Abstract
Experimental studies indicate significant cardioprotective effects of recombinant erythropoietin (Epo) by binding to the Epo receptor (EpoR) and by inducing various molecular mechanisms, including activation of Gata4, a transcription factor that induces anti-apoptotic genes. However, specific molecular mechanisms of EpoR regulation in cardiomyocytes are unknown. We identified a 774 bp regulatory domain in the EpoR 5′ flanking region by reporter gene assays in murine HL-1 cardiomyocytes. The binding sites for Gata and Sp transcription factors both significantly contributed to EpoR promoter activity. DNA-binding studies (EMSA and ChIP assays) identified Gata4 and Sp1 as EpoR promoter-binding proteins in HL1 cardiomyocytes. Although Sp1 alone stimulates EpoR only slightly, forced expression of Gata4 significantly induced EpoR mRNA expression. In addition, knockdown of Gata4 (but also of Sp1) resulted in a significant decrease of EpoR transcript levels in HL-1 cardiomyocytes. Cumulative in vitro data suggest that function of the Sp1 site is essential for the Gata4-mediated transcription. In vivo, analysis of transgenic mice expressing an inducible small-hairpin RNA against Gata4 confirmed suppression of EpoR expression in the heart. Treating mice with high-dose doxorubicin not only resulted in Gata4 protein depletion, but also down-regulated EpoR, followed by up-regulation of EpoR transcripts when Gata4 levels recovered. In conclusion, we identified Gata4 as novel regulator of EpoR transcription in cardiomyocytes. In models of cardiac injury, down-regulation of Gata4 or Sp1 may limit the accessibility of the EpoR for binding of erythropoiesis-stimulating agents (ESA). Thereby our data underline the essential role of Gata4 in mediating cardioprotective effects.
Keywords: cardioprotection, erythropoietin, erythropoietin receptor, GATA4, transcription
Introduction
Animal studies indicate that recombinant erythropoietin (rEpo) or other erythropoiesis-stimulating agents (ESA) exert cardioprotective effects in ischaemia-reperfusion injury and also in non-ischaemic cardiac dysfunction [1, 2]. The in vivo effects of rEpo include enhanced cardiac functional recovery, better left ventricular contractility, decreased infarct size, suppressed myocardial inflammation, reduced apoptosis and decreased remodelling. Although the underlying molecular mechanisms are not entirely clear, recent data indicate that rEpo restores protein levels of the cardiac transcription factor Gata4 in cardiomyocytes treated with doxorubicin [3], a frequently used anthracycline causing cardiomyopathy in cancer treatment, or after cardiac ischaemic-reperfusion injury [4]. Gata4 regulates genes that are relevant for proper cardiomyocyte integrity and function, such as α-myosin heavy chain (α-Mhc), β-Mhc, troponin, atrial natriuretic factor (Anf) and bone morphogenetic protein-4 (Bmp4) [5, 6]. Because Gata4 exhibits direct anti-apoptotic function by regulating genes of the bcl-XL family and other factors [4, 7], rEpo appears to be an attractive pharmaceutical substance to prevent or even to treat cardiomyopathy via Gata4 restoration [8, 9]. However, rEpo or its derivates have not been tested clinically for anthracycline-induced cardiomyopathy, and the first clinical data on rEpo for the protection against ischaemia-reperfusion heart injury are rather disappointing if compared to experimental data [10]. Because Epo’s actions require binding to the extracellular domain of the Epo receptor (EpoR), the question on the regulation of the EpoR gene in cardiomyocytes is of particular interest.
The first evidence for an essential role of Epo and its receptor in the heart resulted from the analysis of transgenic mice with homozygous deletion of the Epo (Epo−/−) or the EpoR (EpoR−/−) genes [11, 12]. Both Epo−/− and EpoR−/− embryos suffer from ventricular hypoplasia, epicardium detachment and vascular abnormalities [11]. In the murine heart, EpoR is expressed in a temporal and cell type-specific manner. From mid-gestation onwards, EpoR expression has been detected in foetal, neonatal and adult cardiomyocytes [13–16]. Organ cultures or primary cell cultures from embryonic heart showed that rEpo acts as mild mitogen for cardiomyocytes [11]. In EpoR−/− mice that have been rescued from the lethal haematopoietic defect, the endogenous EpoR system is relevant for protection against pressure-overload induced cardiac dysfunction and for protection against myocardial ischemia/reperfusion injury [17, 18]. Although low Epo transcript levels have been detected in the murine embryonic and human foetal heart [11, 14], significant Epo expression in cardiomyocytes was not observed under normoxia or hypoxia [15]. This supports the hypothesis that the EpoR has the major implication in mediating the effects of endogenous Epo in cardiac morphogenesis and of rEpo for cardioprotection, even if the EpoR number is low under normal conditions.
The regulatory mechanisms of EpoR expression in cardiomyocytes in health and disease are unknown yet. In haematopoietic cells, transcriptional regulation of EpoR gene is controlled by cis-acting elements [19]. Earlier studies showed that the transcription factors Gata1, Sp1 and Wt1 activate EpoR gene expression in haematopoietic cells by binding to a 452 bp minimal EpoR promoter element [20–23]. However, Gata1 expression is restricted to haematopoietic progenitor cells and Sertoli cells [24]. The analysis of EpoR gene expression in the heart of Wt1-deficient embryos revealed that the transcriptionally active Wt1 protein was not involved in cardiac EpoR expression [23]. Because the transcriptionally active Wt1 protein is expressed exclusively in the epicardium [25], however, further experiments may be required to test whether Wt1 affects EpoR expression in cardiogenic progenitors of the epicardium that are involved in cardiac regeneration.
To improve the translation of experimental work on cardioprotection by rEpo or ESA into future clinical strategies, we aimed to elucidate the regulation of EpoR gene expression in cardiomyocytes. Herein, we provide the first evidence that Gata4 specifically activates EpoR expression in cardiomyocytes.
Material and methods
Cell culture
The murine cardiomyocyte cell line HL-1 was cultured as described [26].
Animal experiments
The investigations conform to the Guide for the Care and Use of Laboratory Animals, published by the US National Institutes of Health (NIH Publication No. 85–23, revised 1996).
Doxorubicin-induced cardiomyopathy
Cardiomyopathy was induced by a single intraperitoneal injection of 15 mg/kg doxorubicin-HCl (Merck, Darmstadt, Germany) in 10-weeks-old male C57BL/6J mice (n = 9). In sham-treated mice (n = 6), the equal volume of saline was intraperitoneally injected. Mice were sacrificed after 24 hrs (n = 6) or 5 days (n = 3), respectively, and hearts were isolated for RNA and protein preparation (IRB Protocol LaGeSo Berlin G0224/06).
Transgenic mice expressing short hairpin RNA (shRNA) against Gata4
The transgenic mouse line (H1:G4/TetR), exhibiting an inducible shRNA construct against Gata4, has been recently developed by us [27]. To induce expression of Gata4 shRNA via the TetR system, mice (n = 4) were fed with 20 mg doxycycline per ml drinking water. The control group (n = 4) was fed with doxycycline at the same concentration. After 38 days mice were sacrificed, and hearts were taken for analysis (IRB Protocol D669, University of Florida, USA).
RT-PCR
For RNA preparation, tissue specimens were homogenized in TRIzol® (Invitrogen, Karlsruhe, Germany) reagent. RNA was isolated according to the manufacturer’s protocol. Subsequently, cDNA was synthesized and transcripts of interest were amplified by using specific primers (Table 1).
Table 1.
Sequences of primers for RT-PCR analysis in murine tissue specimens and HL-1 cells
| Gene | Accession no. | Forward primer | Reverse primer | Product size (bp) |
|---|---|---|---|---|
| β-actin | NM_007393.2 | actgctctggctcctagcac | acatctgctggaaggtggac | 115 |
| EpoR | NM_010149 | agcccgcgtcactactgacc | gtcctgggcatgctcactgc | 386 |
| Epo | NM_007942 | ctgggagctcagaaggaattgatg | ctggagtgtccatgggacagactg | 340 |
| Gata1 | NM_008089 | gagtccagacctcctgacgc | cagtggagtagccgttgctc | 220 |
| Gata4 | NM_008092 | gatgggacgggacactacctg | acctgctggcgtcttagattt | 288 |
| Gata6 | NM_010258 | gcacgctttccctactcgc | cgagtaggtcgggtgatgg | 222 |
| Fog2 | NM_011766 | gcaagctcagacacagagc | ccagctcggacatcttcgc | 276 |
| Sp1 | NM_013672 | ggatgaagtgacagctgtgg | gaagagatgatctgccagcc | 318 |
| Wt1 | NM_144783 | agagccagcctaccatccgc | ggctgcctgtgcaactgtca | 221 |
| α-Mhc | NM_010856.3 | ttagctggaagatcacccgg | aagtcagagaaggaacgcctaga | 783 |
All sequences are shown in 5′–3′ direction.
Quantitative real-time PCR (Q-PCR)
For Q-PCR, 5’-FAM-labelled probes of murine Gata4, EpoR (TaqMan Gene Expression Assays; Mm00484689, Mm00833882) and β-actin (4352933E) from Applied Biosystems (Foster City, CA, USA) were used. PCR reactions were performed as described [28]. The threshold value was calculated using the iCycle iQ Optical System Software, version 3.1. Data were normalized against β-actin.
Plasmids and site-directed mutagenesis
Firefly luciferase reporter gene constructs were generated by cloning various EpoR promoter fragments into the pGL2-basic vector (Promega, Mannheim, Germany). For preliminary experiments, a 5 kb fragment of the 5’-flanking region of the murine EpoR gene (NCBI accession no. NC_000075) was cloned, spanning the sequence from nucleotide −4900 to +100 relative to the transcription start site. This fragment was shortened to generate two additional constructs containing the either an 1842 bp fragment (−1742 to +100) and or a 774 bp fragment (−674 to +100; Fig. S1). The latter construct, named full-length mEpoRpro, was used for detailed analysis. Potential binding sites for transcription factors were mutated within the pGL2-mEpoRpro construct by using a site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA). For details and primer sequences see ‘Supporting information’.
The murine Gata4 cDNA (NM_008092; bp 608–1933) was cloned into the EcoRI site of pcDNA3.1(+) vector (Invitrogen). A XhoI/EcoRI fragment containing the open reading frame of Sp1 cDNA (NM_013672; bp 101–2446) was cloned into the corresponding site of pcDNA3.1(−). The pCMVTag3B/Gata-6 vector was kindly provided by Edward Morrisey [29]. pBS/U6-shScram and pBS/U6-shSp1–2 vectors were kindly provided by Grace Gill [30].
Reporter gene assays
Four micrograms of each reporter gene construct and 20 ng of the Renilla expression plasmid phRL-TK were transiently co-transfected into HL-1 cells performed with Lipofectamine™ 2000 (Invitrogen). After 24 hrs cells were lysed and the luciferase activities were measured (Dual-Luciferase Reporter Assay System, Promega). Relative light units (RLU) of firefly luciferase were normalized against Renilla luciferase-activities and protein concentrations. The amount of total protein was determined using Bradford reagent.
Nuclear extracts, oligo-labelling and electrophoretic mobility shift assay (EMSA)
EMSA oligonucleotides of the EpoR promoter region (NCBI accession no. NC_000075; −74/−45 GATA 5′-gttcccttggtggctgtagcttatctgtcc-3′; −44/−14 GC-rE 5′-ccagcctgcaagctggccccgccccctgga-3′) were end-labelled with 32P and subsequently incubated with 5 μl of HL-1 nuclear extract. Where indicated, the GATA-4, GATA-6 or Sp1 antibodies (all from Santa Cruz Biotechnology, Santa Cruz, CA, USA) were added.
Chromatin immunoprecipitation (ChIP)
ChIP assays were performed with freshly prepared cardiomyocytes from three male 10-week-old C57/BL6 mice by using the ChIP Assay Kit (Millipore, Schwalbach/Ts, Germany) according to manufacturer’s instructions. See ‘Supporting information’ for technical details.
Forced expression of transcription factors
For transient overexpression, 4 μg of each plasmid were transfected into HL-1 cells performed with Lipofectamine™ 2000. After 48 hrs, RNA was extracted, and cDNA was synthesized. The effects of overexpression on EpoR mRNA abundance were quantified by Q-PCR.
Gata4 and Sp1 knockdown in HL-1 cells
pBS/U6-shSp1 or Gata4 siRNA duplexes were transfected into HL-1 performed with Lipofectamine™ 2000. Forty-eight hours past transfection, RNA and protein were extracted and analysed for EpoR mRNA or Sp1 and Gata4 protein, respectively. See ‘Supporting information’ for details.
Western blot analyses
Proteins were extracted from HL-1 cells or murine heart were performed as previously described [28]. The following primary antibodies were used: GATA-4, GATA-6, Sp1 (Santa Cruz Biotechnology) and fl-tubulin (Promega). Signals were visualized using Western Blot ECL reagent (Pierce, Rockford, IL, USA).
Statistical analysis
Data are presented as mean plus S.D. ANOVA with Bonferroni test as post hoc test calculations and Student’s t-test were performed as indicated to reveal statistical significances. P-values < 0.05 were considered statistically significant.
Results
Regulatory elements upstream of the murine EpoR gene
Considering previous studies that identified regulatory domains in the upstream region of the EpoR gene, we first analysed a 5 kb fragment, spanning from the nucleotide −4900 to +100 relative to the transcription start site of the 5’-flanking region of the murine EpoR gene (Fig. S1A). This reporter gene includes the minimal EpoR promoter element as well as an enhancer and an inhibitor element that have previously been identified in haematopoietic progenitors, bone marrow stroma cells or fibroblasts [31, 32]. Removing the 5′-flanking enhancer and inhibitor sequence, which are regulated in a lineage- and temporal-specific manner, led to an increase in reporter gene activity. The shortened 774 bp fragment retained an E-box and the minimal haematopoietic EpoR promoter (Fig. S1B). In the subsequent experiments, we further dissected the 774 bp fragment of the murine EpoR gene (Fig. 1A) [19, 20]. This 774 bp fragment (mEpoRpro), spanning from nucleotides (nt) −674 to +100 relative to the transcription start site (Fig. 1B), contains various putative transcription factor binding sites, including one E-box, several CACCC-motifs, one GATA site and a proximal GC-rich element (GC-rE). For reporter gene analysis, mEpoR promoter constructs were transiently transfected into HL-1 cardiomyocytes. Compared to the full-length mEpoRpro, the truncated reporter construct Δ1mEpoRpro (nt-234/+100) showed a reduction of activity to 60 ± 9.7%, and a further truncation (Δ2mEpoRpro, nt-44/+100) reduced reporter gene activity to 42 ± 2.2%. Δ3mEpoRpro (nt-8/+100), which contained only the 5′-untranslated region, showed very low activity (11 ± 1.9%) compared to mEpoRpro, and was similar to the empty pGL2 basic vector (7 ± 0.8%; Fig. 1C). Mutations of the GC-rE resulted in a significantly reduced activity, compared to that of the full-length mEpoRpro reporter gene (down to 20 ± 6.4%). Mutation of the GATA-site showed a moderate reduction in luciferase activity (down to 72 ± 18.6%), whereas mutations of the E-box or various CACCC-motifs had no effect (Fig. 1C). The combined data highlight the functional implication of the 774 bp fragment and particularly of its GATA-site and GC-rich elements in mediating EpoR promoter activity.
Fig 1.
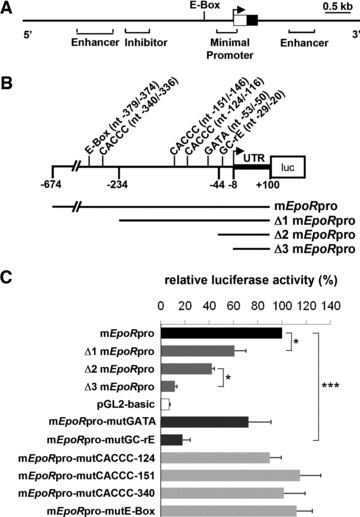
(A) Diagram of the murine EpoR gene locus highlighting the upstream promoter region. The open box displays the 5′-untranslated region and the filled box represents exon 1. Known enhancer and inhibitory elements are indicated [19, 47]. The transcription start site is marked by an arrow. (B) Location of putative cis-regulatory elements in the analysed 774 bp EpoR promoter element and constructs used for reporter gene assays. Numbers indicate the position of the nucleotides (nt) relative to the transcription start site and refer to various lengths of the reporter gene constructs. mEpoRpro represents the 774 bp murine EpoR promoter cloned into the luciferase reporter plasmid pGL2. The truncated promoter regions ΔmEpoRpro, Δ2mEpoRpro and Δ3mEpoRpro are illustrated. Specific DNA-binding motifs for transcription factors are indicated with the nucleotide position numbers: one E-box, three CACCC motifs, the GATA element and one GC-rich element, which represents putative binding sites for Sp1 and Wt1. (C) Reporter gene activities after transfection of wild-type or mutant promoter constructs into HL-1 cells. The full-length EpoR promotor construct (mEpoRpro) and the three truncated forms were compared to the pGL2-basic reporter construct alone. The specific implication of the DNA-binding motifs of interest has been analysed by site-directed mutangenesis of the GATA element, the GC-rich elements, the CACCC motifs and the E-box. Firefly luciferase activities were normalized to the corresponding Renilla luciferase values and protein concentrations (n = 3 independent experiments; ***P < 0.001; *P < 0.05).
Expression pattern of candidate factors for EpoR regulation in the murine heart and HL-1 cardiomyocytes
Next, we analysed the developmental expression profile of Epo, EpoR and transcription factors that could potentially bind to the EpoR promoter as annotated by TRANSFAC analysis or based on previous data on EpoR regulation in haematopoietic cells [20–23, 32, 33]. Although Epo gene expression was silenced in the developing and adult heart, EpoR transcripts were detected at all stages (Fig. S2). Heart tissue specimens were negatively tested for Gata1 mRNA. Similarly, Wt1 mRNA, which is only expressed at low levels in the epicardium [34], was not detectable. Among factors regulating EpoR expression in haematopoietic cells, Sp1 was expressed throughout development and adulthood. As expected according to the literature [35], transcripts of Gata4, Gata6 and their co-factor friend-of-GATA 2 (Fog2) were detected in each specimen. Notably, murine HL-1 cardiomyocytes revealed the same expression profile for the genes expressed in the wild-type murine heart (Fig. S2), suggesting that this cell line is useful and appropriate for later in vivo experiments.
In vitro and in vivo binding of Gata4 and Sp1 to the EpoR promoter in cardiomyocytes
To further explore whether Gata4, Gata6 or Sp1 bind to the EpoR promoter, EMSA experiments were carried out with nuclear extracts from HL-1 cells and end-labelled oligonucleotides, which contain regulatory elements of the EpoR promoter. By using antibodies, we identified a Gata4, but not Gata6-specific protein/DNA complex, as well as a Sp1-specific protein/DNA complex (Fig. 2A). Of note, the Sp1-specific complex was abolished, if the oligonucleotide that contains only the GC-rich element, but not the GATA-site was incubated with an antibody against Gata4 (Fig. 2A). Thus, Gata4 may be a component of the Sp1-specific complex at the GC-rE. ChIP experiments confirmed that Gata4 as well as Sp1 bind to the EpoR promoter in fresh primary cardiomyocytes (Fig. 2B).
Fig 2.
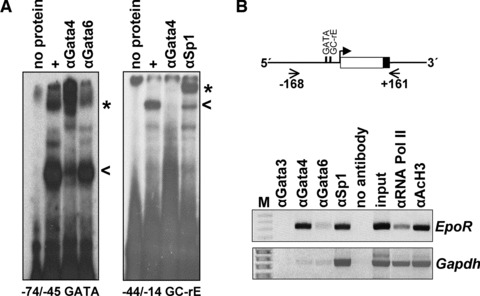
Analysis of Gata4 and Sp1 binding to the murine EpoR promoter. (A) Electrophoretic mobility shift assays demonstrate binding of Gata4 (probe nt-74/−45 GATA) and Sp1 (probe nt-44/−14 GC-rE) to DNA elements of the murine EpoR promoter. Asterisks indicate specific Gata4 and Sp1-mediated supershifts by antibodies directed against Gata4 (αGata4), Gata6 (αGata6) and Sp1 (αSp1). Of note, incubation with an antibody directed against Gata4 totally abolishes the specific complex generated by binding of Sp1 to the −44/−14 GC-rE. (B) Chromatin-immunoprecipitation assays with the EpoR minimal promoter were performed in freshly prepared cardiomyocytes (male C57/BL6). Immunoprecipitation with an antibody against Gata3 (αGata3) served as negative control, whereas that with the antibodies against RNA polymerase II (αRNA Pol II) and against acetylated histon 3 (αAcH3) as positive controls indicate active transcription of the EpoR gene. Precipitated DNA was amplified by PCR with primers spanning the EpoR promoter (nt-168/+161). Amplification of the Gapdh promoter served as control. Gel pictures were inverted.
Enhanced EpoR mRNA expression by forced Gata4 expression
The functional implication of Gata4 and Sp1 binding to the EpoR promoter was tested by transient transfection of HL-1 cells. Forty-eight hours after transient Gata4 overexpression a 3-fold stimulation of endogenous EpoR expression was found (P < 0.05; Fig. 3A), while forced expression of Gata6 had no effect. In contrast to data in haematopoietic cells [22], Sp1 overexpression resulted only in a slight, but not significant increase of EpoR mRNA levels in HL-1 cells. Co-transfection of Sp1 with either Gata4 or Gata6 plasmids did also not significantly (further) enhance EpoR expression (Fig. 3A). These data clearly support the role of Gata4 as a positive regulator of EpoR expression in cardiomyocytes. Unfortunately, the analysis of EpoR protein expression could not be performed due to non-specific cross-reactivity of the currently available EpoR antibodies [36, 37].
Fig 3.
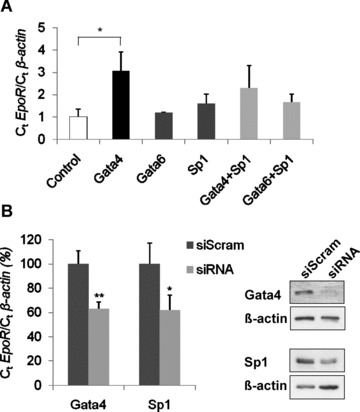
Influence of Gata4 and Sp1 overexpression or suppression on endogenous EpoR expression levels. (A) Increase of EpoR mRNA by forced expression of Gata4. Transcript levels of EpoR and β-actin were quantified by Q-PCR 48 hrs after transient transfection of Gata4, Gata6 or Sp1 (alone or combined) in HL-1 cells. (B) Decrease of EpoR mRNA by Gata4 and Sp1 knockdown. HL-1 cells were transfected with Gata4 siRNA oligonucleotides and Sp1 short hairpin vector, respectively. EpoR values were normalized to β-actin (n = 3 independent experiments; **P < 0.01; *P < 0.05). Analysis of EpoR protein expression could not be performed due to non-specific cross reactivity of currently available EpoR antibodies [36, 37]. Gata4 and Sp1 knockdowns were verified by immunoblot.
Decreased EpoR mRNA expression in Gata4 and Sp1 siRNA experiments
To further study the functional implications of Gata4 and Sp1 on EpoR gene regulation in HL-1 cells, we performed knockdown experiments with siRNA. For Gata4 knockdown experiments, scrambled or Gata4 siRNA oligonucleotides were transfected into HL-1 cells. Forty-eight hours later, protein and RNA were analysed for Gata4 and EpoR expression levels. Western blot analysis showed a distinct reduction of Gata4, which is associated with a significant (P < 0.01) decrease of EpoR mRNA to 63 ± 6% (Fig. 3B). For Sp1 knockdown, we transfected a short hairpin Sp1 plasmid or the corresponding scramble control. The Sp1 knockdown was verified by Western blot. Q-PCR analysis showed a clear reduction in EpoR mRNA to 55 ± 13% (P < 0.05) upon Sp1 knockdown (Fig. 3B). Taken together, these experiments underline the implication of Gata4 and Sp1 in regulating the EpoR gene in HL-1 cardiomyocytes.
Down-regulation of cardiac EpoR expression in transgenic mice with inducible RNA interference directed against Gata4
To study the effect of Gata4 knockdown on endogenous EpoR expression in vivo, we analysed transgenic mice with a stably integrated shRNA against Gata4 under control of the TetR system [27]. To induce the expression of Gata4 shRNA, transgenic mice were fed with doxycycline over 38 days, because this resulted in the most efficient, significant down-regulation of endogenous Gata4 mRNA expression to 10 ± 4.5% of the level in control mice (P < 0.01). In our experiments, reduction of Gata4 protein levels was associated with the down-regulation of selective Gata4-downstream targets, such as the genes encoding Anf and Bmp4, but not αMHC, and is shown elsewhere [27]. Data from us and others (using conditional cardiac Gata4 deletion) indicated that under non-stress conditions Gata4 depletion of up to 80% of normal levels did not result in cardiac failure [6, 27]. However, down-regulation of Gata4 expression was associated with a significant reduction of EpoR mRNA expression to 20 ± 10% of normal levels (Fig. 4). In addition, Gata6 levels remained normal in transgenic mice with inducible Gata4 shRNA [27], confirming our data from DNA-binding analysis and overexpression analysis which indicate that Gata6 is not involved in cardiac EpoR regulation (Figs 2 and 3).
Fig 4.
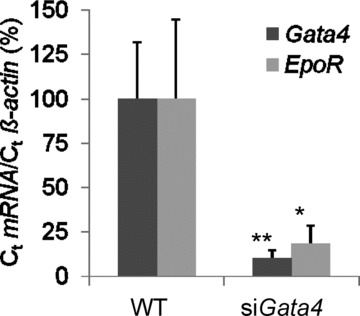
Inhibition of cardiac EpoR mRNA expression in transgenic mice with short hairpin RNA against Gata4. Transgenic mice with a stably integrated, doxycycline-inducible Gata4 shRNA construct and FvB wild-type mice (control) were fed with 20 mg doxycycline per ml drinking water over 38 days. Endogenous Gata4 and EpoR expression levels were quantified by Q-PCR and normalized to β-actin. All samples were measured in duplicates according to the threshold cycle method (**P < 0.01; *P < 0.05).
Gata4 depletion in doxorubicin-induced cardiomyopathy is associated with decreased EpoR expression
In a more physiological and clinical relevant approach, we studied the role of Gata4 in regulating cardiac EpoR expression in a mouse model of doxorubicin-induced cardiomyopathy. This model is characterized by Gata4 depletion accompanied with cardiomyocyte atrophy, degeneration and myocardial fibrosis [3]. After a single intraperitoneal injection of 15 mg/kg doxorubicin hydrochloride or the equivalent volume of saline for controls, animals were sacrificed 24 hrs or 5 days later. Cardiac Gata4, Gata6 and Sp1 levels were analysed by Western blot analysis. EpoR levels were quantified by Q-PCR (Fig. 5A). Of note, 24 hrs after single high-dose doxorubicin injection, Gata4 was depleted, but recovered to almost normal levels after day 5. Coincidentally with Gata4 depletion, EpoR expression significantly decreased (to 19 ± 18% of normal levels) within the first 24 hrs. After 5 days, EpoR mRNA expression recovered in parallel to Gata4 to almost normal levels. Again, Gata6 and Sp1 protein level were unaffected, whereas suppression and later recovery of α-Mhc mRNA expression served as bona fide control for the specific Gata4 effect (Fig. 5B and C). These in vivo data also indicate that Gata4 specifically activates EpoR expression and may render cardiomyocytes more responsive to rEpo or other ESA.
Fig 5.
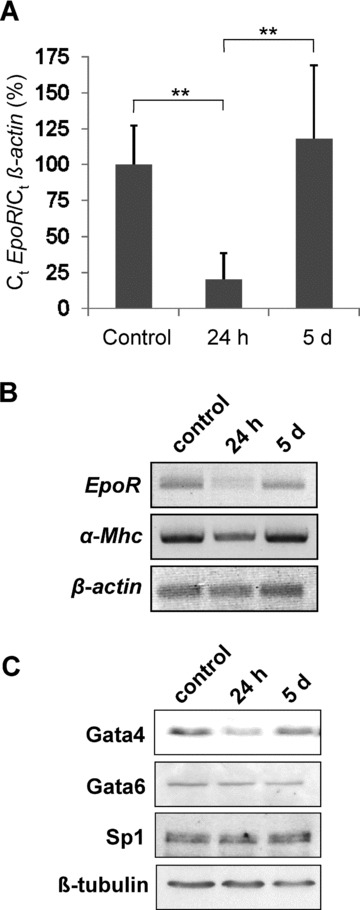
EpoR expression levels in a mouse model of doxorubicin-induced cardiomyopathy following depletion and recovery of Gata4. Cardiomyopathy was induced in 10-week-old male C57BL/6J mice by a single intraperitoneal injection of 15 mg/kg doxorubicin hydrochloride. In sham-treated mice, the equivalent volume of saline was injected. (A) Q-PCR analysis of EpoR expression, normalized to β-actin. All samples were measured in duplicates (**P < 0.01). (B) Analysis of α-Mhc expression as Gata4 downstream target served as bona fide control for the specificity of the effect. Photographies of agarose gels were inverted. (C) Immunoblot of Gata4, Gata6, Sp1 and β-tubulin.
Discussion
Herein, we identified Gata4 as a novel transcriptional regulator of the EpoR gene in cardiomyocytes. This is supported by several findings: Gata4 specifically binds to the EpoR promoter in vitro (EMSA) and in living cells (ChIP Assay; Fig. 2). Furthermore, mutation of the GATA-motif results in a reduced activity of the EpoR promoter (Fig. 1). The combined data from the reporter gene analysis indicate that additional cis-regulatory elements are involved in regulating EpoR promoter activity (Fig. 1). Our data also confirm that DNA elements in the 5′-flanking region of the murine EpoR gene are clearly regulated by both lineage- and temporal-specific mechanisms (Fig. S1) [31]. We also show that Sp1 specifically binds to a GC-rich element within the EpoR promoter in cardiomyocytes (Fig. 2). Although forced expression of Sp1 only slightly stimulates endogenous EpoR expression, Gata4 significantly enhances EpoR mRNA expression (Fig. 3). In an opposite approach, knockdown of Gata4, but also of Sp1 was associated with a significant down-regulation of EpoR mRNA expression (Figs 3 and 4). The high expression of Sp1 in HL-1 cardiomyocytes may explain that forced overexpression of Sp1 did not result in a more pronounced induction of EpoR mRNA levels. Of note, Gata4 is known to interact physically with Sp1 [38], and EMSA experiments indeed indicate that Gata4 binds to the Sp1-specific complex at the GC-rE in the minimal EpoR promoter (Fig. 2A). This suggests that accessibility of Gata4 may be important for Sp1 to activate EpoR expression or vice versa. In haematopoietic progenitor cells, synergistic effects of Sp1 and Gata1 have previously been proposed for EpoR regulation and may also depend on the stage of cellular differentiation [22, 23]. In neuronal cells, Gata2 and Gata3 bind to the same GATA-motif in the EpoR promoter, but this does not result in a significant induction of EpoR expression, neither alone nor in combination with Sp1 [28]. Our in vitro data therefore indicate that regulation of EpoR expression by Gata4 in cardiomyocytes is a very specific mechanism that differs from EpoR regulation in haematopoietic and neuronal cells.
The in vivo models provide further evidence that Gata4 predominantly regulates EpoR expression in the structurally normal murine heart. Analysis of transgenic mice with inducible shRNA against Gata4 shows significant down-regulation of EpoR expression, if Gata4 expression is reduced (Fig. 4). In these transgenic mice, some Gata4 downstream targets, such as the genes encoding Bmp4 and Anf, are expressed at lower levels than in wild-type mice, whereas other Gata4 downstream targets such as the α-Mhc gene are normally expressed [27]. This phenomenon could be due to differences in the affinity of Gata4 to regulatory elements of the target genes. In this context it is important that Gata6, which has often been discussed to compensate for Gata4 deficiency [5, 6, 39], is not modulated in the Gata4 shRNA expressing transgenic mice [27] and does not regulate cardiac EpoR expression (Figs 2, 3 and 5).
Our data confirming the activation of cardiac EpoR expression by Gata4 in the mouse model of doxorubicin-induced cardiomyopathy are very important for the future translation into strategies for cardioprotection. The high clinical impact is given by the fact that GATA4 mRNA and protein levels are about twofold lower in failing human hearts than in healthy cardiac tissues [40]. Doxorubicin, which is broadly used for cancer treatment, causes irreversible degenerative cardiomyopathy and congestive heart failure [41], which is associated with Gata4 depletion in wild-type mice [3, 7, 42]. Indeed, Gata4 depletion resulted in a significant down-regulation of EpoR expression, while during the recovery period EpoR mRNA levels normalized with restored Gata4 levels. The specificity of this mechanism is supported by results on the α-Mhc expression as bona fide control (Fig. 5). Of note, in doxorubicin-induced cardiomyopathy Gata4 can be experimentally restored and Gata4 phosphorylation can be increased by rEpo treatment [3, 4]. This process is associated with the induction of the ErK/MAPK signalling pathway [3]. Activation of this pathway stimulates nuclear translocation of the Gata4 protein and increases Gata4 binding to regulatory DNA elements of downstream target genes [35]. Furthermore, activation of the MAPK cascade leads to the phosphorylation of Gata4, which potentates its activity and direct, Epo-independent, cardioprotective effects [7, 42–44]. Thus, we propose that rEpo may induce EpoR expression in cardiomyocytes by stimulating Gata4 expression and its transcriptional activity, at least under circumstances of Gata4 depletion. For clinical concepts performed with rEpo for cardioprotection it may be important to elucidate whether GATA4 also activates EpoR expression in endothelial cells [10, 45, 46].
In conclusion, we demonstrate that Gata4 is a critical component for normal EpoR expression in cardiomyocytes. Under clinical conditions, such as cardiomyopathy due to cancer treatment with anthracyclines or ischaemia/reperfusion injury [3, 44], down-regulation of Gata4 or Sp1 may limit the accessibility of the EpoR for binding of rEpo. This may explain some limitations in the first clinical trials performed with rEpo or ESA for cardioprotection in human beings [10, 46]. Thus, restoring Gata4 levels under conditions of cardiac failure may be important for future clinical concepts in using rEpo or other ESA for cardioprotection. In general, our study highlights the fundamental role of Gata4 in regulating myocardial function and recovery.
Acknowledgments
We greatly appreciate the gift of the murine cardiomyocyte cell line HL-1 from Dr. William C. Claycomb (New Orleans, LA, USA). The pCMVTag3B/Gata-6 vector was kindly provided by Dr. Edward Morrisey (Philadelphia, PA, USA). pBS/U6-shScram and pBS/U6-shSp1–2 vectors were kindly provided by Dr. Grace Gill (Boston, MA, USA). We also thank Petra Knaus (Freie Universität Berlin) for discussion of the paper. This work was supported by the Roche Foundation for Anemia Research (RoFAR; Award ID 5463534641; cycle IV), Meggen, Switzerland.
Conflict of interest
The authors declare no conflicts of interest.
Supporting Information
(A) EpoR promoter constructs used forreporter gene assays highlighting the minimal promoter as well asenhancer and inhibitor elements as previously identified inhaematopoietic progenitors, bone marrow stroma cells, orfibroblasts (Youssoufian H et al., Mol Cell Biol.1993; 13: 98--104, and Youssoufian H. Blood. 1994; 83:1428--35). Numbers indicate the position of the nucleotidesrelative to the transcription start site and refer to variouslengths of the reporter gene constructs. The -4900/+100 constructsrepresents the 5 kb murine EpoR promoter region cloned intothe luciferase reporter plasmid pGL2. The truncated promoterregions -1742/+100 and mEpoRpro are also illustrated (seealso Fig. 1 of the main manuscript). (B) Reporter geneactivities after transfection of the 5 kb EpoR promoterconstruct, the truncated promoter constructs, or pGL2 into murineHL-1 cardiomyocytes. Firefly luciferase activities were normalizedto the corresponding Renilla luciferase values and proteinconcentration (n = 3 independent experiments; **P< 0.01; *P < 0.05).
Developmental expression profile of Epo,EpoR and candidate factors for EpoR regulation in the heart of wild-type mice and murine HL-1 cardiomyocytes.
References
- 1.Riksen NP, Hausenloy DJ, Yellon DM. Erythropoietin: ready for prime-time cardioprotection. Trends Pharmacol Sci. 2008;29:258–67. doi: 10.1016/j.tips.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Fandrey J. A cordial affair-erythropoietin and cardioprotection. Cardiovasc Res. 2006;72:1–2. doi: 10.1016/j.cardiores.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Li L, Takemura G, Li Y, et al. Preventive effect of erythropoietin on cardiac dysfunction in doxorubicin-induced cardiomyopathy. Circulation. 2006;113:535–43. doi: 10.1161/CIRCULATIONAHA.105.568402. [DOI] [PubMed] [Google Scholar]
- 4.Shan X, Xu X, Cao B, et al. Transcription factor GATA-4 is involved in erythropoietin-induced cardioprotection against myocardial ischemia/reperfusion injury. Int J Cardiol. 2009;134:384–92. doi: 10.1016/j.ijcard.2008.03.043. [DOI] [PubMed] [Google Scholar]
- 5.Bisping E, Ikeda S, Kong SW, et al. Gata4 is required for maintenance of postnatal cardiac function and protection from pressure overload-induced heart failure. Proc Natl Acad Sci USA. 2006;103:14471–6. doi: 10.1073/pnas.0602543103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oka T, Maillet M, Watt AJ, et al. Cardiac-specific deletion of Gata4 reveals its requirement for hypertrophy, compensation, and myocyte viability. Circ Res. 2006;98:837–45. doi: 10.1161/01.RES.0000215985.18538.c4. [DOI] [PubMed] [Google Scholar]
- 7.Aries A, Paradis P, Lefebvre C, et al. Essential role of GATA-4 in cell survival and drug-induced cardiotoxicity. Proc Natl Acad Sci USA. 2004;101:6975–80. doi: 10.1073/pnas.0401833101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorn GW., 2nd Novel pharmacotherapies to abrogate postinfarction ventricular remodeling. Nat Rev Cardiol. 2009;6:283–91. doi: 10.1038/nrcardio.2009.12. [DOI] [PubMed] [Google Scholar]
- 9.Mehta JL. Erythropoietin in cardioprotection: does it have a future or is it all in the past. Cardiovasc Res. 2008;79:549–50. doi: 10.1093/cvr/cvn176. [DOI] [PubMed] [Google Scholar]
- 10.Unger EF, Thompson AM, Blank MJ, et al. Erythiopoiesis-stimulating agents - time for a reevaluation. N Engl J Med. 2010;362:189–92. doi: 10.1056/NEJMp0912328. [DOI] [PubMed] [Google Scholar]
- 11.Wu H, Lee SH, Gao J, et al. Inactivation of erythropoietin leads to defects in cardiac morphogenesis. Development. 1999;126:3597–605. doi: 10.1242/dev.126.16.3597. [DOI] [PubMed] [Google Scholar]
- 12.Yu X, Lin CS, Costantini F, et al. The human erythropoietin receptor gene rescues erythropoiesis and developmental defects in the erythropoietin receptor null mouse. Blood. 2001;98:475–7. doi: 10.1182/blood.v98.2.475. [DOI] [PubMed] [Google Scholar]
- 13.Tramontano AF, Muniyappa R, Black AD, et al. Erythropoietin protects cardiac myocytes from hypoxia-induced apoptosis through an Akt-dependent pathway. Biochem Biophys Res Commun. 2003;308:990–4. doi: 10.1016/s0006-291x(03)01503-1. [DOI] [PubMed] [Google Scholar]
- 14.Juul SE, Yachnis AT, Christensen RD. Tissue distribution of erythropoietin and erythropoietin receptor in the developing human fetus. Early Hum Dev. 1998;52:235–49. doi: 10.1016/s0378-3782(98)00030-9. [DOI] [PubMed] [Google Scholar]
- 15.Cai Z, Manalo DJ, Wei G, et al. Hearts from rodents exposed to intermittent hypoxia or erythropoietin are protected against ischemia-reperfusion injury. Circulation. 2003;108:79–85. doi: 10.1161/01.CIR.0000078635.89229.8A. [DOI] [PubMed] [Google Scholar]
- 16.Wright GL, Hanlon P, Amin K, et al. Erythropoietin receptor expression in adult rat cardiomyocytes is associated with an acute cardioprotective effect for recombinant erythropoietin during ischemia-reperfusion injury. FASEB J. 2004;18:1031–3. doi: 10.1096/fj.03-1289fje. [DOI] [PubMed] [Google Scholar]
- 17.Tada H, Kagaya Y, Takeda M, et al. Endogenous erythropoietin system in non-hematopoietic lineage cells plays a protective role in myocardial ischemia/reperfusion. Cardiovasc Res. 2006;71:466–77. doi: 10.1016/j.cardiores.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 18.Asaumi Y, Kagaya Y, Takeda M, et al. Protective role of endogenous erythropoietin system in nonhematopoietic cells against pressure overload-induced left ventricular dysfunction in mice. Circulation. 2007;115:2022–32. doi: 10.1161/CIRCULATIONAHA.106.659037. [DOI] [PubMed] [Google Scholar]
- 19.Youssoufian H, Longmore G, Neumann D, et al. Structure, function, and activation of the erythropoietin receptor. Blood. 1993;81:2223–36. [PubMed] [Google Scholar]
- 20.Zon LI, Youssoufian H, Mather C, et al. Activation of the erythropoietin receptor promoter by transcription factor GATA-1. Proc Natl Acad Sci USA. 1991;88:10638–41. doi: 10.1073/pnas.88.23.10638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiba T, Ikawa Y, Todokoro K. GATA-1 transactivates erythropoietin receptor gene, and erythropoietin receptor-mediated signals enhance GATA-1 gene expression. Nucleic Acids Res. 1991;19:3843–8. doi: 10.1093/nar/19.14.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chin K, Oda N, Shen K, et al. Regulation of transcription of the human erythropoietin receptor gene by proteins binding to GATA-1 and Sp1 motifs. Nucleic Acids Res. 1995;23:3041–9. doi: 10.1093/nar/23.15.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kirschner KM, Hagen P, Hussels CS, et al. The Wilms’ tumor suppressor Wt1 activates transcription of the erythropoietin receptor in hematopoietic progenitor cells. FASEB J. 2008;22:2690–701. doi: 10.1096/fj.07-097576. [DOI] [PubMed] [Google Scholar]
- 24.Cantor AB, Orkin SH. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 2002;21:3368–76. doi: 10.1038/sj.onc.1205326. [DOI] [PubMed] [Google Scholar]
- 25.Zhou B, Ma Q, Rajagopal S, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–13. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Claycomb WC, Lanson NA, Jr, Stallworth BS, et al. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci USA. 1998;95:2979–84. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thurisch B, Liang SY, Sarioglu N, et al. Transgenic mice expressing small interfering RNA against Gata4 point to a crucial role of Gata4 in the heart and gonads. J Mol Endocrinol. 2009;43:157–69. doi: 10.1677/JME-09-0030. [DOI] [PubMed] [Google Scholar]
- 28.Wallach I, Zhang J, Hartmann A, et al. Erythropoietin-receptor gene regulation in neuronal cells. Pediatr Res. 2009;65:619–24. doi: 10.1203/PDR.0b013e31819ea3b8. [DOI] [PubMed] [Google Scholar]
- 29.Weidenfeld J, Shu W, Zhang L, et al. The WNT7b promoter is regulated by TTF-1, GATA6, and Foxa2 in lung epithelium. J Biol Chem. 2002;277:21061–70. doi: 10.1074/jbc.M111702200. [DOI] [PubMed] [Google Scholar]
- 30.Valin A, Cook JD, Ross S, et al. Sp1 and Sp3 regulate transcription of the cyclin-dependent kinase 5 regulatory subunit 2 (p39) promoter in neuronal cells. Biochim Biophys Acta. 2009;1789:204–11. doi: 10.1016/j.bbagrm.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Youssoufian H, Lodish HF. Transcriptional inhibition of the murine erythropoietin receptor gene by an upstream repetitive element. Mol Cell Biol. 1993;13:98–104. doi: 10.1128/mcb.13.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Youssoufian H. Further characterization of cis-acting regulatory sequences in the genomic locus of the murine erythropoietin receptor: evidence for stage-specific regulation. Blood. 1994;83:1428–35. [PubMed] [Google Scholar]
- 33.Maouche L, Cartron JP, Chretien S. Different domains regulate the human erythropoietin receptor gene transcription. Nucleic Acids Res. 1994;22:338–46. doi: 10.1093/nar/22.3.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scholz H, Wagner KD, Wagner N. Role of the Wilms’ tumour transcription factor, Wt1, in blood vessel formation. Pflugers Arch. 2009;458:315–23. doi: 10.1007/s00424-008-0621-3. [DOI] [PubMed] [Google Scholar]
- 35.Pikkarainen S, Tokola H, Kerkela R, et al. Transcription factors in the developing and adult heart. Cardiovasc Res. 2004;63:196–207. doi: 10.1016/j.cardiores.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 36.Elliott S, Busse L, Bass MB, et al. Anti-Epo receptor antibodies do not predict Epo receptor expression. Blood. 2006;107:1892–5. doi: 10.1182/blood-2005-10-4066. [DOI] [PubMed] [Google Scholar]
- 37.Kirkeby A, van Beek J, Nielsen J, et al. Functional and immunochemical characterisation of different antibodies against the erythropoietin receptor. J Neurosci Methods. 2007;164:50–8. doi: 10.1016/j.jneumeth.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 38.Fluck CE, Miller WL. GATA-4 and GATA-6 modulate tissue-specific transcription of the human gene for P450c17 by direct interaction with Sp1. Mol Endocrinol. 2004;18:1144–57. doi: 10.1210/me.2003-0342. [DOI] [PubMed] [Google Scholar]
- 39.Morrisey EE, Tang Z, Sigrist K, et al. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 1998;12:3579–90. doi: 10.1101/gad.12.22.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hall JL, Grindle S, Han X, et al. Genomic profiling of the human heart before and after mechanical support with a ventricular assist device reveals alterations in vascular signaling networks. Physiol Genomics. 2004;17:283–91. doi: 10.1152/physiolgenomics.00004.2004. [DOI] [PubMed] [Google Scholar]
- 41.Singal PK, Iliskovic N. Doxorubicin-induced cardiomyopathy. N Engl J Med. 1998;339:900–5. doi: 10.1056/NEJM199809243391307. [DOI] [PubMed] [Google Scholar]
- 42.Kim Y, Ma AG, Kitta K, et al. Anthracycline-induced suppression of GATA-4 transcription factor: implication in the regulation of cardiac myocyte apoptosis. Mol Pharmacol. 2003;63:368–77. doi: 10.1124/mol.63.2.368. [DOI] [PubMed] [Google Scholar]
- 43.Liang Q, Wiese RJ, Bueno OF, et al. The transcription factor GATA4 is activated by extracellular signal-regulated kinase 1- and 2-mediated phosphorylation of serine 105 in cardiomyocytes. Mol Cell Biol. 2001;21:7460–9. doi: 10.1128/MCB.21.21.7460-7469.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki YJ, Nagase H, Day RM, et al. GATA-4 regulation of myocardial survival in the preconditioned heart. J Mol Cell Cardiol. 2004;37:1195–203. doi: 10.1016/j.yjmcc.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 45.Anagnostou A, Liu Z, Steiner M, et al. Erythropoietin receptor mRNA expression in human endothelial cells. Proc Natl Acad Sci USA. 1994;91:3974–8. doi: 10.1073/pnas.91.9.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfeffer MA, Burdmann EA, Chen CY, et al. A trial of darbepoetin alfa in type 2 diabetes and chronic kidney disease. N Engl J Med. 2009;361:2019–32. doi: 10.1056/NEJMoa0907845. [DOI] [PubMed] [Google Scholar]
- 47.Heberlein C, Fischer KD, Stoffel M, et al. The gene for erythropoietin receptor is expressed in multipotential hematopoietic and embryonal stem cells: evidence for differentiation stage-specific regulation. Mol Cell Biol. 1992;12:1815–26. doi: 10.1128/mcb.12.4.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) EpoR promoter constructs used forreporter gene assays highlighting the minimal promoter as well asenhancer and inhibitor elements as previously identified inhaematopoietic progenitors, bone marrow stroma cells, orfibroblasts (Youssoufian H et al., Mol Cell Biol.1993; 13: 98--104, and Youssoufian H. Blood. 1994; 83:1428--35). Numbers indicate the position of the nucleotidesrelative to the transcription start site and refer to variouslengths of the reporter gene constructs. The -4900/+100 constructsrepresents the 5 kb murine EpoR promoter region cloned intothe luciferase reporter plasmid pGL2. The truncated promoterregions -1742/+100 and mEpoRpro are also illustrated (seealso Fig. 1 of the main manuscript). (B) Reporter geneactivities after transfection of the 5 kb EpoR promoterconstruct, the truncated promoter constructs, or pGL2 into murineHL-1 cardiomyocytes. Firefly luciferase activities were normalizedto the corresponding Renilla luciferase values and proteinconcentration (n = 3 independent experiments; **P< 0.01; *P < 0.05).
Developmental expression profile of Epo,EpoR and candidate factors for EpoR regulation in the heart of wild-type mice and murine HL-1 cardiomyocytes.


