Abstract
The ubiquitous sequence diverse 4-Toluene Sulfonate Uptake Permease (TSUP) family contains few characterized members and is believed to catalyze the transport of several sulfur-based compounds. Prokaryotic members of the TSUP family outnumber the eukaryotic members substantially, and in prokaryotes, but not eukaryotes, extensive lateral gene transfer occurred during family evolution. Despite unequal representation, homologues from the three taxonomic domains of life share well-conserved motifs. We show that the prototypical eight TMS topology arose from an intragenic duplication of a four TMS unit. Possibly, a two TMS α-helical hairpin structure was the precursor of the 4 TMS repeat unit. Genome context analyses confirmed the proposal of a sulfur-based compound transport role for many TSUP homologues, but functional outliers appear to be prevalent as well. Preliminary results suggest that the TSUP family is a member of a large novel superfamily that includes rhodopsins, integral membrane chaperone proteins, transmembrane electron flow carriers and several transporter families. All of these proteins probably arose via the same pathway: 2 → 4 → 8 TMSs followed by loss of a TMS either at the N- or C-terminus, depending on the family, to give the more frequent 7 TMS topology.
Keywords: Transport proteins, secondary active transport, TSUP, uptake/efflux, evolution, superfamily
1. Introduction
Using functional, phylogenetic, and membrane topological information derived from over 10,000 publications on transport systems, we have classified most recognized transport systems into over 600 families. Our work is summarized in the IUBMB-approved Transporter Classification (TC) Database (TCDB; http://www.tcdb.org; [1, 2]), a carefully curated database presenting the TC system [3], which is analogous to the function-only-based Enzyme Commission (EC) system (http://www.chem.qmul.ac.uk/iubmb/enzyme/). The current study focuses on the putative 4-Toluene Sulfonate Uptake Permease (TSUP) family (TC# 2.A.102; previously TC# 9.A.29).
Transport systems play crucial roles in all processes associated with life. They catalyze nutrient uptake, metabolite excretion, the establishment of electrochemical gradients, macromolecular export, drug and toxin efflux and intercellular communication by transporting signaling molecules [3]. However, their effectiveness can be utilized in ways that are detrimental to humans and other organisms. This is exemplified by multi-drug resistant (MDR) pathogenic microbial strains, arising partially because of the excessive use of antibiotics in meat production and medicine. Characterizing transporters can pave the way for computational modes of drug discovery, which would allow us to more effectively target various MDR pathogens and diseases [4, 5]. The importance of transport proteins, constituting roughly 10% of the proteome of an organism, on the average, cannot be overstated.
The TSUP family includes thousands of currently sequenced members spanning the bacterial, eukaryotic and archaeal domains. Within the prokaryotes, we have found these proteins in virtually all well studied phyla (see below). The occurrence of multiple organismal sources within single phylogenetic clusters implies extensive horizontal transfer of genes encoding the homologues [6]. A majority of prokaryotic protein members are shown to range in size from 240 amino acids (aas) to 280 aas with few exceptions. The archaeal members, on average, are slightly smaller than the bacterial members, but the eukaryotic members are typically 40–50% larger and range from 400–500 amino acyl residues in size [7]. Eukaryotic members often possess N- and C-terminal extensions, which may play regulatory roles [8, 9]. When only the bacterial members were analyzed, an 8 transmembrane segment (TMS) topology proved to predominate [6], although eukaryotic and archaeal homologues may exhibit greater topological variation. We show that some of the prokaryotic members have undergone intragenic duplication of a 4 TMS unit yielding 8 TMS proteins.
Functions for most TSUP family members have not been assigned and cannot be assumed due to the great sequence divergence among homologues. In fact, the few analyses that have been performed with TSUP homologues suggest differing functionalities. One TSUP member, TsaS in Comamonas testosteroni (TC# 2.A.102.1.1), was proposed to catalyze 4-toluene sulfonate uptake via a proton symport mechanism [10, 11]. A cotranscribed protein, TsaT, was proposed to be membrane anion porin [12], but we found that it is homologous to periplasmic binding proteins and therefore propose that, as for TRAP-T family members [13, 14], it is an extracytoplasmic binding receptor that feeds substrates to the TsaS carrier. TsaT is only expressed in the presence of 4-toluene sulfonate, whereas TsaS is expressed following growth in the presence of this and other compounds [12]. TsaS has 7 putative TMSs, and the tight expressional control implied that TsaT might confer specificity to TsaS.
TsaS homologue, TauE (TC# 2.A.102.2.1), was predicted to be a sulfite exporter, but its mechanism of action was not investigated [15]. A putative sulfate uptake porter termed CysZ (TC# 2.A.102.6.1) was also shown to belong to the TSUP family. As for TauE, its mechanism of action is unknown [16]. Yet another homologue, SafE1 (TC# 2.A.102.2.2), was proposed to be a sulfoacetate exporter [17]. A recent study has identified PmpC (TC# 2.A.102.4.2), a TSUP family member, to be part of the PigP regulon in Serratia sp. strain ATCC 39006. It was predicted to be inner-membrane localized along with the DUF395 family proteins PmpA and PmpB. PmpA, B and C were all predicted to transport sulfur-containing compounds [18].
It is not known how TSUP family members arose, and conserved motifs have not been identified. Additionally, structural characteristics such as sidedness and rigorous determination of TMS numbers have not been performed. In this report, we use bioinformatic approaches to correct these deficiencies.
2. Methods
2.1 Obtaining homologues and removing redundancies
Query sequences used to identify TSUP family members were (1) Orf of Pyrococcus abyssi (gi# 74545625; TC# 2.A.102.4.1), (2) YfcA of Escherichia coli (gi# 82592533; TC# 2.A.102.3.1) and (3) Orf of Oryza sativa (gi# 75252893; TC# 2.A.102.5.1). NCBI PSI-BLAST with two iterations were performed with these 3 proteins using default settings, with the output set to 1000 sequences, and with a cutoff of e−4 and e−6 for the first and second iterations, respectively [19, 20]. Due to program restrictions, the corresponding TinySeq XML files were inputted into the MakeTable5 program separately, and a 70% cutoff was used to remove fragments, redundancies, and sequences having greater than 70% identity with a retained sequence [6]. CD-HIT was then used with a 45% cutoff after combining the three files [21]. 216 sequences remained, and MakeTable5 then created a FASTA file for the sequences as well as a table, which included the corresponding abbreviation, sequence description, organismal source, size, gi number, organismal group or phylum, and organismal domain for each protein. The methods used here to establish homology have been shown to exceed other available methods in terms of reliability and sensitivity as documented in [22, 23].
2.2 Multiple alignment and phylogenetic/16S/18S rRNA trees
The ClustalX program was used to create multiple alignments of homologous proteins and a neighbor-joining phylogenetic tree, visualized using the FigTree program [24, 25]. Based on the multiple alignment, 27 fragmented sequences were removed, bringing the total number to 189. 16S/18S rRNA sequences were obtained using the Silva rRNA database.
2.3 Topological analyses
For topological analyses of single protein sequences, the WHAT, TMHMM 2.0, and HMMTOP programs were used [26–30]. The TMHMM 2.0 program was used in TMS count determinations while the HMMTOP program was used for determining protein sidedness [31, 32]. In cases where the TMS count was in agreement, but protein sidedness differed between the two programs, the positive inside rule was used to make educated guesses concerning protein sidedness [33]. Inputting the multiple alignment file generated by ClustalX into the Average Hydropathy, Amphipathicity and Similarity (AveHAS) program facilitated topological assessments of multiple proteins or entire subfamilies [34].
2.4 Establishing internal repeats
Based on visual analysis of AveHAS plots, potential internal repeats were examined using the IntraCompare (IC) program [35]. The best comparison scores, reported in standard deviations (S.D.), were confirmed and analyzed further using the GSAT/GAP program [36]. The GSAT/GAP program was set at default settings with a gap creation penalty of 8 and a gap extension penalty of 2 with 500 random shuffles. A length of 60 amino acyl residues, the average size of a typical protein domain, and 10 S.D., corresponding to a probability of 10−24 that the level of similarity arose by chance, is considered sufficient to prove homology between two proteins or internal repeat units [2, 6, 37, 38]. Optimization of the GSAT/GAP alignment was performed on sequences by maximizing the number of identities, minimizing gaps, and removing non-aligned sequences at the ends. Optimization yields a higher comparison score that better represents the level of similarity between two shorter internal sequences.
2.5 Functional domains and motifs
A search for recognized functional domains in TSUP family members was performed using the conserved domain database (CDD) of NCBI [39]. Protein sequence motifs were identified using the MEME and MAST programs in 2 separate runs due to program restrictions [29, 30]. The most conserved motifs across the 2 runs were analyzed and blended into a single conserved motif based on individual amino acid conservation. The appearance of duplicates of conserved motifs was noted as further evidence of internal repeat elements.
2.6 Genome and operon context analyses
To propose possible related functions, genome context analyses were performed using The SEED-Viewer, which allows the exploration of over 1,500 curated genomes in order to find homologous genes, their operon context, and consequently their known or putative roles in other organisms [40]. This was done alongside RegPrecise and RegPredict, which allow for the identification of transcription factor binding sites [41, 42].
3. Results
3.1 Phylogenetic analysis
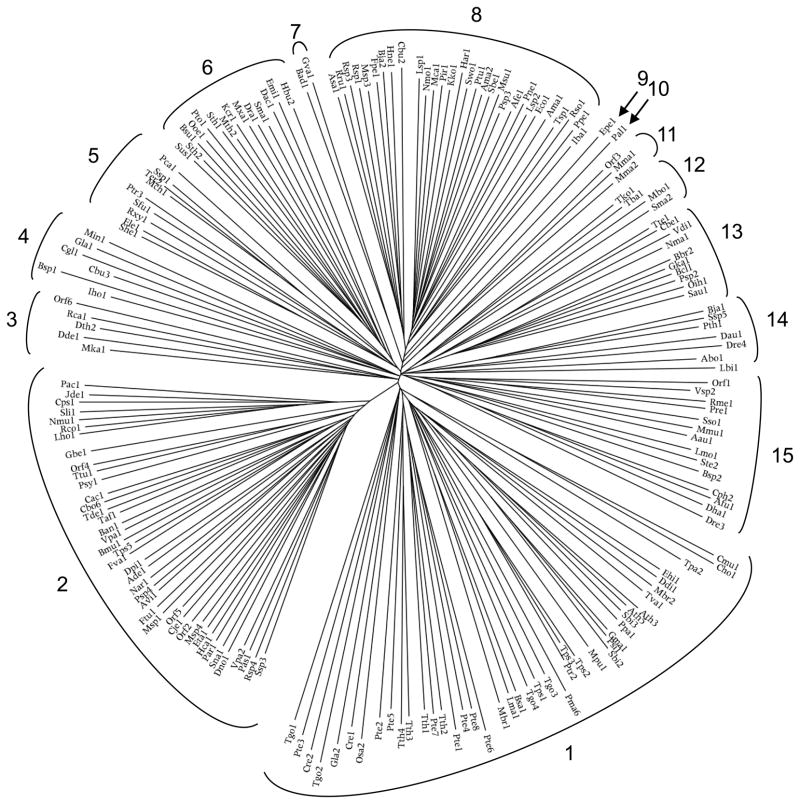
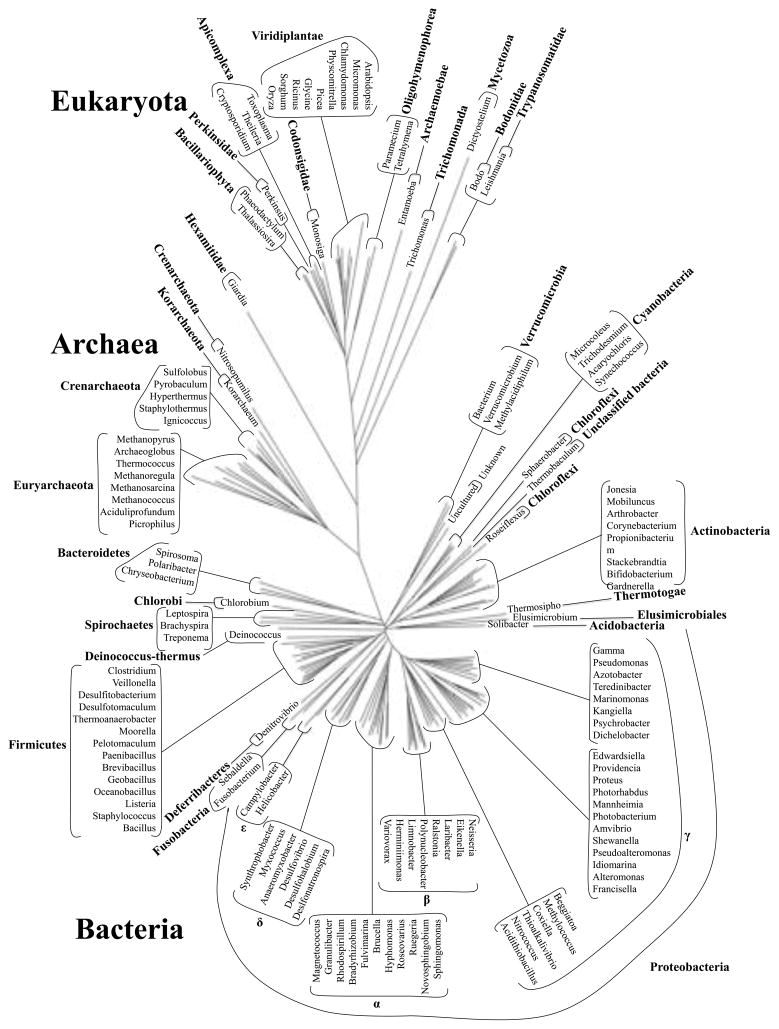
The 189 TSUP family members included in this study fell into 15 phylogenetic clusters as shown in Figures 1 and S1, while the phylogenetic tree for the corresponding 16S/18S rRNAs, representing the genuses from which the proteins derived, is shown in Figures 2 and S2. In Table 1, the proteins with their properties are tabulated according to their positions in the phylogenetic tree in Figure 1 in a clockwise fashion. The Conserved Domain Database (CDD) [39] referred to TSUP family members as DUF81, TauE and COG0730 domains [15, 17]. These proteins will be discussed according to cluster.
Figure 1.
Phylogenetic tree of the 189 TSUP family proteins included in this study. The tree was generated using the ClustalX multiple alignment and FigTree programs. Protein abbreviations and their descriptions are listed in Table 1 in a clockwise fashion starting from cluster 1. The positions of individual proteins within the phylogenetic tree are revealed in Fig. S1.
Figure 2.
16S/18S rRNA phylogenetic tree of genuses represented in this study. The Cloacamonas, Symbiobacterium, Oenococcus, Endoriftia and Desulforudis genera were excluded due to unreliable sequence data.
Table 1.
The 189 TSUP proteins included in this study. Proteins are listed clockwise starting from Cluster 1 in Fig. 1. Protein abbreviations, taxonomic origins, protein sizes (aas), gi numbers, organismal phyla, organismal domains, putative numbers of TMSs, and N-terminal orientation are included (see key below). N-terminal orientation results lacking asterisks signify agreement between programs. Average size and standard deviation values are provided for all clusters with two or more members. Additional average size and SD values are provided after removing the specified outliers.
| Abbreviation | Organism | Protein Size | GenBank No. | Phylum | Domain | No. of TMSs |
|---|---|---|---|---|---|---|
| Cluster 1 | ||||||
| Cmu1 | Cryptosporidium muris RN66 | 525 | 209881434 | Apicomplexa | Eukaryota | 11 |
| Cho1 | Cryptosporidium hominis TU502 | 518 | 67601741 | Apicomplexa | Eukaryota | 10 |
| Tpa2 | Theileria parva strain Muguga | 409 | 71033393 | Apicomplexa | Eukaryota | 9 |
| Ehi1 | Entamoeba histolytica | 460 | 67466247 | none | Eukaryota | 10 |
| Ddi1 | Dictyostelium discoideum AX4 | 549 | 66825573 | none | Eukaryota | 10 |
| Mbr2 | Monosiga brevicollis MX1 | 499 | 167525260 | Codonosigidae | Eukaryota | 8 |
| Tva1 | Trichomonas vaginalis G3 | 448 | 123437805 | Trichomonada | Eukaryota | 9 |
| Ath3 | Arabidopsis thaliana | 491 | 6554197 | Viridiplantae | Eukaryota | 9 |
| Ath5 | Arabidopsis thaliana | 431 | 2911082 | Viridiplantae | Eukaryota | 9 |
| Sbi3 | Sorghum bicolor | 473 | 242058941 | Viridiplantae | Eukaryota | 9 |
| Ppa1 | Physcomitrella patens subsp. patens | 405 | 168065030 | Viridiplantae | Eukaryota | 9 |
| Gma1 | Glycine max | 469 | 83853809 | Viridiplantae | Eukaryota | 9 |
| Psi1 | Picea sitchensis | 505 | 148906357 | Viridiplantae | Eukaryota | 10 |
| Sbi2 | Sorghum bicolor | 383 | 242044420 | Viridiplantae | Eukaryota | 7 |
| Mpu1 | Micromonas pusilla CCMP1545 | 461 | 226458924 | Viridiplantae | Eukaryota | 8 |
| Tps2 | Thalassiosira pseudonana CCMP1335 | 564 | 223992571 | Bacillariophyta | Eukaryota | 11 |
| Tps3 | Thalassiosira pseudonana CCMP1335 | 385 | 223998204 | Bacillariophyta | Eukaryota | 7 |
| Ptr2 | Phaeodactylum tricornutum CCAP 1055/1 | 644 | 219112381 | Bacillariophyta | Eukaryota | 11 |
| Pma6 | Perkinsus marinus ATCC 50983 | 385 | 239878631 | Perkinsidae | Eukaryota | 6 |
| Tgo3 | Toxoplasma gondii GT1 | 482 | 221485444 | Apicomplexa | Eukaryota | 10 |
| Tps1 | Thalassiosira pseudonana CCMP1335 | 522 | 224014684 | Bacillariophyta | Eukaryota | 9 |
| Tgo4 | Toxoplasma gondii VEG | 665 | 221505087 | Apicomplexa | Eukaryota | 11 |
| Bsa1 | Bodo saltans | 526 | 206598109 | Bodonidae | Eukaryota | 10 |
| Lma1 | Leishmania major strain Friedlin | 511 | 157873729 | Trypanosomatidae | Eukaryota | 10 |
| Mbr1 | Monosiga brevicollis MX1 | 512 | 167521960 | Codonosigidae | Eukaryota | 9 |
| Pte6 | Paramecium tetraurelia strain d4-2 | 491 | 145483119 | Oligohymenophorea | Eukaryota | 9 |
| Pte8 | Paramecium tetraurelia strain d4-2 | 473 | 145514235 | Oligohymenophorea | Eukaryota | 9 |
| Pte4 | Paramecium tetraurelia strain d4-2 | 424 | 145501808 | Oligohymenophorea | Eukaryota | 10 |
| Pte1 | Paramecium tetraurelia strain d4-2 | 441 | 145493138 | Oligohymenophorea | Eukaryota | 10 |
| Tth2 | Tetrahymena thermophila | 570 | 118348626 | Oligohymenophorea | Eukaryota | 9 |
| Pte7 | Paramecium tetraurelia strain d4-2 | 430 | 145531341 | Oligohymenophorea | Eukaryota | 10 |
| Tth1 | Tetrahymena thermophila | 505 | 146183328 | Oligohymenophorea | Eukaryota | 11 |
| Tth3 | Tetrahymena thermophila | 503 | 118395416 | Oligohymenophorea | Eukaryota | 10 |
| Tth4 | Tetrahymena thermophila | 1325 | 118401229 | Oligohymenophorea | Eukaryota | 8 |
| Pte5 | Paramecium tetraurelia strain d4-2 | 400 | 145528512 | Oligohymenophorea | Eukaryota | 10 |
| Pte2 | Paramecium tetraurelia strain d4-2 | 406 | 145538953 | Oligohymenophorea | Eukaryota | 9 |
| Osa2 | Oryza sativa Japonica Group | 465 | 222625716 | Viridiplantae | Eukaryota | 5 |
| Cre1 | Chlamydomonas reinhardtii | 929 | 159479540 | Viridiplantae | Eukaryota | 6 |
| Gla2 | Giardia lamblia ATCC 50803 | 748 | 159117352 | Hexamitidae | Eukaryota | 10 |
| Tgo2 | Toxoplasma gondii VEG | 299 | 221501858 | Apicomplexa | Eukaryota | 2 |
| Cre2 | Chlamydomonas reinhardtii | 1854 | 159469083 | Viridiplantae | Eukaryota | 9 |
| Pte3 | Paramecium tetraurelia strain d4-2 | 454 | 145493226 | Oligohymenophorea | Eukaryota | 7 |
| Tgo1 | Toxoplasma gondii GT1 | 1659 | 221487433 | Apicomplexa | Eukaryota | 8 |
| Average Size= 572 +/− 312 (all) | ||||||
| Average Size= 476 +/− 71 (w/out Tth4, Cre1, Gla2, Cre2, Tgo1) | ||||||
| Cluster 2 | ||||||
| Ssp3 | Sphingomonas sp. SKA58 | 259 | 94497264 | Alphaproteobacteria | Bacteria | 7 |
| Rsp4 | Ruegeria sp. TM1040 | 252 | 99082858 | Alphaproteobacteria | Bacteria | 5 |
| Pas1 | Photorhabdus asymbiotica subsp. asymbiotica ATCC 43949 | 260 | 211638062 | Gammaproteobacteria | Bacteria | 7 |
| Vpa2 | Variovorax paradoxus S110 | 270 | 239813891 | Betaproteobacteria | Bacteria | 7 |
| Dno1 | Dichelobacter nodosus VCS1703A | 258 | 146329063 | Gammaproteobacteria | Bacteria | 6 |
| Sna1 | Stackebrandtia nassauensis DSM 44728 | 256 | 229865833 | Actinobacteria | Bacteria | 6 |
| Par1 | Psychrobacter arcticus 273-4 | 251 | 71066392 | Gammaproteobacteria | Bacteria | 5 |
| Hca1 | Helicobacter canadensis MIT 98-5491 | 250 | 224418685 | Epsilonproteobacteria | Bacteria | 7 |
| Eta1 | Edwardsiella tarda | 280 | 158512112 | Gammaproteobacteria | Bacteria | 6 |
| Msp4 | Marinomonas sp. MED121 | 258 | 87120732 | Gammaproteobacteria | Bacteria | 7 |
| Orf2 | gamma proteobacterium | 250 | 90416226 | Gammaproteobacteria | Bacteria | 6 |
| Cje1 | Campylobacter jejuni RM1221 | 254 | 57238492 | Epsilonproteobacteria | Bacteria | 7 |
| Orf5 | bacterium Ellin514 | 268 | 223939838 | Verrucomicrobia | Bacteria | 6 |
| Msp1 | Marinomonas sp. MED121 | 253 | 87118707 | Gammaproteobacteria | Bacteria | 8 |
| Ftu1 | Francisella tularensis subsp. holarctica FSC200 | 366 | 167010238 | Gammaproteobacteria | Bacteria | 8 |
| Avi1 | Azotobacter vinelandii DJ | 289 | 226943937 | Gammaproteobacteria | Bacteria | 7 |
| Psp4 | Psychromonas sp. CNPT3 | 274 | 90407709 | Gammaproteobacteria | Bacteria | 8 |
| Nar1 | Novosphingobium aromaticivorans DSM 12444 | 256 | 87200262 | Alphaproteobacteria | Bacteria | 6 |
| Ade1 | Anaeromyxobacter dehalogenans 2CP-C | 254 | 86157393 | Deltaproteobacteria | Bacteria | 7 |
| Dpi1 | Desulfovibrio piger ATCC 29098 | 259 | 212704568 | Deltaproteobacteria | Bacteria | 7 |
| Fva1 | Fusobacterium varium ATCC 27725 | 276 | 253583632 | Fusobacteria | Bacteria | 7 |
| Tps5 | Thermoanaerobacter pseudethanolicus ATCC 33223 | 253 | 167038325 | Firmicutes | Bacteria | 7 |
| Bmu1 | Brachyspira murdochii DSM 12563 | 255 | 227999578 | Spirochaetes | Bacteria | 6 |
| Vpa1 | Veillonella parvula DSM 2008 | 264 | 227372642 | Firmicutes | Bacteria | 8 |
| Ban1 | Bacillus anthracis str. A2012 | 263 | 65318350 | Firmicutes | Bacteria | 8 |
| Taf1 | Thermosipho africanus TCF52B | 254 | 217077973 | Thermotogae | Bacteria | 7 |
| Tde1 | Treponema denticola ATCC 35405 | 262 | 42525707 | Spirochaetes | Bacteria | 7 |
| Cbo6 | Clostridium bolteae | 251 | 160940895 | Firmicutes | Bacteria | 7 |
| Cac1 | Cloacamonas acidaminovorans | 257 | 218961280 | none | Bacteria | 7 |
| Psy1 | Pseudomonas syringae pv. oryzae | 258 | 237801487 | Gammaproteobacteria | Bacteria | 6 |
| Ttu1 | Teredinibacter turnerae T7901 | 256 | 237685094 | Gammaproteobacteria | Bacteria | 7 |
| Orf4 | uncultured marine bacterium 439 | 252 | 40062756 | none | Bacteria | 7 |
| Gbe1 | Granulibacter bethesdensis CGDNIH1 | 253 | 114328287 | Alphaproteobacteria | Bacteria | 7 |
| Lho1 | Laribacter hongkongensis HLHK9 | 310 | 226942144 | Betaproteobacteria | Bacteria | 6 |
| Rco1 | Ricinus communis | 301 | 223512929 | Viridiplantae | Eukaryota | 8 |
| Nmu1 | Neisseria mucosa ATCC 25996 | 256 | 225367635 | Betaproteobacteria | Bacteria | 7 |
| Sli1 | Spirosoma linguale DSM 74 | 254 | 229867512 | Bacteroidetes | Bacteria | 6 |
| Cps1 | Corynebacterium | 260 | 227490282 | Actinobacteria | Bacteria | 8 |
| pseudogenitalium ATCC 33035 | ||||||
| Jde1 | Jonesia denitrificans DSM 20603 | 269 | 227383462 | Actinobacteria | Bacteria | 7 |
| Pac1 | Propionibacterium acnes KPA171202 | 255 | 50842975 | Actinobacteria | Bacteria | 6 |
| Average Size= 264 +/− 21 (all) | ||||||
| Cluster 3 | ||||||
| Mka1 | Methanopyrus kandleri AV19 | 252 | 20093583 | Euryarchaeota | Archaea | 7 |
| Dde1 | Desulfovibrio desulfuricans | 379 | 220904085 | Deltaproteobacteria | Bacteria | 9 |
| Dth2 | Desulfonatronospira thiodismutans ASO3-1 | 254 | 225199785 | Deltaproteobacteria | Bacteria | 7 |
| Rca1 | Roseiflexus castenholzii | 251 | 156743559 | Chloroflexi | Bacteria | 8 |
| Orf6 | uncultured bacterium | 654 | 239787713 | none | Bacteria | 9 |
| Average Size= 358 +/− 174 (all) | ||||||
| Average Size= 252 +/− 2 (w/out Orf6, Dde1) | ||||||
| Cluster 4 | ||||||
| Iho1 | Ignicoccus hospitalis KIN4/I | 240 | 156936864 | Crenarchaeota | Archaea | 7 |
| Bsp1 | Beggiatoa sp. PS | 787 | 153869281 Gammaproteobacteria | Bacteria | 10 | |
| Cbu3 | Coxiella burnetii Dugway 5J108-111 | 274 | 209364180 Gammaproteobacteria | Bacteria | 8 | |
| Cgl1 | Chryseobacterium gleum ATCC 35910 | 505 | 227369714 | Bacteroidetes | Bacteria | 8 |
| Gla1 | Giardia lamblia ATCC 50803 | 520 | 159115095 | Hexamitidae | Eukaryota | 10 |
| Min1 | Methylacidiphilum infernorum V4 | 267 | 189218632 | Verrucomicrobia | Bacteria | 8 |
| Average Size= 432 +/− 214 (all) | ||||||
| Average Size= 260 +/− 18 (w/out Bsp1, Cgl1, Gla1) | ||||||
| Cluster 5 | ||||||
| She1 | Slackia heliotrinireducens DSM 20476 | 277 | 229879562 | Actinobacteria | Bacteria | 8 |
| Ele1 | Eggerthella lenta DSM 2243 | 307 | 227411437 | Actinobacteria | Bacteria | 8 |
| Rxy1 | Rubrobacter xylanophilus DSM 9941 | 267 | 108803101 | Actinobacteria | Bacteria | 8 |
| Sfu1 | Syntrophobacter fumaroxidans MPOB | 269 | 116750841 | Deltaproteobacteria | Bacteria | 7 |
| Ptr3 | Phaeodactylum tricornutum | 2798 | 219127009 | Bacillariophyta | Eukaryota | 4 |
| Mch1 | Microcoleus chthonoplastes | 267 | 224407624 | Cyanobacteria | Bacteria | 8 |
| Ter2 | Trichodesmium erythraeum | 305 | 113475233 | Cyanobacteria | Bacteria | 8 |
| Ssp1 | Synechococcus sp. JA-3-3Ab | 317 | 86606127 | Cyanobacteria | Bacteria | 8 |
| Pca1 | Pyrobaculum calidifontis JCM 11548 | 244 | 126458964 | Crenarchaeota | Archaea | 8 |
| Average Size= 561 +/− 839 (all) | ||||||
| Average Size= 282 +/− 25 (w/out Ptr3) | ||||||
| Cluster 6 | ||||||
| Sus1 | Candidatus Solibacter usitatus Ellin6076 | 281 | 116624708 | Acidobacteria | Bacteria | 8 |
| Sth2 | Symbiobacterium thermophilum IAM 14863 | 279 | 51892120 | Firmicutes | Bacteria | 8 |
| Bsu1 | Brucella suis 1330 | 289 | 23500891 | Alphaproteobacteria | Bacteria | 8 |
| Ooe1 | Oenococcus oeni PSU-1 | 283 | 116491798 | Firmicutes | Bacteria | 8 |
| Pto1 | Picrophilus torridus DSM 9790 | 333 | 48478318 | Euryarchaeota | Archaea | 8 |
| Sth1 | Sphaerobacter thermophilus | 282 | 229877687 | Chloroflexi | Bacteria | 8 |
| Mth2 | Moorella thermoacetica | 271 | 83589239 | Firmicutes | Bacteria | 8 |
| Kcr1 | Candidatus Korarchaeum cryptofilum OPF8 | 285 | 170290371 | Korarchaeota | Archaea | 8 |
| Mxa1 | Myxococcus xanthus DK 1622 | 260 | 108758495 | Deltaproteobacteria | Bacteria | 7 |
| Dra1 | Deinococcus radiodurans R1 | 255 | 15805571 | Deinococcus-Thermus | Bacteria | 8 |
| Sma1 | Staphylothermus marinus F1 | 250 | 126465319 | Crenarchaeota | Archaea | 8 |
| Dac1 | Denitrovibrio acetiphilus | 274 | 227423788 | Deferribacteres | Bacteria | 7 |
| Emi1 | Elusimicrobium minutum Pei191 | 275 | 187251557 | candidate division TG1 | Bacteria | 7 |
| Hbu2 | Hyperthermus butylicus | 255 | 124028506 | Crenarchaeota | Archaea | 8 |
| Average Size= 277 +/− 20 (all) | ||||||
| Cluster 7 | ||||||
| Bad1 | Bifidobacterium adolescentis ATCC 15703 | 292 | 119026567 | Actinobacteria | Bacteria | 8 |
| Gva1 | Gardnerella vaginalis ATCC 14019 | 267 | 227507357 | Actinobacteria | Bacteria | 8 |
| Average Size= 280 +/− 18 (all) | ||||||
| Cluster 8 | ||||||
| Asa1 | Aliivibrio salmonicida | 279 | 16605593 | Gammaproteobacteria | Bacteria | 8 |
| Rru1 | Rhodospirillum rubrum | 276 | 83592684 | Alphaproteobacteria | Bacteria | 8 |
| Rsp3 | Roseovarius sp. HTCC2601 | 274 | 114764120 | Alphaproteobacteria | Bacteria | 8 |
| Rsp1 | Ruegeria sp. TM1040 | 278 | 99080207 | Alphaproteobacteria | Bacteria | 7 |
| Msp3 | Magnetococcus sp. MC-1 | 265 | 117925601 | Proteobacteria | Bacteria | 6 |
| Fpe1 | Fulvimarina pelagi HTCC2506 | 275 | 114707272 | Alphaproteobacteria | Bacteria | 7 |
| Bja2 | Bradyrhizobium japonicum USDA 110 | 287 | 27375621 | Alphaproteobacteria | Bacteria | 6 |
| Hne1 | Hyphomonas neptunium ATCC 15444 | 314 | 114797241 | Alphaproteobacteria | Bacteria | 9 |
| Cbu2 | Coxiella burnetii RSA 331 | 275 | 161831015 | Gammaproteobacteria | Bacteria | 7 |
| Lsp1 | Limnobacter sp. MED105 | 278 | 149925520 | Betaproteobacteria | Bacteria | 7 |
| Nmo1 | Nitrococcus mobilis Nb-231 | 266 | 88811005 | Gammaproteobacteria | Bacteria | 8 |
| Mca1 | Methylococcus capsulatus str. Bath | 294 | 53802665 | Gammaproteobacteria | Bacteria | 6 |
| Pir1 | Polaribacter irgensii 23-P | 281 | 88803086 | Bacteroidetes | Bacteria | 7 |
| Kko1 | Kangiella koreensis DSM 16069 | 268 | 227997603 | Gammaproteobacteria | Bacteria | 8 |
| Har1 | Herminiimonas arsenicoxydans | 287 | 134096092 | Betaproteobacteria | Bacteria | 7 |
| Swo1 | Shewanella woodyi ATCC 51908 | 268 | 170728324 | Gammaproteobacteria | Bacteria | 8 |
| Ptu1 | Pseudoalteromonas tunicata D2 | 269 | 88860323 | Gammaproteobacteria | Bacteria | 8 |
| Ama2 | Alteromonas macleodii 'Deep ecotype' | 268 | 196158505 | Gammaproteobacteria | Bacteria | 7 |
| Sbe1 | Shewanella benthica KT99 | 267 | 163752420 | Gammaproteobacteria | Bacteria | 8 |
| Msu1 | Mannheimia succiniciproducens MBEL55E | 266 | 52424462 | Gammaproteobacteria | Bacteria | 8 |
| Psp3 | Photobacterium sp. SKA34 | 267 | 89072545 | Gammaproteobacteria | Bacteria | 8 |
| Afe1 | Acidithiobacillus ferrooxidans ATCC 23270 | 264 | 218665563 | Gammaproteobacteria | Bacteria | 8 |
| Pne1 | Polynucleobacter necessarius subsp. asymbioticus QLW-P1DMWA-1 | 272 | 145589361 | Betaproteobacteria | Bacteria | 8 |
| Lsp2 | Limnobacter sp. MED105 | 289 | 149926219 | Betaproteobacteria | Bacteria | 8 |
| Eco1 | Eikenella corrodens ATCC 23834 | 270 | 225024689 | Betaproteobacteria | Bacteria | 7 |
| Ama1 | Acaryochloris marina | 278 | 158336922 | Cyanobacteria | Bacteria | 9 |
| Tsp1 | Thioalkalivibrio sp. K90mix | 268 | 224818668 | Gammaproteobacteria | Bacteria | 8 |
| Rso1 | Ralstonia solanacearum | 273 | 17549483 | Betaproteobacteria | Bacteria | 8 |
| Ppe1 | Proteus penneri ATCC 35198 | 271 | 226330327 | Gammaproteobacteria | Bacteria | 8 |
| Iba1 | Idiomarina baltica OS145 | 264 | 85713215 | Gammaproteobacteria | Bacteria | 8 |
| Average Size= 275 +/− 11 (all) | ||||||
| Cluster 9 | ||||||
| Epe1 | Endoriftia persephone 'Hot96_1+Hot96_2' | 287 | 167948520 | Gammaproteobacteria | Bacteria | 5 |
| Cluster 10 | ||||||
| Pal1 | Providencia alcalifaciens DSM 30120 | 271 | 212712467 | Gammaproteobacteria | Bacteria | 8 |
| Cluster 11 | ||||||
| Orf3 | uncultured archaeon GZfos34A6 | 276 | 52549977 | none | Archaea | 8 |
| Mma1 | Methanosarcina mazei Go1 | 270 | 21228951 | Euryarchaeota | Archaea | 8 |
| Mma2 | Methanococcus maripaludis S2 | 270 | 45358505 | Euryarchaeota | Archaea | 8 |
| Average Size= 272 +/− 3 (all) | ||||||
| Cluster12 | ||||||
| Tko1 | Thermococcus kodakarensis KOD1 | 254 | 57640914 | Euryarchaeota | Archaea | 7 |
| Tba1 | Thermococcus barophilus MP | 251 | 223475524 | Euryarchaeota | Archaea | 8 |
| Mbo1 | Methanoregula boonei 6A8 | 269 | 154149849 | Euryarchaeota | Archaea | 8 |
| Sma2 | Staphylothermus marinus F1 | 265 | 126466107 | Crenarchaeota | Archaea | 8 |
| Average Size= 260 +/− 9 (all) | ||||||
| Cluster 13 | ||||||
| Tte1 | Thermobaculum terrenum ATCC BAA-798 | 255 | 227375491 | none | Bacteria | 8 |
| Cbe1 | Clostridium beijerinckii NCIMB 8052 | 272 | 150017843 | Firmicutes | Bacteria | 8 |
| Vdi1 | Veillonella dispar ATCC 17748 | 264 | 238018311 | Firmicutes | Bacteria | 8 |
| Nma1 | Nitrosopumilus maritimus SCM1 | 257 | 161528556 | Crenarchaeota | Archaea | 7 |
| Bbr2 | Brevibacillus brevis | 274 | 226314422 | Firmicutes | Bacteria | 7 |
| Gka1 | Geobacillus kaustophilus HTA426 | 300 | 56421519 | Firmicutes | Bacteria | 8 |
| Bcl1 | Bacillus clausii KSM-K16 | 272 | 56964722 | Firmicutes | Bacteria | 8 |
| Psp2 | Paenibacillus sp. JDR-2 | 272 | 251794851 | Firmicutes | Bacteria | 8 |
| Oih1 | Oceanobacillus iheyensis HTE831 | 285 | 23099829 | Firmicutes | Bacteria | 8 |
| Sau1 | Staphylococcus aureus subsp. aureus Mu50 | 275 | 15923912 | Firmicutes | Bacteria | 8 |
| Average Size= 273 +/− 13 (all) | ||||||
| Cluster 14 | ||||||
| Bja1 | Bradyrhizobium japonicum USDA 110 | 380 | 27376265 | Alphaproteobacteria | Bacteria | 8 |
| Ssp5 | Sphingomonas sp. SKA58 | 304 | 94498747 | Alphaproteobacteria | Bacteria | 7 |
| Pth1 | Pelotomaculum | 299 | 147678596 | Firmicutes | Bacteria | 7 |
| thermopropionicum SI | ||||||
| Dau1 | Desulforudis audaxviator | 394 | 169832116 | Firmicutes | Bacteria | 8 |
| Dre4 | Desulfotomaculum reducens MI-1 | 426 | 134299284 | Firmicutes | Bacteria | 9 |
| Abo1 | Aciduliprofundum boonei T469 | 254 | 223473124 | Euryarchaeota | Archaea | 8 |
| Lbi1 | Leptospira biflexa serovar Patoc strain 'Patoc 1 (Paris)' | 325 | 183219704 | Spirochaetes | Bacteria | 8 |
| Average Size= 340 +/− 61 (all) | ||||||
| Average Size= 296 +/− 30 (w/out Bja1, Dau1, Dre4) | ||||||
| Cluster 15 | ||||||
| Orf1 | synthetic construct | 284 | 62258462 | none | Unclassified | 8 |
| Vsp2 | Verrucomicrobium spinosum DSM 4136 | 264 | 171915322 | Verrucomicrobia | Bacteria | 8 |
| Rme1 | Ralstonia metallidurans CH34 | 268 | 94311333 | Betaproteobacteria | Bacteria | 8 |
| Pre1 | Providencia rettgeri DSM 1131 | 264 | 223992411 | Gammaproteobacteria | Bacteria | 8 |
| Sso1 | Sulfolobus solfataricus P2 | 293 | 15899038 | Crenarchaeota | Archaea | 8 |
| Mmu1 | Mobiluncus mulieris 35243 | 361 | 227876711 | Actinobacteria | Bacteria | 9 |
| Aau1 | Arthrobacter aurescens TC1 | 300 | 119952309 | Actinobacteria | Bacteria | 8 |
| Lmo1 | Listeria monocytogenes EGD-e | 246 | 16802663 | Firmicutes | Bacteria | 8 |
| Ste2 | Sebaldella termitidis ATCC 33386 | 246 | 229881273 | Fusobacteria | Bacteria | 7 |
| Bsp2 | Bacillus sp. B14905 | 282 | 126650500 | Firmicutes | Bacteria | 8 |
| Cph2 | Chlorobium phaeobacteroides | 408 | 189499528 | Chlorobi | Bacteria | 9 |
| Afu1 | Archaeoglobus fulgidus | 325 | 11499708 | Euryarchaeota | Archaea | 7 |
| Dha1 | Desulfitobacterium hafniense DCB-2 | 312 | 219669180 | Firmicutes | Bacteria | 7 |
| Dre3 | Desulfohalobium retbaense | 569 | 227420936 | Deltaproteobacteria | Bacteria | 7 |
| Average Size= 316 +/− 86 (all) | ||||||
| Average Size= 280 +/− 26 (w/out Mmu1, Cph2, Dre3) | ||||||
Cluster 1 (43 proteins) includes proteins derived from plants, apicomplexa, ciliates and other unicellular eukaryotes (See Figure 1 and Table 1). Considering the organismal diversity of cluster 1 proteins, it is not surprising that these sequences are so diverse. The average size of these proteins is 572 ± 312 amino acids (aas), but five of them (Tth4, Cre1, Gla2, Cre2 and Tgo1) are much larger than the others. Excluding these five proteins, the average size is 476 ± 71 aas. Extra hydrophilic domains were found at the N-termini and between TMSs, but not at the C-termini. However, these regions did not represent domains recognized by CDD. Apicomplexa proteins occur in five distinct positions, plant homologues occur in three positions, and the Bacillariophyta, Codonosigidae and Oligohymenophorea phyla are each present in two positions in cluster 1, suggesting a lack of orthology. Explanations for this diversity include (1) the occurrence of pseudogenes, (2) early arising paralogues and (3) horizontal gene transfer.
Cluster 2 consists of 40 homologues, all much smaller than those in cluster 1 (average size is 264 ± 21 aas). The largest of these (366 aas) is from Franscisella tularensis, a γ-proteobacterium, with a unique N-terminal 110 aa hydrophilic extension. Although individual proteins are predicted to have from 5 to 8 TMSs, based on the TMHMM 2.0 program [31, 32], an AveHAS plot [35] suggested the presence of 8 TMSs.
Cluster 2 proteins derive from (five sub-phyla) Proteobacteria, Actinobacteria, Verrucomicrobia, Fusobacteria, Firmicutes, Spirochetes and Thermotogae plus one plant, Ricinus communis. This last mentioned protein could be a chloroplast protein, explaining its clustering with bacterial homologues. Extensive horizontal gene transfer has evidently occurred during the evolution of this cluster.
The five cluster 3 proteins derive from δ-Proteobacteria, Chloroflexi and a Euryarchaeon. The average size of these proteins is 358 ± 174 aas. Dde1 (379 aas) from Desulfovibrio desulfuricans contains an insertion of unknown function between the first and second TMSs
The larger Orf6 protein contains an N-terminal 160 aa N-terminal degP_htrA_DO domain [43] with probable serine protease and/or chaperone activity [44, 45]. An N-terminal TMS preceding this domain may target it to the cell surface. Members of this serine protease family usually reside in the periplasm of Gram-negative bacteria. It is possible that Orf6 hydrolyzes peptides and transports the products. Although a phylum assignment for Orf6 has not been made, the corresponding 16S rRNA (Fig. 2) clusters with the Verrucomicrobia.
The average protein size of cluster 4 (6 proteins) is 432 ± 214 aas, but when the large Bsp1, Cgl1 and Gla1 proteins were excluded, it was 260 ± 18 aas. Members possess 7, 8 or 10 predicted TMSs. Bsp1 was found to have an extra C-terminal domain with two tandem copies of the Universal Stress Protein (USP_like) domain. USP proteins are upregulated in response to stress-causing agents [46] and possess an ATP-binding alpha/beta fold motif. The presence of a potential ATP-binding or hydrolyzing domain introduces the possibility that this transporter may either be regulated by ATP or be energized by ATP hydrolysis and thus function as a primary active transporter rather than a secondary carrier (see [47, 48]). Bacterial arsenite and ECF transporters provide examples of carriers with superimposed ATPases that allow secondary carriers to function as primary active transporters (see [47, 49] and references cited therein).
The nine proteins comprising cluster 5 derive from all three domains of life. The bacterial proteins are from Actinobacteria, Cyanobacteria and δ-Proteobacteria, while the single eukaryotic and archaeal proteins are from Bacillariophyta and Crenarchaeota, respectively. When the large (2798 aa) eukaryotic Ptr3 homologue was excluded, an average value of 282 ± 25 aas was calculated. Ptr3 was predicted to have 8 TMSs by the WHAT program and 10 TMSs based on visual inspection of the hydropathy profile together with comparison with its 8 TMS homologues. Ptr3 has an N-terminal hydrophobic domain followed by a >2000 aa hydrophilic domain that shows sequence similarity with a hydrophilic protein of 2409 aas from Thalassiosira pseudonana. Based on the 16S rRNA tree, the three Actinobacterial homologues and the three Cyanobacterial homologues may all be orthologous.
The fourteen cluster 6 proteins derive from a diversity of bacterial and archaeal phyla. This fact, plus the observation that the bacterial and archaeal homologues are interspersed, clearly suggests that extensive lateral gene transfer has occurred. The sizes of these proteins range from 250 aas to 333 aas (average, 277 ± 20 aas with 7 or 8 putative TMSs). Pto1 from Picrophilus torridus, a Euryarchaeota, is the largest protein with 333 aas, reflecting a unique 60 aa hydrophilic insert between TMSs 5 and 6.
Cluster 7 contains only 2 proteins derived from Actinobacteria. Bad1 from Bifidobacterium adolescentis and Gva1 from Gardnerella vaginalis are 292 and 267 aas in size, respectively, both with 8 putative TMSs.
Cluster 8 (30 proteins) is the third largest cluster and includes proteins solely from bacteria. These proteins cluster closely together in two sub-clusters. The first sub-cluster derives from α- and γ-Proteobacteria while the second, primarily from β- and γ-Proteobacteria, but including one protein each from Bacteroidetes and Cyanobacteria, are interspersed, suggestive of lateral gene transfer. Very little size variation is observed with the average size being 275 ± 11 aas and the majority possessing 8 TMSs. Each of clusters 9 and 10 consists of a single protein from a γ-Proteobacterium with sizes, and topologies similar to those in cluster 8.
Cluster 11 (3 proteins) and cluster 12 (4 proteins) derive from archaea. These proteins possess 8 putative TMSs with an average size of 266 ± 7 aas. Comparison with the 16S rRNA tree revealed that these proteins cannot be orthologous.
Cluster 13 (10 proteins) derives primarily from Firmicutes, and most of these proteins may be orthologues. However, homologues from a single Crenarchaeota and a bacterium of unknown phylum are also present. These proteins are of uniform size (273 ± 13 aas) and topology (7 to 8 putative TMSs). The bacterial Tte1 homologue 16S rRNA clusters with the Chloroflexi.
Cluster 14 (7 proteins) derives from α-Proteobacteria, Firmicutes, Spirochetes, and Euryarchaeota. Considerable size variation is observed with the Bja1, Dau1 and Dre4 proteins being the largest. The average size for this cluster is 340 ± 61 aas. Despite the size variation and large phylogenetic distances, the topologies are fairly uniform with members possessing 7 to 9 putative TMSs.
Cluster 15 (14 proteins) derives from eight different archaeal and bacterial phyla. Three Firmicute homologues are sandwiched in between those derived from other phyla. The size and sequence divergence of these homologues is tremendous (246 aas to 569 aas), but their predicted topologies range from 7 to 9 TMSs with the majority exhibiting 8 TMSs. The average size for the proteins in this cluster is 316 ± 86 aas.
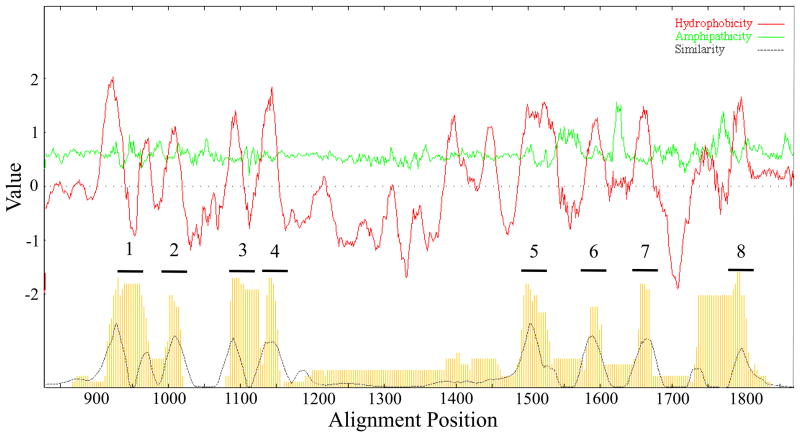
3.2 Topological analyses
The average hydropathy (top, dark lines), amphipathicity (top, light lines) and similarity (bottom) plots for the 189 TSUP family members included in this study are presented in Figures 3 and S3. Eight major peaks of hydrophobicity correlate with 8 major peaks of similarity. The peaks of amphipathicity observed between the TMSs exceed the amphipathicities of the transmembrane domains as is frequently the case for transmembrane permeases. TMSs 1 to 4 cluster together separated from TMSs 5 to 8 which also cluster together, suggestive of intragenic duplication as demonstrated below. Additionally, TMSs 1 and 2 cluster closely together, as do TMSs 3 and 4, consistent with duplication of a primordial 2 TMS hairpin peptide. TMSs 5 to 8 are more distant from each other than are TMSs 1 to 4.
Figure 3.
Portion of average hydropathy, amphipathicity, and similarity (AveHAS) plots for the 189 TSUP family proteins included in this study. A magnification of the TMS-containing region is presented due to the large size of the plot which reveals 8 clear TMSs. However, as many as 12 poorly conserved peaks of hydrophobicity can be seen, suggesting that some homologues have additional TMSs.
The first four TMSs are separated from the last four by a large hydrophilic loop not recognized as a conserved domain by CDD. Two peaks of hydrophobicity and two corresponding peaks of similarity are observed within the loop region prior to TMS 5. These peaks may be due to the 10 TMS proteins in eukaryotes.
3.3 Establishing internal repeats
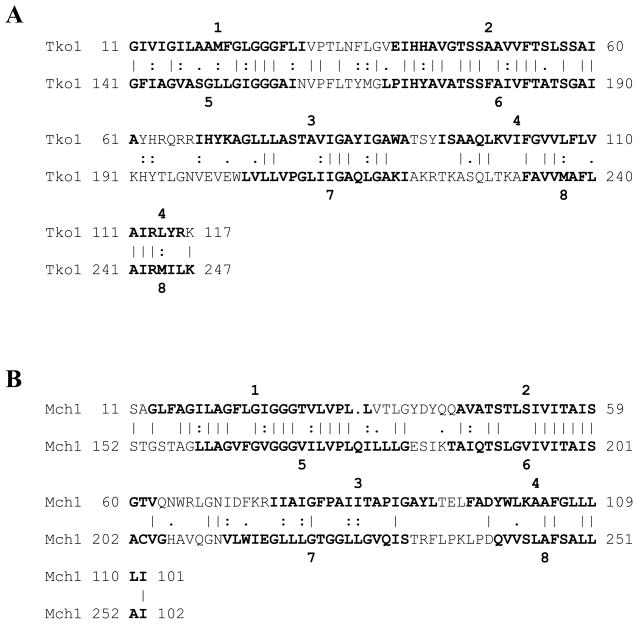
Most TSUP family members possess 8 predicted TMSs, while those that deviate from this topology may have 7 or 9. To establish the evolutionary origin of this family, the IC and GSAT/GAP programs were used to compare putative repeat elements [2, 6]. Comparing TMSs 1–4 of all TSUP proteins with TMSs 5–8 of the same homologues yielded many comparison scores in excess of 10 S.D. For example, comparing TMSs 1 – 4 with TMSs 5–8 of Tko1 resulted in a comparison score of 26 S.D. (Fig. 4A), while the same comparison for Mch1 resulted in a comparison score of 17 S.D. (Fig. 4B). Thus, an intragenic duplication event occurred during the evolution of the TSUP family. The large comparison scores indicate that the duplication event occurred fairly recently in evolutionary time. When comparing 2 TMS hairpin structures with the adjacent hairpins [50], comparison scores were insufficient to establish homology. However, we have demonstrated homology for these 2 TMS repeats in proteins of other families homologous to TSUP proteins (see Discussion).
Figure 4.
Demonstration that 8 TMS TSUP family members arose by intragenic duplication of a primordial 4 TMS encoding genetic element. (A) GAP alignment of TMSs 1–4 of TSUP Tko1 (Thermococcus kodakarensis; gi# 57640914) with TMSs 5–8 of the same protein. Initial identification of repeat units was done using the IC program. The GAP program was run with default settings and 500 random shuffles. Residue identities are signified by vertical lines, while close and more distant similarities are signified by colons or periods, respectively. The numbers at both ends of each line signify the positions of the residues in the proteins. TMS positions were predicted using the TMHMM 2.0 program for this and subsequent comparisons. A comparison score of 26 S.D. was obtained. (B) GAP alignment of TMSs 1–4 of TSUP Mch1 (Microcoleus chthonoplastes; gi# 224407624) with TMSs 5–8 of Mch1. A comparison score of 17 S.D. was obtained.
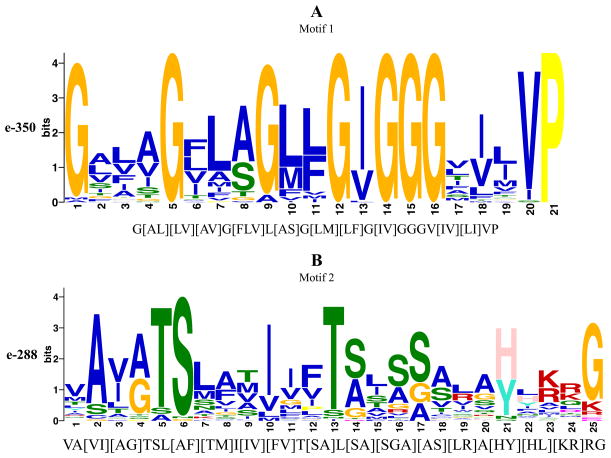
3.4 Conserved motifs
The ClustalX program did not identify fully conserved amino acyl residues [24, 25], but the MEME program [29, 30] revealed two well conserved motifs (Figure 5A–B) derived from a 46 residue stretch that spans the first and second TMSs. Motif 1, spanning the second half of TMS 1, is 21 residues in length (Fig. 5A). Glycines (Gs) at positions 1, 5, 9, 12, and 14–16 are the best conserved. I, V and P at positions 13, 20 and 21, respectively, are also well conserved. The large number of Gs in TMS 1 suggests a high degree of conformational flexibility, possibly important for function. A “reverse” Rossmann fold (GxGxxG) sub-motif (GxxGxG) is observed in motif 1 which resides in TMS 1 and may play a role in regulating transport given its potential for binding nucleotides [49, 51, 52].
Figure 5.
(A–B) The two best conserved motifs found within the TSUP family as predicted by MEME. Corresponding statistical scores are presented on the y-axis. MAST predictions of motifs based on the MEME results are presented on the x-axis below each motif graphic.
Motif 2, the second best conserved motif, is 25 residues in length (Fig. 5B). The sub-motif A(VI)(AG)TSL(AF)(TM) (positions 2–9) is highly conserved. Residues 1–20 of this motif mainly comprise TMS 2 within which I, T, and three Ss are well conserved at positions 10, 13, 14, 16 and 17. Residue 21 (H or Y) marks the beginning of the loop region connecting TMSs 2 and 3, with a conserved G at position 25. Motifs 1 and 2, spanning TMSs 1 and 2, respectively, repeat in TMSs 5 and 6, as expected since the second half of these proteins arose by an intragenic duplication event.
3.5 Genome context analyses for functional predictions
Small gene size, high gene density, intronless coding regions and simple operon organization in bacterial genomes render functional predictions feasible [53]. Our previous molecular genetics studies based on operon context have proven to be successful in identifying the substrates of transporters of unknown function [54–56]. Operon context analyses and identification of transcription factor-controlled regulons were facilitated by the use of the SEED database [40] along with RegPrecise and RegPredict [41, 42]. SEED identifies close homologues using the PSI-BLAST algorithm [19, 20]. Our findings are summarized in Table 2. For detailed analyses by cluster, see Supplementary Material designated “Functional Predictions.”
Table 2.
Summary of functional predictions made for representative members of each phylogenetic cluster (Figure 1).
| Cluster # | Proposed Functions |
|---|---|
| 1 | Transport of sulfur-based compounds; FeS cluster assembly. |
| 2 | Transport of peptides/amino acids/glycerate. Nitrogen-based compound transport; |
| 3 | Arsenate/arsenite resistance; Oxidative, heat and metabolic stress response. |
| 4 | Transport of sulfur-based compounds. |
| 5 | Transport of sulfur-based compounds/ions/peptides/amino acids; FeS cluster assembly; Stress response. |
| 6 | Transport of carbon or sulfur-based compounds. |
| 7 | Stress response and virulence. |
| 8 | Transport of sulfur-based compounds. |
| 9–11 | None identified |
| 12 | Transport of amino acids. |
| 13 | Transport of sulfur-based compounds/Iron/peptides/amino acids. |
| 14 | Transport of sulfur-based compounds. |
| 15 | Transport of sulfur-based compounds; Extrusion of sulfite. |
3.5.1 Functional predictions
Cluster 1 homologues in Mycoplasma penetrans, M. gallisepticum, Staphylococcus aureus and Holdemania filiformis were designated iron-sulfur (FeS) cluster assembly proteins, SufB. The SUF system, encoded by the sufABCDSE operon is one of the three FeS cluster assembly systems with the other two being the iron-sulfur cluster (ISC) and nitrogen fixation (NIF) systems [57]. FeS clusters serve as cofactors mediating electron transfer [58, 59]. SufA may be an iron chaperone and is essential for FeS cluster assembly under aerobic, but not anaerobic conditions [60]. SufS is a cysteine desulfurase, and SufE is a scaffold protein. SufB and the paralogous SufD, both of which are homologous to Ath3 from A. thaliana, function as parts of a cytoplasmic complex along with SufC [61]. It seems likely that Ath3 is chloroplastic, and its bacterial homologues mediate the uptake of sulfur-based compounds (see also cluster 5 analyses).
In cluster 2, the E. coli YfcA protein (TC# 2.A.102.3.1) is one of eight presumed orthologues. One homologue is encoded in an operon with a gene encoding a phosphoserine phosphatase in Silibacter sp. TM1040. Genes encoding phosphoserine aminotransferase, D-3-phosphoglycerate dehydrogenase, serine/threonine phosphatase and L-threonine 3-dehydrogenase are near this operon. These genes encode the serine biosynthetic pathway starting with 3-phosphoglycerate. Glycerate is a known carbon source for E. coli and S. typhimurium although the transporter gene has not been identified [62, 63]. Therefore YfcA could be the long sought glycerate uptake porter.
Pas1 from Photorhabdus asymbiotica and its homologue from Neisseria meningitidis, are encoded in monocistronic operons, but surrounding genes are involved in the methycitrate cycle, acetyl-CoA generation and the propionate-CoA to succinate module. Dno1 is in an operon along with EngB, a GTP-binding protein, and is next to L-asparaginase and a putative protease in Dichelobacter nodosus. In Marinomonas sp. MWYL1, genes involved in glycine and serine utilization and glycerolipid metabolism surround the Dno1 homologue. Par1 from Psychrobacter sp. 273-4 is encoded near a gene for a putative glutamate symporter of the Dicarboxylate/Amino Acid:Cation (Na+ or H+) Symporter (DAACS) family (TC# 2.A.23) and a butyryl-CoA dehydrogenase involved in Ile/Val degradation and Lys fermentation. Its homologue in Mannheimia succiniciproducens is part of an operon along with a murein endopeptidase, in an arrangement similar to that observed for the Dno1 homologue in P. multocida. A function involving peptide/amino acid transport is suggested.
Mka1 from M. kandleri in cluster 3 is part of an operon along with a predicted nucleotide-binding protein related to the universal stress protein, UspA, which is upregulated by metabolic, oxidative and temperature stresses [64]. Genes involved with oxidative stress also surround the Mka1 homologue from Thermococcus kodakarensis. The Mka1 homologues in Pyrococcus furiosus and P. horikoshii are found in operons that appear to function in protein degradation, possibly suggesting an amino acid transport role, and/or representing a part of the stress response. The large Dde1-like proteins of Desulfovibrio desulfurican, D. vulgaris and D. baculatum are encoded adjacently and transcribed divergently from a sigma-54 (σ54)-dependent transcriptional regulator. σ54 or σN often plays a global regulatory role for genes encoding proteins involved in nitrogen metabolism [65]. RegPrecise identified RpoN as the regulator of Dde1 transcription.
Homologues of the Roseiflexus castenholzi Rca1 protein are encoded in operons that are divergently transcribed from a gene encoding a putative efflux pump of the Arsenical Resistance-3 (ACR3) family (TC# 2.A.59). Upstream of this operon is a gene of the putative permease Duf318 family (TC# 9.B.28). Duf318 homologues have been implicated in arsenate/arsenite resistance [66]. Consistent with this finding, closely upstream of the Duf318 genes, genes encoding redox-active disulfide proteins and an ArsR transcriptional regulator are found. ArsR homologues regulate many transporters including Duf318 transporters, and it may also regulate Rca1 [49]. The two transporters may transport arsenate/arsenite, and/or other stress-related substrates.
Cluster 4 orthologues in two Thermus thermophilus strains are located in operons along with genes encoding a ferredoxin-sulfite reductase, a sulfate adenylyltransferase involved in inorganic sulfur assimilation and a (phospho)adenylyl-sulfate reductase involved in cysteine biosynthesis. Several genes homologous to those involved in heme and siroheme biosynthesis are also part of the operon. The siroheme prosthetic group is known to be essential for the function of sulfite-reductases, which convert sulfite (derived from sulfate) to sulfide [67]. Thus, cluster 4 proteins may be involved in the uptake of sulfate or other sulfur-based compounds, a suggestion supported by sequence similarity between cluster 4 proteins and CysZ of Corynebacterium glutamicum, a putative sulfate transporter [16].
Proteins of cluster 5 appear to exhibit functional diversity. One of two Syntrophobacter fumaroxidans paralogues is located adjacent to 3 genes involved in cobalt/zinc/cadmium resistance with sequence similarity to members of the RND superfamily (TC# 2.A.6). It is possible that the second Sfu1 paralogue extrudes anions or mediates the uptake of cations during Co2+/Zn2+/Cd2+-induced stress. The homologue in Meiothermus ruber is surrounded on one side by an operon encoding an acyltransferase and peptidase M19, and on the other side by an ATP-dependent protease, suggestive of a possible amino acid or peptide transport role. However, the homologue in Meiothermus silvanus is located near a gene encoding a scaffold protein for (4Fe-4S) cluster assembly, which makes transport of sulfur-based compounds a reasonable possibility. A Trichodesmium erythraeum-derived Ter2 homologue in Nostoc punctiforme is in an operon also coding for a cysteine desulfurase and the sulfur oxidation molybdopterin C protein. Thus, cluster 5 proteins may transport sulfur-based compounds or amino acids/peptides in response to stress.
Cluster 6 includes Sus1 and its homologues, which all appear in operons along with transcriptional regulators of the GntR superfamily [68–71]. Members of the GntR family usually respond to sugar metabolite effector molecules and control genes involved in carbon metabolism [72, 73]. Unlike its homologue in Chitinophaga pinensis, Sus1 from Solibacter usitatus is not part of an operon with a GntR transcription regulator, but is instead in an operon with two hypothetical proteins and rhodanase (thiosulfate sulfurtransferase; see cluster 15). Roles in carbon or sulfur-based compound transport are implied.
The only protein from cluster 7 to be in SEED was Bad1 from Bifidobacterium adolescentis. Bad1 is found in a monocistronic operon adjacent to operons encoding heat stress and pathogenicity genes. Thus, Bad1 and other cluster 7 proteins could play roles in stress responses and virulence.
In the mainly proteobacterial cluster 8, Rru1 from Rhodospirillum rubrum, Rsp1 and its homologues from Silibacter sp. TM1040 and Magnetococcus sp. MC-1, are found in operons with or nearby genes encoding a γ-glutamyltranspeptidase involved in glutathione and poly-gamma-glutamate biosynthesis as well as enzymes involved in the utilization of glutathione as a sulfur source. Rsp1 and its homologues may transport glutathione or another compound to supply sulfur to supplant the glutathione utilization pathway. TsaS (TC# 2.A.102.1.1) is a cluster 8 protein predicted to be a 4-toluenesulfonate uptake permease, further supporting a role in transport of sulfur-containing compounds [10, 12, 14].
The 2 proteins comprising clusters 9 and 10 are from organisms not included in the SEED database. In cluster 11, Mma2 from Methanococcus maripaludis is in an operon with genes encoding an FMN adenylyltransferase and a putative membrane protein with sequence similarity to members of the Autoinducer-2 Exporter (AI-2E) family (TC# 2.A.86). Tko1 of T. kodakarensis (cluster 12) is encoded within an operon along with a gene encoding glycyl-tRNA synthetase. Therefore, Tko1 may function as a glycine uptake protein. It is likely that Cbe1 from Clostridium beijerincki in cluster 13 functions in sulfur-based compound uptake. Although not present in a polycistronic operon, it is near genes encoding an iron-sulfur-binding protein, the dissimilatory sulfite reductase (desulfoviridin) and the CoA-disulfide reductase. The nma1 gene from Nitrosopumilus maritimus is in a dense gene cluster containing the iron-dependent repressor IdeR of the DtxR family. Deregulation of iron metabolism or superoxide dismutase deficiency can favor the Fenton reaction, which contributes to oxidative stress, DNA damage, mutagenesis and sensitivity to H2O2 [74, 75]. Nma1 may therefore be regulated by IdeR and function in iron uptake. The Nma1 homologue in Thermococcus onnurineus, localized to an operon coding for an aminopeptidase and a dehydrogenase, may be an amino acid/peptide uptake permease.
Pth1 of cluster 14 from Pelotomaculum thermopropionicum appears in an operon with 2 hypothetical proteins and is accompanied by an adjacent transporter, not part of the same operon, with greatest similarity to Sulfate/Tungstate Uptake Transporters (SulT; TC# 3.A.1.6) within the ABC superfamily. Accordingly, Pth1 may transport a sulfur-containing compound.
Homologues in Magnetospirillum magnetotacticum and M. magneticum are located in operons along with genes encoding UspA stress homologues, and next to an operon encoding the NifU protein. The NifU scaffold protein is part of the N2 fixation system [57] (see cluster 1) of FeS cluster assembly and is known to colocalize with the Fe-S center-containing rubrerythrin, a peroxidase involved in hydroperoxide detoxification [76, 77]. Homologues of Pth1 may serve as a means for the uptake of inorganic sulfide [78, 79].
Cluster 15 includes a homologue in Cupriavidus (Ralstonia) metallidurans encoded in a monocistronic operon. It may be co-regulated with the two nearby operons, the first, a large operon encoding enzymes involved in cytochrome c biogenesis and acting on sulfur-based compounds. These proteins include thioredoxin, the thiol:disulfide interchange protein, and a protein-disulfide reductase [80]. Rme1 and its homologues are likely to transport sulfur-based compounds and play a role in metabolic pathways. Sulfolobus solfataricus-derived Sso1, also in a monocistronic operon, is orthologous to a protein in S. acidocaldarius, which is located closely upstream of an operon encoding the various subunits of the CoB-CoM heterodisulfide reductase, further supporting a possible sulfur-based compound transport role.
Aau1 from Arthrobacter aurescens is encoded within an operon with a gene encoding a rhodanase domain-containing protein. It is surrounded by several other small genes encoding a thioredoxin and proteins that also contain rhodanase-like domains. Rhodanase catalyzes the transfer of a sulfur atom from sulfane sulfur-containing compounds (sulfur atoms at the 0 or -1 oxidation state) to sulfur acceptors like cyanide and thiols in order to generate molecules that are less toxic to the cell [81]. An example of a reaction which rhodanase catalyzes is:
Aau1 and its homologues in Corynebacterium glutamicum, C. efficiens, Salinispora tropica and Mycobacterium sp. JLS are all present in operons containing genes encoding hydroxyacylglutathione hydrolase, which may also serve as a polysulfide binding protein. In C. efficiens and S. tropica, hydroxyacylglutathione hydrolase may be encoded in an operon with the Aau1 homologue.
Bsp2 from Bacillus B-14905 and its homologues in B. cereus, Geobacillus kaustophilus and Exiguobacterium sibiricum have essentially the same gene arrangement as Aau1 and its respective homologues, with various rhodanase-like domain proteins and the hydroxyacylglutathione hydrolase joining them in operons. In addition, Bsp2 and its homologues are in an operon with, or are surrounded by a putative sulfide reductase, protein disulfide isomerase (S-S rearrangase) and/or a putative pyridine nucleotide-disulfide oxidoreductase. Aau1 and Bsp2, as well as their respective homologues, may therefore function in sulfur-based compound uptake, or equally likely, sulfite export, the assigned function of TauE (TC# 2.A.102.2.1) [15], a member of cluster 15. Biochemical and genome context analyses suggest that SafE1 is a putative exporter of sulfoacetate (TC# 2.A.102.2.2), while PmpC (TC# 2.A.102.4.2) may be an organo-sulfur compound transporter. Both belong to cluster 15, providing further evidence for a sulfur-based compound transport role for members of this cluster [17, 18].
3.6 The novel Rhodopsin Superfamily
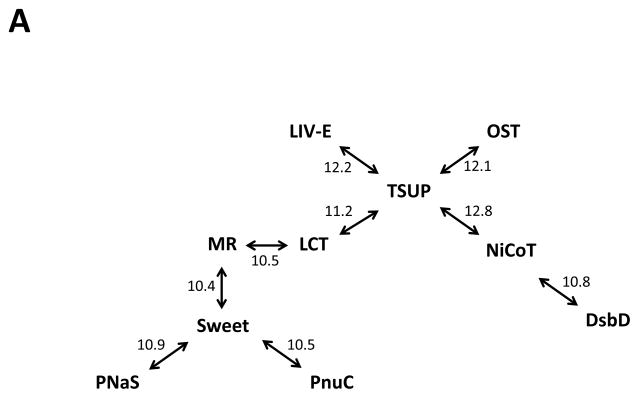
Comparing the TSUP family to TC subclasses 2.A, 3.E, 4.B, 5.A and 9.A proved to be fruitful and led to the identification of a novel superfamily including at least ten previously recognized families of transporters. This novel superfamily includes members mostly of 7 or 8 TMSs (Table 3). Surprisingly, in addition to families of transporters, the superfamily includes vertebrate and invertebrate rhodopsins, G-protein coupled receptors (GPCRs), a variety of hormone receptors, and invertebrate odorant receptors among others (MA Shlykov, DC Yee, VS Reddy, S Aurora, JS Chen, EI Sun and MH Saier Jr., manuscript in preparation). A summary of the comparisons performed is presented in Figure 6A, while specific comparisons between the TSUP, LCT and NiCoT families are presented in Figures 6B and 6C. These comparisons also demonstrate the primordial 4 TMS repeat unit found in the members of all of these families. More details, including phylogenetic and topological analyses of each family as well as proposed evolutionary pathways for the appearance of these proteins will be presented in a forthcoming publication (Shlykov et al., in preparation).
Table 3.
Currently recognized families of transporters included within the Rhodopsin Superfamily. The family names, abbreviations, TC numbers and dominant topologies are presented. This superfamily also includes the non-transporter eukaryotic 7 TMS proteins that include vertebrate and invertebrate rhodopsins, G-protein coupled receptors (GPCRs), a variety of hormone receptors, and invertebrate odorant receptors among others (MA Shlykov, DC Yee, VS Reddy, S Aurora, JS Chen, EI Sun and MH Saier Jr., manuscript in preparation).
| Family Name | Family Abbreviation | TC # | # TMSs |
|---|---|---|---|
| Ion-Translocating Microbial Rhodopsin | MR | 3.E.1 | 7 |
| Sweet | Sweet | 9.A.58 | 3 or 7 |
| Branched Chain Amino Acid Exporter | LIV-E | 2.A.78 | 7 or 8 |
| Nicotinamide Ribonucleotide Uptake Permease | PnuC | 4.B.1 | 7 or 8 |
| 4-Toluene Sulfonate Uptake Permease | TSUP | 2.A.102 | 7–9 |
| Ni2+-Co2+ Transporter | NiCoT | 2.A.52 | 6–8 |
| Organic Solute Transporter | OST | 2.A.82 | 7 |
| Phosphate:Na+ Symporter PNaS | PNaS | 2.A.58 | 8 or 9 |
| Lysosomal Cystine Transporter | LCT | 2.A.43 | 7 |
| Disulfide Bond Oxidoreductase D | DsbD | 5.A.1 | 6–9 |
Figure 6.
(A) Rhodopsin superfamily homology established through the use of GSAT/GAP and the Superfamily Principle. Established Rhodopsin superfamily proteins from TCDB and their homologues were used to establish homology between all members of the ten families. GSAT/GAP scores, adjacent to the arrows, are expressed in terms of standard deviations (S.D.). (B) Homology between members of the TSUP and LCT families. GAP alignment of TMSs 1–3 of TSUP Axy3 (Achromobacter xylosoxidans; gi 311107599) with TMSs 4–6 of LCT Asu1 (Ascaris suum; gi 324511247). A comparison score of 11.2 S.D. was obtained with 43.8% similarity and 28.1% identity. (C) Homology between members of the TSUP and NiCoT families. GAP alignment of TMSs 1–3 of TSUP Bja1 (Bradyrhizobium japonicum; gi 27376265) with TMSs 4–6 of NiCoT Pla1 (Parvibaculum lavamentivorans; gi 154252649). A comparison score of 12.8 S.D. was obtained with 44.7% similarity and 36.4% identity.
3.6.1 The LCT and MR families
The evolutionary pathway of the 7 TMS LCT family has been partially elucidated (Zhai et al., 2001), and LCT family members were found to be homologous to members of the Microbial Rhodopsin (MR) family which includes fungal chaperone proteins (see TC entries under 3.E.1). All of the transporters are light-driven ion pumps or light-activated ion channels. Because MR family members are the best characterized from structural, functional and mechanistic standpoints, we have designated this new superfamily the Rhodopsin Superfamily.
LCT family members range in size from 300 to 400 amino acyl residues (aas) and are generally larger than MR proteins which have ~220 to 300 residues. Eukaryotic homologues within a single family tend to be ~40% larger than their bacterial homologues [7]. Whereas the LCT family is found exclusively in the eukaryotic domain, the MR family is ubiquitous. Despite these differences, both families possess a uniform 7 TMS topology.
TMSs 1–3 in LCT family members duplicated to give rise to TMSs 5–7, with TMS 4 showing insignificant sequence similarity to any one of the other six TMSs [82]. The precursor could have been an 8 TMS protein which generated the present-day 7 TMS proteins by loss of TMS 1 or 8, and strong evidence for this possibility is presented here.
The 8 TMS Axy3 protein of the TSUP family is homologous to the 7 TMS Asu1 protein of the LCT family (Fig. 6B). The repeat regions shown in Fig. 6B include TMSs 1–3 of Axy3 and 4–6 of Asu1 (comparison score = 11.2 S.D.). This demonstrates that TSUP TMSs 1–3 and 5–7 are homologous to LCT/MR TMSs 4–6. TMS 1 of most TSUP members was lost to generate the 7 TMS topology of the LCT/MR families.
3.6.2 The NiCoT family
Members of sub-family 1 within the ubiquitous NiCoT family are typically 300 to 380 aas in size and possess 6–8 putative TMSs [83]. NiCoT sub-family 2 is comprised of distant homologues of great size, sequence and topological variation. NiCoT transporters catalyze the uptake of Ni2+ and Co2+ using a pmf-dependent mechanism; however, a Ni2+ and Co2+ resistance protein that is believed to export the two metals to the external environment has been reported [84, 85].
Comparing TMSs 1–3 of TSUP Bja1 (8 TMSs) with TMSs 4–6 of NiCoT Pla1 (6 TMSs) yielded a comparison score of 12.8 S.D. (Fig. 6C). This comparison establishes homology between members of these two families. Based on this and other alignments, it is likely that the 6 TMS NiCoT proteins arose from the loss of TMSs 1 and 8 after the 4 TMS intragenic duplication event that gave an 8 TMS topology.
4. Discussion
Members of the ubiquitous TSUP family appear to function as secondary carriers primarily for sulfur-based compounds. The vast majority of homologues within the TSUP family possess eight putative TMSs, with some predicted to have seven or nine TMSs, possibly as a result of N- or C-terminal deletions or extensions. Conserved motifs were identified, and their presence in multiple copies within TSUP homologues supports a two-fold symmetry within the proteins (Figs. 5A–B).
The greatest size and topological variation of all phylogenetic TSUP clusters was observed in the diverse cluster 1, which consists solely of eukaryotic members that are in general, 40–50% larger than their prokaryotic counterparts [7], although large homologues were also identified in prokaryotic clusters. These may have been the products of gene fusion events where hydrophilic domains were introduced during their evolutionary histories. Most hydrophilic domains proved to be unrecognizable by CDD, but the degP_htrA_DO domain of Orf6 and the USP_like domain of Bsp1 were identified. Their presence suggests a group-translocation-like function for Orf6 and a stress response role for Bsp1. The known functions of these domains correlate nicely with the predicted functions of the phylogenetic clusters in which they reside.
Comparative analyses of the phylogenetic protein and 16S/18S rRNA trees revealed that lateral gene transfer was common within the bacterial and archaeal domains but much less common within the eukaryotic domain. Lateral gene transfer between bacteria and archaea must have been frequent but rare between prokaryotes and eukaryotes. As a result, orthology was generally not observed within the bacterial domain with the notable exception of the Actinobacterial and Cyanobacterial homologues in cluster 5.
We were able to demonstrate a 4 TMS repeat in homologues from bacteria, eukaryotes and archaea. 2 TMS repeat units have been found in several families of transport proteins, including the Oligopeptide Transporter (OPT; TC# 2.A.67) and CRAC channel/CDF carrier superfamily [23, 50]. However, our methods were unable to detect a 2 TMS repeat unit within TSUP homologues. Sequence divergence may have accounted for this failure.
Comparisons between the TSUP family and other transport systems have revealed superfamily relationships with the Ion-Translocating Microbial Rhodopsins, Sweet sugar porters, Branched Chain Amino Acid Exporters, Nicotinamide Ribonucleotide Uptake Permeases, Ni2+-Co2+ Transporters, Organic Solute Transporters, Disulfide Bond Oxidoreductase D 2-electron carriers, Phosphate:Na+ Symporters and Lysosomal Cystine Transporters (See Table 3). These ten families comprise the novel Rhodopsin Superfamily. Of these proteins, high resolution 3-d structures are available only for the microbial rhodopsins [86]. We predict that all members of these ten families will prove to have similar structures. The functional diversification of this superfamily is unprecedented among transmembrane protein superfamilies (see Superfamily link in TCDB). However, it is worth noting that in some of these families it was possible to demonstrate that duplication of a 2 TMS hairpin structure gave rise to the 4 TMS precursor that duplicated internally in all of the proteins of these families to give the current 8 or 7 TMS proteins. Using the Superfamily Principle, we therefore conclude that the same must have been true for the TSUP family.
Genome context analyses supported the few biochemical assays that have been performed using TSUP homologues. Results for clusters 1, 5, 6, 8 and 12–15 substantiate a transport role for sulfur-based compounds (Table 2). Given the apparent functional diversity of our predictions as well as the sequence diversity inherent in the TSUP family, it may be that members transport a wide range of compounds. These may include (1) amino acids/peptides, (2) nucleotides/nucleosides/nucleobases and (3) carbohydrates. Some TSUP proteins may function as parts of stress responses and/or play roles in cofactor precursor transport. At least some TSUP members may function with auxiliary cytoplasmic and periplasmic proteins. The elucidation of these functions, using the predictions presented here as guides, are likely to open up new fields of study.
Supplementary Material
Dendogram of the 189 TSUP family proteins included in this study corresponding to the phylogenetic tree shown in Figure 1.
Dendogram corresponding to the 16S/18S rRNA phylogenetic tree presented in Figure 2.
Entire average hydropathy, amphipathicity, and similarity (AveHAS) plot for the 189 TSUP family proteins included in this study. Proteins with N- and C-terminal hydrophilic domain extensions contributed to the large size of the plot; their functional roles are discussed in the text.
Highlights.
The 4-toluene sulfonate uptake permease (TSUP) family is ubiquitous in nature.
Uptake of sulfur-containing compounds is mediated by members of the TSUP family.
No bioinformatic characterization of this family of porters has yet been described.
We here demonstrate that these proteins arose by intragenic duplication of 4 TMSs.
Phylogeny, motifs, topologies and functional predictions are described herein.
Acknowledgments
The work reported in this paper was supporter by NIH Grant 2 RO1 GM077402-05A1.
Footnotes
We request that any color figures be reproduced in color on the web only.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Saier MH, Jr, Tran CV, Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34:D181–186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saier MH, Jr, Yen MR, Noto K, Tamang DG, Elkan C. The Transporter Classification Database: recent advances. Nucleic Acids Res. 2009;37:D274–278. doi: 10.1093/nar/gkn862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Busch W, Saier MH., Jr The transporter classification (TC) system, 2002. Crit Rev Biochem Mol Biol. 2002;37:287–337. doi: 10.1080/10409230290771528. [DOI] [PubMed] [Google Scholar]
- 4.Gatti L, Cossa G, Beretta GL, Zaffaroni N, Perego P. Novel Insights into Targeting ATP-Binding Cassette Transporters for Antitumor Therapy. Current Medicinal Chemistry. 2011 doi: 10.2174/092986711797189682. [DOI] [PubMed] [Google Scholar]
- 5.Slavic K, Krishna S, Derbyshire ET, Staines HM. Plasmodial sugar transporters as anti-malarial drug targets and comparisons with other protozoa. Malaria Journal. 2011;10:165. doi: 10.1186/1475-2875-10-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yen MR, Choi J, Saier MH., Jr Bioinformatic analyses of transmembrane transport: novel software for deducing protein phylogeny, topology, and evolution. J Mol Microbiol Biotechnol. 2009;17:163–176. doi: 10.1159/000239667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung YJ, Krueger C, Metzgar D, Saier MH., Jr Size comparisons among integral membrane transport protein homologues in bacteria, Archaea, and Eucarya. J Bacteriol. 2001;183:1012–1021. doi: 10.1128/JB.183.3.1012-1021.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saier MH., Jr Vectorial metabolism and the evolution of transport systems. J Bacteriol. 2000;182:5029–5035. doi: 10.1128/jb.182.18.5029-5035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barabote RD, Tamang DG, Abeywardena SN, Fallah NS, Fu JY, Lio JK, Mirhosseini P, Pezeshk R, Podell S, Salampessy ML, Thever MD, Saier MH., Jr Extra domains in secondary transport carriers and channel proteins. Biochim Biophys Acta. 2006;1758:1557–1579. doi: 10.1016/j.bbamem.2006.06.018. [DOI] [PubMed] [Google Scholar]
- 10.Locher HH, Poolman B, Cook AM, Konings WN. Uptake of 4-toluene sulfonate by Comamonas testosteroni T-2. J Bacteriol. 1993;175:1075–1080. doi: 10.1128/jb.175.4.1075-1080.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tralau T, Cook AM, Ruff J. An additional regulator, TsaQ, is involved with TsaR in regulation of transport during the degradation of p-toluenesulfonate in Comamonas testosteroni T-2. Arch Microbiol. 2003;180:319–326. doi: 10.1007/s00203-003-0594-8. [DOI] [PubMed] [Google Scholar]
- 12.Mampel J, Maier E, Tralau T, Ruff J, Benz R, Cook AM. A novel outer-membrane anion channel (porin) as part of a putatively two-component transport system for 4-toluenesulphonate in Comamonas testosteroni T-2. Biochem J. 2004;383:91–99. doi: 10.1042/BJ20040652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mulligan C, Kelly DJ, Thomas GH. Tripartite ATP-independent periplasmic transporters: application of a relational database for genome-wide analysis of transporter gene frequency and organization. J Mol Microbiol Biotechnol. 2007;12:218–226. doi: 10.1159/000099643. [DOI] [PubMed] [Google Scholar]
- 14.Rabus R, Jack DL, Kelly DJ, Saier MH., Jr TRAP transporters: an ancient family of extracytoplasmic solute-receptor-dependent secondary active transporters. Microbiology. 1999;145(Pt 12):3431–3445. doi: 10.1099/00221287-145-12-3431. [DOI] [PubMed] [Google Scholar]
- 15.Weinitschke S, Denger K, Cook AM, Smits TH. The DUF81 protein TauE in Cupriavidus necator H16, a sulfite exporter in the metabolism of C2 sulfonates. Microbiology. 2007;153:3055–3060. doi: 10.1099/mic.0.2007/009845-0. [DOI] [PubMed] [Google Scholar]
- 16.Ruckert C, Koch DJ, Rey DA, Albersmeier A, Mormann S, Puhler A, Kalinowski J. Functional genomics and expression analysis of the Corynebacterium glutamicum fpr2-cysIXHDNYZ gene cluster involved in assimilatory sulphate reduction. BMC Genomics. 2005;6:121. doi: 10.1186/1471-2164-6-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krejcik Z, Denger K, Weinitschke S, Hollemeyer K, Paces V, Cook AM, Smits TH. Sulfoacetate released during the assimilation of taurine-nitrogen by Neptuniibacter caesariensis: purification of sulfoacetaldehyde dehydrogenase. Arch Microbiol. 2008;190:159–168. doi: 10.1007/s00203-008-0386-2. [DOI] [PubMed] [Google Scholar]
- 18.Gristwood T, McNeil MB, Clulow JS, Salmond GP, Fineran PC. PigS and PigP regulate prodigiosin biosynthesis in Serratia via differential control of divergent operons, which include predicted transporters of sulfur-containing molecules. J Bacteriol. 2011;193:1076–1085. doi: 10.1128/JB.00352-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 20.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 22.Wang B, Dukarevich M, Sun EI, Yen MR, Saier MH., Jr Membrane porters of ATP-binding cassette transport systems are polyphyletic. J Membr Biol. 2009;231:1–10. doi: 10.1007/s00232-009-9200-6. [DOI] [PubMed] [Google Scholar]
- 23.Matias MG, Gomolplitinant KM, Tamang DG, Saier MH., Jr Animal Ca2+ release-activated Ca2+ (CRAC) channels appear to be homologous to and derived from the ubiquitous cation diffusion facilitators. BMC Res Notes. 2010;3:158. doi: 10.1186/1756-0500-3-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 26.Zhai Y, Saier MH., Jr A web-based program (WHAT) for the simultaneous prediction of hydropathy, amphipathicity, secondary structure and transmembrane topology for a single protein sequence. J Mol Microbiol Biotechnol. 2001;3:501–502. [PubMed] [Google Scholar]
- 27.Tusnady GE, Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- 28.Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 29.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 30.Bailey TL, Gribskov M. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 1998;14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- 31.Moller S, Croning MD, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- 32.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 33.von Heijne G, Gavel Y. Topogenic signals in integral membrane proteins. Eur J Biochem. 1988;174:671–678. doi: 10.1111/j.1432-1033.1988.tb14150.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhai Y, Saier MH., Jr A web-based program for the prediction of average hydropathy, average amphipathicity and average similarity of multiply aligned homologous proteins. J Mol Microbiol Biotechnol. 2001;3:285–286. [PubMed] [Google Scholar]
- 35.Zhai Y, Saier MH., Jr A simple sensitive program for detecting internal repeats in sets of multiply aligned homologous proteins. J Mol Microbiol Biotechnol. 2002;4:375–377. [PubMed] [Google Scholar]
- 36.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dayhoff MO, Barker WC, Hunt LT. Establishing homologies in protein sequences. Methods Enzymol. 1983;91:524–545. doi: 10.1016/s0076-6879(83)91049-2. [DOI] [PubMed] [Google Scholar]
- 38.Saier MH., Jr Computer-aided analyses of transport protein sequences: gleaning evidence concerning function, structure, biogenesis, and evolution. Microbiol Rev. 1994;58:71–93. doi: 10.1128/mr.58.1.71-93.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, Gerdes S, Glass EM, Goesmann A, Hanson A, Iwata-Reuyl D, Jensen R, Jamshidi N, Krause L, Kubal M, Larsen N, Linke B, McHardy AC, Meyer F, Neuweger H, Olsen G, Olson R, Osterman A, Portnoy V, Pusch GD, Rodionov DA, Ruckert C, Steiner J, Stevens R, Thiele I, Vassieva O, Ye Y, Zagnitko O, Vonstein V. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Novichkov PS, Laikova ON, Novichkova ES, Gelfand MS, Arkin AP, Dubchak I, Rodionov DA. RegPrecise: a database of curated genomic inferences of transcriptional regulatory interactions in prokaryotes. Nucleic Acids Res. 2010;38:D111–118. doi: 10.1093/nar/gkp894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Novichkov PS, Rodionov DA, Stavrovskaya ED, Novichkova ES, Kazakov AE, Gelfand MS, Arkin AP, Mironov AA, Dubchak I. RegPredict: an integrated system for regulon inference in prokaryotes by comparative genomics approach. Nucleic Acids Res. 2010;38:W299–307. doi: 10.1093/nar/gkq531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krojer T, Garrido-Franco M, Huber R, Ehrmann M, Clausen T. Crystal structure of DegP (HtrA) reveals a new protease-chaperone machine. Nature. 2002;416:455–459. doi: 10.1038/416455a. [DOI] [PubMed] [Google Scholar]
- 44.Lipinska B, Zylicz M, Georgopoulos C. The HtrA (DegP) protein, essential for Escherichia coli survival at high temperatures, is an endopeptidase. J Bacteriol. 1990;172:1791–1797. doi: 10.1128/jb.172.4.1791-1797.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spiess C, Beil A, Ehrmann M. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell. 1999;97:339–347. doi: 10.1016/s0092-8674(00)80743-6. [DOI] [PubMed] [Google Scholar]
- 46.Sousa MC, McKay DB. Structure of the universal stress protein of Haemophilus influenzae. Structure. 2001;9:1135–1141. doi: 10.1016/s0969-2126(01)00680-3. [DOI] [PubMed] [Google Scholar]
- 47.Hebbeln P, Rodionov DA, Alfandega A, Eitinger T. Biotin uptake in prokaryotes by solute transporters with an optional ATP-binding cassette-containing module. Proc Natl Acad Sci U S A. 2007;104:2909–2914. doi: 10.1073/pnas.0609905104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drumm JE, Mi K, Bilder P, Sun M, Lim J, Bielefeldt-Ohmann H, Basaraba R, So M, Zhu G, Tufariello JM, Izzo AA, Orme IM, Almo SC, Leyh TS, Chan J. Mycobacterium tuberculosis universal stress protein Rv2623 regulates bacillary growth by ATP-Binding: requirement for establishing chronic persistent infection. PLoS Pathog. 2009;5:e1000460. doi: 10.1371/journal.ppat.1000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Castillo R, Saier MH. Functional Promiscuity of Homologues of the Bacterial ArsA ATPases. Int J Microbiol. 2010;2010:187373. doi: 10.1155/2010/187373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gomolplitinant KM, Saier MH., Jr Evolution of the oligopeptide transporter family. J Membr Biol. 2011;240:89–110. doi: 10.1007/s00232-011-9347-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iwaki H, Wang S, Grosse S, Bergeron H, Nagahashi A, Lertvorachon J, Yang J, Konishi Y, Hasegawa Y, Lau PC. Pseudomonad cyclopentadecanone monooxygenase displaying an uncommon spectrum of Baeyer-Villiger oxidations of cyclic ketones. Appl Environ Microbiol. 2006;72:2707–2720. doi: 10.1128/AEM.72.4.2707-2720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blander G, Guarente L. The Sir2 family of protein deacetylases. Annu Rev Biochem. 2004;73:417–435. doi: 10.1146/annurev.biochem.73.011303.073651. [DOI] [PubMed] [Google Scholar]
- 53.Ochman H, Davalos LM. The nature and dynamics of bacterial genomes. Science. 2006;311:1730–1733. doi: 10.1126/science.1119966. [DOI] [PubMed] [Google Scholar]
- 54.Harvat EM, Zhang YM, Tran CV, Zhang Z, Frank MW, Rock CO, Saier MH., Jr Lysophospholipid flipping across the Escherichia coli inner membrane catalyzed by a transporter (LplT) belonging to the major facilitator superfamily. J Biol Chem. 2005;280:12028–12034. doi: 10.1074/jbc.M414368200. [DOI] [PubMed] [Google Scholar]
- 55.von Rozycki T, Schultzel MA, Saier MH., Jr Sequence analyses of cyanobacterial bicarbonate transporters and their homologues. J Mol Microbiol Biotechnol. 2004;7:102–108. doi: 10.1159/000078653. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Z, Feige JN, Chang AB, Anderson IJ, Brodianski VM, Vitreschak AG, Gelfand MS, Saier MH., Jr A transporter of Escherichia coli specific for L- and D-methionine is the prototype for a new family within the ABC superfamily. Arch Microbiol. 2003;180:88–100. doi: 10.1007/s00203-003-0561-4. [DOI] [PubMed] [Google Scholar]
- 57.Barras F, Loiseau L, Py B. How Escherichia coli and Saccharomyces cerevisiae build Fe/S proteins. Adv Microb Physiol. 2005;50:41–101. doi: 10.1016/S0065-2911(05)50002-X. [DOI] [PubMed] [Google Scholar]
- 58.Chahal HK, Dai Y, Saini A, Ayala-Castro C, Outten FW. The SufBCD Fe-S scaffold complex interacts with SufA for Fe-S cluster transfer. Biochemistry. 2009;48:10644–10653. doi: 10.1021/bi901518y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saini A, Mapolelo DT, Chahal HK, Johnson MK, Outten FW. SufD and SufC ATPase activity are required for iron acquisition during in vivo Fe-S cluster formation on SufB. Biochemistry. 2010;49:9402–9412. doi: 10.1021/bi1011546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang W, Huang H, Tan G, Si F, Liu M, Landry AP, Lu J, Ding H. In vivo evidence for the iron-binding activity of an iron-sulfur cluster assembly protein IscA in Escherichia coli. Biochem J. 2010;432:429–436. doi: 10.1042/BJ20101507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Iwasaki T. Iron-sulfur world in aerobic and hyperthermoacidophilic archaea Sulfolobus. Archaea. 2010;2010 doi: 10.1155/2010/842639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Umbarger HE, Umbarger MA, Siu PM. Biosynthesis of Serine in Escherichia Coli and Salmonella Typhimurium. J Bacteriol. 1963;85:1431–1439. doi: 10.1128/jb.85.6.1431-1439.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saier MH, Jr, Wentzel DL, Feucht BU, Judice JJ. A transport system for phosphoenolpyruvate, 2-phosphoglycerate, and 3-phosphoglycerate in Salmonella typhimurium. J Biol Chem. 1975;250:5089–5096. [PubMed] [Google Scholar]
- 64.Liu WT, Karavolos MH, Bulmer DM, Allaoui A, Hormaeche RD, Lee JJ, Khan CM. Role of the universal stress protein UspA of Salmonella in growth arrest, stress and virulence. Microb Pathog. 2007;42:2–10. doi: 10.1016/j.micpath.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 65.Zhao B, Yeo CC, Poh CL. Proteome investigation of the global regulatory role of sigma 54 in response to gentisate induction in Pseudomonas alcaligenes NCIMB 9867. Proteomics. 2005;5:1868–1876. doi: 10.1002/pmic.200401081. [DOI] [PubMed] [Google Scholar]
- 66.Wang L, Jeon B, Sahin O, Zhang Q. Identification of an arsenic resistance and arsenic-sensing system in Campylobacter jejuni. Appl Environ Microbiol. 2009;75:5064–5073. doi: 10.1128/AEM.00149-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thomas D, Surdin-Kerjan Y. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1997;61:503–532. doi: 10.1128/mmbr.61.4.503-532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee MH, Scherer M, Rigali S, Golden JW. PlmA, a new member of the GntR family, has plasmid maintenance functions in Anabaena sp. strain PCC 7120. J Bacteriol. 2003;185:4315–4325. doi: 10.1128/JB.185.15.4315-4325.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rigali S, Derouaux A, Giannotta F, Dusart J. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J Biol Chem. 2002;277:12507–12515. doi: 10.1074/jbc.M110968200. [DOI] [PubMed] [Google Scholar]
- 70.Rigali S, Schlicht M, Hoskisson P, Nothaft H, Merzbacher M, Joris B, Titgemeyer F. Extending the classification of bacterial transcription factors beyond the helix-turn-helix motif as an alternative approach to discover new cis/trans relationships. Nucleic Acids Res. 2004;32:3418–3426. doi: 10.1093/nar/gkh673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vindal V, Suma K, Ranjan A. GntR family of regulators in Mycobacterium smegmatis: a sequence and structure based characterization. BMC Genomics. 2007;8:289. doi: 10.1186/1471-2164-8-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hillerich B, Westpheling J. A new GntR family transcriptional regulator in streptomyces coelicolor is required for morphogenesis and antibiotic production and controls transcription of an ABC transporter in response to carbon source. J Bacteriol. 2006;188:7477–7487. doi: 10.1128/JB.00898-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hoskisson PA, Rigali S, Fowler K, Findlay KC, Buttner MJ. DevA, a GntR-like transcriptional regulator required for development in Streptomyces coelicolor. J Bacteriol. 2006;188:5014–5023. doi: 10.1128/JB.00307-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jittawuttipoka T, Sallabhan R, Vattanaviboon P, Fuangthong M, Mongkolsuk S. Mutations of ferric uptake regulator (fur) impair iron homeostasis, growth, oxidative stress survival, and virulence of Xanthomonas campestris pv. campestris. Arch Microbiol. 2010;192:331–339. doi: 10.1007/s00203-010-0558-8. [DOI] [PubMed] [Google Scholar]
- 75.Rodriguez GM, Voskuil MI, Gold B, Schoolnik GK, Smith I. ideR, An essential gene in mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect Immun. 2002;70:3371–3381. doi: 10.1128/IAI.70.7.3371-3381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lumppio HL, Shenvi NV, Summers AO, Voordouw G, Kurtz DM., Jr Rubrerythrin and rubredoxin oxidoreductase in Desulfovibrio vulgaris: a novel oxidative stress protection system. J Bacteriol. 2001;183:101–108. doi: 10.1128/JB.183.1.101-108.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maralikova B, Ali V, Nakada-Tsukui K, Nozaki T, van der Giezen M, Henze K, Tovar J. Bacterial-type oxygen detoxification and iron-sulfur cluster assembly in amoebal relict mitochondria. Cell Microbiol. 2010;12:331–342. doi: 10.1111/j.1462-5822.2009.01397.x. [DOI] [PubMed] [Google Scholar]
- 78.Kiyasu T, Asakura A, Nagahashi Y, Hoshino T. Contribution of cysteine desulfurase (NifS protein) to the biotin synthase reaction of Escherichia coli. J Bacteriol. 2000;182:2879–2885. doi: 10.1128/jb.182.10.2879-2885.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yuvaniyama P, Agar JN, Cash VL, Johnson MK, Dean DR. NifS-directed assembly of a transient [2Fe-2S] cluster within the NifU protein. Proc Natl Acad Sci U S A. 2000;97:599–604. doi: 10.1073/pnas.97.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Missiakas D, Schwager F, Raina S. Identification and characterization of a new disulfide isomerase-like protein (DsbD) in Escherichia coli. Embo J. 1995;14:3415–3424. doi: 10.1002/j.1460-2075.1995.tb07347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wrobel M, Lewandowska I, Bronowicka-Adamska P, Paszewski A. The level of sulfane sulfur in the fungus Aspergillus nidulans wild type and mutant strains. Amino Acids. 2009;37:565–571. doi: 10.1007/s00726-008-0175-x. [DOI] [PubMed] [Google Scholar]
- 82.Zhai Y, Heijne WH, Smith DW, Saier MH., Jr Homologues of archaeal rhodopsins in plants, animals and fungi: structural and functional predications for a putative fungal chaperone protein. Biochim Biophys Acta. 2001;1511:206–223. doi: 10.1016/s0005-2736(00)00389-8. [DOI] [PubMed] [Google Scholar]
- 83.Saier MH, Jr, Eng BH, Fard S, Garg J, Haggerty DA, Hutchinson WJ, Jack DL, Lai EC, Liu HJ, Nusinew DP, Omar AM, Pao SS, Paulsen IT, Quan JA, Sliwinski M, Tseng TT, Wachi S, Young GB. Phylogenetic characterization of novel transport protein families revealed by genome analyses. Biochim Biophys Acta. 1999;1422:1–56. doi: 10.1016/s0304-4157(98)00023-9. [DOI] [PubMed] [Google Scholar]
- 84.Iwig JS, Rowe JL, Chivers PT. Nickel homeostasis in Escherichia coli - the rcnR-rcnA efflux pathway and its linkage to NikR function. Mol Microbiol. 2006;62:252–262. doi: 10.1111/j.1365-2958.2006.05369.x. [DOI] [PubMed] [Google Scholar]
- 85.Rodrigue A, Effantin G, Mandrand-Berthelot MA. Identification of rcnA (yohM), a nickel and cobalt resistance gene in Escherichia coli. J Bacteriol. 2005;187:2912–2916. doi: 10.1128/JB.187.8.2912-2916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schobert B, Cupp-Vickery J, Hornak V, Smith S, Lanyi J. Crystallographic structure of the K intermediate of bacteriorhodopsin: conservation of free energy after photoisomerization of the retinal. J Mol Biol. 2002;321:715–726. doi: 10.1016/s0022-2836(02)00681-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dendogram of the 189 TSUP family proteins included in this study corresponding to the phylogenetic tree shown in Figure 1.
Dendogram corresponding to the 16S/18S rRNA phylogenetic tree presented in Figure 2.
Entire average hydropathy, amphipathicity, and similarity (AveHAS) plot for the 189 TSUP family proteins included in this study. Proteins with N- and C-terminal hydrophilic domain extensions contributed to the large size of the plot; their functional roles are discussed in the text.