Abstract
Microphthalmia, anophthalmia and coloboma (MAC) are structural congenital eye malformations that cause a significant proportion of childhood visual impairments. Several disease genes have been identified but do not account for all MAC cases, suggesting that additional risk loci exist. We used SNP homozygosity mapping (HM) and targeted next-generation sequencing to identify the causative mutation for autosomal recessive isolated colobomatous micro-anophthalmia (MCOPCB) in a consanguineous Irish Traveller family. We identified a double nucleotide polymorphism (g.1157G>A and g.1156G>A; p.G304K) in STRA6 that was homozygous in all of the MCOPCB patients. The STRA6 p.G304K mutation was subsequently detected in additional MCOPCB patients, including one individual with Matthew-Wood syndrome (MWS; MCOPS9). STRA6 encodes a transmembrane receptor involved in vitamin A uptake, a process essential to eye development and growth. We have shown that the G304K mutant STRA6 protein is mislocalised and has severely reduced vitamin A uptake activity. Furthermore, we reproduced the MCOPCB phenotype in a zebrafish disease model by inhibiting retinoic acid synthesis, suggesting that diminished retinoic acid levels account for the eye malformations in STRA6 p.G304K patients. The current study demonstrates that STRA6 mutations can cause isolated eye malformations in addition to the congenital anomalies observed in MWS.
Keywords: STRA6, homozygosity mapping, Matthew-Wood syndrome, MWS
Introduction
Microphthalmia, anophthalmia and coloboma (MAC) are related structural, congenital eye malformations which display a spectrum of severity and can occur in isolation or as part of a syndrome. Microphthalmia refers to a small eye, defined by axial length, while anophthalmia denotes the complete absence of an eye (Morrison, et al., 2002). Both conditions can be present in uni- or bi-lateral form. The third type of structural malformation, coloboma, is a segmental ocular defect resembling a “keyhole” deficiency in the iris. Although the reported prevalence at birth varies greatly, microphthalmia and anophthalmia are estimated to occur in 14 and 3 per 100,000 births respectively (Morrison, et al., 2002), and estimates of the combined prevalence reach 30 per 100,000 births (Verma and Fitzpatrick, 2007). The estimated prevalence of coloboma is 1 in 10,000 (Stoll, et al., 1997). Although individually rare, MAC is reported to cause 14.3% of cases of childhood severe visual loss (Hornby, et al., 2000).
The aetiology of MAC is not well understood. Environmental factors such as maternal vitamin A deficiency and exposure to infections, viruses or toxins during pregnancy have been linked to eye malformations, however, the extent of their contribution to MAC remains to be clarified (Busby, et al., 2005; Hornby, et al., 2002; Jana and Sharma, 2010). Familial clustering of the conditions has implicated a significant genetic component whereby microphthalmia, anophthalmia or coloboma are occasionally found to be present in different eyes of the same individual or within different individuals of the same family (Francois and Haustrate-Gosset, 1976). Inheritance of MAC may be autosomal dominant (Morle, et al., 2000), autosomal recessive (Bessant, et al., 1998) or X-linked (Graham, et al., 1991) and linkage analysis in a small number of families where these disorders segregate in a Mendelian pattern has led to the mapping of loci at 2q37.1, 14q32, 15q12–q15 and Xq27 (Bessant, et al., 1998; Graham, et al., 1991; Hmani-Aifa, et al., 2009; Morle, et al., 2000). Heritable causes of MAC, including chromosomal anomalies and single gene mutations, have also been identified although a high degree of genetic heterogeneity is evident. Several genes that are known to play a role in ocular development have been implicated in MAC including SOX2 (MIM# 184429), CHX10 (MIM# 142993), GDF3 (MIM# 606522), GDF6 (MIM# 601147), MFRP (MIM# 66227), PAX6 (MIM# 607108), PRSS56 (MIM# 613858), OTX2 (MIM# 600037), PAX2 (MIM# 167409), RX (MIM# 601881), SHH (MIM# 600725) and SIX6 (MIM# 606326) (Amiel, et al., 2000; Fantes, et al., 2003; Ferda Percin, et al., 2000; Gal, et al., 2011; Gallardo, et al., 2004; Glaser, et al., 1994; Ragge, et al., 2005a; Schimmenti, et al., 2003; Sundin, et al., 2005; Tassabehji, et al., 2008; Voronina, et al., 2004; Ye, et al., 2010). However causative mutations in these genes do not account for all MAC cases (Ragge, et al., 2005b), suggesting that additional MAC loci are yet to be identified.
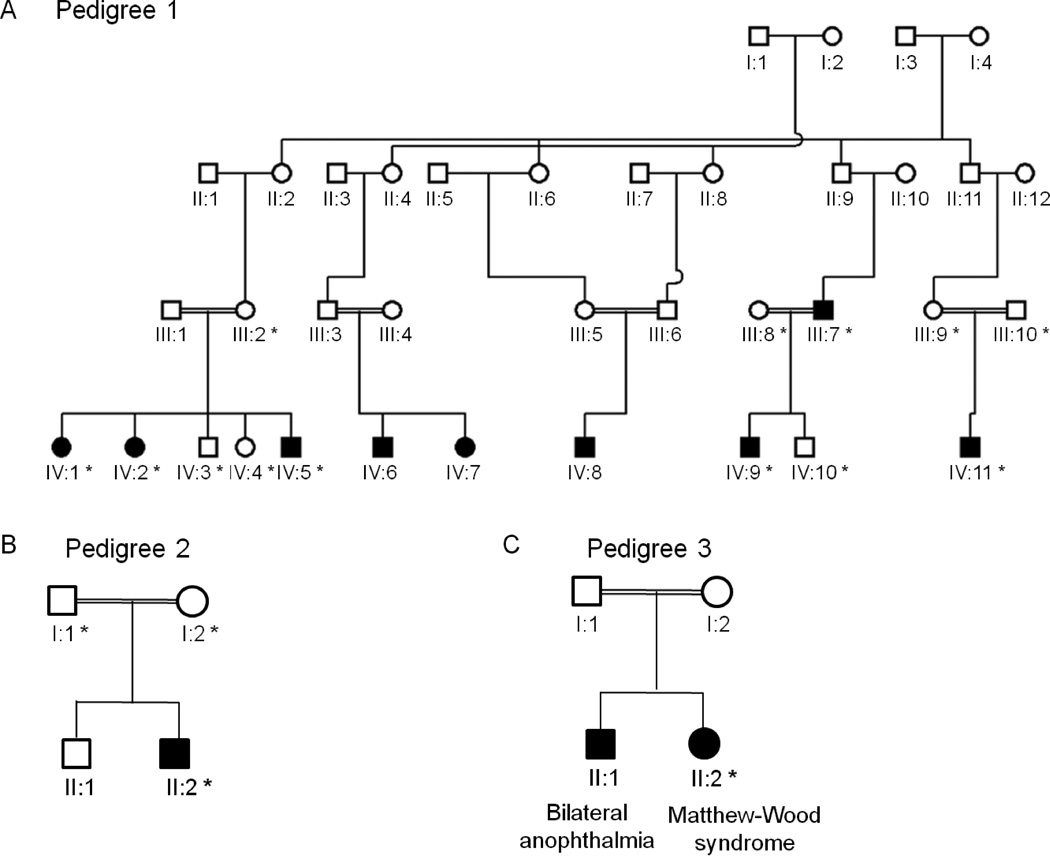
The current study involves a consanguineous Irish Traveller family with nine members who are affected by one or more of microphthalmia, anophthalmia and ocular coloboma of varying severity (pedigree 1; Figure 1A). Three patients have bilateral micro/anophthalmos, two patients have unilateral anophthalmos and contralateral microphthalmos with iris coloboma, two patients have unilateral anophthalmos and contralateral iris coloboma, one patient has unilateral colobomatous microphthalmos and one patient has unilateral microphthalmos (Table 1). Clinical assessment of the affected individuals confirmed that the ocular malformations are non-syndromic. A diagnosis was made of an autosomal recessive non-syndromic colobomatous micro-anophthalmia, referred to as MCOPCB. This is the first report of MCOPCB in the Irish Traveller population, an endogamous group of nomads within the Irish population. Linkage analysis at 14q32, 18q21 and 20p11 had previously excluded OTX2, CHX10, RX and VSX1 (MIM# 605020) as possible candidate genes in this MCOPCB pedigree. We undertook a genome-wide SNP homozygosity mapping analysis to identify the locus responsible for MCOPCB in this family.
Figure 1.
Pedigrees of Irish Traveller families with MCOPCB. (A–C) The inheritance pattern in each pedigree is compatible with an autosomal recessive disease gene. Pedigrees 1–3 are from the Irish Traveller population but are unrelated to each other. A: DNA samples were available for 6 affected and 7 unaffected family members in pedigree 1. B: DNA was available for the affected proband and his parents in pedigree 2. C: The female proband in pedigree 3 was diagnosed with Matthew-Wood syndrome (MWS). Her elder brother had bilateral anophthalmia only and no other clinical features. DNA was available for the MWS patient.
Table 1.
Clinical features of patients with MCOPCB in pedigree 1
| Ind | Sex | Age | Eye malformation |
|---|---|---|---|
| IV:1 | M | 16 | Right microphthalmia, left iris coloboma |
| IV:2* | F | 5 | Right choroidal coloboma, left anophthalmia (small cystic mass) |
| IV:5* | F | 15 | Bilateral microphthalmia |
| IV:6 | M | 4 | Right microphthalmia, left microphthalmia with coloboma |
| IV:7 | F | 2 | Right microphthalmia and choroidal coloboma, left anophthalmia |
| IV:8 | M | 20 | Bilateral anophthalmia |
| III:7* | M | 26 | Right anophthalmia, left microphthalmia with sclerocornea |
| IV:9* | M | 15 mo | Right microphthalmus, left anophthalmia and optic cyst |
| IV:11* | M | 16 mo | Bilateral anophthalmus (poor fornices) |
Nine members of an Irish Traveller family are affected with varying degrees of microphthalmia, anophthalmia and ocular coloboma. DNA from 6 affected individuals (denoted with a *) was available for molecular analysis.
Materials and Methods
Patients
The initial family (pedigree 1) was referred to the clinical genetic service for genetic assessment by at least one of the authors. They are members of the Irish Traveller population with significant consanguinity (average F = 0.063, range 0.032–0.118). Nine family members in two generations are affected by varying degrees of microphthalmia, anophthalmia and coloboma. All patients had normal cranial and abdominal ultrasounds and no extra-ocular defects were observed. The patients had normal intellectual development. A diagnosis was made of an autosomal recessive non-syndromic colobomatous micro-anophthalmia (MCOPCB). Subsequently, another child was seen in the genetics clinic from an apparently unconnected Irish Traveller family (pedigree 2). The child presented with severe bilateral microphthalmia. The optic nerves could not be identified on a CT brain scan but the bony orbits were well developed (Supp. Figure S1A). No significant intracranial pathology was found. The only other congenital anomaly was a dysplastic right kidney. A child from a third unconnected Irish Traveller family later presented at clinic (pedigree 3). The patient had bilateral anophthalmia, absent pulmonary valves, polysplenia, absent uterus and hydronephrosis with ureterocoeles. Brain MRI at age 4 months showed that structure and myelination were normal for the child’s age (Supp. Figure S1B–D). The ventricular system was also normal. Her clinical features were consistent with a diagnosis of Matthew-Wood syndrome (MWS; MIM# 601186). The G banded karyotype of the eight patients was normal at 550 band resolution. Ethical approval for the study was obtained from the ethics committee of Our Lady’s Children’s Hospital Crumlin, Ireland.
SNP genotyping and identification of runs of homozygosity
DNA samples from six patients (IV:1, IV:2, IV:4, III:7, IV:9 and IV:11) and seven unaffected relatives (III:2, IV:3, IV:5, III:8, IV10, III:9 and III:10) in pedigree 1 were available for molecular analysis. Genomic DNA was extracted from peripheral lymphocytes and genotyped for 1 million single nucleotide polymorphisms (SNPs) on an Illumina BeadStation 500GX platform. Runs of homozygosity (ROH) containing a minimum of 50 SNPs were identified using Illumina BeadStudio software. ROH that were common to all six affected individuals but were not homozygous in any of the seven unaffected relatives were identified. Log R ratios and B allele frequencies in ROH of interest were examined to exclude hemizygous deletions (Supp. Figure S2).
Targeted sequence capture
A customised NimbleGen sequence capture array was designed to specifically isolate the four candidate homozygous regions at 15q23–24.1. Whole genome amplified DNA from five individuals, 3 affected (IV:1, IV:2 and IV:9) and 2 unaffected (IV:5 and IV:10), was selected for sequence capture which was performed at Roche NimbleGen.
Single read sequencing and data analysis
Libraries were prepared from the captured DNA according to the Illumina protocol (Illumina Part # 1003806 Rev. B) and sequenced on an Illumina GAII sequencer (Trinity Genome Sequencing Laboratory). The single sequence reads were mapped to the reference human genome (hg18) using BWA version 0.5.7 (Li and Durbin, 2009). Reads of inadequate sequence quality and potential PCR duplicates (those with identical sequences, start and end sites) were discarded. The quality scores for the aligned reads were recalibrated using GATK (McKenna, et al., 2010). Variants and indels were identified using SAMtools (Li, et al., 2009). Loci with inadequate sequence coverage (<3×) were excluded. Homozygous variants that were shared by the 3 patients that were sequenced, but differed to the 2 unaffected relatives, were retained for further analysis. Variants with a population (European) frequency >1%, as reported in dbSNP129, were considered unlikely to be disease-causing and were excluded. Of the remaining novel homozygous variants, non-synonymous substitutions were prioritised. Mutations of interest were validated by Sanger sequencing.
Validation of STRA6 mutations
STRA6 variants were confirmed by PCR in all available family members from pedigrees 1–3. A 248 base-pair region surrounding the NM_001142617.1:g.1157G>A and NM_001142617.1:g.1156G>A mutations in exon 11 of STRA6 was amplified (primers 5’-ctgctggcccttttcctg-3’ and 3’-cctggagagctgggttgg-5’ and an annealing temperature of 62°C.) The PCR products were analysed by Sanger sequencing. Amino acid residues are numbered from the first methionine residue (reference sequence NM_001142617.1(STRA6_v001), according to journal guidelines (www.hgvs.org/mutnomen). The double nucleotide polymorphism is submitted to dbSNP under accession NM_001142617.1:c.910_911delinsAA. A control panel, comprising 100 chromosomes from healthy members of the Irish Traveller population, was also tested for the STRA6 g.1157G>A and g.1156G>A mutations by direct sequencing. The remaining exons of STRA6 (exons 2–10 and 12–19) were amplified and sequenced in the patient diagnosed with MWS (pedigree 3). Primer sequences and reaction conditions are available on request.
Assay for retinol uptake activity using 3H-retinol/RBP
A human STRA6 p.G304K mutant was produced using PCR and confirmed by DNA sequencing. The production of 3H-retinol/RBP was performed as previously described (Kawaguchi, et al., 2007). STRA6 was cotransfected with lecithin retinol acyltransferase (LRAT) for the retinol uptake assay in COS-1 cells. To determine cellular 3H-retinol uptake from 3H-retinol/RBP, cells were washed with Hank’s Buffered Salt Solution (HBSS) before incubation with 3H-retinol/RBP diluted in serum free medium for 1 hour at 37°C. The reactions were stopped by removing the medium, washing the cells with HBSS, and solubilising the cells in 1% Triton X-100 in PBS. Radioactivity was measured with a scintillation counter.
Live and permeabilised cell staining of wild type and mutant STRA6
For live cell staining, a Myc epitope was inserted into an extracellular loop of human STRA6 between residues 132 and 133. Epitope insertion at this position does not affect vitamin A uptake activity or RBP binding activity of STRA6 (Kawaguchi, et al., 2007). STRA6-Myc constructs were transfected into COS-1 cells growing on gelatin-coated coverslips. At 24 hours after transfection, anti-Myc monoclonal antibody was added to the media. After 1 hour incubation at 37°C, the cells were washed with HBSS 3 times and fixed in 4% paraformaldehyde in PBS for 10 minutes at room temperature. After 3 washes with PBS, the fixed cells were incubated with the blocking buffer (5% normal goat serum and 0.3% Triton X-100 in PBS) for 1 hour at room temperature. The cells were then incubated with Alexa Fluor 488-conjugated goat anti-mouse antibody (Molecular Probes) diluted in the blocking buffer for another hour at room temperature. After 3 washes with PBS, the coverslips containing cells were mounted onto slides using VectaShield mounting medium. The cell surface expression of STRA6-Myc proteins was examined by fluorescence microscopy. Permeabilised staining was performed similarly to live cell staining procedures except that cells were incubated with the anti-Myc antibody after they have been fixed in 4% paraformaldehyde in PBS for 10 min.
Pharmacological inhibition of retinoid synthesis in zebrafish
Zebrafish embryos were used to model different levels of retinoic acid using citral dimethyl acetal, a precursor to the retinoic acid synthesis inhibitor citral (Marsh-Armstrong, et al., 1994). Embryos in which the primitive optic primordium had formed were treated with 0.05, 0.5 and 5 µM citral dimethyl acetal at the 6- and 11-somite stage. The drug was added to the embryo media for one hour and then exchanged with embryo media multiple times to wash out the drug. The ratio of eye area to head size was measured in the treated zebrafish at 3 days post fertilisation (dpf). The morphology and eye structure of the treated zebrafish were examined and images taken at 3 and 5 dpf.
Results
SNP Homozygosity mapping
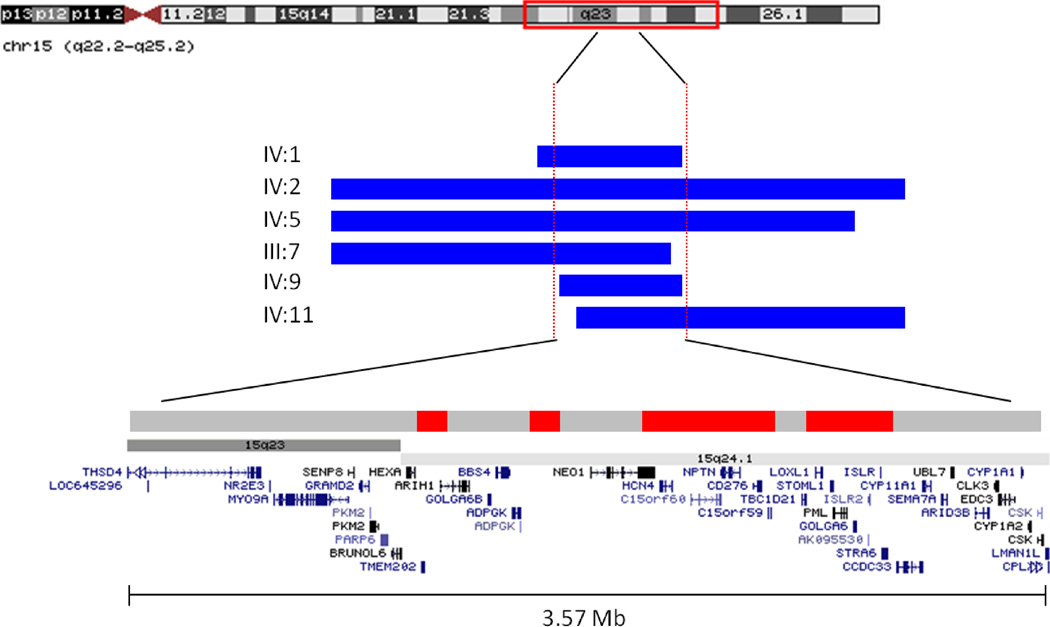
We undertook a genome-wide SNP homozygosity mapping analysis to identify the genetic locus involved in MCOPCB in pedigree 1. The 6 patients affected by MCOPCB shared 73 ROH (17.9 Mb), ranging in size from 12.8 kb to 3.6 Mb. ROH that were exclusively shared by the MCOPCB patients, and were not homozygous in any of the 7 unaffected relatives, were identified and comprised of four consecutive homozygous regions at 15q23–24.1. The candidate loci totalled 0.9 Mb in size and contained 15 genes (Figure 2 and Table 2). All affected individuals share a common 3.6 Mb homozygous haplotype between markers rs4777352 and rs3743487 (chr15:69342218–72909909), a finding that supports a common ancestral origin for this disease locus in pedigree 1.
Figure 2.
Putative MCOPCB disease locus at 15q23–24.1 in pedigree 1. The ROH at 15q23–24.1 in 6 members of the family affected by MCOPCB are shown as horizontal blue/black bars. The ROH vary in size from 4.1 Mb to 11.1 Mb. The maximal shared region is 3.57 Mb containing 43 RefSeq genes. Analysis of the homozygosity patterns in 7 unaffected family members (light grey bars) reduced the ROH of interest to 0.9 Mb in size and 15 genes (red/dark grey bars).
Table 2.
Candidate loci identified by SNP homozygosity mapping
| Genomic position | ROH size (kb) |
RefSeq genes |
|---|---|---|
| 15:70,482,945–70,578,645 | 95.70 | TMEM202, ARIH1 |
| 15:70,900,743–70,985,517 | 84.77 | Intergenic |
| 15 :71,350,548–71,867,728 | 517.18 | NEO1, HCN4, C15orf60, NPTN, CD276, C15orf59 |
| 15:72,004,570–72,306,485 | 301.92 | LOXL1, STOML1, PML, GOLGA6, ISLR2, ISLR, STRA6 |
SNP homozygosity mapping identified four consecutive ROH at 15q24.1 that were shared by the 6 MCOPCB patients but were not homozygous in the 7 unaffected relatives. The four regions total 0.9 Mb and contain 15 RefSeq genes. Genomic positions are based on NCBI build 36.1 (hg18).
Mutation analysis
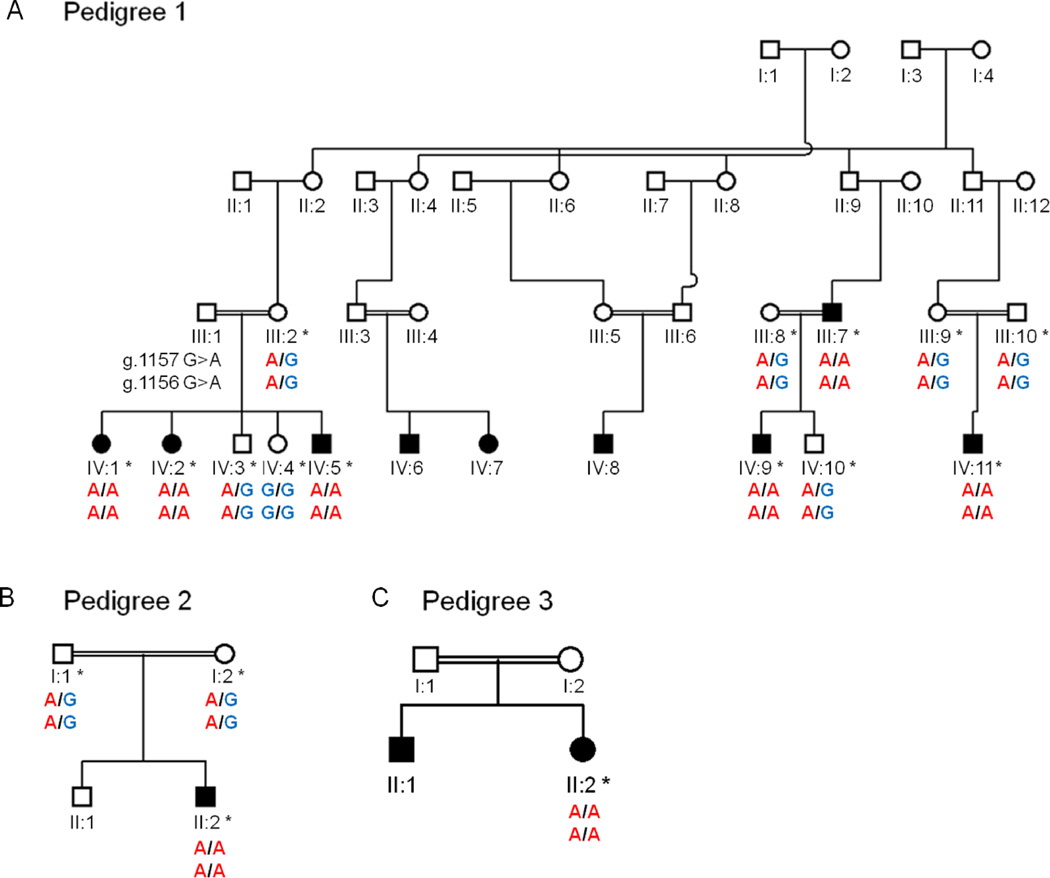
A custom NimbleGen sequence capture array was designed to target the genomic loci of interest at 15q23–24.1 and the captured DNA was sequenced on an Illumina GAII sequencer. We identified 10 homozygous variants that segregated with the MCOPCB phenotype (Supp. Table S1). Four of the variants were reported in dbSNP129 with a population frequency ≥1% and were considered unlikely to be disease-causing (Supp. Table S2). Of the remaining 6 novel mutations, only two resulted in amino acid substitutions (Table 3). Both missense mutations occur in the gene encoding the stimulated by retinoic acid 6 protein (STRA6; MIM# 610745) and are within the same amino acid residue. The MCOPCB patients are homozygous for a double nucleotide polymorphism (NM_001142617.1:c.910_911delinsAA) that induces a non-synonymous change (p.G304K) from glycine (non-polar) to lysine (polar) at a highly conserved residue of STRA6 (Supp. Figure S3). Additional family members were then analysed for G304K by Sanger sequencing. We show complete segregation of the G304K mutation with the disease phenotype in all investigated family members (Figure 3 and Supp. Figure S4). The putative disease mutation was not present in 100 chromosomes of healthy, ethnically matched control samples from the Irish Traveller population.
Table 3.
Novel homozygous mutations at the 15q24.1 locus that segregate with the MCOPCB phenotype
| Position | Gene | Type | IV:1 | IV:2 | IV:9 | IV:5 | IV:10 | Substitution |
|---|---|---|---|---|---|---|---|---|
| 71,382,144 | NEO1 | 3’ utr | CC | CC | CC | CT | CT | - |
| 71,401,251 | HCN4 | 3’ utr | GG | GG | GG | AA | AA | - |
| 71,640,342 | NPTN | Exon | AA | AA | AA | AG | GG | Synonymous |
| 72,270,250 | STRA6 | Exon | TT | TT | TT | CT | CC | G304E* |
| 72,270,251 | STRA6 | Exon | TT | TT | TT | CT | CC | G304R* |
| 72,274,265 | STRA6 | 3’ utr | AA | AA | AA | AG | GG | - |
Sequence analysis of the 15q24.1 candidate locus identified 6 novel homozygous mutations that segregated with the MCOPCB phenotype. Two of the exonic mutations are in STRA6 and result in non-synonymous substitutions.
The mutations at positions 72,270,250 (NM_001142617.1:g.1157G>A) and 72,270,251 (NM_001142617.1:g.1156G>A) are consecutive DNA bases of the same amino acid residue (304) and combine to give a p.G304K substitution in STRA6 (NM_001142617.1:c.910_911delinsAA). The genomic positions refer to build hg18.
Figure 3.
Segregation of STRA6 p.G304K with MCOPCB. Analysis of Sanger sequence confirmed segregation of the G304K mutation with the eye malformation phenotype in pedigrees 1–3 (A–C). The top and bottom genotypes denote the DNA mutations at chromosome 15 positions 72,270,250 (NM_001142617.1:g.1157G>A) and 72,270,251 (NM_001142617.1:g.1156G>A) respectively (hg18).
Identification of STRA6 p.G304K in additional patients
Subsequently, another child from an apparently unconnected consanguineous Irish Traveller family presented at clinic with eye malformations (pedigree 2; Figure 1B). The child had severe bilateral anophthalmia and a dysplastic right kidney. The STRA6 locus was analysed by Sanger sequencing and the proband was also found to be homozygous for the G304K mutation, adding further support for its role in MCOPCB (Figure 3B). A child from a third unconnected consanguineous Irish Traveller family (pedigree 3) later presented at clinic with eye defects and congenital abnormalities (Figure 1C). Her clinical features were consistent with a diagnosis of Matthew-Wood syndrome (MWS; MIM# 601186). Of note, her elder brother was reported to have bilateral anophthalmia with no other malformations but was unavailable for clinical assessment or blood sampling. Sequence analysis confirmed that the patient with MWS was homozygous for the STRA6 p.G304K mutation (Figure 3C). To investigate the possibility of additional STRA6 mutations in the more severely affected MWS patient, the remainder of the STRA6 gene was sequenced. However, no other mutation suspected to be deleterious (missense, nonsense, frameshift or splice site) was identified (Supp. Table S3). SNP genotype analysis showed that the affected individuals from pedigrees 2 and 3 have the same 3.6 Mb haplotype surrounding the STRA6 p.G304K mutation as the patients from pedigree 1, supporting a common ancestral origin for the disease mutation in all three families.
Activity and localisation of mutant STRA6
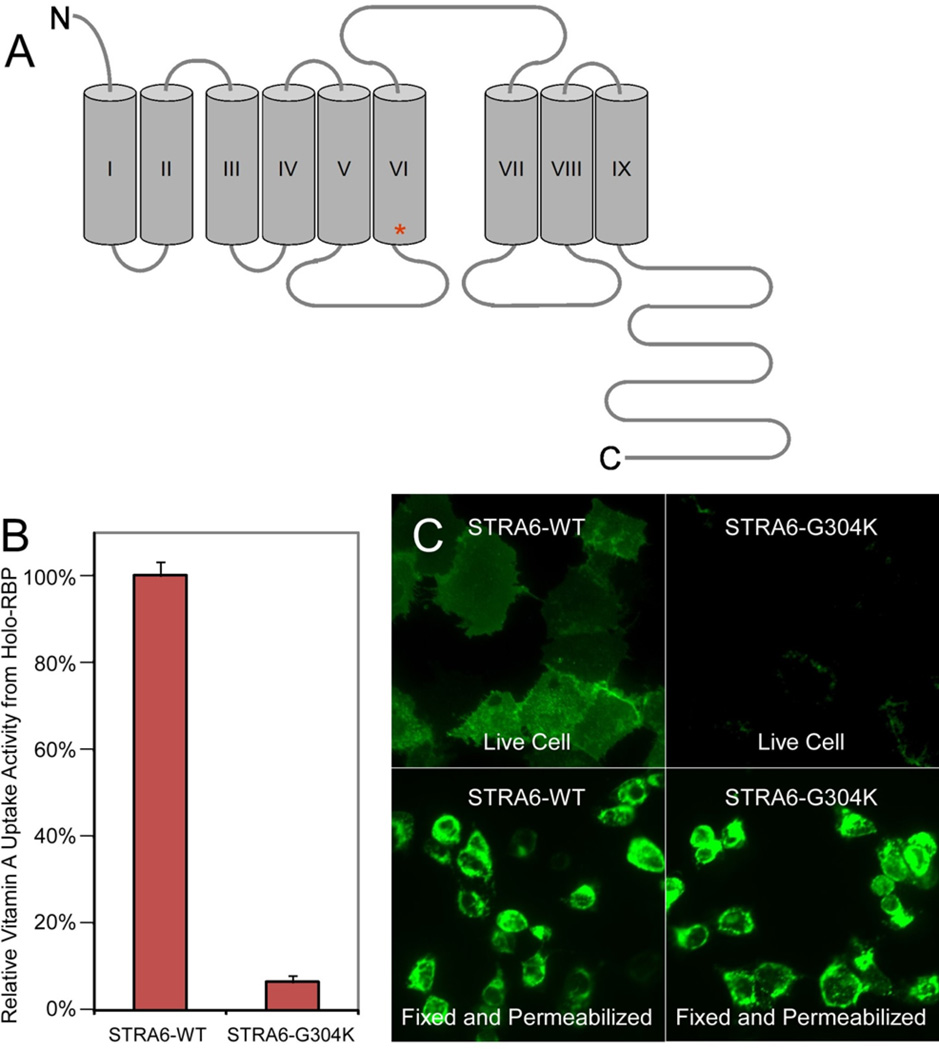
The functional consequences of the STRA6 p.G304K mutation identified in the MCOPCB patients were investigated in vitro by introducing the mutation into COS-1 cells and examining protein activity and localisation. Compared to the wild-type STRA6 protein, we found that the G304K mutation almost completely abolished vitamin A uptake activity (6.4% of wild type activity level) of STRA6 from holo-RBP (Figure 4B). Live cell staining showed that the G304K mutation led to loss of cell surface expression of STRA6 (Figure 4C). Permeabilised immunostaining demonstrated that the G304K mutant was still expressed (Figure 4C) suggesting that the missense mutation either causes misfolding of STRA6 or interferes with its targeting mechanism to prevent cell surface expression, which may be responsible for the loss of vitamin A uptake activity from holo-RBP.
Figure 4.
Examination of vitamin A uptake and localisation of the G304K STRA6 mutant protein in COS-1 cells. A: STRA6 transmembrane topology model based on Kawaguchi et al. 2008 (Mapping the membrane topology and extracellular ligand binding domains of the retinol binding protein receptor. Biochemistry 47:5387–5395). Residue G304 is located at the initial segment of transmembrane domain 6 (red asterisk). B: Retinol uptake assay from holo-RBP comparing wild-type human STRA6 and the G304K STRA6 mutant. Wild-type STRA6’s activity is defined as 100%. The mutant has greatly reduced retinol uptake activity (6.4%). C: The G304K STRA6 mutant is expressed similarly to the wild-type protein as shown in the permeabilised staining but is poorly expressed on the cell surface as shown in the live cell staining.
Pharmacological inhibition of retinoic acid synthesis in zebrafish
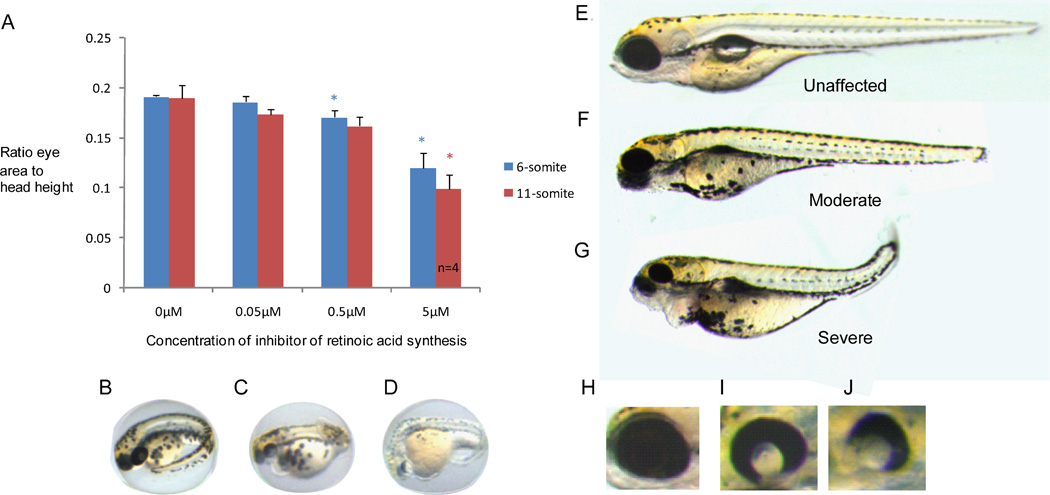
In vitro analyses demonstrated that the G304K STRA6 mutant has a severely diminished ability to transport retinoids into cells. Therefore, it is likely that retinoic acid levels, synthesised intracellularly, will also be diminished in MCOPCB patients homozygous for the missense mutation. Differences in retinoic acid levels within individual patients could account for the varying severity of MAC malformations. We chose a pharmacological approach to reduce retinoic acid levels during zebrafish eye development. Zebrafish embryos were used to model different levels of retinoic acid using citral dimethyl acetal, a precursor to the retinoic acid synthesis inhibitor citral (Marsh-Armstrong, et al., 1994). Inhibition of retinoic acid synthesis produced a dose-dependent microphthalmia at 3 dpf, with an average of 40% reduction in eye size with 5 µM inhibitor (Figure 5A and 5E–G). At 5 dpf, the effect of the retinoic acid inhibitor on eye size was even more pronounced. The inhibitor produced developmental eye defects ranging from mild to severe microphthalmia (Figure 5B–D). Furthermore, retinal pigment epithelium coloboma was also apparent and again with a range of severity (Figure 5H–J). Other developmental defects were visible, including defects in heart morphogenesis, consistent with the role of retinoic acid in multiple developmental processes.
Figure 5.
Pharmacological inhibition of retinoic acid synthesis in zebrafish embryos. Treatment of zebrafish embryos with a retinoic acid inhibitor, citral dimethyl acetal, has a dose-dependent effect on eye development, producing microphthalmic zebrafish. A: The x-axis represents the concentration of citral dimethyl acetal and the y-axis denotes the ratio of eye area to head height. Measurements were taken 3 days post fertilization. The embryos were treated during early eye development at the 6- (blue) or 11- (red) somite stage. P-values ≤0.05 are denoted by a *. Unless otherwise stated, n=5. B, C, D: Representative images of an untreated, moderately affected and severely affected zebrafish respectively at 3 days post fertilisation. E, F, G: Representative images of an unaffected, moderately affected and severely affected zebrafish respectively following treatment with citral dimethyl acetal. The images were taken at 5 days post fertilisation. H, I, J: Images of a wild type zebrafish eye and a moderate and severe retinal pigment epithelium coloboma respectively after inhibition of retinoic acid synthesis.
Discussion
We report a study to determine the causative mutation for autosomal recessive non-syndromic colobomatous micro-anophthalmia (MCOPCB) in a large consanguineous pedigree from an ethnic minority Irish population. Using homozygosity mapping and targeted next-generation sequencing, we identified a novel mutation (G304K) in STRA6 which encodes a transmembrane receptor involved in vitamin A uptake. The STRA6 p.G304K mutation was also found in a patient with Matthew-Wood syndrome, a condition with congenital malformations which includes eye defects. The findings from cellular work and a zebrafish disease model, confirm the importance of vitamin A in normal eye development. The data presented here also shows that the same genetic mutation may be responsible for a single disorder (isolated eye defect) in one individual, but may give rise to a syndromic disorder (MWS) in another. Although 11 patients with true isolated anophthalmia/microphthalmia have been tested for STRA6 mutations (White, et al., 2008), no pathogenic variants were identified. This is the first study to report the identification of causative STRA6 mutations in patients with non-syndromic anophthalmia (6 patients in pedigree 1).
STRA6 and eye development
STRA6 encodes a transmembrane receptor for the retinol-binding protein (RBP) and is responsible for mediating vitamin A uptake from circulation to target organs. The protein is expressed in a number of organs that require vitamin A for normal development, including the eye, brain, placenta and testis (Kawaguchi, et al., 2007). The activity of STRA6 may be especially important for tissues or cell types requiring a large amount of vitamin A, which is supported by the variation in tissue expression of the vitamin A transport system (RBP-STRA6). Retinoic acid (RA), a derivative of vitamin A, has long been known to play a critical role in vision and eye development (Hyatt and Dowling, 1997). Once transported across the membrane by STRA6, retinol is converted to a number of derivatives, termed retinoids, which in turn regulate the expression of vision-related genes. Retinoids can both enhance and suppress gene expression and excessive or deficient retinoid levels can result in severe birth defects (Collins and Mao, 1999). Therefore, tight regulation of retinol transport and maintenance of the correct level of endogenous retinoids is crucial for maintaining normal embryonic development (Zile, 2001). Different tissues vary in their vulnerability to vitamin A deficiency and excess. It is notable that the eye is the organ most dependent on vitamin A (Wilson, et al., 1953) and hence, most sensitive to STRA6 mutations. Therefore it is not surprising that, while there is considerable variability in the clinical presentation of individuals with MWS, eye anomalies are a consistent feature.
Several STRA6 missense mutations resulting in phenotypic effects have been reported to date (Chassaing, et al., 2009). The site of the missense mutation identified in the MCOPCB patients (G304) is located in the initial segment of the sixth transmembrane domain of STRA6 (Kawaguchi, et al., 2008b) (Figure 4A). The insertion of a positively charged polar lysine side chain into the hydrophobic environment of the cell membrane is predicted to have a negative effect on the stability of the protein (White and Wimley, 1999) and the efficiency of protein anchoring (Tsuzuki, et al., 2003). This effect is very likely to cause local unfolding or significant structural rearrangements. Furthermore, due to their small size and backbone flexibility, glycine residues are often found in parts of proteins where a specific geometry is required for the polypeptide backbone (Claessens, et al., 1989). Insertion of the lysine residue, which is more restricted in conformation, is likely to further disrupt the functionally active conformation of STRA6.
Analysis of the G304K mutant STRA6 protein in COS-1 cells
We propose that the G304K mutation may cause misfolding of STRA6 or interfere with its targeting mechanism, leading to loss of vitamin A uptake activity and reduced intracellular retinoid levels. The STRA6 G304K mutation (as identified in the MCOPCB patients) was investigated in vitro. We found that the STRA6 mutation causes a severe reduction in vitamin A uptake activity from holo-RBP (6.4% of wild-type protein activity). Additional expression analysis found that the G304K mutant protein was well expressed but localised to the endoplasmic reticulum instead of the cell surface. Taken together, the cellular work suggested that patients homozygous for STRA6 p.G304K would have severely depleted intracellular retinoid levels. The effect of low intracellular retinoid levels on embryonic development was investigated in a zebrafish model. Mild to severe microphthalmia was observed following inhibition of retinoic acid synthesis in zebrafish embryos which supports the view that diminished retinoic acid levels account for the eye malformation phenotypes in STRA6 p.G304K patients. The functional analyses provide insight into the pathogenesis of the disorder and suggest that the ocular malformations observed in the MCOPCB patients are a result of the inability of the mutant STRA6 to mediate sufficient vitamin A uptake from circulation into the cell, leading to reduced retinoid levels during embryonic development.
Intra-familial phenotypic variability
The affected family members in pedigree 1 are homozygous for the same missense mutation but there is considerable heterogeneity in the phenotype of their eye malformations. The intra-familial phenotype heterogeneity may be attributed to the variability in vitamin A intake of the affected individuals (environmental factor) and/or differences in the degree of loss of STRA6 function (genetic factor). RBP/STRA6 independent mechanisms of retinoid delivery, including random diffusion and transport of retinyl esters on plasma lipoproteins, can provide cellular retinoids in the absence of RBP or STRA6. However, these random mechanisms have many limitations that made it necessary to evolve the RBP/STRA6 system (Smith and Goodman, 1976). Firstly, random retinoid distribution is associated with mild to severe toxic side effects (Adams, 1993; Nau, 2001; Nau, et al., 1994). The RBP/STRA6 system is designed to achieve specific delivery of vitamin A to target cells. The specificity is achieved through the high affinity and low off-rate of RBPs binding to retinol and the specific binding of RBP to its high-affinity receptor STRA6 on specific target cells that can store vitamin A. Secondly, random retinoid distribution requires constant retinoid intake to supply target tissues and cannot provide a stable supply during times of insufficiency. RBP plays the essential role of mobilizing vitamin A stored in the liver and serving as a buffer to maintain stable vitamin A concentration in the blood (Blomhoff, et al., 1990; Quadro, et al., 2005). The buffering function is important given the adverse effect of both low and high retinoid levels on the growth and function of diverse organs. Thirdly, random retinoid distribution may not satisfy tissues that need a large amount of retinoid for proper function such as the eye. The combined actions of these RBP/STRA6-independent mechanisms of vitamin A uptake mean that the intracellular retinol levels will differ between individuals and may contribute to the variation in phenotypic severity.
Matthew Wood syndrome or isolated eye defects
STRA6 has previously been implicated in MWS, a multisystem disorder that includes microphthalmia and anophthalmia (Chassaing, et al., 2009; Golzio, et al., 2007; Pasutto, et al., 2007; West, et al., 2009; White, et al., 2008). MWS patients typically present with various combinations of eye, heart, lung and diaphragmatic defects. Of note, all of the 20 MWS patients reported in the literature to date had eye anomalies in the form of anophthalmia or microphthalmia, with heart malformations being the second most consistent feature. Currently, seventeen different STRA6 mutations (8 missense, 3 nonsense and 6 frameshift) have been found in patients with MWS (Chassaing, et al., 2009). The clinical spectrum associated with STRA6 mutations is extremely variable and no correlations between the nature of a STRA6 mutation and phenotypic severity have been found (Chassaing, et al., 2009). The current study has shown that patients homozygous for the STRA6 p.G304K mutation can develop an isolated eye phenotype (pedigrees 1 and 2) or the more severe MWS (pedigree 3), raising the question of how the G304K mutation produces an isolated eye malformation of variable severity in some patients but a multi-system disorder in others? Several mechanisms are proposed.
Firstly, one of the simplest explanations for the observed phenotypic differences may relate to the nature and location of the pathogenic STRA6 mutation and its impact on STRA6 function. It is plausible that MWS patients have more severe STRA6 mutations which completely abolish vitamin A uptake activity, while the STRA6 mutation observed in the MCOPCB patients may result in only a partial reduction in vitamin A uptake activity. STRA6 is highly enriched in the retinal pigment epithelium in the adult eye (Bouillet, et al., 1997) suggesting a high requirement of tightly regulated vitamin A uptake in this organ. Accordingly, even small deficiencies in vitamin A are sufficient to produce an ocular malformation. Therefore, although the STRA6 mutation observed in the MCOPCB patients results in an eye defect, there may still be sufficient uptake of vitamin A for proper functioning of the non-ocular tissues, such as the heart and brain, which do not have as high a dependency on vitamin A. In contrast, the more severe MWS mutations may result in complete loss of STRA6 activity in all tissues, leading to the multi-system malformations which extend beyond eye defects. However, the G304K mutation identified in the MCOPCB patients reduces STRA6 activity to a level similar to that observed for MWS mutations (~6%) (Kawaguchi, et al., 2008a) suggesting that the variation in phenotype arises through an alternate mechanism.
Secondly, differences in the timing and tissue expression of the mutation may explain why some patients with STRA6 mutations have a phenotype restricted to the eye, but others present with multi-organ syndromes. Thirdly, a second as yet unidentified disease gene may contribute to the broad spectrum of symptoms associated with MWS, similar to the tri-allelic inheritance model proposed in Bardet-Biedl syndrome (BBS) (Katsanis, et al., 2001). However, Isken and colleagues demonstrated that knock-down of stra6 in zebrafish produced a MWS phenotype whereby STRA6 deficiency resulted in craniofacial and cardiac defects and microphthalmia (Isken, et al., 2008). Although these observations need to be confirmed in germline mutants, they indicate that loss of STRA6 alone is sufficient to produce the MWS phenotype. Finally, the STRA6 p.G304K mutation may exhibit variable expressivity, which is not uncommon for MAC disease genes (Zenteno, et al., 2006). Similar to BBS, the inter- and intra-familial clinical variability in MCOPCB could be explained, in part, by the presence of second-site mutations that serve as modifiers of expressivity (Beales, et al., 2003; Leitch, et al., 2008; Stoetzel, et al., 2006). Recent work by Yeyati and colleagues has demonstrated that functional reduction of a second gene, HSP90, in two zebrafish eye mutants significantly influenced the expressivity of the eye defect (Yeyati, et al., 2007). Therefore it is possible that unidentified modifier loci may contribute to the variable expression of STRA6 mutations.
It is also possible that while genetic factors predispose to eye malformations, the interaction of environmental risk factors may contribute to the development of multiple congenital anomalies, such as those observed in MWS. It is well established that vitamin A intake alone (either insufficient or excessive) can lead to severe developmental defects without any genetic contribution (Wilson, et al., 1953). The evidence from teratogen- and dietary-induced animal models of congenital diaphragmatic hernia (a feature of MWS) is suggestive of environmental and/or nutritional factors also being involved and more research in this area is warranted (Clugston, et al., 2006; Holder, et al., 2007).
We identified a missense mutation (STRA6 p.G304K) that segregates with recessive MCOPCB and MWS in families from the Irish Traveller population. We have shown that substituting a highly conserved glycine at residue 304 with a lysine essentially abolishes vitamin A uptake activity of STRA6, most likely due to mislocalisation of the mutant protein. Inhibition of retinoid synthesis in a zebrafish disease model reproduced the MCOPCB phenotype suggesting that individuals homozygous for the G304K allele are unable to attain the intracellular levels of vitamin A and retinoids required to support normal eye development during embryogenesis. To our knowledge, this is the first report of STRA6 mutations associated with isolated eye malformations. Currently, diagnostic genetic testing of the STRA6 gene is standard for patients presenting with MWS but is not typically recommended for patients with non-syndromic eye anomalies. However, our study shows that STRA6 mutations can be responsible for non-syndromic eye malformations and we suggest that patients with isolated eye defects such as anophthalmia/microphthalmia should be considered for STRA6 screening.
Supplementary Material
Acknowledgments
We sincerely thank the participating families for the use of genetic samples and clinical information. We would also like to thank Dr. Ethna Phelan (Department of Radiology at Our Lady’s Children’s Hospital Crumlin) for her help in reporting on the patient’s CT and MRI images and Roche NimbleGen for providing the sequence capture service. This work was supported by the National Children’s Research Centre Our Lady’s Children’s Hospital Crumlin Ireland (grant SAC/95/07), the Health Research Board Ireland (HRA_HSR/2010/3) and Science Foundation Ireland (07/SRC/B1156). The Trinity Genome Sequencing Laboratory is funded by Science Foundation Ireland. Jillian Casey is supported by an EMBARK postgraduate award from the Irish Research Council for Science, Engineering and Technology (IRCSET).
Footnotes
Supporting Information for this preprint is available from the Human Mutation editorial office upon request (humu@wiley.com)
References
- Adams J. Structure-activity and dose-response relationships in the neural and behavioral teratogenesis of retinoids. Neurotoxicol Teratol. 1993;15(3):193–202. doi: 10.1016/0892-0362(93)90015-g. [DOI] [PubMed] [Google Scholar]
- Amiel J, Audollent S, Joly D, Dureau P, Salomon R, Tellier AL, Auge J, Bouissou F, Antignac C, Gubler MC, et al. PAX2 mutations in renal-coloboma syndrome: mutational hotspot and germline mosaicism. Eur J Hum Genet. 2000;8(11):820–826. doi: 10.1038/sj.ejhg.5200539. [DOI] [PubMed] [Google Scholar]
- Beales PL, Badano JL, Ross AJ, Ansley SJ, Hoskins BE, Kirsten B, Mein CA, Froguel P, Scambler PJ, Lewis RA, et al. Genetic interaction of BBS1 mutations with alleles at other BBS loci can result in non-Mendelian Bardet-Biedl syndrome. Am J Hum Genet. 2003;72(5):1187–1199. doi: 10.1086/375178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessant DA, Khaliq S, Hameed A, Anwar K, Mehdi SQ, Payne AM, Bhattacharya SS. A locus for autosomal recessive congenital microphthalmia maps to chromosome 14q32. Am J Hum Genet. 1998;62(5):1113–1116. doi: 10.1086/301843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomhoff R, Green MH, Berg T, Norum KR. Transport and storage of vitamin A. Science. 1990;250(4979):399–404. doi: 10.1126/science.2218545. [DOI] [PubMed] [Google Scholar]
- Bouillet P, Sapin V, Chazaud C, Messaddeq N, Decimo D, Dolle P, Chambon P. Developmental expression pattern of Stra6, a retinoic acid-responsive gene encoding a new type of membrane protein. Mech Dev. 1997;63(2):173–186. doi: 10.1016/s0925-4773(97)00039-7. [DOI] [PubMed] [Google Scholar]
- Busby A, Dolk H, Armstrong B. Eye anomalies: seasonal variation and maternal viral infections. Epidemiology. 2005;16(3):317–322. doi: 10.1097/01.ede.0000158817.43037.ab. [DOI] [PubMed] [Google Scholar]
- Chassaing N, Golzio C, Odent S, Lequeux L, Vigouroux A, Martinovic-Bouriel J, Tiziano FD, Masini L, Piro F, Maragliano G, et al. Phenotypic spectrum of STRA6 mutations: from Matthew-Wood syndrome to non-lethal anophthalmia. Hum Mutat. 2009;30(5):E673–E681. doi: 10.1002/humu.21023. [DOI] [PubMed] [Google Scholar]
- Claessens M, Van Cutsem E, Lasters I, Wodak S. Modelling the polypeptide backbone with 'spare parts' from known protein structures. Protein Eng. 1989;2(5):335–345. doi: 10.1093/protein/2.5.335. [DOI] [PubMed] [Google Scholar]
- Clugston RD, Klattig J, Englert C, Clagett-Dame M, Martinovic J, Benachi A, Greer JJ. Teratogen-induced, dietary and genetic models of congenital diaphragmatic hernia share a common mechanism of pathogenesis. Am J Pathol. 2006;169(5):1541–1549. doi: 10.2353/ajpath.2006.060445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MD, Mao GE. Teratology of retinoids. Annu Rev Pharmacol Toxicol. 1999;39:399–430. doi: 10.1146/annurev.pharmtox.39.1.399. [DOI] [PubMed] [Google Scholar]
- Fantes J, Ragge NK, Lynch SA, McGill NI, Collin JR, Howard-Peebles PN, Hayward C, Vivian AJ, Williamson K, van Heyningen V, et al. Mutations in SOX2 cause anophthalmia. Nat Genet. 2003;33(4):461–463. doi: 10.1038/ng1120. [DOI] [PubMed] [Google Scholar]
- Ferda Percin E, Ploder LA, Yu JJ, Arici K, Horsford DJ, Rutherford A, Bapat B, Cox DW, Duncan AM, Kalnins VI, et al. Human microphthalmia associated with mutations in the retinal homeobox gene CHX10. Nat Genet. 2000;25(4):397–401. doi: 10.1038/78071. [DOI] [PubMed] [Google Scholar]
- Francois J, Haustrate-Gosset MF. Genetic counseling in cases of microphthalmos and anophthalmos. J Genet Hum. 1976;24(Suppl):35–41. [PubMed] [Google Scholar]
- Gal A, Rau I, El Matri L, Kreienkamp HJ, Fehr S, Baklouti K, Chouchane I, Li Y, Rehbein M, Fuchs J, et al. Autosomal-recessive posterior microphthalmos is caused by mutations in PRSS56, a gene encoding a trypsin-like serine protease. Am J Hum Genet. 2011;88(3):382–390. doi: 10.1016/j.ajhg.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo ME, Rodriguez De Cordoba S, Schneider AS, Dwyer MA, Ayuso C, Bovolenta P. Analysis of the developmental SIX6 homeobox gene in patients with anophthalmia/microphthalmia. Am J Med Genet A. 2004;129A(1):92–94. doi: 10.1002/ajmg.a.30126. [DOI] [PubMed] [Google Scholar]
- Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, Maas RL. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7(4):463–471. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- Golzio C, Martinovic-Bouriel J, Thomas S, Mougou-Zrelli S, Grattagliano-Bessieres B, Bonniere M, Delahaye S, Munnich A, Encha-Razavi F, Lyonnet S, et al. Matthew-Wood syndrome is caused by truncating mutations in the retinol-binding protein receptor gene STRA6. Am J Hum Genet. 2007;80(6):1179–1187. doi: 10.1086/518177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham CA, Redmond RM, Nevin NC. X-linked clinical anophthalmos. Localization of the gene to Xq27–Xq28. Ophthalmic Paediatr Genet. 1991;12(1):43–48. doi: 10.3109/13816819109023084. [DOI] [PubMed] [Google Scholar]
- Hmani-Aifa M, Ben Salem S, Benzina Z, Bouassida W, Messaoud R, Turki K, Khairallah M, Rebai A, Fakhfekh F, Soderkvist P, et al. A genome-wide linkage scan in Tunisian families identifies a novel locus for non-syndromic posterior microphthalmia to chromosome 2q37.1. Hum Genet. 2009;126(4):575–587. doi: 10.1007/s00439-009-0688-8. [DOI] [PubMed] [Google Scholar]
- Holder AM, Klaassens M, Tibboel D, de Klein A, Lee B, Scott DA. Genetic factors in congenital diaphragmatic hernia. Am J Hum Genet. 2007;80(5):825–845. doi: 10.1086/513442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornby SJ, Gilbert CE, Rahi JK, Sil AK, Xiao Y, Dandona L, Foster A. Regional variation in blindness in children due to microphthalmos, anophthalmos and coloboma. Ophthalmic Epidemiol. 2000;7(2):127–138. [PubMed] [Google Scholar]
- Hornby SJ, Ward SJ, Gilbert CE, Dandona L, Foster A, Jones RB. Environmental risk factors in congenital malformations of the eye. Ann Trop Paediatr. 2002;22(1):67–77. doi: 10.1179/027249302125000193. [DOI] [PubMed] [Google Scholar]
- Hyatt GA, Dowling JE. Retinoic acid. A key molecule for eye and photoreceptor development. Invest Ophthalmol Vis Sci. 1997;38(8):1471–1475. [PubMed] [Google Scholar]
- Isken A, Golczak M, Oberhauser V, Hunzelmann S, Driever W, Imanishi Y, Palczewski K, von Lintig J. RBP4 disrupts vitamin A uptake homeostasis in a STRA6-deficient animal model for Matthew-Wood syndrome. Cell Metab. 2008;7(3):258–268. doi: 10.1016/j.cmet.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana M, Sharma S. Bilateral anophthalmia with septo-optic dysplasia. Oman J Ophthalmol. 2010;3(2):86–88. doi: 10.4103/0974-620X.64233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsanis N, Ansley SJ, Badano JL, Eichers ER, Lewis RA, Hoskins BE, Scambler PJ, Davidson WS, Beales PL, Lupski JR. Triallelic inheritance in Bardet-Biedl syndrome, a Mendelian recessive disorder. Science. 2001;293(5538):2256–2259. doi: 10.1126/science.1063525. [DOI] [PubMed] [Google Scholar]
- Kawaguchi R, Yu J, Honda J, Hu J, Whitelegge J, Ping P, Wiita P, Bok D, Sun H. A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science. 2007;315(5813):820–825. doi: 10.1126/science.1136244. [DOI] [PubMed] [Google Scholar]
- Kawaguchi R, Yu J, Wiita P, Honda J, Sun H. An essential ligand-binding domain in the membrane receptor for retinol-binding protein revealed by large-scale mutagenesis and a human polymorphism. J Biol Chem. 2008a;283(22):15160–15168. doi: 10.1074/jbc.M801060200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R, Yu J, Wiita P, Ter-Stepanian M, Sun H. Mapping the membrane topology and extracellular ligand binding domains of the retinol binding protein receptor. Biochemistry. 2008b;47(19):5387–5395. doi: 10.1021/bi8002082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch CC, Zaghloul NA, Davis EE, Stoetzel C, Diaz-Font A, Rix S, Alfadhel M, Lewis RA, Eyaid W, Banin E, et al. Hypomorphic mutations in syndromic encephalocele genes are associated with Bardet-Biedl syndrome. Nat Genet. 2008;40(4):443–448. doi: 10.1038/ng.97. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh-Armstrong N, McCaffery P, Gilbert W, Dowling JE, Drager UC. Retinoic acid is necessary for development of the ventral retina in zebrafish. Proc Natl Acad Sci U S A. 1994;91(15):7286–7290. doi: 10.1073/pnas.91.15.7286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morle L, Bozon M, Zech JC, Alloisio N, Raas-Rothschild A, Philippe C, Lambert JC, Godet J, Plauchu H, Edery P. A locus for autosomal dominant colobomatous microphthalmia maps to chromosome 15q12–q15. Am J Hum Genet. 2000;67(6):1592–1597. doi: 10.1086/316894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison D, FitzPatrick D, Hanson I, Williamson K, van Heyningen V, Fleck B, Jones I, Chalmers J, Campbell H. National study of microphthalmia, anophthalmia, and coloboma (MAC) in Scotland: investigation of genetic aetiology. J Med Genet. 2002;39(1):16–22. doi: 10.1136/jmg.39.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nau H. Teratogenicity of isotretinoin revisited: species variation and the role of all-trans-retinoic acid. J Am Acad Dermatol. 2001;45(5):S183–S187. doi: 10.1067/mjd.2001.113720. [DOI] [PubMed] [Google Scholar]
- Nau H, Chahoud I, Dencker L, Lammer EJ, Scott WJ. Teratogenicity of Vitamin A and Retinoids. Vitamin A in Health and Disease: Marcel Dekker, Inc. 1994 [Google Scholar]
- Pasutto F, Sticht H, Hammersen G, Gillessen-Kaesbach G, Fitzpatrick DR, Nurnberg G, Brasch F, Schirmer-Zimmermann H, Tolmie JL, Chitayat D, et al. Mutations in STRA6 cause a broad spectrum of malformations including anophthalmia, congenital heart defects, diaphragmatic hernia, alveolar capillary dysplasia, lung hypoplasia, and mental retardation. Am J Hum Genet. 2007;80(3):550–560. doi: 10.1086/512203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadro L, Hamberger L, Gottesman ME, Wang F, Colantuoni V, Blaner WS, Mendelsohn CL. Pathways of vitamin A delivery to the embryo: insights from a new tunable model of embryonic vitamin A deficiency. Endocrinology. 2005;146(10):4479–4490. doi: 10.1210/en.2005-0158. [DOI] [PubMed] [Google Scholar]
- Ragge NK, Brown AG, Poloschek CM, Lorenz B, Henderson RA, Clarke MP, Russell-Eggitt I, Fielder A, Gerrelli D, Martinez-Barbera JP, et al. Heterozygous mutations of OTX2 cause severe ocular malformations. Am J Hum Genet. 2005a;76(6):1008–1022. doi: 10.1086/430721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragge NK, Lorenz B, Schneider A, Bushby K, de Sanctis L, de Sanctis U, Salt A, Collin JR, Vivian AJ, Free SL, et al. SOX2 anophthalmia syndrome. Am J Med Genet A. 2005b;135(1):1–7. doi: 10.1002/ajmg.a.30642. discussion 8. [DOI] [PubMed] [Google Scholar]
- Schimmenti LA, de la Cruz J, Lewis RA, Karkera JD, Manligas GS, Roessler E, Muenke M. Novel mutation in sonic hedgehog in non-syndromic colobomatous microphthalmia. Am J Med Genet A. 2003;116A(3):215–221. doi: 10.1002/ajmg.a.10884. [DOI] [PubMed] [Google Scholar]
- Smith FR, Goodman DS. Vitamin A transport in human vitamin A toxicity. N Engl J Med. 1976;294(15):805–808. doi: 10.1056/NEJM197604082941503. [DOI] [PubMed] [Google Scholar]
- Stoetzel C, Laurier V, Faivre L, Megarbane A, Perrin-Schmitt F, Verloes A, Bonneau D, Mandel JL, Cossee M, Dollfus H. BBS8 is rarely mutated in a cohort of 128 Bardet-Biedl syndrome families. J Hum Genet. 2006;51(1):81–84. doi: 10.1007/s10038-005-0320-2. [DOI] [PubMed] [Google Scholar]
- Stoll C, Alembik Y, Dott B, Roth MP. Congenital eye malformations in 212,479 consecutive births. Ann Genet. 1997;40(2):122–128. [PubMed] [Google Scholar]
- Sundin OH, Leppert GS, Silva ED, Yang JM, Dharmaraj S, Maumenee IH, Santos LC, Parsa CF, Traboulsi EI, Broman KW, et al. Extreme hyperopia is the result of null mutations in MFRP, which encodes a Frizzled-related protein. Proc Natl Acad Sci U S A. 2005;102(27):9553–9558. doi: 10.1073/pnas.0501451102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassabehji M, Fang ZM, Hilton EN, McGaughran J, Zhao Z, de Bock CE, Howard E, Malass M, Donnai D, Diwan A, et al. Mutations in GDF6 are associated with vertebral segmentation defects in Klippel-Feil syndrome. Hum Mutat. 2008;29(8):1017–1027. doi: 10.1002/humu.20741. [DOI] [PubMed] [Google Scholar]
- Tsuzuki D, Hichiya H, Okuda Y, Yamamoto S, Tamagake K, Shinoda S, Narimatsu S. Alteration in catalytic properties of human CYP2D6 caused by substitution of glycine-42 with arginine, lysine and glutamic acid. Drug Metab Pharmacokinet. 2003;18(1):79–85. doi: 10.2133/dmpk.18.79. [DOI] [PubMed] [Google Scholar]
- Verma AS, Fitzpatrick DR. Anophthalmia and microphthalmia. Orphanet J Rare Dis. 2007;2:47. doi: 10.1186/1750-1172-2-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina VA, Kozhemyakina EA, O'Kernick CM, Kahn ND, Wenger SL, Linberg JV, Schneider AS, Mathers PH. Mutations in the human RAX homeobox gene in a patient with anophthalmia and sclerocornea. Hum Mol Genet. 2004;13(3):315–322. doi: 10.1093/hmg/ddh025. [DOI] [PubMed] [Google Scholar]
- West B, Bove KE, Slavotinek AM. Two novel STRA6 mutations in a patient with anophthalmia and diaphragmatic eventration. Am J Med Genet A. 2009;149A(3):539–542. doi: 10.1002/ajmg.a.32682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SH, Wimley WC. Membrane protein folding and stability: physical principles. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- White T, Lu T, Metlapally R, Katowitz J, Kherani F, Wang TY, Tran-Viet KN, Young TL. Identification of STRA6 and SKI sequence variants in patients with anophthalmia/microphthalmia. Mol Vis. 2008;14:2458–2465. [PMC free article] [PubMed] [Google Scholar]
- Wilson JG, Roth CB, Warkany J. An analysis of the syndrome of malformations induced by maternal vitamin A deficiency. Effects of restoration of vitamin A at various times during gestation. Am J Anat. 1953;92(2):189–217. doi: 10.1002/aja.1000920202. [DOI] [PubMed] [Google Scholar]
- Ye M, Berry-Wynne KM, Asai-Coakwell M, Sundaresan P, Footz T, French CR, Abitbol M, Fleisch VC, Corbett N, Allison WT, et al. Mutation of the bone morphogenetic protein GDF3 causes ocular and skeletal anomalies. Hum Mol Genet. 2010;19(2):287–298. doi: 10.1093/hmg/ddp496. [DOI] [PubMed] [Google Scholar]
- Yeyati PL, Bancewicz RM, Maule J, van Heyningen V. Hsp90 selectively modulates phenotype in vertebrate development. PLoS Genet. 2007;3(3):e43. doi: 10.1371/journal.pgen.0030043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenteno JC, Perez-Cano HJ, Aguinaga M. Anophthalmia-esophageal atresia syndrome caused by an SOX2 gene deletion in monozygotic twin brothers with markedly discordant phenotypes. Am J Med Genet A. 2006;140(18):1899–1903. doi: 10.1002/ajmg.a.31384. [DOI] [PubMed] [Google Scholar]
- Zile MH. Function of vitamin A in vertebrate embryonic development. J Nutr. 2001;131(3):705–708. doi: 10.1093/jn/131.3.705. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.