Abstract
The human transforming growth factor-β1 (TGF-β1) gene, namely TGFB1, contains several single-nucleotide polymorphisms (SNPs) and some of the polymorphic variants were shown to affect the TGF-β1 protein levels. A number of studies reported the association between 915G/C polymorphism and susceptibility to chronic hepatitis C virus (HCV) infection. However, the results were inconsistent. This meta-analysis was conducted to assess the association of TGFB1 915G/C polymorphism with susceptibility to chronic HCV infection. PubMed, ISI Web of Knowledge, ScienceDirect and Google Scholar databases were systematically searched up to August, 2013 to identify relevant studies. The pooled odds ratios (ORs) with their corresponding 95% confidence intervals (95% CIs) were calculated in 5 genetic comparison models (C vs. G, CC vs. GG, GC vs. GG, CC vs. GG+GC and CC+GC vs. GG). The Galbraith plot and subgroup analyses based on ethnicity, genotyping methods, sample size and fibrosis were performed to investigate possible sources of heterogeneity. A sensitivity analysis and assessment of publication bias were also conducted. Finally, 8 eligible case-control studies on TGFB1 915G/C polymorphism, including a total of 910 cases and 632 controls, were included in this meta-analysis. Overall, there was no evidence of any gene-disease association obtained from the subgroup analyses. Therefore, this meta-analysis demonstrated that there is no association between TGFB1 915G/C polymorphisms and susceptibility to chronic HCV infection.
Keywords: transforming growth factor β1, polymorphism, 915G/C, hepatitis C virus, meta-analysis
Introduction
Hepatitis C virus (HCV) infection is one of the major causes of chronic liver disease, with ~170 million HCV carriers worldwide (1). Chronic infection develops in 80% of the infected patients, leading to a higher risk of cirrhosis, hepatocellular carcinoma and end-stage liver disease (2). Although several factors affect the outcome of HCV infection, immunological and genetic factors may play important roles (3).
Transforming growth factor-β1 (TGF-β1), one of the three isoforms of TGF-β, is a multifunctional cytokine that is involved in cell growth and differentiation, angiogenesis, extracellular matrix formation, immune response regulation and development of cirrhosis (4–6). The concentration of TGF-β1 in the plasma has been associated with the progression of HCV-induced liver fibrosis (7,8). Changes in the secretion or function of TGF-β may cause a deregulation of the host immune response in chronic HCV patients (9). Therefore, we hypothesized that abnormal plasma levels of TGF-β1 may be associated with HCV persistence.
The human TGF-β1 gene, namely TGFB1, which is located on chromosome 19q13 (10), contains several single-nucleotide polymorphisms (SNPs) and some of the polymorphic variants were shown to affect the TGF-β1 protein levels (11). We focused on one SNP that was associated with gene expression and susceptibility to disease in previous studies (12,13). The G→C transition at position 915 of the TGF-β1 signal sequence causes the amino acid sequence to change from arginine to proline in codon 25. A number of previous studies reported the association between this SNP and susceptibility to chronic HCV infection (14–21). However, the currently available results are controversial. In order to determine whether TGFB1 915G/C is associated with susceptibility to chronic HCV infection, we undertook a meta-analysis to provide a quantitative assessment of the collective information.
Materials and methods
Search strategy and selection criteria
We conducted a comprehensive literature search in PubMed, ISI Web of Knowledge, ScienceDirect and Google Scholar databases up to March, 2013. The search terms used were as follows: transforming growth factor, hepatitis C, TGF, HCV and polymorphism. The reference lists of the retrieved reviews and articles were also hand-searched in order to identify additional relevant studies.
Studies were included in this meta-analysis if they met the following selection criteria: i) articles published in English; ii) studies designed as case-control studies investigating the association between the TGFB1 915G/C polymorphism and chronic HCV infection; iii) the studies provided the number of chronic HCV infection cases and controls; iv) the genotype or allele frequencies were available for estimating an odds ratio (OR) with its 95% confidence interval (95% CI); and v) if more than one article was published using the same case series, only the study with the largest sample size was selected. The major exclusion criteria were as follows: i) no proper controls; ii) duplicates; and iii) no usable data reported.
Data extraction
Data extraction was performed by two independent investigators and a consensus was reached on all items through discussion. The following information was extracted from each included study: name of first author, year of publication, country of the source of cases and controls, ethnicity of study population (Asian or Caucasian), genotyping methods, number of cases and controls, cases with and without fibrosis, genotype distribution, mean age, male percentage and source of control group.
Statistical analysis
Hardy-Weinberg equilibrium (HWE) was asessed in the control group of each study with the Chi-square method to assess the latent bias resulting from the deviation of genotype distribution (22). Genotype frequencies were considered as being consistent with HWE when the exact P-value of the Chi-square method was >0.05. OR with 95% CI was used to assess the strength of the association between 915G/C polymorphism and susceptibility to chronic HCV infection. The significance of the pooled OR was determined by the Z test and a P<0.05 was considered to indicate a statistically significant difference. The following genetic comparison models were used in this meta-analysis (23): allele contrast (C vs. G), homozygous comparison (CC vs. GG), heterozygous comparison (GC vs. GG), recessive model (CC vs. GG+GC) and dominant model (CC+GC vs. GG). Two meta-analysis models for dichotomous outcomes were used: the fixed effects model, using the Mantel-Haenszel method (24) and the random effects model, using the DerSimonian and Laird method (25). The Chi-square-based Q statistic test (Cochran’s Q statistic) and the I2 statistic were calculated to determine between-study heterogeneity. Heterogeneity was considered significant when P<0.10 for Cochran’s Q statistic (26) or I2>50% for I2 statistic (27) and the random effects model was adopted as the pooling method; otherwise, the fixed effects model was used (P>0.10 and I2<50%). When heterogeneity was observed, the Galbraith plot was used to detect the possible sources of heterogeneity (28). A combined analysis was performed by excluding the studies that possibly caused the heterogeneity to confirm the robustness of the pooled OR. In addition, subgroup analyses based on ethnicity, genotyping methods, sample size and fibrosis, were also performed to investigate the sources of heterogeneity. A sensitivity analysis was performed to investigate the effect of each individual study on the overall meta-analysis OR by omitting a single study each time. Any individual study is suspected to have excessive influence if the point estimate of its omitted analysis lies outside the 95% CI of the combined analysis (omitting none of the studies). Publication bias was assessed with Begg’s funnel plot and Egger’s test and P<0.05 indicated the presence of publication bias. All the statistical analyses were performed using STATA software, version 10 (StataCorp LP, College Station, TX, USA) with two-sided P-values.
Results
Characteristics of the studies
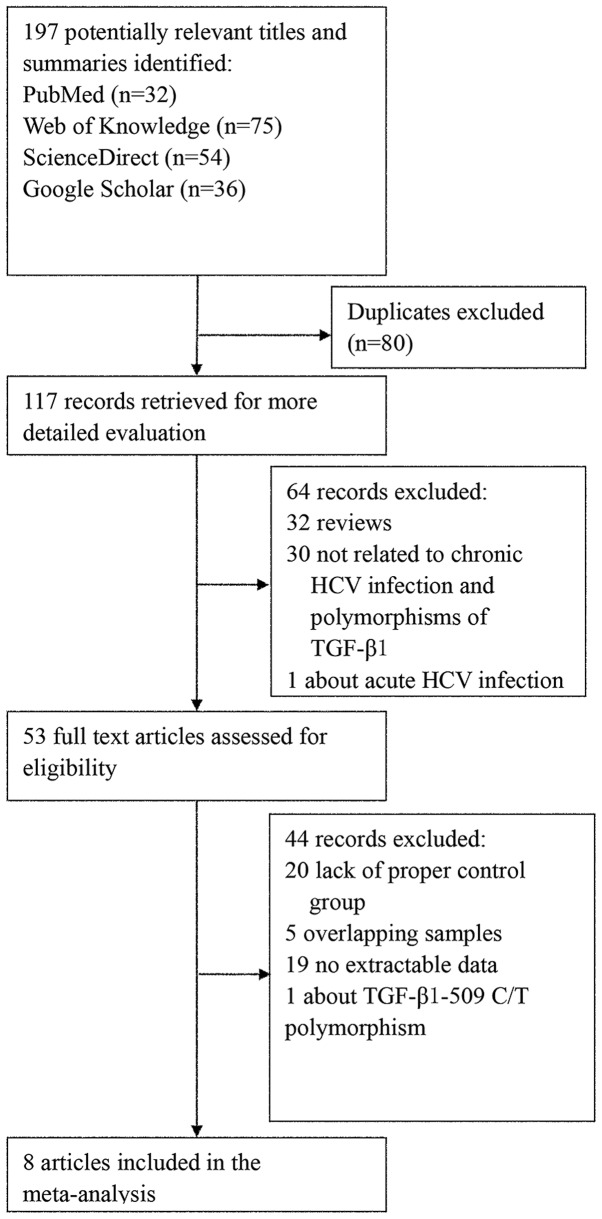
The combined search yielded 117 references after excluding duplicates. The study selection process is shown in Fig. 1. Finally, 8 eligible studies including a total of 1,542 participants (910 cases and 632 controls) were identified (14–21). A summary of the characteristics of each study, including first author, year of publication, ethnicity of cases and controls, genotyping methods, number of cases and controls, cases with and without fibrosis, genotype distribution and P-value for HWE, is shown in Table I. We considered the study of Vidigal et al (14) and the Caucasian arm of the study of Zein et al (20) (there are two studies in this paper) to be reduplicates after comparing the authors and the information of the subjects in the case and control groups. Subsequently, only the study of Vidigal et al (14) was selected according to the selection criteria. All the cases were of chronic HCV infection; there were no other infections, such as HBV and HIV, no report of excessive alcohol consumption and all the controls were healthy individuals. Although some information on mean age and gender percentage was not available, the majority of the studies involved age- and gender ratio-matched cases and controls. For most studies, the mean age of the case or control group was ~45 years and the male percentage was >50%. The distribution of genotypes in the controls was consistent with HWE, except for one study (21).
Figure 1.
Flowchart of the study selection process.
Table I.
Characteristics of studies included in the meta-analysis.
| First author | Year | Ethnicity | Methods | No. (cases/controls) | Fibrosis (Y/N) | Genotype (cases/controls) | P-value for HWE | Refs. | ||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| CC | GC | GG | ||||||||
| Vidigal et al | 2002 | Caucasian | Sequencing | 80/37 | Y | 1/0 | 11/3 | 68/34 | 1.000 | (14) |
| Suzuki et al | 2003 | Asian | PCR-RFLP | 206/101 | Y | 0/0 | 0/0 | 206/101 | NA | (21) |
| Zein et al | 2004 | African | Sequencing | 24/45 | N | 0/0 | 3/4 | 21/41 | 1.000 | (20) |
| Wang et al | 2005 | Caucasian | LightCycler | 210/50 | Y | 0/0 | 32/8 | 178/42 | 1.000 | (19) |
| Pereira et al | 2008 | Mixeda | PCR-SSP | 128/94 | N | 1/1 | 14/29 | 113/64 | 0.452 | (16) |
| Armendariz-Borunda et al | 2008 | Caucasian | ARMS-PCR | 13/30 | Y | 0/6 | 0/13 | 13/11 | 0.706 | (18) |
| Fang et al | 2008 | Asian | ARMS-PCR | 85/106 | N | 3/3 | 9/15 | 73/88 | 0.060 | (17) |
| Romani et al | 2011 | Caucasian | PCR-RFLP | 164/169 | N | 1/2 | 18/16 | 145/151 | 0.101 | (15) |
Caucasian and African.
Y, yes, cases with fibrosis; N, no, cases without fibrosis; HWE, Hardy-Weinberg equilibrium; PCR, polymerase chain reaction; RFLP, restriction fragment length polymorphism; NA, not available; SSP, sequence-specific primers; ARMS, amplification-refractory mutation system.
Meta-analysis for TGFB1 915G/C polymorphism
The results of the combined analysis for the association between TGFB1 915G/C polymorphism and susceptibility to chronic HCV infection are presented in Table II. For the C vs. G model, OR=0.77 and 95% CI: 0.43–1.40; for the CC vs. GG model, OR=0.52 and 95% CI: 0.20–1.35; for the GC vs. GG model, OR=0.73 and 95% CI: 0.38–1.40; for the CC vs. GG+GC model, OR=0.64 and 95% CI: 0.24–1.71; and for the CC+GC vs. GG model, OR=0.73 and 95% CI: 0.38–1.41. Overall, there was no association between the TGFB1 915G/C polymorphism and susceptibility to chronic HCV infection in none of the genetic comparison models. There was heterogeneity in the C vs. G, GC vs. GG and CC+GC vs. GG models. Therefore, the random effects model was used for these genetic comparison models.
Table II.
Combined analysis under all genetic models.
| Heterogeneity | ||||
|---|---|---|---|---|
|
|
||||
| Genetic modela | OR (95% CI) | PZ | PQ | I2 (%) |
| C vs. G | 0.77 (0.43–1.40) | 0.40 | 0.01 | 63.5 |
| CC vs. GG | 0.52 (0.20–1.35) | 0.18 | 0.51 | 0.0 |
| GC vs. GG | 0.73 (0.38–1.40) | 0.34 | 0.01 | 65.7 |
| CC vs. GG+GC | 0.64 (0.24–1.71) | 0.37 | 0.74 | 0.0 |
| CC+GC vs. GG | 0.73 (0.38–1.41) | 0.35 | 0.01 | 66.2 |
Certain studies had to be excluded in some genetic comparison models for 915G/C, as they contained no individuals carrying these genotypes. For C vs. G, GC vs. GG and CC+GC vs. GG, the study by Suzuki et al (21) was excluded; for CC vs. GG and CC vs. GG+GC, Suzuki et al (21), Zein et al (20) and Wang et al (19) were excluded.
OR, odds ratio; 95% CI, 95% confidence interval; PZ, P-value of Z test; PQ, P-value of Cochran’s Q statistic; I2, value of I2 statistic.
Sources of between-study heterogeneity for TGFB1 915G/C polymorphism
The studies potentially causing between-study heterogeneity were identified in the C vs. G, GC vs. GG and CC+GC vs. GG genetic comparison models by the Galbraith plot (Fig. 2). When these studies were excluded, heterogeneity disappeared and the results of the combined analyses after excluding these studies still showed no association between TGFB1 915G/C polymorphism and chronic HCV infection susceptibility (Table III).
Figure 2.
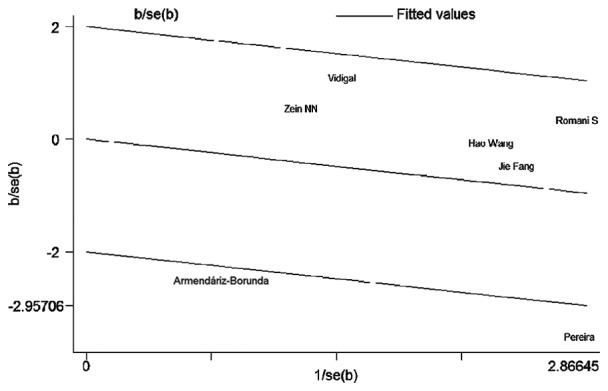
Identification of studies acting as sources of heterogeneity by the Galbraith plot under the CC+GC vs. GG genetic model. Each name represents a separate study for the indicated association. The random effects model was used.
Table III.
Combined analysis after excluding sources of between-study heterogeneity.
| Heterogeneity | |||||
|---|---|---|---|---|---|
|
|
|||||
| Genetic model | Excluded studies (refs.) | OR (95% CI) | PZ | PQ | I2 (%) |
| C vs. G | Pereira et al (16) and Armendariz-Borunda et al (18) | 1.06 (0.73–1.54) | 0.76 | 0.81 | 0.0 |
| GC vs. GG | Pereira et al (16) and Armendariz-Borunda et al (18) | 1.06 (0.70–1.61) | 0.78 | 0.80 | 0.0 |
| CC+GC vs. GG | Pereira et al (16) and Armendariz-Borunda et al (18) | 1.06 (0.71–1.59) | 0.77 | 0.81 | 0.0 |
OR, odds ratio; 95% CI, 95% confidence interval; PZ, P-value of Z test; PQ, P-value of Cochran’s Q statistic; I2, value of I2 statistic.
The results of the subgroup analyses (Table IV) revealed no association between TGFB1 915G/C polymorphism and chronic HCV infection susceptibility. As regards ethnicity, there was only one study on Asian (17), African (20) and Caucasian and African (mixed) populations (16), respectively. Therefore, we could only calculate pooled OR with its 95% CI for a Caucasian population and there was no association between TGFB1 915G/C polymorphism and susceptibility to chronic HCV infection in that population. As regards genotyping methods, there was also only one study using polymerase chain reaction (PCR)-restriction fragment length polymorphism (15), LightCycler (Roche, Basel, Switzerland) (19) and PCR sequence-specific primers (16), respectively. As regards the methods of PCR-direct sequencing and amplification-refractory mutation system-PCR, a subgroup analysis was conducted without any effect on the overall conclusion. In addition, no association was revealed by the subgroup analyses by sample size and fibrosis.
Table IV.
Subgroup analyses based on ethnicity, genotyping methods, sample size and fibrosis under three genetic models with heterogeneity.
| Heterogeneity | |||||
|---|---|---|---|---|---|
|
|
|||||
| Genetic model | Subgroup | OR (95% CI) | PZ | PQ | I2 (%) |
| C vs. G | Caucasian | 0.87 (0.35–2.19) | 0.77 | 0.03 | 65.4 |
| Sequencing | 1.82 (0.69–4.84) | 0.23 | 0.71 | 0.0 | |
| ARMS-PCR | 0.19 (0.00–8.84) | 0.40 | 0.01 | 85.7 | |
| Smalla | 0.66 (0.26–1.67) | 0.38 | 0.01 | 71.9 | |
| Largeb | 1.00 (0.61–1.65) | 1.00 | 0.87 | 0.0 | |
| Fibrosisc | 0.60 (0.10–3.50) | 0.57 | 0.01 | 78.0 | |
| Non-fibrosisd | 0.74 (0.39–1.38) | 0.34 | 0.05 | 61.1 | |
| GC vs. GG | Caucasian | 0.93 (0.39–2.18) | 0.86 | 0.08 | 55.4 |
| Sequencing | 1.68 (0.61–4.66) | 0.32 | 0.83 | 0.0 | |
| ARMS-PCR | 0.20 (0.01–4.85) | 0.32 | 0.04 | 77.6 | |
| Smalla | 0.58 (0.22–1.53) | 0.27 | 0.02 | 67.6 | |
| Largeb | 1.07 (0.62–1.85) | 0.80 | 0.70 | 0.0 | |
| Fibrosisc | 0.65 (0.14–3.10) | 0.54 | 0.04 | 69.9 | |
| Non-fibrosisd | 0.70 (0.32–1.54) | 0.89 | 0.02 | 68.2 | |
| CC+GC vs. GG | Caucasian | 0.86 (0.33–2.23) | 0.76 | 0.04 | 64.2 |
| Sequencing | 1.78 (0.65–4.89) | 0.26 | 0.77 | 0.0 | |
| ARMS-PCR | 0.17 (0.00–7.15) | 0.35 | 0.01 | 84.0 | |
| Smalla | 0.59 (0.22–1.62) | 0.31 | 0.01 | 72.2 | |
| Largeb | 1.04 (0.61–1.77) | 0.90 | 0.78 | 0.0 | |
| Fibrosisc | 0.57 (0.10–3.35) | 0.62 | 0.01 | 76.5 | |
| Non-fibrosisd | 0.71 (0.34–1.48) | 0.92 | 0.03 | 67.0 | |
Small sample of subgroup analysis based on sample size (n≤250).
Large sample of subgroup analysis based on sample size (n>250).
Cases with chronic hepatitis C virus infection with fibrosis.
Cases with chronic hepatitis C virus infection without fibrosis.
OR, odds ratio; 95% CI, 95% confidence interval; PZ, P-value of Z test; PQ, P-value of Cochran’s Q statistic; I2, value of I2 statistic; ARMS-PCR, amplification-refractory mutation system-polymerase chain reaction.
Sensitivity analysis and publication bias for TGFB1 915G/C polymorphism
The sensitivity analysis revealed that no single study exerted a significant effect on the combined results in all the genetic comparison models (Fig. 3). Begg’s funnel plots and Egger’s tests indicated that there were no publication biases (for the C vs. G, CC vs. GG, GC vs. GG, CC vs. GG+GC and CC+GC vs. GG genetic models, Egger’s tests P=0.856, 0.375, 0.872, 0.371 and 0.825, respectively; Begg’s funnel plots not shown). However, two studies were outside the expected 95% CI of Begg’s funnel plot in the C vs. G, GC vs. GG and CC+GC vs. GG models. After excluding these two studies contributing to heterogeneity (16,18), there was no obvious asymmetry of Begg’s funnel plot in the C vs. G, GC vs. GG and CC+GC vs. GG models, with Egger’s test P=0.110, 0.147 and 0.204, respectively.
Figure 3.
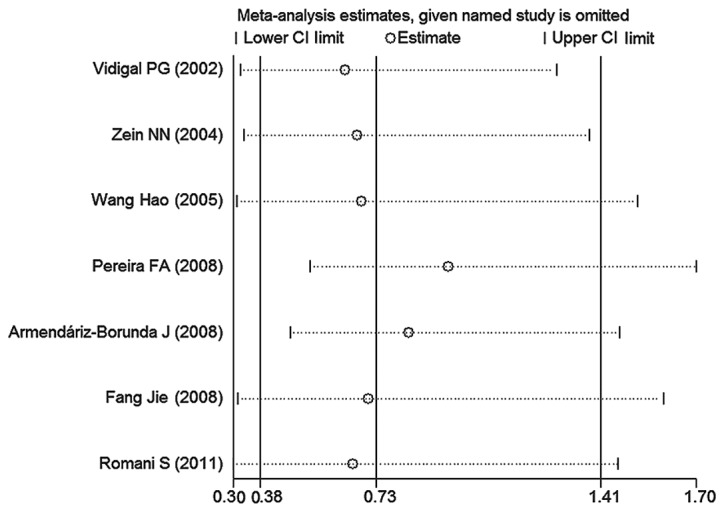
Sensitivity analysis through exclusion of one study at a time to reflect the effect of individual datasets on the pooled ORs under the CC+GC vs. GG genetic model.
Discussion
Individuals infected with HCV have two possible outcomes, clearance or persistent infection. The majority of the infected patients fail to clear HCV and some individuals progress to chronic hepatitis, liver cirrhosis and eventually hepatocellular carcinoma (29). Several factors, either virus- or host-related, have been investigated in an attempt to elucidate the mechanism underlying chronic HCV infection. As regards the virus-related factors, the HCV genotypes do not appear to be correlated with the activity of HCV infection (30) or the outcome (31), although an Italian study reported that children infected with HCV genotype 3 had the highest chance of spontaneous viremia clearance early in life (32). As regards host-related factors, the age at infection, male gender and race were not found to be statistically associated with HCV clearance (32,33). In addition to the abovementioned factors, host genetic factors are considered to exert an effect on the outcome of HCV infection (34). TGF-β1, the expression product of TGFB1, is a polypeptide that is mainly secreted by regulatory T cells (Tregs) (35,36) and was previouly associated with the development of chronic HCV infection (37). Tregs have been reported to suppress T-cell immune responses through the secretion of cytokines, including TGF-β1 (16). HCV infection is characterized by the impairment of HCV-specific effector T-cell responses, suggesting that TGF-β1, as an effector cytokine, possibly contributes to the long-term persistence of HCV infection (16). Furthermore, Presser et al (38) demonstrated that TGF-β1 positively regulates HCV RNA replication. The 915G/C polymorphism in TGFB1 is associated with TGF-β1 levels. Stimulating the leucocytes of patients with the GG genotype in vitro was shown to produce significantly more TGF-β1 compared to individuals with the GC genotype (39). Therefore, the association between the SNP in TGFB1 and susceptibility to chronic HCV infection was investigated by several researchers, yielding, however, inconsistent conclusions (14–21). To the best of our knowledge, this is the first meta-analysis performed to assess the association between TGFB1 915G/C polymorphism and susceptibility to chronic HCV infection.
Overall, no association was detected between TGFB1 915G/C polymorphism and susceptibility to chronic HCV infection. The Galbraith plot and subgroup analyses based on ethnicity, genotyping method, sample size and fibrosis were performed to identify the possible sources of heterogeneity. As a result, the studies of Armendariz-Borunda et al (18) and Pereira et al (16) were considered to cause heterogeneity that may be attributed to genotyping methods and sample size. After excluding these two studies, the pooled ORs with 95% CIs did not change distinctly without the heterogeneity. In addition, no association between TGFB1 915G/C polymorphism and susceptibility to chronic HCV infection was identified by the subgroup analyses.
There were some limitations to our meta-analysis. First, the number of studies and sample size was not adequate. As a result, certain subgroup analyses, particularly by ethnicity, could not be conducted. Second, there were several HCV genotypes. However, the included studies did not provide relevant information. Therefore, we were not able to identify the association between TGFB1 915G/C polymorphism and chronic HCV infection by subgroup analysis of the HCV genotype. Pereira et al (16) reported no significant difference in the frequency of TGFB1 915G/C polymorphism according to HCV genotype distribution. Third, our meta-analysis only included published studies. Therefore, publication bias may have occurred, alhough it was not identified by statistical tests.
It was previously demonstrated that TGF-β1 is associated with the progression of fibrosis in chronic HCV infection (40,41). The TGFB1 915G/C polymorphism in codon 25 was also implicated in this process (42). In the meta-analysis, some cases included in certain studies (14,18,19,21) exhibited chronic HCV infection that progressed to fibrosis. Therefore, a subgroup analysis by fibrosis was performed; however, no association was observed, although the studies causing heterogeneity (16,18) were excluded (data not shown). All the evidence confirmed the robustness of the overall conclusion.
In summary, the association of TGFB1 915G/C polymorphism with susceptibility to chronic HCV infection was not proven in this meta-analysis. However, further well-designed studies including larger sample size and providing more details are required to confirm our conclusions.
References
- 1.Lavanchy D. The global burden of hepatitis C. Liver Int. 2009;29(Suppl 1):74–81. doi: 10.1111/j.1478-3231.2008.01934.x. [DOI] [PubMed] [Google Scholar]
- 2.Lauer GM, Walker BD. Hepatitis C virus infection. N Engl J Med. 2001;345:41–52. doi: 10.1056/NEJM200107053450107. [DOI] [PubMed] [Google Scholar]
- 3.Boyer N, Marcellin P. Pathogenesis, diagnosis and management of hepatitis C. J Hepatol. 2000;32(Suppl 1):98–112. doi: 10.1016/s0168-8278(00)80419-5. [DOI] [PubMed] [Google Scholar]
- 4.Clarke DC, Liu X. Decoding the quantitative nature of TGF-beta/Smad signaling. Trends Cell Biol. 2008;18:430–442. doi: 10.1016/j.tcb.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li MO, Sanjabi S, Flavell RA. Transforming growth factor-beta controls development, homeostasis, and tolerance of T cells by regulatory T cell-dependent and -independent mechanisms. Immunity. 2006;25:455–471. doi: 10.1016/j.immuni.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 6.Nelson DR, Gonzalez-Peralta RP, Qian K, et al. Transforming growth factor-beta 1 in chronic hepatitis C. J Viral Hepat. 1997;4:29–35. doi: 10.1046/j.1365-2893.1997.00124.x. [DOI] [PubMed] [Google Scholar]
- 7.de Andrade DR, Jr, de Andrade DR. The influence of the human genome on chronic viral hepatitis outcome. Rev Inst Med Trop Sao Paulo. 2004;46:119–126. [PubMed] [Google Scholar]
- 8.Tsushima H, Kawata S, Tamura S, et al. Reduced plasma transforming growth factor-beta1 levels in patients with chronic hepatitis C after interferon-alpha therapy: association with regression of hepatic fibrosis. J Hepatol. 1999;30:1–7. doi: 10.1016/s0168-8278(99)80001-4. [DOI] [PubMed] [Google Scholar]
- 9.Kondo Y, Ueno Y, Shimosegawa T. Dysfunction of immune systems and host genetic factors in hepatitis C virus infection with persistent normal ALT. Hepat Res Treat. 2011;2011:713216. doi: 10.1155/2011/713216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujii D, Brissenden JE, Derynck R, Francke U. Transforming growth factor beta gene maps to human chromosome 19 long arm and to mouse chromosome 7. Somat Cell Mol Genet. 1986;12:281–288. doi: 10.1007/BF01570787. [DOI] [PubMed] [Google Scholar]
- 11.Grainger DJ, Heathcote K, Chiano M, et al. Genetic control of the circulating concentration of transforming growth factor type beta1. Hum Mol Genet. 1999;8:93–97. doi: 10.1093/hmg/8.1.93. [DOI] [PubMed] [Google Scholar]
- 12.Ben-Ari Z, Pappo O, Druzd T, et al. Role of cytokine gene polymorphism and hepatic transforming growth factor beta1 expression in recurrent hepatitis C after liver transplantation. Cytokine. 2004;27:7–14. doi: 10.1016/j.cyto.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 13.Falleti E, Fabris C, Toniutto P, et al. TGF-beta1 genotypes in cirrhosis: relationship with the occurrence of liver cancer. Cytokine. 2008;44:256–261. doi: 10.1016/j.cyto.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 14.Vidigal PG, Germer JJ, Zein NN. Polymorphisms in the interleukin-10, tumor necrosis factor-alpha, and transforming growth factor-beta1 genes in chronic hepatitis C patients treated with interferon and ribavirin. J Hepatol. 2002;36:271–277. doi: 10.1016/s0168-8278(01)00243-4. [DOI] [PubMed] [Google Scholar]
- 15.Romani S, Azimzadeh P, Mohebbi SR, et al. Investigation of transforming growth factor-beta1 gene polymorphisms among Iranian patients with chronic hepatitis C. Hepat Mon. 2011;11:901–906. doi: 10.5812/kowsar.1735143X.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pereira FA, Pinheiro da Silva NN, Rodart IF, Carmo TM, Lemaire DC, Reis MG. Association of TGF-beta1 codon 25 (G915C) polymorphism with hepatitis C virus infection. J Med Virol. 2008;80:58–64. doi: 10.1002/jmv.21011. [DOI] [PubMed] [Google Scholar]
- 17.Fang J, Liu ZW, Han QY. Polymorphism of codon25 in signal peptide region of transforming growth factor beta 1 and its association with chronic hepatitis C virus infection. Chin J Hepatol. 2008;6:586–589. (In Chinese) [PubMed] [Google Scholar]
- 18.Armendariz-Borunda J, Rincon AR, Munoz-Valle JF, et al. Fibrogenic polymorphisms (TGF-beta, PAI-1, AT) in Mexican patients with established liver fibrosis. Potential correlation with pirfenidone treatment. J Investig Med. 2008;56:944–953. doi: 10.2310/JIM.0b013e3181891512. [DOI] [PubMed] [Google Scholar]
- 19.Wang H, Mengsteab S, Tag CG, et al. Transforming growth factor-beta1 gene polymorphisms are associated with progression of liver fibrosis in Caucasians with chronic hepatitis C infection. World J Gastroenterol. 2005;11:1929–1936. doi: 10.3748/wjg.v11.i13.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zein NN, Germer JJ, El-Zayadi AR, Vidigal PG. Ethnic differences in polymorphisms of tumor necrosis factor-alpha, interleukin-10, and transforming growth factor-beta1 genes in patients with chronic hepatitis C virus infection. Am J Trop Med Hyg. 2004;70:434–437. [PubMed] [Google Scholar]
- 21.Suzuki S, Tanaka Y, Orito E, et al. Transforming growth factor-beta-1 genetic polymorphism in Japanese patients with chronic hepatitis C virus infection. J Gastroenterol Hepatol. 2003;18:1139–1143. doi: 10.1046/j.1440-1746.2003.03161.x. [DOI] [PubMed] [Google Scholar]
- 22.Thakkinstian A, McElduff P, D’Este C, Duffy D, Attia J. A method for meta-analysis of molecular association studies. Stat Med. 2005;24:1291–1306. doi: 10.1002/sim.2010. [DOI] [PubMed] [Google Scholar]
- 23.Attia J, Thakkinstian A, D’Este C. Meta-analyses of molecular association studies: methodologic lessons for genetic epidemiology. J Clin Epidemiol. 2003;56:297–303. doi: 10.1016/s0895-4356(03)00011-8. [DOI] [PubMed] [Google Scholar]
- 24.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 25.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 26.Paul SR, Donner A. Small sample performance of tests of homogeneity of odds ratios in K 2×2 tables. Stat Med. 1992;11:159–165. doi: 10.1002/sim.4780110203. [DOI] [PubMed] [Google Scholar]
- 27.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 28.Bax L, Ikeda N, Fukui N, Yaju Y, Tsuruta H, Moons KG. More than numbers: the power of graphs in meta-analysis. Am J Epidemiol. 2009;169:249–255. doi: 10.1093/aje/kwn340. [DOI] [PubMed] [Google Scholar]
- 29.Ascione A, Tartaglione T, Di Costanzo GG. Natural history of chronic hepatitis C virus infection. Dig Liver Dis. 2007;39(Suppl 1):S4–S7. doi: 10.1016/s1590-8658(07)80003-x. [DOI] [PubMed] [Google Scholar]
- 30.Roffi L, Ricci A, Ogliari C, et al. HCV genotypes in Northern Italy: a survey of 1368 histologically proven chronic hepatitis C patients. J Hepatol. 1998;29:701–706. doi: 10.1016/s0168-8278(98)80249-3. [DOI] [PubMed] [Google Scholar]
- 31.Zeuzem S, Franke A, Lee JH, Herrmann G, Ruster B, Roth WK. Phylogenetic analysis of hepatitis C virus isolates and their correlation to viremia, liver function tests, and histology. Hepatology. 1996;24:1003–1009. doi: 10.1002/hep.510240505. [DOI] [PubMed] [Google Scholar]
- 32.Bortolotti F, Resti M, Marcellini M, et al. Hepatitis C virus (HCV) genotypes in 373 Italian children with HCV infection: changing distribution and correlation with clinical features and outcome. Gut. 2005;54:852–857. doi: 10.1136/gut.2004.053744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehmann M, Meyer MF, Monazahian M, Tillmann HL, Manns MP, Wedemeyer H. High rate of spontaneous clearance of acute hepatitis C virus genotype 3 infection. J Med Virol. 2004;73:387–391. doi: 10.1002/jmv.20103. [DOI] [PubMed] [Google Scholar]
- 34.Bellanti F, Vendemiale G, Altomare E, Serviddio G. The impact of interferon lambda 3 gene polymorphism on natural course and treatment of hepatitis C. Clin Dev Immunol. 2012;2012:849373. doi: 10.1155/2012/849373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Birerdinc A, Afendy A, Stepanova M, Younossi I, Baranova A, Younossi ZM. Gene expression profiles associated with depression in patients with chronic hepatitis C (CH-C) Brain Behav. 2012;2:525–531. doi: 10.1002/brb3.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hall CH, Kassel R, Tacke RS, Hahn YS. HCV+hepatocytes induce human regulatory CD4+T cells through the production of TGF-beta. PloS One. 2010;5:e12154. doi: 10.1371/journal.pone.0012154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dooley S, ten Dijke P. TGF-beta in progression of liver disease. Cell Tissue Res. 2012;347:245–256. doi: 10.1007/s00441-011-1246-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Presser LD, Haskett A, Waris G. Hepatitis C virus-induced furin and thrombospondin-1 activate TGF-beta1: role of TGF-beta1 in HCV replication. Virology. 2011;412:284–296. doi: 10.1016/j.virol.2010.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Awad MR, El-Gamel A, Hasleton P, Turner DM, Sinnott PJ, Hutchinson IV. Genotypic variation in the transforming growth factor-beta1 gene: association with transforming growth factor-beta1 production, fibrotic lung disease, and graft fibrosis after lung transplantation. Transplantation. 1998;66:1014–1020. doi: 10.1097/00007890-199810270-00009. [DOI] [PubMed] [Google Scholar]
- 40.Presser LD, McRae S, Waris G. Activation of TGF-beta1 promoter by hepatitis C virus-induced AP-1 and Sp1: role of TGF-beta1 in hepatic stellate cell activation and invasion. PloS One. 2013;8:e56367. doi: 10.1371/journal.pone.0056367. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Mehmedovic A, Mesihovic R, Prnjavorac B, et al. Non-invasive liver fibrosis markers: use of serum levels of cytokines IL 1α and TGF β1 in management of chronic liver diseases. Med Glas (Zenica) 2013;10:20–27. [PubMed] [Google Scholar]
- 42.Powell EE, Edwards-Smith CJ, Hay JL, et al. Host genetic factors influence disease progression in chronic hepatitis C. Hepatology. 2000;31:828–833. doi: 10.1053/he.2000.6253. [DOI] [PubMed] [Google Scholar]