Abstract
Considerable interest, speculation and controversy have been generated utilising surface-enhanced laser desorption/ionization in conjunction with mass spectrometry (SELDI-MS) for the diagnosis, prognosis and therapeutic monitoring of cancer and offers an attractive approach to cancer biomarker discovery from tissues and biological fluids. This technology utilises a combination of mass spectrometry and chromatography to facilitate protein profiling of complex biological mixtures. Compared to some other more traditional proteomic platforms, such as 2D polyacrylamide gel electrophoresis, it has a high-throughput capability and can resolve low-mass proteins. However, a considerable number of challenging issues related to the design of studies, including reproducibility, sensitivity, specificity, variation in sample collection, processing and storage, have been reported as problematic with this technology; albeit some of these concerns could perhaps also be lauded against other proteomic approaches that have attempted to address complex protein mixtures, such as plasma. Applications, successes and limitations of SELDI-MS in both clinical and basic science arenas will be reviewed in this article.
Keywords: SELDI-MS, cancer, diagnosis, prognosis, therapeutics
Introduction
Despite the recent advances in molecular medicine, genomics, proteomics and translational research, our efforts to alleviate cancer have been ill reputed. According to the World Health Organisation, it is estimated that there will be 16 million new cases every year by 2020. Cancer accounts for seven million deaths every year or 12.5% of deaths worldwide. Current strategies to combat cancer include early diagnosis and administration of effective treatment and monitoring patients after treatment response. Cancer biomarkers that are currently used for the diagnosis, prognosis, monitoring of patients and prediction of therapeutic response include CA125 (ovarian), CA15.3 (breast), CA19.9 (gastrointestinal) and β human chorionic gonadotropin and serum α-fetoprotein (testicular cancer). Due to the lack of early detection methods and specific and sensitive biomarkers, both scientist and clinicians are moving towards using proteomics as a means to (i) discover and validate new biomarkers or ensembles of biomarkers that have better specificity and sensitivity characteristics than existing biomarker assays or cytology [1] and to(ii) improve our understanding of cancer initiation and progression. Herein, we will discuss the applications and limitations of surface-enhanced laser desorption/ionization-mass spectrometry (SELDI-MS) as a diagnostic and cancer biomarker discovery tool in both a clinical and scientific setting.
Surface-enhanced laser desorption/ionization-mass spectrometry
SELDI-MS, also known as ProteinChip®, is a high-throughput pro-teomics technique that facilitates multiple biomarker discovery, purification and identification. The process involves binding of a crude sample to a ProteinChip array which is subsequently washed several times to insure complete removal of unbound proteins and other interfering substances such as contaminants or buffers (Fig. 1; Table 1). As in matrix-assisted laser desorp-tion/ionization (MALDI) time-of-flight (TOF) mass spectrometry, an energy absorbing matrix (EAM), such as sinapinic acid (SPA) or α-cyano-4-hydroxycinnamic acid matrix (CHCA), is applied to the protein spots to facilitate ionization and desorption of proteins from the surface. The mass-to-charge ratios for desorbed molecules are analysed as they fly down the TOF tube and an individual protein spectrum is generated for each sample tested. Differentially expressed proteins are determined from the protein profiles by comparing the peak intensities of spectra.
1.
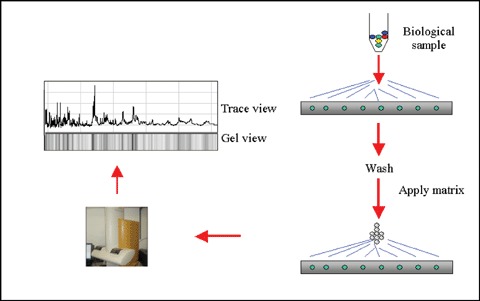
Schematic demonstrating ProteinChip technology
1.
ProteinChip surfaces and their applications
| Chemical surfaces Chromatographic surfaces | Biological surfaces Pre-activated surfaces | Applications |
|---|---|---|
| Hydrophobic (i.e.H50) CM10, WCX2) | PS10, PS20, | Protein profiling, purification, antibody–antigen interaction, phosphorylation/signal transduction, toxicity markers, clinical trials, glycosylation analysis and epitope mapping. |
| Weak cation exchange (i.e. Q10, SAX2) | RS100, PG20 | |
| Strong anionic exchange (i.e. Q10, SAX2) |
Sampling
A wide variety of sample types can be used to detect biomarkers from crude samples, such as blood serum or plasma, intestinal fluid, cellular extracts (from microdissected cells, cell culture or xenografts), cellular secretion products, fine needle aspirates, tissue, urine and cerebrospinal fluids (CSFs). According to the literature (Table 3), the majority of clinical studies use blood plasma or serum, as it is a readily accessible protein-rich body fluid. Moreover, it perfuses all other tissues of the body, so it carries not only plasma-specific proteins but also proteins derived from other tissues. However, there are a number of drawbacks associated with blood serum or plasma, one of which is the highly abundant proteins such as albumin and immunoglobulin. These proteins, which account for 97% of all proteins, suppress middle and low-abundance proteins in the sample. Another disadvantage is the contamination with peptides produced by the activation of platelets or derived from the coagulation process during serum collection. Serum fractionation or protein depletion assays kits have been employed to remove these high-abundant proteins. However, these methodologies generate new difficulties such as the loss of small abundant proteins that bind to albumin. Removal of albumin could result in the loss of potential biomark-ers. The use of tissue derived from organs also faces a number of challenges, one of which is the loss of potentially important spatial, thus anatomical, information or context during sample preparation as the result of homogenisation and extraction procedures.
3.
Examples of SELDI-MS studies performed on specimens from patients with cancer
| Study | Specimen | Cancer type | Proteins or peaks | Patient number | Clinical perspective |
|---|---|---|---|---|---|
| Fung et al.[65] | Serum | Ovarian, breast, colon | 3 | 142 | Diagnosis |
| Li et al.[66] | Serum | Breast | 3 | 169 | Diagnosis |
| Pawlik et al.[9] | *NAF | Breast | 27 | 28 | Diagnosis |
| Sauter et al.[67] | NAF | Breast | 7 | 114 | Diagnosis, Prognosis |
| Nakagawa et al.[32] | Tissue | Breast | 2 | 65 | Diagnosis, Prognosis |
| Shi et al.[55] | Plasma | Breast | 1 | 122 | Diagnosis |
| Becker et al.[7] | Serum | Breast | 4/8? | 62 | Diagnosis |
| Chen et al[17] | Serum | Colon | 4 | Diagnosis, Prognosis | |
| Liu et al.[68] | Serum | Colon | 2 | 99 | Diagnosis |
| De Bont et al.[24] | †CSF | Brain | 1 | 102 | Diagnosis |
| M Roesch-Ely et al. | Tissue | ‡HNSCC | 48 | 303 | Diagnosis, Prognosis |
| [36] | |||||
| Soltys et al.[69] | Plasma | HNSCC | 65 | 217 | Diagnosis |
| Wang et al.[70] | Serum | Thyroid | 2 | 80 | Diagnosis |
| Yang et al.[10] | Serum | Lung | 5 | 208 | Diagnosis |
| Zhukov et al.[11] | Tissue | Lung | 3 | 45 | Diagnosis, Detection |
| Rosty et al.[71] | Pancreatic juices | Pancreas | 2 | 91 | Diagnosis |
| Lin et al.[14] | Plasma | Ovarian | 4 | 65 | Diagnosis |
| Zhang et al.[15] | Serum | Ovarian | 3 | 503 | Diagnosis |
| Yu et al.[72] | Serum | Pancreas | 6 | 100 | Diagnosis, Prognosis |
| Koopmann et al.[73] | Serum | Pancreas | 2 | 180 | Diagnosis |
| Qian et al.[74] | Serum | Gastric | 16 | 130 | Diagnosis |
| Su et al.[16] | Serum | Gastric | 3 | 245 | Diagnosis |
| Soltys et al.[69] | Plasma | HNSCC | 65 | 109 | Diagnosis |
| Ye, et al.[77] | Serum | Ovarian | 1 | 266 | Diagnosis |
| Moshkovskii et al.[78] | Serum | Ovarian | 1 | 61 | Diagnosis |
| Scarlett et al.[23] | Serum | Cholangiocarcinoma | 14/16 | 22 | |
| Wilson et al.[79] | Serum | Melanoma | 3 | 49 | Diagnosis |
| Engwegan et al.[18] | Serum | Colorectal | 2 | 157 | Detection |
| Chen et al.[17] | Serum | Colorectal | 147 | 4 | Diagnosis |
| Zhu et al.[80] | Serum | Endometrial | 13 | 40 | Detection |
NAF; nipple aspirate fluid; †CSF; cerebrospinal fluid; ‡HNSCC; head and neck squamous carcinoma.
Clinical applications
Detection and diagnosis
Despite significant investment in cancer research over the past few decades, advances in cancer treatment and improvements in cancer outcomes are modest. A great deal of research has been invested in improving the treatment modality for advanced disease as a lot of patients are diagnosed at the latter stages of the disease. With a few notable exceptions (mostly childhood cancers), survival rates for people diagnosed with advanced cancer have changed little over the past 20 years [2]. According to Cancer Statistics 2007, a total of 1,444,920 new cancer cases and 559,650 deaths for cancers are projected to occur in the United States [3]. High-throughput non-invasive or minimally invasive tests to diagnose cancer at early stages are now essential. Currently, both scientist and clinicians are moving towards using tumour protein expression (proteomics) as a means to identify novel diagnostic and therapeutic targets in oncological malignancies [4].
Several laboratories have demonstrated the feasibility of using SELDI mass spectrometry for the diagnosis and detection of various cancers (Tables 2 and 3). Breast cancer is the most common form of cancer worldwide and is responsible for 502,000 deaths per year (World Health Organisation [WHO]). Early detection is crucial to reduce breast cancer mortality rates. Current screening methods include mammography, clinical breast examination and self-examination. Whilst mammography has been shown to reduce breast cancer mortality by about 20% to 35% in women aged 50 to 69 years and slightly less in women aged 40 to 49 years at 14 years of follow-up, the predictive value of mammogra-phy declines in patients with dense breast tissue and smaller lesions as well as in pre-menopausal women [5–7]. The potential for an increase in successful treatment based on early diagnosis has driven the search for diagnostic biomarkers [5, 8]. Currently, SELDI-MS has been used for the detection of changes in protein expression pattern in breast cancer patients with stage I or II unilateral invasive breast carcinoma. Nipple aspirate fluid (NAF) collected from cancerous and non-cancerous breast of patients were applied to SELDI-MS ProteinChip arrays and protein expression was analysed using time of flight mass spectrometry. Of the 463 peaks detected, 17 peaks were overexpressed in breast cancer patients compared to breasts of healthy volunteers (P< 0.0005).
2.
Percentage of SELDI-MS studies performed to date (Mid Jan 2007)
| Cancer type | % of studies |
|---|---|
| Breast | 16.3% |
| Colon | 3.6% |
| Colorectal | 8.2% |
| Glioma | 2.8% |
| Hepatocellular/liver | 8.9% |
| Kidney/renal | 4.1% |
| Laryngeal | 0.4% |
| Leukaemia/CLMC | 2% |
| Ovarian | 10.2% |
| Thyroid | 1.2% |
| Bladder | 4.1% |
| Pancreas | 2.8% |
| Lung | 2.8% |
| Head and neck | 4% |
| Endometrial | 1.6% |
| Brain | 1.2% |
| Melanoma | 1.6% |
SELDI-MS was able to detect differences in the phenotypic pro-teomic profile of NAF samples taken from patients with early stage breast cancer and healthy women [9]. In 2004, Becker et al. profiled serum from BRCA-1 (breast cancer 1) breast cancer patients, BRCA-1 mutation carriers, patients with sporadic breast cancer and normal controls using IMAC-Cu ProteinChip arrays. Differentially expressed peaks distinguished between BRCA-1 breast cancer patients and sporadic breast cancer patients with sensitivity and specificity of 94% and 100% respectively. BRCA-1 mutation carriers and BRCA-1 breast cancer patients were correctly detected with a sensitivity and specificity of 87% each. Thus, SELDI-MS may be able to identify patients with the BRCA-1 mutations that have occult disease and those who do not [7].
Lung cancer is the second leading killer after cardiovascular disease and is responsible for 1.3 million deaths per year (WHO). Current screening tools for lung cancer include chest X-ray, low-dose computed tomography, bronchoscopy, sputum cytology and tumour markers. Due to the lack of adequate sensitivity and/or specificity of these approaches, there is an urgent need for new screening tools. Yang et al. (2005) used SELDI-MS to screen the sera of 158 lung cancer patients and 50 healthy individuals which had been randomly divided into two sets: training set (including 11 sera from patients with stages I/II lung cancer, 63 from patients with stages III/IV lung cancer and 20 from healthy controls) and blind set (43 sera from patients with stages I/II lung cancer, 41 from patients with stages III/IV lung cancer and 30 from healthy controls). Five protein peaks at 11,493, 6429, 8245, 5335 and 2538 Da were detected in the training set. The blind test yielded a sensitivity of 91.4% in the detection of non-small cell lung cancers, which was significantly higher than that in the detection of small cell lung cancers (P< 0.05) [10]. Similarly, Zhukov et al. (2002) assayed 45 laser capture microdissected samples from malignant/pre-malignant peripheral lung lesions and normal lung tissue by SELDI-MS. Three peaks at 17,250, 17,930 and 22,250 Da were increased in lung tumour cells when compared with normal cells. The 17,250 Da peak, which was not detected in any of the normal cells, was present at low levels in the atypical cell samples [11].
Ovarian cancer is the fifth commonest form of cancer amongst women and presents at an advanced stage with poor outcome. Cancer antigen 125 (CA125) and ultrasound are the main screening tools for the detection of ovarian cancer. Recent studies suggest that SELDI-MS offers exciting opportunities for the detection of novel biomarkers or patterns of markers that will have greater sensitivity and lead time for pre-clinical disease than CA125 [12, 13]. Lin et al. (2006) used SELDI in combination with mass spectrometry to identify new plasma biomarkers. Plasma from 35 ovarian cancer patients and 30 control patients was used in this study. Four protein peaks with molecular masses of 6190, 5147, 11523 and 11538 Da were identified in ovarian cancer patients, but not in controls. Two peaks, with molecular masses of 5296 and 8780 Da were present in control patients but not in ovarian cancer patients [14]. In a similar study, Zhang et al. (2004) detected and identified three peaks with molecular masses of 12,828 (form of transthyretin), 28,043 (apolipoprotein A1) and 3272 (a fragment of human inter-a trypsin inhibitor, heavy chain H4) Da in the sera of 153 patients with invasive epithelial ovarian cancer, 42 with other ovarian cancers, 166 with benign pelvic masses and 142 healthy women. Transthyretin and apolipoprotein A1 were down-regulated in the cancer group whilst human inter-α trypsin inhibitor fragment was up-regulated in the cancer group [15]. These findings suggest that these biomarkers have the potential to improve the detection of early stage ovarian cancer.
Gastric cancer is another form of cancer where specific and sensitive biomarkers that can be used for its diagnosis are still unavailable [16]. Using SELDI-MS, Su et al. (2006) analysed 245 serum samples from individuals with gastric cancer, age and sex-matched healthy individuals and benign and colorectal cancer patients. Three proteins with a m/z ratio of 1468 (fibrinopeptide A), 3935 and 7560 were detected as potential biomarkers for the diagnosis of gastric cancer [16]. In a similar study, Chen et al. (2004) and Engwegan et al. (2006) detected serum proteins with potential as biomarkers for the detection and diagnosis of colorectal cancer [17, 18].
Cervical cancer is the third most common form of cancer affecting women aged 25 years or older. Human papillomavirus (HPV) has been shown to play a major role in cervical cancer development [19]. Current screening strategies include cervical smear test or liquid-based cytology followed by a colposcopy, large-loop excision of the transformation zone or cone biopsy if abnormal cells are detected. Wong et al. (2004) used SELDI analysis to differentiate cervical cancer from non-cancer patients. Sixty-two samples microdissected from 35 invasive cervical cancer and 27 age-matched normal cervix tissue were applied to WCX2 ProteinChip arrays. A training set comprising of 20 cervical cancer and 15 normal cervix tissue specimens was used to develop a classification scoring system and a blind test set of 27 samples was used to evaluate this scoring systems ability to distinguish cervical cancer from non-cancer [20]. Using this model, a sensitivity of 87%, a specificity of 100%, a positive predictive value of 100% and a negative predictive value of 86% for the test population were obtained. Seven proteins were down-regulated in cervical cancer cells compared to normal cervical epithelial cells [20]. Similarly, Von Eggeling et al. (2001) used SELDI-MS analysis to differentiate cervical cancer tissue from pre-cancerous tissue. He found that a number of differentially expressed proteins (10–15.5 kDa) that were detected in the pre-cancerous tissue were not present in the tumour samples. These findings suggest that SELDI mass spectrometry may play a role in distinguishing cervical cancer from normal cervical cells [21].
Cholangiocarcinoma (CC) is a rare cancer that has a poor prognosis. The diagnosis is often made on imaging and a high index of suspicion. Confirming the diagnosis histologically can be difficult due to the poor representativity of the biopsy procedures [22].
There are two tumour biomarkers in clinical use for CC, carci-noembryonic antigen (CEA) and CA 19.9. These have a sensitivity of 70% and 50% respectively. Scarlett et al. (2006) used SELDI-MS to detect additional potential protein biomarkers of CC [23]. Twenty-two resected CC samples were compared with non-involved bile duct tissue. In addition, serum from patients with CC, benign disease and normal healthy controls were profiled. Fourteen differentially expressed peaks between the CC and normal bile duct tissue were detected. Serum profiling detected four peaks that differentiated CC from benign stricture and 12 peaks that distinguished CC from normal healthy controls [23].
SELDI-MS has also been used successfully for the detection of brain tumours. De Mont et al. (2006) detected 123 differentially expressed proteins in CSF collected from patients with or without brain tumours. From this listing, apolipoprotein A-II was identified in CSF taken from brain tumour patients [24]. In summary, these studies highlight the potential of using SELDI-MS to detect bio-markers in cancer.
Pre-operative staging of cancer
The ability to accurately stage cancers pre-operatively is desirable. It can dictate the most appropriate treatment strategy for the patient, that is neoadjuvant versus adjuvant therapy. Currently definitive colorectal cancer staging is based on histopathological features, such as the grade of the tumour, the depth of bowel wall invasion, lymphatic spread and lymphovascular invasion. Despite this pathological staging system, there is debate as to whether stage II colorectal cancer patients benefit from adjuvant chemotherapy. Xu et al. (2006) examined the serum SELDI-MS proteomic patterns of 76 patients with stage I–IV colorectal cancer. The spectra were generated using a weak cation exchange ProteinChip array. They were able to create various stage models with the data. These models had the ability to distinguish between the various stages of colorectal cancer. In particular, they were able to differentiate between stage II and III disease with an accuracy of 86%[25]. This technology may be able to generate a molecular classification of colorectal disease which may be able to correctly identify those high-risk patients with stage II disease who will benefit from adjuvant treatment [25].
Predicting response to therapy
Accurate prediction of chemosensitivity in cancer therapy is particularly desirable in the clinic to avoid toxic side effects and to eliminate the use of any ineffective agent. Therefore, new biomarkers for predicting chemosensitivity are highly sought after to improve the current clinical capabilities. Using SELDI-MS analysis, Menard et al. (2006) detected proteomic changes in serum protein composition in patients with various forms of cancer before and during radiotherapy in an effort to discover clinical biomarkers of ionizing radiation exposure. SELDI-MS analysis of pooled sera taken from 68 patients detected 23 protein/peptide fragments including interleukin-6 precursor protein in the radiation exposure group. In addition, proteomic profiles were able to distinguish unexposed from radiation-exposed patients with 91–100% sensitivity and 97–100% specificity; and could distinguish high from low-dose volume radiation exposure [26]. Similarly, Wibom et al. (2006) detected changes in the protein expression profile in brain tumour tissues following radiotherapy. Seventy-seven peaks whose intensity significantly changed after radiotherapy were detected [27].
In chemotherapeutic studies, proteomic changes in plasma were examined in patients with stage I–III breast carcinoma who had received paclitaxel or 5-fluorouracil, doxorubicin and cyclophosphamide (FAC) chemotherapy. Plasma collected from 69 patients' pre- and post-treatment or from 15 healthy individuals every 3 days were used to generate protein profiles viaSELDI-MS. A single chemotherapy-inducible SELDI-MS peak (m/z ratio of 2790) and five other peaks that distinguished plasma obtained from breast cancer patients and healthy individuals were detected. These proteins are candidate markers of micrometastatic disease after surgery [28].
Smith et al. (2007) used SELDI-MS analysis to predict histolog-ical response of locally advanced rectal cancer to neoadjuvant radiochemotherapy. From 230 spectra generated representing all available time points from nine good responders (tumour regression grade [TRG] 1 + 2) and 11 poor responders (TRG 3–5), a cohort of 14 protein peaks were detected that collectively differentiated between good and poor responders, with 87.5% sensitivity and 80% specificity [29]. In a Phase I clinical trial, Baker et al. 2006 used SELDI-MS to measure plasma Thioredoxin-1 from patients treated with 1-methylpropyl 2-imidazolyl disulfide (PX-12). Four peaks, 13.86 kD, 22.20 kD, 44.46 kD and 110.53 kD, showed a significant decrease after 4 hrs of PX-12 treatment, independent of pre-treatment peak intensity. The 13.86 kD peak was identified using liquid chromatography (LC) tandem MS as a variant of transthyretin [30].
In oesophageal cancer, the ability to predict chemoradiosensi-tivity may have an impact on the treatment strategies patients receive. Using SELDI-MS analysis, Hayashida et al. (2005) detected four peaks at 7420, 9112, 17,123 and 12,867 m/z. These peaks could distinguish responders from non-responders and could correctly diagnosed chemoradiosensitivity in 93.3% of the test group of 15 independent samples [31]. In short, these studies highlight the potential of using SELDI-MS to predict responses to therapy.
Metastasis and disease progression
A number of studies have focused on the ability of SELDI-MS to determine if protein signatures in primary cancers can predict metastasis. Nakagawa et al. (2006) showed that protein profiling can detect differential protein peaks in primary breast cancers that predict the presence and number of axillary lymph node (ALN) metastases and non-sentinel lymph node (SLN) status [32].
Sixty-five laser capture microdissected samples from patients undergoing resection with SLN or level I and II ALN dissection were subjected to SELDI-MS. ProteinChip array analysis identified two metal-binding polypeptides at 4871 and 8596 D as significant risk factors for nodal metastasis. Lymphovascular invasion was the only clinicopathologic factor predictive of ALN metastasis. In a similar study, Goncalves et al. (2006) used SELDI-MS to detect a protein signature correlating with metastatic relapse in early postoperative serum from 81 high-risk early breast cancer patients. Using various bioinformatic approaches, a multi-protein model that correctly predicted outcome in 83% of patients was created. The 5-year metastasis-free survival in ‘good prognosis' and ‘poor prognosis' patients were strikingly different (83 and 22%, respectively). In a multivariate Cox regression model including conventional pathological factors and multi-protein index, the poor prognosis patients retained the strongest independent prognostic significance for metastatic relapse. Major components of the multi-protein index included haptoglobin, C3a complement fraction, transferrin, apolipoprotein C1 and apolipoprotein A1 [33].
In another study, Wu et al. (2002) identified and validated metastasis-associated proteins in head and neck squamous cell carcinomas (HNSCC) using SELDI-MS in conjunction with 2D-gel electrophoresis. Three proteins namely annexin I, annexin II and enolase a were identified [34]. Wadsworth et al. (2004) used SELDI-MS analysis to detect sera changes between HNSCC patients versus control patients [35]. Using protein peak clustering and classification analyses of SELDI-MS spectral data, several proteins with masses ranging from 2778 to 20,800 D, were differentially expressed between HNSCC and the healthy controls. In particularly, a protein peak at 5064 D was under-expressed in the HNSCC sera when compared with the control sera. Similarly Roesch-Ely et al. (2007) analysed 303 biopsies (113 HNSCCs, 73 healthy, 99 tumour-distants and 18 tumour-adjacent squamous mucosae) in order to detect changes in protein expression occurring at different stages of tumourigenesis. Forty-eight protein peaks were differentially expressed between healthy mucosa and HNSCC. Calgizarrin (S100A11), Cystatin A, Acyl-CoA-binding protein, Stratifin (14–3-3 sigma), Histone H4, a- and β-Haemoglobin, a C-terminal fragment of β-haemoglobin and the α-defensins 1–3 were identified by mass spectrometry [36]. These results suggest that these proteins may play a pivotal role in head and neck cancer invasion and metastasis.
Schwegler et al. (2005) generated a list of 38 differentially expressed peaks across four separate patient groups including patients with no liver disease, liver disease without cirrhosis, cirrhosis and hepatocellular carcinoma. The SELDI-MS peak data could distinguish hepatitis C viral infection from hepatitis C viral-associated hepatocellular carcinoma with a sensitivity and specificity of 61% and 76% respectively. When the known values of serum markers a fetoprotein, des-γ carboxyprothrombin and GP73 were used in conjunction with the SELDI-MS peak data, the sensitivity and specificity increased to 75% and 92% respectively [37]. These findings suggest that serum proteomic profiles may predict metastasis in primary cancers and may be used to detect the progression of disease.
Drug resistance
Platinum compounds are the most effective antineoplastic agents in the treatment against ovarian cancer. Development of resistance to cisplatin during chemotherapy is common and hence a 5-year survival rate of women afflicted with this disease is just 18%[38]. Due to continued absence of effective early detection test for ovarian cancer, detailed knowledge of factors that confer tumour cell resistance to platinum compounds is a necessity. Britten et al. (2005) used SELDI-MS to identify low-mass proteins that are uniquely expressed in cisplatin-resistant OAW42 and 2780 cell lines. Two polypeptide peaks (m/z 5041 and 7324) were identified in these cisplatin-resistance ovarian cancer cells [38]. These findings suggest that SELDI-MS may be useful in monitoring the emergence of cisplatin-resistant tumour cell clones [38].
Similarly, Lauten et al. (2006) identified vasosin-containing protein as a putative marker for glucocorticoid resistance to human leukaemia cells from prednisone good responders and prednisone poor responders using SELDI-MS [39]. Taken together, these findings suggest that SELDI-MS may serve as an important diagnostic tool in identifying protein signatures that confer drug resistance.
In vitro applications
Cell culture models
Cell culture models are commonly used to study a multitude of diseases including cancer due to the lack of clinical models. The advantage of using cell lines over other conventional methods is that they are readily accessible and do not require excessive extraction procedures prior to usage. A number of cell culture models have been assessed by SELDI-MS to identify protein signatures for the detection, prognosis and the treatment of cancer (Table 4). Nakamura et al. (2006) used SELDI-MS analysis to detect proteins showing unique peaks in the renal cell carcinoma cell lines with different interferon (IFN) susceptibility. Five proteins with molecular masses of 8049, 3157, 3993, 8959 and 1623 D were detected. Comparison of these proteins may help to identify the IFN sensitivity [40].
4.
SELDI-MS studies performed on in vitromodels of cancer
| Study | Specimen | Cancer type | Proteins/peaks identified/detected |
|---|---|---|---|
| Akashi et al.[44] | Cell lines | Lung, colorectal, gastric, breast, ovarian, glioma, renal, melanoma | 2 |
| Britten et al.[38] | Cell lines | Ovarian | 2 |
| Wu, et al.[34] | Cell lines | Head and neck | 4 |
| Currid et al.[41] | HT-1080 | Fibrosarcoma | 3 |
| Nakamura et al.[40] | RCC | Kidney | 5 |
| Traub et al.[81] | HMEC and MCF-7 cell lines | Breast | 140 |
| Le et al.[43] | Human FaDu cells | ‡HNSCC | 1 |
| Chen et al.[82] | Myofibroblasts HT29 KM20 | Colon | 40 |
| Yim et al.[83] | Cell lines | Cervical | 8 |
‡HNSCC, head and neck squamous carcinoma.
Currid et al. (2006) employed SELDI-MS technology to characterise, at a proteomic level, factors released from HT-1080 human fibrosarcoma cells displaying inducible p21 expression. Three p21-regulated proteins were observed at 10.2, 11.7 and 13.4 kD. SDS-PAGE and MS analysis of tryptic digests identified the 13.4 kD protein as cystatin C, the 10.2 kD protein as pro-platelet basic protein (PPBP) and the 11.7 kD protein as β-2-microglobulin [41].
Using SELDI analysis in conjunction with MALDI-TOF/TOF MS, Dowling et al. (2007) detected and identified a 7.6 kD protein (fragment of bovine transferring) in serum-free conditioned media from paclitaxel-resistant superinvasive variant (MDA-MB-435S-F/Taxol10p4pSI) and human cancer drug-sensitive and invasive cell line (MDA-MB-435S-F) [42]. In another study, Le et al. (2005) used SELDI-MS analysis to detect a novel hypoxia-induced secreted protein in cell culture media derived from FaDu cells (HNSCC) that may have a role in malignant progression. A 15 kDa peak was identified by MS analysis to be galectin-1 [43].
Akashi et al. (2007) used SELDI-MS analysis to detect novel biomarkers for predicting sensitivity to a PI3K inhibitor, LY294002. Using SELDI-MS, a protein expression database was generated for 39 cancer cell lines. This database was combined with a previously determined chemosensitivity database (obtained by measuring the growth inhibition parameters of LY294002 for the cells in the 39 cancer cell lines). From this data, ribosomal P2 was identified. This study also showed that the phosphorylation status of ribosomal P2 was responsible for determining the sensitivity to PI3K inhibitors, especially LY294002, in the 39 cancer cell lines [44].
Melle et al. (2006) used SELDI-MS to identify molecular changes occurring in hepatocellular carcinomas (HCC). Microdissected cells from control liver tissue and hepatic tumour tissues were used in this study. Using peptide fingerprint mapping and SELDI-MS, 53 proteins were identified in the tumour tissue. Ferritin light subunit (FLS) and adenylate kinase 3 α-like 1 (AK3) showed decreased expressions in hepatic tumour, whilst biliverdin reductase B (BVRB) was up- regulated in HCC [45].
SELDI-MS technology has also allowed the discovery of 40 differentially expressed protein peaks between wild-type mice and mice treated with peroxisome proliferators to induce cancer in liver cells [46]. Taken together, these findings suggest that SELDI-MS in combination with mass spectrometry can be used to identify protein signatures for the detection, prognosis and the treatment of cancer from cell culture models.
Protein interactions
To further understand the biological function of a protein within a broader cellular context, the identification of protein–protein interactions is an important component [47]. Recent studies have shown that SELDI-MS technology can also be used to detect and identify protein–protein, protein/DNA or protein/metabolite interactions from crude biological samples. For example, Lehmann et al. (2007) studied the interaction of S100 proteins, known to be involved in several human diseases such as rheumatoid arthritis and cancer [47]. Using specific immunoaffinity beads and SELDI analysis, Lehmann (2007) was able to detect specific interactions between different S100 proteins [47]. In a similar study, Hegedus et al, 2007 carried out serum proteome analysis of individuals with different IL6–174G.C genotypes would provide insight on genotype–phenotype associations of this polymorphism and its role in disease susceptibility [48]. Specimens were obtained from healthy control individuals in an ongoing study of non-Hodgkin lymphoma. Using SELDI-MS analysis, an association of the -174C allele with increased apolipoprotein C-I was reported. Additionally, Hegedus et al. (2007) confirmed previous findings by others of an association of the -174C allele with lower autoantibodies to heat shock protein 60 and the absence of any association between the IL6–174G.C genotype and serum IL-6 levels [48]. Using SELDI-MS analysis in conjunction with 2D gel electrophoresis and ELISA, Escher et al. (2007) was able to detect and confirm posttranslational modifications of transthyretin in the sera of patients with mycosis fungoides [49]. These findings suggest that SELDI-MS can be used for the detection of protein interactions and modifications.
Advantages over conventional technologies
SELDI-MS offers a more efficient way to directly analyse proteins from crude biological samples compared to conventional laser desorption ionization (approaches such as MALDI) and/ionization MALDI approaches. Complex biological samples can be applied directly (do not require complicated treatment prior to analysis) due to specific retention of target proteins. Additionally, samples can be applied in minuscule amounts. Another major advantage of SELDI-MS is its rapid transition from discovery to assay on a single platform [50]. No pre-selection markers are required as many molecules are screened in a single experiment. Additionally, no biological knowledge about the pathophysiology of the disease is required. To date, it is the only proteomics platform available for the complete process of biomarker discovery/protein expression and identification, validation, purification, characterisation and assay development. Moreover, SELDI-MS provides a rapid and low-cost possibility to compare two samples. Another benefit is its greater specificity with the discovery of a larger amount of significant peaks [38]. The significant peaks/proteins highlighted throughout this review can be used to design predictive or diagnostic tests for different areas of oncology. Quality data analysis for this purpose is required but it is dependant on quality data production.
Limitations of SELDI-MS
Biomarker discovery using SELDI-MS relies on the consistency of the mass of proteins and peptides between samples and over time. Improper sample handling and storage, such as freeze thaw can lead to protein degradation, reducing the power of a study to detect differences between samples. The addition of protease inhibitors and/or bacterial inhibitors to samples and proper storage at –70°C not only circumvents these problems but also provides a better recovery of proteins. Extrinsic and intrinsic factors can have an impact on protein profiling [51]. For example, the complexity of biological samples can negatively influence the detection of biomarkers with the ProteinChip Array. The observed peak intensity of a particular protein may not reflect the actual amount present in the sample but rather result from the amount of protein actually bound to the ProteinChip array. ProteinChip arrays have limited binding capacity such that components present in complex samples compete for binding on the chip surface and and/or ionization energy [52]. Diamandis et al. (2002) argued that the ProteinChip surfaces preferentially bind high abundance proteins and that this technology will not be able to identify small alternations in the proteome caused by an early cancer [53].
Natural variability, such as age, gender and diet of the donor and changes in the sample preparation protocol, can influence the protein concentration in the sample and thus alter the resulting spectra. Another issue relates to the identification of discriminatory peaks (peptides, proteins or protein fragments) that have originated ex vivo due to samples standing at room temperature. Statistical analysis of spectra generated under such influences can produce artificial scores [12]. Another important methodological artefact is data analysis of protein profiles. Instrument settings such as calibration and matrix composition can all have an impact on protein profiling. Poor instrument resolution, ProteinChip variability, drift and noise of the spectrometer and the stability of this technology when constantly applied in a diagnostic mode contribute to poor reproducibility of spectra [54]. Users need to be aware of the issues surrounding analysis in order to produce high quality and biologically accurate spectra. The pre-processing of the data is the key step in SELDI-MS analysis. Pre-processing includes calibration, baseline correction, noise calculation, normalisation and peak detection. Calibration uses well-characterised peptides with known molecular weights to derive the calibration equation for a particular instrument. This application improves the molecular weight determination accuracy values of unknown peptides. The noise from the matrix is removed by baseline correction and removing the lower end of the spectrum from analysis. Baseline correction means removing the signal that is collected from the noise of the EAM and the signal made intrinsically from the SELDI-MS profiling process from the SELDI-MS spectra. The outcome should be the correct peak height and area for that spectrum and ideally the baseline of a spectrum should lie on the zero horizontal line. The m/z values between 0 and 2000 are generally removed from further analysis; however this range can vary from as low as 1000 D [16] to as stringent as 3000 D [55] depending on the individual experiment. Normalisation removes systematic variation between spectra. This helps to compensate for variation in the data including levels of sample concentration loaded onto the chip, levels of total protein in the sample and ion detection. Peak detection is a significant problem with SELDI-MS spectra as there is variation between peaks along the horizontal axis with m/z values and along the vertical axis with peak intensities. The peak detection settings with CiphergenExpress software (available from Ciphergen Biosystems) are user-defined, therefore the number of detected peaks can be easily altered by the user and therefore it is necessary to use parameter settings that are suitable for the biological samples under investigation [56]. Pre-processing SELDI-MS data can be viewed as a limitation as it essential but challenging and the analysis is individual for every experiment under investigation.
Another critical limitation of using SELDI-MS analysis is its inability to identify proteins or peptides [57]. Identified proteins and peptides not only provide biological information about the pathophysiology of the disease but may also contribute or lead to the identification of novel, therapeutic targets—this information may not be available if the SELDI-MS approach is used [58]. However, if an identified biomarker has been validated, then the SELDI-MS approach could then be used for a high-throughput, rapid-screening programme [58].
Another concern is the reproducibility of results between host institutions [59–63]. Recent studies have shown discrepancies between research groups in terms of data handling, instrument settings, study designs, experimental protocols and, bioinformatics tools used to generate data. Inter- and intratumour heterogeneity, inappropriate diagnosis and unexpected changes of cell line properties are also a concern. Other issues relate to the inability to identify classical cancer biomarkers, that is prostate specific antigen (PSA) is due to the low sensitivity of the SELDI-MS approach [64]. These obstacles must be overcome if SELDI-MS is to be routinely used for clinical diagnostic purposes.
Conclusion
This review highlights the potential use of SELDI-MS in identifying new diagnostic, prognostic and predictive protein signatures from biological samples obtained both in vitro and in vivo and will help in developing new molecularly targeted antineoplastic drugs. Several concerns with regard to reproducibility, data handling and interpretation across institutions remain to be addressed before this proteomics platform can enter clinical practice.
Acknowledgments
Funding is acknowledged from Science Foundation Ireland and the Health Research Board of Ireland, the latter under the auspices of the HRB Programme Grant “Breast Cancer Metastasis: Biomarkers and Functional Mediators”. UCD Conway Institute is funded by the Programme for Third Level Institutions (PRTLI), as administered by the Higher Education Authority (HEA) of Ireland.
References
- 1.Pisitkun T, Johnstone R, Knepper MA. Discovery of urinary biomarkers. Mol Cell Proteomics. 2006;5:1760–71. doi: 10.1074/mcp.R600004-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Etzioni R, Urban N, Ramsey S, McIntosh M, Schwartz S, Reid B, Radich J, Anderson G, Hartwell L. The case for early detection. Nat Rev Cancer. 2003;3:243–52. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 4.Weissinger EM, Mischak H, Ganser A, Hertenstein B. Value of proteomics applied to the follow-up in stem cell transplantation. Ann Hematol. 2006;85:205–11. doi: 10.1007/s00277-005-0057-1. [DOI] [PubMed] [Google Scholar]
- 5.Belluco C, Petricoin EF, Mammano E, Facchiano F, Ross-Rucker S, Nitti D, Maggio CD, Liu C, Lise M, Liotta LA, Whiteley G. Serum proteomic analysis identifies a highly sensitive and specific discriminatory pattern in stage 1 breast cancer. Ann Surg Oncol. 2007;14:2470–6. doi: 10.1245/s10434-007-9354-3. [DOI] [PubMed] [Google Scholar]
- 6.Elmore JG, Armstrong K, Lehman CD, Fletcher SW. Screening for breast cancer. Jama. 2005;293:1245–56. doi: 10.1001/jama.293.10.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker S, Cazares LH, Watson P, Lynch H, Semmes OJ, Drake RR, Laronga C. Surfaced-enhanced laser desorp-tion/ionization time-of-flight (SELDI-TOF) differentiation of serum protein profiles of BRCA-1 and sporadic breast cancer. Ann Surg Oncol. 2004;11:907–14. doi: 10.1245/ASO.2004.03.557. [DOI] [PubMed] [Google Scholar]
- 8.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–67. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 9.Pawlik TM, Fritsche H, Coombes KR, Xiao L, Krishnamurthy S, Hunt KK, Pusztai L, Chen JN, Clarke CH, Arun B, Hung MC, Kuerer HM. Significant differences in nipple aspirate fluid protein expression between healthy women and those with breast cancer demonstrated by time-of-flight mass spectrometry. Breast Cancer Res Treat. 2005;89:149–57. doi: 10.1007/s10549-004-1710-4. [DOI] [PubMed] [Google Scholar]
- 10.Yang SY, Xiao XY, Zhang WG, Zhang LJ, Zhang W, Zhou B, Chen G, He DC. Application of serum SELDI proteomic patterns in diagnosis of lung cancer. BMC Cancer. 2005;5:83. doi: 10.1186/1471-2407-5-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhukov TA, Johanson RA, Cantor AB, Clark RA, Tockman MS. Discovery of distinct protein profiles specific for lung tumors and pre-malignant lung lesions by SELDI mass spectrometry. Lung Cancer. 2003;40:267–79. doi: 10.1016/s0169-5002(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs IJ, Menon U. Progress and challenges in screening for early detection of ovarian cancer. Mol Cell Proteomics. 2004;3:355–66. doi: 10.1074/mcp.R400006-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Petricoin EF, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, Steinberg SM, Mills GB, Simone C, Fishman DA, Kohn EC, Liotta LA. Use of proteomic patterns in serum to identify ovarian cancer. Lancet. 2002;359:572–7. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 14.Lin YW, Lin CY, Lai HC, Chiou JY, Chang CC, Yu MH, Chu TY. Plasma proteomic pattern as biomarkers for ovarian cancer. Int J Gynecol Cancer. 2006;16:139–46. doi: 10.1111/j.1525-1438.2006.00475.x. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Z, Bast RC, Jr, Yu Y, Li J, Sokoll LJ, Rai AJ, Rosenzweig JM, Cameron B, Wang YY, Meng XY, Berchuck A, Van Haaften-Day C, Hacker NF, De Bruijn HW, Van Der Zee AG, Jacobs IJ, Fung ET, Chan DW. Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res. 2004;64:5882–90. doi: 10.1158/0008-5472.CAN-04-0746. [DOI] [PubMed] [Google Scholar]
- 16.Su Y, Shen J, Qian H, Ma H, Ji J, Ma H, Ma L, Zhang W, Meng L, Li Z, Wu J, Jin G, Zhang J, Shou C. Diagnosis of gastric cancer using decision tree classification of mass spectral data. Cancer Sci. 2006;98:37–43. doi: 10.1111/j.1349-7006.2006.00339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen YD, Zheng S, Yu JK, Hu X. Artificial neural networks analysis of surface-enhanced laser desorption/ionization mass spectra of serum protein pattern distinguishes colorectal cancer from healthy population. Clin Cancer Res. 2004;10:8380–5. doi: 10.1158/1078-0432.CCR-1162-03. [DOI] [PubMed] [Google Scholar]
- 18.Engwegen JY, Helgason HH, Cats A, Harris N, Bonfrer JM, Schellens JH, Beijnen JH. Identification of serum proteins discriminating colorectal cancer patients and healthy controls using surface-enhanced laser desorption ionisation-time of flight mass spectrometry. World J Gastroenterol. 2006;12:1536–44. doi: 10.3748/wjg.v12.i10.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bosch FX, Lorincz A, Munoz N, Meijer CJ, Shah KV. The causal relation between human papillomavirus and cervical cancer. Journal of Clinical Pathology. 2002;55:244–65. doi: 10.1136/jcp.55.4.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong YF, Cheung TH, Lo KW, Wang VW, Chan CS, Ng TB, Chung TK, Mok SC. Protein profiling of cervical cancer by protein-biochips: proteomic scoring to discriminate cervical cancer from normal cervix. Cancer Letters. 2004;211:227–34. doi: 10.1016/j.canlet.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 21.Von Eggeling F, Junker K, Fiedle W, Wollscheid V, Durst M, Claussen U, Ernst G. Mass spectrometry meets chip technology: a new proteomic tool in cancer research? Electrophoresis. 2001;22:2898–902. doi: 10.1002/1522-2683(200108)22:14<2898::AID-ELPS2898>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 22.Abu-Hamda EM, Baron TH. Endoscopic management of cholangiocarcinoma. Seminars in Liver Disease. 2004;24:165–75. doi: 10.1055/s-2004-828893. [DOI] [PubMed] [Google Scholar]
- 23.Scarlett CJ, Saxby AJ, Nielsen A, Bell C, Samra JS, Hugh T, Baxter RC, Smith RC. Proteomic profiling of cholangiocarcinoma: diagnostic potential of SELDI-TOF MS in malignant bile duct stricture. Hepatology. 2006;44:658–66. doi: 10.1002/hep.21294. [DOI] [PubMed] [Google Scholar]
- 24.De Bont JM, Den Boer ML, Reddingius RE, Jansen J, Passier M, Van Schaik RH, Kros JM, Sillevis Smitt PA, Luider TH, Pieters R. Identification of apolipoprotein A-II in cerebrospinal fluid of pediatric brain tumor patients by protein expression profiling. Clin Chem. 2006;52:1501–9. doi: 10.1373/clinchem.2006.069294. [DOI] [PubMed] [Google Scholar]
- 25.Xu WH, Chen YD, Hu Y, Yu JK, Wu XG, Jiang TJ, Zheng S, Zhang SZ. Preoperatively molecular staging with CM10 ProteinChip and SELDI-TOF-MS for colorectal cancer patients. Journal of Zhejiang University. Science. 2006;7:235–40. doi: 10.1631/jzus.2006.B0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menard C, Johann D, Lowenthal M, Muanza T, Sproull M, Ross S, Gulley J, Petricoin E, Coleman CN, Whiteley G, Liotta L, Camphausen K. Discovering clinical biomarkers of ionizing radiation exposure with serum proteomic analysis. Cancer Res. 2006;66:1844–50. doi: 10.1158/0008-5472.CAN-05-3466. [DOI] [PubMed] [Google Scholar]
- 27.Wibom C, Pettersson F, Sjostrom M, Henriksson R, Johansson M, Bergenheim AT. Protein expression in experimental malignant glioma varies over time and is altered by radiotherapy treatment. Br J Cancer. 2006;94:1853–63. doi: 10.1038/sj.bjc.6603190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pusztai L, Gregory BW, Baggerly KA, Peng B, Koomen J, Kuerer HM, Esteva FJ, Symmans WF, Wagner P, Hortobagyi GN, Laronga C, Semmes OJ, Wright GL, Jr, Drake RR, Vlahou A. Pharmacoproteomic analysis of prechemotherapy and postchemotherapy plasma samples from patients receiving neoadjuvant or adjuvant chemotherapy for breast carcinoma. Cancer. 2004;100:1814–22. doi: 10.1002/cncr.20203. [DOI] [PubMed] [Google Scholar]
- 29.Smith FM, Gallagher WM, Fox E, Stephens RB, Rexhepaj E, Petricoin EF, 3rd, Liotta L, Kennedy MJ, Reynolds JV. Combination of SELDI-TOF-MS and data mining provides early-stage response prediction for rectal tumors undergoing multi-modal neoadjuvant therapy. Ann Surg. 2007;245:259–66. doi: 10.1097/01.sla.0000245577.68151.bd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baker AF, Dragovich T, Tate WR, Ramanathan RK, Roe D, Hsu CH, Kirkpatrick DL, Powis G. The antitumor thioredoxin-1 inhibitor PX-12 (1-methyl-propyl 2-imidazolyl disulfide) decreases thioredoxin-1 and VEGF levels in cancer patient plasma. J. Lab. Clin. Med. 2006;147:83–90. doi: 10.1016/j.lab.2005.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hayashida Y, Honda K, Osaka Y, Hara T, Umaki T, Tsuchida A, Aoki T, Hirohashi S, Yamada T. Possible prediction of chemoradiosensitivity of esophageal cancer by serum protein profiling. Clin Cancer Res. 2005;11:8042–7. doi: 10.1158/1078-0432.CCR-05-0656. [DOI] [PubMed] [Google Scholar]
- 32.Nakagawa T, Huang SK, Martinez SR, Tran AN, Elashoff D, Ye X, Turner RR, Giuliano AE, Hoon DS. Proteomic profiling of primary breast cancer predicts axillary lymph node metastasis. Cancer Res. 2006;66:11825–30. doi: 10.1158/0008-5472.CAN-06-2337. [DOI] [PubMed] [Google Scholar]
- 33.Goncalves A, Esterni B, Bertucci F, Sauvan R, Chabannon C, Cubizolles M, Bardou VJ, Houvenaegel G, Jacquemier J, Granjeaud S, Meng XY, Fung ET, Birnbaum D, Maraninchi D, Viens P, Borg JP. Postoperative serum proteomic profiles may predict metastatic relapse in high-risk primary breast cancer patients receiving adjuvant chemotherapy. Oncogene. 2006;25:981–9. doi: 10.1038/sj.onc.1209131. [DOI] [PubMed] [Google Scholar]
- 34.Wu W, Tang X, Hu W, Lotan R, Hong WK, Mao L. Identification and validation of metastasis-associated proteins in head and neck cancer cell lines by two-dimensional electrophoresis and mass spectrometry. Clin Exp Metastasis. 2002;19:319–26. doi: 10.1023/a:1015515119300. [DOI] [PubMed] [Google Scholar]
- 35.Wadsworth JT, Somers KD, Cazares LH, Malik G, Adam BL, Stack BC, Jr, Wright GL, Jr, Semmes OJ. Serum protein profiles to identify head and neck cancer. Clin Cancer Res. 2004;10:1625–32. doi: 10.1158/1078-0432.ccr-0297-3. [DOI] [PubMed] [Google Scholar]
- 36.Roesch-Ely M, Nees M, Karsai S, Ruess A, Bogumil R, Warnken U, Schnolzer M, Dietz A, Plinkert PK, Hofele C, Bosch FX. Proteomic analysis reveals successive aberrations in protein expression from healthy mucosa to invasive head and neck cancer. Oncogene. 2007;26:54–64. doi: 10.1038/sj.onc.1209770. [DOI] [PubMed] [Google Scholar]
- 37.Schwegler EE, Cazares L, Steel LF, Adam BL, Johnson DA, Semmes OJ, Block TM, Marrero JA, Drake RR. SELDI-TOF MS profiling of serum for detection of the progression of chronic hepatitis C to hepato-cellular carcinoma. Hepatology Baltimore, Md. 2005;41:634–42. doi: 10.1002/hep.20577. [DOI] [PubMed] [Google Scholar]
- 38.Britten RA, Hardy C, Vlahou A, Gregory B, Giri PS, Drake R. Identification of reproducible low mass SELDI protein profiles specific to cisplatin resistance in human ovarian cancer cells. Oncol Rep. 2005;14:1323–30. [PubMed] [Google Scholar]
- 39.Lauten M, Schrauder A, Kardinal C, Harbott J, Welte K, Schlegelberger B, Schrappe M, Von Neuhoff N. Unsupervised proteome analysis of human leukaemia cells identifies the Valosin-containing protein as a putative marker for glucocorticoid resistance. Leukemia. 2006;20:820–6. doi: 10.1038/sj.leu.2404162. [DOI] [PubMed] [Google Scholar]
- 40.Nakamura K, Yoshikawa K, Yamada Y, Saga S, Aoki S, Taki T, Tobiume M, Shimazui T, Akaza H, Honda N. Differential profiling analysis of proteins involved in anti-proliferative effect of inter-feron-alpha on renal cell carcinoma cell lines by protein biochip technology. Int J Oncol. 2006;28:965–70. [PubMed] [Google Scholar]
- 41.Currid CA, O’Connor DP, Chang BD, Gebus C, Harris N, Dawson KA, Dunn MJ, Pennington SR, Roninson IB, Gallagher WM. Proteomic analysis of factors released from p21-overexpressing tumour cells. Proteomics. 2006;6:3739–53. doi: 10.1002/pmic.200500787. [DOI] [PubMed] [Google Scholar]
- 42.Dowling P, Maurya P, Meleady P, Glynn SA, Dowd AJ, Henry M, Clynes M. Purification and identification of a 7.6-kDa protein in media conditioned by superinvasive cancer cells. Anticancer Research. 2007;27:1309–17. [PubMed] [Google Scholar]
- 43.Le QT, Shi G, Cao H, Nelson DW, Wang Y, Chen EY, Zhao S, Kong C, Richardson D, O’Byrne KJ, Giaccia AJ, Koong AC. Galectin-1: a link between tumor hypoxia and tumor immune privilege. J Clin Oncol. 2005;23:8932–41. doi: 10.1200/JCO.2005.02.0206. [DOI] [PubMed] [Google Scholar]
- 44.Akashi T, Nishimura Y, Wakatabe R, Shiwa M, Yamori T. Proteomics-based identification of biomarkers for predicting sensitivity to a PI3-kinase inhibitor in cancer. Biochem Biophys Res Commun. 2007;352:514–21. doi: 10.1016/j.bbrc.2006.11.052. [DOI] [PubMed] [Google Scholar]
- 45.Melle C, Ernst G, Scheibner O, Kaufmann R, Schimmel B, Bleul A, Settmacher U, Hommann M, Claussen U, Eggeling FV. Identification of specific protein markers in microdissected hepatocellular carcinoma. J Proteome Res. 2007;6:306–15. doi: 10.1021/pr060439b. [DOI] [PubMed] [Google Scholar]
- 46.Chu R, Zhang W, Lim H, Yeldandi AV, Herring C, Brumfield L, Reddy JK, Davison M. Profiling of acyl-CoA oxidase-deficient and peroxisome proliferator Wy14,643-treated mouse liver protein by surface-enhanced laser desorption/ionization ProteinChip Biology System. Gene Expr. 2002;10:165–77. doi: 10.3727/000000002783992460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lehmann R, Melle C, Escher N, Von Eggeling F. Detection and identification of protein interactions of S100 proteins by ProteinChip technology. J Proteome Res. 2005;4:1717–21. doi: 10.1021/pr050163s. [DOI] [PubMed] [Google Scholar]
- 48.Hegedus CM, Skibola CF, Bracci P, Holly EA, Smith MT. Screening the human serum proteome for genotype-phenotype associations: an analysis of the IL6 -174G>C polymorphism. Proteomics. 2007;7:548–57. doi: 10.1002/pmic.200600366. [DOI] [PubMed] [Google Scholar]
- 49.Escher N, Kaatz M, Melle C, Hipler C, Ziemer M, Driesch D, Wollina U, Von Eggeling F. Posttranslational modifications of transthyretin are serum markers in patients with mycosis fungoides. Neoplasia. 2007;9:254–9. doi: 10.1593/neo.06805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liotta LA, Ferrari M, Petricoin E. Clinical proteomics: written in blood. Nature. 2003;425:905. doi: 10.1038/425905a. [DOI] [PubMed] [Google Scholar]
- 51.Schaub S, Wilkins J, Weiler T, Sangster K, Rush D, Nickerson P. Urine protein profiling with surface-enhanced laser-desorption/ionization time-of-flight mass spectrometry. Kidney Int. 2004;65:323–32. doi: 10.1111/j.1523-1755.2004.00352.x. [DOI] [PubMed] [Google Scholar]
- 52.Van Breemen MJ, Bleijlevens B, De Koster CG, Aerts JM. Limitations in quantitation of the biomarker CCL18 in Gaucher disease blood samples by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry. Biochim Biophys Acta. 2006;1764:1626–32. doi: 10.1016/j.bbapap.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 53.Diamandis EP. Proteomic patterns in serum and identification of ovarian cancer. Lancet. 2002;360:170. doi: 10.1016/s0140-6736(02)09390-x. author reply 70–1. [DOI] [PubMed] [Google Scholar]
- 54.Everley PA, Zetter BR. Proteomics in tumor progression and metastasis. Ann N Y Acad Sci. 2005;1059:1–10. doi: 10.1196/annals.1339.001. [DOI] [PubMed] [Google Scholar]
- 55.Shi Q, Harris LN, Lu X, Li X, Hwang J, Gentleman R, Iglehart JD, Miron A. Declining plasma fibrinogen alpha fragment identifies HER2-positive breast cancer patients and reverts to normal levels after surgery. J Proteome Res. 2006;5:2947–55. doi: 10.1021/pr060099u. [DOI] [PubMed] [Google Scholar]
- 56.Beyer S, Walter Y, Hellmann J, Kramer PJ, Kopp-Schneider A, Kroeger M, Ittrich C. Comparison of software tools to improve the detection of carcinogen induced changes in the rat liver proteome by analyzing SELDI-TOF-MS spectra. J Proteome Res. 2006;5:254–61. doi: 10.1021/pr050279o. [DOI] [PubMed] [Google Scholar]
- 57.Merchant M, Weinberger SR. Recent advancements in surface-enhanced laser desorption/ionization-time of flight-mass spectrometry. Electrophoresis. 2000;21:1164–77. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1164::AID-ELPS1164>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 58.Downes MR, Byrne JC, Dunn MJ, Fitzpatrick JM, Watson RW, Pennington SR. Application of proteomic strategies to the identification of urinary biomarkers for prostate cancer: a review. Biomarkers. 2006;11:406–16. doi: 10.1080/13547500600799821. [DOI] [PubMed] [Google Scholar]
- 59.Diamandis EP. Analysis of serum proteomic patterns for early cancer diagnosis: drawing attention to potential problems. J Natl Cancer Inst. 2004;96:353–6. doi: 10.1093/jnci/djh056. [DOI] [PubMed] [Google Scholar]
- 60.Diamandis EP. Mass spectrometry as a diagnostic and a cancer biomarker discovery tool: opportunities and potential limitations. Mol Cell Proteomics. 2004;3:367–78. doi: 10.1074/mcp.R400007-MCP200. [DOI] [PubMed] [Google Scholar]
- 61.Diamandis EP. Re: diagnostic potential of serum proteomic patterns in prostate cancer. J Urol. 2004;171:1244–5. doi: 10.1097/01.ju.0000112784.51142.bd. author reply 124–5-64. [DOI] [PubMed] [Google Scholar]
- 62.Sorace JM, Zhan M. A data review and re-assessment of ovarian cancer serum proteomic profiling. BMC Bioinformatics. 2003;4:24. doi: 10.1186/1471-2105-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baggerly KA, Edmonson SR, Morris JS, Coombes KR. High-resolution serum proteomic patterns for ovarian cancer detection. Endocr Relat Cancer. 2004;11:583–4. doi: 10.1677/erc.1.00868. [DOI] [PubMed] [Google Scholar]
- 64.Banez LL, Prasanna P, Sun L, Ali A, Zou Z, Adam BL, McLeod DG, Moul JW, Srivastava S. Diagnostic potential of serum proteomic patterns in prostate cancer. J Urol. 2003;170:442–6. doi: 10.1097/01.ju.0000069431.95404.56. [DOI] [PubMed] [Google Scholar]
- 65.Fung ET, Yip TT, Lomas L, Wang Z, Yip C, Meng XY, Lin S, Zhang F, Zhang Z, Chan DW, Weinberger SR. Classification of cancer types by measuring variants of host response proteins using SELDI serum assays. Int J Cancer. 2005;115:783–9. doi: 10.1002/ijc.20928. [DOI] [PubMed] [Google Scholar]
- 66.Li J, Zhang Z, Rosenzweig J, Wang YY, Chan DW. Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clin Chem. 2002;48:1296–304. [PubMed] [Google Scholar]
- 67.Sauter ER, Shan S, Hewett JE, Speckman P, Du Bois GC. Proteomic analysis of nipple aspirate fluid using SELDI-TOF-MS. Int J Cancer. 2005;114:791–6. doi: 10.1002/ijc.20742. [DOI] [PubMed] [Google Scholar]
- 68.Liu XP, Shen J, Li ZF, Yan L, Gu J. A serum proteomic pattern for the detection of colorectal adenocarcinoma using surface enhanced laser desorption and ionization mass spectrometry. Cancer Invest. 2006;24:747–53. doi: 10.1080/07357900601063873. [DOI] [PubMed] [Google Scholar]
- 69.Soltys SG, Le QT, Shi G, Tibshirani R, Giaccia AJ, Koong AC. The use of plasma surface-enhanced laser desorp-tion/ionization time-of-flight mass spectrometry proteomic patterns for detection of head and neck squamous cell cancers. Clin Cancer Res. 2004;10:4806–12. doi: 10.1158/1078-0432.CCR-03-0469. [DOI] [PubMed] [Google Scholar]
- 70.Wang JX, Yu JK, Wang L, Liu QL, Zhang J, Zheng S. Application of serum protein fingerprint in diagnosis of papillary thyroid carcinoma. Proteomics. 2006;6:5344–9. doi: 10.1002/pmic.200500833. [DOI] [PubMed] [Google Scholar]
- 71.Rosty C, Christa L, Kuzdzal S, Baldwin WM, Zahurak ML, Carnot F, Chan DW, Canto M, Lillemoe KD, Cameron JL, Yeo CJ, Hruban RH, Goggins M. Identification of hepatocarcinoma-intestine-pancreas/pancreatitis-associated protein I as a biomarker for pancreatic ductal adenocarci-noma by protein biochip technology. Cancer Res. 2002;62:1868–75. [PubMed] [Google Scholar]
- 72.Yu Y, Chen S, Wang LS, Chen WL, Guo WJ, Yan H, Zhang WH, Peng CH, Zhang SD, Li HW, Chen GQ. Prediction of pancreatic cancer by serum biomarkers using surface-enhanced laser desorption/ionization-based decision tree classification. Oncology. 2005;68:79–86. doi: 10.1159/000084824. [DOI] [PubMed] [Google Scholar]
- 73.Koopmann J, Zhang Z, White N, Rosenzweig J, Fedarko N, Jagannath S, Canto MI, Yeo CJ, Chan DW, Goggins M. Serum diagnosis of pancreatic adenocarcinoma using surface-enhanced laser desorption and ionization mass spectrometry. Clin Cancer Res. 2004;10:860–8. doi: 10.1158/1078-0432.ccr-1167-3. [DOI] [PubMed] [Google Scholar]
- 74.Qian HG, Shen J, Ma H, Ma HC, Su YH, Hao CY, Xing BC, Huang XF, Shou CC. Preliminary study on proteomics of gastric carcinoma and its clinical significance. World J Gastroenterol. 2005;11:6249–53. doi: 10.3748/wjg.v11.i40.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lehrer S, Roboz J, Ding H, Zhao S, Diamond EJ, Holland JF, Stone NN, Droller MJ, Stock RG. Putative protein markers in the sera of men with prostatic neoplasms. BJU Int. 2003;92:223–5. doi: 10.1046/j.1464-410x.2003.04341.x. [DOI] [PubMed] [Google Scholar]
- 76.Malik G, Ward MD, Gupta SK, Trosset MW, Grizzle WE, Adam BL, Diaz JI, Semmes OJ. Serum levels of an isoform of apolipoprotein A-II as a potential marker for prostate cancer. Clin Cancer Res. 2005;11:1073–85. [PubMed] [Google Scholar]
- 77.Ye B, Cramer DW, Skates SJ, Gygi SP, Pratomo V, Fu L, Horick NK, Licklider LJ, Schorge JO, Berkowitz RS, Mok SC. Haptoglobin-alpha subunit as potential serum biomarker in ovarian cancer: identification and characterization using proteomic profiling and mass spectrometry. Clin Cancer Res. 2003;9:2904–11. [PubMed] [Google Scholar]
- 78.Moshkovskii SA, Serebryakova MV, Kuteykin-Teplyakov KB, Tikhonova OV, Goufman EI, Zgoda VG, Taranets IN, Makarov OV, Archakov AI. Ovarian cancer marker of 11.7 kDa detected by proteomics is a serum amyloid A1. Proteomics. 2005;5:3790–7. doi: 10.1002/pmic.200401205. [DOI] [PubMed] [Google Scholar]
- 79.Wilson LL, Tran L, Morton DL, Hoon DS. Detection of differentially expressed proteins in early-stage melanoma patients using SELDI-TOF mass spectrometry. Ann N Y Acad Sci. 2004;1022:317–22. doi: 10.1196/annals.1318.047. [DOI] [PubMed] [Google Scholar]
- 80.Zhu LR, Zhang WY, Yu L, Zheng YH, Zhang JZ, Liao QP. Serum proteomic features for detection of endometrial cancer. Int J Gynecol Cancer. 2006;16:1374–8. doi: 10.1111/j.1525-1438.2006.00561.x. [DOI] [PubMed] [Google Scholar]
- 81.Traub F, Feist H, Kreipe HH, Pich A. SELDI-MS-based expression profiling of ductal invasive and lobular invasive human breast carcinomas. Pathol Res Pract. 2005;201:763–70. doi: 10.1016/j.prp.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 82.Chen AL, Soman KV, Rychahou PG, Luxon BA, Evers BM. Proteomic analysis of colonic myofibroblasts and effect on colon cancer cell proliferation. Surgery. 2005;138:382–90. doi: 10.1016/j.surg.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 83.Yim EK, Lee KH, Namkoong SE, Um SJ, Park JS. Proteomic analysis of ursolic acid-induced apoptosis in cervical carcinoma cells. Cancer letters. 2006;235:209–20. doi: 10.1016/j.canlet.2005.04.007. [DOI] [PubMed] [Google Scholar]


