Abstract
Background
Internalin A (InlA) facilitates the invasion of Listeria monocytogenes into a host cell. Some strains of Listeria monocytogenes express truncated forms of InlA, which reduces invasiveness. However, few virulence-related genes other than inlA have been analyzed in InlA-truncated strains. In the present study, we sequenced the draft genome of strain 36-25-1, an InlA-truncated strain, with pyrosequencing and compared 36 major virulence-related genes in this strain and a clinical wild-type strain.
Results
Strain 36-25-1 possessed all of the virulence-related genes analyzed. Of the analyzed genes, only 4 genes (dltA, gtcA, iap, and inlA) differed when the nucleotide sequences of strain 36-25-1 and the clinical wild-type strain were compared. Analysis of the deduced amino acid sequences found no mutations that significantly influenced virulence in genes other than inlA.
Conclusions
The virulence-associated genes in strain 36-25-1 differ little from those of the clinical wild-type strain, indicating that a slight mutation in the nucleotide sequence determines the virulence of the InlA-truncated strain. In addition, the results suggest that, aside from InlA-mediated cell invasiveness, there is almost no difference between the virulence of strain 36-25-1 and that of the clinical wild-type strain.
Keywords: Listeria monocytogenes, Internalin A, Next-generation-sequencing
Background
Listeria monocytogenes is a gram-positive, non-sporulating foodborne infectious pathogen. The pathogen can cause serious diseases, such as septicemia and meningitis, especially in high-risk groups (pregnant women, neonates, and immunocompromised people) with a high mortality rate of 20–30% [1]. L. monocytogenes is ubiquitous in nature; it can survive under conditions of high salt and low pH. Because, it can grow even at low temperatures, the bacterium can be found in many kinds of foods during storage [2]. In particular, ready-to-eat (RTE) foods, which do not require heat cooking, are a main source of foodborne listeriosis cases [3-5].
Internalin A (InlA) plays an important role in L. monocytogenes invasion by attaching to host cells [6-9]. However, some L. monocytogenes strains express a truncated form of InlA, which lowers the invasion rate [10-14]. Truncated InlA, caused by a premature stop codon (PMSC) in inlA, often lacks the LPXTG motif that anchors InlA to the surface of the pathogen, leading to a decrease in invasiveness [12,13]. Some reports have shown that InlA-truncated strains account for 35–45% of L. monocytogenes in RTE foods [15,16]. However, aside from invasiveness, the virulence of these mutated strains has not been studied.
Our previous study showed that an InlA-truncated strain had wild-type PrfA, which regulates the expression of virulence related genes [11]. On the other hand, Tèmoin et al. (2008) reported that all of 5 InlA-truncated strains analyzed had the same amino acid sequence mutations in the migration factor plcA and the invasion factor inlB[17]. However, approximately 50 genes are related to the L. monocytogenes infection cycle [18], and most of them have not been investigated in strains with truncated InlA. A small number of studies have investigated virulence-related genes in InlA-truncated strains [11,17]; the studies do not completely explain the virulence of the strains.
This study aimed to identify the major virulence-related gene sequences present in InlA-truncated strains. In recent years, the analysis of bacterial whole genomes has become faster and easier with the development of next-generation sequencing methods such as pyrosequencing. In this study, we used pyrosequencing to construct a draft sequence of strain 36-25-1, and we compared 36 main virulence-related genes in the InlA-truncated strain and a clinical wild-type strain.
Results
Presence of virulence-related genes
After de novo assembly of the reads for strain 36-25-1, the total contig length was 2,957,538 bp with a peak depth of 11.0. The contigs aligned to 2,861,194 bp of the EGDe whole genome sequence, and showed 99.84% identity (Table 1).
Table 1.
The draft sequences results
| 36-25-1 | EGDe | |
|---|---|---|
| Total bases (bp) |
2,957,538 (11.0*) |
2,944,528 |
| Alignment length (bp) |
2,861,194 |
|
| Similarity | 99.84 % |
*The number in the parentheses shows the peak depth of de novo assembly results.
In strain 36-25-1, 36 open reading frames (ORFs) showed high similarity with the 36 EGDe virulence-related genes, indicating that strain 36-25-1 has all of these genes.
Comparison of the nucleotide and amino acid sequences of the virulence-related genes
Nucleotide mutations were found in 4 genes (dltA, gtcA, inlA,and iap) of strain 36-25-1, when compared to EGDe (Table 2). Substitutions of 1 bp were found in dltA, gtcA, and inlA. The mutation in iap was an insertion of 12 bp (Figure 1).
Table 2.
The alignment results of 36-25-1 and EGDe
| Gene | Mutation type | Mutation loci | EGDe allelic type * | 36-25-1 allelic type * | Function of mutation loci |
|---|---|---|---|---|---|
|
actA |
N/A |
|
|
|
|
|
ami |
N/A |
|
|
|
|
|
aut |
N/A |
|
|
|
|
|
ctaP |
N/A |
|
|
|
|
|
dltA |
Silence |
891 |
T (N) |
C (N) |
AMP binding site |
|
fbpA |
N/A |
|
|
|
|
|
fri |
N/A |
|
|
|
|
|
gap |
N/A |
|
|
|
|
|
gtcA |
Missense |
200 |
T (F) |
C (S) |
Function unknown |
|
inlA |
Nonsense |
1578 |
A (K) |
T (*) |
Listeria-bacteroides repeat domain |
|
inlB |
N/A |
|
|
|
|
|
inlC |
N/A |
|
|
|
|
|
inlH |
N/A |
|
|
|
|
|
inlJ |
N/A |
|
|
|
|
|
lap |
N/A |
|
|
|
|
|
lgt |
N/A |
|
|
|
|
|
hly |
N/A |
|
|
|
|
|
lntA |
N/A |
|
|
|
|
|
lpeA |
N/A |
|
|
|
|
|
lplA1 |
N/A |
|
|
|
|
|
lsp |
N/A |
|
|
|
|
|
mpl |
N/A |
|
|
|
|
|
mprF |
N/A |
|
|
|
|
|
murA |
N/A |
|
|
|
|
|
oppA |
N/A |
|
|
|
|
|
iap |
Insertion |
982 |
N/A |
ACAAATACAAAT (TNTN) |
Non coding region |
|
pgdA |
N/A |
|
|
|
|
|
pgl |
N/A |
|
|
|
|
|
plcA |
N/A |
|
|
|
|
|
plcB |
N/A |
|
|
|
|
|
prsA2 |
N/A |
|
|
|
|
|
pycA |
N/A |
|
|
|
|
|
recA |
N/A |
|
|
|
|
|
sipZ |
N/A |
|
|
|
|
|
sod |
N/A |
|
|
|
|
| svpA | N/A |
* In the allelic type columns, the amino acid residues are described in the parenthesis.
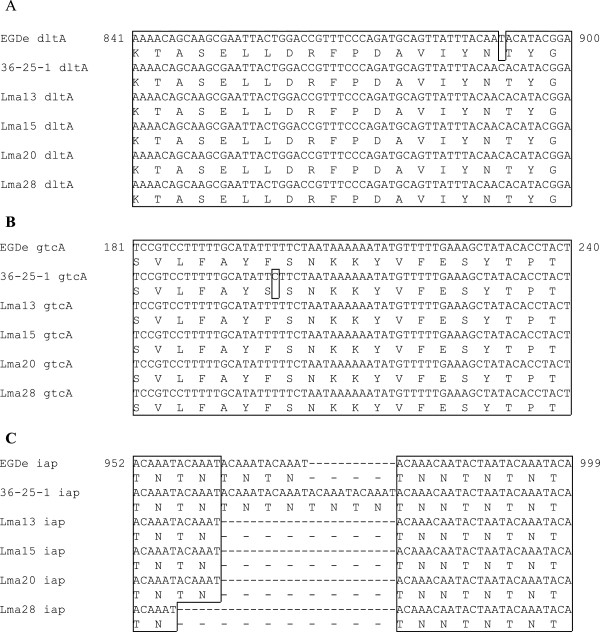
Figure 1.
The alignment of mutation loci in EGDe and InlA-truncated strains. Nucleotide sequences and amino acid sequences are shown for each strain. The numbers shown on the both sides mean the nucleotide sequence positions in the ORF of strain EGDe. The frames show identical sequences among the strains. (A) The alignment dltA. (B) The alignment gtcA. (C) The alignment iap.
In dltA, thymine was changed to cytosine at position 891 of the ORF. This mutation is a silent mutation, which does not cause an amino acid sequence change (Figure 1A). On the other hand, the mutation in gtcA is a missense mutation, which affects the amino acid sequence; substitution of thymine with cytosine position 200 changed the phenylalanine in EGDe to serine in strain 36-25-1 (Figure 1B). The inserted region in iap is a tandem repeat sequence. Whereas EGDe has 5 repeats of the ACAAAT motif, strain 36-25-1 has 7 repeats, resulting in 2 additional threonine-asparagine (TN) repeats (Figure 1C). Among the genes analyzed, a nonsense mutation was found only in inlA (Additional file 1).
Mutation of virulence-related genes in other InlA-truncated strains
The 4 genes, in which the nucleotide sequences differed between strain 36-25-1 and EGDe were also sequenced in other InlA-truncated strains (Lma13, Lma15, Lma20, and Lma28). The silent mutation in dltA was found in all of the InlA-truncated strains (Figure 1A). On the other hand, the missense mutation in gtcA found in 36-25-1 was not found in the other InlA-truncated strains (Figure 1B). As for the tandem repeat of the ACAAAT motif in iap, strains Lma13, Lma15, and Lma20 had 3 repeats; strain Lma28 had 2 repeats (Figure 1C).
Discussion
Virulence-related genes in strain 36-25-1
The contig of the 36-25-1 strain constructed by de novo assembly showed similarity as high as 99.84% in the regions that aligned with the EGDe strain. In addition, this strain possessed all of the 36 virulence-associated genes analyzed. The genus Listeria is considered to have lost virulence-associated genes as it differentiated from ancestors that showed virulence [19]. Multiple virulence-associated genes are missing in strain 4a, a serotype of L. monocytogenes showing no virulence [20]. Because strain 36-25-1 possesses all of the 36 genes investigated in the present study, we conclude that the InlA-truncated strain has not undergone changes that have resulted in any major loss of regions present in the clinical wild-type strain.
Virulence-related genes with mutations
Among the 36 virulence-associated genes in strain 36-25-1, 32 genes possess a sequence identical to that of the corresponding gene in the EGDe strain. Therefore, we conclude that the virulence of these genes is the same in the 36-25-1 and EGDe strains. Nucleotide sequence differences were found in only 4 genes (dltA, gtcA, iap, and inlA).
dltA is a part of the dlt operon, which is composed of 4 genes that function in the addition of alanine to lipoteichoic acid (LTA) [21]. Experiments using a strain in which dltA was artificially inactivated suggest that dltA influences the electric charge of the bacterial surface to increase adhesiveness to host cells [22]. The dltA mutation found in strain 36-25-1 is present in all other InlA-truncated strains examined in this study. Whether or not this mutation is characteristic of InlA-truncated strains requires investigation of other clinical wild-type strains. However, this mutation does not influence the phenotype of these strains as a silent mutation.
Similar to dltA, gtcA is involved in the addition of a saccharide to LTA [21]. In the present study, the nucleotide sequence of gtcA in strain 36-25-1 differed from that in the EGDe strain, and the encoded amino acid sequence differed as well. However, this mutation is not common to InlA-truncated strains: the mutation was not found in the other InlA-truncated strains examined.
The mutation in iap is in the tandem repeat region, in which the number of repeats has been reported to vary even among clinical wild-type strains [23-26]. p60, encoded by iap, is involved in the movement of L. monocytogenes inside a host cell and in cell-to-cell propagation [24]. p60 possesses multiple LysM motifs at its C-terminus, which are used to bind to the cell wall of L. monocytogenes, and an SH3 structure at the N-terminus exposed outside the cell [27,28]. The iap mutation in strain 36-25-1 occurred between the LysM domain and the SH3 structure. Therefore, we conclude that this mutation has almost no influence on the functions of these domains.
Pathogenicity of InlA truncated strain
The results of the present study indicate that, except for InlA-mediated cell invasiveness, virulence in the InlA-truncated 36-25-1 strain is almost equivalent to that in a clinical wild-type strain. However, these results differ from those of a study by Témoin et al. (2008), which demonstrated that 3 virulence genes, including inlA, were simultaneously truncated [17]. In addition, Van Stelten et al. (2011), after orally administering an InlA-truncated strain to guinea pigs, reported that only the translocation rate to the spleen was lower in the truncated strain, when compared to a clinical wild-type strain [14]. Moreover, in the study by Olier et al. (2005) using a strain that originally showed virulence but was genetically modified to express truncated InlA, the mortality of chicken embryos infected with the transformed strain was lower than that of chickens infected with a clinical wild-type strain [13]. These reports indicate that aspects of virulence other than cell invasiveness differ between InlA-truncated strains and clinical wild-type strains; this cannot be explained by the results of the present study. Therefore, we expect that mutations in the virulence-associated genes of InlA-truncated strains will exhibit heterogeneity. The results from InlA-truncated strains other than strain 36-25-1 in the present study support this hypothesis. In addition, many L. monocytogenes genes have functions that are not yet elucidated. Hence, we cannot exclude the possibility that genes not analyzed in the present study contribute to the differences between InlA-truncated strains and clinical wild-type strains. Analyzing other InlA-truncated strains and determining the unknown functions of L. monocytogenes genes will resolve these questions.
Conclusions
In the present study, we analyzed the major virulence-associated genes in strain 36-25-1, an InlA-truncated strain. With the exception of inlA, the virulence-associated genes in the InlA-truncated strain are almost identical to those in a clinical wild-type strain. The results indicate that a slight mutation in the nucleotide sequence, such as a PMSC, determines the virulence of InlA-truncated strains. In addition, post-translational analysis of each gene indicated that, except for InlA-mediated cell invasiveness, the virulence of the 36-25-1 strain is equivalent to that of clinical wild-type strains. However, this result does not completely explain the results of previous studies on InlA-truncated strains. To clarify the relationship between virulence and genes in InlA-truncated strains, analyses of more InlA-truncated strains, including pyrosequencing, and studies of gene expression are required to understand the virulence of InlA-truncated strains deeper.
Methods
Bacterial strains used in this study
L. monocytogenes strain 36-25-1, with truncated InlA, was sequenced by whole genome shot gun sequencing to analyze virulence-related genes. The low invasiveness of the strain compared to that of the wild-type strain was shown in our previous study [11].
In addition, four InlA-truncated strains (Lma13, Lma15, Lma20, and Lma28) isolated from raw meat products were sequenced by Sanger sequencing for reference [29].
The whole genome sequence of EGDe, a clinical wild-type strain, was obtained from GenBank (GenBank accession no. NC 003210).
Genome extraction
All L. monocytogenes strains were cultured overnight in brain heart infusion broth (Eiken Chemical, Tokyo, Japan) at 37°C. The bacterial DNA was extracted using the phenol-chloroform and ethanol precipitation method [30]. One milliliter of enriched culture was centrifuged at 10,000 × g for 10 min, and bacterial cells were incubated in 567 μL of Tris-EDTA buffer containing lysozyme (2 mg/mL) for 1 h at 37°C. Cells were lysed by the addition of 30 μL of 10% (wt/vol) sodium dodecyl sulfate and 3 mL of 20 mg/mL proteinase K, with incubation for 1 h at 37°C. Next, 100 μL of 5 M NaCl was added, and DNA was extracted with chloroform–isoamyl alcohol (24:1) followed by phenol–chloroform–isoamyl alcohol (25:24:1). DNA was then precipitated with isopropanol, washed with 70% ethanol, and dried. Purified DNA was dissolved in Tris-EDTA buffer and used as the DNA template for whole genome shot gun sequencing and Sanger sequencing.
Whole genome shot gun sequencing and de novo assembly
For whole genome shot gun sequencing, a Roche GS Junior platform (Roche, Basel, Schweiz) was employed using a GS Junior Rapid Library Preparation kit and GS Junior emPCR kit (Lib-L) according to the manufacture’s protocol. The read sequences were used to construct a contig without a reference sequence by de novo assembly using the GS De Novo Assembler (Roche, Basel, Schweiz). In this assembly, the program parameters were set to: seed step, 12; seed length, 16; seed count, 1; minimum overlap, 10; and minimum identity, 90.
Extraction of virulence-related gene loci and comparison analysis
The contigs of strain 36-25-1 and the EGDe whole genome sequence were aligned using NUCmer, an application of MUMmer 3.0 (http://mummer.sourceforge.net/).
The virulence-related gene loci of strain 36-25-1 were extracted from the contigs using GenomeTraveler (In Silico Biology, Kanagawa, Japan). Briefly, among the ORFs extracted from the contigs, those that showed high identity with EGDe virulence-related genes were selected for further analysis.
The extracted gene sequences were aligned with the EGDe sequences by GENETYX ver11.0.0 (Genetyx, Tokyo, Japan) to identify nucleotide mutations. When a genomic mutation was found, the corresponding amino acid sequences were also compared.
Sanger sequencing at the mutation loci
When gene mutations were identified in strain 36-25-1, Sanger sequencing was conducted for confirmation. Other InlA-truncated strains (Lma13, Lma15, Lma20, and Lma28) were sequenced at the same loci. The primers for PCR amplification and Sanger sequencing were designed using ABI PRISM Primer Express v2.0.0 (Life Technologies, Carlsbad, CA, USA) (Table 3). The PCR reaction mixture, prepared in 50 μL, contained 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 50 pmol of each primer, 2.5 mM of each dNTP, 25 ng template DNA, and 1.25 U Takara Taq DNA polymerase (Takara Bio, Shiga, Japan). The PCR program consisted of 94°C for 4 min; 30 cycles of 94°C for 30 sec, annealing temperature for 30 sec, and 72°C 2 min; and 72°C for 4 min (Table 3). The PCR products were purified using Agencourt AMPure (Beckman Coulter, Brea, CA, USA). Sanger sequencing was conducted using an Applied Biosystems 3130 Genetic Analyzer (Life Technologies, Carlsbad, CA, USA) using a Big-Dye Terminator ver3.1 Cycle Sequencing Kit (Life Technologies, Carlsbad, CA, USA) and Agencourt CleanSEQ (Beckman Coulter, Brea, CA, USA), according to each manufacturer’s protocol.
Table 3.
The primers used in PCR amplification and sequencing for confirmation of the mutation
| Gene | Forward | Reverse | Annealing temperature |
|---|---|---|---|
|
dltA |
AAGTAGTGCAGTTTAGGAGAGGA |
AGATTGTACCACCGGATGTC |
58.0 |
|
gtcA |
TTGAGCTCTTAGTAGAACCTGAC |
CTGGTTTCGCTATCTCATTAG |
54.5 |
| iap | CAAAATGCTACTACACACGCT | GTCAAAGAATACTAAATCACCAGC | 56.5 |
Availability of supporting data
The draft genome sequences of L. monocytogenes strain 36-25-1 are available in DDBJ/EMBL/GenBank under accession number BASN01000001-BASN01000122. The gene sequences of other strains Lma13, Lma15, Lma20 and Lma28 are available under accession number AB845328-845343.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
Conception and design of this study: HT, KB. Laboratory work and data analysis: DK, HT. Manuscript writing, review and revision: DK, HT, SM, TK. All authors read and approved the final manuscript.
Supplementary Material
The alignment of inlA in EGDe and InlA truncated strains. Nucleotide sequences and amino acid sequences are shown for each strain. The numbers shown on the both sides mean the nucleotide sequence positions in the ORF of strain EGDe. The frames show identical sequences among the strains.
Contributor Information
Daisuke Kyoui, Email: d131006@kaiyodai.ac.jp.
Hajime Takahashi, Email: hajime@kaiyodai.ac.jp.
Satoko Miya, Email: s-miya@kaiyodai.ac.jp.
Takashi Kuda, Email: kuda@kaiyodai.ac.jp.
Bon Kimura, Email: kimubo@kaiyodai.ac.jp.
Acknowledgements
This work was supported by a Grant-in-Aid for Scientific Research (B 24380115) from the Ministry of Education, Science, Sports, Culture and Technology in Japan.
References
- Swaminathan B, Gerner-Smidt P. The epidemiology of human listeriosis. Microbes Infect. 2007;9:1236–1243. doi: 10.1016/j.micinf.2007.05.011. [DOI] [PubMed] [Google Scholar]
- Ivanek R, Gröhn YT, Wiedmann M. Listeria monocytogenes in multiple habitats and host populations: review of available data for mathematical modeling. Foodborne Pathog Dis. 2006;3:4. doi: 10.1089/fpd.2006.3.4. [DOI] [PubMed] [Google Scholar]
- Rocourt J, BenEmbarek P, Toyofuku H, Schlundt J. Quantitative risk assessment of Listeria monocytogenes in ready-to-eat foods: the FAO/WHO approach. FEMS Immunol Med Microbiol. 2003;33:263–267. doi: 10.1016/S0928-8244(02)00468-6. [DOI] [PubMed] [Google Scholar]
- U.S. Department of Health and Human Service, U.S. Department of Agriculture Food Safety and Inspection service, and Centers for Disease Control. Quantitative assessment of the relative risk to public health from foodborne Listeria monocytogenes among selected categories of ready-to-eat foods. Washington, D.C: United States Department of Health and Human Services and United States Department of Agriculture; 2003. http://www.fda.gov/Food/FoodScienceResearch/RiskSafetyAssessment/ucm183966.htm. [Google Scholar]
- World Health Organization, Food and Agriculture Organization of the United Nations. Risk assessment of Listeria monocytogenes in ready-to-eat foods. Microbiological risk assessment series 5. Roma, Italy: Food and Agriculture Organization of the United Nations and World Health Organization; 2004. http://www.fao.org/docrep/010/y5394e/y5394e00.HTM. [Google Scholar]
- Gaillard JL, Berche P, Frehel C, Gouin E, Cossart P. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell. 1991;65:1127–1141. doi: 10.1016/0092-8674(91)90009-N. [DOI] [PubMed] [Google Scholar]
- Mengaud J, Ohayon H, Gounon P, Mège RM, Cossart P. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell. 1996;84:923–932. doi: 10.1016/S0092-8674(00)81070-3. [DOI] [PubMed] [Google Scholar]
- Miya S, Takahashi H, Ishikawa T, Fujii T, Kimura B. Risk of Listeria monocytogenes contamination of raw ready-tp-eat seafood products available at retail outlets in Japan. Appl Environ Microbiol. 2010;76:3383–3386. doi: 10.1128/AEM.01456-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert WD, Urbanke C, Ziehm T, Beier V, Machner MP, Domann E, Wehland J, Chakraborty T, Heinz DW. Structure of internalin, a major invasion protein of Listeria monocytogenes, in complex with its human receptor E-cadherin. Cell. 2002;111:825–836. doi: 10.1016/S0092-8674(02)01136-4. [DOI] [PubMed] [Google Scholar]
- Chen Y, Ross WH, Whiting RC, Van Stelten A, Nightingale KK, Wiedmann M, Scott VN. Variation in Listeria monocytogenes dose responses in relation to subtypes encoding a full-length or truncated internalin A. Appl Environ Microbiol. 2011;77:4. doi: 10.1128/AEM.01564-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa-Miya S, Kimura B, Takahashi H, Sato M, Ishikawa T, Igarashi K, Fujii T. Nonsense-mutated inlA and prfA not widely distributed in Listeria monocytogenes isolates from ready-to-eat seafood products in Japan. Int J Food Microbiol. 2007;117:312–318. doi: 10.1016/j.ijfoodmicro.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Jonquières R, Bierne H, Mengaud J, Cossart P. The inlA gene of Listeria monocytogenes LO28 harbors a nonsense mutation resulting in release of internalin. Infect Immun. 1998;66:7. doi: 10.1128/iai.66.7.3420-3422.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olier M, Garmyn D, Rousseaux S, Lemaître JP, Piveteau P, Guzzo J. Truncated internalin A and asymptomatic Listeria monocytogenes carriage: in vivo investigation by allelic exchange. Infect Immun. 2005;73:644–648. doi: 10.1128/IAI.73.1.644-648.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Stelten A, Simpson JM, Chen Y, Scott VN, Whiting RC, Ross WH, Nightingale KK. Significant shift in median guinea pig infectious dose shown by an outbreak-associated Listeria monocytogenes epidemic clone strain and a strain carrying a premature stop codon mutation in inlA. Appl Environ Microbiol. 2011;77:2479–2487. doi: 10.1128/AEM.02626-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet C, Doumith M, Gordon JI, Martin PMV, Cossart P, Lecuit M. A molecular marker for evaluating the pathogenic potential of foodborne Listeria monocytogenes. J Infect Dis. 2004;189:2094–2100. doi: 10.1086/420853. [DOI] [PubMed] [Google Scholar]
- Van Stelten A, Simpson JM, Ward TJ, Nightingale KK. Revelation by single-nucleotide polymorphism genotyping that mutations leading to a premature stop codon in inlA are common among Listeria monocytogenes isolates from ready-to-eat foods but not human listeriosis cases. Appl Environ Microbiol. 2010;76:2783–2790. doi: 10.1128/AEM.02651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Témoin S, Roche SM, Grépinet O, Fardini Y, Velge P. Multiple point mutations in virulence genes explain the low virulence of Listeria monocytogenes field strains. Microbiol. 2008;154:939–948. doi: 10.1099/mic.0.2007/011106-0. [DOI] [PubMed] [Google Scholar]
- Camejo A, Carvalho F, Reis O, Leitão E, Sousa S, Cabanes D. The arsenal of virulence factors deployed by Listeria monocytogenes to promote its cell infection cycle. Virulence. 2011;2:379–394. doi: 10.4161/viru.2.5.17703. [DOI] [PubMed] [Google Scholar]
- Bakker HC, Cummings CA, Ferreira V, Vatta P, Orsi RH, Degoricija L, Barker M, Petrauskene O, Furtado MR, Wiedmann M. Comparative genomics of the bacterial genus Listeria: genome evolution is characterized by limited gene acquisition and limited gene loss. BMC Genomics. 2010;11:688. doi: 10.1186/1471-2164-11-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hain T, Ghai R, Billion A, Kuenne CT, Steinweg C, Izar B, Mohamed W, Mraheil MA, Domann E, Schaffrath S, Kärst U, Goesmann A, Oehm S, Pühler A, Merkl R, Vorwerk S, Glaser P, Garrido P, Rusniok C, Buchrieser C, Goebel W, Chakraborty T. Comparative genomics and transcriptomics of lineages I, II, and III strains of Listeria monocytogenes. BMC Genomics. 2012;13:144. doi: 10.1186/1471-2164-13-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H, Cossart P. Listeria monocytogenes surface proteins: from genome predictions to function. Microbiol Mol Biol Rev. 2007;71:377–397. doi: 10.1128/MMBR.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abachin E, Poyart C, Pellegrini E, Milohanic E, Fiedler F, Berche P, Trieu-Cuot P. Formation of D-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol Microbiol. 2002;43:1–14. doi: 10.1046/j.1365-2958.2002.02723.x. [DOI] [PubMed] [Google Scholar]
- Bubert A, Kuhn M, Goebel W, Köhler S. Structural and functional properties of the p60 proteins from different Listeria species. J Bacteriol. 1992;174:8166–8171. doi: 10.1128/jb.174.24.8166-8171.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilgrim S, Kolb-Mäurer A, Gentschev I, Goebel W, Kuhn M. Deletion of the gene encoding p60 in Listeria monocytogenes leads to abnormal cell division and loss of actin-based motility. Infect Immun. 2003;71:3473–3484. doi: 10.1128/IAI.71.6.3473-3484.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen OF, Skouboe P, Dons L, Rossen L, Olsen JE. Listeria monocytogenes exists in at least three evolutionary lines: evidence from flagellin, invasive associated protein and listeriolysin O genes. Microbiol. 1995;141:2053–2061. doi: 10.1099/13500872-141-9-2053. [DOI] [PubMed] [Google Scholar]
- Schmid M, Walcher M, Bubert A, Wagner M, Wagner M, Schleifer KH. Nucleic acid-based, cultivation-independent detection of Listeria spp. and genotypes of L. monocytogenes. FEMS Immunol Med Microbiol. 2003;35:215–225. doi: 10.1016/S0928-8244(02)00456-X. [DOI] [PubMed] [Google Scholar]
- Cabanes D, Dehoux P, Dussurget O, Frangeul L, Cossart P. Surface proteins and the pathogenic potential of Listeria monocytogenes. Trends Microbiol. 2002;10:5. doi: 10.1016/S0966-842X(01)02281-8. [DOI] [PubMed] [Google Scholar]
- Köhler S, Leimeister-Wächter M, Chakraborty T, Lottspeich F, Goebel W. The gene coding for protein p60 of Listeria monocytogenes and its use as a specific probe for Listeria monocytogenes. Infect Immun. 1990;58:1943–1950. doi: 10.1128/iai.58.6.1943-1950.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Handa-Miya S, Kimura B, Sato M, Yokoi A, Goto S, Watanabe I, Koda T, Hisa K, Fujii T. Development of multilocus single strand conformation polymorphism (MLSSCP) analysis of virulence genes of Listeria monocytogenes and comparison with existing DNA typing methods. Int J Food Microbiol. 2007;118:274–284. doi: 10.1016/j.ijfoodmicro.2007.07.047. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2. Cold Spring HarborCold: Spring Harbor Laboratory Press; 1989. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The alignment of inlA in EGDe and InlA truncated strains. Nucleotide sequences and amino acid sequences are shown for each strain. The numbers shown on the both sides mean the nucleotide sequence positions in the ORF of strain EGDe. The frames show identical sequences among the strains.