Abstract
A recent study showed cardioprotective effects of resveratrol on the diabetic heart. The present study sought to compare the protein profiles of the normal versus diabetic hearts after resveratrol treatment using differential proteomic analysis. Rats were randomly divided into two groups: control and diabetic. Both groups of rats were fed resveratrol (2.5 mg/kg/day) for 7 days, and then the rats were sacrificed, hearts were isolated and cytoplasmic fraction from left ventricular tissue was collected to carry out proteomic profiling as well as immunoblotting. Compared to normal hearts, diabetic hearts show increased myocardial infarct size and cardiomy-ocyte apoptosis upon ex vivo global ischaemia of 30 min. followed by 2 hrs of reperfusion. Resveratrol reduced infarct size and apop-totic cell death for both the groups, but the extent of infarct size and apoptosis remained higher for the diabetic group compared to the normal group. The left ventricular cytoplasmic proteins were analysed by 2D-DIGE and differentially displayed bands were further analysed by nano Liquid Chromatography-Mass Spectroscopy (LC-MS/MS). The results showed differential regulation of normal versus diabetic hearts treated with resveratrol of many proteins related to energy metabolism of which several were identified as mitochondrial proteins. Of particular interest is the increased expression of several chaperone proteins and oxidative stress and redox proteins in the diabetic group including Hsc70, HSPp6, GRP75, peroxiredoxin (Prdx)-1 and Prdx-3 whose expression was reversed by resveratrol. Western blot analysis was performed to validate the up- or down-regulation of these stress proteins. The results indicate the differential regulation by resveratrol of stress proteins in diabetic versus normal hearts, which may explain in part the beneficial effects of resveratrol in diabetic induced cardiovascular complications.
Keywords: diabetes, proteomics, stress proteins, heat shock proteins, redox proteins, oxidative stress-inducible proteins
Introduction
Consumption of red wine reduces oxidative stress caused by increased blood glucose levels after meals or in diabetic conditions [1–5]. Resveratrol, the principal effector constituent of red wine, can also alleviate diabetes-induced complications [6–11]. Since diabetes is the major cardiovascular risk factor [12] and since resveratrol can reduce diabetes-induced cardiovascular complications [7], the mechanisms of action of resveratrol-medi-ated reduction of cardiovascular dysfunction need to be better understood. Several studies from our laboratory revealed that resveratrol provides cardioprotection through the modulation of several stress proteins [13–14] as well as redox-regulated proteins [15, 16]. Interestingly, the same redox regulated and stress proteins also play a significant role in the diabetic hearts [17–19]. There was an increased association between plasma levels of heat shock protein 60 and cardiovascular disease in patients with diabetes mellitus [17]. Phosphorylated heat shock protein 27 was found to be involved in enhanced heart tolerance to ischaemia in short-term type 1 diabetic rats [19].
To further explore the role of stress proteins and redox proteins in resveratrol-mediated cardioprotection in the diabetic heart, we sought to examine differential proteomic profiling of the effects of resveratrol on the normal versus diabetic hearts. A recent proteomics study showed the role of several stress proteins including HSP 27 and αβ-crystallin on resveratrol pre-conditioning of the ischaemic myocardium [16]. We undertook a modified approach to assess the proteomic profiling of the effects of resveratrol on the diabetic hearts. The results of our study provide valuable information on the regulation of stress-and redox- regulated proteins in the resveratrol-mediated protection of diabetic hearts.
Materials and methods
Animals
All animals used in this study were treated in compliance with the principles of the laboratory animal care formulated by the National Society for Medical Research and Guide for the Care and Use of Laboratory Animals prepared by the National Academy of Sciences and published by the National Institutes of Health (Publication Number NIH 85-23, revised 1985). Sprague Dawley male rats weighing between 250–300 g were fed regular rat chow ad libitum with free access to water until the start of the experimental procedure. The rats were randomly assigned to one of the two groups: normal and diabetic. Diabetes was induced by one intraperi-toneal administration of streptozotocin (65 mg/kg) dissolved in citrate buffer (pH 4.5). The rats whose blood glucose was more than 400 mg/dl after 7 days were considered diabetic. Both normal and diabetic rats were fed resveratrol (2.5 mg/kg) dissolved in 30% ethanol by gavaging for 7 days. Then hearts were isolated and perfused with Krebs Henseleit buffer in Langendorff apparatus for 15 min. Hearts were removed and stored for proteomic and immunoblot analysis. A group of hearts was subjected to global ischaemia for 30 min. followed by reperfusion for 2 hrs. At the end of reperfusion, hearts were stored for infarct size or apoptosis analysis.
Infarct size estimation
At the end of reperfusion, the left ventricle was cut into transverse slices. The slices were incubated in 1% triphenyl tetrazolium solution in phosphate buffer (Na2HPO4 88 mM, NaH2PO4 1.8 mM, pH 7.4) for 20 min. at 37°C. This procedure distinguishes necrotic tissue from viable myocardium. The slices were stored for 48 hrs in 10% buffered formalin. The heart slices were photographed and the weights of the slices were monitored. Digital images of the slices were magnified, and the area of necrosis in each slice was quantified by computerized planimetry. The risk and infarct volumes in cm3 of each slice were then calculated on the basis of slice weight to remove the introduction of any errors due to non-uniformity of heart slice thickness. The risk volumes and infarct volumes of each slice were summed to obtain the risk and infarct volumes for the whole heart. Infarct size was taken to be the percent infarct volume of risk volume for any one heart.
TUNEL assay for assessment of apoptotic cell death
Immunohistochemical detection of apoptotic cells was carried out using TUNEL. Left ventricular tissue sections were incubated with mouse monoclonal antibody recognizing cardiac a-myosin heavy chain to specifically recognize apoptotic cardiomyocytes. The fluorescence staining was viewed with a confocal laser microscope. The number of apoptotic cardiomyocytes was counted and expressed as a percentage of total myocyte population.
Analysis of cytoplasmic proteins by 2D-DIGE
About 100 mg of the left ventricular tissue was excised from non-diabetic and diabetic resveratrol-treated groups (n = 6 each) and homogenized in an equal volume of 40mM Tris-HCl, 1mM ethylenediaminetetraacetic acid (EDTA), pH 8.5 supplemented with protease inhibitor cocktail (GE Healthcare Bio-Sciences, Piscataway NJ, USA) with a microdismembrator (Braun, Melsungen, Germany) at liquid nitrogen temperature. Frozen powder was collected and two volumes of Tris-buffer and one volume of 5% Triton X-100 in Tris-buffer both supplemented with protease inhibitor were added on ice. Samples were sonicated in a Bioruptor (Diagenode, Liège, Belgium) for 15 min. on ice water at setting ‘high’ with 15 sec. on/off cycles and incubated on ice for another 15 min. After subsequent centrifu-gation at 4°C for 15 min. at 13,400 g, the first supernatant containing the cytosolic fraction was collected. The pellet (myofilament fraction) was re-suspended in two volumes of 1% Triton X-100 in Tris-buffer supplemented with protease inhibitors and re-centrifuged. The combined super-natants completed the cytosolic fraction. Protein amount was determined with the DC protein assay (BioRad, Hercules, CA, USA). Extracts were stored at −80°C.
A pool sample was prepared containing equal protein amounts of all samples. Aliquots of 50 μg of this pool were labelled with CyDye DIGE Fluor Cy2 (GE Healthcare Bio-Sciences, Piscataway, NJ, USA) by incubation with 400 pmol of the dye. Other 50 μg aliquots of individual samples were either labelled with CyDye DIGE Fluor Cy3 or Cy5 (GE Healthcare Bio-Sciences, Piscataway, NJ, USA) according to manufacturer's instructions (minimal labelling). Bias towards a certain dye was prevented by labelling half of the samples of a group with Cy3 and the other half of that group with Cy5 (dye swap). One Cy3 labelled sample and one Cy5 labelled sample of the other group were combined with a Cy2 labelled pool sample in re-hydration buffer supplemented with 69 mM 1, 4 Dithiothreitol (DTT), 0.6% v/v Biolytes (BioRad, Hercules, CA, USA) and a few grains of bromophenol blue in a total volume of 450 μl. An additional 100 μg of unlabelled pool sample was added previously to increase the amount of protein available for spot picking and further analysis. Isoelectric focusing was performed using 24 cm IPG strips (GE Healthcare Bio-Sciences, Piscataway, NJ, USA) with a non-linear pH range of 3–11. Samples were transferred to a re-swelling tray and dry IPG strips were placed on top for overnight re-hydration of the strips at room temperature. Next, the proteins in the strips were focused in a Protean IEF Cell (BioRad, Hercules, CA, USA) according to the following protocol: 300V for 3 hrs followed by a linear increase to 8000 V in 9 hrs and 8000 V steady until 55kVh. Next the strips were equilibrated in SDS-equilibration buffer as described [16]. SDS-PAGE was performed using the EttanDalt Twelve system (GE Healthcare Bio-Sciences, Piscataway, NJ, USA) and 12% gels fixed to the large glass plates by Bindsilane treatment. Strips were sealed in place with a 1% agarose solution in running buffer. The 2-D elec-trophoresis was carried out at 5 mA/gel for 1 hr followed by 12.5 mA/gel at 25°C until the bromophenol blue front reached the bottom of the glass plate. After the 2-DE separation the gels were immediately scanned sequentially for Cy2, Cy3 and Cy5 fluorescence on a Typhoon 9410 imag-er (GE Healthcare Bio-Sciences, Piscataway, NJ, USA) at 100-μm resolution. Thereafter two of the gels were fixed overnight in 40% ethanol, 10% acetic acid and post stained with Sypro Ruby fluorescent protein stain and scanned in a Typhoon 9410 imager. 2D-DIGE images were analysed by DeCyder 6.5 software (GE Healthcare Bio-Sciences, Piscataway, NJ, USA). Spots were picked from a matched Sypro Ruby post-stained gel with an Ettan Spot Picker (GE Healthcare Bio-Sciences, Piscataway, NJ, USA) equipped with a 1.4 mm picker head.
Identification of differentially expressed proteins
2D gel plugs selected for further analysis after the Decyder analysis were destained and subjected to in-gel reduction with dithiothreitol, alkylation with iodoacetamide and digestion with trypsin (sequencing grade, Promega, Madison, WI, USA), essentially as described by Wilm et al. [20] NanoLC-MS/MS was performed on an 1100 series capillary LC system (Agilent Technologies, Santa Clara, CA, USA) coupled to an LTQ ion trap mass spectrometer (Thermo Scientific, Waltham, MA, USA) operating in positive mode and equipped with a nanospray source. Peptide mixtures were trapped on a 1.5 cm x 100 μm in-house packed 5 μm ReproSil C18 reversed phase column (Dr Maisch GmbH, Ammerbuch-Entringen, Germany) at a flow rate of 8 μl/min. Peptide separation was performed on a 20 cm x 50 μm in-house packed 3 μm ReproSil C18 reversed phase column using a linear gradient from 0 to 30% B in A (A = 2% (v/v) acetoni-trile, 0.1% (v/v) formic acid; B = 80% (v/v) acetonitrile, 0.1% (v/v) formic acid) in 70 min. and at a constant flow rate of 200 nl/min using a splitter. The column eluent was directly sprayed into the ESI source of the mass spectrometer. Mass spectra were acquired in continuum mode. Fragmentation of the peptides was performed in data-dependent mode. Peak lists were automatically created from raw data files using the Mascot Distiller software (version 2.0; Matrix Science, Boston, MA, USA). The Mascot search algorithm (version 2.0, Matrix Science) was used for searching against the NCBInr database (release date: January 20, 2006; taxonomy Rattus). The peptide tolerance was typically set to 2 Da and the fragment ion tolerance to 0.8 Da. Only double and triple charged peptides were searched for. A maximum number of two missed cleavages by trypsin were allowed and carbamidomethylated cysteine and oxidized methionine were set as fixed and variable modifications respectively. The Mascot score cut-off value for a positive protein hit was set at 40. Individual peptide MS/MS spectra with Mowse scores below 40 were checked manually and either interpreted as valid identifications or discarded.
Immunoblot analysis
For the confirmative immunoanalysis of differentially displayed stress-related proteins, about 100 mg of left ventricular tissue was homogenized in a buffer containing 25 mM Tris, 25mM NaCl, 1 mM sodium orthovanadate, 10 mM sodium fluoride, 10 mM sodium pyrophosphate, 10 nM okadaic acid, 0.5 mM EDTA, 1 mM PMSF and protease inhibitors cocktail (leupeptin, aprotinin and pepstatin). Homogenates were cen-trifuged at 3000 rpm for 10 min. at 4°C, and the supernatant was again centrifuged at 10,000 rpm for 20 min. at 4°C. The supernatant was saved as cytosolic extract. The resultant pellet was re-suspended in an equal volume of the above-mentioned buffer containing 0.1% Triton-X100, and incubated on ice for 1 hr with intermittent tapping. The supernatant after centrifugation at 14,000 rpm for 10 min. at 4°C was collected as mito-chondrial extract. Total protein concentration in both cytosolic and mito-chondrial extract was determined using BCA Protein Assay Kit (Pierce, Rockford, IL, USA).
Cytosolic or mitochondrial proteins (50 μg of total protein) were separated in SDS-PAGE and transferred to nitrocellulose filters. Filters were blocked in 5% non-fat dry milk, and probed overnight with a primary antibody. Antibodies against heat shock protein β6 (1:20,000 dilution), glutathione S transferase μ2 (1:3000), peroxiredoxin1(Prdx)-1 (1;10000) and Prdx-3 (1:3000) were obtained from Abcam, Cambridge, MA. C-Crk I/II, GRP 75, catalase and Hsc70 antibodies were obtained from Santa Cruz Biotechnology Santa Cruz, CA, and used at the dilution of 1:1000. Hsp70 antibody was obtained from Cell Signaling Technology, Danvers, MA, and used at the dilution of 1:1000. Protein bands were identified with horseradish peroxidase-conjugated secondary antibody (1:2000 dilution) and Western Blotting Luminol Reagent (Santa Cruz Biotechnology, Santa Cruz, CA, USA). The resulting blots were digitized, subjected to densitometric scanning using a standard NIH image program, and normalized against loading control.
Statistical analysis
The differential analysis software DeCyder 6.5 was primarily used for the identification of up- and down- regulated spots comparing the duplicate left ventricular samples obtained from each animal group. For the statistical analysis of heart function and images of the immunoblots the statistical software program SPSS (version 10.1 for windows, Microsoft) was used to re-evaluate the Mann-Whitney statistics and calculate the exact P-values. Data are presented as means ± SEM. A P-value of <0.05 was considered statistically significant.
Results
Effects of resveratrol on the infarct size and cardiomyocyte apoptosis on diabetic and normal hearts
Hearts were excised from diabetic and normal rats and subjected to ex vivo 30 min. of global ischaemia followed by 2 hrs of reper-fusion. The hearts from the diabetic rats had greater infarct size and increased number of apoptotic cardiomyocytes compared to those from the non-diabetic control animals. As shown in (Fig. 1A), the amount of myocardial infarct size expressed as per cent of area at risk was increased by about 20%. Number of apop-totic cardiomyocytes in the diabetic hearts was increased by 35% compared to that in the control hearts (Fig. 1B and C). Feeding resveratrol to the animals for a period of 7 days lowered the myocardial infarct size by about 35% and apoptotic number of cardiomyocytes by about 45%. This effect of resveratrol was similar in control and diabetic animals.
1.
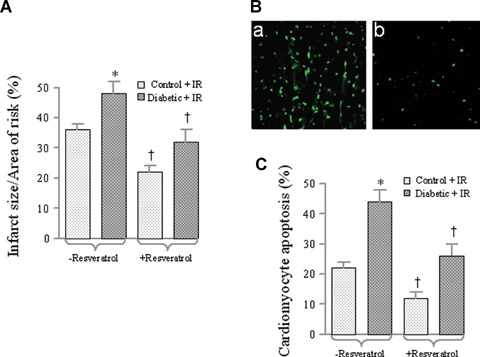
Effect of ex vivo myocardial infact size and cardiomyocyte apoptosis. Normal and diabetic rats were treated for 7 days with or without resveratrol (Control) treatment. Thereafter, hearts were isolated and perfused with KHB in Langendorff apparatus for 15 min. followed by 30 min. of global ischaemia and 2 hrs of reperfusion. At the end of reperfusion, hearts were either prepared for infarct size measurement by Triphenyl tetrazolium chloride (TTC) staining (A) or cardiomyocyte apoptosis by TUNEL method (B and C). (B) Representative images of TUNEL staining with (b) or without (a) resveratrol treatment.
Analysis of the 2D-DIGE images
Per gel up to 2100 Cye-labelled spots were detected (compared with 6000 for Sypro Ruby) and up to 1400 spots were matched to the master gel (the 2D-DIGE gel containing most spots). One gel was excluded from the analysis due to lack of resolution. Differentially expressed protein spots were selected for spot picking when P < 0.05 (t-test), the average ratio was <−1.3 or >1.3, and the spot was present in at least eight out of the 17 images. All 84 spots that met these criteria were visually checked. Spots that were not detected in the Sypro Ruby post-stained gel or poor quality spots were omitted. Finally, 41 spots were selected. Some blank spots and spots containing known proteins from previous experiments were added for validation purposes. A representative image of a Sypro Ruby post stained 2D-DIGE gel is shown in Figure 2.
2.
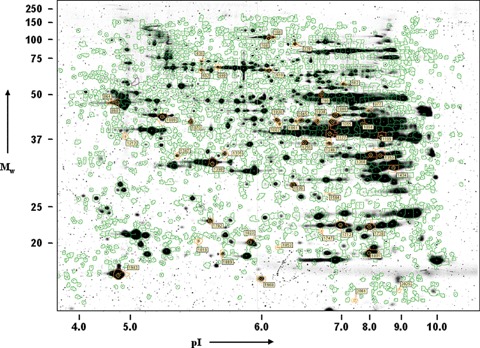
Two-dimensional differential gel elec-trophoresis. Representative image of a Sypro Ruby post-stained 2D-DIGE gel of the cyto-plasmic subfraction of rat left ventricle. Green lines represent spot boundaries of spots matched to the master gel and the label shows the DeCyder master gel spot number of the selected spots for MS analysis.
Identification of differentially expressed proteins
The patterns of proteins in resveratrol-treated diabetic hearts versus normal hearts are shown in Figures 3–4. Figure 3 shows an increased spot (compared to normal) in diabetic group while Figure 4 depicts a reduced spot (compared to normal) in the same group. The identity of the proteins in these spots was clarified by LC-MS/MS followed by SwissProt database search. The search output was filtered to eliminate irrelevant proteins and comparison was made between theoretical and actual pI and MW. Many proteins related to glycolysis, citric acid cycle, fatty acid oxidation were down-regulated, as well as creatine kinases (Table 1) in the diabetic group compared to control group while a few within this group were up-regulated (Table 2). Interestingly, the up-regulated proteins from these groups include mitochondrial proteins for example, delta (3,5)-delta (2,4)-dienoyl-CoA isomerase, or dihydrolipoylly-sine-residue acetyl transferase component of pyruvate dehydroge-nase E1 α subunit.
3.
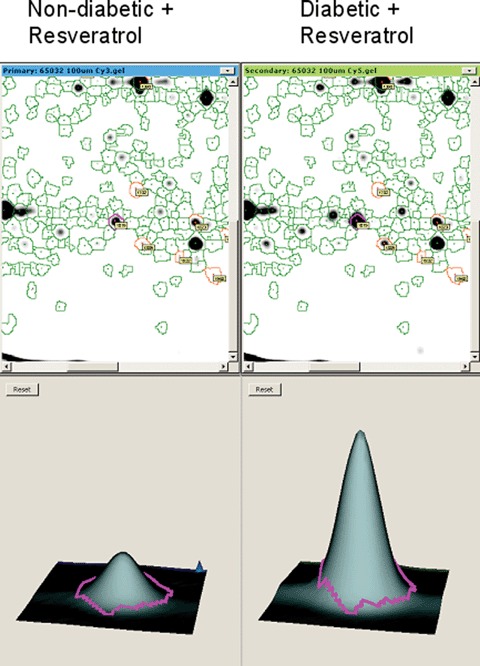
Up-regulated proteins in resvera-trol-treated diabetic rats. A representative DeCyder picture of part of a 2D-DIGE gel showing the up-regulation of a protein (apolipoprotein A-1 precursor, spot number 1819) in diabetic compared to non-diabetic rat heart, both treated with resveratrol.
4.
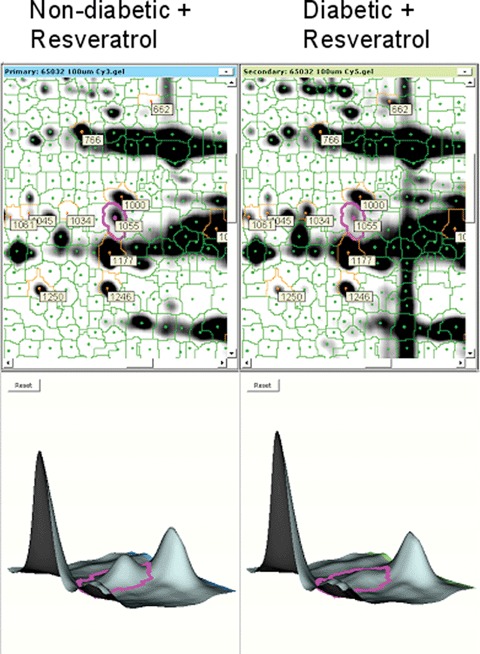
Down-regulated proteins in resvera-trol-treated diabetic rats. A representative DeCyder picture showing the down-regulation of a protein (pyruvate dehydroge-nase E1 component, alpha subunit, spot number 1055) in resveratrol treated diabetic versus non-diabetic hearts.
1.
Proteins which were downregulated in diabetic when compared to non-diabetic rat hearts both treated with resveratrol
| Spot number | Protein | Function | Mowse score | Mw (theor., kD) | pI (theor.) | Mw (on gel, est. kD) | pI (on gel, est.) | Relative abundance | P-value |
|---|---|---|---|---|---|---|---|---|---|
| 1199 | Fructose-bisphosphate aldolase A (Muscle-type aldolase) | Glycolysis | 1374 | 40 | 8.3 | 40 | 8.4 | −2.1 | 0.000324 |
| 1055 | Pyruvate dehydrogenase E1 component alpha subunit, somatic form, mitochondrial precursor | Citric acid cycle | 852 | 44 | 8.5 | 45 | 6.8 | −2.0 | 0.001255 |
| 1070 | Creatine kinase B-type | Energy metabolism | 858 | 43 | 5.4 | 45 | 5.4 | −2.0 | 0.0038 |
| 168 | 2-oxoglutarate dehydrogenase E1 component, mitochondrial precursor | Citric acid cycle | 2430 | 117 | 6.3 | 110 | 6.1 | −1.8 | 0.003655 |
| 1000 | Beta-enolase (2-phospho-D-glycerate hydro-lyase) (Muscle-specific enolase) Glycolysis | Glycolysis | 1247 | 47 | 7.1 | 45 | 6.9 | −1.7 | 0.000578 |
| 1434 | Malate dehydrogenase, mito-chondrial precursor | Citric acid cycle | 1451 | 36 | 8.9 | 30 | 8.8 | −1.6 | 0.01374 |
| 1368 1350 | Glyceraldehyde-3-phosphate dehydrogenase | Glycolysis | 1065 1350 | 36 | 8.1 | 35 35 | 8.0 8.4 | −1.4 −1.6 | 0.001473 8.75E-05 |
| 1347 | Proto-oncogene C-crk (p38) (Adapter molecule crk) | Interacts with insulin receptor | 409 | 34 | 5.4 | 35 | 5.3 | −1.5 | 0.04918 |
| 1335 | Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial precursor | Citric acid cycle | 609 | 40 | 6.5 | 35 | 5.6 | −1.5 | 0.00018 |
| 1968 | Heat-shock protein beta-6 (HspB6) (Heat-shock 20 kDa-like protein p20) | Chaperone | 302 | 18 | 6.1 | 17 | 6.0 | −1.5 | 0.01284 |
| 1250 | Short-chain specific acyl-CoA dehydrogenase, mitochondrial precursor | Fatty acid oxidation | 1134 | 45 | 8.5 | 37 | 6.4 | −1.5 | 0.04758 |
| 1094 | Creatine kinase, sarcomeric mitochondrial precursor | Energy metabolism | 1249 | 48 | 8.6 | 45 | 7.8 | −1.5 | 0.01682 |
| 1009 | Ubiquinol-cytochrome-c reduc-tase complex core protein 1 | Oxidative phos-phorylation | 1066 | 54 | 5.6 | 45 | 5.2 | −1.5 | 0.03161 |
| 766 | Dihydrolipoyl dehydrogenase, mitochondrial precursor | Citric acid cycle | 1232 | 55 | 8.0 | 50 | 6.7 | −1.4 | 0.000489 |
| 1891 | Peroxiredoxin-1 (Thioredoxin peroxidase 2) | Oxidative stressVanti oxi-dant | 476 | 22 | 8.3 | 19 | 8.0 | −1.4 | 0.00058 |
| 1730 | 3-hydroxyacyl-CoA dehydrogenase type-2 | Fatty acid oxidation | 476 | 27 | 8.9 | 23 | 8.0 | −1.4 | 0,04545 |
| 518 | Succinate dehydrogenase [ubiquinone] flavoprotein sub-unit, mitochondrial precursor | Citric acid cycle | 1647 | 73 | 6.8 | 60 | 6.1 | −1.4 | 0.01509 |
| 1177 | Creatine kinase M-type | Energy metabolism | 1236 | 43 | 6.6 | 40 | 6.8 | −1.4 | 0.004462 |
| 1712 | Triosephosphate isomerase (TIM) (Triose-phosphate isomerase) | Glycolysis | 640 | 27 | 6.9 | 23 | 7.1 | −1.4 | 0.001169 |
| 1702 | NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial precursor | Oxidative phos-phorylation | 499 | 30 | 6.4 | 23 | 5.5 | −1.4 | 0.004078 |
| 1246 | Isocitrate dehydrogenase [NAD] subunit beta, mitochondrial precursor | Citric acid cycle | 1059 | 43 | 8.9 | 37 | 6.8 | −1.4 | 0.00435 |
| 1823 | Thioredoxin-dependent peroxide reductase, mitochondrial precursor (Peroxiredoxin-3) | Oxidative stress/anti oxidant | 513 | 29 | 7.1 | 21 | 5.9 | −1.4 | 0.006387 |
2.
Proteins which were upregulated in diabetic when compared to non-diabetic rat hearts both treated with resveratrol
| Spot number | Protein | Function | Mowse score | Mw (theor., kDa) | pI (theor.) | Mw (on gel, est. kDa) | pI (on gel, est.) | Relative abundance | P-value |
|---|---|---|---|---|---|---|---|---|---|
| 1536 | Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial precursor | Fatty acid oxidation | 935 | 36 | 8.1 | 27 | 6.3 | 1.4 | 3.00E-05 |
| 406 | Stress-70 protein, mitochondrial precursor (75 kDa glucose-regulated protein) (GRP 75) | Proliferation, aging, chaperone | 830 | 74 | 6.0 | 70 | 5.5 | 1.4 | 0.01418 |
| 502 | Dihydrolipoyllysine-residue acetyltransferase comp. of pyruvate dehydr. complex, mitochondrial precursor | Citric acid cycle | 914 | 68 | 8.8 | 60 | 5.5 | 1.5 | 0.000653 |
| 228 | Glycogen phosphorylase, brain form | Glycogenolysis | 1418 | 97 | 6.3 | 100 | 6.4 | 1.6 | 0.01427 |
| 440 | Heat shock cognate 71 kDa protein (Heat shock 70 kDa protein 8) | Chaperone | 416 | 71 | 5.4 | 65 | 5.6 | 1.7 | 0.02859 |
| 1889 | Adenine phosphoribosyltrans-ferase (APRT) | Nucleotide salvage pathway | 371 | 20 | 6.2 | 19 | 5.6 | 1.8 | 0.003842 |
| 1034 1038 1045 | Pyruvate dehydrogenase E1 component alpha subunit, somatic form, mitochondrial precursor | Citric acid cycle | 686 785 758 | 44 | 8.5 | 45 45 45 | 6.6 6.1 6.4 | 1.9 1.6 1.6 | 0.001224 0.03529 0.0356 |
| 1747 | Glutathione S-transferase Mu 2 (GSTM2–2) (Glutathione S-transferase Yb-2) | Oxidative stress/anti oxidant | 464 | 26 | 6.9 | 22 | 6.7 | 2.0 | 0.005773 |
| 662 | Catalase | Oxidative stress/anti oxidant | 1330 | 60 | 7.1 | 55 | 7.1 | 2.1 | 0.00077 |
| 977 | α-enolase (2-phospho-D-glyc-erate hydro-lyase) (Non-neural enolase) (NNE) | Glycolysis | 736 | 47 | 6.2 | 45 | 8.0 | 2.6 | 0.000602 |
| 1212 | Myosin-7 (Myosin heavy chain 7) (Myosin heavy chain, cardiac muscle beta isoform) | Structural protein | 1203 | 224 | 5.6 | 40 | 4.9 | 2.7 | 0.000241 |
| 1061 1082 | Elongation factor Tu, mitochondrial precursor | Protein synthesis | 769 661 | 50 | 7.2 | 45 45 | 6.3 6.2 | 1.5 3.5 | 0.001967 4.42E-05 |
| 1584 | Carbonyl reductase [NADPH] 1 (NADPH-dependent carbonyl reductase 1) | Oxidoreductase | 742 | 31 | 8.2 | 26 | 6.8 | 3.6 | 0.004523 |
| 1819 | Apolipoprotein A-I precursor (Apo-AI) | Fat metabolism | 550 | 30 | 5.5 | 21 | 5.5 | 7.4 | 4.49E-05 |
Of particular interest is the increased expression of several chaperone proteins and oxidative stress related proteins in the diabetic group. As apparent from the proteomics analysis, heat shock protein (HSP) β6, Prdx1 and Prdx-3 expression levels were lower in the diabetic hearts compared to normal hearts. In contrast, HSP 70, GRP 75, heat shock cognate 71 kDa protein, glutathione S-transferase and catalase were up-regulated in the diabetic group.
Effect of resveratrol on stress protein expression in diabetic versus non-diabetic rats
Since it is well-known that oxidative stress plays a crucial role in the pathophysiology of diabetes, we decided to focus primarily on the stress-related proteins identified to be differentially expressed in diabetic versus non-diabetic rat hearts in the present study. Western blot analysis was performed to determine the effect of resveratrol on expression of several stress-related proteins. As shown in Figure 6A and B, glutathione S-transferase μ2, catalase, Hsc70, Hsp70 and Prdx-1 and Prdx-3 proteins were up-regulated in streptozotocin-induced diabetic rats when compared to normal rats. Treatment with resveratrol reduced the expression of glutathione S-transferase μ2, Hsp70, Hsc 70, Prdx-1 and Prdx-3 in diabetic rats to a level similar to untreated normal hearts, or to even lower levels (HSP70 or Prdx-3. Protein expression of GRP75 and HSP β6 was down-regulated in diabetic rats relative to normal rats, and this effect was reversed by resveratrol treatment. The expression of GRP 75 was significantly higher in resveratrol-treated diabetic groups than in the other groups, which is in accordance with the proteomic data. However, protein expression of catalase was elevated in diabetic rats, and its expression was further enhanced in resveratrol-treated diabetic rats. Our immunoblotting results demonstrate that resveratrol reversed the diabetes-induced changes in stress protein expression except for catalase, which was further induced.
6.
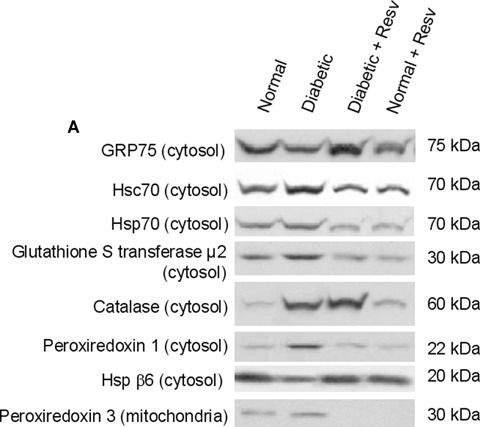
Effect of resveratrol on oxidative stress proteins in diabetic and control hearts. (A) Cytosolic and mitochondrial total protein was extracted from the left ventricular tissue, as described in methods. Equal amounts of total protein were loaded and separated by SDS-PAGE, and protein expression analyses was carried out by immunoblotting. Figures are representative images of three different samples. (B) Quantification of immunoblots obtained as mentioned in Figure 6A using Scion Image software. All proteins are expressed as arbitrary units relative to normal myocardium. Resv, resveratrol. *P < 0.05 versusdiabetic group.
Discussion
The results of the present study extend our previous observation that resveratrol could protect the diabetic heart from diabetes-mediated cardiac dysfunction [7] and proteomic profiling of the cardiac pharmacological pre-conditioning by resveratrol [16]. This study was specifically designed to compare the resveratrol-induced protein profiles on the control versus diabetic hearts. As expected, most of the proteins that differ between resveratrol-treated diabetic and control hearts included the proteins related to fatty acid oxidation, glycolysis, citric acid cycle, oxidative phos-phorylation and creatine kinases. The majority of these proteins are nuclear-encoded mitochondrial proteins. The salient features of the proteomic analysis also include down-regulation of proto-oncogene Crk (p38) (adapter molecule Crk) and up-regulation of carbonyl reductase (NADPH)-1 and elongation factor Tu. c-Crk is known to interact with the insulin receptor [21] while elongation factor Tu is involved in mitochondrial protein synthesis. Of particular interest, however, is differential expression of stress-related proteins in diabetic versus normal hearts in response to resvera-trol (Fig. 6). The stress proteins include HSPs as well as oxidative stress-inducible proteins. Although the effects of resveratrol on normal versus diabetic hearts have never been studied previously, the role of stress proteins in diabetes is known. For example, a recent study demonstrated down-regulation of HSC70 in diabetic myocardium secondary to insulin deficiency suggesting that insulin plays a major role in maintaining adequate expression of HSC70 in cardiac muscle [22]. Another related study showed reduced HSP60 in diabetic myocardium [23]. In the latter study, HSP60 was found to augment IGF-1 receptor signalling in cardiac muscle cells. Unlike HSP60, HSP 70 and HSC 70 cannot modulate IGF-1 receptor signalling in the diabetic heart [22]. Another study suggested a key role of HSP70 in the natural resistance of human b cells against nitric oxide induced injury by preserving mitochon-drial function [24].
Reactive oxygen species (ROS) have been known for a long time to play a crucial role in the pathogenesis of diabetes [25]. The antioxidants such as superoxide dismutase (SOD) and catalase can reduce the complications due to diabetes. For example, cata-lase was found to protect sarcomere function in diabetic myocytes [26]. Elevating MnSOD provided extensive protection to diabetic mitochondria and provided overall protection to the diabetic heart [27]. Mitochondrial glutathione level was shown to be more sensitive to diabetes than whole heart glutathione [28].
Similar to the antioxidant proteins, many redox-regulated proteins are modulated in diabetes. For example, Prdxs 1, 4 and 6 as well as glutathione reductase were found to be affected by dia-betogenic stress [29]. In a recent study using cDNA microarray, several thiol antioxidant genes including glutathione peroxidase 1, Prdx 6 and thioredoxin 2 were found to be up-regulated by high glucose [30]. Another study demonstrated induction of Prdxs by oxidative and nitrosative stress in pancreatic beta cells [31]. Consistent with these findings, our study showed modulation of Prdxs in diabetes and further demonstrated that resveratrol differentially regulate Prdx-1 and Prdx-3.
In summary, we found that resveratrol regulates many genes in the diabetic heart involved in oxidative stress, etc. Notably, resver-atrol reverses several of the diabetes-induced changes in stress protein expression to the level of control, untreated hearts. This may indicate that resveratrol reduces the oxidative stress induced by diabetes. In contrast, resveratrol further up-regulated catalase expression in the diabetic hearts, which may suggest that the drug increased protection against oxidative stress. These results suggest that resveratrol treatment improve cardiac dysfunction in streptozocin-induced diabetic rats at least in part via regulation of stress-induced redox proteins. The ability of resveratrol to trigger gene and protein expression was demonstrated in many earlier studies including our own study [16] where we showed induction of the expression of proteins related to preconditioning. Similar to the results of the present study, resveratrol was found to modulate expression of genes related to redox metabolism and cell proliferation in non-small-cell lung carcinoma cells [32]. Another related study demonstrated that treatment of cells with resveratrol potentiated chromatin-associated SIRT1 protein on the cIAP promoter region, an effect that correlated with a loss of NF-kB-regulated gene expression [33].
Finally, the present study involves treatment with resveratrol for 7 days, the long-term effects of resveratrol in diabetic condition are not known. Most of the studies described in the literature found cardioprotective effects within 1–2 weeks of treatment. However, our results supply valuable information on resveratrol's ability to differentially regulate several redox-regulated stress proteins in diabetic versus normal hearts suggesting that these beneficial proteins are expressed in the diabetic hearts to protect the tissue from cellular injury. It is tempting to speculate that resver-atrol may be used as nutritional supplement to prevent metabolic disorders related to Syndrome X.
Acknowledgments
This study was supported in part by NIH HL 34360, HL22559 and HL 33889. The authors have no conflicting financial interests
References
- 1.Ceriello A, Bortolotti N, Motz E, Lizzio S, Russo A, Selmo V, Catone B, Tonutti L, Taboga C. Meal-generated oxidative stress in diabetes. The protective effect of red wine. Diabetes Care. 1999;22:2084–5. doi: 10.2337/diacare.22.12.2084. [DOI] [PubMed] [Google Scholar]
- 2.Montilla P, Barcos M, Munoz MC, Bujalance I, Munoz-Castaneda JR, Tunez I. Red wine prevents brain oxidative stress and nephropa-thy in streptozotocin-induced diabetic rats. J Biochem Mol Biol. 2005;38:539–44. doi: 10.5483/bmbrep.2005.38.5.539. [DOI] [PubMed] [Google Scholar]
- 3.Napoli R, Cozzolino D, Guardasole V, Angelini V, Zarra E, Matarazzo M, Cittadini A, Sacca L, Torella R. Red wine consumption improves insulin resistance but not endothelial function in type 2 diabetic patients. Metabolism. 2005;54:306–13. doi: 10.1016/j.metabol.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Caimi G, Carollo C, Lo Presti R. Diabetes mellitus: oxidative stress and wine. Curr Med Res Opin. 2003;19:581–6. doi: 10.1185/030079903125002324. [DOI] [PubMed] [Google Scholar]
- 5.Ceriello A, Bortolotti N, Motz E, Lizzio S, Catone B, Assaloni R, Tonutti L, Taboga C. Red wine protects diabetic patients from meal-induced oxidative stress and thrombosis activation: a pleasant approach to the prevention of cardiovascular disease in diabetes. Eur J Clin Invest. 2001;31:322–8. doi: 10.1046/j.1365-2362.2001.00818.x. [DOI] [PubMed] [Google Scholar]
- 6.Su HC, Hung LM, Chen JK. Resveratrol, a red wine antioxidant, possesses an insulinlike effect in streptozotocin-induced diabetic rats. Am J Physiol Endocrinol Metab. 2006;290:E1339–46. doi: 10.1152/ajpendo.00487.2005. [DOI] [PubMed] [Google Scholar]
- 7.Thirunavukkarasu M, Penumathsa SV, Koneru S, Juhasz B, Zhan L, Otani H, Bagchi D, Das DK, Maulik N. Resveratrol alleviates cardiac dysfunction in streptozo-tocin-induced diabetes: Role of nitric oxide, thioredoxin, and heme oxygenase. Free Radic Biol Med. 2007;43:720–9. doi: 10.1016/j.freeradbiomed.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar A, Kaundal RK, Iyer S, Sharma SS. Effects of resveratrol on nerve functions, oxidative stress and DNA fragmentation in experimental diabetic neuropathy. Life Sci. 2007;80:1236–44. doi: 10.1016/j.lfs.2006.12.036. [DOI] [PubMed] [Google Scholar]
- 9.Chi TC, Chen WP, Chi TL, Kuo TF, Lee SS, Cheng JT, Su MJ. Phosphatidylinositol-3-kinase is involved in the antihyperglycemic effect induced by resveratrol in streptozo-tocin-induced diabetic rats. Life Sci. 2007;80:1713–20. doi: 10.1016/j.lfs.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Sharma S, Anjaneyulu M, Kulkarni SK, Chopra K. Resveratrol, a polyphenolic phytoalexin, attenuates diabetic nephropa-thy in rats. Pharmacology. 2006;76:69–75. doi: 10.1159/000089720. [DOI] [PubMed] [Google Scholar]
- 11.Aribal-Kocaturk P, Kavas GO, Buyukkagnici DI. Pretreatment effect of resveratrol on streptozotocin-induced diabetes in rats. Biol Trace Elements Res. 2007;118:244–9. doi: 10.1007/s12011-007-0031-y. [DOI] [PubMed] [Google Scholar]
- 12.Bartnik M, Norhammar A, Ryden L. Hyperglycaemia and cardiovascular disease. J Intern Med. 2007;262:145–56. doi: 10.1111/j.1365-2796.2007.01831.x. [DOI] [PubMed] [Google Scholar]
- 13.Sato M, Maulik N, Das DK. Cardioprotection with alcohol: role of both alcohol and polyphenolic antioxidants. Ann N Y Acad Sci. 2002;957:122– 35. doi: 10.1111/j.1749-6632.2002.tb02911.x. [DOI] [PubMed] [Google Scholar]
- 14.Das S, Tosaki A, Bagchi D, Maulik N, Das DK. Potentiation of a survival signal in the ischemic heart by resveratrol through p38 mitogen-activated protein kinase/mitogen-and stress-activated protein kinase 1/cAMP response element-binding protein signaling. J Pharmacol Exp Ther. 2006;317:980–8. doi: 10.1124/jpet.105.095133. [DOI] [PubMed] [Google Scholar]
- 15.Juric D, Wojciechowski P, Das DK, Netticadan T. Prevention of concentric hypertrophy and diastolic impairment in aortic-banded rats treated with resveratrol. Am J Physiol Heart Circ Physiol. 2007;292:H2138–43. doi: 10.1152/ajpheart.00852.2006. [DOI] [PubMed] [Google Scholar]
- 16.Bezstarosti K, Das S, Lamers JM, Das DK. Differential proteomic profiling to study the mechanism of cardiac pharmacological preconditioning by resveratrol. J Cell Mol Med. 2006;10:896–907. doi: 10.1111/j.1582-4934.2006.tb00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Shamaei-Tousi A, Stephens JW, Bin R, Cooper JA, Steptoe A, Coates ARM, Henderson B, Humphries SE. Association between plasma levels of heat shock protein 60 and cardiovascular disease in patients with diabetes mellitus. European Heart J. 2006;27:1565–70. doi: 10.1093/eurheartj/ehl081. [DOI] [PubMed] [Google Scholar]
- 18.Hayden MR, Tyagi SC. Myocardial redox stress and remodeling in metabolic syndrome, type 2 diabetes mellitus, and congestive heart failure. Med Sci Monit. 2003;9:47–74. [PubMed] [Google Scholar]
- 19.Chen H, Wu XJ, Lu XY, Zhu L, Wang LP, Yang HT, Chen HZ, Yuan WJ. Phosphorylated heat shock protein 27 is involved in enhanced heart tolerance to ischemia in short-term type 1 diabetic rats. Acta Pharmacol Sinica. 2005;26:806–812. doi: 10.1111/j.1745-7254.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- 20.Wilm M, Shevchenko A, Houthaeve T, Breit S, Schweigerer L, Fotsis T, Mann M. Femtomole sequencing of proteins from polyacrylamide gels by nano-electro-spray mass spectrometry. Nature. 1996;379:466–9. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- 21.Karas M, Koval AP, Zick Y, LeRoith D. The insulin-like growth factor I receptor-induced interaction of insulin receptor substrate-4 and Crk-II. Endocrinology. 2001;142:1835–40. doi: 10.1210/endo.142.5.8135. [DOI] [PubMed] [Google Scholar]
- 22.Chen HS, Jia J, Su HF, Lin HD, Chen JW, Lin SJ, Yang JY, Lai HC, Mestril R, Wang PH. Downregulation of the constitutively expressed HSC70 in diabetic myocardium is mediated by insulin deficiency. J Endocrinol. 2006;190:433–40. doi: 10.1677/joe.1.06692. [DOI] [PubMed] [Google Scholar]
- 23.Shan Y, Yang T, Mestril R, Wang PH. HSP10 and HSP60 suppresses ubiquitination of IGF1 receptor and augments IGF1 receptor signaling in cardiac muscle-Implications on decreased myocardial protection in diabetic cardiomyopathy. J Biol Chem. 2003;278:45492–8. doi: 10.1074/jbc.M304498200. [DOI] [PubMed] [Google Scholar]
- 24.Burkart V, Liu H, Bellmann K, Wissing D, Jaattela M, Cavallo MG, Pozzilli P, Briviba K, Kolb H. Natural resistance of human beta cells toward nitric oxide is mediated by heat shock protein 70. J Biol Chem. 2000;275:19521–8. doi: 10.1074/jbc.M002265200. [DOI] [PubMed] [Google Scholar]
- 25.Baynes JW. Role of oxidative stress in development of complications in diabetes. Diabetes. 1991;40:405–12. doi: 10.2337/diab.40.4.405. [DOI] [PubMed] [Google Scholar]
- 26.Ye G, Metreveli NS, Donthi RV, Xia S, Xu M, Carison EC, Epstein PN. Catalase protects cardiomyocyte function in models of type 1 and type 2 diabetes. Diabetes. 2004;53:1336–43. doi: 10.2337/diabetes.53.5.1336. [DOI] [PubMed] [Google Scholar]
- 27.Shen X, Zheng S, Metreveli NS, Epstein PN. Protection of cardiac mitochondria by overexpression of MnSOD reduces diabetic cardiomyopathy. Diabetes. 2006;55:798–805. doi: 10.2337/diabetes.55.03.06.db05-1039. [DOI] [PubMed] [Google Scholar]
- 28.Liang Q, Carlson EC, Donthi RV, Kralik PM, Shen X, Epstein PN. Overexpression of metallothionein reduces diabetic car-diomyopathy. Diabetes. 2002;51:174–81. doi: 10.2337/diabetes.51.1.174. [DOI] [PubMed] [Google Scholar]
- 29.Nagaoka Y, Iuchi Y, Ikeda Y, Fujii J. Glutathione reductase is expressed at high levels in pancreatic islet cells. Redox report. 2004;9:321–4. doi: 10.1179/135100004225006812. [DOI] [PubMed] [Google Scholar]
- 30.Morrison J, Knoll K, Hessner MJ, Liang M. Effect of high glucose on gene expression in mesangial cells: upregulation of the thiol pathway is an adaptational response. Physiological Genomics. 2004;17:271–82. doi: 10.1152/physiolgenomics.00031.2004. [DOI] [PubMed] [Google Scholar]
- 31.Bast A, Wolf G, Obrbaumer I, Walther R. Oxidative and nitrosative stress induces peroxiredoxins in pancreatic beta cells. Diabetologia. 2002;45:867–76. doi: 10.1007/s00125-002-0846-1. [DOI] [PubMed] [Google Scholar]
- 32.Hu Y, Rahlfs S, Mersch-Sundermann V, Becker K. Resveratrol modulates mRNA transcripts of genes related to redox metabolism and cell proliferation in non-small-cell lung carcinoma cells. Biol Chem. 2007;388:207–19. doi: 10.1515/BC.2007.023. [DOI] [PubMed] [Google Scholar]
- 33.Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–80. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]


