Abstract
The term riboswitch usually refers to small molecule sensing regulatory modules in the 5′ untranslated regions of a mRNA. They are typically comprised of separate ligand binding and regulatory domains. The T box riboswitch is unique from other identified riboswitches because its effector is an essential macromolecule, tRNA. It senses the aminoacylation state of tRNA to regulate genes involved in a variety of functions relating to amino acid metabolism and tRNA aminoacylation. T box riboswitches performs an intuitively simple process using a complex structured RNA element and, until recently, the underlying mechanisms were poorly understood. Only two sequence-specific contacts had been previously identified: (1) between the specifier sequence (codon) and the tRNA anticodon and (2) between an anti-terminator stem loop and the tRNA acceptor arm CCA tail. tRNA aminoacylation blocks the latter interaction and therefore serves as the switch between termination and anti-termination. Outside of these two contacts, the structure and functions of T box riboswitches have come to light in some recent studies. We recently described the X-ray crystal structure of the highly conserved T box riboswitch distal Stem I region and demonstrated that this region interacts with the tRNA elbow to anchor it to the riboswitch. Independently, Lehmann et al. used sequence homology search to arrive at a similar model for Stem I-tRNA interactions. The model was further supported by two recent structures of the Stem I-tRNA complex, determined independently by our group and by Zhang and Ferré-D’Amaré. This article highlights some of these contributions to synthesize an updated model for tRNA recognition by the T box riboswitch.
Keywords: RNA, structure, T-loop, interlocking T-loop, RNA-RNA complex
RNA molecules perform diverse functions in cells, from coding to catalysis. Riboswitches are widespread structured RNAs located in the 5′ untranslated region of some mRNAs where they bind their target metabolites to affect expression.1 Riboswitches are typically modular with a ligand-binding (or aptamer) domain and an effector domain that alters either transcription elongation or translation initiation. The T box mRNA leader was discovered approximately 20 y ago in pioneering work by Tina Henkin’s laboratory.2,3 These studies provided some of the first conclusive demonstrations that structured RNA played crucial gene regulatory roles in biology. Since then, significant strides have been made to understand the T box leader mechanisms (for a recent review, see Green et al.4). T box mRNA leader sequences do not fit into the classic definition of a riboswitch because instead of sensing a small molecule, they bind another macromolecule, tRNA; however, they containing both aptamer and effector domains and due to their functional similarities, here we refer to them as T box riboswitches.
The transcriptional regulatory T box riboswitch is comprised of several highly conserved motifs that typically form three complex stems (Stems I–III, though Stem II is absent from some T box riboswitches) and a fourth effector stem that is stabilized as an anti-terminator or terminator depending on the aminoacylation state of the bound tRNA (Fig. 1).5,6 Stem I is approximately 100 nucleotides (nts) long and contains several conserved motifs. From proximal to distal, these include: the GA/k-turn motif, specifier loop, L3/4, AG bulge, and the distal loop, linked by five short helical segments (P1–5). Only the role of the specifier loop in tRNA binding was well established. It contains a specifier sequence that directly recognizes the cognate tRNA through codon–anticodon pairing.2,7-10 An NMR structure of the base of Stem I reveals the GA motif forms a k-turn that could be important in orienting the 3′ region of the T box riboswitch, relative to the bound tRNA.11,12 Otherwise, the functions of the highly conserved distal elements were largely unknown prior to recent structural and biochemical analysis performed in our group,13,14 by Lehmann et al.,15 and by Zhang and Ferré-D’Amaré.16 Through the combined contribution of these studies, a much more lucid model for tRNA binding has taken shape.
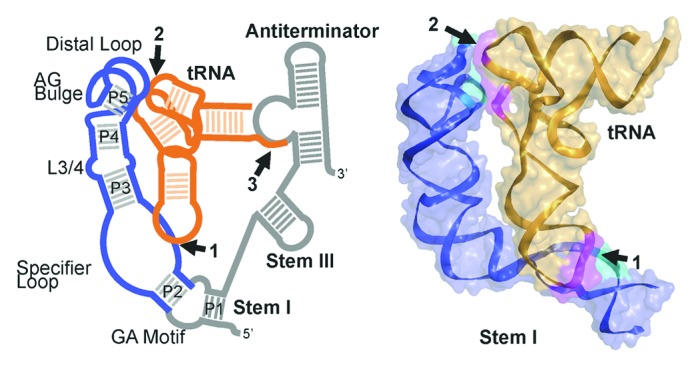
Figure 1. Structure of the T box riboswitch tRNA complex. (A) Schematic of the T box riboswitch (blue/gray)—tRNA (orange) complex with intermolecular interactions highlighted and numbered: (1) specifier sequence-anticodon base pairs, (2) distal Stem I base stacking against the tRNA elbow, (3) acceptor-antiterminator base pairs. Regions of the T box riboswitch shown in the crystal structure in panel B are highlighted in blue. (B) The overall 3.2 Å crystal structure of the Stem I-tRNA complex (PDB ID: 4MGN), shown as ribbons with a semi-transparent molecular surface. The two intermolecular contacts highlighted in cyan (Stem I) and magenta (tRNA) and numbered, as in (A).
Structure-to-Function Discovery of tRNA Binding
It has long been established that tRNA is recognized by the T box riboswitch through sequence-specific base-pairing interactions between: (1) the specifier sequence-anticodon and (2) the anti-terminator-acceptor arm.2,7 Additional highly conserved motifs in the T box riboswitch have been identified, but their roles in tRNA anchoring are largely unknown.9 In our recent work, presented in the Proceedings of the National Academy of Sciences, we used a structure-to-function approach to understand the tRNA recognition mechanism by the T box riboswitch.13 A series of T box riboswitch truncations were used to identify essential structural and sequence elements required for tRNA binding. The tRNA binding affinity was narrowed down to a fragment that contains 86-nts of Stem I, excluding the GA motif/k-turn (denoted Stem I86, Fig. 1). Further division of Stem I86 into the distal half and specifier half completely abolished tRNA binding; however, outside of the specifier-anticodon interaction, it was unclear how the distal half of Stem I contributed to binding. An NMR structure of the tRNA-free specifier half of Stem I was previously determined,11,12 but the structure of ~50 nts in the Stem I tip was unknown. We therefore began by determining the 2.65 Å crystal structure of the distal half of Stem I (57 nucleotides, Stem I57), including the highly conserved L3/4, AG bulge, and distal loop motifs. Fortuitously, the structure provided a tremendous amount of information. For example, the L3/4 motif (CUC bulge in Stem I57) imparts a kink between P3 and P4 that changes the arch of Stem I and could foreseeably provide flexibility in the stem by altering the helical register or kinking between P3 and P4. Given its conservation, it seems likely that the main role for this mid-stem motif is to provide essential flexibility to the stem for tRNA binding.
The most significant observation revealed by the structure was the extensive tertiary interactions between the AG bulge and distal loop. The bases from both motifs stack together in an interwoven zipper-like motif, referred to as a loop–loop interaction in our work, or more precisely, an interlocking T-loop (or head-to-tail double T-loop).13,15,17 T-loops are five-nucleotide motifs that form a compact U-turn, receive an intercalating base between its fourth and fifth nucleotides, and close the motif with a base pair between the first and fifth nucleotides (for an excellent review, see Chan et al. 201317). Interlocked T-loops have been describe in RNase P18,19 and the ribosome L1 stalk15 and are comprised of two compact, pseudo-symmetrically oriented T-loops that interact across two interwoven stacked base triples. The result is a rigid structure with a well-formed base-stacking surface suited to dock against RNA bases. The ability of the motif to form an interaction surface is clearly reflected in the prominent Stem I57 crystal lattice contacts, formed across both ends of the interlocking T-loop, resulting in a continuously stacked structure through the crystal.13 We constructed a hypothetical structural model of the T box-tRNA complex by first assembling a T box Stem I model from the crystal structure of the distal region13 and the NMR structure of the specifier region,12 then modeling bound tRNA by overlaying the mRNA-petidyl site tRNA complex from the Thermus thermophilus ribosome structure20 with the T box riboswitch specifier sequence (Fig. 1). The theoretical models revealed a possible second tRNA contact, ~60 Å away from the specifier-anticodon interface, where the apical interlocking T-loop structure stacks against the tRNA D/T loop. This observation is supported by previous work that demonstrated altering the tRNA anticodon arm length or D/T-loop interactions was detrimental to T box function.21,22 Given these observations, we proposed that the tRNA anticodon is specifically bound at the specifier sequence with the anticodon arm extending up along Stem I to dock in a structurally specific fashion against the Stem I apex (Fig. 1B). Lehmann et al. independently derived and classified the distal structure as a head to tail double T-loop module (or interlocking T-loops) based on the striking sequence similarity between 23S rRNA L1 stalk. They correctly predicted the secondary structure of the T box Stem I and proposed a similar structure model for the T box Stem I-tRNA complex.15
Biochemical Evidence for tRNA Stacking against the Stem I Apex
Several predictions can be made based on our hypothetical structural model of the T box-tRNA complex. For example, our model predicts that the G62-C43•G55 base triple in the interlocking T-loop module stacks against the D/T loops of tRNA.13 This is tested using several independent techniques. First, individual point mutations of the distal base triple nucleotides were analyzed for tRNA binding by electrophoretic mobility shift assays, revealing that a substitution of any base in the base triple disrupts tRNA binding, to various extent. To more accurately probe nucleotides involved, selective 2’-hydroxyl acylation analyzed by primer extension (SHAPE) was used to identify reactivity changes due to T box-tRNA complex formation. Key complex-induced stabilization was observed in both strands of the specifier loop and at G55 in from the distal base triple from Stem I and in the anticodon and the D-loop of tRNA. Previous in-line probing of the T box riboswitch–tRNA interaction revealed contacts in both strands of the specifier loop and the tRNA anticodons and D-loop;22,23 however, this was the first direct observation of an induced change in the distal Stem I region. The mutagenesis and SHAPE data provided indirect means of assessing interactions, so to examine the intermolecular contacts more directly, the product of intermolecular UV cross-linking was analyzed by reverse transcription. Using this method, intermolecular cross-links were identified at C43 and G55 in Stem I and U19, U32, and G55 in tRNA. Together, these biochemical analyses provide strong supporting evidence that two contacts are formed between Stem I and tRNA: one between the specifier sequence and the tRNA anticodon and the second between the apical base triple and the tRNA elbow. With this model in hand we used small-angle X-ray scattering on the Stem I-tRNA complex and demonstrated that the hypothetical model generated by us13 (and foreseeably the similar model by Lehmann et al.15) closely resembles the unbiased reconstructions of the complex in solution.
A High-Resolution Snapshot of the Stem I-tRNA Complex
Using our well-defined minimal functional tRNA binding constructs, we determined the co-crystal structure of the G. kaustophilus (Gkau) glyQ Stem I86-tRNAGly complex.14 Independently, Zhang and Ferré-D’Amaré determined the full-length Oceanobacillus iheyensis (Oihe) glyQ Stem I-tRNAGly complex by stabilizing the basal GA/Kink-turn motif by co-crystallization with a Kink-turn binding protein, YbxF.16 Both structures were determined at 3.2 Å resolution, providing key insights into tRNA recognition. Stem I forms a structurally similar, ~110 Å long, arched stem in both cases. Indeed, Stem I packs against tRNA at the specifier-anticodon interaction, and as predicted, at the Distal Stem I interlocking T-loop-tRNA elbow region. The distal stacking interface is structurally analogous, but is a C-G•G base triple in Gkau and C-G•A in Oihe. Despite the G/A variation in the base triple, the purine seems to be essential for binding as replacing it with a pyrimidine disrupted binding.13,16 This requirement seems to be driven by optimal base stacking of the tRNA T-loop (elbow) base, G56. The structures beautifully revealed the extensive tertiary contacts throughout the specifier loop-anticodon loop interaction. The expected base pairs CGG/GCC base pairs provide sequence-specificity, but the are preceded by several conserved features for binding: An S-loop motif sets up the interface by inducing a sharp bend in the stem that is required for robust tRNA binding10,14,16 and several tertiary interactions between both strands of the Stem I specifier loop and the tRNA anticodon loop, including two Type II A-minor motifs, stabilize docking. These structures provide both confirmation for the models proposed by us13 and Lehman et al.,15 but more importantly, they reveal high resolution molecular level details for the interaction.
A Model for T box Riboswitch Regulation and Perspective
Based on these experimental data, we propose an updated model for tRNA sensing. Stem I forms a long, arched stem that curves around tRNA to dock its specifier sequence against the tRNA anticodon and wrap up to the tRNA elbow, where it stacks the exposed apical base triple from the interlocking T-loop against the tRNA elbow (D/T-loops). These two contacts securely anchor tRNA to the T box riboswitch. Intuitively, since Stem I is the first element transcribed, tRNA anchoring could be performed while downstream components are being transcribed. The function of Stems II and III are unclear at this time, but they are generally conserved suggesting they either provide supporting contacts to the tRNA or modulate the rate of polymerase progression to the terminator/antiterminator. tRNA binding kinetics are not well-established, but a major polymerase pause site was identified in Stem III24 and this or other pausing sites could foreseeably allow sufficient time for tRNA to bind to Stem I prior to terminator transcription. With tRNA anchored to Stem I, the aminoacylation status of tRNA further influences the downstream T box structure formation. The antiterminator stem-loop is stabilized by an uncharged tRNA through base-pairing between the tRNA CCA tail and a bulged UCC sequence in the T box anti-terminator, allowing full-length transcription. The same interaction is disfavored if tRNA is aminoacylated, disfavoring the anti-terminator over the terminator stem-loop in T box and causing its premature transcription termination. The model that comes to light from these studies provides significant refinements to the previous model, but many questions remain. For instance, what is the architecture of the remaining T box riboswitch (Stems I–III and the anti-terminator/terminator)? Are transcriptional pause sites required for regulation or are the kinetics of tRNA docking on Stem I rapid enough without it? Do binding kinetics correlate with tRNA abundance? Once anchored, how does tRNA aminoacylation destabilize the anti-terminator structure? These and other questions remain the focus of our continued work on this system.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by an NIH grant GM-086766 to Ke A and a Postdoctoral Fellowship from Canadian Institute of Health Research to Grigg JC. We thank Frank Grundy and Tina Henkin for introducing the G. kau glyQS T box system to us.13
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/26996
References
- 1.Breaker RR. Riboswitches and the RNA world. Cold Spring Harb Perspect Biol. 2012;4:4. doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grundy FJ, Henkin TM. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993;74:475–82. doi: 10.1016/0092-8674(93)80049-K. [DOI] [PubMed] [Google Scholar]
- 3.Henkin TM. tRNA-directed transcription antitermination. Mol Microbiol. 1994;13:381–7. doi: 10.1111/j.1365-2958.1994.tb00432.x. [DOI] [PubMed] [Google Scholar]
- 4.Green NJ, Grundy FJ, Henkin TM. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett. 2010;584:318–24. doi: 10.1016/j.febslet.2009.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gutiérrez-Preciado A, Henkin TM, Grundy FJ, Yanofsky C, Merino E. Biochemical features and functional implications of the RNA-based T-box regulatory mechanism. Microbiol Mol Biol Rev. 2009;73:36–61. doi: 10.1128/MMBR.00026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vitreschak AG, Mironov AA, Lyubetsky VA, Gelfand MS. Comparative genomic analysis of T-box regulatory systems in bacteria. RNA. 2008;14:717–35. doi: 10.1261/rna.819308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grundy FJ, Hodil SE, Rollins SM, Henkin TM. Specificity of tRNA-mRNA interactions in Bacillus subtilis tyrS antitermination. J Bacteriol. 1997;179:2587–94. doi: 10.1128/jb.179.8.2587-2594.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grundy FJ, Winkler WC, Henkin TM. tRNA-mediated transcription antitermination in vitro: codon-anticodon pairing independent of the ribosome. Proc Natl Acad Sci U S A. 2002;99:11121–6. doi: 10.1073/pnas.162366799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rollins SM, Grundy FJ, Henkin TM. Analysis of cis-acting sequence and structural elements required for antitermination of the Bacillus subtilis tyrS gene. Mol Microbiol. 1997;25:411–21. doi: 10.1046/j.1365-2958.1997.4851839.x. [DOI] [PubMed] [Google Scholar]
- 10.Chang AT, Nikonowicz EP. Solution NMR determination of hydrogen bonding and base pairing between the glyQS T box riboswitch Specifier domain and the anticodon loop of tRNA LID - S0014-5793(13)00673-X [pii] LID - 10.1016/j.febslet.2013.09.003 []. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Henkin TM, Nikonowicz EP. NMR structure and dynamics of the Specifier Loop domain from the Bacillus subtilis tyrS T box leader RNA. Nucleic Acids Res. 2010;38:3388–98. doi: 10.1093/nar/gkq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J, Nikonowicz EP. Solution structure of the K-turn and Specifier Loop domains from the Bacillus subtilis tyrS T-box leader RNA. J Mol Biol. 2011;408:99–117. doi: 10.1016/j.jmb.2011.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grigg JC, Chen Y, Grundy FJ, Henkin TM, Pollack L, Ke A. T box RNA decodes both the information content and geometry of tRNA to affect gene expression. Proc Natl Acad Sci U S A. 2013;110:7240–5. doi: 10.1073/pnas.1222214110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grigg JC, Ke A. Structural determinants for geometry and information decoding of tRNA by T box leader RNA. Structure (London, England: 1993) 2013:In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehmann J, Jossinet F, Gautheret D. A universal RNA structural motif docking the elbow of tRNA in the ribosome, RNAse P and T-box leaders. Nucleic Acids Res. 2013;41:5494–502. doi: 10.1093/nar/gkt219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Ferré-D’Amaré AR. Co-crystal structure of a T-box riboswitch stem I domain in complex with its cognate tRNA. Nature. 2013;500:363–6. doi: 10.1038/nature12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan CW, Chetnani B, Mondragón A. Structure and function of the T-loop structural motif in noncoding RNAs. Wiley Interdiscip Rev RNA. 2013;4:507–22. doi: 10.1002/wrna.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krasilnikov AS, Xiao Y, Pan T, Mondragón A. Basis for structural diversity in homologous RNAs. Science. 2004;306:104–7. doi: 10.1126/science.1101489. [DOI] [PubMed] [Google Scholar]
- 19.Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragón A. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature. 2010;468:784–9. doi: 10.1038/nature09516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Selmer M, Dunham CM, Murphy FV, 4th, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–42. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 21.Grundy FJ, Collins JA, Rollins SM, Henkin TM. tRNA determinants for transcription antitermination of the Bacillus subtilis tyrS gene. RNA. 2000;6:1131–41. doi: 10.1017/S1355838200992100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yousef MR, Grundy FJ, Henkin TM. tRNA requirements for glyQS antitermination: a new twist on tRNA. RNA. 2003;9:1148–56. doi: 10.1261/rna.5540203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yousef MR, Grundy FJ, Henkin TM. Structural transitions induced by the interaction between tRNA(Gly) and the Bacillus subtilis glyQS T box leader RNA. J Mol Biol. 2005;349:273–87. doi: 10.1016/j.jmb.2005.03.061. [DOI] [PubMed] [Google Scholar]
- 24.Grundy FJ, Henkin TM. Kinetic analysis of tRNA-directed transcription antitermination of the Bacillus subtilis glyQS gene in vitro. J Bacteriol. 2004;186:5392–9. doi: 10.1128/JB.186.16.5392-5399.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


