, Heck AJ, Jiskoot W. Mass spectrometric analysis of intact human monoclonal antibody aggregates fractionated by size-exclusion chromatography. Pharm Res. 2010;27:2197–204. doi: 10.1007/s11095-010-0224-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Zhang Y, Rempel DL, Zhang J, Sharma AK, Mirica LM, Gross ML. Pulsed Hydrogen-Deuterium Exchange Mass Spectrometry Probes Conformational Changes in Amyloid Beta Peptide Aggregation. PNAS. 2013;110:14604–14609. doi: 10.1073/pnas.1309175110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Zhang A, Qi W, Singh SK, Fernandez EJ. A new approach to explore the impact of freeze-thaw cycling on protein structure: hydrogen/deuterium exchange mass spectrometry (HX-MS) Pharm Res. 2011;28:1179–93. doi: 10.1007/s11095-011-0383-z. [DOI] [PubMed] [Google Scholar]
- [110].Zhang A, Singh SK, Shirts MR, Kumar S, Fernandez EJ. Distinct aggregation mechanisms of monoclonal antibody under thermal and freeze-thaw stresses revealed by hydrogen exchange. Pharm Res. 2012;29:236–50. doi: 10.1007/s11095-011-0538-y. [DOI] [PubMed] [Google Scholar]
- [111].Iacob R, Bou-Assaf G, Makowski L, Engen J, Berkowitz S, D H. Investigating Monoclonal Antibody Aggregation Using a Combination of H/DX-MS and Other Biophysical Measurements. J. Pharm. Sci. 2013 doi: 10.1002/jps.23754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Hambly DM, Gross ML. Cold chemical oxidation of proteins. Anal. Chem. 2009;81:7235–42. doi: 10.1021/ac900855f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Deperalta G, Alvarez M, Bechtel C, Dong K, McDonald R, Ling V. Structural analysis of a therapeutic monoclonal antibody dimer by hydroxyl radical footprinting. Mabs. 2013;5:86–101. doi: 10.4161/mabs.22964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Zhao A, Hao G, Gu J. Chemical crosslinking and mass spectrometric identification of interaction sites within soluble aggregate of protein therapeutics. J Pharm Biomed Anal. 2013;73:99–102. doi: 10.1016/j.jpba.2012.05.006. [DOI] [PubMed] [Google Scholar]
- [115].Hawiet AE, Shoemaker GK, Daneshfar R, Kitova EN, Klassen JS. Analytical chemistry. 2012;84:50–58. doi: 10.1021/ac202760e. [DOI] [PubMed] [Google Scholar]
- [116].Obungu VH, Gelfanova V, Rathnachalam R, Bailey A, Sloan-Lancaster J, Huang L. Determination of the mechanism of action of anti-FasL antibody by epitope mapping and homology modeling. Biochemistry. 2009;48:7251–60. doi: 10.1021/bi900296g. [DOI] [PubMed] [Google Scholar]
- [117].Malito E, et al. Defining a protective epitope on factor H binding protein, a key meningococcal virulence factor and vaccine antigen. Proc Natl Acad Sci U S A. 2013;110:3304–9. doi: 10.1073/pnas.1222845110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Coales SJ, Tuske SJ, Tomasso JC, Hamuro Y. Epitope mapping by amide hydrogen/deuterium exchange coupled with immobilization of antibody, on-line proteolysis, liquid chromatography and mass spectrometry. Rapid Commun Mass Spectrom. 2009;23:639–47. doi: 10.1002/rcm.3921. [DOI] [PubMed] [Google Scholar]
- [119].Pandit D, et al. Mapping of discontinuous conformational epitopes by amide hydrogen/deuterium exchange mass spectrometry and computational docking. J Mol Recognit. 2012;25:114–24. doi: 10.1002/jmr.1169. [DOI] [PubMed] [Google Scholar]
- [120].Jones LM, J BS, J AC, Gross ML. Fast photochemical oxidation of proteins for epitope mapping. Anal Chem. 2011;83:7657–61. doi: 10.1021/ac2007366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Mayne L, Paterson Y, Cerasoli D, Englander SW. Effect of antibody binding on protein motions studied by hydrogen-exchange labeling and two-dimensional NMR. Biochemistry. 1992;31:10678–85. doi: 10.1021/bi00159a006. [DOI] [PubMed] [Google Scholar]
- [122].Rose RJ, Damoc E, Denisov E, Alexander M, Heck AJR. High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies. Nature Methods. 2012;9:1084–1086. doi: 10.1038/nmeth.2208. [DOI] [PubMed] [Google Scholar]
- [123].Thompson NJ, Rosati S, Rose RJ, Heck AJ. The Impact of Mass Spectrometry on the Study of Intact Antibodies: From Post-Translational Modifications to Structural Analysis. Chem. Comm. 2013;49:538–548. doi: 10.1039/c2cc36755f. [DOI] [PubMed] [Google Scholar]
- [124].Shaw JB, et al. Complete protein characterization using top-down mass spectrometry and ultraviolet photodissociation. J Am Chem Soc. 2013;135:12646–51. doi: 10.1021/ja4029654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Fernandez-Lima FA, Kaplan DA, Park MA. Note: Integration of trapped ion mobility spectrometry with mass spectrometry. Rev Sci Instrum. 2011;82:126106. doi: 10.1063/1.3665933. [DOI] [PMC free article] [PubMed] [Google Scholar]