Abstract
Disrupted-in-schizophrenia 1 (DISC1) is a gene disrupted by a translocation, t(1;11) (q42.1;q14.3), that segregates with major psychiatric disorders, including schizophrenia, recurrent major depression and bipolar affective disorder, in a Scottish family. Here we report that mammalian DISC1 endogenously expressed in oligodendroglial lineage cells negatively regulates differentiation of oligodendrocyte precursor cells into oligodendrocytes. DISC1 expression was detected in oligodendrocytes of the mouse corpus callosum at P14 and P70. DISC1 mRNA was expressed in primary cultured rat cortical oligodendrocyte precursor cells and decreased when oligodendrocyte precursor cells were induced to differentiate by PDGF deprivation. Immunocytochemical analysis showed that overexpressed DISC1 was localized in the cell bodies and processes of oligodendrocyte precursor cells and oligodendrocytes. We show that expression of the myelin related markers, CNPase and MBP, as well as the number of cells with a matured oligodendrocyte morphology, were decreased following full length DISC1 overexpression. Conversely, both expression of CNPase and the number of oligodendrocytes with a mature morphology were increased following knockdown of endogenous DISC1 by RNA interference. Overexpression of a truncated form of DISC1 also resulted in an increase in expression of myelin related proteins and the number of mature oligodendrocytes, potentially acting via a dominant negative mechanism. We also identified involvement of Sox10 and Nkx2.2 in the DISC1 regulatory pathway of oligodendrocyte differentiation, both well-known transcription factors involved in the regulation of myelin genes.
Introduction
DISC1 gene is specifically disrupted by a t(1;11) (q42.1;q14.3) balanced translocation, in a large Scottish pedigree, which leads to several major mental illnesses, such as schizophrenia (SZ), bipolar affective disorder and recurrent major depression [1]–[3]. Many subsequent genetic studies indicated that DISC1 is not only implicated in schizophrenia and mood disorders, but also in autism spectrum disorders, Asperger syndrome, attention deficit and hyperactivity disorder (ADHD) and agenesis of the corpus callosum [4]–[9]. Biological studies have shown that DISC1 plays a role in multiple process of brain development such as neuronal proliferation, migration, differentiation, and modulation of DISC1 gene in rodents causes behavioral changes [10]–[21].
Multiple lines of evidence, obtained by brain imaging, studies in postmortem brains and genetic association studies, have implicated oligodendrocytes and myelin dysfunction in SZ, major depressive disorder (MDD), autism and ADHD [22]–[24]. Specifically, compromised white matter (WM)/myelin integrity, a reduced number and/or altered morphology of oligodendrocytes, and the aberrant expression and genetic association of oligodendrocytes/myelin-related genes have been identified by a number of studies [25]–[32]. It can also be inferred from the higher than chance co-occurrence of WM-diseases, such as multiple sclerosis (MS), leukodystrophies and velocardiofacial syndrome, with SZ-like psychoses [33]–[35], that oligodendrocytes and myelin dysfunction may play a key role in the pathophysiology of mental illness.
Despite substantial evidence indicating the role of oligodendrocyte abnormalities in pathophysiology of psychosis, neurobiological studies have predominantly focused on neurons. Accordingly, a large number of studies have shown the key role of DISC1 in neurons [10]–[19], while only a handful of studies have addressed a possible role of DISC1 in oligodendrocytes [36]–[38]. DISC1 expression in human brain and primary cultured rat cortical oligodendrocytes was shown by Seshadri et al. [37] and a critical requirement for DISC1 in oligodendroglial development, by promoting specification of olig2-positive cells in the hindbrain and other brain regions of zebrafish, was reported by Wood et al [36]. Nevertheless, no study to date, has directly addressed the functional role of DISC1 expressed in a mammalian cell of glial lineage.
By examining the effect of RNA interference (RNAi) on endogenous DISC1, and also overexpression of either truncated DISC1 or full length DISC1, we here show for the first time that endogenous DISC1 expressed in glial cells negatively regulates mammalian oligodendrocyte development in vitro, acting upstream of Sox10 and/or Nkx2.2.
Materials and Methods
Ethics Statement
The study protocol was approved by the Institutional Animal Care and Use Committee of Osaka University (No. 20-138-006).
Antibodies
Antibodies used in this study are as follows: anti-CNPase (Sigma, St. Louis, MO, USA); anti-APC (Millipore, Billerica, MA, USA); anti-MBP (Millipore); anti-β-tubulin (Sigma); anti-GFP (Abcam, Cambridge, MA, USA); anti-GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA, USA); HRP-linked anti-IgG (Cell Signaling, Danvers, MA, USA); Alexa-Flour568-anti-IgG and Alexa-Flour488-anti-IgG (Molecular Probes). DAPI was purchased from Invitrogen.
Plasmids and Adenovirus
Complementary DNAs coding full length human DISC1 and truncated human DISC1 (1–598) were cloned into pEGFP-C1. A recombinant adenovirus expressing either GFP, human DISC1 with GFP fused to the C-terminus, or truncated human DISC1 (1–598) with GFP fused to the C-terminus (GFP-Adv, DISC1-GFP-Adv and trDISC1-GFP-Adv, respectively) was generated using the ViraPower Adenoviral Expression System (Invitrogen), according to the manufacturer’s instructions.
In situ Hybridization-immunohistochemistry
cDNA fragments of mouse DISC1 were amplified by reverse-transcribed-PCR using the sense/antisense primer set of 5′- ATGCAGGGCGGGGGTCCCCGG -3′/5′-TCAGGCCTCGGTTTCCTGAG-3′, and used as templates for probe synthesis. Probe was hydrolyzed and in situ hybridization of coronal mouse brain sections with DIG-labeled RNA probes was performed as described previously [39]. The slides were washed thoroughly in PBS following a colorimetric reaction. Next, the slides were incubated overnight at 4°C with the primary antibody (monoclonal mouse anti-APC antibody) at 1∶50 in PBS. After washing in PBS, the slides were incubated for 30 min at RT with the secondary antibody (biotinylated anti-mouse IgG antibody from Vector Laboratories). After amplification with the avidin-biotin complex using ABC kit (Vector Laboratories), reaction products were visualized with 50 mM Tris-HCl buffer (pH 7.6) containing 0.02% diaminobenzidine tetrahydrochloride (Sigma) and 0.01% hydrogen peroxide. After dehydration, the sections were sealed using Entellan.
siRNA Design and RNA Interference
The targeted sequences of rat DISC1 were: 5′-GGCTACATGAGAAGCACAG-3′ (DISC1-siRNA-1) and 5′-CTGGCTGATGCGAGAGAAA-3′ (DISC1-siRNA-2). The DISC1-siRNA-1 was previously shown to knockdown mouse and rat DISC1 [15], [40], [41]. The DISC1-siRNA-2 was designed using online software tool siDirect. We validated knockdown of endogenous rat DISC1 transcripts by transfecting DISC1-siRNA-1 or DISC1-siRNA-2 in oligodendrocyte precursor cells. The targeted sequences of rat Sox10 and Nkx2.2 siRNAs were: 5′-CTGTGTCACTGTCCTAAA-3′ (Sox10-siRNA) and 5′-GTTTGTGTGAGTAGCGATA-3′ (Nkx2.2-siRNA). A scrambled sequence (5′-GCGCGCTTTGTAGGATTCGT-3′) was used as a control. Transfection of these siRNAs was carried out using RNAiMAX (Invitrogen), according to the manufacturer’s protocol. To assess the transfection efficiency of siRNA, cells were transfected with Block-iT Alexa Fluor Red Fluorescent Oligo (Invitrogen) and examined under fluorescence microscope 24 hours later.
In vitro Oligodendrocyte Differentiation and Transfection
Primary cultures of rat oligodendrocyte precursor cells were established and induced to differentiate into oligodendrocytes according to the method of Chen et al., with some modification [42]. Single cell suspensions of P1 rat cortex was prepared in MEM supplemented with 0.292 g/l L-glutamine, 4 g/l D-glucose, 3.2 g/l NaHCO3 and 10% FBS, and plated on poly-L-lysine (PLL) coated culture flasks (Nunc). These mixed brain cell cultures were cultured in humidified CO2 incubators for 10 to 14 days with the medium changed every 3 days. Twelve hours after the last medium change, the flask was rotated at 200 rpm for 20 hours to dislodge glial lineage cells. Dislodged cells were plated on non-coated dishes and incubated for 1 hour to allow astrocytes and microglia to adhere to the dish. Oligodendrocyte precursor cells were collected as non-adherent cells, re-suspended in proliferation medium (PM: Neurobasal medium (Invitrogen) supplemented with 5 ng/ml insulin (Sigma), 5 ng/ml NT3 (Pepro Tech Inc., Rocky Hill, NJ, USA), 10 ng/ml PDGF (Wako, Osaka, Japan), 2 mM L-glutamine (Sigma) and B27 (Invitrogen)). Oligodendrocyte precursor cells plated on PLL coated flasks were maintained for 3 days with half-medium-changes with PM every second day. Differentiation of oligodendrocyte precursor cells to oligodendrocytes was induced by replacing the whole medium with PM deprived of PDGF (0 hours). After the induction of differentiation, cells were maintained with half-medium-changes with PDGF deprived PM. Cells were infected with adenovirus expressing GFP, DISC1-GFP or trDISC1-GFP, 12 hours before PDGF deprivation at 0 hours. We confirmed that proportion of cells positive for oligodendrocyte precursor cells marker was 91.8±2.4% of the whole cell population at 0 hours by immunostaining with anti-NG2 antibody. Transfection of siRNAs was performed at 0 hours and the whole medium was changed with PM 4 hours after transfection. In rescue experiments, cells were infected with adenovirus expressing GFP or DISC1-GFP 24 hours after the siRNA transfection and the whole medium was changed with PM 12 hours after the infection.
Quantitative Real-time PCR (qRT-PCR)
Total RNA was reverse-transcribed using High-Capacity cDNA Reverse Transcription Kits (Applied Biosystems, Warrington, UK), and analyzed by RT-PCR to determine expression levels of DISC1, CNPase, MBP, Sox10, Nkx2.2, β-actin and GAPDH. For DISC1 gene, two sets of forward/reverse primers were used: DISC1 5′- TGGCTGTCCCTAGAACACCC-3′/5′- CTCATGCCTATGGCTTCGC-3′ (DISC1 primer-1) or 5′-TGATGCGAGAGAAAGAGCAA-3′/5′- AGCATCTCCTGATCCTCCAA-3′ (DISC1 primer-2). DISC1 primer-1 and DISC1 primer-2 target to exon 11 to 12 and exon 5 to 6 of rat DISC1 gene, respectively. The following sets of forward/reverse primers were used for other genes: CNPase 5′-CAACAGGATGTGGTGAGGA-3′/5′-CTGTCTTGGGTGTCACAAAG-3′, MBP 5′-CACACACAAGAACTACCCA-3′/5′-CACACACAAGAACTACCCA-3′, Sox10 5′-AGCCCAGGTGAAGACAGAGA-3′/5′-CCCCTCTAAGGTCGGGATAG-3′ and Nkx2.2 5′-CGGGCTGAGAAAGGTATGGA-3′/5′-TGTGCTGTCGGGTACTGGG-3′. To standardize the experiments, we designed primer sets (5′-GCCTTCTCTTGTGACAAAGTGG-3′/5′-ATTCTCAGCCTTGACTGTGCC-3′) and (5′-CCTGTATGCCTCTGGTCGTA-3′/5′-CCATCTCTTGCTCGAAGTCT-3′) to amplify a portion of the rat GAPDH and β-actin gene respectively. RT-PCR was set up using Power SYBR Green PCR Master Mix (Applied Biosystems). Comparison of specific ratios (gene of interest/GAPDH or β-actin) was used to assess differences in expression levels between groups.
Western Blotting
Cells were lysed with lysis buffer (50 mM Tris-HCl, pH 7.4, containing 50 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate and protease inhibitor mixture). Western blotting was performed as described previously [43].
Immunocytochemical Analysis
Immunocytochemistry was performed as described [44]. Mouse anti-β-tubulin, rabbit anti-GFP and mouse anti-CNPase antibodies were all used at dilutions of 1∶200. Confocal microscopy was performed using an LSM-510 laser scanning microscope (Carl Zeiss, Oberkochen, Germany). For the analysis of morphological differentiation, cells were classified to one of the following morphological categories: simple, bipolar or stellate cells having short primary branches; intermediate, cells having very long primary branches and/or secondary branches; or complex morphology, cells with tertiary branches [45].
Statistical Analyses
Unless otherwise mentioned, all statistical comparisons were determined by Student’s t-test, with significant differences indicated by p<0.05.
Results
DISC1 is Expressed in Oligodendrocytes in the Mouse Corpus Callosum
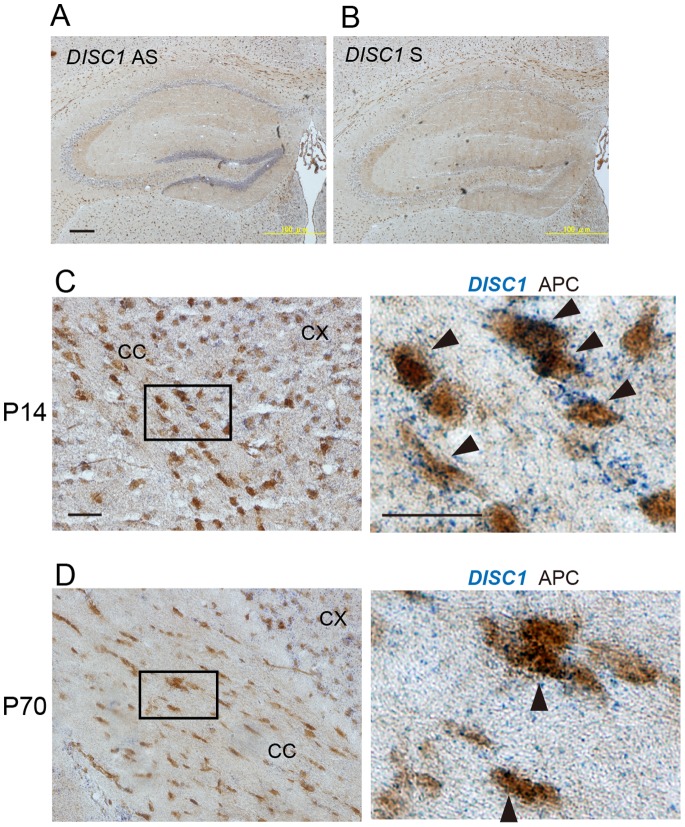
Although a previous study reported DISC1 expression in oligodendrocytes in human brain, DISC1 expression in oligodendroglial lineage cells in mouse brain had not been investigated [37]. To investigate whether DISC1 transcripts are expressed in oligodendrocytes of mouse brain, we performed in situ hybridization-immunohistochemistry using DIG-labeled RNA probe for mouse DISC1 and anti-APC antibody. Consistent with previous studies [41], [46], DISC1 mRNA was highly expressed in the hippocampus at P70 (Fig. 1 A). No signal was detected in sections hybridized with the sense probe confirming the specificity of our in situ hybridization probe (Fig. 1 B). DISC1 mRNA was found in cells expressing APC, an oligodendrocyte marker, both at P14 (Fig. 1 C) and P70 (Fig. 1 D). Furthermore, higher expression of DISC1 mRNA in oligodendrocytes at P14 than P70 was suggested. These results show that DISC1 is expressed in oligodendrocytes of mouse brain.
Figure 1. DISC1 mRNA is expressed in oligodendrocytes in the corpus callosum of mouse brain.
A Double In situ hybridization-immunohistochemistry analysis of hippocampal sections from P70 mice with the antisense RNA probe to DISC1 and antibody against APC. Scale bar, 200 µm. H: hippocampus AS: antisense B As controls, adjacent sections were hybridized with DIG-labeled sense RNA probe. S: sense C, D, Double In situ hybridization-immunohistochemistry analysis of brain sections from P14 and P70 mice demonstrates the expression of DISC1 mRNA in APC expressing oligodendrocytes in the corpus callosum of mouse at P14 (C) and P70 (D). High magnified images of the squared region in the left panels are shown in the adjacent right panels. Arrowheads indicate DISC1+/APC+ cells. Scale bars, 50 µm.
DISC1 Expression Decreases in the Course of in vitro Oligodendrocyte Differentiation
As a developmental decrease of DISC1 mRNA in the mouse corpus callosum was suggested, we investigated DISC1 expression during in vitro differentiation of oligodendrocyte precursor cells to oligodendrocytes. Primary cultured rat oligodendrocyte precursor cells were induced to differentiate to oligodendrocytes by depriving PDGF from the culture medium. Quantitative PCR analysis using two sets of primers for DISC1 showed that DISC1 mRNA expression was reduced after PDGF deprivation (Fig. 2 A). The decrease of DISC1 expression was confirmed using DISC1 primer-1 and another reference gene (β-actin) (100% for 0 h; 48.9±11.3% for 48 h; 26.0±4.8% for 96 h; 36.4±13.5% for 120 h; 22.6±3.8% for 144 h). These results suggest that DISC1 is involved in differentiation of oligodendrocyte lineage cells. Next, we examined the subcellular localization of overexpressed DISC1 in primary cultured oligodendrocyte precursor cells and oligodendrocytes by immunocytochemistry. Overexpressed DISC1 was preferentially expressed in the cell soma and cytoplasmic processes of oligodendrocytes, with marginal nuclear expression. In oligodendrocyte precursor cells, subcellular localization of DISC1 was similar to oligodendrocytes (Fig. 2 B). These results demonstrate that DISC1 expression is more abundant in oligodendrocyte precursor cells and DISC1 protein is expressed in cell soma and processes in oligodendrocyte precursor cells and oligodendrocytes.
Figure 2. DISC1 expression decreases in the course of oligodendrocyte differentiation.
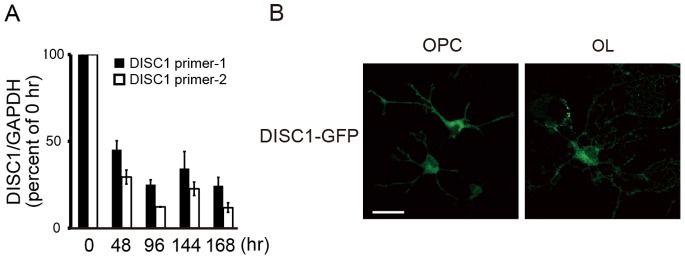
A, Primary cultured cells were harvested at indicated times after PDGF deprivation and mRNA was quantified by qRT-PCR. Data are expressed as the mean±s.e.m. of at least three independent experiments. DISC1 mRNA level at 0 hr is higher than later time-points with p<0.01 by one-way ANOVA followed by Tukey’s test. B, Intracellular localization of overexpressed DISC1-GFP in cultured oligodendrocyte precursor cells and oligodendrocytes. Cells were immunostained with anti-GFP antibody. Scale bar = 50 µm.
DISC1 Overexpression Retards Oligodendrocyte Differentiation
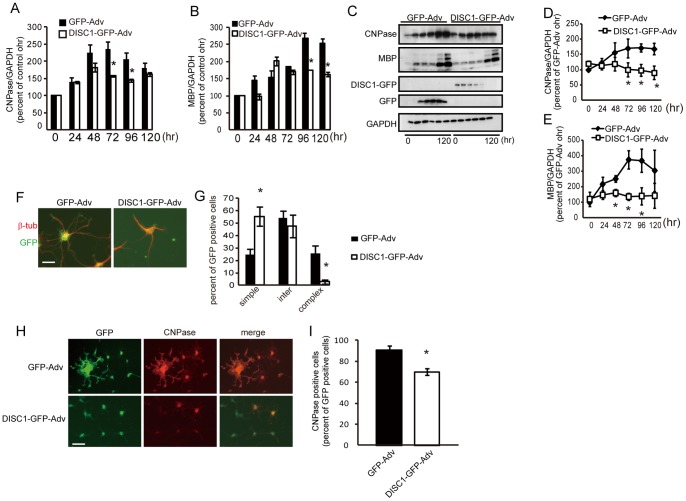
To explore the functional role of decreased expression of DISC1 in the course of oligodendrocytes differentiation (Fig. 2A), DISC1 was overexpressed in oligodendrocyte precursor cells using a DISC1 expressing adenovirus (DISC1-GFP-Adv). In cells induced to differentiate 12 hours after DISC1-GFP-Adv infection, expression of the myelin genes, CNPase (Fig. 3 A, C, D) and MBP (Fig. 3 B, C, E), were decreased at both the mRNA and protein level, compared with control (GFP-Adv) infected cells. To confirm the reduced CNPase expression in DISC1 overexpressing cells, we determined the ratio of GFP expressing cells with CNPase expression in GFP-Adv or DISC1-GFP-Adv infected cells by immunostaining with anti-CNPase antibody. The percentage of CNPase immuno-positive cells decreased 96 hours after PDGF deprivation, following infection of DISC1-GFP-Adv (70.1±3.3%) compared with GFP-Adv (90.6±2.5%) (Fig. 3 H, I).
Figure 3. DISC1 overexpression inhibits oligodendrocyte differentiation.
A, B, Cells infected with GFP-Adv or DISC1-GFP-Adv were harvested at indicated times after PDGF deprivation and mRNA levels of CNPase (A) and MBP (B) were quantified by qRT-PCR. Data are expressed as mean ± s.e.m. of at least three independent experiments. *p<0.05 vs. GFP-Adv. C, Cells infected with GFP-Adv or DISC1-GFP-Adv were lysed at 0, 24, 48, 72, 96 and 120 hours after PDGF deprivation and subjected to western blot analysis. D, E, Quantitation of relative band densities for CNPase (D) and MBP (E) were performed by scanning densitometry. Data are expressed as mean ± s.e.m. of at least three independent experiments. *p<0.05 vs. GFP-Adv. F–I, Oligodendrocyte precursor cells were infected with GFP-Adv or DISC1-GFP-Adv for 12 hours and induced to differentiate by PDGF deprivation for 96 hours then fixed for immunostaining. F, G, Cells were immunostained with anti-GFP and anti-β-tubulin antibodies for morphological observation. Infected cells from three independent cultures were classified according to their morphology (simple, intermediate, or complex) and quantified. The percentage of cells within each category, relative to the total number of GFP positive cells, is shown. *p<0.05 vs. GFP-Adv. Scale bar = 50 µm. H, I, Cells were immunostained with anti-GFP and anti-CNPase antibodies. The percentage of CNPase positive cells relative to the total number of GFP positive cells is shown. Infected cells from three experiments were analyzed. *p<0.05 vs. GFP-Adv. Scale bar = 100 µm.
Morphological transformation of cells infected by GFP-Adv or DISC1-GFP-Adv was examined 96 hours after PDGF deprivation. Infected cells were identified by GFP expression and their morphology was assessed by immunostaining with anti-β-tubulin. The morphology of infected cells was classified as simple, intermediate or complex, as described previously [45]. Of 238 GFP-Adv infected cells, 24.9±3.4% showed a complex morphology, characterized by the presence of several interlaced fine branches indicative of advanced differentiation. Undifferentiated cells, showing a simple morphology defined by primary branches and the absence of secondary and tertiary processes, comprised 23.6±2.2% of the total. In contrast, of 191 DISC1-overexpressing cells, only 2.5±0.8% displayed a complex morphology and 53.3±3.9% showed a simple morphology (Fig. 3 F, G). Thus, DISC1 overexpression results in a reduction of cells with a complex morphology, and an increase in cells with a simple morphology, suggesting that decrease in endogenous DISC1 expression upon PDGF deprivation in oligodendrocyte precursor cells has a functional role to promote oligodendrocyte differentiation.
DISC1 Knockdown Promotes Oligodendrocyte Differentiation
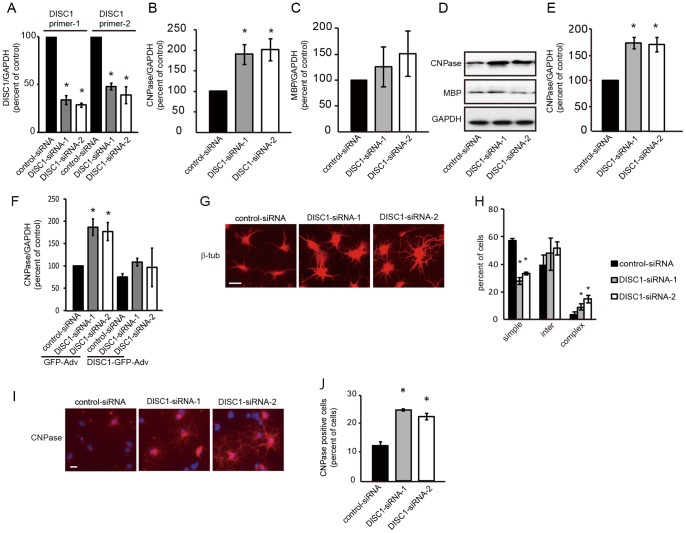
To further investigate the role of endogenous DISC1 in oligodendrocyte differentiation, we treated oligodendrocyte precursor cells with DISC1 specific siRNA (DISC1-siRNA) and examined mRNA or protein expression levels of CNPase and MBP 48 or 72 hours after siRNA transfection. To isolate the effect of DISC1 knockdown induced by DISC1-siRNA, cells were maintained in medium with PDGF during the course of the experiment. The proportion of siRNA-transfected oligodendrocyte precursor cells determined using Block-iT Alexa Fluor Red Fluorescent Oligo was 93.6±1.2% of total cell population. Two DISC1-siRNAs (DISC1-siRNA-1 and DISC1-siRNA-2) targeting exon2 and exon5 of the DISC1 gene respectively have already been shown to effectively suppress rat DISC1 protein expression [15], [40], [41]. Suppression of DISC1 expression by these siRNAs was confirmed by qRT-PCR with two different primer sets for rat DISC1 (DISC1 primer-1 (100% for control siRNA; 33.7±4.8% for DISC1-siRNA-1; 28.5±2.1% for DISC1-siRNA-2); DISC1 primer-2 (100% for control siRNA; 47.9±3.6% for DISC1 siRNA-1; 39.0±8.8% for DISC1 siRNA-2)) (Fig. 4 A). Effective knockdown of DISC1 expression was also confirmed using another reference gene (β-actin) (data not shown). Transfection of either of two siRNAs for DISC1 resulted in an increase of CNPase, at both the mRNA and protein level, compared with control-siRNA treated cells (Fig. 4 B, D, E). Although not statistically significant, we also observed a trend towards increased expression of MBP (Fig. 4 C, D). Immunostaining of in vitro oligodendroglial lineage cells with anti-CNPase antibody revealed that DISC1-siRNA treatment increased the percentage of CNPase positive cells (DISC1-siRNA-1: 24.6±0.7%; DISC1-siRNA-2: 22.3±1.9%), compared with control-siRNA treated cells (12.2±2.3%) (Fig. 4 I, J). To confirm the effect of DISC1 knockdown on expression of CNPase, we performed rescue experiment by co-expression of DISC1 siRNAs and human DISC1-GFP. Human DISC1 rescued the increased CNPase expression by DISC1 siRNA (Fig. 4 F). Morphological transformation of cells transfected with control-siRNA or DISC1-siRNAs was examined by immunostaining for β-tubulin 72 hours after siRNA transfection. Of 225 control-siRNA transfected cells, 57.4±0.9% showed a simple morphology and only 3.5±0.7% of cells displayed a complex morphology. In contrast, DISC1 knockdown decreased the cells with a simple morphology (DISC1-siRNA-1: 27.9±4.6%; DISC1-siRNA-2: 33.5±1.3%), and increased cells with a complex morphology (DISC1-siRNA-1: 6.2±2.7%; DISC1-siRNA-2: 14.7±1.7%) (Fig. 4 G, H). Our results suggest that endogenous DISC1 in an oligodendrocyte lineage cell negatively regulates oligodendrocyte differentiation.
Figure 4. DISC1 knockdown promotes oligodendrocyte differentiation.
A–C, Cells transfected with control-siRNA, DISC1-siRNA-1 or DISC1-siRNA-2 were cultured for 24 (A)or 48 (B,C) hours in medium containing PDGF and mRNA levels of DISC1 (A), CNPase (B) and MBP (C) were quantified by qRT-PCR. Data are expressed as mean ± s.e.m. of at least three independent experiments. *p<0.05 vs. control-siRNA. D, Cells transfected with control-siRNA, DISC1-siRNA-1 or DISC1-siRNA-2 were lysed 72 hours after siRNA transfection and analyzed by western blotting. E, Quantitation of CNPase was performed by scanning densitometry. Data are expressed as mean ± s.e.m. of at least three independent experiments. *p<0.05 vs. control-siRNA. F, DISC1 knockdown mediated increase of CNPase mRNA was rescued by overexpression of DISC1. Cells were infected with GFP- or DISC1-GFP-Adv 24 hours after control- or DISC1-siRNA transfection. Forty-eight hours after the infection, mRNA level of CNPase was quantified by qRT-PCR. Data are expressed as mean ± s.e.m. of at least three independent experiments. *p<0.05 vs. control-siRNA and GFP-Adv. G–J Oligodendrocyte precursor cells were transfected with control-siRNA, DISC1-siRNA-1 or DISC1-siRNA-2 and cultured in medium containing PDGF for 72 hours then fixed for immunostaining. Cells were immunostained with anti-β-tubulin antibody (G, H) or anti-CNPase-antibody (I, J) and analyzed as described in figure legend 3. *p<0.05 vs. control-siRNA. Scale bars = 50 µm.
Overexpressed Truncated-DISC1 Promotes Oligodendrocyte Differentiation
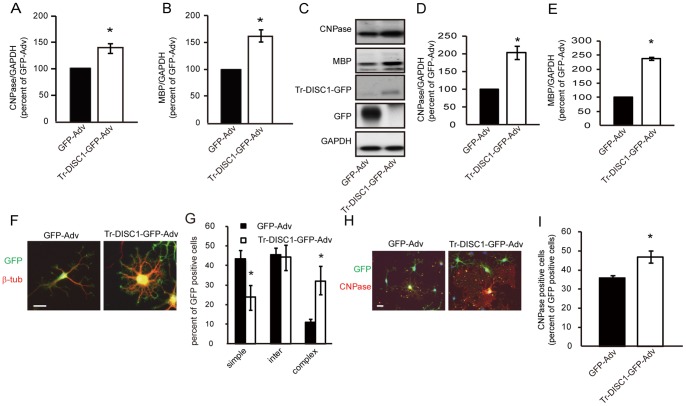
To further confirm the negative regulatory role of DISC1 in oligodendrocyte differentiation, we overexpressed truncated DISC1, which has been suggested to be generated when the DISC1 gene is disrupted by the balanced translocation of chromosome 1. Truncated DISC1 is predicted to function in a dominant negative fashion, possibly by competing with the full-length form for interacting proteins [12]. When oligodendrocyte precursor cells infected with a truncated DISC1 expressing adenovirus (trDISC1-GFP-Adv) for 12 hours, were deprived of PDGF, levels of CNPase and MBP, both mRNA and protein levels, were significantly increased compared with control (GFP-Adv) adenovirus infected cells (Fig. 5 A–E). Furthermore, immunostaining analysis with anti-CNPase antibody showed a higher proportion of CNPase positive cells in trDISC1-GFP-Adv, compared with control adenovirus, infected cells (GFP-Adv: 36.0±1.2%; tr-DISC1-GFP-Adv: 46.9±3.1%) (Fig. 5 H, I).
Figure 5. Overexpressed truncated DISC1 promotes oligodendrocyte differentiation.
A–E Effect of truncated DISC1 overexpression on CNPase and MBP expression. Oligodendrocyte precursor cells were infected with GFP-Adv or trDISC1-GFP-Adv for 12 hours and induced to differentiate by PDGF deprivation for 36 hours. mRNA levels of CNPase (A) and MBP (B) were quantified by qRT-PCR. Data are expressed as mean ± s.e.m. of at least three independent experiments. *p<0.05 vs. GFP-Adv. C, Cells infected with GFP-Adv or trDISC1-GFP-Adv for 12 hours were lysed 60 hours after PDGF deprivation and subjected to western blot analysis. D, E, Quantitation of relative band densities for CNPase (D) and MBP (E) was performed by scanning densitometry. Data are expressed as mean ± s.e.m. of at least three independent experiments. *p<0.05 vs. GFP-Adv. F–I Oligodendrocyte precursor cells were infected with GFP-Adv or trDISC1-GFP-Adv for 12 hours and induced to differentiate by depriving PDGF for 60 hours then fixed for immunostaining. Cells were immunostained for GFP and β-tubulin (F, G) or for GFP and CNPase (H, I) and analyzed as described in figure legend 3. *p<0.05 vs. GFP-Adv. Scale bar = 50 µm.
Immunostaining of trDISC1-GFP-Adv infected cells with anti-β-tubulin antibody showed that upon differentiation, there was a higher ratio of cells with a complex morphology compared with control adenovirus infected cells (GFP-Adv: 11.0±0.8%; trDISC1-GFP-Adv: 32.0±4.9%) (Fig. 5 F, G). Conversely, the ratio of cells displaying a simple morphology in trDISC1-GFP-Adv, compared to GFP-Adv, infected cultures was significantly lower (GFP-Adv: 43.4±2.1%; trDISC1-GFP-Adv: 23.9±4.2%). These results suggest that truncated DISC1 expression promotes differentiation of oligodendroglial lineage cells, and provides additional supporting evidence for negative regulation of oligodendrocyte differentiation by endogenous DISC1.
Involvement of Sox10 and/or Nkx2.2 in the Regulatory Pathway of Oligodendrocyte Differentiation by DISC1
Transcription factors involved in oligodendrocyte specification and differentiation include, but are not limited to, the basic helix-loop-helix (bHLH) family members Olig1 and Olig2, the inhibitor of DNA binding (Id) family of proteins Id2 and Id4, SRY box containing (Sox) family members Sox10, and the homeobox containing (Hox) transcription factor Nkx2.2 [47], [48].
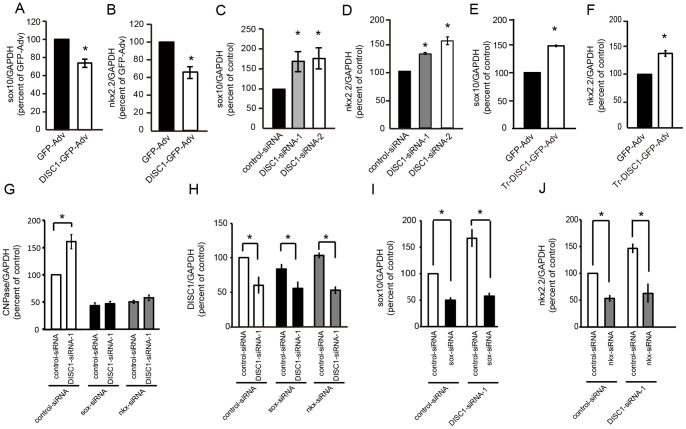
To examine the potential involvement of transcription factors in the regulatory pathway of oligodendroglial lineage cells by DISC1, we investigated the effects of manipulating DISC1 expression on the mRNA expression of transcription factors. Of the transcription factors examined (sox10, nkx2.2, mash1, olig1, olig2, Id2 and Id4), mRNA expression of Sox10 and Nkx2.2 were significantly decreased in DISC1 overexpressing cells (Fig. 6 A, B) while expression level of other transcription factors were not significantly changed compared with control cells (mash1; 104±0.3%, olig1; 107±0.2%, olig2; 99.5±1.6%, Id2; 111.8±6.0%; Id4; 112.0±8.2%). In contrast, knockdown of endogenous DISC1 resulted in enhanced expression of Sox10 and Nkx2.2 (Fig. 6 C, D). Furthermore, truncated form of DISC1 also increased Sox10 and Nkx2.2 expression (Fig. 6 E, F).
Figure 6. Involvement of Sox10 and/or Nkx2.2 in the regulatory pathway of oligodendrocyte differentiation by DISC1.
A, B, Expression of Sox10 and Nkx2.2 mRNA were decreased by DISC1 overexpression. Oligodendrocyte precursor cells were infected with GFP-Adv or DISC1-GFP-Adv for 12 hours and induced to differentiate by depriving PDGF for 36 hours. C, D, Expression of Sox10 and Nkx2.2 mRNA were increased by DISC1 knockdown. Oligodendrocyte precursor cells were transfected with control-siRNA, DISC1-siRNA-1 or DISC1-siRNA-2 and cultured in medium containing PDGF for 48 hours. E, F, Expression of Sox10 and Nkx2.2 mRNA were increased by truncated DISC1 overexpression. Oligodendrocyte precursor cells were infected with GFP-Adv or trDISC1-GFP-Adv for 12 hours and induced to differentiate by PDGF deprivation for 36 hours. G–J DISC1 knockdown mediated increase of CNPase mRNA was inhibited by a simultaneous knockdown of either Sox10 or Nkx2.2. Oligodendrocyte precursor cells were co-transfected with control-siRNA or DISC1-siRNA-1 and sox10-siRNA or nkx2.2-siRNA and cultured in medium containing PDGF for 24 (H) or 48 hours (G, I, J). mRNA quantification was performed 48 hours after adenovirus infection (A, B, E, F) or siRNA transfection (C, D, G, I, J) or 24 h hours after siRNA transfection (H) by qRT-PCR. Data are expressed as mean ± s.e.m. of at least three independent experiments. *p<0.05 vs. GFP-Adv (A, B), *p<0.05 vs. control-siRNA (C, D), *p<0.05 (E–H).
We examined CNPase expression levels in cells co-transfected with DISC1-siRNA-1 and siRNAs targeting either Sox10 or Nkx2.2 (sox-siRNA or nkx-siRNA) to elucidate if DISC1 is acting upstream or downstream of these transcription factors. When either Sox10 or Nkx2.2 was simultaneously knocked-down with DISC1-siRNA-1 treatment, promotion of oligodendrocyte differentiation by DISC1 knockdown was inhibited (Fig. 6 G). The effect of DISC1-siRNA-1 on its target mRNA was not altered when either sox-siRNA or nkx-siRNA was co-transfected (Fig. 6 H). Similarly, DISC1-siRNA-1 co-transfection did not alter the effect of sox-siRNA or nkx-siRNA (Fig. 6 I, J). Therefore, these results suggest DISC1 negatively regulates oligodendrocyte differentiation by acting upstream of Sox10 and/or Nkx2.2 to suppress their expression.
Discussion
The major findings of this study are as follows. First, we show that DISC1 is expressed in oligodendrocytes in the corpus callosum of postnatal mouse brain (Fig. 1). Second, DISC1 expression is decreased during in vitro differentiation of oligodendrocyte precursor cells to oligodendrocytes (Fig. 2 A). Third, DISC1 endogenously expressed in cells of an oligodendroglial lineage, negatively regulates differentiation of oligodendrocyte precursor cells to oligodendrocytes, as shown by promotion of oligodendrocyte differentiation by either DISC1 knockdown or overexpression of truncated DISC1 (Fig. 4, 5), while overexpression of full length DISC1 inhibits oligodendrocyte differentiation (Fig. 3). Finally, we have implicated Sox10 and/or Nkx2.2 in the DISC1 regulatory pathway of oligodendrocyte differentiation, with knockdown of endogenous DISC1 increasing, and DISC1 overexpression decreasing, expression of Sox10 and Nkx2.2. Additionally, promotion of oligodendrocyte differentiation by DISC1 knockdown was prevented by simultaneous knockdown of either Sox10 or Nkx2.2 (Fig. 6).
DISC1 Expression in Oligodendrocytes
Our in situ hybridization analysis shows that DISC1 mRNA is expressed not only in oligodendrocytes of mouse brain but also in primary cultured rat oligodendrocytes and oligodendrocyte precursor cells (Fig. 1, 2). These results are consistent with previous report by Seshadri et al. showing colocalization of DISC1 with the oligodendrocyte marker in human brain and primary cultured rat cortical oligodendrocytes [37]. Furthermore, DISC1 expression in oligodendrocytes in the corpus callosum was higher in developing stage than in adulthood, similar to the developmental expression pattern of DISC1 in neurons [20], [46]. These results suggest that DISC1 has a developmental role in oligodendrocyte lineage cells as well as in neurons. Overexpressed DISC1 localized preferentially in cell bodies and processes of oligodendrocyte precursor cells and oligodendrocytes in vitro, with marginal nuclear localization (Fig. 2 B). The subcellular localization of DISC1 in oligodendrocyte precursor cells and oligodendrocytes is similar to that in neurons, suggesting the possibility that DISC1 has common functional roles between neurons and oligodendrocyte lineage cells.
DISC1 Function in Oligodendrocytes
DISC1 has been shown to play an important role in immature neurons, regulating their differentiation, migration and proliferation [12], [17], [49]. Thus our findings, namely, decrease of DISC1 expression during the course of oligodendrocyte differentiation (Fig. 2 A), and higher DISC1 expression in oligodendrocytes in the mouse corpus callosum at P14 (Fig. 1 C), suggest DISC1 may also have a developmental role in immature oligodendroglial lineage cells as well. Supporting evidence is discussed below.
Overexpressed DISC1 disrupts not only induction of CNPase and MBP expression, but also transformation of oligodendrocytes to a complex morphology (Fig. 3), indicating a negative regulatory role of DISC1 in differentiation of oligodendroglial lineage cells in vitro. Conversely, both expression of CNPase and the number of matured oligodendrocytes, were increased when endogenously expressed DISC1 was knocked-down by siRNA, even if the cells were maintained in medium containing PDGF (Fig. 4). Although MBP mRNA levels were increased by DISC1 knockdown, the result did not reach statistical significance. This is likely due to both the transient nature of DISC1 knockdown by siRNA, compared to the more stable adenovirus overexpression system, and also that MBP expression increases at a later stage of differentiation than CNPase [50]. More robust increases of MBP expression may be observed at later time-points, with stronger and more continuous inhibition of DISC1. Truncated DISC1 is predicted to function in a dominant negative fashion, potentially by competing with full length DISC1 for interacting proteins. Thus promotion of oligodendrocyte differentiation by truncated DISC1 overexpression suggests a negative regulatory role for DISC1 in oligodendrocyte differentiation (Fig. 5). Further studies are needed to determine if DISC1 interacts with other proteins in oligodendrocyte lineage cells, as in neurons [51], [52]. Nevertheless, our results do indicate that decreased level of endogenous DISC1 promotes differentiation of oligodendrocyte precursor cells to oligodendrocytes.
To date, a functional role for endogenous DISC1 expressed in mammalian oligodendrocyte lineage cells has not been reported. A critical requirement for DISC1 in oligodendroglial development, by promoting specification of olig2-positive cells in the hindbrain and other brain regions of zebrafish, was reported by Wood et al [36]. Although this report also shows regulation of oligodendroglial development by DISC1, the low homology between zebrafish DISC1 and mammalian DISC1 (homologies between zebrafish and rat, mouse or human are 31, 32, 36% respectively), highlights the necessity of our study. Furthermore, it is not clear if neuronal or glial expressed DISC1 is responsible for oligodendroglial development. Katsel et al., showed that oligodendrocyte-associated gene/protein expression was changed in the forebrain of transgenic mice with forebrain restricted expression of mutant human DISC1 (ΔhDISC1) at embryonic, neonatal and adulthood stages [38]. The transgenic mice show neuron-specific overexpression of ΔhDISC1, therefore the observed alterations in oligodendrocyte-associated gene/protein expression are caused by mutant DISC1 expressed in neurons. However, the results of our study show that glial expressed DISC1 regulates oligodendrocyte differentiation. To further determine the role of glial expressed DISC1 on oligodendroglial development in vivo, studies using transgenic mice with glia-specific expression of mutant DISC1 are needed.
Sox10, Nkx2.2 Regulation by DISC1
Each step of oligodendrocyte differentiation is under tight transcriptional control. Among many transcription factors acting at different stages, Sox10 and Nkx2.2 play a major role in the transition from oligodendrocyte precursor cells to pre-myelinating oligodendrocytes [48], [53]. Sox10 is a high-mobility group transcriptional regulator restricted in the central nervous system to myelin-forming oligodendrocytes. In Sox10-deficient mice, oligodendrocyte precursor cells develop but terminal differentiation is disrupted [54]. A critical role for the homeodomain-containing protein Nkx2.2 in the development of oligodendroglial lineage cells has also been reported. The number of MBP-positive and proteolipid protein (PLPDM-20)-positive oligodendrocytes are dramatically reduced in the brain of Nkx2.2 null mice [55]. Positive regulatory roles for Sox10 and Nkx2.2 in oligodendrocyte differentiation are consistent with our result that DISC1 regulates oligodendrocyte differentiation via upstream regulation of Sox10 and/or Nkx2.2 (Fig. 6). Negative regulation of Sox10 expression by DISC1 has previously been reported by Drerup et al., although they examined cranio-neural crest cells, which become glial precursors at later stages of development [56]. How DISC1 regulates these transcription factors is not yet known, but intracellular signaling pathways involving molecules such as Akt, cAMP, CREB and MAPK are likely candidates, as neuronal DISC1 regulates these pathways, and moreover, these signaling pathways have functional roles in differentiation of oligodendroglial lineage cells [57]–[62].
The pathophysiological role of Sox10 in SZ has been suggested by a report showing a correlative relationship between the DNA methylation status of the Sox10 gene and oligodendrocyte dysfunction in SZ [63]. In addition, a significant association in the genotype and allelic frequency of a single-nucleotide polymorphism of the Sox10 gene, between schizophrenic patients and controls has been reported [64]. Nkx2.2 is known to form a transcriptional network with Pet1, a molecule involved in differentiation of serotonergic neurons [65]. It is well known that serotonergic neurons are both a relevant pathophysiological factor and therapeutic target in several psychiatric diseases, including SZ, bipolar disorder, major depression and autism. Therefore our finding that DISC1, a key psychiatric disease susceptibility gene, controls Sox10 and/or Nkx2.2 expression is intriguing.
Pathophysiological Role of DISC1 Regulated Oligodendrocyte Differentiation
Overall, our findings suggest that DISC1 dysfunction may cause impaired differentiation of oligodendrocytes by affecting Sox10 and/or Nkx2.2 expression, and consequently contribute to the pathophysiology of psychiatric disorders. Improper myelination of neuronal axons, resulting from impaired oligodendrocyte differentiation, may lead to defective neuronal communication, a likely component in the mechanistic background of “structural disconnectivity”, suggested in the pathophysiology of psychiatric disorders [24], [66]. Therefore, it would be of interest to investigate if WM abnormalities are a feature of the Scottish DISC1 pedigree that harbors the disrupted DISC1 gene. Furthermore, given that SZ-like psychosis co-occurs frequently in demyelinating diseases [67], it is also warranted to investigate the role of DISC1 in pathophysiology of demyelinating diseases such as MS, leukodystrophies and velocardiofacial syndrome.
Acknowledgments
We would like to thank Ms. Moriya and Ms. Ohashi for preparing our experiments and Ms. Nakajima and Ms. Ikuma for excellent technical assistance.
Funding Statement
This work was partially supported by Grant-in-Aid for Scientific Research on Innovative Areas “Neural Diversity and Neocortical Organization”, from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan (No. 23123512 and No. 23700415), and by a grant from Dainippon Sumitomo Pharma Co., Ltd. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Millar JK, Christie S, Anderson S, Lawson D, Hsiao-Wei Loh D, et al. (2001) Genomic structure and localisation within a linkage hotspot of Disrupted In Schizophrenia 1, a gene disrupted by a translocation segregating with schizophrenia. Mol Psychiatry 6: 173–178. [DOI] [PubMed] [Google Scholar]
- 2. Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, et al. (2000) Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet 9: 1415–1423. [DOI] [PubMed] [Google Scholar]
- 3. St Clair D, Blackwood D, Muir W, Carothers A, Walker M, et al. (1990) Association within a family of a balanced autosomal translocation with major mental illness. Lancet 336: 13–16. [DOI] [PubMed] [Google Scholar]
- 4. Callicott JH, Straub RE, Pezawas L, Egan MF, Mattay VS, et al. (2005) Variation in DISC1 affects hippocampal structure and function and increases risk for schizophrenia. Proc Natl Acad Sci U S A 102: 8627–8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kilpinen H, Ylisaukko-Oja T, Hennah W, Palo OM, Varilo T, et al. (2008) Association of DISC1 with autism and Asperger syndrome. Mol Psychiatry 13: 187–196. [DOI] [PubMed] [Google Scholar]
- 6. Hennah W, Varilo T, Kestila M, Paunio T, Arajarvi R, et al. (2003) Haplotype transmission analysis provides evidence of association for DISC1 to schizophrenia and suggests sex-dependent effects. Hum Mol Genet 12: 3151–3159. [DOI] [PubMed] [Google Scholar]
- 7. Hodgkinson CA, Goldman D, Jaeger J, Persaud S, Kane JM, et al. (2004) Disrupted in schizophrenia 1 (DISC1): association with schizophrenia, schizoaffective disorder, and bipolar disorder. Am J Hum Genet 75: 862–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Song W, Li W, Feng J, Heston LL, Scaringe WA, et al. (2008) Identification of high risk DISC1 structural variants with a 2% attributable risk for schizophrenia. Biochem Biophys Res Commun 367: 700–706. [DOI] [PubMed] [Google Scholar]
- 9. Osbun N, Li J, O’Driscoll MC, Strominger Z, Wakahiro M, et al. (2011) Genetic and functional analyses identify DISC1 as a novel callosal agenesis candidate gene. Am J Med Genet A 155A: 1865–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang Q, Charych EI, Pulito VL, Lee JB, Graziane NM, et al. (2011) The psychiatric disease risk factors DISC1 and TNIK interact to regulate synapse composition and function. Mol Psychiatry 16: 1006–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hayashi-Takagi A, Takaki M, Graziane N, Seshadri S, Murdoch H, et al. (2010) Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nat Neurosci 13: 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kamiya A, Kubo K, Tomoda T, Takaki M, Youn R, et al. (2005) A schizophrenia-associated mutation of DISC1 perturbs cerebral cortex development. Nat Cell Biol 7: 1167–1178. [DOI] [PubMed] [Google Scholar]
- 13. Roberts RC (2007) Schizophrenia in translation: disrupted in schizophrenia (DISC1): integrating clinical and basic findings. Schizophr Bull 33: 11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mao Y, Ge X, Frank CL, Madison JM, Koehler AN, et al. (2009) Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK3beta/beta-catenin signaling. Cell 136: 1017–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Duan X, Chang JH, Ge S, Faulkner RL, Kim JY, et al. (2007) Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell 130: 1146–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Porteous DJ, Millar JK, Brandon NJ, Sawa A (2011) DISC1 at 10: connecting psychiatric genetics and neuroscience. Trends Mol Med 17: 699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ishizuka K, Kamiya A, Oh EC, Kanki H, Seshadri S, et al. (2011) DISC1-dependent switch from progenitor proliferation to migration in the developing cortex. Nature 473: 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Niwa M, Jaaro-Peled H, Tankou S, Seshadri S, Hikida T, et al. (2013) Adolescent stress-induced epigenetic control of dopaminergic neurons via glucocorticoids. Science 339: 335–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaaro-Peled H, Niwa M, Foss CA, Murai R, de Los Reyes S, et al. (2013) Subcortical dopaminergic deficits in a DISC1 mutant model: a study in direct reference to human molecular brain imaging. Hum Mol Genet. [DOI] [PMC free article] [PubMed]
- 20. Kuroda K, Yamada S, Tanaka M, Iizuka M, Yano H, et al. (2011) Behavioral alterations associated with targeted disruption of exons 2 and 3 of the Disc1 gene in the mouse. Hum Mol Genet 20: 4666–4683. [DOI] [PubMed] [Google Scholar]
- 21. Hikida T, Jaaro-Peled H, Seshadri S, Oishi K, Hookway C, et al. (2007) Dominant-negative DISC1 transgenic mice display schizophrenia-associated phenotypes detected by measures translatable to humans. Proc Natl Acad Sci U S A 104: 14501–14506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Takahashi N, Sakurai T, Davis KL, Buxbaum JD (2011) Linking oligodendrocyte and myelin dysfunction to neurocircuitry abnormalities in schizophrenia. Prog Neurobiol 93: 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Edgar N, Sibille E (2012) A putative functional role for oligodendrocytes in mood regulation. Transl Psychiatry 2: e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fields RD (2008) White matter in learning, cognition and psychiatric disorders. Trends Neurosci 31: 361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Uranova NA, Vostrikov VM, Orlovskaya DD, Rachmanova VI (2004) Oligodendroglial density in the prefrontal cortex in schizophrenia and mood disorders: a study from the Stanley Neuropathology Consortium. Schizophr Res 67: 269–275. [DOI] [PubMed] [Google Scholar]
- 26. Bernstein HG, Steiner J, Bogerts B (2009) Glial cells in schizophrenia: pathophysiological significance and possible consequences for therapy. Expert Rev Neurother 9: 1059–1071. [DOI] [PubMed] [Google Scholar]
- 27. Feng Y (2008) Convergence and divergence in the etiology of myelin impairment in psychiatric disorders and drug addiction. Neurochem Res 33: 1940–1949. [DOI] [PubMed] [Google Scholar]
- 28. Sokolov BP (2007) Oligodendroglial abnormalities in schizophrenia, mood disorders and substance abuse. Comorbidity, shared traits, or molecular phenocopies? Int J Neuropsychopharmacol 10: 547–555. [DOI] [PubMed] [Google Scholar]
- 29. Tkachev D, Mimmack ML, Ryan MM, Wayland M, Freeman T, et al. (2003) Oligodendrocyte dysfunction in schizophrenia and bipolar disorder. Lancet 362: 798–805. [DOI] [PubMed] [Google Scholar]
- 30. Aston C, Jiang L, Sokolov BP (2004) Microarray analysis of postmortem temporal cortex from patients with schizophrenia. J Neurosci Res 77: 858–866. [DOI] [PubMed] [Google Scholar]
- 31. Aston C, Jiang L, Sokolov BP (2005) Transcriptional profiling reveals evidence for signaling and oligodendroglial abnormalities in the temporal cortex from patients with major depressive disorder. Mol Psychiatry 10: 309–322. [DOI] [PubMed] [Google Scholar]
- 32. Hof PR, Haroutunian V, Friedrich VL Jr, Byne W, Buitron C, et al. (2003) Loss and altered spatial distribution of oligodendrocytes in the superior frontal gyrus in schizophrenia. Biol Psychiatry 53: 1075–1085. [DOI] [PubMed] [Google Scholar]
- 33. Walterfang M, Wood SJ, Velakoulis D, Copolov D, Pantelis C (2005) Diseases of white matter and schizophrenia-like psychosis. Aust N Z J Psychiatry 39: 746–756. [DOI] [PubMed] [Google Scholar]
- 34. Baumann N, Turpin JC, Lefevre M, Colsch B (2002) Motor and psycho-cognitive clinical types in adult metachromatic leukodystrophy: genotype/phenotype relationships? J Physiol Paris 96: 301–306. [DOI] [PubMed] [Google Scholar]
- 35. Kosmidis MH, Giannakou M, Messinis L, Papathanasopoulos P (2010) Psychotic features associated with multiple sclerosis. Int Rev Psychiatry 22: 55–66. [DOI] [PubMed] [Google Scholar]
- 36. Wood JD, Bonath F, Kumar S, Ross CA, Cunliffe VT (2009) Disrupted-in-schizophrenia 1 and neuregulin 1 are required for the specification of oligodendrocytes and neurones in the zebrafish brain. Hum Mol Genet 18: 391–404. [DOI] [PubMed] [Google Scholar]
- 37. Seshadri S, Kamiya A, Yokota Y, Prikulis I, Kano S, et al. (2010) Disrupted-in-Schizophrenia-1 expression is regulated by beta-site amyloid precursor protein cleaving enzyme-1-neuregulin cascade. Proc Natl Acad Sci U S A 107: 5622–5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Katsel P, Tan W, Abazyan B, Davis KL, Ross C, et al. (2011) Expression of mutant human DISC1 in mice supports abnormalities in differentiation of oligodendrocytes. Schizophr Res 130: 238–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Koyama Y, Fujiwara T, Kubo T, Tomita K, Yano K, et al. (2008) Reduction of oligodendrocyte myelin glycoprotein expression following facial nerve transection. J Chem Neuroanat 36: 209–215. [DOI] [PubMed] [Google Scholar]
- 40. Maher BJ, LoTurco JJ (2012) Disrupted-in-schizophrenia (DISC1) functions presynaptically at glutamatergic synapses. PLoS One 7: e34053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Faulkner RL, Jang MH, Liu XB, Duan X, Sailor KA, et al. (2008) Development of hippocampal mossy fiber synaptic outputs by new neurons in the adult brain. Proc Natl Acad Sci U S A 105: 14157–14162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen Y, Balasubramaniyan V, Peng J, Hurlock EC, Tallquist M, et al. (2007) Isolation and culture of rat and mouse oligodendrocyte precursor cells. Nat Protoc 2: 1044–1051. [DOI] [PubMed] [Google Scholar]
- 43. Hattori T, Baba K, Matsuzaki S, Honda A, Miyoshi K, et al. (2007) A novel DISC1-interacting partner DISC1-Binding Zinc-finger protein: implication in the modulation of DISC1-dependent neurite outgrowth. Mol Psychiatry 12: 398–407. [DOI] [PubMed] [Google Scholar]
- 44. Miyoshi K, Honda A, Baba K, Taniguchi M, Oono K, et al. (2003) Disrupted-In-Schizophrenia 1, a candidate gene for schizophrenia, participates in neurite outgrowth. Mol Psychiatry 8: 685–694. [DOI] [PubMed] [Google Scholar]
- 45. Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P (2002) Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci 22: 10333–10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Honda A, Miyoshi K, Baba K, Taniguchi M, Koyama Y, et al. (2004) Expression of fasciculation and elongation protein zeta-1 (FEZ1) in the developing rat brain. Brain Res Mol Brain Res 122: 89–92. [DOI] [PubMed] [Google Scholar]
- 47. Emery B (2010) Regulation of oligodendrocyte differentiation and myelination. Science 330: 779–782. [DOI] [PubMed] [Google Scholar]
- 48. Nicolay DJ, Doucette JR, Nazarali AJ (2007) Transcriptional control of oligodendrogenesis. Glia 55: 1287–1299. [DOI] [PubMed] [Google Scholar]
- 49. Ozeki Y, Tomoda T, Kleiderlein J, Kamiya A, Bord L, et al. (2003) Disrupted-in-Schizophrenia-1 (DISC-1): mutant truncation prevents binding to NudE-like (NUDEL) and inhibits neurite outgrowth. Proc Natl Acad Sci U S A 100: 289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Baumann N, Pham-Dinh D (2001) Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol Rev 81: 871–927. [DOI] [PubMed] [Google Scholar]
- 51. Brandon NJ (2007) Dissecting DISC1 function through protein-protein interactions. Biochem Soc Trans 35: 1283–1286. [DOI] [PubMed] [Google Scholar]
- 52.Camargo LM, Wang Q, Brandon NJ (2008) What can we learn from the disrupted in schizophrenia 1 interactome: lessons for target identification and disease biology? Novartis Found Symp 289: 208–216; discussion 216–221, 238–240. [DOI] [PubMed]
- 53. Emery B (2010) Transcriptional and post-transcriptional control of CNS myelination. Curr Opin Neurobiol 20: 601–607. [DOI] [PubMed] [Google Scholar]
- 54. Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, et al. (2002) Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev 16: 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Qi Y, Cai J, Wu Y, Wu R, Lee J, et al. (2001) Control of oligodendrocyte differentiation by the Nkx2.2 homeodomain transcription factor. Development 128: 2723–2733. [DOI] [PubMed] [Google Scholar]
- 56. Drerup CM, Wiora HM, Topczewski J, Morris JA (2009) Disc1 regulates foxd3 and sox10 expression, affecting neural crest migration and differentiation. Development 136: 2623–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kim JY, Duan X, Liu CY, Jang MH, Guo JU, et al. (2009) DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron 63: 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Millar JK, Pickard BS, Mackie S, James R, Christie S, et al. (2005) DISC1 and PDE4B are interacting genetic factors in schizophrenia that regulate cAMP signaling. Science 310: 1187–1191. [DOI] [PubMed] [Google Scholar]
- 59.Sawamura N, Ando T, Maruyama Y, Fujimuro M, Mochizuki H, et al. (2008) Nuclear DISC1 regulates CRE-mediated gene transcription and sleep homeostasis in the fruit fly. Mol Psychiatry 13: 1138–1148, 1069. [DOI] [PMC free article] [PubMed]
- 60. Johnson MA, Ables JL, Eisch AJ (2009) Cell-intrinsic signals that regulate adult neurogenesis in vivo: insights from inducible approaches. BMB Rep 42: 245–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hashimoto R, Numakawa T, Ohnishi T, Kumamaru E, Yagasaki Y, et al. (2006) Impact of the DISC1 Ser704Cys polymorphism on risk for major depression, brain morphology and ERK signaling. Hum Mol Genet 15: 3024–3033. [DOI] [PubMed] [Google Scholar]
- 62. Taveggia C, Feltri ML, Wrabetz L (2010) Signals to promote myelin formation and repair. Nat Rev Neurol 6: 276–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Iwamoto K, Bundo M, Yamada K, Takao H, Iwayama-Shigeno Y, et al. (2005) DNA methylation status of SOX10 correlates with its downregulation and oligodendrocyte dysfunction in schizophrenia. J Neurosci 25: 5376–5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Maeno N, Takahashi N, Saito S, Ji X, Ishihara R, et al. (2007) Association of SOX10 with schizophrenia in the Japanese population. Psychiatr Genet 17: 227–231. [DOI] [PubMed] [Google Scholar]
- 65. Alenina N, Bashammakh S, Bader M (2006) Specification and differentiation of serotonergic neurons. Stem Cell Rev 2: 5–10. [DOI] [PubMed] [Google Scholar]
- 66. Spencer KM, Nestor PG, Niznikiewicz MA, Salisbury DF, Shenton ME, et al. (2003) Abnormal neural synchrony in schizophrenia. J Neurosci 23: 7407–7411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Walterfang M, Velakoulis D, Whitford TJ, Pantelis C (2011) Understanding aberrant white matter development in schizophrenia: an avenue for therapy? Expert Rev Neurother 11: 971–987. [DOI] [PubMed] [Google Scholar]