Abstract
Upon transformation into Escherichia coli or Cos-1 monkey cells, heteroduplex DNA made of two sequences containing many nucleotide mismatches yields a wide array of different molecules, some with a patchwork structure. Thus, complex heteroduplexes can be processed to generate many genetic variants.
Full text
PDF
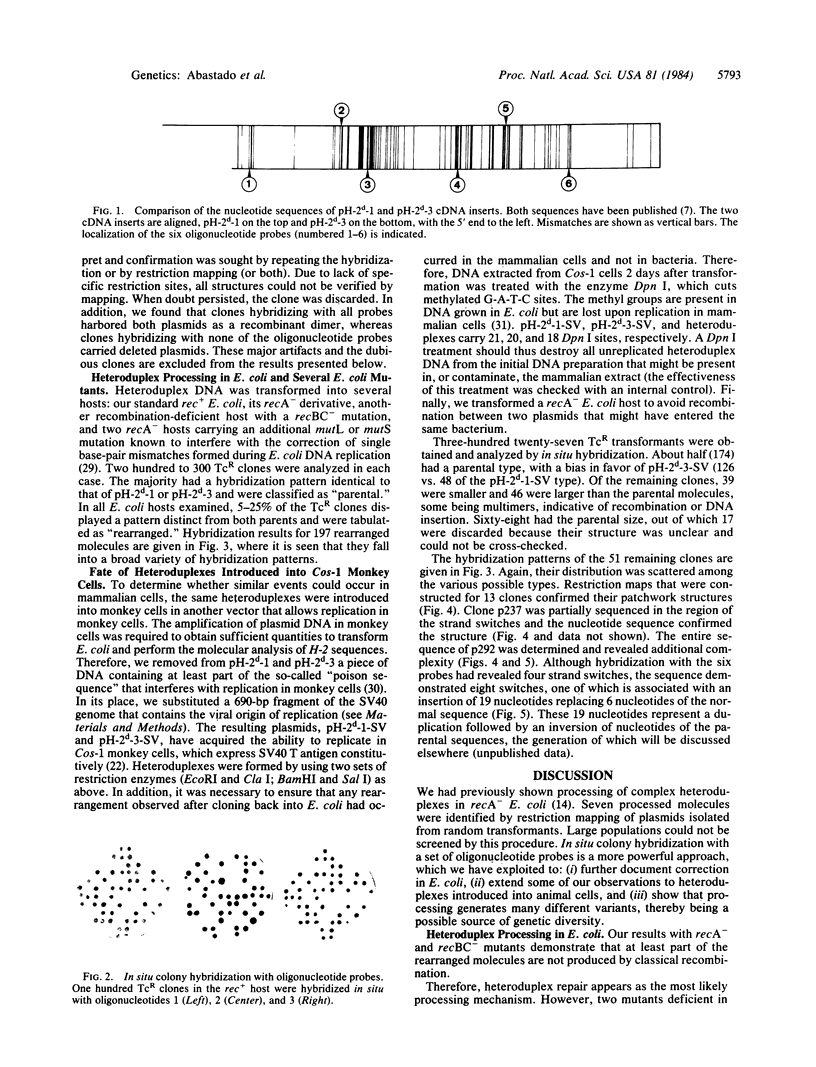

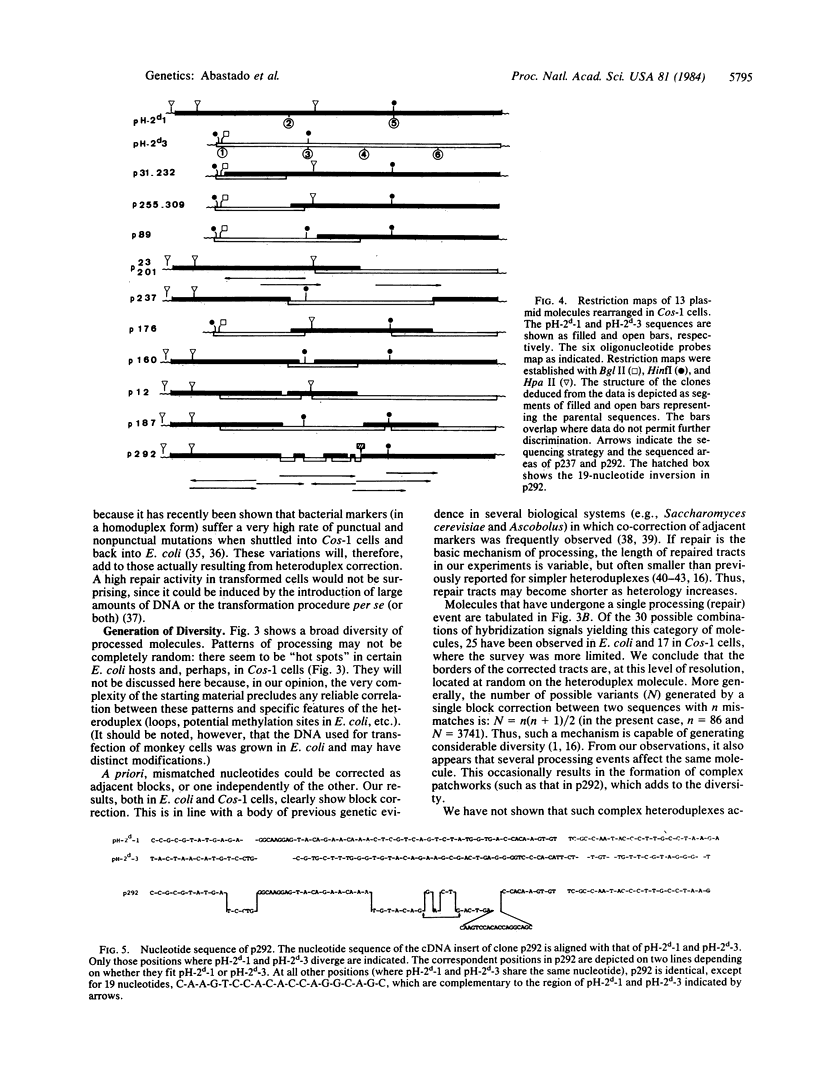

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baltimore D. Gene conversion: some implications for immunoglobulin genes. Cell. 1981 Jun;24(3):592–594. doi: 10.1016/0092-8674(81)90082-9. [DOI] [PubMed] [Google Scholar]
- Bourguignon-Van Horen F., Brotcorn A., Caillet-Fauquet P., Diver W. P., Dohet C., Doubleday O. P., Lecomte P., Maenhaut-Michel G., Radman M. Conservation and diversification of genes by mismatch correction and SOS induction. Biochimie. 1982 Aug-Sep;64(8-9):559–564. doi: 10.1016/s0300-9084(82)80087-4. [DOI] [PubMed] [Google Scholar]
- Calos M. P., Lebkowski J. S., Botchan M. R. High mutation frequency in DNA transfected into mammalian cells. Proc Natl Acad Sci U S A. 1983 May;80(10):3015–3019. doi: 10.1073/pnas.80.10.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cami B., Chambon P., Kourilsky P. Correction of complex heteroduplexes made of mouse H-2 gene sequences in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1984 Jan;81(2):503–507. doi: 10.1073/pnas.81.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman B. W., Radman M. Escherichia coli mutator mutants deficient in methylation-instructed DNA mismatch correction. Proc Natl Acad Sci U S A. 1980 Feb;77(2):1063–1067. doi: 10.1073/pnas.77.2.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981 Jan;23(1):175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967 Jun 14;26(2):365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- Hood L., Steinmetz M., Malissen B. Genes of the major histocompatibility complex of the mouse. Annu Rev Immunol. 1983;1:529–568. doi: 10.1146/annurev.iy.01.040183.002525. [DOI] [PubMed] [Google Scholar]
- Kourilsky P. Genetic exchanges between partially homologous nucleotide sequences: possible implications for multigene families. Biochimie. 1983 Feb;65(2):85–93. doi: 10.1016/s0300-9084(83)80178-3. [DOI] [PubMed] [Google Scholar]
- Lai C. J., Nathans D. A map of temperature-sensitive mutants of simian virus 40. Virology. 1975 Jul;66(1):70–81. doi: 10.1016/0042-6822(75)90179-8. [DOI] [PubMed] [Google Scholar]
- Lalanne J. L., Bregegere F., Delarbre C., Abastado J. P., Gachelin G., Kourilsky P. Comparison of nucleotide sequences of mRNAs belonging to the mouse H-2 multigene family. Nucleic Acids Res. 1982 Feb 11;10(3):1039–1049. doi: 10.1093/nar/10.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichten M., Fox M. S. Effects of nonhomology on bacteriophage lambda recombination. Genetics. 1983 Jan;103(1):5–22. doi: 10.1093/genetics/103.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusky M., Botchan M. Inhibition of SV40 replication in simian cells by specific pBR322 DNA sequences. Nature. 1981 Sep 3;293(5827):79–81. doi: 10.1038/293079a0. [DOI] [PubMed] [Google Scholar]
- Meselson M. S., Radding C. M. A general model for genetic recombination. Proc Natl Acad Sci U S A. 1975 Jan;72(1):358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L. K., Cooke B. E., Fried M. Fate of mismatched base-pair regions in polyoma heteroduplex DNA during infection of mouse cells. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3073–3077. doi: 10.1073/pnas.73.9.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray N. E., Brammar W. J., Murray K. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol Gen Genet. 1977 Jan 7;150(1):53–61. doi: 10.1007/BF02425325. [DOI] [PubMed] [Google Scholar]
- Nicolas A., Rossignol J. L. Gene conversion: point-mutation heterozygosities lower heteroduplex formation. EMBO J. 1983;2(12):2265–2270. doi: 10.1002/j.1460-2075.1983.tb01733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollo R., Rougeon F. Gene conversion and polymorphism: generation of mouse immunoglobulin gamma 2a chain alleles by differential gene conversion by gamma 2b chain gene. Cell. 1983 Feb;32(2):515–523. doi: 10.1016/0092-8674(83)90471-3. [DOI] [PubMed] [Google Scholar]
- Pays E., Van Assel S., Laurent M., Darville M., Vervoort T., Van Meirvenne N., Steinert M. Gene conversion as a mechanism for antigenic variation in trypanosomes. Cell. 1983 Sep;34(2):371–381. doi: 10.1016/0092-8674(83)90371-9. [DOI] [PubMed] [Google Scholar]
- Pease L. R., Schulze D. H., Pfaffenbach G. M., Nathenson S. G. Spontaneous H-2 mutants provide evidence that a copy mechanism analogous to gene conversion generates polymorphism in the major histocompatibility complex. Proc Natl Acad Sci U S A. 1983 Jan;80(1):242–246. doi: 10.1073/pnas.80.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden K. W., Pipas J. M., Pearson-White S., Nathans D. Isolation of mutants of an animal virus in bacteria. Science. 1980 Sep 19;209(4463):1392–1396. doi: 10.1126/science.6251547. [DOI] [PubMed] [Google Scholar]
- Petes T., Fink G. R. Gene conversion between repeated genes. Nature. 1982 Nov 18;300(5889):216–217. doi: 10.1038/300216a0. [DOI] [PubMed] [Google Scholar]
- Pukkila P. J., Peterson J., Herman G., Modrich P., Meselson M. Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli. Genetics. 1983 Aug;104(4):571–582. doi: 10.1093/genetics/104.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radding C. M. Genetic recombination: strand transfer and mismatch repair. Annu Rev Biochem. 1978;47:847–880. doi: 10.1146/annurev.bi.47.070178.004215. [DOI] [PubMed] [Google Scholar]
- Radding C. M. Homologous pairing and strand exchange in genetic recombination. Annu Rev Genet. 1982;16:405–437. doi: 10.1146/annurev.ge.16.120182.002201. [DOI] [PubMed] [Google Scholar]
- Razzaque A., Mizusawa H., Seidman M. M. Rearrangement and mutagenesis of a shuttle vector plasmid after passage in mammalian cells. Proc Natl Acad Sci U S A. 1983 May;80(10):3010–3014. doi: 10.1073/pnas.80.10.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A. A., Johnson M. J., Schöld M., Ito H., Ike Y., Morin C., Itakura K., Wallace R. B. Identification of an H-2Kb-related molecule by molecular cloning. Immunogenetics. 1981;14(5):383–392. doi: 10.1007/BF00373318. [DOI] [PubMed] [Google Scholar]
- Roman H., Fabre F. Gene conversion and associated reciprocal recombination are separable events in vegetative cells of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6912–6916. doi: 10.1073/pnas.80.22.6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougeon F., Kourilsky P., Mach B. Insertion of a rabbit beta-globin gene sequence into an E. coli plasmid. Nucleic Acids Res. 1975 Dec;2(12):2365–2378. doi: 10.1093/nar/2.12.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman J. G., Leder A., Nau M., Norman B., Leder P. Antibody diversity. Science. 1978 Oct 6;202(4363):11–17. doi: 10.1126/science.99815. [DOI] [PubMed] [Google Scholar]
- Slightom J. L., Blechl A. E., Smithies O. Human fetal G gamma- and A gamma-globin genes: complete nucleotide sequences suggest that DNA can be exchanged between these duplicated genes. Cell. 1980 Oct;21(3):627–638. doi: 10.1016/0092-8674(80)90426-2. [DOI] [PubMed] [Google Scholar]
- Stoeckert C. J., Jr, Collins F. S., Weissman S. M. Human fetal globin DNA sequences suggest novel conversion event. Nucleic Acids Res. 1984 Jun 11;12(11):4469–4479. doi: 10.1093/nar/12.11.4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suggs S. V., Wallace R. B., Hirose T., Kawashima E. H., Itakura K. Use of synthetic oligonucleotides as hybridization probes: isolation of cloned cDNA sequences for human beta 2-microglobulin. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6613–6617. doi: 10.1073/pnas.78.11.6613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccaro K. K., Siegel E. C. The frameshift mutability of polA1 and recA1 derivatives of mutator strains of Escherichia coli. Mutat Res. 1977 Mar;42(3):443–446. doi: 10.1016/s0027-5107(77)80048-1. [DOI] [PubMed] [Google Scholar]
- Wagner R., Jr, Meselson M. Repair tracts in mismatched DNA heteroduplexes. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4135–4139. doi: 10.1073/pnas.73.11.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss E. H., Mellor A., Golden L., Fahrner K., Simpson E., Hurst J., Flavell R. A. The structure of a mutant H-2 gene suggests that the generation of polymorphism in H-2 genes may occur by gene conversion-like events. Nature. 1983 Feb 24;301(5902):671–674. doi: 10.1038/301671a0. [DOI] [PubMed] [Google Scholar]
- Wigler M., Pellicer A., Silverstein S., Axel R., Urlaub G., Chasin L. DNA-mediated transfer of the adenine phosphoribosyltransferase locus into mammalian cells. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1373–1376. doi: 10.1073/pnas.76.3.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildenberg J., Meselson M. Mismatch repair in heteroduplex DNA. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2202–2206. doi: 10.1073/pnas.72.6.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]




