Summary
Cell surface growth factor receptors couple environmental cues to the regulation of cytoplasmic homeostatic process including autophagy, and aberrant activation of such receptors is a common feature of human malignancies. Here, we defined the molecular basis by which the epidermal growth factor receptor (EGFR) tyrosine kinase regulates autophagy. Active EGFR binds to the autophagy protein Beclin 1, leading to its multisite tyrosine phosphorylation, enhanced binding to inhibitors, and decreased Beclin 1-associated Class III phosphatidylinositol-3 kinase activity. EGFR tyrosine kinase inhibitor (TKI) therapy disrupts Beclin 1 tyrosine phosphorylation and binding to its inhibitors, and restores autophagy in non-small cell lung carcinoma (NSCLC) cells with a TKI-sensitive EGFR mutation. In NSCLC tumor xenografts, the expression of a tyrosine phosphomimetic Beclin 1 mutant leads to reduced autophagy, enhanced tumor growth, tumor dedifferentiation, and resistance to TKI therapy. Thus, oncogenic receptor tyrosine kinases directly regulate the core autophagy machinery, which may contribute to tumor progression and chemoresistance.
Introduction
Epidermal growth factor receptor (EGFR), an oncogenic receptor tyrosine kinase, links extracellular signals to cellular homeostasis. In normal cells, EGFR signaling is triggered by the binding of growth factors, such as epidermal growth factor (EGF), leading to homodimerization or heterodimerization with other EGFR family members (such as HER2/neu) and autophosphorylation of the intracellular domain (Lemmon and Schlessinger, 2010). The phosphotyrosines formed serve as a docking site for adaptor molecules, which results in the activation of signaling pathways including the Ras/MAPK pathway, the PI3K/Akt pathway, and STAT signaling pathways. In tumor cells, the tyrosine kinase activity of EGFR may be dysregulated by EGFR gene mutation, increased EGFR gene copy number, or EGFR protein overexpression, leading to aberrant EGFR signaling and increased tumor cell survival, proliferation, invasion and metastasis (Ciardiello and Tortora, 2008). EGFR signaling is deregulated in many human cancers, including those of the lung, head and neck, colon, pancreas, and brain.
The deregulation of EGFR in human cancers has led to the development of anticancer agents that target EGFR, including: (1) anti-EGFR antibodies that inhibit ligand binding: and (2) small molecule receptor tyrosine kinase inhibitors (TKIs), erlotinib and gefitinib, that block EGFR intracellular tyrosine kinase activity. Although the EGFR TKIs have shown limited clinical benefit in the majority of solid tumors, they are effective in non-small lung carcinomas (NSCLCs) that harbor specific mutations in the tyrosine kinase domain of EGFR (most commonly, in-frame deletion in exon 19 around codons 746-750 or single-base substitution, L858R, in exon 21) (Ciardiello and Tortora, 2008; Lynch et al., 2004; Pao and Chmielecki, 2010). Most patients with NSCLCs with EGFR mutations initially respond favorably to erlotinib or gefitinib, suggesting these mutations drive tumorigenesis. However, among tumors that initially respond to EGFR TKIs, most eventually acquire resistance, often due to the emergence of a secondary mutation, T790M, in the kinase domain of EGFR (Pao and Chmielecki, 2010).
Several studies have shown that EGFR signaling regulates autophagy, a lysosomal degradation pathway that functions in cellular homeostasis and protection against a variety of diseases, including cancer (Levine and Kroemer, 2008). The downstream targets of EGFR – PI3K, Akt, and mTOR – are well-established negative regulators of autophagy (Botti et al., 2006). Moreover, EGFR inhibitors induce autophagy in NSCLCs (Gorzalczany et al., 2011; Han et al., 2011) and other cancer cells (Fung et al., 2012). However, the links between EGFR signaling and autophagy remain poorly understood, particularly (1) the molecular mechanisms by which EGFR signaling suppresses autophagy; (2) the role of EGFR suppression of autophagy in lung cancer pathogenesis; and (3) the role of autophagy induction in the response to TKI therapy. EGFR inhibitor-induced autophagy in lung cancer cells has been postulated to exert either cytoprotective (Han et al., 2011) or cytotoxic (Gorzalczany et al., 2011) effects.
Conflicting results regarding the role of autophagy in the response or resistance to EGFR TKI treatment reflects broader uncertainties in the role of autophagy in cancer therapy (Rubinsztein et al., 2012). It is not understood in what contexts autophagy induction contributes to tumor progression or suppression and to tumor chemoresistance or chemosensitivity. There is a general consensus that autophagy prevents tumor initiation, as loss-of-function mutations of several different autophagy genes results in spontaneous tumorigenesis (beclin 1, Atg5, Atg7) and/or increased chemical-induced tumorigenesis (Atg4C) in mice (Rubinsztein et al., 2012). Despite this inhibitory role in tumor initiation, it has been proposed that autophagy may promote the growth of established tumors and contribute to chemoresistance, principally through its actions to prolong the survival of metabolically-stressed neoplastic cells (Rubinsztein et al., 2012).
To understand the relationship between oncogenic signaling, autophagy, and distinct stages of tumorigenesis, it is important to define the molecular mechanisms by which oncogenic signaling regulates autophagy. We recently showed that the oncogene Akt inhibits autophagy independently of mTOR signaling via serine phosphorylation of the essential autophagy protein, Beclin 1 (Wang et al., 2012), a haploinsufficient tumor suppressor protein frequently monoallelically deleted in human breast and ovarian cancer (Levine and Kroemer, 2008). Moreover, Akt-mediated phosphorylation of Beclin 1 contributes to Akt-dependent fibroblast transformation, supporting the concept that inactivation of Beclin 1-dependent autophagy plays a role in tumor initiation. However, it is not known whether oncogenic inactivation of Beclin 1 (or other autophagy proteins) influences progression of established tumors and/or their response to therapy.
Here we identify the molecular basis by which EGFR tyrosine kinase activity regulates autophagy. We show that active EGFR binds to Beclin 1, leading to its tyrosine phosphorylation, alteration of its interactome, and inhibition of its autophagy function. A mutant of Beclin 1 containing phosphomimetic mutations in the EGFR-dependent tyrosine phosphorylation sites enhances autophagy suppression in EGFR-mutated NSCLC cells, resulting in enhanced tumor progression, altered tumor cell differentiation, and partial tumor resistance to EGFR TKI therapy. These findings demonstrate a heretofore unknown link between oncogenic receptor tyrosine kinases and the autophagy machinery, which may contribute to tumor progression and resistance to targeted therapy.
Results
EGFR Activation Promotes Autophagy Inhibition and EGFR/Beclin 1 Complex Formation
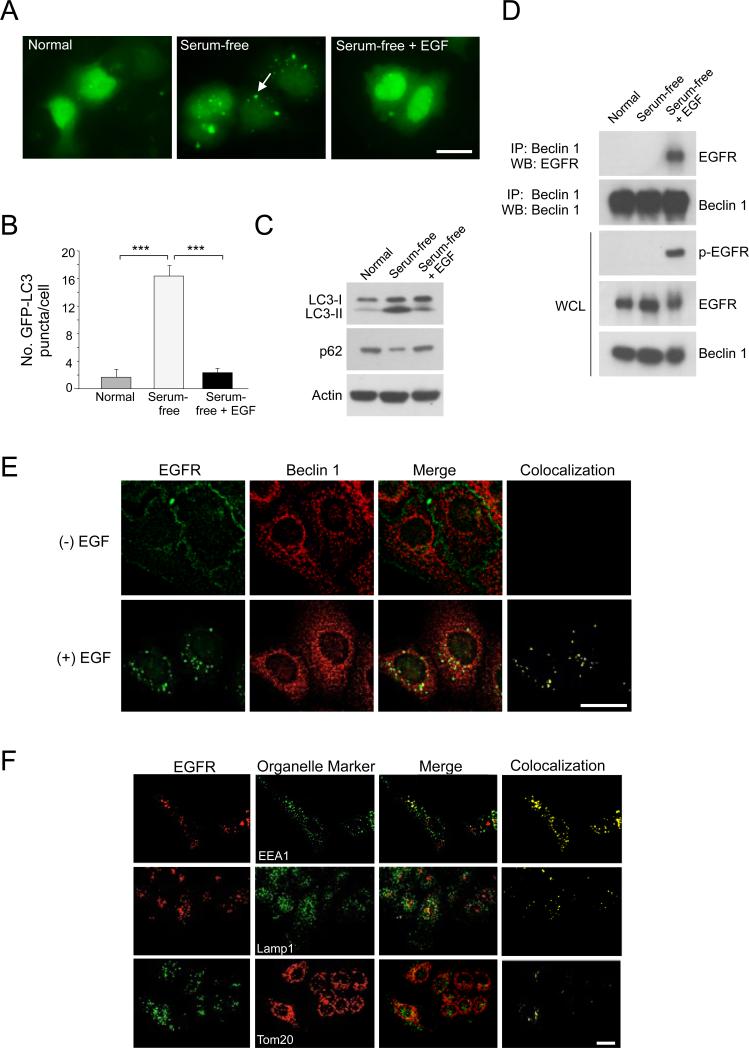
To evaluate whether EGFR activation inhibits autophagy, we used a human non-small cell lung carcinoma (NSCLC) cell line, A549, that express wild-type (WT) EGFR (VanMeter et al., 2008). We measured autophagy by examining the subcellular localization of a green fluorescent autophagy reporter protein, GFP-LC3, and by measuring levels of LC3-II, the autophagosome-associated lipidated form of LC3, and p62, an autophagic substrate (Mizushima et al., 2010). Serum depletion resulted in marked autophagy induction, as evidenced by an increase in GFP-LC3 puncta (autophagosomes) per cell, increased LC3-II conversion, and increased p62 degradation (Figure 1A-C). EGF addition to the serum-starved cells for 30 min partially reversed autophagy induction.
Figure 1. EGFR Activation Inhibits Autophagy and Promotes EGFR/Beclin 1 Complex Formation.
(A) Representative images of GFP-LC3 puncta (autophagosomes) in A549 NSCLC cells cultured O/N in normal medium, serum-free medium, or serum-free medium plus EGF (50 ng/ml, 30 min).
(B) Quantitation of GFP-LC3 puncta in conditions shown in (A). Bars are mean ± SEM of triplicate samples (≥50 cells analyzed per sample). Similar results were observed in 3 independent experiments. ***, P < 0.001, one-way ANOVA.
(C) LC3-I/II and p62 western blot analysis in A549 cells in conditions shown in (A).
(D) Immunoprecipitation of EGFR with Beclin 1 in A549 NSCLC cells in conditions shown in (A).
(E) Colocalization of EGFR and Beclin 1 in EGF-treated A549 cells stably expressing Flag-Beclin 1 (A549/Flag-Beclin 1). Cells were cultured O/N in serum-free medium +/− EGF (50 ng/ml, 30 min), fixed, and immunostained with anti-EGFR (green) and anti-Flag to detect Flag-Beclin 1 (red). Yellow indicates colocalization.
(F) Colocalization of active EGFR with cellular organelles. Cells from (E) were cultured O/N in serum-free medium + EGF (50 ng/ml, 30 min), stained with anti-EGFR and antibodies to detect early endosomes (EEA1), late endosomes/lysosomes (LAMP1) or mitochondria (Tom20). Yellow indicates colocalization.
Scale bars, 20 μm.
See also Figure S1.
To explore the mechanism by which EGFR activation inhibits autophagy, we investigated whether the EGFR can interact with Beclin 1, a component of the Class III phosphatidylinositol 3-kinase (PI3K) (VPS34) autophagy-inducing complex. During growth in normal or serum-free media when no EGFR phosphorylation was observed, EGFR did not co-immunoprecipitate with Beclin 1 (Figure 1D). However, when cells were treated with EGF and EGFR was phosphorylated, EGFR co-immunoprecipitated with Beclin 1. Thus, active phosphorylated EGFR interacts with a key component of the autophagy machinery, Beclin 1, in human NSCLC cells in a ligand-dependent manner.
The EGFR is a cell surface receptor that undergoes ligand-dependent dimerization, phosphorylation of tyrosine residues within its cytoplasmic tail, and endocytosis (Lemmon and Schlessinger, 2010). We examined the colocalization of WT EGFR and Beclin 1 in A549 cells stably expressing Flag-Beclin 1 (Figure 1E). In the absence of EGF stimulation, almost all EGFR localized to the cell surface and did not colocalize with Flag-Beclin 1. In contrast, after EGF stimulation, EGFR was observed in punctate intracellular vesicles and colocalized with Flag-Beclin 1. This colocalization required EGFR endocytosis, and not just receptor homodimerization, as inhibition of EGFR endocytosis by clathrin siRNA blocked EGFR and Beclin 1 colocalization in EGF-treated A549 cells (Figure S1). To determine the identity of the intracellular vesicles with EGFR immunostaining, we evaluated the colocalization of EGFR with different organelle markers, including EEA1 (an early endosome marker), LAMP1 (a late endosome/lysosome marker) and Tom20, a mitochondrial marker (Figure 1F). Internalized EGFR colocalized with EEA1, indicating that the majority of active EGFR resides in endosomes. We also observed weak colocalization of EGFR with LAMP1 and Tom20, consistent with the lysosomal trafficking of endocytosed wild-type EGFR (Carpenter, 1987) and its reported mitochondrial localization (Yao et al., 2010).
Together, these data indicate that ligand-dependent EGFR activation leads to the interaction of EGFR and Beclin 1, which likely occurs primarily in endosomes.
Active EGFR Mutants Interact with Beclin 1 and Inhibit Autophagy Independently of mTOR
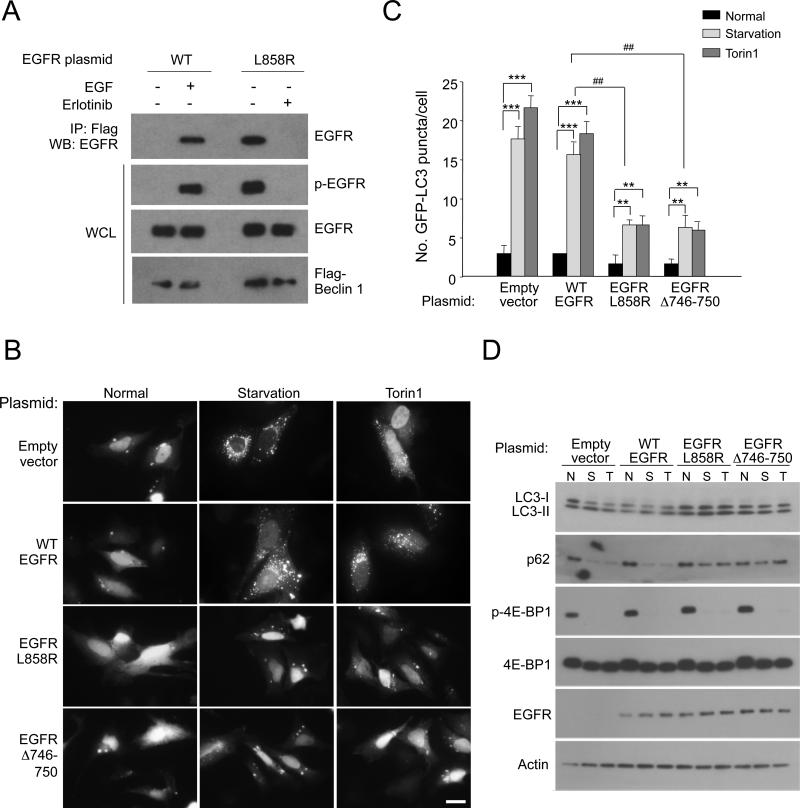
To further evaluate whether ligand-dependent and -independent activation of EGFR results in the formation of an EGFR/Beclin 1 complex and autophagy suppression, we used HeLa cells which express low levels of endogenous EGFR (Figure S2A). HeLa cells were transfected either with WT EGFR, which requires EGF stimulation for EGFR phosphorylation and activation, or with constitutively active mutants of EGFR. WT EGFR co-immunoprecipitated with Flag-Beclin 1 only when activated (tyrosine phosphorylated) by EGF stimulation (Figure 2A). In contrast, an active EGFR mutant common in NSCLCs, EGFR L858R, co-immunoprecipitated with Flag-Beclin 1 in the absence of EGF stimulation. The TKI, erlotinib, which binds to the EGFR ATP binding site, dephosphorylated EGFR and abolished the EGFR/Beclin 1 interaction. Similar results were also observed with another active mutant EGFR, EGFRΔ746-750, also commonly found in NSCLC patients (data not shown). Thus, both ligand-dependent stimulation of WT EGFR and activating mutations in EGFR associated with NSCLC promote the formation of an EGFR/Beclin 1 complex.
Figure 2. Active EGFR Mutants Interact with Beclin 1 and Inhibit Autophagy Independently of mTOR.
(A) Immunoprecipitation of EGFR with anti-Flag in HeLa cells co-transfected with Flag-Beclin 1 and indicated EGFR plasmid. After O/N serum starvation, they were treated +/− EGF (50 ng/ml, 30 min). Cells transfected with EGFR L858R were either treated with DMSO (Erlotinib −) or (1 μM, 4 h) (Erlotinib +).
(B) Representative images of GFP-LC3 puncta in HeLa cells transfected with indicated plasmid, and grown in normal medium, subjected to amino acid starvation (2 h), or treated with Torin1 (50 ng/ml, 2 h). Scale bar, 20 μm.
(C) Quantification of GFP-LC3 puncta in conditions shown in (B). Bars are mean ± SEM of triplicate samples (≥50 cells analyzed per sample). Similar results were observed in 3 independent experiments. **P < 0.01, ***P < 0.001, one-way ANOVA for indicated comparison; ##P<0.01, two-way ANOVA for comparison of magnitude of changes between different groups.
(D) Western blot detection of p62 and LC3-I/II (to measure autophagy) and p-4E-BP1 (to measure mTOR activity).
See also Figure S2.
We investigated the effects of active EGFR mutants that constitutively bind Beclin 1 on autophagy. Both amino acid starvation and the ATP-competitive inhibitor of mTOR, Torin1, induced autophagy in HeLa cells transfected with empty vector or WT EGFR (Figure 2B, 2C, 2D). In cells transfected with the active EGFR mutants, L858R or Δ746-750, starvation and Torin1 induced lower levels of autophagy. However, active EGFR did not block either starvation- or Torin1-induced mTOR deactivation (Figure 2D). Thus, active EGFR mutants suppress autophagy in a manner that is partially independent of mTOR.
Active EGFR Mutants and EGF-Stimulation of Wild-Type EGFR Alter the Beclin 1 Interactome
The results above suggested that the EGFR/Beclin 1 interaction may alter the Beclin 1-containing autophagy-inducing Class III PI3K complex. We mapped the EGFR binding domains of Beclin 1 in HeLa cells co-transfected with the active EGFR L858R mutant and Beclin 1 truncation mutants (Figure S3). Amino acids 1-135 but not 1-115 of Beclin 1 co-immunoprecipitated with active EGFR, suggesting that amino acids 115-135 containing the BH3 domain contribute to the interaction between Beclin 1 and EGFR. In addition, the evolutionarily conserved domain (ECD) of Beclin 1 spanning from amino acids 244-377 was sufficient to bind EGFR. Thus, Beclin 1 contains at least two domains (the BH3 domain and the ECD domain) capable of binding to EGFR.
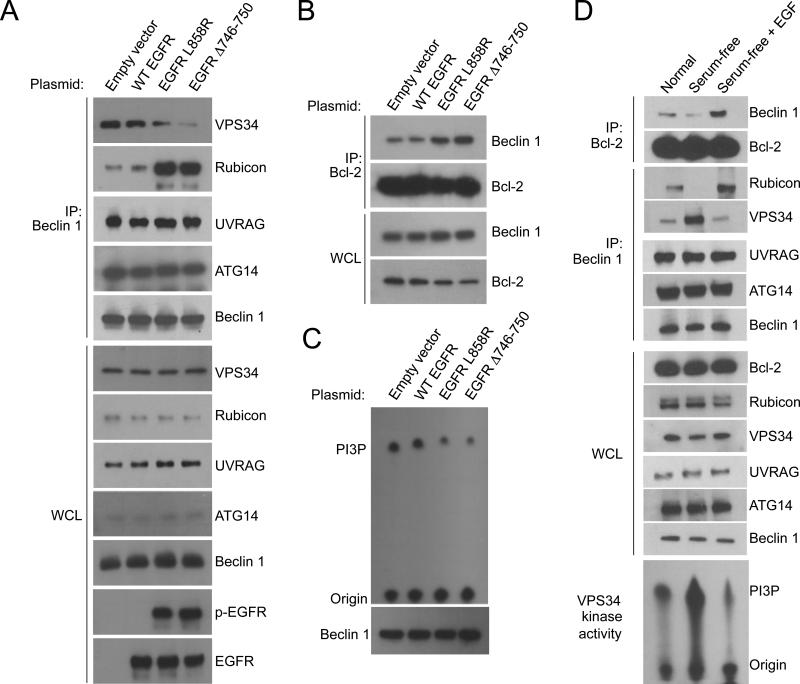
We hypothesized that EGFR binding enhances the interaction of Beclin 1 with negative regulators such as Bcl-2 (which binds to the Beclin 1 BH3 domain) and Rubicon (which binds to the Beclin 1 ECD) and diminishes the interaction of Beclin 1 with VPS34, the Class III PI3K involved in autophagosome initiation (which binds to the Beclin 1 ECD) (He and Levine, 2010). Therefore, we examined the effects of expression of WT and active mutant EGFRs in HeLa cells on the interaction between Beclin and its binding partners, Bcl-2, Rubicon, VPS34, UVRAG and ATG14. Active EGFR did not influence the interaction between Beclin 1 and ATG14 or Beclin 1 and UVRAG. However, expression of either of the active EGFR mutants (L858R or Δ746-750) increased co-immunoprecipitation of Beclin 1 and Rubicon (Figure 3A) and of Beclin 1 and Bcl-2 (Figure 3B). Conversely, cells expressing either of the active EGFR mutants had decreased co-immunoprecipitation of Beclin 1 and VPS34 (Figure 3A). This EGFR-regulated alteration in the Beclin 1 interactome was associated with decreased Beclin 1-associated VPS34 kinase activity (Figure 3C). EGF stimulation resulted in similar alterations in the Beclin 1 interactome and a similar decrease in Beclin 1-associated VPS34 kinase activity in A549 NSCLC cells that express WT EGFR (Figure 3D). Together, these data indicate that binding of EGFR to Beclin 1 (either via ligand-dependent EGFR activation or via activating mutations in EGFR) suppresses autophagy by regulating the Beclin 1 interactome.
Figure 3. Active EGFR Mutants and EGF Ligand Stimulation of WT EGFR Alter the Beclin 1 Interactome.
(A) Immunoprecipitation of indicated proteins with Beclin 1 in Hela cells transfected with indicated EGFR constructs.
(B) Immunoprecipitation of Beclin 1 with Bcl-2 in HeLa cells transfected with indicated EGFR constructs.
(C) Beclin 1-associated VPS34 in vitro kinase assay in anti-Beclin 1 immunoprecipitates of Hela cells transfected with indicated EGFR constructs.
(D) EGF effects on the Beclin 1 interactome and Beclin 1-associated VPS34 kinase activity in A549 cells. Cells cultured as in Figure 1A were subjected to immunoprecipitation with anti-Bcl-2 followed by western blot detection of Beclin 1 or immunoprecipitation with anti-Beclin 1 followed by either western blot detection of indicated proteins or a VPS34 in vitro kinase assay.
See also Figure S3.
EGFR-Dependent Regulation of Autophagy and Beclin 1 Complex Formation in NSCLC Cells with Active EGFR Mutants
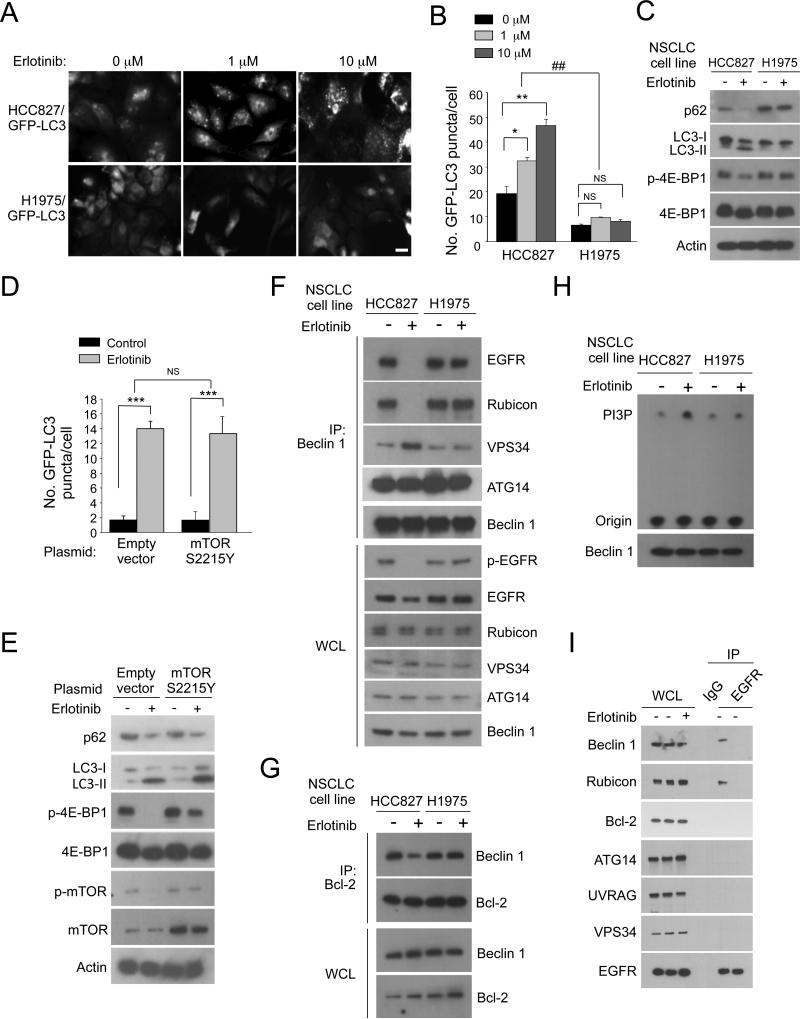
We investigated whether this mechanism underlies EGFR regulation of autophagy in NSCLC cells with activating mutations in EGFR. We used two NSCLC lines: HCC827 cells have the Δ746-750 EGFR activating mutation and are TKI-sensitive, and H1975 cells have the L858R EGFR activating mutation and are TKI-resistant due to a T790M mutation. First, we evaluated if TKI treatment induces autophagy; erlotinib blocked the Beclin 1/EGFR interaction and increased GFP-LC3 puncta in a dose-dependent manner in TKI-sensitive HCC827 cells stably transfected with GFP-LC3 but not in TKI-resistant H1975 cells stably transfected with GFP-LC3 (Figure 4A, 4B, 4F). Furthermore, HCC827 cells but not H1975 cells demonstrated LC3-II conversion and p62 degradation after erlotinib treatment (Figure 4C). Thus, TKI therapy disrupts the EGFR/Beclin 1 complex and induces autophagy in TKI-sensitive but not TKI-resistant cells. This association between TKI sensitivity and autophagy induction was also observed in vivo in tumor xenografts formed by HCC827/GFP-LC3 and H1975/GFP-LC3 cells (Figure S4A-C); HCC827/GFP-LC3 xenografts had increased autophagosomes after TKI treatment and underwent complete regression within several days, whereas H1975/GFP-LC3 xenografts did not have increased autophagosomes and failed to respond to TKI treatment.
Figure 4. Erlotinib-Induced Autophagy in NSCLC Cells is Associated with Regulation of the Beclin 1 Interactome.
(A) Representative images of GFP-LC3 puncta in NSCLC cell lines treated with indicated concentration of erlotinib (4 h). Scale bar, 20 μm.
(B) Quantitation of GFP-LC3 puncta in NSCLC cell lines treated as in (A). Bars are mean ± SEM of triplicate samples (≥ 50 cells analyzed per sample). Similar results were observed in 3 independent experiments.
(C) Western blot detection of p62, LC3-I/II, and p-4E-BP1 in indicated cells +/− erlotinib (1 μM, 2 h).
(D) Quantitation of GFP-LC3 puncta in HCC827 cells transiently co-transfected with GFP-LC3 and control vector or a constitutively active mTOR mutant (S2215Y) and incubated +/− DMSO (control) or erlotinib (1 μM, 2 h). Bars are mean ± SEM of triplicate samples (≥ 50 cells analyzed per sample). Similar results were observed in 3 independent experiments.
(E) Western blot detection of p62, LC3-I/II, and p-4E-BP1 in HCC827 cells treated as in (D).
(F) Immunoprecipitation of indicated Beclin 1 binding partners with Beclin 1 in HCC827 and H1975 cell lines incubated +/− erlotinib (1 μM, 4 h) prior to immunoprecipitation.
(G) Immunoprecipitation of Beclin 1 with Bcl-2 in the conditions in (F).
(H) Beclin 1-associated VPS34 in vitro kinase assay after immunoprecipitation of Beclin 1 complexes from indicated cells +/− erlotinib (1 μM, 4 h).
For (B) and (D), NS, not significant, *P < 0.05, **P < 0.01, ***P<0.001; one-way ANOVA for indicated comparison. ##P < 0.01; two-way ANOVA for comparison of magnitude of changes between different groups.
See also Figure S4.
Since erlotinib decreased mTOR activity in HCC827 cells, as measured by phosphorylation of the mTOR substrate, 4E-BP1 (Figure 4C), we asked if erlotinib-induced autophagy is dependent on suppression of mTOR. We transfected HCC827 cells with a constitutively active mTOR mutant S2215Y (Sato et al., 2010) that blocks starvation-induced mTOR deactivation and dephosphorylation of 4E-BP1. mTOR S2215Y expression in HCC827 cells decreased levels of erlotinib-induced mTOR and 4E-BP1 dephosphorylation but had no effect on erlotinib-induced autophagy (Figure 4D, 4E). Thus, TKI-induced autophagy in a NSCLC cell line with an active EGFR mutation is independent of mTOR deactivation.
We investigated whether TKI-induced autophagy in HCC827 cells involves regulation of the Beclin 1 interactome. In TKI-sensitive HCC827 cells (but not in TKI-resistant H1975 cells), erlotinib led to EGFR dephosphorylation, disruption of EGFR/Beclin 1 binding, disruption of Beclin 1/Rubicon binding, increased Beclin 1/VPS34 binding, decreased Beclin 1/Bcl-2 binding, and increased Beclin 1-associated VPS34 kinase activity (Figure 4F-H). Thus, TKI-induced autophagy in NSCLCs with active EGFR is associated with increased Beclin 1-associated VPS34 kinase activity and disruption of the interaction between Beclin 1 and EGFR and between Beclin 1 and negative regulators of autophagy such as Bcl-2 and Rubicon. The effect of active EGFR on Beclin 1/Bcl-2 and Beclin 1/VPS34 interactions is likely indirect, as only Beclin 1 and Rubicon (but not Bcl-2, ATG14, UVRAG, or VPS34) co-immunoprecipitate with active EGFR in HCC827 cells (Figure S4D).
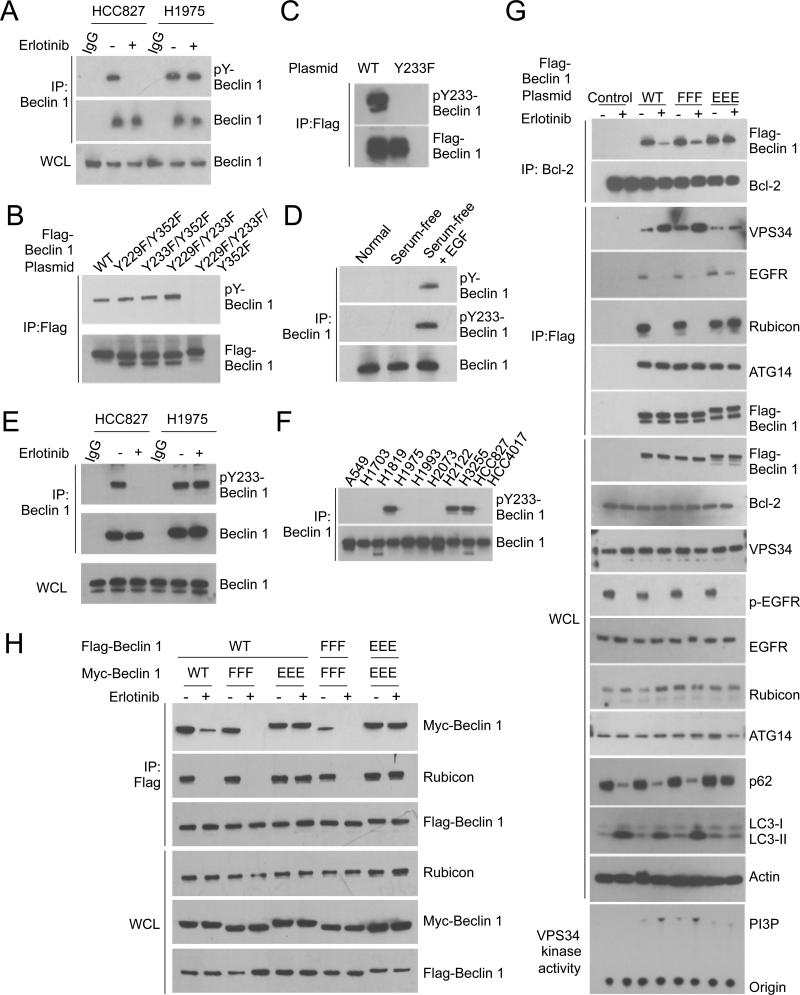
Mutational Activation of EGFR and Ligand Stimulation of Wild-Type EGFR Results in Beclin 1 Tyrosine Phosphorylation
We investigated whether regulation of Beclin 1 function by active EGFR involves Beclin 1 tyrosine phosphorylation. Beclin 1 was tyrosine phosphorylated in NSCLC cells with WT EGFR after EGF stimulation (Figure 5D) and in NSCLC cells with active EGFR mutations (HCC827 and H1975 cells) (Figure 5A). This was decreased by erlotinib in TKI-sensitive HCC827 cells but not TKI-resistant H1975 cells. Inhibition of c-Met, another oncogenic receptor tyrosine kinase activated in HCC827 cells, did not block Beclin 1 tyrosine phosphorylation, and Beclin 1 did not co-immunoprecipitate with active c-Met (Figure S5A, S5B). This suggests that active EGFR (but not other receptor tyrosine kinases) is responsible for Beclin 1 tyrosine phosphorylation in HCC827 cells.
Figure 5. EGFR-Mediated Tyrosine Phosphorylation of Beclin 1 Regulates the Beclin 1 Interactome.
(A) Western blot detection of Beclin 1 tyrosine phosphorylation in NSCLC cells +/− erlotinib (1 μM, 2 h). Lysates were immunoprecipitated with an anti-Beclin 1 followed by western blot analysis with an anti-phosphotyrosine antibody (pY99).
(B) Beclin 1 phosphotyrosine site identification. HCC827 cells transfected with WT or indicated mutant Flag-Beclin 1 constructs were immunoprecipitated with an anti-Flag followed by western blot analysis with anti-pY99.
(C) Specificity of phospho-Beclin 1 Y233 antibody. HCC827 cells transfected with WT or mutant Flag-Beclin 1 constructs were immunoprecipitated with anti-Flag followed by western blot analysis with anti-Beclin 1 pY233.
(D) Western blot detection of phospho-Beclin 1 Y233 in A549 cells cultured as in Figure 3D. Lysates from same experiment were immunoprecipitated with anti-Beclin 1 followed by western blot analysis with anti-pY99 or anti-phospho-Beclin 1 Y233.
(E) Western blot detection of phospho-Beclin 1 Y233 in NSCLC cells +/− erlotinib (1 μM, 4 h). Lysates were immunoprecipitated with anti-Beclin 1 followed by western blot analysis with anti phospho-Beclin 1 Y233.
(F) Western blot detection of phospho-Beclin 1 Y233 in NSCLC cell lines with or without active EGFR mutations. Phospho-Beclin 1 Y233 was detected as in (E). See text for information on cell lines.
(G) Immunoprecipitation of Beclin 1 binding partners with WT and indicated Flag-Beclin 1 mutant constructs in stably transfected HCC827 cells +/− erlotinib (1 μM, 2 h). Lysates were immunoprecipitated with anti-Bcl-2 or anti-Flag and subjected to western blot analysis with indicated antibodies. Beclin 1 immunoprecipitates were also used to measure Beclin 1-associated VPS34 in vitro kinase activity (lower panel).
(H) Effect of Beclin 1 tyrosine phosphorylation site mutations on EGFR-regulated Beclin 1 dimerization and binding to Rubicon. Immunoprecipitation with anti-Flag and western blot analysis with indicated antibodies of HCC827 cells co-transfected with indicated Flag epitope-tagged Beclin 1 constructs and Myc epitope-tagged Beclin 1 constructs +/− erlotinib (1 μM, 2 h). WT, wild-type Flag-Beclin 1; FFF, Flag-Beclin 1 Y229F/Y233F/Y352F; Flag-Beclin 1 Y229E/Y233E/Y352E.
See also Figure S5.
We identified sites of EGFR-mediated in vitro Beclin 1 phosphorylation using recombinant active EGFR L858R/T790M and two synthetic peptides derived from regions of Beclin 1 containing three database-identified candidate tyrosine phosphorylation sites, Y229, Y233 and Y352. Active EGFR phosphorylated the Beclin 1 peptide spanning amino acids 223-239 in a concentration-dependent manner (Figure S5C), while control tyrosine kinases, including PDGFRβ, mutationally active PDGFRα T674I and SRMS, did not (data not shown). Mutation of both Y229 and Y233 was required to block EGFR-dependent phosphorylation of the Beclin 1 amino acid 223-239 peptide. The peptide spanning Y352 (Beclin 1 amino acids 345-358) underwent low levels of phosphorylation at the highest peptide concentration examined. Thus, Beclin 1 Y229, Y233, and possibly Y352 may be substrates of EGFR-mediated Beclin 1 tyrosine phosphorylation.
To evaluate whether Beclin 1 Y229, Y233 and/or Y352 are required for Beclin 1 tyrosine phosphorylation in HCC827 NSCLC cells, we expressed Flag epitope-tagged WT and mutant Beclin 1 constructs. While the peptide spanning Beclin 1 Y352 was only mildly phosphorylated by active EGFR in vitro, simultaneous mutation of all three candidate tyrosine phosphorylation sites (Y229F, Y233F and Y352F) in full-length Beclin 1 was required to block Beclin 1 tyrosine phosphorylation (Figure 5B, Figure S5D). Thus, in NSCLC cells with active EGFR, three Beclin 1 tyrosine residues — Y229, Y233 and Y352 — are phosphorylated. These residues are conserved in Beclin 1 throughout metazoan evolution (data not shown).
Flag-Beclin 1 Y233 phosphorylation was detected in HCC827 cells transfected with WT Flag-Beclin 1 but not with mutant Flag-Beclin 1 Y233F using a phosphospecific antibody (Figure 5C). Endogenous Beclin 1 Y233 phosphorylation was detected in A549 cells expressing WT EGFR after EGF stimulation (Figure 5D) and in HCC827 and H1975 cells with active EGFR mutations (Figure 5E). This phosphorylation was blocked by erlotinib in HCC827 but not in H1975 cells. Thus, Beclin 1 Y233 phosphorylation is regulated by ligand-dependent and ligand-independent activation of EGFR. Furthermore, NSCLC cells with activating mutations in EGFR (H1975, H3255 and HCC827) had detectable Beclin 1 Y233 phosphorylation, whereas cells with amplified wild-type EGFR (A549, H1703, H1819 and H2073), with amplified c-Met (H1933) or with K-Ras mutation (HCC4017, H2122) did not (Figure 5F), indicating that Beclin 1 Y233 phosphorylation may be a specific marker of mutationally active EGFR in NSCLC.
To evaluate the functional consequences of Beclin 1 tyrosine phosphorylation, we generated NSCLC cells that stably express WT Flag-Beclin 1 or Flag-Beclin 1 containing three non-phosphorylatable mutations, Y229F/Y233F/Y352F (FFF) or three phosphomimetic mutations, Y229E/Y233E/Y352E (EEE) (Figure 5G). Flag-Beclin 1 FFF interacted with active EGFR and its interactome was regulated by EGFR inhibition in a manner similar to that of WT Flag-Beclin 1. This most likely reflects the presence of endogenous Beclin 1 in these cells (which is tyrosine phosphorylated when EGFR is active) and the homodimerization of Beclin 1 FFF with WT Beclin 1 (Figure 5H), as Beclin 1 homodimerization is predicted to favor binding to Rubicon and Bcl-2 and disfavor binding to VPS34 (see model, Figure S5E). In contrast, the phosphomimetic Beclin 1 EEE mutant escaped regulation by TKI treatment; erlotinib resulted in dephosphorylation of EGFR (indicating preserved receptor sensitivity to TKIs) but did not disrupt Beclin 1 EEE/EGFR, Beclin 1 EEE/Rubicon, or Beclin 1/Bcl-2 interactions, and did not increase the Beclin 1 EEE/VPS34 interaction or increase Beclin 1 EEE-associated VPS34 kinase activity. In addition, unlike WT Beclin 1 or Beclin 1 FFF, the Beclin 1 EEE mutant dimerized with itself or with WT Beclin 1 in HCC827 cells in the setting of EGFR inhibition (Figure 5H). Together, these findings indicate that the Beclin 1 EEE mutant mimics tyrosine phosphorylated Beclin 1.
Perhaps via constitutive dimerization with endogenous WT Beclin 1 (Figure 5H), Beclin 1 EEE functioned as a dominant negative mutant of TKI-induced autophagy. Despite the presence of endogenous Beclin 1 in NSCLC cells, overexpression of Beclin 1 EEE suppressed erlotinib-induced autophagy (Figure 5G). Thus, Beclin 1 tyrosine dephosphorylation (and disruption of Beclin 1 homodimerization) may be essential for TKI-induced autophagy. Moreover, a lower level of basal autophagy and Beclin 1-associated VPS34 kinase activity was observed in non-TKI-treated HCC827 cells expressing Beclin 1 EEE. This suggests that constitutive Beclin 1 tyrosine phosphorylation enhances autophagy suppression in cells with active EGFR mutation.
A Beclin 1 Tyrosine Phosphomimetic Mutant Enhances Autophagy Suppression, Tumor Growth, and Tumor Dedifferentiation of NSCLC Xenografts
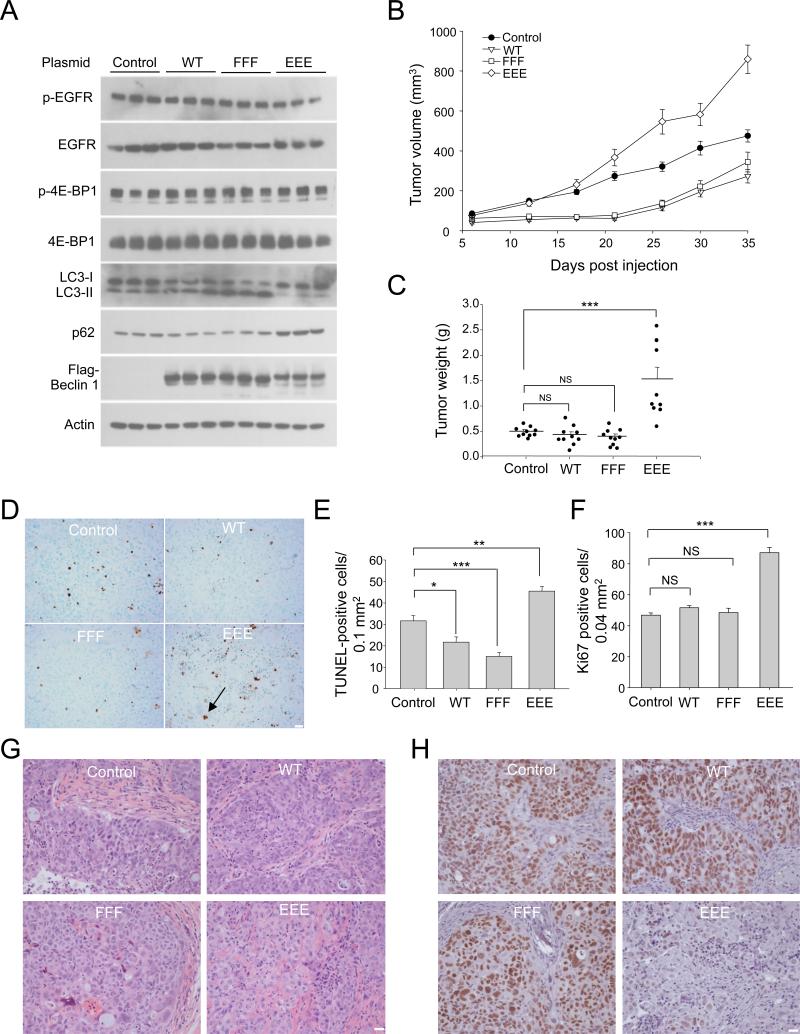
As Beclin 1 EEE expression in NSCLC cells enhanced autophagy suppression in vitro, we evaluated the effects of Beclin 1 EEE expression on NSCLC autophagy and tumor progression in vivo, using a SCID/NOD mouse xenograft model. Beclin 1 EEE-expressing NSCLC xenografts had lower levels of autophagy (Figure 6A), and grew at a faster rate than those derived from NSCLCs transfected with empty vector control, WT Beclin 1, or Beclin 1 FFF (Figure 6B). Conversely, although there was no difference in tumor volume on day 35 (Figure 6C), xenografts from NSCLCs expressing either WT Beclin 1 or Beclin 1 FFF grew at a slower rate than control xenografts (Figure 6B) and had biochemical evidence of increased autophagy (Figure 6A). Thus, in human NSCLC xenografts with EGFR mutation, changes in autophagic activity are associated with inverse changes in the rates of tumor growth.
Figure 6. Beclin 1 Tyrosine Phosphorylation is Associated with Enhanced Tumorigenesis in vivo.
(A) Western blot analysis of autophagy (p62 and LC3), EGFR activity (p-EGFR) and mTOR activity (p-4E-BP1) in indicated xenografts (3 randomly chosen samples per condition; similar results observed in all 10 samples per condition).
(B) Tumor volume of xenografts formed after subcutaneous injection of NOD SCID® mice with HCC827 cells stably transfected with indicated Beclin 1 constructs. Results are mean volume ± SEM for 10 mice per group per time point. P<0.0001 for EEE vs. control; P=0.0006 for WT vs. control; P=0.0074 for FFF vs. control; linear-mixed effect model.
(C) Tumor weights from experiment in (B) upon autopsy at day 35.
(D) Representative images of TUNEL-labeling of indicated HCC827 xenograft tumor genotype. Black arrow denotes representative TUNEL-positive cell.
(E-F) Quantitation of TUNEL-positive nuclei (E) and Ki67-positive nuclei (F) in indicated xenograft tumor genotype. Bars are mean + SEM for all tumors in each genotype (10 per group), from 30 randomly selected fields for each tumor.
(G) Representative H & E images of indicated HCC827 xenograft tumor genotypes. See text for description.
(H) Representative images of TTF-1 immunostaining in indicated HCC827 xenograft tumor genotype.
For (C), (E), (F), NS, not significant, *P < 0.05, **P < 0.01, ***P<0.001; one-way ANOVA. Scale bars, 20 μm.
See also Figure S6.
This inverse relationship seemingly contradicts the paradigm that autophagy fosters the growth of established tumors (Amaravadi et al., 2011; Rubinsztein et al., 2012). This pro-tumor function of autophagy is thought to be related to its role in promoting cell survival in the metabolically stressed tumor microenvironment, and indeed, we found that NSCLC xenografts with increased autophagy had decreased cell death. NSCLC xenografts with overexpression of WT Flag-Beclin 1 or mutant Flag-Beclin 1 FFF (which increased steady-state levels of autophagy) had decreased numbers of TUNEL-positive cells, whereas NSCLC xenografts with overexpression of mutant Flag-Beclin 1 EEE (which suppressed steady-state levels of autophagy) had increased numbers of TUNEL-positive cells (Figure 6D, 6E). Thus, although autophagy exerts pro-survival effects in established tumors, the amount of autophagy-dependent cell survival may be insufficient to determine tumor growth in NSCLC.
We observed other differences that may account for increased tumor growth in NSCLC xenografts expressing the Beclin 1 tyrosine phosphomimetic mutant. NSCLC xenografts expressing Beclin 1 EEE exhibited increased cell proliferation (Figure 6F). Consistent with this increased proliferation, NSCLC xenografts expressing Beclin 1 EEE also had an increase in variance of nuclear DNA intensity, perimeter, and area (Figure S6A-C). More entotic (cell-in-cell structures) and more multinucleated cells were also observed in NSCLC xenografts expressing Beclin 1 EEE (Figure S6D-F). This increase in entotic and multinucleated cells was not associated with an increase in variance in DNA ploidy in cells with single nuclei, although NSCLC xenografts expressing WT Beclin 1 or Beclin 1 FFF (which had increased levels of autophagy) had a decreased variance in DNA ploidy (Figure S6G).
In addition to the aforementioned features, histopathologic analyses revealed significant morphologic differences in NSCLCs xenografts expressing Beclin 1 EEE (Figure 6G, 6H). HCC827 cells were derived from 39 year-old never-smoking woman with primary lung adenocarcinoma (Gazdar and Minna, 1996), and xenografts from control HCC827 cells, HCC827/Flag-Beclin 1, and HCC/Flag-Beclin 1 FFF cells displayed characteristic features of lung adenocarcinomas with mutated EGFR (Shim et al., 2011). They manifested varying degrees of gland formation and cohesive clusters of pleomorphic epithelial cells (admixed with clusters of smaller ovoid or spindle-shaped cells) (Figure 6G). In contrast, HCC827/Flag-Beclin 1 EEE xenografts appeared more poorly differentiated; they lacked overt glandular differentiation, had more infiltrative features, and tumor infiltrates often exhibited a solid growth pattern with morphologic characteristics suggesting squamous differentiation i.e. large cells with centrally placed nuclei and increased amounts of pale pink cytoplasm (low nucleus-cytoplasmic ratio). No differences were observed among the NSCLC xenograft groups in terms of immunostaining for the markers of epithelial to mesenchymal transition, e-cadherin and vimentin (data not shown).
To confirm the change in differentiation status of HCC827/Flag-Beclin 1 EEE tumors, we stained all xenografts for TTF-1, p63, and cytokeratin 5 (CK5), which are used clinically to distinguish between lung adenocarcinoma and squamous carcinoma (Rekhtman et al., 2011). All xenografts had diffuse low expression of p63 (a marker of squamous differentiation, but also expressed by many lung adenocarcinomas) and lacked expression of CK5 (a marker of squamous cell carcinoma) (data not shown). In contrast, marked differences were observed in expression of TTF-1, a homeodomain protein that is highly expressed in lung adenocarcinomas and whose loss of expression is associated with aggressive tumor behavior and decreased median survival in patients with NSCLC (Saad et al., 2004; Tang et al., 2011). All control HCC827, HCC827/Flag-Beclin 1, and HCC827/Flag-Beclin 1 FFF xenografts displayed strong TTF-1 staining homogenously in nearly all tumor cells, while the majority of tumor cells in HCC827/Flag-Beclin 1 EEE xenografts displayed either weak or undetectable TTF-1 staining (Figure 6H). Using an established pathological score for grading TTF-1 staining (Saad et al., 2004), 10/10 xenografts were strongly positive in the control HCC827, HCC827/Flag-Beclin 1, and HCC827/Flag-Beclin 1 FFF groups, whereas 7/10 xenografts were weakly positive and 3/10 xenografts were moderately positive in the HCC827/Flag-Beclin 1 EEE group.
Thus, expression of a Beclin 1 tyrosine phosphomimetic in NSCLC xenografts in mice results in decreased autophagy, increased cellular proliferation, accelerated tumor growth, and dedifferentiation from TTF-1-positive adenocarcinomas to TTF-1-negative poorly differentiated tumors with features of squamous differentiation. This phenotype is analogous to human adenosquamous carcinoma, an NSCLC subtype associated with a worse prognosis. Similar findings were observed in NSCLC xenografts from three independent Beclin 1 EEE clones (data not shown), indicating that their aggressive features do not represent a clonal artifact.
Inhibition of TKI-Induced Autophagy in NSCLC Xenografts Results in Resistance to TKI Therapy
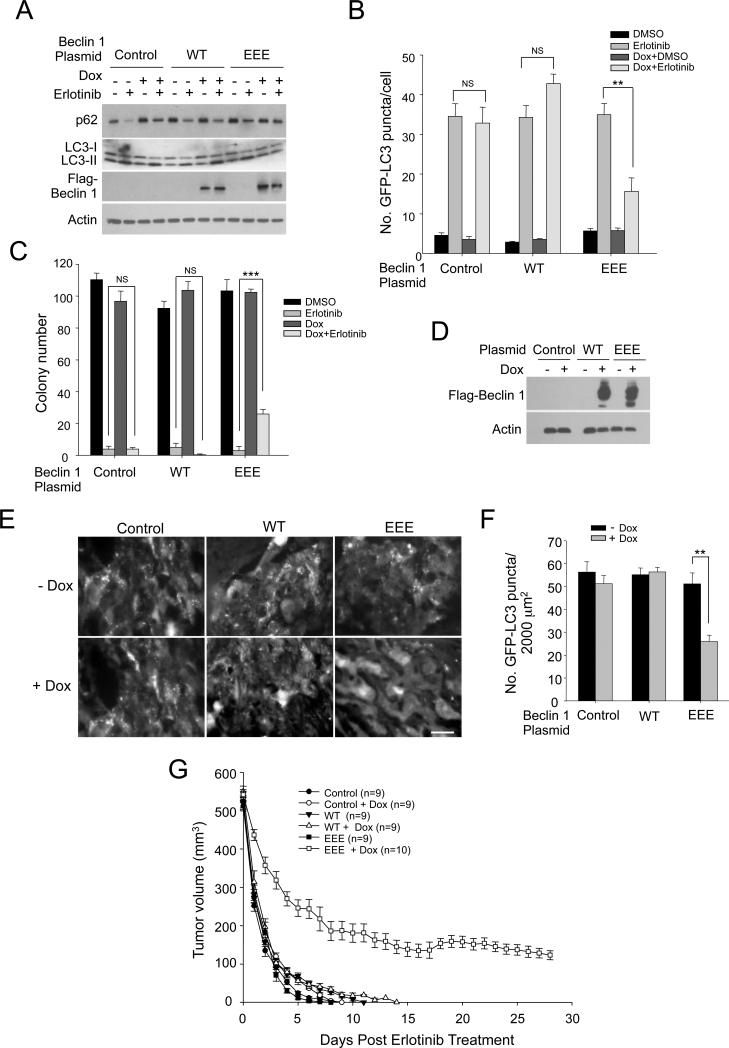
We evaluated the effects of blocking TKI-mediated autophagy on the response of NSCLCs to TKI therapy. Since constitutive Beclin 1 EEE expression alters the natural history of HCC827 xenografts, we generated HCC827 cell lines that express tetracycline-inducible WT Beclin 1 or Beclin 1 Y229E/Y233E/Y352 (EEE) to induce protein expression in established tumors immediately prior to initiation of TKI therapy. We confirmed that doxycycline induction of Beclin 1 EEE expression (but not of WT Beclin 1) in HCC827 cells reduced levels of erlotinib-induced autophagy (Figure 7A, 7B). The suppression of erlotinib-induced autophagy by Beclin 1 EEE expression was associated with an increase in clonogenic survival of HCC827 cells after treatment with erlotinib (Figure 7C). A similar increase in clonogenic survival was also observed in erlotinib-treated HCC827 cells with ATG7 siRNA knockdown (Figure S7A, S7B), suggesting that autophagy suppression, rather than other potential effects of Beclin 1 EEE expression, contributes to TKI-resistance. Similarly, selective inhibition of autophagy (without inhibition of apoptosis) by a mutant viral Bcl-2 (M11 AAA) protein that binds to Beclin 1 but not Bax increased HCC827 clonogenic survival following treatment with erlotinib (Figure S7C-G).
Figure 7. Beclin 1 Tyrosine Phosphorylation Leads to Erlotinib Resistance in NSCLC Xenografts.
(A) Western blot detection of Flag-Beclin 1, p62 and LC3-I/II in indicated HCC827/GFP-LC3 cells with doxycycline-inducible expression of WT Beclin 1 or Beclin 1 Y229E/Y233E/Y352E (EEE). Cells were incubated in +/− doxycycline (1 μg ml−1, 4 days) to induce Flag-Beclin 1 expression and then treated +/− erlotinib (0.2 μM, 2 h).
(B) Quantification of GFP-LC3 puncta for cells treated as in (A). Bars are mean ± SEM of triplicate samples (≥50 cells analyzed per sample). Similar results were observed in 3 independent experiments.
(C) Clonogenic survival of indicated HCC827/GFP-LC3/Beclin 1 cell lines grown +/− erlotinib (0.2 μM, 14 days). Beclin 1 expression was induced by doxycycline (1 μg ml−1, 4 days) prior to erlotinib treatment. Bars are mean ± SEM of triplicate wells per treatment condition. Similar results were observed in 3 independent experiments.
(D) Effect of doxycycline treatment of mice on expression of indicated Flag-Beclin 1 constructs in tumor xenografts. Shown are western blot analyses of tumors 3 days after doxycycline or control treatment.
(E) Representative images of GFP-LC3 staining in indicated NSCLC xenografts treated with 12.5 mg kg−1 erlotinib for 1 day. Scale bar, 20 μm.
(F) Quantitation of number of GFP-LC3 dots per unit area of xenograft. Bars are mean ± SEM for 3 mice per genotype, with 50 randomly selected fields for each tumor.
(G) Effect of erlotinib on tumor response of xenografts formed by indicated HCC827/GFP-LC3/Beclin 1 cell lines. Mice bearing HCC827/GFP-LC3/Beclin 1 xenografts were treated with or without doxycline for 3 days to induce Flag-Beclin 1 expression when the tumor volume reached ~400 mm3, and then treated with 12.5 mg kg−1 erlotinib daily. Values are mean tumor volume ± SEM per time point for 9-10 mice per group. P<0.0001 for EEE vs. EEE+dox; NS for WT vs. WT+dox and FFF vs. FFF+dox; linear-mixed effect model.
For (B), (C), (F), NS, not significant, **P < 0.01, ***P<0.001; one-way ANOVA.
See also Figure S7.
We confirmed that these effects of autophagy suppression in vitro on NSCLC resistance to TKI therapy also occurred in vivo, using xenografts derived from HCC827 cells expressing tetracycline-inducible Beclin 1 EEE and from HCC827 cells expressing the mutant viral Bcl-2 protein. In HCC827 xenografts, induced expression of Beclin 1 EEE (Figure 7D) decreased numbers of autophagosomes one day after initiation of erlotinib (Figure 7E, 7F) and resulted in partial resistance to erlotinib (Figure 7G). In untreated control mice and in mice with induced expression of WT Beclin 1, there was complete regression of all tumor xenografts within 14 days (Figure 7G). In contrast, xenografts with induced expression of Beclin 1 EEE had a slower rate of tumor regression and all tumors remained at the end of the 28-day observation period. Similarly, xenografts derived from HCC827 cells expressing the viral Bcl-2 inhibitor of autophagy had decreased erlotinib-induced autophagy and were partially resistant to erlotinib (Figure S7H-J). Together, these results suggest that Beclin 1 tyrosine phosphorylation and autophagy suppression contribute to TKI resistance in NSCLCs with EGFR mutations.
Discussion
Active EGFR Suppresses Autophagy through Beclin 1 Tyrosine Phosphorylation
The Beclin 1/VPS34 complex plays a crucial role in autophagosome formation. Moreover, Beclin 1 is emerging as a central node of autophagy regulation via cross-talk with diverse cellular and viral autophagy stimulatory or inhibitory proteins; such cross-talk allows the integration of diverse environmental cues with levels of cellular autophagy. Although it was known that activation of cell surface receptors involved in growth factor signaling suppresses autophagy, the precise mechanisms underlying such suppression have been poorly understood. Here we demonstrate that upon activation, EGFR interacts with Beclin 1, promoting its tyrosine phosphorylation and inactivation. This effect occurs in cells expressing WT EGFR upon EGF binding as well as in human NSCLC cells with activating cancer-driving mutations in EGFR. Thus, EGFR signaling suppresses autophagy via its interaction with Beclin 1 during normal mitogenic signaling as well as during aberrant cell proliferation in cancer cells.
These findings directly link receptor tyrosine kinases involved in cell growth control and autophagy suppression. Several downstream molecules in the EGFR signaling pathway, including PI3K, Akt and mTOR, are known to negatively regulate autophagy (Botti et al., 2006). Our results uncover a new mechanism of EGFR suppression of autophagy that is mTOR independent – involving an interaction between EGFR and the Beclin 1 autophagy protein – which underscores the importance of autophagy suppression by EGFR signaling. While we did not find evidence of EGFR-independent Beclin 1 tyrosine phosphorylation in NSCLC cells with active EGFR mutations, other members of the EGFR family (e.g. HER2, HER3, HER4) and/or other oncogenic receptor tyrosine kinases may also inactivate Beclin 1 and the autophagy pathway in other cell types.
The mechanism by which EGFR suppresses Beclin 1 function involves EGFR interaction with two domains (BH3 and ECD) of Beclin 1; EGFR-mediated multisite tyrosine phosphorylation of Beclin 1 on residues Y229, Y233 and Y352; and EGFR-mediated alterations in the Beclin 1 interactome (increased binding to the negative regulators, Bcl-2 and Rubicon, and decreased binding to the VPS34 lipid kinase). The interaction between EGFR and Beclin 1 most likely occurs in endosomes, the site of EGFR signaling following ligand-dependent internalization (with WT EGFR) and of kinase domain mutants that promote constitutive activation and constant endocytic internalization (Wang et al., 2002). Active EGFR interacts with Beclin 1 and Rubicon, but not other Beclin 1 binding partners, such as Bcl-2, VPS34, UVRAG, or ATG14. Thus, the regulation of the Beclin 1 interactome (increased Beclin 1/Bcl-2 binding and decreased Beclin 1/VPS34 binding) by EGFR-mediated phosphorylation is indirect. Our data showing that Beclin 1 tyrosine phosphorylation promotes Beclin 1 homodimerization suggests a model (supported by the crystal structures of the Beclin 1 coiled coil region and evolutionarily conserved domain) in which EGFR-mediated Beclin 1 tyrosine phosphorylation favors the formation of Beclin 1 dimers that are unable to bind VPS34 and promote autophagy (Figure S5E). In addition to blocking autophagy, we speculate that EGFR-mediated Beclin 1 tyrosine phosphorylation may also block its own endolysosomal degradation by promoting the binding of Rubicon to tyrosine phosphorylated Beclin 1, as Rubicon inhibits EGF-stimulated EGFR degradation in A549 cells (Matsunaga et al., 2009). Thus, activating mutations in EGFR may exert multiple effects on Beclin 1; EGFR-dependent Beclin 1 tyrosine phosphorylation may both suppress its autophagy activity as well as its role in endolysosomal trafficking.
EGFR Suppression of Beclin 1 May Contribute to Tumor Progression in Non-Small Cell Lung Carcinoma
Several lines of evidence indicate that decreased Beclin 1 function contributes to tumor initiation: allelic loss of beclin 1 increases the incidence of spontaneous malignancies in mice; Akt-mediated phosphorylation of Beclin 1 contributes to Akt-fibroblast transformation; and allelic loss of beclin 1 in immortalized mammary epithelial cell lines enhances their tumorigenesis in vivo (Levine and Kroemer, 2008; Wang et al., 2012). However, to date, it has been unclear whether loss of Beclin 1 function contributes to the progression of established tumors. Our present findings suggest that enhanced Beclin 1 inactivation (via constitutive expression of a tyrosine phosphomimetic mutant) augments tumor growth of existing NSCLCs with active EGFR mutation. In addition to accelerated growth and increased cell proliferation, tumors with expression of the Beclin 1 tyrosine phosphomimetic have histopathologic features indicative of a de-differentiated, more aggressive neoplasm, including squamous differentiation, increased infiltration, and loss of TTF-1 expression. Since activating EGFR mutations in NSCLC result in constitutive Beclin 1 tyrosine phosphorylation, we postulate that endogenous Beclin 1 tyrosine phosphorylation in NSCLCs may similarly contribute to tumor progression.
We cannot definitively conclude that inactivation of the autophagy function of Beclin 1, rather than its other potential membrane-trafficking functions, contributes to tumor progression in NSCLCs expressing the Beclin 1 tyrosine phosphomimetic mutant. As Beclin 1-associated VPS34 kinase activity is likely essential for both autophagy and other membrane-trafficking events, it is difficult to genetically separate these functions. Nonetheless, we observed a strong relationship between Beclin 1 tyrosine phosphorylation and autophagy suppression in NSCLCs. EGF stimulation suppressed autophagy in NSCLCs expressing WT EGFR; cell lines with constitutive mutations in EGFR had constitutive Beclin 1 tyrosine phosphorylation and autophagy suppression; cell lines with EGFR TKI-sensitive mutations, but not with EGFR TKI-resistant mutations, decreased Beclin 1 tyrosine phosphorylation and autophagy suppression following TKI treatment; and overexpression of a Beclin 1 tyrosine phosphomimetic mutant in TKI-sensitive EGFR mutant NSCLC cells enhanced autophagy suppression.
Regardless of whether Beclin 1 tyrosine phosphorylation functions to enhance tumor progression via suppression of autophagy or some other Beclin 1/VPS34-dependent trafficking event(s), our results reveal an important principle regarding the interrelationship between autophagy, cell survival and tumor progression. We observed decreased cell death in NSCLCs with increased autophagy (i.e. those overexpressing WT Beclin 1 or a non-phosphorylatable mutant of Beclin 1, Beclin 1 Y229F/Y233F/Y352F), and we observed increased cell death in NSCLCs with decreased autophagy (i.e. those overexpressing a tyrosine phosphomimetic mutant of Beclin 1, Beclin 1 Y229E/Y233E/Y352E). These results are consistent with previous studies demonstrating that autophagy functions as a pro-survival pathway in the metabolically stressed tumor microenvironment (Amaravadi et al., 2011; Rubinsztein et al., 2012). However, at least in NSCLCs, the amount of cell death may not be the primary determinant of tumor progression. Other previously described anti-tumor functions of autophagy such as preventing DNA damage and chromosomal instability, and limiting cell proliferation (Levine and Kroemer, 2008) and/or other effects of Beclin 1 such as the prevention of tumor dedifferentiation, may prevail over the pro-survival effects of autophagy in limiting tumor progression. Our observation that tumors with the least autophagy and the greatest amount of cell death also were the most aggressive (in terms of growth rate and histopathology) raise important questions about the strategy of inhibiting autophagy in cancer simply because it can function as a pro-survival pathway. Although such strategies may increase tumor cell death, they may promote tumor progression by blocking other autophagy-dependent functions.
NSCLC Response to EGFR TKI Therapy May Involve Activation of Beclin 1 and Autophagy
Our results have direct implications for the treatment of patients with active EGFR mutations and non-small cell lung carcinoma. Based on previous data that EGFR TKIs induce autophagy (Gorzalczany et al., 2011; Han et al., 2011) and the belief that autophagy induction may lead to chemoresistance (Amaravadi et al., 2011), there are currently several NIH-sponsored clinical trials that combine autophagy inhibitory agents (e.g. chloroquine and hydoxychloroquine) with EGFR inhibitors in the treatment of NSCLC. Earlier studies have either claimed that autophagy induction can enhance or limit the response to EGFR TKI therapy, but they suffer from experimental limitations, including (1) the use of NSCLC cell lines with WT EGFR or overexpressed EGFR rather than NSCLCs with TKI-sensitive mutations in EGFR (which are the tumors that clinically respond best to TKI therapy); (2) induction of autophagy with the use of high doses of TKIs, which are not relevant to those prescribed to patients; (3) the use of assays based on mitochondrial activity (which reflect both cell proliferation and death) rather than clonogenic survival assays to determine the effect of autophagy manipulation on NSCLC survival; and most importantly (4) the lack of in vivo studies that directly assess the effects of modulating autophagy on the response of NSCLC to TKIs. By contrast, we assessed clonogenic survival of NSLCs cells with an active mutation in EGFR after treatment with a clinically relevant concentration of erlotinib using three complementary approaches to inactivate erlotinib-induced autophagy: expression of a Beclin 1 tyrosine phosphomimetic, expression of a viral Bcl-2 mutant that selectively inhibits autophagy but not apoptosis, and siRNA knockdown of an autophagy gene ATG7 that functions downstream of Beclin 1. In all these studies, autophagy inhibition was associated with increased clonogenic survival and TKI resistance in vitro. Moreover, in tumor xenografts formed by TKI-sensitive NSCLC cells, TKI resistance was conferred either by expression of a viral Bcl-2 autophagy inhibitor or inducible expression of the Beclin 1 tyrosine phosphomimetic mutant that blocks autophagy. Together, our data suggest that (1) autophagy induction contributes to EGFR TKI responses in NSCLCs with active EGFR mutations; and (2) the use of autophagy inhibitors in patients receiving EGFR TKIs may adversely (rather than favorably) affect their clinical course.
Experimental Procedures
Cell Culture and Transfection
HeLa cells and the NSCLC tumor cell lines were grown in RPMI-1640 medium (Invitrogen) with 10% fetal bovine serum (FBS) (Invitrogen). A549, HCC827, and H1975 cell lines stably transfected with GFP-LC3 and/or Beclin 1 expression constructs were generated as described in the Extended Experimental Procedures.
Antibodies, Plasmids and Reagents
pTyr233-Beclin 1 antibody was custom-produced by PhosphoSolutions Inc. See Extended Experimental Procedures for details of other antibodies, plasmids and reagents.
Co-immunoprecipitations and Western Blot Analyses
Beclin 1, EGFR, or Bcl-2 complexes were immunoprecipitated in HeLa cells, A549 cells, HCC827 cells in the presence or absence of 50 ng/ml EGF or 1 μM erlotinib as described in the Extended Experimental Procedures and western blot analyses were performed on the immunoprecipitates to detect Beclin 1, EGFR, phospho-EGFR, phospho-Beclin 1, Bcl-2, VPS34, ATG14, Rubicon, and UVRAG.
VPS34 and EGFR in vitro Kinase Assays
Beclin 1-VPS34 complexes were immunoprecipitated with anti-Beclin 1 from A549, HeLa or HCC827 cells and used as substrates for a VPS34 in vitro kinase assay (Wang et al., 2012). To identify the tyrosine phosphorylation sites of Beclin 1, an in vitro Peptide Kinase Assay wasperformed by ProQinase GmbH (Freiburg, Germany) as described in the Extended Experimental Procedures.
Autophagy Assays
Autophagy was measured by quantitation of GFP-LC3 puncta using fluorescence microscopy in cells and tumor xenografts by an observed blinded to experimental condition, and by western blot analysis of LC3 and p62 in cell and tissue lysates.
Immunostaining
Immunofluorescence staining of HeLa cells was performed to detect EGFR, Flag-Beclin 1, EEA1, Lamp1, and Tom20 and the images were analyzed on a Zeiss AxioImager Z2 microscope. Immunuoperoxidase staining to detect TTF-1 and Ki67 expression and TUNEL staining to detect apoptotic cells was performed on tumor xenograft sections. See Extended Experimental Procedures for details.
Tumor Xenograft Studies
Six week-old NOD SCID® mice were injected subcutaneously in the flank region with 107 NSCLC (HCC827) tumor cells, and tumor volume was measured daily using the formula (tumor volume = ½(L × W2)) for 35 days. To measure tumor response to erlotinib therapy, mice were randomized after tumors reached ~400 mm3 to receive water containing either 2 mg ml−1 doxycycline or no doxycycline and beginning three days later, treated daily with 12.5 mg kg−1 i.p erlotinib until tumors were no longer detectable or for a maximum of 28 days. All animal procedures were performed in accordance with institutional guidelines and with approval from the Institutional Animal Care and Use Committee. See Extended Experimental Procedures for details.
Supplementary Material
Research Highlights.
EGFR negatively regulates autophagy by binding to Beclin 1
Active EGFR phosphorylates Beclin 1 and alters its interactome
EGFR suppression of Beclin 1 may contribute to tumor progression in lung cancer
Lung cancer responses to EGFR inhibitors may involve activation of Beclin 1
ACKNOWLEDGEMENTS
We thanks Noboru Mizushima, Tamotsu Yoshimori, and Sandra Schmid for supplying critical reagents; Haley Harrington for assistance with manuscript preparation; and Lori Nguyen for technical assistance. This work was supported by NIH grants NCI RO1 84254 and NCI RO1 109618 to B.L. and the Lung Cancer SPORE P50CA70907 to J.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, DiPaola RS, Lotze MT, White E. Principles and current strategies for targeting autophagy for cancer treatment. Clin. Cancer Res. 2011;17:654–666. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botti J, Djavaheri-Mergny M, Pilatte Y, Codogno P. Autophagy signaling and the cogwheels of cancer. Autophagy. 2006;2:67–73. doi: 10.4161/auto.2.2.2458. [DOI] [PubMed] [Google Scholar]
- Carpenter G. Receptors for epidermal growth factor and other polypeptide mitogens. Annu. Rev. Biochem. 1987;56:881–914. doi: 10.1146/annurev.bi.56.070187.004313. [DOI] [PubMed] [Google Scholar]
- Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N. Engl. J. Med. 2008;358:1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- Fung C, Chen X, Grandis JR, Duvvuri U. EGFR tyrosine kinase inhibition induces autophagy in cancer cells. Cancer Biol. Ther. 2012;13 doi: 10.4161/cbt.22002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar AF, Minna JD. NCI series of cell lines: an historical perspective. J. Cell Biochem. Suppl. 1996;24:1–11. doi: 10.1002/jcb.240630502. [DOI] [PubMed] [Google Scholar]
- Gorzalczany Y, Gilad Y, Amihai D, Hammel I, Sagi-Eisenberg R, Merimsky O. Combining an EGFR directed tyrosine kinase inhibitor with autophagy-inducing drugs: a beneficial strategy to combat non-small cell lung cancer. Cancer Lett. 2011;310:207–215. doi: 10.1016/j.canlet.2011.07.002. [DOI] [PubMed] [Google Scholar]
- Han W, Pan H, Chen Y, Sun J, Wang Y, Li J, Ge W, Feng L, Lin X, Wang X, et al. EGFR tyrosine kinase inhibitors activate autophagy as a cytoprotective response in human lung cancer cells. PLoS One. 2011;6:e18691. doi: 10.1371/journal.pone.0018691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Levine B. The Beclin 1 interactome. Curr. Opin. Cell Biol. 2010;22:140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell. Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat. Rev. Cancer. 2010;10:760–774. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rekhtman N, Ang DC, Sima CS, Travis WD, Moreira AL. Immunohistochemical algorithm for differentiation of lung adenocarcinoma and squamous cell carcinoma based on large series of whole-tissue sections with validation in small specimens. Mod. Path. 2011;24:1348–1359. doi: 10.1038/modpathol.2011.92. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat. Rev. Drug Discov. 2012;11:709–730. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad RS, Liu YL, Han H, Landreneau RJ, Silverman JF. Prognostic significance of thyroid transcription factor-1 expression in both early-stage conventional adenocarcinoma and bronchioloalveolar carcinoma of the lung. Hum. Pathol. 2004;35:3–7. doi: 10.1016/j.humpath.2003.08.011. [DOI] [PubMed] [Google Scholar]
- Sato T, Nakashima A, Guo L, Coffman K, Tamanoi F. Single amino-acid changes that confer constitutive activation of mTOR are discovered in human cancer. Oncogene. 2010;29:2746–2752. doi: 10.1038/onc.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim HS, Lee da H, Park EJ, Kim SH. Histopathologic characteristics of lung adenocarcinomas with epidermal growth factor receptor mutations in the International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society lung adenocarcinoma classification. Arch. Pathol. Lab Med. 2011;135:1329–1334. doi: 10.5858/arpa.2010-0493-OA. [DOI] [PubMed] [Google Scholar]
- Tang X, Kadara H, Behrens C, Liu DD, Xiao Y, Rice D, Gazdar AF, Fujimoto J, Moran C, Varella-Garcia M, et al. Abnormalities of the TITF-1 lineage-specific oncogene in NSCLC: implications in lung cancer pathogenesis and prognosis. Clin. Cancer Res. 2011;17:2434–2443. doi: 10.1158/1078-0432.CCR-10-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanMeter AJ, Rodriguez AS, Bowman ED, Jen J, Harris CC, Deng J, Calvert VS, Silvestri A, Fredolini C, Chandhoke V, et al. Laser capture microdissection and protein microarray analysis of human non-small cell lung cancer: differential epidermal growth factor receptor (EGPR) phosphorylation events associated with mutated EGFR compared with wild type. Mol. Cell. Proteomics. 2008;7:1902–1924. doi: 10.1074/mcp.M800204-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RC, Wei Y, An Z, Zou Z, Xiao G, Bhagat G, White M, Reichelt J, Levine B. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science. 2012;338:956–959. doi: 10.1126/science.1225967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Pennock S, Chen X, Wang Z. Endosomal signaling of epidermal growth factor receptor stimulates signal transduction pathways leading to cell survival. Mol. Cell. Biol. 2002;22:7279–7290. doi: 10.1128/MCB.22.20.7279-7290.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Wang G, Li Z, Yan B, Guo Y, Jiang X, Xi Z. Mitochondrially localized EGFR is independent of its endocytosis and associates with cell viability. Acta Biochim. Biophys. Sin. (Shanghai) 2010;42:763–770. doi: 10.1093/abbs/gmq090. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.