Abstract
microRNAs are a newly discovered class of short (∼22 nt) naturally occurring single-stranded RNA molecules that regulate the expression of target genes post-transcriptionally. Despite only being discovered 7 years ago, microRNAs have been implicated as key regulatory molecules in nearly every biological process examined so far and abnormal expression of microRNAs have been linked to many forms of disease including cancer where they can function as both tumour-suppressors and oncogenes. So why are microRNAs causing so much excitement? And will this excitement translate into new medical breakthroughs? This review attempts to answer these questions in the wider context of cancer, focusing on the role that microRNAs play in normal lymphoid development and malignancy.
Keywords: microRNA, lymphoma, leukaemia, lymphocyte, B cell, T cell
Introduction
The microRNA field has only existed for 7 years, yet it has become one of the hottest topics in biological sciences and there now well over 3000 microRNA publications (PubMed-June 2008). microRNAs have been found to play key roles in nearly every biological process examined so far and abnormal expression of these molecules have been linked with many disease types including infectious diseases, genetic disorders and cancer. So why are microRNAs causing so much excitement? And will this excitement translate into new medical breakthroughs? This review attempts to answer these questions in the wider context of cancer, focusing on the role that microRNAs play in normal lymphoid development and malignancy.
A concise history of small RNA
Despite the fundamental role that microRNAs appear to play in biology, these molecules were unknown to the scientific world until 1993 when lin-4, a C. elegans developmental regulator was identified [1, 2]. The significance of this finding was not however realised until 7 years later when another worm microRNA, let-7 was discovered [3]. Unlike lin-4, the sequence of let-7 was found to be highly conserved in almost all organisms [4]. It was soon realised that similar sequences were scattered throughout eukary-otic genomes that were first called microRNAs in 2001 [5]. Since then, over 6000 microRNAs have been identified from a huge range of both prokaryotic and eukaryotic organisms [6]. There are currently over 600 human microRNA sequences annotated in the miRBase database (http://microrna.sanger.ac.uk/sequences/), although it is believed that the true figure is closer to one thousand [7, 8]. Despite the relatively small number of microRNAs, because a single microRNA can target several hundred genes, and conversely a single target gene can cooperatively bind multiple microRNAs [9], it is currently believed that between 10% and 30% of all human genes are a target for microRNA regulation [10, 11]. microRNAs are expressed in a tissue-/cell-specific manner, some expressed ubiquitously whilst others appear to be restricted to single cell types [12].
microRNA biosynthesis and function
Mature microRNAs are 19–24 nucleotide non-coding single-stranded RNA molecules which regulate the expression of target genes through perfect (in plants) or imperfect (in animals) binding to the 3′-UTR (un-translated region) and possibly 5′-UTR [13] of mRNA. The majority of human microRNAs are encoded within introns, exgenically, within the exons of non-coding mRNAs, or within the 3′UTR sequence of mRNA [14]. With the exception of microRNAs encoded within Alu repeat sequences, which are transcribed by Pol-III [15], microRNAs are transcribed as 5′-capped polyadenylated transcripts (pri-microRNA) in a Pol II-dependent manner. Approximately 40% of human microRNAs are co-transcribed as clusters encoding up to eight distinct microRNA sequences in a single transcript that can be longer than 1kb [16, 17]. Pri-microRNAs are cleaved by the microprocessor complex consisting of a nulease Drosha, and a co-factor, DGCR8 in human beings, Pasha in Drosophila[18]. The resulting 60–70 nucleotide hairpin structure (pre-microRNA) encodes a single microRNA sequence that is exported to the cytoplasm by Exportin5 in a Ran-GTP dependent manner [19]. Cytoplasmic pre-microRNAs are further cleaved, by Dicer associated with TRBP and PACT co-factors (in human beings), to remove the loop sequence forming a shortlived asymmetric duplex intermediate (microRNA: microRNA*) [20]. This intermediate is in turn loaded into the miRISC complex containing Argonaut (Ago) proteins [21]. The strand that becomes the active mature microRNA is dependent upon which has the lowest free energy 5′ end, whilst the other strand is degraded by an unknown nuclease [22, 23]. The loaded miRISC is guided by the mature microRNA sequence to its cognate recognition sequence in the UTR of the target mRNA.
Although repression of translation without mRNA degradation appears to be the modus operandi of animal microRNAs, the situation appears to be more complex than previously thought, as there is now compelling evidence that microRNAs also effect transcriptional levels through de-adenylation and/or degradation [24] and may even positively affect translation in some instances [25]. How translational repression occurs remains unclear. It has been suggested that mRNA bound to the microRNA-miRISC complex may be sequestered away from the translational machinery in P-bodies that additionally act in concert with enzymes to remove the 5′-cap hence preventing translation [26, 27]. Alternatively it has been suggested that microRNAs may prevent recognition of the 5′cap by translation factors [28].
Aberrant expression of microRNA is a common feature of cancer
There is now compelling evidence that dysfunctional expression of microRNAs is a common feature of malignancy [29]. Aberrant expression of specific microRNAs has now been associated with all cancer types including solid and haematopoietic tumours (Table 1). Currently there are >650 publications relating to microRNA involvement in cancer (source PubMed database (http://www.pubmed.gov)). Moreover, it has been suggested that microRNA expression profiling can distinguish cancers according to diagnosis and developmental stage of the tumour to a greater degree of accuracy than traditional gene expression analysis [30]. microRNAs are proposed to play a direct role in oncogenesis as they can function as both oncogenes (e.g., miR-155 and members of miR-17–92 cluster) and tumour suppressor molecules (e.g., miR-15a and miR-16) (see below).
1.
Common aberrantly expressed microRNAs in lymphoid malignancies
| microRNA | Expression | Target | Regulation | Reference |
|---|---|---|---|---|
| miR-15a/16-1 | ↓CLL; APL | BCL2 | 13q14 deletion (CLL) | [55–62, 64–67, 125] |
| miR-17-92 cluster | DLBCL, BL, MCL, lymphoma | E2F1, CDKN1A, Bim | E2F3, c-myc, 13q31 (c13orf25) amplification | [49, 99–102, 104, 107–109] |
| miR-155 | DLBCL (ABC), FL CLL, HL, PMBL, PTLD, pediatric BL; ↓adult BL | AGTR1, FADD, RIPK1, BACH1, PU.1 | FOXP3, AP-1 | [49, 64, 71, 76, 93, 94, 96] |
| miR-29 | ↑ CLL; ↓SMZL | TCL1 | [68, 69] | |
| miR-181a | ↑ CLL, AML(M1/M2); ↓APL, AML(M3/M4) | TCL1, HOXA11, BCL2 | [64, 65, 68, 116, 126] | |
| miR-143/145 | ↓CLL, DLBCL, MALT, BL | ERK5 | [83] | |
| miR-21 | ↑ CLL, FL, DLBCL (ABC), HL, myeloma | PTEN, BCL2 | [64, 70, 76, 82] | |
| miR-221 | DLBCL (ABC), FL | c-KIT, p27 | [76] | |
| miR-196a | HL | ERK5 | [49,72] |
Abbreviations: CLL, chronic lymphocytic leukaemia; APL, acute promyelocytic leukaemia; AML, acute myeloid leukaemia; DLBCL, diffuse large B-cell lymphoma; HL, Hodgkin's lymphoma; PMBL, primary mediastinal B-cell lymphoma; FL, follicular lymphoma; PTLD, post-trasplantation lymphopro-liferative disorder; BL, Burkitt lymphoma; MALT, extranodal marginal zone B-cell lymphoma of mucosa-associated lymphoid tissue; SMZL, splenic marginal zone lymphoma.
What causes microRNAs to be aberrantly expressed in cancer is fundamental to understanding their mode of action and hence identifying clinically useful properties. There are currently understood to be at least three mechanisms whereby microRNAs are de-regulated in cancer; (i) chromosomal lesions at regions encoding microRNAs, (ii) defects in the microRNA biosynthetic pathway machinery and (iii) epigenetic regulation.
The majority of human and mouse microRNAs have been found to be encoded at cancer-associated genomic regions such as fragile sites, minimal regions of loss of heterozygosity, minimal regions of amplification and common break point regions [31, 32]. Using array comparative genomic hybridization of 283 microRNA loci in solid tumours, a large proportion were found to be associated with DNA copy number alterations [33]. These data suggest that dysregulation of microRNA expression by genomic alterations is probably a widespread phenomenon in cancer.
Global effects on microRNA expression can be exerted through aberrant expression/activity of components of the microRNA biosynthetic machinery. For example, reduced levels of Dicer but not Drosha in lung cancer have been associated with poor prognostic outcome [34]. Dicer was also found to be down-regulated in B-cell lymphomas (Lawrie- unpublished data). Conversely up-regulation of Dicer has been reported in lung adenocarcinoma [35] and prostate adenocarcinoma [36] and Drosha levels in cervical squamous cell carcinoma are up-regulated [37]. When endogenous microRNA processing was silenced in both cell lines and mice, they displayed enhanced cellular transformation and tumourgenesis, giving a pathological significance to the down-regulation of tumour suppressor microRNAs in cancer [38].
More recently, it has been found that microRNA expression can be influenced by treatment with either de-methylating agents or histone modifying agents (reviewed by [39]), suggesting that epigenetic control of microRNA expression may also be involved in malignancy.
Whilst these three mechanisms may, in part, help explain aberrant microRNA expression in cancer, a paucity of understanding of how microRNAs are regulated means that in the majority of cases, the causes behind dysfunctional microRNA expression remains unknown.
microRNAs in lymphocyte development
The first study to suggest a role for microRNAs in normal lymphocyte development was by Chen et al. who cloned ∼150 microRNAs from murine bone marrow and found that miR-181a, miR-223 and miR-142 were preferentially expressed in haematopoietic tissue [40]. miR-181a, more highly expressed in B-cell lineage samples, when expressed ectopically, resulted in a dramatic increase in the proportion of B-lineage cells produced. Expression of miR-181a was also found to be up-regulated in CD4+/CD8+ double positive (DP) thymocytes where it was shown to target BCL2, CD69 and TCRα in vitro, although levels of BCL2 were unexpectedly decreased in Dicer-deficient mice [41]. Furthermore, the inhibition of miR-181a in DP cells by antagomirs was found to impair sensitivity to antigens and to inhibit positive and negative selection in T-cell development [42] pointing to a fundamental role for this microRNA in normal T-cell function.
Another microRNA found to be lymphocyte associated, miR-150 was shown to be down-regulated in response to T-cell stimulation in both Th1 or Th2 subsets [43]. The role of miR-150 in lymphocyte maturation was explored further by Zhou et al. who observed that ectopic expression of this microRNA in haematopoietic stem cells transplanted into mice had no effect on T cells, macrophages or granulocytes but greatly impaired formation of mature B cells [44]. More detailed investigation revealed that over-expression of miR-150 reduced numbers through increased apop-totic rates in different developmental stages of B cells with the exception of pro-B cells suggesting that this microRNA inhibits the transition from pro- to pre-B cell. miR-150 was recently shown to control B-cell differentiation in vivo though specific targeting of c-Myb levels [45].
Mir-155, originally identified as being abnormally expressed in lymphoma (see below), has more recently been found to play a pivotal role in normal lymphocyte development using knockout mice. Rodriguez et al. found that miR-155-deficient mice were also immunodeficient and displayed increased lung airway remodelling [46]. These mice when challenged with the pathogen Salmonella typhimurium were not protected by immunization unlike wild-type mice. Furthermore, B cells from these mice produced significantly reduced levels of immunoglobulins in response to antigen treatment, whilst T cells produced reduced levels of IL-2 or IFN-γ and dendritic cells failed to efficiently activate T cells. Gene expression analysis of CD4+ cells from miR-155-deficient mice showed that many of the genes aberrantly expressed in these cells were computationally predicted as targets for this microRNA. Another study by Thai et al. used both BIC/miR-155-deficient and mice overexpressing miR-155 to investigate germinal centre (GC) B cell responses [47]. They found a reduction in total numbers of GC B cells in deficient mice and an increase in numbers in mice overexpressing miR-155. In vitro activation of wild type B cells resulted in strong up-regulation of miR-155 expression as did activation of CD4+ T cells. Similar to the study of Rodriguez et al., immunization resulted in decreased antibody titres in miR-155-deficient mice but the authors observed no difference in B-cell proliferation levels or somatic hypermutation when stimulated in vitro. It was also observed that activated miR-155-deficient B cells expressed about a third of normal levels of TNF leading to the suggestion that miR-155 controls B-cell activation through control of cytokine production. Further research on miR-155-deficient revealed that miR-155 is required for B-cell responsiveness to both thymus-dependent and independent antigens and that B cells lacking this microRNA generated reduced extrafollicular and GC responses and failed to generate high affinity IgG antibodies [48]. This phenotype was reproduced by overexpressing a target gene of miR-155, the transcription factor PU.1.
Microarray analysis of in vitro B-cell activation by IL-2 or IgM resulted in changes to the expression of eleven microRNAs (including miR-155), the majority (9/11) of which were up-regulated. In contrast, plasma cell development was associated with the down-regulation of (10/11) of microRNAs [49]. Three microRNAs implicated in both B-cell developmental stages (miR-17, miR-20b and miR-106a) form part of the miR-17–92 cluster leading to the proposition that this cluster played a role in normal B-cell development. Subsequently, this has been found that the miR-17–92 cluster is indeed essential for normal B-cell development as its deletion in a murine model resulted in increased levels of the pro-apoptotic protein Bim and inhibited B-cell development at the stage of pro- to pre-B-cell transition [50].
In addition, to the role of specific microRNAs, the general necessity of microRNAs in lymphocyte development was investigated by selective deletion of Dicer in the thymus at both an early stage of T-cell development and at a much later stage [51, 52]. Surprisingly, this deficiency, in both experiments, did not inhibitT-cell differentiation but did cause a severe block in peripheral CD8+ development and reduced numbers of CD4+ cells which when stimulated underwent increased apoptosis and proliferated poorly.
microRNAs in lymphoid malignancy
Lymphoma is the fifth most common cancer type in the UK with approximately 12,000 cases per annum. The age-adjusted incidence of non-Hodgkin's lymphoma (NHL) in the United States has increased 74% between 1976 and 2001 [53]. The WHO classification divides NHL into either B-cell (85%) or T- and NK-cell (15%) neoplasms. Compared with solid tumours the identity and role of microRNAs in lymphomas remains little studied with the exception perhaps of chronic lymphocytic leukaemia (CLL) (Fig. 1).
1.
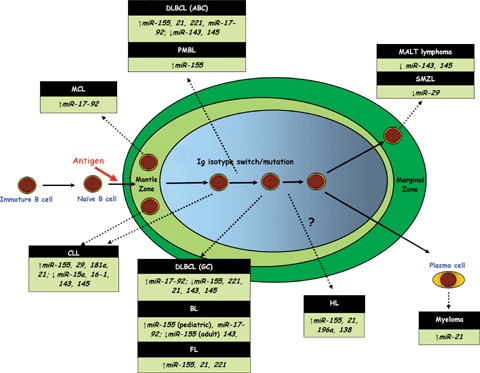
microRNA involvement in lymphoid malignancy shown relative to proposed counterpart B-cell developmental stage in the context of the lymph node germinal centre. Abbreviations used: CLL, chronic lymphocytic leukaemia; DLBCL, diffuse large B-cell lymphoma; HL, Hodgkin's lymphoma; PMBL, primary mediastinal B-cell lymphoma; FL, follicular lymphoma; BL, Burkitt lymphoma; MALT, extranodal marginal zone B-cell lymphoma of mucosa-associated lymphoid tissue; SMZL, splenic marginal zone lymphoma.
Chronic lymphocytic leukaemia (CLL)
CLL is the most common leukaemia among adults in the western world and despite its name is classified by WHO as a B-cell lymphoma being characterized by clonal expansion of CD5+ mature B lymphocytes. Although generally an indolent disease some patients develop a more aggressive form with consequent poor prognostic outcome. Several factors have emerged as having prognostic power in CLL including certain genetic aberrations, somatic IgVH mutation status and levels of ZAP-70 protein (reviewed by Chiorazzi et al. [54]).
The first report of aberrant microRNA expression in CLL or indeed in any cancer was by Calin et al. in 2002, who found that that two microRNAs, miR-15a and miR-16–1 encoded at the 13q14 locus, a region deleted in the majority of CLL cases, were down-regulated in 68% of cases that harboured this deletion [55]. These microRNAs were subsequently shown to target BCL2 and to induce apoptosis in vitro, suggesting they have a tumour-suppressor role in CLL [56]. Consistent with this role, it has recently been demonstrated that miR-16 negatively regulates cellular growth and cell cycle progression [57] and exogenous delivery of miR-16 to a mouse model of CLL with low endogenous levels of miR-16 resulted in altered cell-cycle and increased apoptosis [58]. In addition, the potential effect of miR-15a/miR-16–1 expression in a leukaemic model was investigated in vitro by restoring endogenous expression levels in an acute megakaryocytic leukaemia cell line (MEG-01) containing the 13q14 deletion [59]. Gene expression and proteomic profiling identified a signature enriched for AU-rich elements (ARE) as well as cancer genes as ETS1, JUN, BCL2 and MCL1. A previous involvement of miR-16 in ARE-medi-ated mRNA decay has also been reported [60]. Ectopic expression of miR-15a and miR-16 in Dicer-deficient cell lines led to a decrease in cell growth and down-regulation of genes that caused an accumulation at G0/G1 [57].
However, a number of factors call into question the importance of miR-15a/16–1 in the pathogenesis of CLL. Firstly, the link between the 13q14 deletion and miR-15a/16–1 levels is unclear. Recently, it was found that ∼85% of 171 CLL cases displayed a wide expression range of these microRNAs that was independent of 13q14 status [61] and in a study of 56 patients although all cases with 13q14 homozygous deletions were significantly down-regulated, those with a heterozygous deletion were not significantly different from controls [62]. Additionally, although an inverse relationship between miR-15a/miR-16–1 and BCL2 was reported in 26 CLL cases [56], other reports using much larger patients cohorts did not find a correlation [61, 62]. Secondly, a homologous cluster to miR-15a/16–1 is encoded at 3q25 (miR-15b/16–2), a region not commonly deleted in CLL. It is possible that consequent residual expression of these microRNAs might explain the more favourable prognosis observed in CLL patients with 13q14 deletions compared to other common CLL chromosomal abnormalities such as 17p13 or 11q23. Consistent with this idea it has recently been reported that CLL patients with a monoallelic 13q14 deletion had slower lymphocyte growth kinetics than those with a biallelic deletions [63].
Asides from miR-15a/miR-16–1 involvement several groups have investigated the microRNA expression profile of CLL using a variety of techniques. Microarrays were used by Calin et al. to identify microRNA signatures that distinguished between karyotype, the presence of ZAP-70 and IgVH mutation status in a series of 38 CLL patients [64]. A follow-up study by the same authors extended the profiling studies to 94 CLL cases and defined a prognostically significant 13-microRNA signature [65]. Fulci et al. used a combination of cloning and qRT-PCR technologies to profile 56 CLL patients [62]. They found that miR-21 and miR-155 were highly overexpressed in CLL compared to controls whilst miR-92 was down-regulated and miR-223, miR-29b and miR-29c were up-regulated in cases with IgVH mutations. Consistent with these findings high expression of miR-16, miR-21 and miR-150 were reported in 37 CLL cases by qRT-PCR [66]. Furthermore, in situ hybridization (ISH) was used to show that CLL proliferation centres were characterized by low miR-150 but high miR-155 expression. A cloning strategy was used by Marton et al. to clone pools of patients (n= 3) with either mutated (M) or non-mutated (NM) IgVH from which they obtained 287 and 237 clones respectively of which 32 and 24 clones represented 13 previously annotated microRNAs as well as the identification of 5 novel microRNAs [67].
A further indication of the role that microRNAs could play in the pathogenesis of CLL was provided by Pekarsky et al. who found that the TCL1oncogene, previously shown to be linked with an aggressive CLL phenotype, was regulated by miR-29 and miR-181b in vitroand that levels of these microRNAs inversely correlated with levels of TCL1 expression in clinical samples of CLL [68]. Interestingly a similar finding was reported in cases of splenic marginal zone lymphoma (SMZL) [69].
Hodgkin's lymphoma (HL)
Hodgkin's lymphoma (HL) accounts for approximately 1% of cancers worldwide and is characterized by the presence of relatively few Hodgkin/Reed-Sternberg (HRS) tumour cells surrounded by a non-neoplastic cellular infiltrate. Navarro et al. investigated the microRNA profile of 49 classical HL (cHL) cases along with 10 reactive lymph nodes (RLN) by qRT-PCR [70]. They identified a 25-microRNA signature associated with disease and 36 microRNAs differentially expressed between two histological subtypes of cHL (nodular sclerosing and mixed cellularity). However, only a subset of these microRNAs were also expressed in cHL cell lines suggesting the remainder of microRNAs were associated with non-neoplastic cells. Of note miR-138 expression was associated with clinical stage in both training and validation sets and miR-21, miR-134 and miR-138 expression was visualized in HRS cells by ISH. Although this study did not identify miR-155 expres-sion as being highly expressed in cHL cases, at odds with other publications [71], they did visualize miR-155 expression in HRS cells by ISH. Microarray analysis of four cHL cell lines identified seven differentially expressed microRNAs [49]. As well as miR-155 this study found that miR-196a was up-regulated, a microRNA that was shown in another study to be up-regulated along with neighbouring homeobox gene HOXB9 in cHL cell lines as a result of constitutively active ERK5 pathway [72].
Non-Hodgkin's lymphoma
Diffuse large B-cell lymphoma (DLBCL)
Diffuse large B-cell lymphoma (DLBCL) is the most common form of adult lymphoma accounting for nearly 40% of all lymphoid tumours [73]. Gene expression and immunohistochemical studies of this clinically heterogeneous disease have revealed the presence of at least two prognostically distinct subtypes of DLBCL; those with a good prognostic outcome that are germinal centre B cell-like (GCB) and those with a worse prognosis that are activated B cell-like (ABC) [74, 75]. microRNA profiling of five cell lines representing these molecular subtypes identified differentially expressed microRNAs [76]. Three of the microRNAs associated with ABC-type cell lines (miR-155, miR-221 and miR-21) were found to be up-regulated in clinical cases of follicular lymphoma (FL), transformed DLBCL and were more highly expressed in cases of de novoDLBCL that had an ABC-type immunophenotype than those with a GCB-immunophenotype. Furthermore, miR-21 expression level was shown to be an independent prognostic indicator in these patients.
Recently, miR-155 levels measured in 22 DLBCL cell lines revealed that although high expression was seen in the ABC-type cell lines OCI-Ly3 and OCI-Ly10, expression was also high in RC-K8 and Farage cell lines that are GC-type. The authors observed a positive correlation between miR-155 and NFkB levels in eight of these cell lines [77]. Using data mining of existing microarray data from 176 DLBCL cases that contained pri-miR-155 probe data it was found that miR-155 levels were indeed higher in ABC-type cases than GC-type (or type III) cases but not in other schemes that sub-classify DLBCL. Forty-two genes were found to be correlated inversely with miR-155 expression, nine of which were targets of miR-155 by multiple predictive algorithms.
Primary effusion lymphoma (PEL) is a rare form of post-germinal centre DLBCL caused by Kaposi sarcoma-associated herpesvirus (KSHV) that is also associated with EBV infection in about 80% of cases. Nine cases of PEL were subject to DNA copy-number, pre-microRNA and mature microRNA profiling by qRT-PCR [78]. A 68-microRNA specific signature was identified as well as microRNAs associated with KSHV and EBV infection. KSHV itself encodes 12 microRNAs, including KSHV-miR-K12–11, which shares 100% seed sequence homology with miR-155. The viral microRNA was shown to function as an orthologue of miR-155[79, 80] and in PEL, KSHV-miR-K12–11 but not miR-155 is expressed. Both microRNAs were expressed in 293T cells, leading to the identification of 66 genes that were commonly down-regulated, 20 of which had 3′-UTR seed matches that were involved in cell signalling, cell division and T-cell activation. This suggests that the overexpression of KSHV-miR-K12–11 caused by KSHV infection in B cells mirrors the malignant phenotype of miR-155 overexpression (see below) in non-virally induced DLBCL.
Other non-Hodgkin's lymphoma
Mantle cell lymphoma (MCL) accounts for about 6% of NHLs and is an aggressive disease of poor prognosis with a median survival of only 3 years. The majority of patients harbour a t(11;14) translocation that leads to truncation and overexpression of cyclin D1 (CCND1). Recently it has been found that the truncation of the CCDN1 gene in the t(11;14) translocation alters miR-16–1 binding sites and that miR-16–1 alters CCDN1 mRNA expression [81], suggesting a profound role for microRNAs in the pathogenesis of this disease.
Myeloma is a plasma cell neoplasm that is essentially incurable with a median survival of 3 years accounting for nearly 2% of all deaths from cancer and about 20% of deaths from haematologi-cal cancers. Using myeloma cell lines Löffler et al.found that miR-21 expression is controlled by an up-stream enhancer that binds Stat3 and that miR-21 expression by IL-6 induction was dependent on Stat3 and that ectopically expressing miR-21 without IL-6 reduced apoptosis [82].
Several B-cell malignancies including CLL, DLBCL, BL and EBV-transformed lymphoblastoid cell lines were associated with the down-regulation of miR-143 and miR-145[83]. Expression of these microRNAs in the BL cell line Raji inhibited growth, which was associated with ERK5 protein levels.
microRNA expression in T-cell lymphoma
T cell and natural killer cell lymphoma account for about 15% of total lymphomas and represent a heterogeneous group of diseases that compared with their B-cell counterparts are little understood. The direct role of microRNAs in these lymphomas has not yet been studied; rather the little research that has been carried out suggests a role for microRNAs in these diseases. Recently the T lymphoma causing murine leukaemia virus (MLV), was used in a screen to identify common proviral integration sites and found a high density of integrations upstream of the miR-106a cistron [84]. In tumours containing this integration, the miR-106 acistron, as well as component mature microRNA sequences, was found to be overexpressed. Using the same technique, the same group reported that the miR-17–92 cluster was also a common integration site [85]. Additionally, microRNAs have been found to be encoded in the avian T-cell lymphoma-causing virus, Marek's disease virus (MDV) [86–88]. EBV-associated microRNAs were recently profiled in a peripheral T-cell lymphoma case, which were consistent with a type II latency infection [89].
Lymphoma-associated oncomirs (miR-155 and the miR-17–92 cluster)
The non-coding BIC locus was originally identified as a common retrovirus integration site for avian leukosis virus (ALV) that despite being poorly conserved between avian, mouse and human genomes, enhanced lymphogenesis in a mouse myc model [90, 91]. Commenting on the observation by van den Berg et al. that BIC was highly expressed in over 90% of Hodgkin's lymphoma (HL) cases [92], Metzler and colleagues proposed that a phylogenically conserved region of 138 nucleotides in the BIC gene encoded a functional precursor sequence of miR-155[93]. Subsequently, both BIC and miR-155 transcript levels were found to be up-regulated 10–30 fold in DLBCL cases with higher levels of expression observed in cases with an ABC-immunophenotype than those with a GCB- immunophenotype [94]. Similar findings were reported by Kluiver et al. who also found BIC overexpression in HL, primary mediastinal B-cell lymphoma (PMBL), but not other non-Hodgkin's lymphomas [71]. Overexpression of miR-155, in BL and post-transplantation lymphoproliferative disorder (PTLD) at least, appears to be associated with EBV latency type-III infections [95, 96]. Consistent with this hypothesis high miR-155 expression was reported in latency type III infected lymphoblas-toid, Raji and Granta-519 cell lines but not EBV-negative BL cell line Ramos or latency type II infected cell YT [49].
Transgenic mice that overexpressed myc carrying the miR-155 precursor sequence under control of a VH promoter-Ig heavy chain Eμ enhancer, which becomes active at the late pro-B cell stage of B-cell development, were found to develop initially pre-B-cell proliferation in spleen and bone marrow followed by a frank B-cell malignancy resembling high-grade lymphoma after 6 months [97]. It should be noted however that these mice developed a polyclonal lymphoproliferation, suggesting additional factors are necessary for oncogenesis in this model. Intriguingly, overexpression of miR-155in mouse haematopoietic stem cells resulted in a myeloproliferative disorder that resembled acute myeloid leukaemia [98].
A commonly found amplification in B-cell lymphomas, the 13q31 locus encodes a functional precursor microRNA sequence, the miR-17–92 cluster, that itself encodes seven mature microRNA sequences [99]. Levels of the miR-17–92 cluster were found to be elevated in cell lines harbouring this amplification compared to cell lines, which did not. Additionally, amplification of 13q31 has been linked to overexpression of miR-17–92 in BL, mantle cell lymphoma (MCL) and lung cancer [100–102]. However, the link between overexpression of the miR-17–92 cluster and 13q31 amplification is not clear. Recently, it has been suggested that that overexpression of these microRNAs is a more common occurrence in haematological malignancies than previously thought as high expression levels were observed ubiquitously in 40 haematological cell lines examined including cell lines lacking the 13q31 amplification [49]. Although He et al. reported that 65% of 46 lymphoma cases, including 13 DLBCL, overexpressed miR-17–92, comparative genomic hybridisation suggests that just over quarter of DLBCL cases have the 13q31 amplification [103]. In addition, overexpression of miR-17–92 has been described as a common feature of solid tumours irrespective of 13q31 amplification status [104, 105], and indeed loss-of-heterozygosity covering the 13q31 locus is frequently observed in these tumours [33].
Similar to BIC[90, 91], He et al. found that expression of components of the miR-17–92 cluster in mice also overexpress-ing myc greatly accelerated lymphogenesis [99]. Compared to the Eμ-Myc mice, mice also overexpressing the miR-17–92 cluster showed reduced levels of apoptosis, suggesting that the main effect of these microRNAs was to suppress cell death. Increased lymphogenesis was only observed however when these microRNAs were expressed together, but not as individual microRNAs, suggesting a cooperative effect. In contrast, a tumour suppressor role has been proposed for this cluster as expression of miR-17 in breast cancer cell lines resulted in reduced proliferation [106].
O'Donnell and colleagues found that the miR-17–92 cluster itself was up-regulated through direct c-myc binding [107] and it has been suggested that the two factors act synergistically in carcinogenesis [101]. They also reported that components of the cluster, miR-17–5-p and miR-20, negatively regulate expression of E2F1, a pro-apoptotic transcription factor. Conversely it was recently found that the promoter sequence of the miR-17–92 cluster contains two functional E2F binding elements and chromatin immunoprecipitation analysis demonstrated that E2F3 binds this region, forming a negative regulatory loop between the pro-apoptotic E2F1 and the proliferative E2F3 [108]. Recently, the cluster has been shown to control cell cycle progression through targeting of CDKN1A (p21) [109]. In some cell lines, over-expression or inhibition of members of the cluster promoted or delayed entry into S-phase respectively and overexpression inhibited DNA-damage mediated cell cycle arrest. Overexpression of these microRNAs in gastric cancer cells made them insensitive to TGFβ-mediated cell cycle arrest, attributable in part at least to their effect on p21 [104].
MicroRNA: a new class of diagnostic and therapeutic agents for lymphoma?
MicroRNAs as diagnostic/prognostic bio-markers
Many studies have shown that microRNA expression levels have potential diagnostic and/or prognostic significance in cancer [30, 34, 65, 76, 110–114]. Specifically in haematological malignancies, a 13-microRNA prognostic signature was identified from a study of 94 CLL patients, which was associated with other prognostic markers including IgVH status and ZAP70 expression [65]. More recently, a 27-microRNA signature was identified that could distinguish ALL from AML with 97–99% accuracy [115], whilst miR-181a expression was found to discriminate between AML M1/M2 and M3/M3 stages [116]. Additionally, the expression of miR-21 was found to be an independent indicator of overall survival in DLBCL patients [76]. With the advent of more standardised and widely available high throughput technologies to identify microRNAs, this list can only increase in the coming years.
MicroRNAs as non-invasive diagnostic tools
The search for non-invasive tools for the diagnosis and management of cancer has long been a goal of cancer research that has led to great interest in the field of circulating nucleic acids in plasma and serum [117]. Many studies have shown that specific cancer characteristics, both genetic and epigenetic, are detectable in the plasma and serum of cancer patients and may be useful as a tool for early detection, diagnosis and follow-up of cancer patients [117]. microRNAs due to their small size are relatively resistant to RNase degradation and unlike mRNA can be successfully recovered intact from archival formalin-fixed paraffin-embedded (FFPE) material [76, 118, 119], suggesting they would be eminently suitable candidates for detection in biological fluids. We were able to show the presence of circulating microRNAs for the first time and found that tumour-associated microRNAs (miR-21, 155 and 210) were present at significantly higher levels in DLBCL patient sera (n= 60) than in healthy controls (n= 43) [120]. Furthermore, we found that expression levels of serum miR-21, like that of tumoural material [76], were associated with relapse-free survival times for these patients. More recently, placental microRNAs have also been detected in maternal plasma [121]. These findings open a new diagnostic potential for microRNAs that will undoubtedly grow in the future.
microRNAs as therapeutic agents
The association between aberrant expression of microRNAs and malignancy is now so compelling that research is rapidly focusing on the therapeutic use of these molecules. As microRNAs can function as both tumour-suppressor molecules and oncogenes, the delivery of exogenous microRNAs and silencing of microRNAs can be envisaged as possible therapeutic scenarios. The power of the first scenario is perhaps best illustrated by Lim and colleagues who expressed miR-124 and miR-1 in HeLa cells and saw that the gene expression profiles of transfected cells shifted towards that typically seen in brain and skeletal muscle respectively, the organs where these microRNAs are preferentially expressed [122].
Several researchers have explored the potential of microRNA-based therapeutics using an antisense approach to inhibit specific microRNAs. Both in vitro and more recently in vivo studies have successfully used antisense technologies to inhibit specific microRNAs (reviewed by Hammond [123]). Most dramatically Krutzfeldt et al. injected modified 2-O-methyl antisense molecules (antagomirs) into mice and found remarkably efficient and long-lasting inhibition of specific microRNAs in all tissues except the brain [124]. Furthermore, inhibition of liver-specific miR-122 resulted in a 40% decrease in plasma cholesterol levels.
Perhaps one of the biggest problems facing the use of microRNAs and/or associated agents as therapeutics is their pleiotropic mode of action. As a single microRNA can target several hundred genes their perturbation may be expected to give rise to a complex phenotype that may not be readily predictable. It is therefore imperative that such therapeutics is precisely delivered in order to mitigate effects on non-targeted cells.
What is clear however is that microRNAs promise a great deal in furthering understanding of malignancy and of biology in general and can be expected to deliver further therapeutic tools in the not too distant future, although equally, a great deal is yet to be discovered before this vision becomes reality.
Acknowledgments
This work was funded by grants from the Leukaemia Research Fund and the Julian Starmer-Smith Memorial Fund.
References
- 1.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the hete-rochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–62. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 3.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 4.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–9. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 5.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–4. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 6.Griffiths-Jones S, Grocock RJ, Van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–70. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 8.Berezikov E, Guryev V, Van De Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–4. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 9.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 10.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 11.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA. 2007;104:9667–72. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–10. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 16.Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hertel J, Lindemeyer M, Missal K, Fried C, Tanzer A, Flamm C, Hofacker IL, Stadler PF. The expansion of the metazoan microRNA repertoire. BMC Genomics. 2006;7:25. doi: 10.1186/1471-2164-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeom KH, Lee Y, Han J, Suh MR, Kim VN. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res. 2006;34:4622–9. doi: 10.1093/nar/gkl458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng Y. Principles of microRNA production and maturation. Oncogene. 2006;25:6156–62. doi: 10.1038/sj.onc.1209908. [DOI] [PubMed] [Google Scholar]
- 20.Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–32. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim VN. microRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–85. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 22.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–16. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 23.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 24.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–9. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 25.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–4. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. microRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–23. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol. 2005;7:633–6. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 28.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 microRNA in human cells. Science. 2005;309:1573–6. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 29.Calin GA, Croce CM. microRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–4. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 30.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. microRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 31.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genom-ic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sevignani C, Calin GA, Nnadi SC, Shimizu M, Davuluri RV, Hyslop T, Demant P, Croce CM, Siracusa LD. microRNA genes are frequently located near mouse cancer susceptibility loci. Proc Natl Acad Sci USA. 2007;104:8017–22. doi: 10.1073/pnas.0702177104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, Yao G, Medina A, O'Brien-Jenkins A, Katsaros D, Hatzigeorgiou A, Gimotty PA, Weber BL, Coukos G. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA. 2006;103:9136–41. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, Yatabe Y, Takamizawa J, Miyoshi S, Mitsudomi T, Takahashi T. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–5. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiosea S, Jelezcova E, Chandran U, Luo J, Mantha G, Sobol RW, Dacic S. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67:2345–50. doi: 10.1158/0008-5472.CAN-06-3533. [DOI] [PubMed] [Google Scholar]
- 36.Chiosea S, Jelezcova E, Chandran U, Acquafondata M, McHale T, Sobol RW, Dhir R. Up-regulation of dicer, a component of the microRNA machinery, in prostate adenocarcinoma. Am J Pathol. 2006;169:1812–20. doi: 10.2353/ajpath.2006.060480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muralidhar B, Goldstein LD, Ng G, Winder DM, Palmer RD, Gooding EL, Barbosa-Morais NL, Mukherjee G, Thorne NP, Roberts I, Pett MR, Coleman N. Global microRNA profiles in cervical squamous cell carcinoma depend on Drosha expression levels. J Pathol. 2007;212:368–77. doi: 10.1002/path.2179. [DOI] [PubMed] [Google Scholar]
- 38.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–7. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 39.Yang N, Coukos G, Zhang L. microRNA epigenetic alterations in human cancer: one step forward in diagnosis and treatment. Int J Cancer. 2008;122:963–8. doi: 10.1002/ijc.23325. [DOI] [PubMed] [Google Scholar]
- 40.Chen CZ, Li L, Lodish HF, Bartel DP. microRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–6. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 41.Neilson JR, Zheng GX, Burge CB, Sharp PA. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 2007;21:578–89. doi: 10.1101/gad.1522907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, Klein LO, Davis MM, Chen CZ. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–61. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 43.Monticelli S, Ansel KM, Xiao C, Socci ND, Krichevsky AM, Thai TH, Rajewsky N, Marks DS, Sander C, Rajewsky K, Rao A, Kosik KS. microRNA profiling of the murine hematopoietic system. Genome Biol. 2005;6:R71. doi: 10.1186/gb-2005-6-8-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci USA. 2007;104:7080–5. doi: 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–59. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 46.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, Van Dongen S, Grocock RJ, Das PP, Miska EA, Vetrie D, Okkenhaug K, Enright AJ, Dougan G, Turner M, Bradley A. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–11. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–8. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 48.Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, Smith KG, Rada C, Enright AJ, Toellner KM, Maclennan IC, Turner M. microRNA-155 regulates the generation of immunoglobu-lin class-switched plasma cells. Immunity. 2007;27:847–59. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lawrie CH, Saunders NJ, Soneji S, Palazzo S, Dunlop HM, Cooper CDO, Brown PJ, Troussard X, Mossafa H, Enver T, Pezzella F, Boultwood J, Wainscoat JS, Hatton CS. microRNA expression in lymphocyte development and malignancy. Leukemia. 2008;22:1440–6. doi: 10.1038/sj.leu.2405083. [DOI] [PubMed] [Google Scholar]
- 50.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–86. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cobb BS, Nesterova TB, Thompson E, Hertweck A, O'Connor E, Godwin J, Wilson CB, Brockdorff N, Fisher AG, Smale ST, Merkenschlager M. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201:1367–73. doi: 10.1084/jem.20050572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–9. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.SEER. Cancer Statistics Review, 1975-2002. National Cancer Institute. 2005 [Google Scholar]
- 54.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352:804–15. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 55.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce CM. Frequent deletions and down-regulation of microRNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–9. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond CK, Dai H, Chau N, Cleary M, Jackson AL, Carleton M, Lim L. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–52. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raveche ES, Salerno E, Scaglione BJ, Manohar V, Abbasi F, Lin YC, Fredrickson T, Landgraf P, Ramachandra S, Huppi K, Toro JR, Zenger VE, Metcalf RA, Marti GE. Abnormal microRNA-16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood. 2007;109:5079–86. doi: 10.1182/blood-2007-02-071225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, Taccioli C, Zanesi N, Garzon R, Aqeilan RI, Alder H, Volinia S, Rassenti L, Liu X, Liu CG, Kipps TJ, Negrini M, Croce CM. MiR-15a and miR-16–1 cluster functions in human leukemia. Proc Natl Acad Sci USA. 2008;105:5166–71. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di Padova F, Lin SC, Gram H, Han J. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–34. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 61.Ouillette P, Erba H, Kujawski L, Kaminski M, Shedden K, Malek SN. Integrated genomic profiling of chronic lymphocytic leukemia identifies subtypes of deletion 13q14. Cancer Res. 2008;68:1012–21. doi: 10.1158/0008-5472.CAN-07-3105. [DOI] [PubMed] [Google Scholar]
- 62.Fulci V, Chiaretti S, Goldoni M, Azzalin G, Carucci N, Tavolaro S, Castellano L, Magrelli A, Citarella F, Messina M, Maggio R, Peragine N, Santangelo S, Mauro FR, Landgraf P, Tuschl T, Weir DB, Chien M, Russo JJ, Ju J, Sheridan R, Sander C, Zavolan M, Guarini A, Foa R, Macino G. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109:4944–51. doi: 10.1182/blood-2006-12-062398. [DOI] [PubMed] [Google Scholar]
- 63.Pfeifer D, Pantic M, Skatulla I, Rawluk J, Kreutz C, Martens UM, Fisch P, Timmer J, Veelken H. Genome-wide analysis of DNA copy number changes and LOH in CLL using high-density SNP arrays. Blood. 2007;109:1202–10. doi: 10.1182/blood-2006-07-034256. [DOI] [PubMed] [Google Scholar]
- 64.Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, Dell'Aquila ML, Alder H, Rassenti L, Kipps TJ, Bullrich F, Negrini M, Croce CM. microRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA. 2004;101:11755–60. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 66.Wang M, Tan L, Dijkstra M, Van Lom K, Robertus JL, Harms G, Blokzijl T, Kooistra K, Van T'veer M, Rosati S, Visser L, Jongen-Lavrencic M, Kluin P, Van Den Berg A. miRNA analysis in B-cell chronic lymphocytic leukaemia: proliferation centres characterized by low miR-150 and high BIC/miR-155 expression. J Pathol. 2008;215:13–20. doi: 10.1002/path.2333. [DOI] [PubMed] [Google Scholar]
- 67.Marton S, Garcia MR, Robello C, Persson H, Trajtenberg F, Pritsch O, Rovira C, Naya H, Dighiero G, Cayota A. Small RNAs analysis in CLL reveals a deregulation of miRNA expression and novel miRNA candidates of putative relevance in CLL pathogenesis. Leukemia. 2008;22:330–8. doi: 10.1038/sj.leu.2405022. [DOI] [PubMed] [Google Scholar]
- 68.Pekarsky Y, Santanam U, Cimmino A, Palamarchuk A, Efanov A, Maximov V, Volinia S, Alder H, Liu CG, Rassenti L, Calin GA, Hagan JP, Kipps T, Croce CM. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66:11590–3. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 69.Ruiz-Ballesteros E, Mollejo M, Mateo M, Algara P, Martinez P, Piris MA. microRNA losses in the frequently deleted region of 7q in SMZL. Leukemia. 2007;21:2547–9. doi: 10.1038/sj.leu.2404853. [DOI] [PubMed] [Google Scholar]
- 70.Navarro A, Gaya A, Martinez A, Urbano-Ispizua A, Pons A, Balague O, Gel B, Abrisqueta P, Lopez-Guillermo A, Artells R, Montserrat E, Monzo M. microRNA expression profiling in classic Hodgkin lymphoma. Blood. 2008;111:2825–32. doi: 10.1182/blood-2007-06-096784. [DOI] [PubMed] [Google Scholar]
- 71.Kluiver J, Poppema S, De Jong D, Blokzijl T, Harms G, Jacobs S, Kroesen BJ, Van Den Berg A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lym-phomas. J Pathol. 2005;207:243–9. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 72.Nagel S, Burek C, Venturini L, Scherr M, Quentmeier H, Meyer C, Rosenwald A, Drexler HG, MacLeod RA. Comprehensive analysis of homeobox genes in Hodgkin lymphoma cell lines identifies dysregulated expression of HOXB9 mediated via ERK5 signaling and BMI1. Blood. 2007;109:3015–23. doi: 10.1182/blood-2006-08-044347. [DOI] [PubMed] [Google Scholar]
- 73.Coiffier B. Diffuse large cell lymphoma. Curr Opin Oncol. 2001;13:325–34. doi: 10.1097/00001622-200109000-00003. [DOI] [PubMed] [Google Scholar]
- 74.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–11. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 75.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, Muller-Hermelink HK, Campo E, Braziel RM, Jaffe ES, Pan Z, Farinha P, Smith LM, Falini B, Banham AH, Rosenwald A, Staudt LM, Connors JM, Armitage JO, Chan WC. Confirmation of the molecular classification of diffuse large B-cell lym-phoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–82. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 76.Lawrie CH, Soneji S, Marafioti T, Cooper CDO, Palazzo S, Paterson JC, Cattan H, Enver T, Mager R, Boultwood J, Wainscoat JS, Hatton CSR. microRNA expression distinguishes between germinal center B celllike (GCB) and activated B cell-like (ABC) subtypes of diffuse large B cell lymphoma. Int J Cancer. 2007;121:1156–61. doi: 10.1002/ijc.22800. [DOI] [PubMed] [Google Scholar]
- 77.Rai D, Karanti S, Jung I, Dahia PL, Aguiar RC. Coordinated expression of microRNA-155 and predicted target genes in diffuse large B-cell lymphoma. Cancer Genet Cytogenet. 2008;181:8–15. doi: 10.1016/j.cancergencyto.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.O'Hara AJ, Vahrson W, Dittmer DP. Gene alteration and precursor and mature microRNA transcription changes contribute to the miRNA signature of primary effusion lymphoma. Blood. 2008;111:2347–53. doi: 10.1182/blood-2007-08-104463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, Braich R, Manoharan M, Soutschek J, Ohler U, Cullen BR. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–9. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, Baker HV, Renne R. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol. 2007;81:12836–45. doi: 10.1128/JVI.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen RW, Bemis LT, Amato CM, Myint H, Tran H, Birks DK, Eckhardt SG, Robinson WA. Truncation in CCND1 mRNA alters miR-16–1 regulation in mantle cell lymphoma. Blood. 2008;112:822–9. doi: 10.1182/blood-2008-03-142182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Loffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermuller J, Kretzschmar AK, Burger R, Gramatzki M, Blumert C, Bauer K, Cvijic H, Ullmann AK, Stadler PF, Horn F. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–3. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 83.Akao Y, Nakagawa Y, Kitade Y, Kinoshita T, Naoe T. Downregulation of microRNAs-143 and -145 in B-cell malignancies. Cancer Sci. 2007;98:1914–20. doi: 10.1111/j.1349-7006.2007.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lum AM, Wang BB, Li L, Channa N, Bartha G, Wabl M. Retroviral activation of the mir-106a microRNA cistron in T lymphoma. Retrovirology. 2007;4:5. doi: 10.1186/1742-4690-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang CL, Wang BB, Bartha G, Li L, Channa N, Klinger M, Killeen N, Wabl M. Activation of an oncogenic microRNA cistron by provirus integration. Proc Natl Acad Sci USA. 2006;103:18680–4. doi: 10.1073/pnas.0609030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yao Y, Zhao Y, Xu H, Smith LP, Lawrie CH, Sewer A, Zavolan M, Nair V. Marek's disease virus type 2 (MDV-2)-encoded microRNAs show no sequence conservation to those encoded by MDV-1. J Virol. 2007;81:7164–70. doi: 10.1128/JVI.00112-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Burnside J, Bernberg E, Anderson A, Lu C, Meyers BC, Green PJ, Jain N, Isaacs G, Morgan RW. Marek's disease virus encodes microRNAs that map to meq and the latency-associated transcript. J Virol. 2006;80:8778–86. doi: 10.1128/JVI.00831-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yao Y, Zhao Y, Xu H, Smith LP, Lawrie CH, Watson M, Nair V. microRNA profile of Marek's disease virus-transformed T-cell line MSB-1: predominance of virus-encoded microRNAs. J Virol. 2008;82:4007–15. doi: 10.1128/JVI.02659-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jun SM, Hong YS, Seo JS, Ko YH, Yang CW, Lee SK. Viral microRNA profile in Epstein-Barr virus-associated peripheral T cell lymphoma. Br J Haematol. 2008 doi: 10.1111/j.1365-2141.2008.07186.x. DOI May 22. [DOI] [PubMed] [Google Scholar]
- 90.Tam W, Ben-Yehuda D, Hayward WS. bic, a novel gene activated by proviral insertions in avian leukosis virus-induced lym-phomas, is likely to function through its noncoding RNA. Mol Cell Biol. 1997;17:1490–502. doi: 10.1128/mcb.17.3.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Clurman BE, Hayward WS. Multiple proto-oncogene activations in avian leukosis virus-induced lymphomas: evidence for stage-specific events. Mol Cell Biol. 1989;9:2657–64. doi: 10.1128/mcb.9.6.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Van Den Berg A, Kroesen BJ, Kooistra K, De Jong D, Briggs J, Blokzijl T, Jacobs S, Kluiver J, Diepstra A, Maggio E, Poppema S. High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosomes Cancer. 2003;37:20–8. doi: 10.1002/gcc.10186. [DOI] [PubMed] [Google Scholar]
- 93.Metzler M, Wilda M, Busch K, Viehmann S, Borkhardt A. High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma. Genes Chromosomes Cancer. 2004;39:167–9. doi: 10.1002/gcc.10316. [DOI] [PubMed] [Google Scholar]
- 94.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102:3627–32. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jiang J, Lee EJ, Schmittgen TD. Increased expression of microRNA-155 in Epstein-Barr virus transformed lymphoblastoid cell lines. Genes Chromosomes Cancer. 2006;45:103–6. doi: 10.1002/gcc.20264. [DOI] [PubMed] [Google Scholar]
- 96.Kluiver J, Haralambieva E, De Jong D, Blokzijl T, Jacobs S, Kroesen BJ, Poppema S, Van Den Berg A. Lack of BIC and microRNA miR-155 expression in primary cases of Burkitt lymphoma. Genes Chromosomes Cancer. 2006;45:147–53. doi: 10.1002/gcc.20273. [DOI] [PubMed] [Google Scholar]
- 97.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024–9. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.O'Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–94. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human onco-gene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, Yatabe Y, Kawahara K, Sekido Y, Takahashi T. A polycistronic microRNA cluster, miR-17–92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–32. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 101.Tagawa H, Karube K, Tsuzuki S, Ohshima K, Seto M. Synergistic action of the microRNA-17 polycistron and Myc in aggressive cancer development. Cancer Sci. 2007;98:1482–90. doi: 10.1111/j.1349-7006.2007.00531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rinaldi A, Poretti G, Kwee I, Zucca E, Catapano CV, Tibiletti MG, Bertoni F. Concomitant MYC and microRNA cluster miR-17–92 (C13orf25) amplification in human mantle cell lymphoma. Leuk Lymphoma. 2007;48:410–2. doi: 10.1080/10428190601059738. [DOI] [PubMed] [Google Scholar]
- 103.Karnan S, Tagawa H, Suzuki R, Suguro M, Yamaguchi M, Okamoto M, Morishima Y, Nakamura S, Seto M. Analysis of chromosomal imbalances in de novo CD5-positive diffuse large-B-cell lymphoma detected by comparative genomic hybridization. Genes Chromosomes Cancer. 2004;39:77–81. doi: 10.1002/gcc.10298. [DOI] [PubMed] [Google Scholar]
- 104.Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, De Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M, Cavazzini L, Volinia S, Alder H, Ruco LP, Baldassarre G, Croce CM, Vecchione A. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272–86. doi: 10.1016/j.ccr.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 105.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hossain A, Kuo MT, Saunders GF. Mir-17–5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol. 2006;26:8191–201. doi: 10.1128/MCB.00242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–43. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 108.Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic microRNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–4. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 109.Ivanovska I, Ball AS, Diaz RL, Magnus JF, Kibukawa M, Schelter JM, Kobayashi SV, Lim L, Burchard J, Jackson AL, Linsley PS, Cleary MA. microRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol Cell Biol. 2008;28:2167–74. doi: 10.1128/MCB.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, Calin GA, Volinia S, Liu CG, Scarpa A, Croce CM. microRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24:4677–84. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 111.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–6. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 112.Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, Fedele V, Ginzinger D, Getts R, Haqq C. Optimized high-throughput microRNA expression profiling provides novel bio-marker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ, Schmittgen TD. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–54. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. microRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 115.Mi S, Lu J, Sun M, Li Z, Zhang H, Neilly MB, Wang Y, Qian Z, Jin J, Zhang Y, Bohlander SK, Le Beau MM, Larson RA, Golub TR, Rowley JD, Chen J. microRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc Natl Acad Sci USA. 2007;104:19971–6. doi: 10.1073/pnas.0709313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Debernardi S, Skoulakis S, Molloy G, Chaplin T, Dixon-McIver A, Young BD. microRNA miR-181a correlates with morphological sub-class of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia. 2007;21:912–6. doi: 10.1038/sj.leu.2404605. [DOI] [PubMed] [Google Scholar]
- 117.Tsang JC, Lo YM. Circulating nucleic acids in plasma/serum. Pathology. 2007;39:197–207. doi: 10.1080/00313020701230831. [DOI] [PubMed] [Google Scholar]
- 118.Xi Y, Nakajima G, Gavin E, Morris CG, Kudo K, Hayashi K, Ju J. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA. 2007;13:1668–74. doi: 10.1261/rna.642907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Li J, Smyth P, Flavin R, Cahill S, Denning K, Aherne S, Guenther SM, O'Leary JJ, Sheils O. Comparison of miRNA expression patterns using total RNA extracted from matched samples of formalin-fixed paraffin-embedded (FFPE) cells and snap frozen cells. BMC Biotechnol. 2007;7:36. doi: 10.1186/1472-6750-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, Hatton CSR, Harris AL. Detection of elevated tumor-associated microRNAs in serum of patients with diffuse large B cell lymphoma. Br J Haematol. 2008;141:672–5. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 121.Chim SS, Shing TK, Hung EC, Leung TY, Lau TK, Chiu RW, Lo YM. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem. 2008;54:482–90. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- 122.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 123.Hammond SM. microRNA therapeutics: a new niche for antisense nucleic acids. Trends Mol Med. 2006;12:99–101. doi: 10.1016/j.molmed.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 124.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 125.Garzon R, Pichiorri F, Palumbo T, Visentini M, Aqeilan R, Cimmino A, Wang H, Sun H, Volinia S, Alder H, Calin GA, Liu CG, Andreeff M, Croce CM. microRNA gene expression during retinoic acid-induced differentiation of human acute promyelocytic leukemia. Oncogene. 2007;26:4148–57. doi: 10.1038/sj.onc.1210186. [DOI] [PubMed] [Google Scholar]
- 126.Naguibneva I, Ameyar-Zazoua M, Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, Cuvellier S, Harel-Bellan A. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol. 2006;8:278–84. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]


