Abstract
Summary
Competition between microbial species is a product of, yet can lead to a reduction in, the microbial diversity of specific habitats. Microbial habitats can resemble ecological battlefields where microbial cells struggle to dominate and/or annihilate each other and we explore the hypothesis that (like plant weeds) some microbes are genetically hard-wired to behave in a vigorous and ecologically aggressive manner. These ‘microbial weeds’ are able to dominate the communities that develop in fertile but uncolonized – or at least partially vacant – habitats via traits enabling them to out-grow competitors; robust tolerances to habitat-relevant stress parameters and highly efficient energy-generation systems; avoidance of or resistance to viral infection, predation and grazers; potent antimicrobial systems; and exceptional abilities to sequester and store resources. In addition, those associated with nutritionally complex habitats are extraordinarily versatile in their utilization of diverse substrates. Weed species typically deploy multiple types of antimicrobial including toxins; volatile organic compounds that act as either hydrophobic or highly chaotropic stressors; biosurfactants; organic acids; and moderately chaotropic solutes that are produced in bulk quantities (e.g. acetone, ethanol). Whereas ability to dominate communities is habitat-specific we suggest that some microbial species are archetypal weeds including generalists such as: Pichia anomala, Acinetobacter spp. and Pseudomonas putida; specialists such as Dunaliella salina, Saccharomyces cerevisiae, Lactobacillus spp. and other lactic acid bacteria; freshwater autotrophs Gonyostomum semen and Microcystis aeruginosa; obligate anaerobes such as Clostridium acetobutylicum; facultative pathogens such as Rhodotorula mucilaginosa, Pantoea ananatis and Pseudomonas aeruginosa; and other extremotolerant and extremophilic microbes such as Aspergillus spp., Salinibacter ruber and Haloquadratum walsbyi. Some microbes, such as Escherichia coli, Mycobacterium smegmatis and Pseudoxylaria spp., exhibit characteristics of both weed and non-weed species. We propose that the concept of nonweeds represents a ‘dustbin’ group that includes species such as Synodropsis spp., Polypaecilum pisce, Metschnikowia orientalis, Salmonella spp., and Caulobacter crescentus. We show that microbial weeds are conceptually distinct from plant weeds, microbial copiotrophs, r-strategists, and other ecophysiological groups of microorganism. Microbial weed species are unlikely to emerge from stationary-phase or other types of closed communities; it is open habitats that select for weed phenotypes. Specific characteristics that are common to diverse types of open habitat are identified, and implications of weed biology and open-habitat ecology are discussed in the context of further studies needed in the fields of environmental and applied microbiology.
Introduction
As the collective metabolism and ecological activities of microorganisms determine the health and sustainability of life on Earth it is essential to understand the function of both individual microbes and in their communities. The cellular systems of an insignificant portion of microbial species have been intensively characterized at the levels of biochemistry, cell biology, genomics and systems biology; and a substantial body of (top-down) studies has been published in the field of microbial ecology in relation to species interactions, community succession and environmental metagenomics. There are, however, some fundamental questions that remain unanswered in relation to the behaviour of microbial species within communities: what type of biology, for instance, enables microbes to dominate entire communities and their habitats and thereby determine levels of biodiversity in specific environments?
Plant species that primarily inhabit freshly disturbed habitats – known as weeds – are characterized by vigorous growth; tolerance to multiple stresses; exceptional reproduction, dispersal and survival mechanisms; a lack of specific environmental requirements; production of phytotoxic chemicals; and/or other competitive strategies (Table 1). The biology of plant weeds, including their phenotypic and genetic traits, has been the subject of extensive study over the past 100 years and is now relatively well characterized (Table 1; Long, 1932; Anderson, 1952; Salisbury, 1961; Hill, 1977; Mack et al., 2000; Schnitzler and Bailey, 2008; Bakker et al., 2009; Chou, 2010; Wu et al., 2011; Liberman et al., 2012). We hypothesize that weed biology is also pertinent to microbial species, but that the defining characteristic of microbial weeds is their ability to dominate their respective communities. In the field of plant ecology it is open habitats (such as freshly exposed fertile soil) that facilitate the emergence of weed species both within specific ecosystems and across evolutionary timescales. We propose that weed behaviour is equally prevalent in some microbial habitats, that open microbial habitats promote the emergence of microbial weed species, and that microbial weed biology represents a potent ecological and evolutionary mechanism of change for some microbial species and their communities.
Table 1.
A comparison of plant weeds and microbial weeds.a
| Plant weeds | Microbial weeds | References (plants/microbes) | |
|---|---|---|---|
| Definition | Plant species that primarily grow in open (plant) habitatsb | Microbial species able to dominate communities that develop in open (microbial) habitatsc | Baker, 1965; Hill, 1977/current article; Cordero et al., 2012 |
| Ability to rapidly occupy available space | Important trait | Essential trait | Hill, 1977/current article |
| Overall vigourd and competitive ability | Typically vigorous; competitive ability usually superior to that of crop plants | Exceptional vigour and competitive ability | Hill, 1977/Figs 1 and 2; Tables S1 and 2–5 |
| Primary competitors | Intra-specific competition can be stronger than inter-specific competition | Intra-and inter-specific competition are likely to be stronger if dominance results from efficient resource acquisition or antimicrobial substances respectively | Hill, 1977/current article |
| Resource acquisition and storage | Acquisition and/or storage of water, light, nutrients etc. typically excellent | Acquisition and/or storage of nutrients, water and/or light typically excellent | Hill, 1977; Oatham, 1997; Sadeghi et al., 2007/Table5 |
| Reproduction and dispersal | Exceptionale | May not differ from non-weed species | Hill, 1977/current article |
| Obligatory associations or interactions with other organisms | Uncommon/atypical | Uncommon/atypical | Hill, 1977/current article |
| Ubiquity | Many species are environmentally ubiquitous | Not necessarily widespread outside the habitat which they dominate | Hill, 1977/current article |
| Germination | No special requirements; high tolerance to diverse stresses (see below) | No special requirements; high tolerance to diverse stresses (see below) | Hill, 1977/current article |
| Longevity of dormant propagules | Exceptional | May not differ from non-weeds | Hill, 1977/current article |
| Able to regenerate from fragments | Yes, especially root fragments | Not relevant | Hill, 1977/current article |
| Maintenance of growth rate during nutrient limitation | Some species, e.g. black-grass (Alopecurus mysuroides) on low-potassium soils | Many (photosynthetic as well as some heterotrophic) species | Hill, 1977/Table5; Benning, 1998 |
| Capable of creating a zone-of-inhibition | Not known | Yes, in some instances | [Not known]/Fleming, 1929 |
| Secretion of toxic substances | Some species produce phytotoxic inhibitors (exuded from leaves, roots, and/or necromass) | All species produce antimicrobial toxins (secreted into the extracellular environment)f | Chou, 2010/Table6 |
| Production of volatile organic compounds (VOCs) that act as stressors | Some hydrophobic and highly chaotropic compounds act as phytotoxic stressors (e.g. β-caryophellene) | Many species produce a spectrum of VOCs that can inhibit metabolic activity at concentrations in the nanomolar or low millimolar rangef | Wang et al., 2010; Inderjit et al., 2011; Tsubo et al., 2012/Table6 |
| Secretion of biosurfactants that act as stressors | Not known | Some species produce biosurfactants; substances that can solubilize hydrophobic substrates and act as cellular stressorsf | [Not known]/Tables3 and e; Nitschke et al., 2011 |
| Bulk production, and secretion, of moderately chaotropic substances | Not known | Some species produce ethanol, butanol, acetone etc. that are potent cellular stressors at millimolar and molar concentrationsf | [Not known]/Table6 |
| Saprotrophic activity resulting in toxic/stressful breakdown products | Not known | Some species degrade macromolecules to release inhibitory catabolic products (e.g. chaotropic stressors such as phenol, catechol and vanilling) | [Not known]/Park et al., 2004; 2007; Zuroff and Curtis, 2012 |
| Acidification of habitat | Not known | Some species, via bulk production of organic acidsf | [Not known]/Table6 |
| Secretion of aggressive enzymes | Unlikely | Some speciesf | [Not known]/Table6; Walker, 2011 |
| Tolerance to environmental stressesh | Tolerant to multiple, habitat-relevant stress parameters | Tolerant to multiple, habitat-relevant stress parameters | Hill, 1977/Fig. 2, Tables2–4 and 6 |
| Resistance to toxins and/or stressors of biotic originh | Likely | Yes, most species | Hill, 1977/Fig. 2; Tables2–4 and 6 |
| Phenotypic plasticity and intra-specific variation | Greater than that of crop plants | Likely to be above average (e.g. in relation to stress biology) | Hill, 1977/current article; see references in Todd et al., 2004 |
| Resistance to viruses, predators, and/or grazers | Some species can tolerate, resist, and/or repel viruses, sucking insects and grazing animals; and may contain poisonous substances – or have morphological adaptations such as spines – that minimize losses through grazing | Some strains/species can tolerate, resist, repel and/or avoid viruses, grazing plankton and arthropods etc; may contain toxic metabolites or have behavioural adaptations – e.g. formation of large microcolonies – that minimize losses through grazing/predation; and may inhabit environments that are too extreme for predatory species | Thurston et al., 2001; Ralphs, 2002; Shukurov et al., 2012/Table6 |
| Karyotype | Commonly of hybrid origin, polyploid or aneuploid | Frequency of hybrid origin, polyploidy and aneuploidy not yet tested; may contain plasmids that contribute to weediness | Anderson, 1952/Tables3 and e |
| Proportion of total species | Minute (several hundred species) | Not yet established (likely to be minute) | Hill, 1977/current article |
| Association with human activity | Evolution and ecology have been strongly favoured by land disturbance that creates and open (plant) habitatsi | Evolution is likely to have occurred primarily in nature; but habitat dominance is commonplace in both natural and manmade environments | Anderson, 1952; Hill, 1977/Fig. 2; Table S1 and g |
| Problematic for human endeavours | Yes, due to loss of crop-plant yieldsj; toxicity/harm to livestock; role as hosts for crop pests and diseases, etc. | Yes, due to spoilage of foods, drinks; some species are pathogens of crop plants, livestock or humans; may emerge to take over industrial fermentations and other processesk | Hill, 1977/Table S1 |
| Value to humankind | Disproportionately important (origin of many crop-plant species; plant weeds have food value for humans or livestock; model organisms for research) | Disproportionately important (key sources of bulk chemicals and antimicrobialsl; for food and drinks production, and other biotechnological applications) | Hill, 1977; Koornneef and Meinke, 2010/; see Concluding remarks |
Typical traits.
Plant weeds may not be the dominating plant species.
Not necessarily the primary habitat of the microbial weed species.
Vigour can result from a combination of traits such as fast growth, stress tolerance, and immunity to toxins and diseases (see Tables 2–6).
Plant weeds typically have a high output of seeds under both favourable and poor growth-conditions, and can produce seed early (prior to maturity of the parent plant; Hill, 1977).
Each microbial weed species may secrete diverse antimicrobial substances which can be the primary factor in habitat dominance (see Table 7; Hallsworth, 1998; Walker, 2011). For examples of the concentrations at which moderately chaotropic stressors, potent chaotropes and hydrophobic stressors can inhibit cellular systems see Bhaganna and colleagues (2010). Toxins such as antibiotics, and VOCs, biosurfactants, and other stressors can have additional key roles in the cellular biology and ecology of microbes.
Some stressors of biotic origin are structurally identical to and/or exert the same stress mechanisms as stressors of abiotic origin (e.g. see Fig. 2; Tables 2–4 and 6).
Agricultural practices and crop-plant life-cycle can also select for early seed production and other weed traits (Hill, 1977).
Crop-yield losses caused by plant weeds are equivalent to those caused by plant pests or plant diseases (Cramer, 1967).
Open (microbial) habitats are not only the starting point for food and drinks fermentations, but many foodstuffs themselves represent open habitats and it is frequently microbial weed species that cause spoilage and therefore determine shelf-life. Open habitats located on and within host organisms can be readily invaded by microbial weed species including those with pathogenic activities (Cerdeño-Tárraga, et al., 2005). This has given rise to the use of probiotics (Molly et al., 1996; Bron et al., 2012) and a practice of preserving the skin microflora in babies and infants by avoiding use of soaps or detergents (Capone et al., 2011).
Antimicrobial substances are used for various applications; as biofuels, biocides, pharmaceuticals, flavour compounds, food preservatives, etc.
This article focuses on the following questions: (i) what types of substrate or environment facilitate the emergence of dominant microbial species, (ii) are there archetypal weed species that can consistently dominate microbial communities, (iii) what are the stress biology, nutritional strategies, energy-generating capabilities, antimicrobial activities and other competitive strategies of microbial weeds, (iv) which components and characteristics of microbial cells form the mechanics of weed biology, (v) what are the properties of open (microbial) habitats that promote the emergence of weed species, and (vi) how are microbial weeds conceptually distinct from plant weeds, and other ecophysiological groups of microorganism?
Dominance within microbial communities
The microbial diversity of specific habitats, as well as that of Earth's entire biosphere, is an area of intensive scientific interest that has implications within the fundamental sciences and for drug discovery, biotechnology and environmental sustainability. Biodiversity provides the material basis for competition between microbial taxa, species succession in communities, and other aspects of microbial ecology that collectively impact the evolution of microbial species (Cordero et al., 2012). We believe that certain microbes are genetically, and thus, metabolically hard-wired to dominate communities (monopolizing available resources and space) and thereby reduce biodiversity within the habitat. Microbial communities can reach a stationary-phase or climax condition with a relatively balanced species composition (McArthur, 2006). However whether tens or thousands of microbial species are initially present (Newton et al., 2006) the community can become dominated by a single – or two to three – species (Fig. 1; Table S1; Randazzo et al., 2010; Cabrol et al., 2012). For example the indigenous microflora of olives is comprised of significant quantities of phylogenetically diverse species such as Aureobasidium pullulans, Candida spp., Debaryomyces hansenii, Metschnikowia pulcherrima, Pichia guilliermondii, Pichia manshurica, Rhodotorula mucilaginosa and Zygowilliopsis californica (Fig. 1Bi; Nisiotou et al., 2010). Nisiotou and colleagues (2010) found that species succession and the stationary-phase community during olive fermentations are determined by one dominating genus, Pichia, three species of which account for almost 100% of the community by 35 days (see Fig. 1Biii). The most prevalent of these, Pichia anomala, is a microbial generalist that is not only able to inhabit ecologically and nutritionally distinct environments (e.g. palm sugar, cereals, silage, oil-contaminated soils, insects, skin, various marine habitats; see Walker, 2011) but can also dominate the communities that develop in diverse habitat types (Table S1; Tamang, 2010; Walker, 2011). Studies of Pecorino Crotonese cheese fermentations reveal that, even though the curd contains > 300 bacterial strains, the community is eventually dominated by Lactobacillus rhamnosus (Randazzo et al., 2010).
Figure 1.
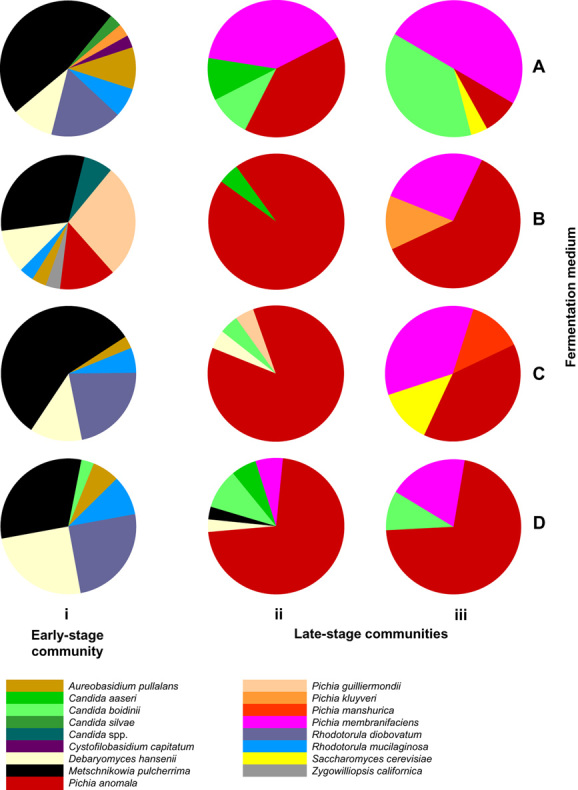
Percentage species composition during olive fermentations at (i) 2 days, (ii) 17 days and (iii) 35 days over a range of conditions: (A) with added NaCl (6% w/v); (B) added NaCl and glucose (6% and 0.5% w/v respectively); (C) added NaCl and lactic acid (6% w/v and 0.2% v/v respectively); and (D) NaCl, glucose and lactic acid (6% and 0.5% w/v, and 0.2% v/v respectively). Data (from Nisiotou et al., 2010) were obtained by DGGE.
Species that are able to dominate communities that develop in open habitats are not restricted to copiotrophs, generalists or any established ecophysiological grouping of microorganism (Table S1). Furthermore dominating species span the Kingdoms of life and, collectively, can be observed in most segments of the biosphere; from bioaerosols to solar salterns, oceans and freshwater to sediments and soils, the phyllosphere and rhizosphere, to those with molar concentrations of salts or sugars (see Table S1). The phenomenon can also be observed during food-production, food-spoilage and waste-decomposition processes where resource-rich habitats are available for colonization (see Table S1). The materials present in and the microbes living on these substrates are ultimately derived from the natural environment so community successions resulting in single-species dominance are likely to be commonplace in comparable open habitats within natural ecosystems (Table S1; Senthilkumar et al., 1993; Nisiotou et al., 2007; Reynisson et al., 2009; Rajala et al., 2011). Microbial weed species are able to dominate open habitats that progress to a closed condition (e.g. fermenting fruit juice) as well as those that remain in a more or less perpetually open state (e.g. the surface-fluid film of sphagnum moss and rhizosphere of plants growing in moist soils); see Table S1. Weeds emerge in habitats where competing cells are in close proximity (such as biofilms; Table S1; Rao et al., 2005) as well as those where there is limited mechanical contact between cells, such as the aqueous habitats of planktonic species (Table S1; Collado-Fabbri et al., 2011; Trigal et al., 2011; Leão et al., 2012); and in both extreme and non-extreme environments (Table S1). Some microbes emerge as a/the dominating species in diverse types of habitat including specialists such as Lactobacillus (that do so, in part, by acidifying their environment) and generalists such as Aspergillus, Pichia and Pseudomonas species (Table S1; Steinkraus, 1983; Tamang, 2010). For weed species such as Saccharomyces cerevisiae (high-sugar environments), Haloquadratum walsbyi (aerobic, hypersaline brines), and Gonyostomum semen and Microcystis aeruginosa (eutrophic freshwater) ability to dominate can be restricted to a specific type of habitat. Most of these microbes do not populate their respective habitat as a one-off or chance event; they can consistently dominate their microbial communities and do so regardless of their initial cell number (Table S1; for P. anomala see Fig. 1; for S. cerevisiae see Pretorius, 2000; Renouf et al., 2005; Raspor et al., 2006; for L. rhamnosus see Randazzo et al., 2010). It is therefore intriguing to consider what combination(s) of traits can give rise to a weed phenotype.
Cellular biology of microbial weeds
Stress tolerance and energy generation
During habitat colonization, even in apparently stable, homogenous environments such as sugar-based solutions, there are dynamic/drastic changes in stressor concentrations, antimicrobials and stress parameters (D'Amore et al., 1990; Hallsworth, 1998; Eshkol et al., 2009; Fialho et al., 2011; see also below). As a result of these fluctuations microbes are exposed to multiple stresses some of which present potentially lethal challenges to the cell. In soil and sediments, for example the water activit can oscillate from low to high and high to low; these substrates typically cycle between desiccation and rehydration and are subjected to changes in solar radiation, temperature, and even freezing and thawing. Soils and sediments can be physically heterogeneous such that water availability, solute concentrations and other stress parameters exhibit profound spatial fluctuations. In addition, intracellular metabolites and extracellular substances (of both biotic and abiotic origin) impose stresses due to ionic, osmotic, chaotropic, hydrophobic and other activities of solutes (see Brown, 1990; Hallsworth et al., 2003a; 2007; Lo Nostro et al., 2005; Bhaganna et al., 2010; Chin et al., 2010). On the one hand resilience to stress undoubtedly contributes to competitive ability but on the other a vigorous growth phenotype and reinforced stress biology also require extraordinary levels of energy generation. Furthermore, microbes that can achieve a greater net gain in energy than competing species will have an ecological advantage (Pfeiffer et al., 2001). Here we suggest that species able to dominate microbial communities are exceptionally well equipped to resist the effects of multiple stress parameters, and that many have high-efficiency energy-generation systems.
We tested the hypothesis that microbial weeds are exceptionally tolerant to habitat-relevant stresses for two species; the soil bacterium Pseudomonas putida (Table 2) and the specialist yeast S. cerevisiae (see below). Environments in which P. putida is a major ecological player (Table S1) typically contain an array of chemically diverse aromatics and hydrocarbons that induce chaotropicity-mediated stresses (Hallsworth et al., 2003a; Bhaganna et al., 2010; Cray et al., 2013). This bacterium is not only renowned for its metabolic versatility, but is also known for its tolerance to specific solutes and solvents (Hallsworth et al., 2003a; Domínguez-Cuevas et al., 2006; Bhaganna et al., 2010). We compared the biotic windows for growth of P. putida for eight mechanistically distinct classes of stress using 37 habitat-relevant substances (Table 2). This bacterium was able to tolerate all substances within each group of stressor; this is the first data set to demonstrate that a mesophilic bacterium can tolerate the following combination of stresses: up to 4–11 mM hexane, toluene and benzene; 8–115 mM concentrations of aromatic, chaotropic solutes; 300–825 mM chaotropic salts; 1300–2160 mM sugars or polyols; 200–750 mM kosmotropic salts; and considerable levels of matric stress (Brown, 1990) generated by kosmotropic polysaccharides (Table 2). The P. putida tolerance limits to highly chaotropic substances and hydrophobic compounds were superior to those of other microbes (including extremophiles and polyextremophiles; Table 2; Hallsworth et al., 2007; Bhaganna et al., 2010). Whereas prokaryotes are generally less tolerant to solute stress than eukaryotes (Brown, 1990), the resilience to this array of mechanistically diverse solute stresses (Table 2) has neither been equalled nor been surpassed by any microbial species to our knowledge.
Table 2.
Growth windows for Pseudomonas putida under habitat-relevant stresses.a
| Stressor concentration (mM) causing P. putida growth-rate inhibition of: |
||||
|---|---|---|---|---|
| Type of stressor | Specific examples | 50%b | 100%b | Notes and references |
| Hydrophobic compoundsc | 1,2,3-Trichlorobenzened | 0.20 | 0.40 | Pseudomonas putida can be exposed to hydrophobic stressors in plant exudates, hydrocarbon-contaminated environments, and as hydrophobic catabolites and antimicrobials from P. putida or other microbes (see Table6; Timmis, 2009; Bhaganna et al., 2010) |
| γ-Hexachlorocyclohexaned,e | 0.00034 | 0.00072 | ||
| 2,5-Dichlorophenold | 0.70 | 0.90 | ||
| n-hexaned | 0.45 | 0.71 | ||
| Toluened | 4.0 | 4.6 | ||
| Benzened | 8.4 | 11.0 | ||
| Surfactants and aromatic solutes (highly chaotropic) | Tween® 80f,g | 108 | 164 | Cells come into contact with biosurfactants of microbial origin, and with chaotropic aromatics from the sources listed above (see also Hallsworth et al., 2003a; Timmis, 2009; Bhaganna et al., 2010) |
| Phenolh | 9.0 | 13.0 | ||
| o-cresole | 3.82 | 8.10 | ||
| Benzyl alcoholh | 20.0 | 32.7 | ||
| Sodium benzoatee | 69.2 | 115 | ||
| Chaotropic salts | CaCl2e | 210 | 302 | Chaotropic salts occur in soils, sediments, evaporite deposits, the Dead Sea, brine lakes (including the Discovery Basin located beneath the Mediterranean Sea and the Don Juan Pond in the Antarctic) and marine-associated habitats including bitterns (see Table S1); can be highly stressful for, and lethal to, microbial systems (Hallsworth et al., 2003a; 2007; Duda et al., 2004) |
| MgCl2d | 182 | 311 | ||
| Guanidine-HClh | 125 | 640 | ||
| LiClh | 595 | 825i | ||
| Alcohols and amides (moderately chaotropic) | Butanold | 158 | 208 | Alcohols are produced by some species as antimicrobials (Table6); ethanol is used as a biocide and can occur as an anthropogenic pollutant. Urea may be produced as an antimicrobial by some bacteria (Table6) and is used as an agricultural fertilizer to which soil bacteria are exposed |
| Ethanolh | 600 | 993i | ||
| Ureah | 650 | 950 | ||
| Formamided | 168 | 1080i | ||
| Relatively neutral substances | Methanold | 970 | 1440 | Some organic compounds have little chao- or kosmotropic activity at biologically relevant concentrations. At low concentrations glycerol is relatively neutral but, despite its role as a stress protectant, can act as a chaotropic stressor at molar concentrations (Williams and Hallsworth, 2009) |
| Ethylene glycolh | 1300 | 2160i | ||
| Glucosej | 840 | 1400 | ||
| Glycerolj | 770 | 2120i | ||
| Maltosej | 326 | 1300 | ||
| Kosmotropic compatible solutes | Prolinej | 800 | 2000 | The majority of microbial compatible solutes are kosmotropic, including other compatible solutes found in P. putida and other bacteria such as mannitol, betaine and ectoine. Pseudomonas may also come into contact with some of these compounds via exposure to plant exudates |
| Sorbitolj | 503 | 1200 | ||
| Dimethyl sulfoxidej | 353 | 1300 | ||
| Trehalosej | 474 | 771 | ||
| Glycinej | 99 | 370 | ||
| Betainej | 1570 | 2560 | ||
| Kosmotropic salts | NaClj | 650 | 750 | NaCl is environmentally ubiquitous, and is the dominant salt in most marine habitats including bioaerosols (Table S1); ammonium sulfate and KH2PO4 are commonly applied to soils as fertilizers. In many habitats kosmotropic salts are the primary osmotic stressors, but stresses imposed on the cell are determined by the net combination of solute activities of ions and other substances present (see Hallsworth et al., 2007) |
| KH2PO4j | 118 | 200 | ||
| Ammonium sulfatej | 198 | 350 | ||
| Sodium citratej | 53 | 500 | ||
| Polysaccharides (kosmotropic) | Dextran 40000j | 10 | 17 | Pseudomonas putida is exposed to the kosmotropic activities of and/or matric stress induced by polysaccharides such as microbe- and plant-derived extracellular polymeric substances, humic substances, etc. |
| Polyethylene glycol 3350j | 43 | 120 | ||
| Polyethylene glycol 6000j | 20 | 40 | ||
Pertinent to the phyllosphere, rhizosphere, hydrocarbon-polluted environments, and other habitats (see Table S1), including catabolic products of hydrocarbon degradation, antimicrobial substances (see Table 6), compatible solutes, and other P. putida metabolites. At sufficient concentrations solutes such as glucose, maltose, proline, sorbitiol, glycine, betaine, NaCl and KH2PO4 induce osmotic stress; whereas dextran and high Mr polyethylene glycols induce matric stress (see Brown, 1990); hydrophobic substances typically induce a (chaotropicity-mediated) hydrocarbon-induced water stress (Bhaganna et al., 2010; Cray et al., 2013); and other aromatics, alcohols, amides and specific salts cause chaotrope-induced water stress (Hallsworth et al., 2003a; Cray et al., 2013).
Relative to no-added stressor controls (see Hallsworth et al., 2003a).
These data were taken from Bhaganna and colleagues (2010); P. putida KT2440 (DSMZ 6125) was grown in a minimal mineral-salt broth (with glucose and NH4Cl as the sole carbon and nitrogen substrates respectively; see Hartmans et al., 1989; Bhaganna et al., 2010) at 30°C. The media for control treatments had no stressors added. Some P. putida strains can tolerate benzene at up to 20 mM (Volkers et al., 2010).
The pesticide γ-hexachlorocyclohexane (γ-HCH) is also known as lindane.
Pseudomonas putida KT2440 was grown in modified Luria–Bertani broth at 30°C; stressors were incorporated into media prior to inoculation, and growth rates calculated, as described by Hallsworth and colleagues (2003a). The media for control treatments had no stressors added.
Tween® 80 was used as a model substitute for biosurfactants (Cray et al., 2013).
These data were taken from Hallsworth and colleagues (2003a); P. putida KT2440 was grown in modified Luria–Bertani broth at 30°C as described in footnote (f).
Extrapolated values.
Glycerol is a notable exception (Williams and Hallsworth, 2009).
Pseudomonas putida is able to avoid contact with or prevent accumulation of stressors by using solvent-efflux pumps and synthesizing extracellular polymeric substances to form a protective barrier (Table 3). This bacterium also utilizes highly efficient macromolecule-protection systems that are upregulated in response to multiple environmental stresses including between 10 and 20 protein-stabilization proteins, oxidative-stress responses, and upregulation of energy generation and overall protein synthesis (Table 3). Furthermore, P. putida can synthesize an array of compatible solutes including betaine, proline, N-acetylglutaminylglutamine amide, glycerol, mannitol and trehalose (Table 3). Individual compatible solutes are unique in their physicochemical properties, interactions with macromolecular systems and functional efficacy for specific roles within the cell (e.g. see Chirife et al., 1984; Crowe et al., 1984; Brown, 1990; Hallsworth and Magan, 1994a; 1995; Yancey, 2005; Bhaganna et al., 2010; Bell et al., 2013). Microbes that are able to selectively utilize/deploy any substance(s) from a range of diverse compatible solutes have enhanced tolerance to diverse stresses (e.g. Hallsworth and Magan, 1995; Bhaganna et al., 2010). The range of compatible solutes utilized by P. putida may be unique for a bacterium (see Brown, 1990) and can confer tolerance to diverse stresses including those imposed by chaotropes and hydrocarbons (Bhaganna et al., 2010).
Table 3.
Stress tolerance and energy generation: cellular characteristics that contribute to weed-like behaviour.a
| Metabolites, proteins, molecular characteristics, and other traits | Function(s) | Phenotypic traits | Notes and references |
|---|---|---|---|
| Tolerances to multiple environmental stresses | |||
| Ability to select from an array of functionally diverse compatible solutes in response to specific stress parameters | Specific compatible solutes can protect against osmotic stress, temperature extremes, freezing, dehydration and rehydration, chaotropicity, and/or hydrophobic stressors (Crowe et al., 1984; Brown, 1990; Hallsworth et al., 2003b; Bhaganna et al., 2010; Chin et al., 2010; Bell et al, 2013) | Ability to maintain functionality of macromolecular systems and/or cell turgor under diverse stresses | Some microbes utilize a wide array of compatible solutes depending on the type and severity of stress; e.g. Pseudomonas putida is exceptional among bacteria in its ability to synthesize and accumulate protectants as diverse as betaine, proline, N-acetylglutaminylglutamine amide, glycerol, mannitol and trehalose (D'Souza-Ault et al., 1993; Kets et al., 1996; Bhaganna et al., 2010); Saccharomyces cerevisiae can accumulate proline, glycerol and trehalose (see Table6; Kaino and Takagi, 2008). Like other fungi, Aspergillus species utilize a range of polyols as well as trehalose (Hallsworth and Magan, 1994c; Hallsworth et al., 2003b) but their exceptional ability to generate energy under stress (see below) may aid glycerol retention and thereby extending the growth window at low water activity (Hocking, 1993; Williams and Hallsworth, 2009) |
| Upregulation of protein-stabilization proteins in combinations tailored to specific stressesb | In addition to stressors (including those of biotic origin; Table6), protein-stabilization proteins also protect against perturbations induced by stresses such as temperature extremes and desiccation (Arsène et al., 2000; Ferrer et al., 2003) | Maintenance of protein structure and function during environmental challenges | Several studies show highly efficient responses of P. putida to chaotropic and hydrophobic stressors via upregulation of between 10 and 20 protein-stabilizing proteins (Hallsworth et al., 2003a; Santos et al., 2004; Segura et al., 2005; Domínguez-Cuevas et al., 2006; Tsirogianni et al., 2006; Volkers et al., 2006; Ballerstedt et al., 2007). The PalA protein in Aspergillus nidulans induces a rapid upregulation of protein-stabilization proteins in response to extreme pH (Freitas et al., 2011); high production rates of protein-stabilization proteins enhance tolerance to chaotropic salts and alcohols in S. cerevisiae and Lactobacillus plantarum, to hydrophobic stressors in S. cerevisiae and Escherichia coli (for references, see Bhaganna et al., 2010), and low temperature, salt and other stresses in Rhodotorula mucilaginosa (Lahav et al., 2004) |
| Intracellular accumulation of ions for osmotic adjustment; intracellular proteins structurally adapted to high ionic strength | The utilization of ions for osmotic adjustment avoids the energy-expensive synthesis of organic compatible solutes (Kushner, 1978; Oren, 1999) | Ability to maintain metabolic activity at high extra- and intracellular ionic strength | Haloquadratum walsbyi, and Salinibacter ruber (unusually for a bacterium), utilize the so-called ‘salt-in’ strategy (Oren et al., 2002; Bardavid and Oren, 2012; Saum et al., 2012) |
| Diverse proteins and pathways conferring tolerance to solvents, chaotropic solutes and hydrophobic stressors | Solvent-tolerant species such as P. putida can function at high concentrations of chaotropic and hydrophobic stressors due to the collective activities of solvent pumps that expel solvent molecules, pathways that catabolize the solvent, membrane-stabilization proteins (see below), compatible solutes (see above), and additional chaotrope- and solvent-stress responses (Ramos et al., 2002; Timmis, 2002; Hallsworth et al., 2003a; Volkers et al., 2006; Bhaganna et al., 2010; Fillet et al., 2012) | High tolerance to chaotropic and hydrophobic solvent stressors | Due to its exceptional solvent tolerance P. putida is the organism of choice for many industrial applications including biotransformation in two-phase systems and bioremediation (Timmis, 2002). |
| Membrane-stabilization proteins; e.g. cis/trans-isomerase (CTI) | Membrane stabilization under stress | Enhanced tolerance to stresses that impact membrane structure | Several proteins are involved in maintaining bilayer rigidity in P. putida during exposure to toluene and other solvents or hydrophobic stressors (Bernal et al., 2007) |
| Proteins involved in the moderation of membrane fluidity | Maintenance of membrane fluidity and membrane-associated processes under conditions such as low temperature (Turk et al., 2011) | Enhanced tolerance to diverse, physicochemical stress parameters | For example Aureobasidium pullulans, Cryptococcus liquefaciens and R. mucilaginosa (Turk et al., 2011) |
| Promoter(s) that simultaneously control multiple stress-response genes, e.g. stress-response elements (STREs) | Activation of multiple cellular stress-responses | Enhanced tolerance to multiple stress-parameters | The transcription factors (Msn2p and Msn4p) involved in STRE-mediated stress responses in S. cerevisiae accumulate in the nucleus upon heat-, ethanol- (chaotrope-), and osmotically induced stresses, and during nutrient limitation. The STRE is upstream of genes for protein-stabilization proteins (see above) metabolic enzymes, and cellular organization proteins (Görner et al., 1998; Moskvina et al., 1998) |
| Proteins involved in DNA protection under stress; e.g. DNA-binding protein (Dps) | Formation of a protein:chromosome complex that protects DNA | Enhanced DNA stability and reduced risk of mutation in relation to multi-farious stresses | The role of Dps has been well-characterized in E. coli in relation to diverse environmental challenges including reactive oxygen species, iron and copper toxicity, thermal stress and extremes of pH (Nair and Finkel, 2004) |
| Pathways for synthesis of carotenoids (e.g. β-carotene in Dunaliella salina; torularhodin in Rhodotorula spp.) | Carotenoids absorb light and thereby protect cellular macromolecules and organelles from exposure to ultraviolet | Tolerance of exposure to ultraviolet irradiation | Synthesis of β-carotene is upregulated in response to light in D. salina (Chen and Jiang, 2009); carotenoids are concentrated in the plasma membrane of S. ruber (Antón et al., 2002); studies of R. mucilaginosa show that torularhodin enhances tolerance to ultraviolet B exposure (Moliné et al., 2010) |
| Preferential utilization of a chaotropic compatible solute at high NaCl concentration and/or low temperature | Rigidification of macromolecular structures at high NaCl and low temperature can induce metabolic inhibition (Brown, 1990; Ferrer et al., 2003; Chin et al., 2010) that can be compensated for by glycerol and/or fructose (Brown, 1990; Chin et al., 2010) | Enhanced tolerance to extreme conditions that rigidify macromolecular systems | The high intracellular concentrations of glycerol in D. salina under salt stress and glycerol and/or fructose in and Aspergillus and Eurotium spp. and R. mucilaginosa at low temperature are able to extend growth windows under these extreme conditions (Brown, 1990; Lahav et al., 2002; Chin et al., 2010). Dunaliella salina has membranes with reduced glycerol permeability thereby aiding retention of this compatible solute (Bardavid et al., 2008) |
| Efficient system to eliminate free radicals (see above) | Elimination of reactive oxygen species (see also Table5) | Various substances and stress parameters can induce oxidative stress | Diverse environmental substances and parameters induce oxidative stress, either directly or indirectly (e.g. via lipid peroxidation). For example, chaotropic stressors and heat shock have been associated with the induction of oxidative stress responses in S. cerevisiae and P. putida (Costa et al., 1997; Hallsworth et al., 2003a) |
| Proteins associated with regulation of cytosolic pH; e.g. the lysine decarboxylase system (coded for by the cadBA operon); the Na+/H+ antiporter | Conversion and export of lysine/cadaverine that results in increased pH of the cytosol in bacteria (Álvarez-Ordóñez et al., 2011); the Na+/H+ antiporter Nha1 decreases intracellular hydrogen ion concentration | Tolerance of low-pH environments | Decarboxylase systems that increase intracellular pH also exist for other amino acids (Álvarez-Ordóñez et al., 2011); Nha1 greatly enhances viability of Saccharomyces boulardii at pH 2 (dos Santos Sant'Ana et al., 2009) |
| Multidrug efflux pumps (for details see Table6) | |||
| Enzymes for synthesis of extracellular polymeric substances | Production and secretion of polymeric substances | Enhanced tolerance to multiple stressors and stress parameters | Extracellular polymeric substances have been shown to confer protection to E. coli cells exposed to high temperature, low pH, salt and oxidative stress (Chen et al., 2004), and are associated with stress protection in Clostridium spp., Pantoea ananatis, P. putida and other Pseudomonas spp. (Chang et al., 2007; Morohoshi et al., 2011; see also Table 6) |
| Upregulation of proteins involved in protein synthesis under stress | To enhance or maintain the quantity, type, and functionality of cellular proteins under stress | Maintenance of the cellular system and its metabolic activity under stress | Can be upregulated in response to stress induced by high temperature, chaotropic solutes, hydrophobic stressors and other stress parameters (e.g. P. putida; Hallsworth et al., 2003a; Volkers et al., 2009) |
| Polyploidy, aneuploidy, association with plasmids, and/or hybridization that enhance(s) stress tolerance | Whereas hybridization, polyploidy, aneuploidy and association with plasmids can be ecologically disadvantageous, such traits can enhance the vigour, stress tolerance and competitive ability of some microbes such as Pichia spp., S. cerevisiae, P. putida and S. ruber (Table1; Rainieri et al., 1999; Naumov et al., 2001; Mongodin et al., 2005; Tark et al., 2005; Dhar et al., 2011; Chen et al., 2012) | Enhanced stress tolerance | Aneuploid cells of S. cerevisiae have enhanced tolerance to oxidative stress and diverse inhibitors of biochemical processes (Chen et al., 2012); polyploidy can enhance vigour and stress tolerance in some species (see also Tables1 and c); the stress tolerance of some microbial weeds is enhanced by their association with plasmids (e.g. the P. putida tol-plasmid that enhances solvent tolerance; Tark et al., 2005; Domínguez-Cuevas et al., 2006; the S. ruber plasmid contains a gene involved in ultraviolet protection; Mongodin et al., 2005) |
| Polyextremophilic growth-phenotype | Ability to grow optimally under multiple, extreme conditions; e.g. extremely acid and alkali (pH 2–10), high-salt (2.5 M NaCl), and low-temperature conditions (Lahav et al., 2002; Libkind and Sampaio, 2010) | Polyextremophile | Rhodotorula mucilaginosa is capable of growth over an incredible range of physicochemical conditions (including almost the entire pH range for microbial life) and is able to colonize a correspondingly wide diversity of environments (for examples see main text) |
| Genes associated with salt stress | Sugar catabolism (e.g. RmPGM2, RmGPA2, RmACK1, RmMCP1 and RmPET9), protein glycosylation (e.g. RmSEC53), aromatic amino acid synthesis (e.g. RmARO4), regulation of protein synthesis (e.g. RmANB1) | Maintenance of protein synthesis and energy generation under salt stress | Between 10 and 20 genes are involved in increased tolerance to NaCl and/or LiCl in R. mucilaginosa (Lahav et al., 2004; Gostinčar et al., 2012) |
| Metabolites associated with resistance to antimicrobial plant metabolites and survival in phyllosphere habitats | Metabolites involved in quorum sensing may enhance competitive ability on the leaf surface | Enhanced survival and competitive ability | By contrast to other phyllosphere microbes, P. ananatis is resistant to the inhibitory effects of plant alkaloids and this correlates with the production of metabolites involved in quorum sensing such as N-acyl-homoserine lactone (Enya et al., 2007) |
| High-efficiency energy-generation systems | |||
| Light-activated proton pumps (e.g. xanthorhodopsin) | To generate a proton-motive force, utilizing light as an energy source | Utilization of light for transmembrane proton transport | Xanthorhodopsin, found in S. ruber, is a proton pump made up of two chromophores; a light-absorbing carotenoid and a protein (Balashov et al., 2005); a light-activated proton pump was also recently described in H. walsbyi (Lobasso et al., 2012) |
| High expression of genes involved in energy generation (e.g. fba, pykF, atpA and atpDc) | Rapid catabolism of carbon substrates leading to enhanced synthesis of ATP | Efficient energy-generation | High expression of E. coli genes fba, pykF, atpA and atpD can be associated with ability to grow rapidly, enhanced stress tolerance and – by implication – greater competitive ability (Karlin et al., 2001) |
| Enhanced energy-efficiency at low nutrient concentrations (e.g. see glutamate dehydrogenase entry in Table5) | |||
| Upregulation of proteins involved in energy generation under chaotropic and solvent-induced (hydrocarbon) stresses (Hallsworth et al., 2003a; Volkers et al., 2006). | Maintenance of energy generation close to the point of system failure under hostile conditions | Efficient energy generation during potentially lethal challenges | The exceptional ability of P. putida to respond and adapt to chaotropic, solvent and hydrocarbon stressors has, in part, been attributed to considerable increases in energy- and NAD(P)H-generating systems |
| Idiosyncratic genome modifications associated with reinforced energy metabolism | Enhanced primary metabolism and energy-generation systems | Enhanced vigour and energy production | Aspergillus species have numerous genome modifications, such as gene duplications for pyruvate dehydrogenase, citrate synthase, aconitase and malate dehydrogenase that enhance primary metabolism including glycolysis and the TCA cycle (Flipphi et al., 2009) |
| Efficient utilization of intracellular reserves for energy generation | Catabolism of stored substances to boost growth, stress tolerance, production of secondary metabolites, motility and/or other key cellular functions under stress | Ability to maintain energy production during nutrient limitation and under hostile conditions | Efficient storage (see Table5) and utilization of energy-generating substances such as trehalose, glycogen and polyhydroxyalkanoates confers a competitive advantage in S. cerevisiae and other species (Castrol et al., 1999; Kadouri et al., 2005) |
| Concomitant energy generation and synthesis of antimicrobials with no loss of ATP-generation efficiency | Via aerobic fermentation S. cerevisiae can simultaneously generate energy while synthesizing the chaotropic stressor ethanol (see Table6) | Energy generation linked to synthesis of an antimicrobial | Each glucose molecule fermented by S. cerevisiae produces two ethanol molecules and two molecules of ATP (Piškur et al., 2006) |
The current table provides examples of proteins, genes or other characteristics that can be associated with and give rise to weediness; the categories of traits are not mutually exclusive. Individual traits may not be unique to microbial weeds; however, weeds species are likely to have a number of these types of characteristics.
For example chaperonins, heat-shock proteins and cold-shock proteins.
That code for fructose-1,6-bisphosphate aldolase, pyruvate kinase and ATP synthase subunits F1 α and F1 β respectively.
The exceptionally wide growth-windows of Aspergillus species in relation to a number of stress parameters correlate with their ability to dominate communities in both extreme and non-extreme (but physicochemically dynamic) environments including soils, plant necromass, saline and very high-sugar habitats (Table S1). At least 20 species of Aspergillus and the closely related genus Eurotium are xerotolerant (Pitt, 1975; Brown, 1990). Aspergillus penicillioides is capable of hyphal growth and conidial germination down to water activities of 0.647 and 0.68 respectively (Pitt and Hocking, 1977; Williams and Hallsworth, 2009); Aspergillus echinulatus can germinate down to 0.62 (Snow, 1949). Halotolerant species such as Aspergillus wentii can grow on the highly chaotropic salt guanidine-HCl at a higher concentration than any other microbe; up to 930 mM (≡ 26.5 kJ g−1 chaotropic activity; Hallsworth et al., 2007) and, along with Aspergillus oryzae, up to ∼ 4 M NaCl or KCl (Hallsworth et al., 1998). Aspergillus (and Eurotium) strains are capable of reasonable growth rates close to 0°C (Chin et al., 2010), and some reports suggests that the biotic windows for some Aspergillus species ultimately fail at ≤ −18°C (Vallentyne, 1963; Siegel and Speitel, 1976). Whereas a number of comparable fungal genera show moderate levels of stress tolerance, Aspergillus species generally out-perform their competitors and top numerous league tables for ascomycete stress tolerance (see Pitt, 1975; Wheeler and Hocking, 1993; Williams and Hallsworth, 2009).
Studies of seven Aspergillus species have revealed a number of genome modifications within this genus that reinforce both primary metabolism and energy generation (Table 3; Flipphi et al., 2009). These include gene duplications for key enzymes that control metabolic flux, such as pyruvate dehydrogenase, citrate synthase, aconitase, and malate dehydrogenase that enhance glycolysis and the TCA cycle (Flipphi et al., 2009). There is evidence that, at the low water activities (high osmotic pressures) associated with very high-salt and high-sugar habitats, the biotic window of xerophilic fungi fail due to the prohibitive energy expenditure required to prevent the high levels of intracellular glycerol from leaking out of the membrane (Hocking, 1993). The adaptability and competitive ability of Aspergillus species (most of which can also proliferate under non-extreme conditions) may be attributed, in part, from enhanced energy-generating capability. Aspergillus species (like many fungi) can utilize a range of polyols – as well as trehalose – as compatible solutes based on their individual strengths as protectants against osmotic stress, desiccation rehydration, chaotropes etc. (Crowe et al., 1984; Brown, 1990; Hallsworth et al., 2003b). Aspergillus and Eurotium strains can also accumulate chaotropic sugars from the medium, or preferentially synthesize and accumulate chaotropic glycerol, in order to enhance growth at temperatures below ∼ 10°C (Chin et al., 2010; see also below).
Salinibacter ruber is a halophilic bacterium found in high numbers at molar salt concentrations in aerobic aquatic habitats that are otherwise more densely populated by archaeal than bacterial species. Unusually for a bacterium, S. ruber takes in ions to use as compatible solutes and thereby avoids synthesis of organic compatible solutes that would be energetically unfavourable in environments of > 1 M NaCl (Oren, 1999) and has evolved intracellular proteins that are both structurally stable and catalytically functional in the cytosol at high ionic strength (Table 3). Salinibacter ruber (as well as the alga Dunaliella salina and yeast R. mucilaginosa) accumulates carotenoids in response to light which can protect against oxidative stress that is a potential hazard in salterns exposed to high levels of ultraviolet (Table 3). Haloquadratum walsbyi, an Archaeon that is also a dominant player in saltern communities (Table S1), also uses the ‘salt-in’ compatible-solute strategy, and – along with S. ruber – has light-activated protein pumps that generate proton-motive force and thereby boost energy generation (Table 3). In addition, H. walsbyi is unusually tolerant to the chaotropic salt MgCl2 that can reach high concentrations in evaporate ponds (Hallsworth et al., 2007; Bardavid et al., 2008). Dunaliella salina is also highly prevalent in saline habitats that (unusually for an alga) can successfully compete with the multitude of halophilic Archaea found in 5 M NaCl environments (see later; Table S1). This alga preferentially utilizes glycerol as a compatible solute, is able to accumulate glycerol to molar concentrations (6–7 M; see Bardavid et al., 2008), and has low membrane permeability to this compatible-solute thus aiding retention (Bardavid et al., 2008).
Glycerol is unique in its ability to maintain the flexibility of macromolecular structures under conditions that would otherwise cause excessive rigidification, e.g. molar concentrations of NaCl, or low temperature (Back et al., 1979; Hallsworth et al., 2007; Williams and Hallsworth, 2009; Chin et al., 2010). Species able to grow at low or subzero temperatures (e.g. Aspergillus, Cryptococcus, Rhodotorula species, including R. mucilaginosa) either primarily use glycerol as a compatible solute or preferentially accumulate this chaotrope at low temperature (Table 3; Lahav et al., 2004; Chin et al., 2010). Rhodotorula mucilaginosa is able to colonize – and frequently dominates – diverse substrates/environments including saline and non-saline soils, aquatic habitats (both liquid- and ice-associated), the acidic surface-film of sphagnum moss (as well as other plant tissues) and various surfaces within the human body and other animals (Table S1; Egan et al., 2002; Trindade et al., 2002; Vital et al., 2002; Lahav et al., 2002; 2004; Lisichkina et al., 2003; Buzzini et al., 2005; Perniola et al., 2006; Goyal et al., 2008; Turk et al., 2011; Kachalkin and Yurkov, 2012). This ability is coupled with an exceptional level of tolerance to multiple stresses: R. mucilaginosa is (highly unusual for a yeast) a polyextremophile that is psychrophilic, highly salt-tolerant, and can growth over the majority of the pH range known to support microbial life (i.e. < 2 and > 10, see Table 3; Lahav et al., 2002).
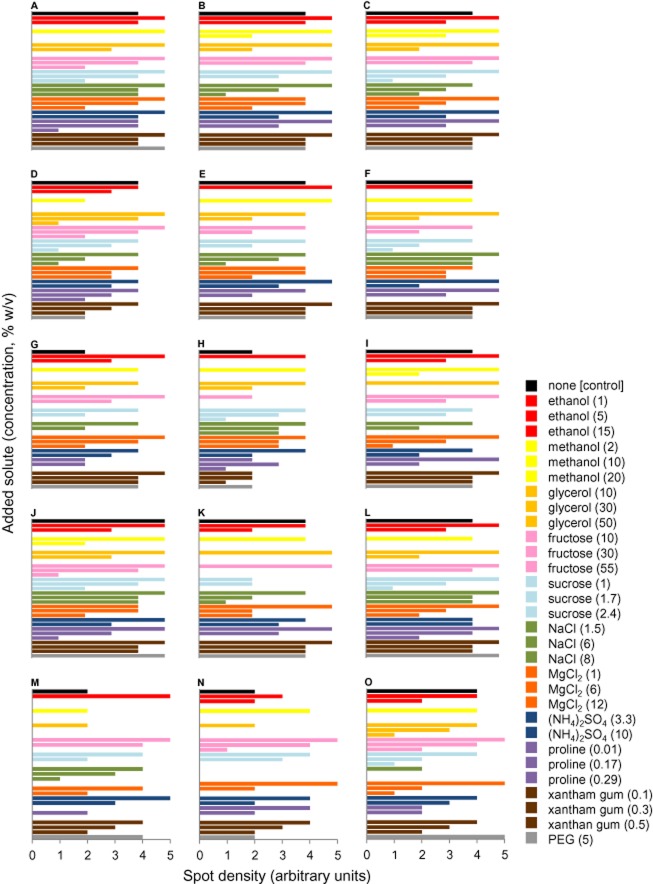
Some microbial habitats such as fruits that have been disconnected from the vascular system (and immune responses) of the parent plant have a favourable water activity and high nutrient availability and are therefore potentially habitable by a wide range of microbes (Ragaert et al., 2006). As the microbial community develops on a detached fruit (or in apple or grape musts; see Table S1) there can be changes in the composition and concentration of sugars, osmolarity, water activity, ethanol concentration (and that of other antimicrobials), pH and other environmental factors that are severe enough to inhibit or eliminate most members of the original microflora (see Brown, 1990; Hallsworth, 1998; de Pina and Hogg, 1999; Pretorius, 2000; Bhaganna et al., 2010). The yeast S. cerevisiae that is relatively scarce in natural habitats (and even when present forms a quantitatively insignificant component of the natural community; Renouf et al., 2005; Raspor et al., 2006) is unsurpassed in its ability to emerge to dominate high-sugar habitats at water activities above 0.9 (Table S1; Hallsworth, 1998; Pretorius, 2000). We therefore compared its tolerance towards a range of habitat-relevant stressors with those of 12 other yeasts that occur at the insect–flower interface, on plant tissues, and/or are environmentally widespread (Fig. 2; Table 4). This experiment revealed that S. cerevisiae has a more vigorous growth phenotype and more robust stress tolerance than the other species tested with the exception of two species of Pichia, another genus characterized by weed-like traits (see Fig. 2; Table 4). Stress phenotypes of the yeasts assayed were compared on the basis of stressor type and mechanistically distinct stress-parameters in order to assess resilience to both the degree and diversity of environmental challenges (Table 4). Under the assay conditions it was the three S. cerevisiae strains, Pichia (Kodamaea) ohmeri and Pichia sydowiorum that exhibited the greatest tolerance to the widest range of solute-imposed stresses (Table 4). Other species studied were slow growers, have specific environmental requirements (e.g. the halotolerant D. hansenii and Hortaea werneckii), and/or otherwise lacked the capacity for vigorous growth on the high-sugar substrate at 30°C (see Fig. 2). Generally S. cerevisiae strains were fast-growing, xerotolerant, and highly resistant to chaotrope-induced, osmotic, matric and kosmotrope-induced stresses and are known to be acidotolerant and capable of growth over a wide temperature range, from ∼0°C to 45°C (Fig. 2A and B; Table 4). Although Pichia species can produce ethanol (P. anomala can produce up to 0.36 M ethanol; Djelal et al., 2005) when growing side by side in high-sugar habitats with S. cerevisiae, Pichia species are unable to tolerate the molar ethanol concentrations that S. cerevisiae can produce (Lee et al., 2011).
Figure 2.
Growth of Saccharomyces cerevisiae strains: (A) CCY-21-4-13, (B) Alcotec 8 and (C) CBS 6412 and other yeast species: (D) Candida etchellsii (UWOPS 01–168.3), (E) Cryptococcus terreus (PB4), (F) Debaryomyces hansenii (UWOPS 05-230.3), (G) Hansenula (Ogataea) polymorpha (CBS 4732), (H) Hortaea werneckii (MZKI B736), (I) Kluyveromyces marxianus (CBS 712), (J) Pichia (Kodamaea) ohmeri (UWOPS 05-228.2), (K) Pichia (Komagataella) pastoris (CBS 704), (L) Pichia sydowiorum (UWOPS 03-414.2), (M) Rhodotorula creatinivora (PB7), (N) Saccharomycodes ludwigii (UWOPS 92–218.4) and (O) Zygosaccharomyces rouxii (CBS 732) on a range of media at 30°C. These were: malt-extract, yeast-extract phosphate agar (MYPiA) without added solutes (control), and MYPiA supplemented with diverse stressors – ethanol, methanol, glycerol, fructose, sucrose, NaCl, MgCl2, ammonium sulfate, proline, xantham gum and polyethylene glycol (PEG) 8000 – over a range of concentrations as shown in key (values indicate %, w/v). A standard Spot Test was carried out (see Chin et al., 2010; Toh et al., 2001) that was modified from Albertyn and colleagues (1994) for stress phenotype characterization (see Table 5). Colony density was assessed after an incubation time of 24 h on a scale of 0–5 arbitrary units (Chin et al., 2010). Cultures were obtained from (for strain A) the Culture Collection of Yeasts (CCY, Slovakia); (strain B) Hambleton Bard Ltd, Chesterfield, UK; (strains C, G, I, K, O) the Centraalbureau voor Schimmelcultures (CBS, the Netherlands); (strains D, F, J, L, N) the University of Western Ontario Plant Sciences Culture Collection (UWOPS, Canada); (strains E, M) were obtained from Dr Rosa Margesin, Institute of Microbiology, Leopold Franzens University, Austria; and (strain H) the Microbial Culture Collection of National Institute of Chemistry (MZKI, Slovenia). All Petri plates containing the same medium were sealed in a polythene bag to maintain water; all experiments were carried out in duplicate, and plotted values are the means of independent treatments.
Table 4.
Stress phenotypes and ecophysiological profiles of Saccharomyces cerevisiae relative to those of other yeast species
| Tolerance to diverse stressorsb |
Tolerance to distinct stress mechanismsb |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yeast speciesa | Alcohols | Glycerol | Sugars | Salts | Others | Chaotropic | Osmotic | Matric | Kosmotropic | Additional notes on stress and growth phenotypesc | Typical habitat | Ecophysiological descriptiond |
| Saccharomyces cerevisiae strain | ||||||||||||
| CCY-21-4-13 | Higher | Mid | Higher | Higher | Higher | Higher | Higher | Higher | Higher | Topt 30–35°C; Tmin ∼0.5°C; Tmax 45°C pHopt 3–7; pHmin ∼2.5e; fast grower | Fruits; other fleshy plant tissues; nectar | Multiple stress tolerances including sugar and ethanolf; stress phenotype of a xerotolerant, acidotolerant, vigorous weed |
| Alcotec 8 | Higher | Mid | Mid | Mid | Higher | Higher | Mid | Higher | Mid | As above | As above | As above |
| CBS 6412 | Higher | Mid | Higher | Mid | Higher | Higher | Mid | Higher | Mid | As above | As above | As above |
| Other species (strain) | ||||||||||||
| Candida etchellsii (UWOPS 01–168.3) | Lower | Higher | Mid | Higher | Mixed | Higher | Higher | Mid | Mid | Slow growerg | Pollinating insects; flowers; fermented foods | Intolerant to specific stressors; slow-growing |
| Cryptococcus terreus (PB4) | Mid | Mid | Higher | Mid | Higher | Mid | Mid | Higher | Mid | Topt 10°C; Tmin 1°Ch; slow grower | Soils and sediments; subsurface environments | A slow-growing psychrophilei |
| Debaryomyces hansenii (UWOPS 05–230.3) | Mid | Mid | Higher | Higher | Higher | Mid | Mid | Higher | Lower | Topt 30–32°C; pHopt 5.5j; fast grower | High-salt environments and foods | Halotolerant; a fast-growing high-salt specialist |
| Hansenula (Ogataea) polymorpha (CBS 4732) | Higher | Mid | Mid | Mid | Higher | Higher | Lower | Higher | Higher | Topt 37–43°C; pHopt 4.5–5.5h | Soil, insect gut, fruit juice | Neither fast-growing nor highly stress tolerant; a high-temperature specialist |
| Hortaea werneckii (MZKI B736) | Mid | Mid | Higher | Higherj | Lower | Mid | Mid | Lower | Mid | Topt ∼29°Ch; slow grower | Salty environments; animal and plant surfaces | Intolerance to matric stress; a slow-growing halophile |
| Kluyveromyces marxianus (CBS 712) | Higher | Lower | Mid | Lower | Higher | Mid | Lower | Higher | Lower | Topt ∼38–40°C; Tmax 65°Ch; not fast-growing | Milk; plant surfaces; natural fermentations | Thermotolerant, solute-intolerant and xero-intolerantj |
| Pichia (Kodamaea) ohmeri (UWOPS 05–228.2) | Higher | Mid | Higher | Higher | Higher | Higher | Higher | Higher | Mid | Topt ∼33°C; Tmax > 50°Ch; fast- grower | Beetles; fruits and other plant | Multiple stress tolerances including ethanol and sugar; stress phenotype of a xerotolerant, vigorous weed |
| Pichia (Komagataella) pastoris (CBS 704) | Mid | Lower | Lower | Mid | Higher | Mid | Lower | Higher | Mid | pHmin ∼2.2h; not fast-growing | Rotting wood; slimes | Intolerant to osmotic stress, glycerol and sugar; a xero-intolerant yeast |
| Pichia sydowiorum (UWOPS 03–414.2) | Higher | Mid | Higher | Higher | Higher | Higher | Mid | Higher | Higher | pHopt ∼6–8h; fast grower | Polluted environments; waste water | Multiple stress tolerances including ethanol and sugar; stress phenotype of a xerotolerant, vigorous weed |
| Rhodotorula creatinivora (PB7) | Lower | Lower | Mid | Mid | Lower | Lower | Lower | Mid | Higher | Topt ∼10°C; Tmin ∼1°Ch; slow grower | Subsurface; soil | Intolerant to alcohols, glycerol; xerotolerant a psychrophilic slow grower |
| Saccharomycodes ludwigii (UWOPS 92–218.4) | Lower | Mid | Higher | Lower | Lower | Lower | Lower | Lower | Lower | pHmin ∼2h; slow grower | Grapes; plant tissues and fermentations | Intolerant to glycerol and NaCl; tolerant to sugars; an osmophile |
| Zygosaccharomyces rouxii (CBS 732) | Mid | Higher | Higher | Lower | Mixed | Higher | Mid | Mid | Mid | pHopt 3–7; pHmin 1.5–2h; slow grower | Fruits and other high-sugar habitats | Intolerant to NaCl; tolerant to glycerol and sugars; an osmophile |
Sources of strains are given in Fig. 2.
Based on data for growth on MYPiA at 30°C (see Fig. 2). Alcohols were ethanol and methanol, sugars were fructose and sucrose, salts were NaCl, MgCl2 and ammonium sulfate, the other stressors were proline, xantham gum and PEG 8000 (Fig. 2). Chaotrope stress is induced by ethanol, fructose, glycerol and MgCl2; osmotic stress by fructose, sucrose, NaCl and proline; matric stress by xantham gum and PEG 8000; and kosmotrope stress by ammonium sulfate (Fig. 2; Brown, 1990; Hallsworth, 1998; Hallsworth et al., 2003a; Hallsworth et al., 2007; Williams and Hallsworth, 2009; Bhaganna et al., 2010; Chin et al., 2010; Cray et al., 2013). Stress-tolerance assessments (higher, mid-range, lower) are relative to the range of those of the other species assayed, and based on values in Fig. 2.
Temperature optima, minima and/or maxima (Topt, Tmin, Tmax) and pH optima, minima and/or maxima (pHopt, pHmin, pHmax) for growth were obtained from cited publications. Extreme growth phenotypes (fast or slow growers) were listed according to growth rates on MYPiA at 30°C (see also Fig. 2). Species designated as slow growers did not have a vigorous growth phenotype under these conditions; i.e. all strains listed are able to grow rapidly within the assay period on/at their favoured media/temperature. The designation slow growing did not relate to an extended lag phase (data not shown).
Primarily based on data presented in the current table as well as Fig. 2 (unless otherwise stated) so is most pertinent to high-sugar habitats at ∼30°C. Habitat dominance will ultimately be influenced by antimicrobial activities of species present within a community; see Table 6). Ecophysiological descriptions of weed-like species are underlined in bold.
Values for temperature- and pH-tolerances were taken from: (for S. cerevisiae) Praphailong and Fleet (1997), Yalcin and Ozbas (2008) and Salvadó and colleagues (2011); (for C. terreus) Krallish and colleagues (2006); (for D. hansenii) Domínguez and colleagues (1997); (for H. polymorpha) Levine and Cooney (1973); (for H. werneckii) Díaz Muñoz and Montalvo-Rodríguez (2005); (for K. marxianus) Aziz and colleagues (2009); Salvadó and colleagues (2011); (for P. ohmeri) Zhu and colleagues (2010); (for P. pastoris) Chiruvolu et al. (1998); (for P. sydowiorum) Kanekar and colleagues (2009); (for R. creatinivora) Krallish and colleagues (2006); (for S. ludwigii) Stratford (2006); and (for Z. rouxii) Restaino and colleagues (1983) and Praphailong and Fleet (1997). Tolerance windows for Candida etchellsii are yet to be established.
Under some conditions strains of S. cerevisiae can tolerate up to 28% v/v (20% w/v) ethanol (see Hallsworth, 1998).
See also Krallish and colleagues (2006).
In high-salt habitats, H. werneckii can grow at molar concentrations of NaCl (see Gunde-Cimerman et al., 2000) but was less salt tolerant under the conditions of this assay.
This species appears to have a requirement for high water activity.
Saccharomyces cerevisiae is metabolically wired to concomitantly synthesize an antimicrobial chaotrope (ethanol), accumulate a compatible solute (glycerol) that affords protection against this chaotrope (see below), and at the same time generate energy without any loss of ATP-generation efficiency (Table 3): a remarkable phenotype that is coupled with the deployment of other compatible solutes under specific conditions (trehalose and proline). Trehalose, which can reach concentrations of up to 13% dry weight in S. cerevisiae (Aranda et al., 2004), is highly effective at stabilizing macromolecular structures that are exposed to chaotropic substances (e.g. ethanol, 2-phenylethanol) and enabling cells to survive desiccation–rehydration cycles (Crowe et al., 1984; Mansure et al., 1994). The highest reported tolerance to ethanol stress was in S. cerevisiae cells capable of growth and metabolism up to 28% v/v (≡ 20% w/v or 4.3 M; chaotropic activity = 25.3 kJ g−1; Hallsworth, 1998; Hallsworth et al., 2007) when the medium contained high concentrations of a compatible solute, proline (Thomas et al., 1993); furthermore of the seven microbes reported to have the highest resistance to other chaotropic substances (including urea, phenol) the majority are weed species including A. wentii, S. cerevisiae, Escherichia coli and P. putida (Hallsworth et al., 2007).
The stress mechanisms for numerous types of cellular stress operate at the level of water:macromolecule interactions (Crowe, et al., 1984; Brown, 1990; Sikkema et al., 1995; Casadei et al., 2002; Ferrer et al., 2003; Hallsworth et al., 2003a; 2007; McCammick et al., 2009; Bhaganna et al., 2010; Chin et al., 2010). As front-line stress protectants, compatible solutes (within limits) can therefore mollify stress the inhibitory effects of an indefinite number of stress parameters (see Crowe, et al., 1984; Brown, 1990; Hallsworth et al., 2003b; Bhaganna et al., 2010; Chin et al., 2010; Bell et al., 2013) but cells also upregulate other protection systems (see also above; Table 3). In S. cerevisiae, an extensive number of genes that code for protein-stabilizing proteins, metabolic enzymes and cellular-organization proteins are regulated by a single promoter – the stress response element (STRE) – which is activated under cellular stress. This regulatory system enables the coordination of an array of stress responses in an energy-efficient manner (Table 3). Numerous studies show that the S. cerevisiae responses to osmotic stress [via the high-osmolarity glycerol-response (HOG) pathway], oxidative stress, and challenges to protein and membrane stability are exceptionally efficient (Table 3; Brown, 1990; Costa et al., 1997; Hallsworth, 1998; Rodríguez-Peña et al., 2010; Szopinska and Morsomme, 2010). In addition to these stress responses, there is evidence that the dynamics of polyphosphate and glycogen metabolism in S. cerevisiae are wired in such a way to boost ATP production (Castrol et al., 1999), and that the ploidy levels of many strains enhance tolerance towards oxidative and other stresses (Table 3). The energy-generation and energy-conservation strategies employed by S. cerevisiae have apparent parallels in other weed species; for example, analyses of synonymous codon bias in the genomes of fast-growing bacteria including E. coli and Bacillus subtilis suggest that genes associated with energy generation are highly expressed (Karlin et al., 2001; Table 5; see also above). The enhanced energy efficiency that is apparent in some weed species not only underpins their robust stress biology, but is required to sustain a vigorous growth phenotype and/or to out-grow competitors.
Table 5.
Growth, nutritional versatility and resource acquisition and storage: cellular characteristics that contribute to weed-like behaviour.a
| Metabolites, proteins, molecular characteristics, and other traits | Function(s) | Phenotypic traits | Notes and references |
|---|---|---|---|
| Ability to out-grow competitors | |||
| Multiple copies of ribosomal DNA operons | Enhanced production of ribosomes | Rapid protein synthesis | Soil bacteria able to grow rapidly on high-nutrient media have an average of ∼ 5.5 copies of ribosomal DNA operons (compared with ∼ 1.4 copies for slow-growing species; Klappenbach et al., 2000) |
| Key roles of di-/tripeptide-transport protein (dtpT) and oligopeptide transport ATP-binding protein (Opp) | Permease (dtpT) and ABC-type transporter (Opp) that enable efficient scavenging of diverse peptides (Altermann et al., 2005) | Efficient uptake of diverse peptides | These proteins enable species such as Lactobacillus acidophilus to efficiently scavenge peptides and thereby minimize the need for de novo synthesis of amino acids (Altermann et al., 2005) |
| High-efficiency DNA polymerase | Apparent correlation with short generation-time in fast-growing bacteria (Couturier and Rocha, 2006) | Rapid genome replication | The relatively fast-growing Mycobacterium smegmatis (generation time ∼ 3 h) has a DNA polymerase processing rate of 600 nucleotide s−1, compared with 50 nucleotide s−1 for Mycobacterium tuberculosis (generation time ∼ 24 h; Straus and Wu, 1980; Couturier and Rocha, 2006) |
| Overlapping replication cycles | Bacteria can initiate a new round of chromosomal DNA replication before the completion of an ongoing round of synthesis | Rapid genome replication | Escherichia coli can operate up to eight origins of replication simultaneously (see Nordström and Dasgupta, 2006) |
| Polyploidy, aneuploidy, association with plasmids, and/or hybridization that enhance(s) vigour | Multiple copies of genomic DNA can lead to increased vitality, enhanced growth rates, increased production of antimicrobials, and enhanced energy generation and stress tolerance (see also Table3) | Enhanced vigour | Shorter generation times in polyploid bacteria may arise from a number of factors including the increased number of ribosomal DNA operons (Cox, 2004; see also above) and gene localization within the genome (Pecoraro et al., 2011). A recent study of F1 hybrids produced by crossing 16 wild-type strains of S. cerevisiae showed that many F1 strains displayed hybrid vigour (Timberlake et al., 2011)b> |
| Traits for high-efficiency energy generation (see Table3) | |||
| Efficient system to eliminate free radicals (see also Table3) | Elimination of reactive oxygen species that can react with and damage macromolecular and cellular structures | Ability to cope with the oxidative stress associated with high growth rates | The enhanced rates of oxidative phosphorylation required for rapid growth result in the increased generation of free radicals; a phenomenon that has been observed in plant weeds (Wu et al., 2011). Aspergillus mutants that lack superoxide dismutase genes are unable to grow under environmental conditions that favour a high metabolic rate (Lambou et al., 2010). Strains of Pseudomonas aeruginosa have an auxiliary manganese-dependent version of superoxide dismutase that does not require iron and is upregulated under iron-limiting conditions (Britigan et al., 2001) |
| Nutritional and metabolic versatilityc | |||
| Genes for catabolism of a wide spectrum of hydrocarbons; e.g. those coding for the catechol-, homogentisate-, phenylacetate- and protocatechuate-catabolic pathways (cat, hmg, pha and pca gene families respectively) | Facilitate the synthesis of proteins that enable the utilization of hydrocarbons (including polycyclic aromatic hydrocarbons) | Ability to utilize diverse hydrocarbons as a carbon and/or energy source | Such compounds can be utilized as a sole carbon source by a small number of metabolically versatile microbes such as Pantoea spp., Pseudomonas putida and Pichia anomala (Timmis, 2002; Pan et al., 2004; Vasileva-Tonkova and Gesheva, 2007) but are also known to exert cellular stress even at low concentrations (Bhaganna et al., 2010). Catabolism of hydrocarbons will therefore alleviate stress at the same time as providing for energy generation and growth (Jiménez et al., 2002; Domínguez-Cuevas et al., 2006) |
| Catabolite-repression system for energy-efficient utilization of complex substrates | Advanced system to regulate utilization of diverse substrates | Energy-efficient growth in nutritionally complex environments | A cAMP-independent catabolite-repression system has been identified in P. putida that enables the ordered assimilation of carbon, nitrogen and sulfur substrates (most energy-favourable first; Daniels et al., 2010) |
| Production and secretion of biosurfactants (e.g. rhamnolipids in Pantoea, P. aeruginosa and P. putida; glycolipoproteins in Aspergillus spp.) | Solubilization of hydrophobic molecules (and antimicrobial activity; see Table6) | Ability to enhance the bioavailability of hydrophobic substrates | In vitro studies of Pantoea and P. aeruginosa demonstrate that biosurfactants enhance the uptake of hydrophobic substrates (Al-Tahhan et al., 2000; Vasileva-Tonkova and Gesheva, 2007; Reis et al., 2011) and thereby facilitate colonization of hydrocarbon-containing habitats (see Table S1). Aspergillus species can produce copious amounts of biosurfactant (Desai and Banat, 1997; Kiran et al., 2009) |
| Diverse saprotrophic enzymes (e.g. cellulose, ligninase) able to function over a range of conditions | Extracellular degradation of polymeric organic molecules for use as a carbon and energy source | Ability to grow saprotrophically | Saprotrophic capabilities enhance the range of substrates from which nutrition can be obtained. Saprotrophic enzymes from Aspergillus species can function over a wide range of environmental stresses (Hong et al., 2001) |
| Proteins for oxidation of methane and/or methanol (e.g. alcohol oxidase in Pichia and Kodamaea) | Oxidation of one-carbon substrates that are used for energy generation/growth | Ability to utilize one-carbon substrates | One-carbon substrates are environmentally commonplace and their utilization can facilitate rapid growth (van der Klei et al., 1991; McDonald and Murrell, 1997; Pacheco et al., 2003) |
| Pathways to utilize a broad range of nitrogen substrates (e.g. D-amino acids, urea, ammonium salts, and macromolecules such as collagen and elastin) | Assimilation of nitrogen into anabolic pathways for de novo synthesis of amino acids and nucleic acids | Ability to utilize a wide spectrum of nitrogen sources | Versatility in the utilization of nitrogen substrates can enable proliferation in open habitats by, and opportunistic pathogenicity for, Aspergillus, Pseudomonas putida and other weed species (Krappmann and Braus, 2005; Daniels et al., 2010; see also Table S1). Rhodotorula spp., including metabolically versatile weed species, can grow on D-amino acids as a sole carbon and nitrogen source (Simonetta et al., 1989; Pollegioni et al., 2007) |
| Genome-mediated responses to maintain growth under low-nutrient conditions (e.g. upregulation of glutamate dehydrogenase in P. putida) | Glutamate dehydrogenase plays key roles in nitrogen assimilation and metabolism | Ability to adapt metabolism for growth at high- or low-nutrient concentrations | Expression of gdh was between 5- and 26-fold greater in P. putida cells under low-nutrient conditions compared with a high-nutrient control (Syn et al., 2004). The synthesis of glutamate via glutamate dehydrogenase, rather than glutamate synthase, consumes 20% less ATP (Syn et al., 2004) |
| Enzyme systems that enable metabolism under nutrient-limiting conditions by avoiding dependence on specific cofactors | Utilization of substitute cofactor, or replacement of enzyme system(s) that would require a cofactor that is environmentally scarce | Maintenance of essential enzyme functions at low nutrient concentrations | For example arginase in S. cerevisiae (Middelhoven et al., 1969) and phospholipase C in P. aeruginosa (Stinson and Hayden, 1979) |
| Pathways to synthesize phosphate-free membrane lipids | Maintenance of membrane functions under phosphate-limited conditions | Ability to remain active, and grow, in phosphate-depleted environments | Pseudomonas spp. can synthesize membranous glycolipids as an alternative to phospholipids under low-phosphate conditions (Minnikin et al., 1974). Salinibacter ruber and a number of phytoplankton species utilize sulfur in membrane glycolipids that have a sulfoquinovose-containing (non-phosphorus) head group and are synthesized in response to low phosphate availability (Benning, 1998; Corcelli et al., 2004; Bellinger and Van Mooy, 2012) |
| High glucosylglycerate under low-nitrogen conditions | Control of nitrogen homeostasis to enable growth under nitrogen-limiting conditions | Nitrogen oligotrophy | Intracellular glucosylglycerate is increased in M. smegmatis under low nitrogen conditions. Mutants unable to synthesize glucosylglycerate showed reduced uptake of ammonium and a reduced growth rate (Behrends et al., 2012) |
| Efficient acquisition and storage of resources | |||
| Transport proteins to scavenge peptides and/or other macromolecules | Efficient scavenging of biomacromolecules avoiding the need for de novo synthesis | Efficient uptake of substrates | For example in L. acidophilus (see also above; Altermann et al., 2005) |
| Biosurfactants to access hydrophobic substrates (see above) | |||
| Production of high quantities of high- affinity saproptrophic enzymes | Efficient degradation of complex extracellular substances | Efficient utilization of high-Mr substrates | For example. xylanases produced by Acinetobacter junii and Aspergillus spp. (Meagher et al., 1988; Lo et al., 2010) |
| High-affinity hexose-transport proteins, e.g. Hxt2p (HXT gene family) | Highly efficient uptake of diverse carbon substrates | Rapid uptake of hexose sugars | These proteins are upregulated in S. cerevisiae at low sugar concentrations (Wendell and Bisson, 1994) |
| Phytase, inorganic phosphate transport proteins and polyphosphate kinase | Extracellular degradation of organic phosphate and uptake and intracellular polymerization of inorganic phosphate | Sequestration and storage of inorganic phosphate | Pichia kudriavzevii has a high rate of inorganic phosphate production from phytate (Hellström et al., 2012). Reduced survival of polyphosphate kinase-deficient bacterial mutants has been observed at low pH, high salinity, and under oxidative stress (Jahid et al., 2006). Other weed species, such as P. putida, Dunaliella salina, Microcystis aeruginosa and Ostreococcus tauri are efficient accumulators of polyphosphate (Bental et al., 1991; Shi et al., 2003; Tobin et al., 1997; 2007; Le Bihan et al., 2011) |
| Production and secretion of siderophores that chelate iron | To scavenge iron from the extracellular milieu which is then removed once the siderophores have been transported back into the cell | Efficient scavenging of iron | The competitive ability of some bacterial weeds (e.g. pseudomonads, Pantoea ananatis) may be enhanced by siderophore production (Duiff et al., 1993; Loaces et al., 2011). Rhodotorula mucilaginosa and M. aeruginosa upregulate siderophore production under iron-limiting conditions (Andersen et al., 2003; Xing et al., 2007); in contrast, Saccharomyces spp. are not known to produce siderophores (Lesuisse et al., 2001) |
| Enhanced efficiency of iron-binding proteins (e.g. transferrin-like protein, TTf, in D. salina) | Binding and uptake of extracellular iron | Ability to accumulate iron under iron-limiting conditions | Dunaliella salina cells undergo a fourfold increase in iron-binding affinity under low-iron conditions. Iron is then taken up stored in vacuoles (Paz et al., 2007) |
| Arginine biosynthesis proteins | Bulk synthesis of arginine as a nitrogen substrate store | Amino acid accumulation and storage | Saccharomyces cerevisiae stores arginine (in vacuoles) that can be utilized when the extracellular environment becomes nitrogen substrate limited (Westenberg et al., 1989) |
| Trehalose-synthesis proteins (e.g. TPS, TPP) and glycogen synthase (e.g. GSY1 and GSY2) | Synthesis of disaccharide- and polysaccharide-storage compounds (e.g. trehalose and glycogen respectively) | Effective synthesis and storage of carbohydrates | Trehalose synthesis can be upregulated prior to the stationary growth phase and during stress (Parrou et al., 1997), and glycogen synthesis is upregulated during periods of nutrient limitation, in S. cerevisiae (Lillie and Pringle, 1980; Farkas et al., 1990). Starch is accumulated by D. salina (Stoynova-Bakalova and Toncheva-Panova, 2003) |
| Polyhydroxybutyrate-synthesis proteins (and those producing other storage lipids) | Synthesis of lipid stores (see also Table3) | Bulk storage of lipids | Polyhydroxybutyrate, in the form of intracellular granules, dominates the cytoplasm of Haloquadratum walsbyi (Saponetti et al., 2011); Dunaliella spp. also store lipids (Chen et al., 2011); recent studies of P. putida suggest that carbon and energy can be channelled into storage of polyhydroxybutyrate without a reduction of growth rate (Escapa et al., 2012) |
| Morphological and physiological adaptations to optimize light capture and absorption of oxygen and nutrients | Flat cell shape gives a high surface-to-volume ratio; gas vesicles help to maintain the horizontal orientation of planktonic cells | Optimized light capture, and oxygen and nutrient absorption | The remarkable morphology of H. walsbyi cells optimizes acquisition of resources in highly competitive high-salt habitats (Bolhuis et al., 2006; Bardavid et al., 2008) |
| Efficient chlorophyll a and chloroplast arrangement | Optimization of light harvesting and high photosynthetic rate | High photosynthetic rate | Habitat dominance by Gonyostomum semen can be attributed, in part, to having a higher photosynthetic rate than other autotrophic species (Coleman and Heywood, 1981; Peltomaa and Ojala, 2010) |
| Genes related to gas synthesis and gas vesicle formation (e.g. gvp family in M. aeruginosa) | Gas vesicles provide buoyancy to optimize access to light and high oxygen concentrations (close to the water:atmosphere interface of aquatic habitats) | Ability to access resources required for photosynthesis | Twelve genes involved in gas synthesis and storage have been identified in M. aeruginosa (Mlouka et al., 2004); models suggest regulation of buoyancy in M. aeruginosa has a key role in habitat dominance (Bonnet and Poulin, 2002) |
The current table provides examples of proteins, genes or other characteristics that can be associated with and give rise to weediness; the categories of traits are not mutually exclusive. Individual traits may not be unique to microbial weeds; however, weeds species are likely to have a number of these types of characteristics.
Whereas polyploidy can enhance vigour (via increased growth rates, enhanced stress tolerance etc.) increased chromosome number can also result in decreased growth rates; the consequences of polyploidy can vary with species and environmental context.
Growth, nutritional versatility, and resource acquisition and storage
Efficient cell division and an ability to out-grow competitors are implicit to the emergence of weed species. Species that grow relatively slowly or remain quiescent, and those with specific requirements for a host, a symbiotic partner, or obligate interactions with other microbial species are disadvantaged during community development in open habitats: dominance ultimately results from vigour and – in many habitats – a degree of nutritional versatility (Tables S1 and5). Ability to out-grow competitors is equally important (possibly even more so) for weed species in nutrient-poor and physicochemically extreme habitats that can both select for slow-growing species and limit overall biomass development. Many traits that give rise to an efficient growth phenotype can be found in weed species (Table 5): multiple copies of ribosomal DNA operons that can enhance rates of protein synthesis; transport proteins to scavenge macromolecules thus negating the need for de novo synthesis; high-efficiency DNA polymerase for rapid genome replication; efficient systems to eliminate reactive oxygen species that are a by-product of rapid growth; and polyploidy, aneuploidy and plasmids that can be associated with enhanced stress tolerance, higher growth-rates and increased overall vigour (Table 5). We believe that another key aspect of weed biology for some microbial species is their ability to sequester and store available nutrient and substrate molecules with a high efficiency as an auxiliary energy reserve (see below), to be able to supply daughter cells with nutrients, and simultaneously deprive other microbes of nutritional resources (e.g. Duiff et al., 1993; Castrol et al., 1999; Loaces et al., 2011; Renshaw et al., 2002; Jahid et al., 2006). Microbial weeds are highly efficient at nutrient-sequestration and substrate utilization, as well as intracellular storage of these substances (Table 5). For example, fungal siderophores that remove iron from the habitat have been shown to suppress growth of other microorganisms due to iron limitation (Renshaw et al., 2002).
Intermittent nutrient availability and low nutrient concentrations are characteristic of many aquatic environments so the ability to scavenge and store, circumvent the requirement for, or strategically utilize scarce and potentially limiting nutrients can greatly enhance the competitive ability of planktonic species. Under phosphate-limiting conditions some aquatic species, including S. ruber, are able to use sulfur-containing glycolipids in place of phospholipids during low phosphate availability (Table 5). The cyanobacterium M. aeruginosa has high-efficiency phosphate-transport systems and accumulates intracellular polyphosphate which is used to maintain activity once extracellular phosphate is depleted (Table 5; Shi et al., 2003). It is likely that this trait contributes to its weed-like phenotype in Lake Taihu (China) that is a well-studied, phosphate-limited habitat frequently dominated by M. aeruginosa (Shi et al., 2003). Iron is another potentially limiting nutrient in aquatic habitats. Dunaliella salina utilizes a highly efficient iron-binding protein to take up iron which is then stored in vacuoles (Paz et al., 2007). Species such as D. salina and H. walsbyi cope with oligotrophic conditions by hyperaccumulating starch and/or lipids such as polyhydroxybutyrate especially when nutrients other than carbon are limiting (Table 5; Bardavid et al., 2008). One weed species with adaptations to endure low-nitrogen conditions is Mycobacterium smegmatis. This species (as well as other mycobacteria) which can be prevalent in relatively low-nutrient aquatic and non-aquatic habitats (Koch, 2001; Kazda and Falkinham III, 2009) have biochemical adaptations to enable growth and metabolic activity even when nitrogen concentrations are an order of magnitude lower than those usually required for structural growth (i.e. a C : N ratio of 8:1; Roels, 1983; Behrends et al., 2012). In addition to nitrogen oligotrophy, M. smegmatis has a DNA polymerase capable of nucleotide processing rates and generation times that are an order of magnitude faster than those of comparable Mycobacterium species (Table 5). Light and/or oxygen are key resources for many planktonic species; 12 genes have been identified that are involved in gas synthesis and storage in species such as M. aeruginosa and H. walsbyi (Table 5). Modelling approaches indicate that buoyancy, which enables the cell to access the upper (well-illuminated, well-oxygenated) section of the water column, plays a key role in habitat dominance by M. aeruginosa (Bonnet and Poulin, 2002). Studies of H. walsbyi suggest that its peculiar, flat cell-shape optimizes the uptake of nutrients and oxygen and its intracellular gas vesicles help to maintain a horizontal orientation to capture light (Table 5). In freshwater habitats dominance of algal blooms by G. semen has been attributed in part to a high photosynthetic rate that results from a combination of its high-efficiency chlorophyll a and a highly efficient chloroplast arrangement (Table 5).
In open habitats that are nutritionally dynamic and/or complex (Table S1) metabolic versatility in nutrient utilization can enhance the competitive ability of microbes (for examples see Table 5). Pseudomonas putida and related species are remarkable in their range of nutritional strategies (Table 5; Timmis, 2002; 2009): they are able to catabolize an indeterminate number of hydrocarbons (e.g. decanoate, toluene) and other organic molecules (e.g. amino acids, and organic acids such as acetic, citric and lactic acid) for use as carbon and energy sources; amino acids, dipeptides, ammonium chloride, urea, ammonium nitrate, and other substances as nitrogen sources; amino acids, organic acids, sodium sulfite, thiouracil, agmatine sulfate, etc. as sulfur sources; as well as accumulating diverse substances as intracellular reserves (Chakrabarty and Roy, 1964; Daniels et al., 2010; Table 5) enabling a switch to oligotrophic growth at low nutrient concentration (Table 5; Timmis, 2002). In addition, P. putida and closely related species have evolved a complex, cAMP-independent catabolite repression system which incorporates five different signals and enables an ordered assimilation of carbon, nitrogen and sulfur substrates from the most to least energetically favourable (Daniels et al., 2010). This may contribute to the ability of many pseudomonads to sustain vigorous growth in nutritionally complex habitats. Pseudomonads, and closely related bacteria such as Acinetobacter, are not only environmentally ubiquitous but can dominate microbial communities in numerous types of open habitat including the plant phyllosphere and rhizosphere, bioaerosols, oil sands, and other hydrocarbon-containing environments (see Table S1; Ogino et al., 2001; Röling et al., 2004; Louvel et al., 2011). Pseudomonas species are able to enhance the availability of hydrophobic substrates by secreting biosurfactants and, at the same time, tolerate the stress induced by the latter (see below). Despite their copiotrophic phenotype, pseudomonads utilize a number of strategies to maintain growth and metabolism under phosphate-, iron- or nitrogen-depleted conditions (Table 5), can synthesize glutamate via glutamate dehydrogenase rather than glutamate synthase (thereby consuming 20% less ATP; Syn et al., 2004), and have even been observed growing in distilled water (Favero et al., 1971; Hirsch, 1986). Specific enzymes of Pseudomonas aeruginosa are catalytically active even when cofactor nutrients are scarce, and some strains have a manganese-dependent superoxide dismutase which does not require iron and is upregulated at low iron concentrations (Table 5).
Weeds such as lactic acid bacteria and S. cerevisiae that lack saprotrophic capabilities can dominate communities in habitats with high nutrient concentrations (Tables S1 and5); they possess high-affinity transport proteins that enable rapid and efficient uptake of substrate molecules to support their vigorous growth phenotypes (see Table 5). Cells of S. cerevisiae are not only highly efficient at storing carbohydrates (combined trehalose and glycogen may reach 25% dry weight; Aranda et al., 2004; Guillou et al., 2004), but they can also accumulate high concentrations of arginine and polyphosphate and have adaptations to retain activity under low-nitrogen conditions (Table 5). Plant weeds that excel in open habitats have distinct genetic properties; they are often of hybrid origin and/or polyploid, traits that give rise to increased growth rates and stress tolerance, and have specific genes that confer their vigorous phenotype (Anderson, 1952; Hill, 1977; Schnitzler and Bailey, 2008; Bakker et al., 2009). There is also evidence that the multiple copies of genomic material found in vigorous microbes that enhanced vigour have arisen via hybridization and/or increased ploidy levels in bacterial weed species, as well as Pichia and Saccharomyces (Table 5; Cox, 2004). A recent study by Timberlake and colleagues (2011) reported vigorous growth for the majority of F1 hybrids produced by crossing 16 wild-type S. cerevisiae strains (although hybridization and/or polyploidy may not always be advantageous; see Table 5). Many microbial weed species are phenotypically variable (like their plant counterparts; Table 1; Hill, 1977) and may form part of a species complex due to polyploidization and hybridization (e.g. Acinetobacter, Pseudomonas, Mycobacterium, Saccharomyces sensu stricto; Table S1; Rainieri et al., 1999; Todd et al., 2004; Ballerstedt et al., 2007; Parkinson et al., 2011; Sicard and Legras, 2011); such factors may contribute to the vigour and weediness of such species.
High-Mr macromolecules are the primary microbial substrates available in many habitats: e.g. hydrocarbons (in bioaerosols, oil-containing environments, and the rhizosphere), and polysaccharides (within or derived from plant necromass such as cellulose, lignin and humic substances; see Table S1). In order to out-compete other species in such habitats, microbes must utilize these macromolecular substrates that typically have a low bioavailability, require specialist enzymes to degrade, can be toxic, and/or are energy-expensive to catabolize. Facultatively saprotrophic microbes such as Aspergillus, Pichia, Rhodotorula and Acinetobacter species are able to access and catabolize recalcitrant polysaccharides; some weed species can produce exceptional amounts, or high-affinity versions, of inducible saprotrophic enzymes, such as xylanases (Table 5). Several saprotrophic enzymes produced by Aspergillus species (that can dominate physicochemically diverse habitats; Table S1) can retain activity over a wide range of environmental stresses (Hong et al., 2001); and Aspergillus, and Rhodotorula spp. are able to utilize a broad range of nitrogen substrates thereby avoiding the need for de novo synthesis (Table 5). Whereas nutritionally complex habitats can select for metabolic versatility (see below) and thereby favour the growth of a great number of generalist species, only a minute fraction of such microbes are able to dominate what were (initially) phylogenetically diverse microbial communities (Table S1).
Antimicrobial strategies and trophic interactions
Grazing, predation and viral infections adversely impact the growth dynamics of and/or incur massive losses on microbial populations and so exert a powerful selective pressure, and this can be well illustrated by the ecologies of many planktonic species in aquatic habitats (Matz et al., 2004; Jüttner et al., 2010; Berdjeb et al., 2011; Thomas et al., 2011; Yang and Kong, 2012). The competitive ability of some aquatic bacteria and eukaryotes (both marine and freshwater species) is associated with their ability to resist or tolerate virus infection (see Table 6). Species with a halophilic growth phenotype, especially those which have high growth rates in salt-saturated environments such as S. ruber and H. walsbyi minimize losses to predators because these salt concentrations are hostile to protozoa due to their low water activity, high ionic strength, and/or (for high-MgCl2 and high-CaCl2 brines) high chaotropicity (Table 6; Hallsworth et al., 2007). Gonyostomum semen has evolved vertical migration patterns through the water column that are out of phase with those of grazing species and thereby minimizes losses (Table 6). Other weeds, including P. aeruginosa and M. aeruginosa, are capable of forming multicellular structures – micrcolonies – that are sufficiently large to reduce losses by many species of grazers and predators (Table 6). Species such as M. aeruginosa accumulate toxic metabolites, and M. aeruginosa and Aspergillus spp. are known to synthesize secondary metabolites that repel to grazing plankton and arthropods respectively (Table 6).
Table 6.
Antimicrobial activities and trophic interactions: cellular characteristics that contribute to weed-like behaviour.a
| Metabolites, proteins, molecular characteristics, and other traits | Function(s) | Phenotypic traits | Notes and references |
|---|---|---|---|
| Avoidance of/resistance to viral infection, predators, and grazers | |||
| Metabolites that repel grazers and/or predators, e.g. aldehydes and other volatile organic compounds in plankton and fungi | Secondary metabolites produced by Aspergillus spp. repel (and can reduce the fecundity of) grazing arthropods such as Folsomia candida; those produced by Microcystis aeruginosa act as a repellent to planktonic grazers | Ability to repel grazers and predators | Aspergillus nidulans mutants lacking the laeA gene, a global regulator of secondary metabolite synthesis, are consumed by F. candida in preference to the wild-type strain (Rohlfs et al., 2007). Aldehydes produced by plankton repel crustacean grazers (Jüttner, 2005); β-cyclocitral from M. aeruginosa has been shown to repel Daphnia magna (Jüttner et al., 2010) |
| Synthesis of extracellular polymeric substances (e.g. M. aeruginosa), or use of flagella (e.g. Pseudomonas aeruginosa), that facilitate microcolony formation | Formation of multicellular structures that are sufficiently large to minimize grazing and predation by single-celled protozoa, zooplankton etc. | Microcolony formation | The role of polymeric substances in colony formation has been established using M. aeruginosa as a model organism (Yang and Kong, 2012). Functional analysis of P. aeruginosa mutants that were unable to form flagella, or were alginate-deficient, demonstrated an inability to form microcolonies that can confer resistance to grazing (Matz et al., 2004) |
| Biochemical adaptations to resist viral infection | Diverse mechanisms including modification of cell surface to prevent binding, intracellular suppression or viral genome replication, and degradation of viral DNA by restriction endonucleases | Resistance to or tolerance of viral infection | There are a number of mechanisms by which dominating species of bacteria and eukaryotic plankton (e.g. P. aeruginosa, Ostreococcus tauri; Bathycoccus and Micromonas spp.) may acquire and maintain resistance to viral infection (see Thomas et al., 2011; Selezska et al., 2012) |
| Toxic metabolites (e.g. microcystin in M. aeruginosa) | Toxicity against grazing or predatory zooplankton | Ability to protect against excessive grazing | Microcystin synthesis in M. aeruginosa can be induced by exposure to zooplankton; concentrations of the toxin in cells exposed to D. magna, Daphnia pulex and Moina macrocopa were up to fivefold those found in control cells (Jang et al., 2003) |
| Halophilic growth phenotype | Colonization of salt-saturated habitats that are hostile to predatory protozoa minimizes losses from grazing and predation | Ability to inhabit environments hostile to predators and grazers | Halophiles that dominate high-salt habitats (e.g. D. salina, S. ruber) minimize interactions with grazers and predators most of which cannot tolerate molar salt concentrations (Bardavid et al., 2008) |
| Biochemical and cellular infrastructure that give rise to idiosyncratic migration patterns in the water column | Vertical migration within the water column that minimizes loss through predation and grazing | Avoidance of predators and grazers | The predominance of Gonyostomum semen in eutrophic habitats has been associated in part with migration patterns that minimize loss through grazing (Salonen and Rosenberg, 2000) |
| Bulk production of highly soluble, moderately chaotropic stressorsb | |||
| Ethanol produced via a high-Km alcohol dehydrogenase (e.g. Adh1 in Saccharomyces cerevisiae) | Efficient conversion of acetaldehyde to ethanol; in contrast, low-Km Adh typically converts ethanol to acetaldehyde under in vivo conditions (Piškur et al., 2006) | Production and secretion of ethanol (up to molar concentrations) | Ethanol, secreted in quantity by species such as S. cerevisiae and Zymomonas mobilis, is a moderately chaotropic substance that is nevertheless a potent stressor at millimolar to molar concentrations (Hallsworth, 1998; Bhaganna et al., 2010; Cray et al., 2013) |
| Enzymes for acetone, isopropanol and butanol synthesis | Synthesis of chaotropic substances in the anaerobic bacterium Clostridium acetobutylicum | Bulk production, and secretion, of multiple chaotropic stressors | Acetone, isopropanol and butanol that are produced by C. acetobutylicum (George et al., 1983) are more chaotropic ethanol – when compared on a molar basis – and are therefore more potent in their inhibitory activity as cellular stressors (Bhaganna et al., 2010; Cray et al., 2013; Bell et al., 2013) |
| Production of high concentrations of urea and ammonia | Synthesis of low-Mr, nitrogenous chaotropes | Production and secretion of urea and ammonia at inhibitory concentrations | Urea and ammonia are produced by phylogenetically diverse bacteria such as Pseudomonas stutzeri and Clostridium aminophilum respectively (Therkildsen et al., 1997; Watanabe et al., 1998; Anderson et al., 2010) |
| Release of VOCs as low-solubility antimicrobial stressors | |||
| Highly volatile metabolites that act as hydrophobic stressors (log P > 1.95)c | Inhibition of microbial metabolism; typically via hydrocarbon-induced water stress (Bhaganna et al., 2010) | Production and release of volatile antimicrobials | Examples are: isoamyl acetate (produced by Pichia anomala; Passoth et al., 2006), ethyl octanoate (S. cerevisiae; Fialho et al., 2011), β-cyclocitral (M. aeruginosa and Dunaliella salina; Herrero et al., 2006; Ozaki et al., 2008), pentane (D. salina; Muñoz et al., 2004); and diverse substances from Aspergillus (see above), R. mucilaginosa (Buzzini et al., 2005), P. putida and other Pseudomonas species (Morales et al., 2005) |
| Highly volatile, strongly chaotropic metabolites (log P typically in the range 1–1.9)d | Inhibition of microbial metabolism via chaotrope-induced water stress (Hallsworth, 1998; Hallsworth et al., 2003a; Bhaganna et al., 2010) | Production and release of volatile antimicrobials | Examples are: ethyl acetate (produced by Pichia anomala; Walker, 2011), dichloromethane (Dunalialla spp.; Colomb et al., 2008) and 2-phenylethanol (Mycobacterium bovis, Mycobacterium smegmatis and S. cerevisiae; Eshkol et al., 2009; McNerney et al., 2012); and a range of chaotropic stressors by R. mucilaginosa (Buzzini et al., 2005), P. putida and other Pseudomonas species (Morales et al., 2005) |
| Volatile catabolites produced from the degradation of macromolecular substrates | Inhibition of microbial metabolism typically via chaotrope-induced or hydrocarbon-induced water stress (Hallsworth et al., 2003a; Bhaganna et al., 2010) | Production and release of volatile antimicrobialsg | Antimicrobial stressors can be released during the saprotrophic degradation of lignin, catabolism of hydrocarbons etc. (e.g. catechol, vanillin and biphenyl in P. putida; Zuroff and Curtis, 2012) |
| Acidification of the extracellular environment | |||
| Secretion of organic acids | Acidification of the extracellular milieu | Ability to acidify the habitat | Acinetobacter rhizosphaerae and S. cerevisiae secrete a range of substances that can acidify the environment (Kotyk et al., 1999; Gulati et al., 2010); Aspergillus spp. produces high levels of citric acid (up to 200 g l−1 in 5–7 days; Ward et al., 2006) |
| High-activity l-lactate dehydrogenase (e.g. ldhL in Lactobacillus plantarum) | Intracellular conversion of pyruvate to lactate | Production and secretion of lactic acid | The specific activity of IdhL from a lactic acid produced by L. plantarum was found to be 2460 U mg−1; compared with 250 U mg−1 for Escherichia coli (Garvie, 1980). Habitat acidification by lactic acid inhibits the growth of many microbial competitors (Table S1; Guillot et al., 2003). Lactobacillus efficiently acidifies its environment even at low sugar concentrations (Klinke et al., 2009) |
| Enzymes for acetic and butyric acid synthesis (e.g. phosphotransacetylase and acetate kinase, and phosphate butyryltransferase and butyrate kinase respectively) | Conversion of acetyl-CoA to acetate; and butyryl-CoA to butyrate | Production/secretion of acetic acid and butyric acid | Clostridium spp. produce acetic, butyric and lactic acids; fermentations carried out in media with an initial pH of ∼ 6.5 resulted in a final pH of 4–4.5 (Wiesenborn et al., 1989; Boynton et al., 1996; Liu et al., 2011) |
| Production of biosurfactants, enzymes, and/or toxins with antimicrobial activities | |||
| Biosurfactants (e.g. rhamnolipids in Pseudomonas spp.; glycolipoproteins in Aspergillus spp.) | Solubilization of hydrophobic substrates; inhibitory to the growth of many microbial competitors | Production of inhibitory biosurfactants | Lipopeptides from Pseudomonas sp. DF 41 were shown to inhibit Sclerotinia sclerotiorum (Berry et al., 2010); purified biosurfactants from Pseudomonas fluorescens inhibit growth of Botrytis cinerea and Rhizoctonia solani at low concentrations (25 μg ml−1) and can lyse zoospores of Phytophthora capsici (Kruijt et al., 2009). Studies of P. aeruginosa rhamnolipid suggests a key role in competitive ability against other bacteria (Wecke et al., 2011). Acinetobacter spp. produces a potent biosurfactant (phosphatidylethanolamine; Desai and Banat, 1997) which may act as an antimicrobial. Biosurfactants from Aspergillus ustus are inhibitory to phylogenetically diverse microbes (Kiran et al., 2009) |
| Panomycocin (an exo-β-1,3-glucanase) | Hydrolysis of glucans such as laminarin | Antimicrobial and cytocidal activity via degradation of cell wall components | Panomycocin is produced by P. anomala and is inhibitory or lethal to many species of yeasts and fungi such as B. cinerea (Izgü et al., 2005) |
| Ochratoxin and other mycotoxins (e.g. in Aspergillus spp.) | Inhibition of other microbial species/strains | Secretion of antimicrobials | Mycotoxins can inhibit phylogenetically diverse microbes (including fungal and bacterial species) and thereby enhance competitive ability (Boutibonnes et al., 1984; Magan et al., 2003) |
| Antifungal peptide (Anafp; in Aspergillus niger) | Mode-of-action not yet resolved | Secretion of an antifungal peptide | Anafp inhibits growth of yeasts and fungi including some Aspergillus and Fusarium spp., Candida albicans, S. cerevisiae and Trichosporon beigelii (Lee et al., 1999) |
| Mycocins (e.g. salt-mediated killer toxin) | Inhibitory to many fungi, yeasts and other microbes | Production of antibiotic peptides | A range of killer toxins produced by Pichia farinose, Rhodotorula spp. and S. cerevisiae that are inhibitory to phylogenetically diverse taxa (Golubev and Churkina, 1993;; Golubev et al., 1996; Suzuki et al., 2001; Trindade et al., 2002; Albergaria et al., 2010) |
| Bacteriocins e.g. plantarcins (pln gene family), lactacin-B (La1791–La1803), plant lectin-like bacteriocin (LlpA) | Modification of membrane structure causing leakage and dissipation of proton-motive force (Anderssen et al., 1998; Parret et al., 2003; Altermann et al., 2005) | Production of antibacterial peptides | For example L. plantarum, Lactobacillus acidophilus and Pseudomonas spp. |
| Pyoluteorin (in P. putida) | Inhibitory to growth of oomycetes and fungi; mode-of-action not yet resolved | Antifungal activity | Pyoluteorin-producing P. putida strains have potential as biocontrol agents of pathogens such as Glomerella tucumanensis (sugarcane red rot; Nowak-Thompson et al., 1999; Hassan et al., 2011) |
| Phenazine and phenazine-derived antimicrobials | Probable antimicrobial activity | Antimicrobial activity | Produced by species such as M. aeruginosa and P. aeruginosa (Mavrodi et al., 2006; Lifshits and Carmeli, 2012) |
| Mechanisms to induce cell lysis | Elimination of potential competitors | Algicidal activity | Gonyostomum semen is able to lyse microbial cells (e.g. Rhodomonas lacustris) on contact; mechanism not yet resolved (Rengefors et al., 2008) |
| Resistance to antimicrobial stressors and toxins | |||
| Coordinated production of compatible solutes and ethanol | Protection against ethanol stress | Resistance to ethanol | The synthesis of glycerol (known to afford some protection against ethanol stress; Hallsworth, 1998; Hallsworth et al., 2003b) and ethanol production are coupled in S. cerevisiae; accumulation of trehalose (an effective protectant against ethanol; Mansure et al., 1994) coincides with the growth phase when ethanol concentrations are highest |
| Compatible-solute accumulation in response to solvent and hydrophobic stressors (see above) | Protection of macromolecular systems challenged by chaotropic and hydrophobic stressors (see studies of A. nidulans, S. cerevisiae and P. putida: Mansure et al., 1994; Hallsworth et al., 2003b; Park et al., 2007; Bhaganna et al., 2010; Bell et al., 2013) | Ability to maintain functionality of macromolecular systems | Kosmotropic compatible-solutes and other kosmotropic substances can reduce stress induced by chaotropic solutes such as MgCl2, glycerold and ethanol (Hallsworth, 1998; Hallsworth et al., 2007; Williams and Hallsworth, 2009). Tryptophan and trehalose are upregulated in cells of both eukaryotic and bacterial species in response to ethanol (Parrou et al., 1997; Purvis et al., 2005; Hirasawa et al., 2007; Horinouchi et al., 2010). S. cerevisiae could tolerate 28% v/v ethanol when protected by the kosmotropic compatible-solute proline (Thomas et al., 1993; Hallsworth, 1998) |
| Upregulation of protein-stabilization proteins (see Table3) | Protection of macromolecular systems from perturbations induced by antimicrobial stressors | Maintenance of protein structure and function during stressor-induced challenges | Saccharomyces cerevisiae upregulates synthesis of up to 10 heat-shock proteins under ethanol stress (Ma and Liu, 2010). Pseudomonas putida synthesizes diverse protein-stabilization proteins in response to chaotropic and hydrophobic stressors (see above; Hallsworth et al., 2003a; Bhaganna et al., 2010) |
| Membrane-stabilization proteins; solvent pumps (see Table3) | |||
| Multidrug efflux pumps (e.g. AcrAB-TolC in E. coli; Pdr5p in S. cerevisiae; ABC transporter proteins in Aspergillus spp.) | Expulsion of chemically diverse stressors and toxins | Tolerance to a wide array of stressors and toxins | Including chaotropic solutes (e.g. phenol, benzyl alcohol), hydrophobic stressors (e.g. n-hexane, p-xylene) and antibiotics (e.g. chlorampenicol, tetracycline, novobiocin) by AcrAB-TolC; fluorescent dyes (e.g. daunorubicin, rhodamine), drugs (e.g. tamoxifen); and ionophoric peptides (e.g. nigericin) by Pdr5p (Kolaczkowski et al., 1996); and antifungals (cilofungin and echinocandin B) by the AfuMDR1 gene product (Tobin et al., 1997; Nishino and Yamaguchi, 2001; Ramos et al., 2002) |
| Proteins for expulsion of, or resistance to, bacteriocins; e.g. lincomycin-resistance protein (lmrB), undercraprenyl pyrophosphate phosphatise (bacA) | Removal of bacteriocins from the cell | Resistance to bacteriocins | LmrB is an ATP-binding cassette-type transporter in Lactococcus lactis (Gajic et al., 2003); BacA is a membrane-associated protein that in E. coli for (El Ghachi et al., 2004) |
| Enzymes that inactivate antimicrobial substances | Detoxification and/or catabolism of inhibitory stressors and toxins | Ability to detoxify or degrade inhibitory antimicrobials | Examples include degradation of β-lactam antibiotics in Acinetobacter spp. and P. aeruginosa (Langaee et al., 2000; Sinha et al., 2007) and degradation of volatile stressors such as dichloromethane (e.g. Bacillus spp.; Wu et al., 2009) |
| Enhanced tolerance to a chaotropic volatile | Resistance to 2-phenylethanol | Resistance to VOCs | One strain of S. cerevisiae was found to be capable of growth at up to ∼ 24 mM 2-phenylethanol (Eshkol et al., 2009). For a chaotropic stressor, log P 1.52, almost as chaotropic as phenol, this is an exceptional level of tolerance (see Bhaganna et al., 2010) |
| Cell wall composition that confers resistance to specific antibiotics | Pathways targeted by specific antibiotics may be absent; cell wall can act as a barrier to antimicrobials | Resistance to antibiotics | The Haloquadratum walsbyi cell wall is not composed of peptidoglycan so this species has reduced sensitivity to β-lactam antibiotics (Bardavid and Oren, 2008); the M. smegmatis cell envelope contains mycolic acids that reduce permeability, and therefore enhance resistance, to stressors and antibiotics (Liu and Nikaido, 1999) |
| Synthesis of extracellular polymeric substances (see Table3) | Production and secretion of polymeric substances | Resistance to diverse antimicrobials | For example production of alginate by P. aeruginosa increases resistance to antibiotics such as tobramycin (Hentzer et al., 2001) |
The current table provides examples of proteins, genes or other characteristics that can be associated with and give rise to weediness; the categories of traits are not mutually exclusive. Individual traits may not be unique to microbial weeds; however, weeds species are likely to have a number of these types of characteristics.
Chaotropic activity of > 5 and < 40 kJ kg−1 mole−1 (Cray et al., 2013).
Volatile organic compounds are unlikely to exceed micromolar concentrations in the cytosol of the weed cell.
A number of agricultural (plant) weeds are able to dominate their habitats due largely to the production of phytotoxic inhibitors that can be exuded from leaves, roots and plant necromass and which inhibit the germination and/or growth of other plant species (Hill, 1977; Xuan et al., 2006; Alsaadawi and Salih, 2009; Chou, 2010; Meksawat and Pornprom, 2010; Inderjit et al., 2011). It is likely that most microbial weed species also secure their competitive advantage – in part at least – via the deployment of antimicrobial substances. Assuming that the examples listed in Table S1 are representative of the microbial biosphere then the proportion of habitats dominated by eukaryotic species is approximately 40%. If a short generation time was sufficient to facilitate habitat domination then prokaryotic species would populate almost all these habitats. The fact that species of algae, fungi and yeast are also able to do so underscores the importance of factors other than generation time in the biology of habitat dominance. Saccharomyces cerevisiae provides an illustration of how one weed species combines the synthesis of a potent antimicrobial, energy generation, and coordinated synthesis of stress protectants in a single process, i.e. aerobic production of the chaotropic alcohol, ethanol (see above; Table 6). Ethanol can be produced and secreted at concentrations > 4 M that cause a reduction of water activity (to 0.896) and exert an exceptionally high chaotropic activity (see above) sufficient to prevent the growth or metabolism of any living system (Hallsworth, 1998; Hallsworth and Nomura, 1999; Hallsworth et al., 2007); S. cerevisiae simultaneously produces glycerol (which can reduce ethanol stress in yeast and fungi; Hallsworth, 1998; Hallsworth et al., 2003b) during ethanol fermentation and is metabolically wired to synthesize high concentrations of the protectant trehalose at the end of its growth cycle when ethanol concentration is highest (see Table 6).
We believe that the bulk production and secretion of chaotropic solutes and organic acids (that can reach molar concentrations) is unique to microbial weed species (Tables 1 and 2). Whereas chaotropic substances can promote growth and expand the biotic windows of microbes under specific conditions (Hallsworth and Magan, 1994b; 1995; Williams and Hallsworth, 2009; Bhaganna et al., 2010; Chin et al., 2010), in other circumstances chaotropicity can be highly inhibitory or even lethal (Hallsworth, 1998; Hallsworth et al., 2003a; 2007; Duda et al., 2004; Bhaganna et al., 2010). For plant weeds it is the early stages of habitat colonization that are key to their success (Hill, 1977), and is may be that the efficient synthesis of antimicrobials is frequently as important as much as the type of substance produced (Hallsworth, 1998; Pretorius, 2000; Eshkol et al., 2009; Fialho et al., 2011). Unlike plant weeds, where phytotoxic inhibitors tend to have the greatest impact on neighbouring plants, microbes can also produce volatile organic compounds (VOCs) that inhibit the growth of both proximal and distal cells via their dispersion in both aqueous and gaseous milieu (Table 6).
Volatile organic compounds – including aldehydes, furones, terpenes and many alcohols – can be powerful inhibitors of microbial metabolism and growth (Table 6; Jorge and Livingston, 1999; Passoth et al., 2006; Minerdi et al., 2009; Fialho et al., 2011; Corcuff et al., 2011). Despite their volatility they are to some degree also water-soluble, and are sufficiently inhibitory to prevent metabolic activity and growth, even in aqueous solution at subsaturated solute-concentrations (Bhaganna et al., 2010). VOCs with a log Poctanol-water < 1.95 typically partition into the aqueous phase of cellular macromolecules and can act as potent chaotropic stressors (Table 6; Hallsworth et al., 2003a; Bhaganna et al., 2010; Cray et al., 2013). Those stressors with a log P of > 1.95 preferentially partition into the hydrophobic domains of membranes and other macromolecular systems, and thereby act as hydrophobic stressors that induce a distinct type of chaotropicity-mediated water stress (Bhaganna et al., 2010). We believe that the primary ecophysiological value of VOCs for microbial cells pertains to competition, species succession and (for weed species) habitat dominance. Saccharomyces cerevisiae synthesizes both hydrophobic and chaotropic volatiles that have antimicrobial activities, including ethyl octanoate and isoamyl acetate, and ethyl acetate and 2-phenylethanol (respectively, see Table 6; Fialho et al., 2011). Ethanol and 2-phenylethanol are synergistic in their antimicrobial activity (Ingram and Buttke, 1984; Seward et al., 1996), and this is consistent with their common chaotropicity-mediated mode-of-action. The more potent chaotrope of the two (when compared on a molar basis) is 2-phenylethanol which prevents growth of microbes in the range 10–40 mM (Berrah and Konetzka, 1962; Eshkol et al., 2009), values consistent with a chaotropic mode-of-action for a stressor with a log P of 1.36 (see Bhaganna et al., 2010). Saccharomyces cerevisiae (like P. anomala, Acinetobacter and lactic acid bacteria) also inhibits the growth of competing species by acidifying the environment which it achieves via the production of a number of organic acids (Table 6; Kotyk et al., 1999; Lowe and Arendt, 2004; Gulati et al., 2010); it also secretes peptides with fungicidal and fungistatic activities (Albergaria et al., 2010); and some strains contain inheritable virus-like particles containing genes that code for proteins inhibitory to other strains of the same species (Pretorius, 2000). Not only does S. cerevisiae employ multiple types of antimicrobial substance that act in concert, it is also has a correspondingly wide range of stressor-protection measures that minimize metabolic inhibition by antimicrobial stressors (those synthesized by either S. cerevisiae or other species), including efflux pumps (Pretorius, 2000), and highly effective compatible-solute strategies (see above), protein-stabilization proteins and adaptations to acid stress (Table 6) and is therefore highly tolerant towards chaotropic and hydrophobic stressors (Fig. 2; Tables 4 and 6). Saccharomyces cerevisiae acid tolerance (as well as that of lactic acid bacteria) is conferred by an array of mechanisms (Table 3) that collectively contribute to the weed phenotype by removing the hydrogen ions and/or by promoting the refolding of partially denatured biomacromolecules.
Pichia anomala utilizes an array of antimicrobials that have synergistic effects as inhibitors and collectively target a wide array of taxa (including fungi, other yeasts, bacteria and viruses; Walker, 2011): chaotropic stressors such as ethanol and ethyl acetate; hydrophobic stressors such as isoamyl acetate; biosurfactants (see below); organic acids; an exo-β-1,3-glucanase known to degrade glucans in fungal cell walls; and diverse toxins that have specific modes-of-action (see Table 6; Passoth et al., 2006; Walker et al., 2011). A semi-quantitative analysis of the antifungal activities of P. anomala suggests that inhibition results primarily from a combination of (in order of importance): killer toxins, hydrolytic enzymes and volatile substances (Table 6), as well as more minor factors such as nutrient competition and substrate acidification (Walker, 2011). Rhodotorula mucilaginosa produces an array of volatile chaotropic and hydrophobic stressors as well as killer toxins (Table 6). Many fungi also produce mycotoxins; for example Aspergillus ochraceus growing on maize grain can produce ochratoxins at up to 1.4 mg g−1 (Magan et al., 2003). Indeed a number of Aspergillus species are notorious for the potent activity of their ochratoxins against other fungal strains and as well as bacteria (Table 6).
Like S. cerevisiae, some other microbial species are able to produce moderately chaotropic antimicrobials in bulk quantities: Clostridium acetobutylicum produces high concentrations of butanol, ethanol and acetone in anaerobic habitats, and Zymomonas mobilis produces high concentrations of ethanol (Tables S1 and6). Butanol is a particularly potent chaotrope that is highly inhibitory at millimolar concentrations (Bhaganna et al., 2010; Ezeji et al., 2010; Cray et al., 2013). We suspect that Clostridium spp. that can dominate chaotropic environments such as ammonia-rich animal slurries (Table S1) have an unusually high resistance to the net chaotropicity of the ammonia, ethanol, butanol, acetone and other chaotropic substances in their habitat (a hypothesis that has not yet been investigated to our knowledge; Williams and Hallsworth, 2009; Cray et al., 2013). The chaotropicity of urea and ammonia is greater than that of ethanol when compared at equivalent molar concentrations (Hallsworth et al., 2003a; Williams and Hallsworth, 2009) and there is evidence that some bacteria that secrete these compounds, including Clostridium aminophilum and Pseudomonas stutzeri, benefit from chaotropicity-induced inhibition of their competitors (Table 6). There is no absolute resistance to the inhibitory/lethal activities of chaotropic and hydrophobic stressors (Sikkema et al., 1995; Hallsworth, 1998; Hallsworth et al., 2003a; 2003b; 2007; Duda et al., 2004; Bhaganna et al., 2010; Bell et al., 2013) so the production of such antimicrobials coupled with reasonably effective self-protection mechanisms (see Tables S1 and6) can sometimes be the ultimate means to achieve community dominance (see also Table S1).
Pseudomonas putida can generate and/or secrete diverse antimicrobial substances including catabolites produced from degradation of macromolecular substrates, VOCs and biosurfactants (Table 6). Biosurfactants that solubilize hydrocarbons and other hydrophobic substrates can also be highly inhibitory to other microbial species. Studies of Pseudomonas rhamnolipids suggest a key role in competitive ability (Wecke et al., 2011); surfactants not only disorder plasma membranes and solubilize lipids, but can be chaotropic even for macromolecules that lack hydrophobic domains (Table 2; Cray et al., 2013). The membrane-permeabilizing activities of rhamnolipids are particularly detrimental to Gram-positive bacteria, and are lethal to Phytophthora zoospores (Kruijt et al., 2009, Wecke et al., 2011). Bacteriocins produced by pseudomonads, like those of lactic acid bacteria, modify cellular membranes increasing permeability, causing leakage, disrupting transport processes and causing a dissipation of proton-motive force (Table 6). Catabolites such as phenol, catechol, vanillin, biphenyl, and sodium benzoate that are produced from the degradation of high-Mr hydrocarbons have inhibitory activity as chaotropic and hydrophobic stressors (Table 6; Hallsworth et al., 2003a; Bhaganna et al., 2010). Pseudomonas putida and other pseudomonads produce a wide variety of VOCs that collectively impact microbial competitors (Table 6). As outlined above, P. putida has multiple layers of defence against the potential stressful impact of solvent, chaotropic, and hydrophobic stressors via a versatile compatible solute metabolism (e.g. trehalose; Park et al., 2007; Bhaganna et al., 2010), protein-stabilization proteins, and production of extracellular polymeric substances etc. (Tables 3 and 6). In addition, and arguably the most important feature for stressor tolerance, P. putida cells are equipped with efflux pumps (for example the plasmid-encoded TtGHI) that are highly effective at expulsion of hydrocarbons such as toluene as well as many other stressors and toxins (Ramos et al., 2002; Fillet et al., 2012).
Whereas the aquatic habitats of planktonic species such as D. salina, G. semen and M. aeruginosa may serve to dilute secreted antimicrobials, there is evidence that VOCs produced by D. salina, M. aeruginosa, and other aquatic species (including compounds such as β-cyclocitral and dichloromethane) are inhibitory to competing species (Table 6). Gonyostomum semen is able to lyse the cells of competing algal species on contact although the mechanism by which it does so has yet to be resolved (Table 6); and M. aeruginosa synthesizes a diverse range of volatile antimicrobials including phenazines and phenazine derivatives (Table 6; Lifshits and Carmeli, 2012). Lactic acid bacteria, in addition to bacteriocins, produce significant quantities of organic acids and secrete a vast array of inhibitory VOCs (Table 6). The analysis of volatiles produced, primarily by L. rhamnosus, during the production of a ewe's-milk cheese (Table S1) gives an insight into the complexity of the microbial warefare that takes place in open habitats: 57 compounds were detected including terpenes, alcohols, ketones and esters (Randazzo et al., 2010) Although it is plausible that some of these substances exert specific toxic effects, all of them have the properties of either chao- or hydrophobic stressors in microbial cells (Table 6; Bhaganna et al., 2010), and their roles in habitat dominance has rarely received attention.
Microbial weed ecology
Open habitats
The concept of an open habitat is a fundamental, well-established principle in the fields of plant and animal ecology. For microorganisms, open habitats can be defined according to the diversity, interactions and community succession of their inhabitants in contrast to other types of microbial habitat that are defined by physicochemical characteristics, substrate type or nutrient concentration (Table 7). Hydrocarbons are the primary carbon substrate in approximately one-third of the open habitats listed in Table S1, and are generally derived from dense, non-aqueous phase liquids (e.g. oil) or plant metabolites secreted into the rhizosphere, phyllosphere or atmosphere (bioaerosols). A distinguishing feature of hydrocarbon-based systems is that they can remain perpetually open because, as dissolved hydrocarbons are assimilated, the substrate reservoir may be continually replenished by molecules from the oil entering the aqueous phase or (for the rhizosphere and phyllosphere) because the plant continues to produce and secrete hydrocarbons and other organics (Nunes et al., 2005; Kazda and Falkinham III, 2009; McCammick et al., 2009; Hunter et al., 2010; Kachalkin and Yurkov, 2012). These habitats are commonly dominated by species such as Acinetobacter johnsonii, Alkanivorax borkumensis, Pantoea ananatis and P. putida. Whereas these organisms – all members of the gammaproteobacteria – may share many molecular and ecophysiological characteristics (see Tables 3) in most cases it is P. putida that emerges as the most prevalent species (Table S1). This is a product of its metabolic versatility, exceptional tolerance to hydrocarbons and other solutes, and antimicrobial activities (Tables 5 and 6). Other weed species, however, have greater acid tolerance than the gammaproteobacteria: in the acidic surface film of the sphagnum-moss grey layer it is Mycobacterium (including Mycobacterium terrae, M. smegmatis and Mycobacterium sphagni) that emerges as a dominant genus; and in the green layer at lower pH (2–2.5), R. mucilaginosa has the competitive advantage (Table S1). Phyllosphere, rhizosphere and other habitats can also remain continuously open if they are replenished with organic substances from ions released from soil particles, extracellular polymeric substances and compatible solutes from microbial cells, root diffusates, animal wastes, diverse sources of necromass, etc.
Table 7.
Properties of open (microbial) habitats
| Notes and examples |
|||
|---|---|---|---|
| Typical characteristic(s) | Value as a descriptor for an open habitata | For open habitats | For habitats that are not open |
| Microbial ecology | |||
| Open to population by a multitude of microbial species | Primary | Potentially habitable by an indefinite number of species which may be from diverse Kingdoms of life (e.g. solar salterns see Table S1) | The fracture water of a South-African goldmine (2.8 km deep) was found to contain a single-species ecosystem (Chivian et al., 2008) |
| Promote intense competition between microbial species | Primary | Conditions favour strong growth of many species and may select for ability to produce antimicrobial substances (see below) | A climax community, single-species habitat, and symbiotic partnership are characterized by low levels of inter-species competition |
| Can facilitate dominance of the developing community by one or more species (though this may not always take place) | Primary | Diverse types of substrate/environment can accommodate community progression towards dominance by one or more microbial species (see Table S1) | The antithesis of an open habitat is one that is occupied by a climax community that is no longer capable of further evolution (e.g. Paul et al., 2006) |
| Transient and self-terminating; unless maintained in an ‘open’ condition | Primary | Frequently leading to the development of a closed community dominated by a single species (see Fig. 1; Table S1); unless ‘re-opened’ due to grazing for example, or replenished (e.g. by fresh inputs of nutrients) | Closed habitats typically lack conditions that facilitate biomass formation and species succession |
| Resident microbes | |||
| Strong selection for microbes able to out-grow competitors | Primary | For example, habitats rich in nutrients may favour species of bacterial copiotroph which may merge as dominating species if they possess other weed traits | A habitat with limited available resources would not would not support growth (the latter is associated with competition/species succession) |
| Open to manipulation by species able to modify environmental conditions and/or secrete antimicrobials | Primary | Dynamic biomass formation makes open habitats available for/vulnerable to manipulation by species that can introduce growth-limiting substances (Table6; Cordero et al., 2012) | In closed habitats the community is already restricted in terms of space, nutrients and/or physicochemical constraints etc. |
| Select for microbes that lack specific requirements (such as rare trace elements, or a specific host) | Secondary | Microbes such as S. cerevisiae and Lactobacillus spp. typically grow without idiosyncratic requirements (such as a requirement for interactions with other species) | Specialist microbes are found in niche habitats, such as those that form lichens (Hoffland et al., 2004) |
| Chemistry and properties of the cellular environment | |||
| Not excessively stressful for resistant microbes in terms of water activity, chaotropicity, pH, temperature etc.b | Primary | Open (microbial) habitats are, by definition, open to population by a large number and high diversity of microbial species so are typically not physically or chemically extremec | Extreme habitats that are occupied by a small number of obligate extremophiles rarely support the high growth rates or promote the intense competition that typify open habitatsc |
| Have the potential to support a substantial microbial biomass and so do not constrain ecosystem development | Primary | Open habitats have sufficient space and resources to facilitate the ecosystem development (Table S1; Fig. 1) | Non-open habitats can be constrained by nutritional and/or physicochemical parameters (Chivian et al., 2008) |
| Not contaminated with inhibitory substances such as heavy metals or other potent toxins | Secondary | Some polluted, hydrocarbon-rich environments may also support the development of microbial communities | High levels of toxic substances will prevent high growth rates and minimize diversity |
Those designated as descriptors of primary importance are considered essential to the definition of an open (microbial) habitat.
Not so extreme that there is limited diversity or possibility for biomass formation: i.e. typically > 0.75 water activity (see Pitt, 1975; Williams and Hallsworth, 2009), < 20–25 kJ kg−1 mole−1 chaotropic activity (Hallsworth et al., 2007), < pH 11 or > −15°C.
Whereas NaCl-saturated habitats are generally considered extreme, their water activity is 3 0.75 – well above that which ultimately limits microbial life (i.e. 0.61–0.71: Pitt and Christian, 1968; Brown, 1990; Grant, 2004; Williams and Hallsworth, 2009; Leong et al., 2011) – and typically supports a remarkably high microbial diversity and biomass (Antón et al., 2002; Daffonchio et al., 2006; Baati et al., 2008; Bardavid et al., 2008; Khemakhem et al., 2010).
Habitats which facilitate weed emergence are typically not so extreme that there is a negligible microbial diversity or little capacity for community development/biomass formation; i.e. > 0.75 water activity, < 20–25 kJ kg−1 mole−1 chaotropic activity, < pH 11 or > −15°C (Table 7). Open habitats have available nutrients, electron acceptors etc and C : N ratios that favour microbial growth, and do not contain high levels of inhibitory substances such as heavy metals or toxic pollutants. They will therefore promote intense competition between species assuming that a diversity of microbial species is present (Tables S1 and7). Whereas high-sugar habitats with a water activity below 0.9 are too stressful for the vast majority of microbes, including S. cerevisiae, there is still a considerable diversity of xerotolerant and xerophilic species that are highly active between water activities of 0.84 and 0.9 (Pitt, 1975; Brown, 1990). Similarly, crystallizer ponds or solar salterns are known as extreme environments and only a small fraction of microbial species can survive and grow at saturated NaCl. Nevertheless, it is the limited solubility of NaCl (and not the limitations of salt-adapted macromolecular and cellular systems) that determines the limit for microbial growth in high-NaCl habitats: a number of obligate halophiles grow optimally at 35% w/v NaCl (0.755 water activity) and different types of NaCl-saturated environment are biodiverse, highly competitive, biomass-rich microbial habitats (Antón et al., 2002; Daffonchio et al., 2006; Baati et al., 2008; Bardavid et al., 2008; Khemakhem et al., 2010). Salterns (and also algal mats) may under some conditions function as continually open habitats if nutrients are not limiting; D. salina for example can release sufficient glycerol into the system to satisfy the carbohydrate requirement of the wider community (Borowitzka, 1981; Bardavid et al., 2008).
Open habitats favour microbes with shorter generation-times and relatively vigorous growth-phenotypes and in this way may drive the evolution of faster-growing and more stress-tolerant species (as well as those with effective antimicrobial strategies; Tables 6 and 7). That the ecology and evolution of plant weeds are intimately and inextricably linked with open habitats has been established for some time (Anderson, 1952); in contrast there is little information about open habitats of microorganisms (see Table 1; Hallsworth, 1998). Open habitats can be spontaneously created (e.g. due to removal of microbial biomass by predators and grazers; McArthur, 2006), dynamic (e.g. due to desiccation–rehydration cycles or nutrient limitation), and/or transient (Tables S1 and7). As is true for open habitats of plants (Anderson, 1952), open habitats of microbes can also become closed if they becomes water-constrained or otherwise resource-depleted; and/or the microbial community reaches stationary phase (Paul et al., 2006): and these changes may be accompanied by – or even a consequence of – domination by weed species. Environments can even progress to become sterile; for example those with high ethanol concentrations (Hallsworth, 1998; Pretorius, 2000). However, a dualistic concept of open habitats on the one hand and closed habitats on the other would be simplistic; in reality there is a seemless continuum between the two extremes of fertile but uncolonized and hostile/uninhabitable environment. Habitats with a favourable water activity for most bacteria (≥ 0.94) and high concentrations of carbon and nitrogen substrates such as milk, cheese and crushed peanuts (Table S1) typically progress to a closed condition (and are dominated by lactic acid bacteria), as do most high-sugar habitats (Table S1). A number of the open habitats listed in Table S1 that are resource-rich, and stressful, temporally or spatially heterogenous, and/or require saprotrophic nutrition were dominated by Pichia (e.g. moist barley grains; salted fermenting olives) or Aspergillus species (e.g. decomposing leaf litter, sugarcane juice). The stress biology, energy-generating capabilities and antimicrobial activities of these species undoubtedly contribute to their success (Tables 3 and 6).
Ecophysiological classification
Microorganisms have been previously categorized based on the way in which their phenotype impacts ecological behaviour: as autotrophs and heterotrophs; generalists and specialists; r- and K-strategists, aerobes and anaerobes; copiotrophs and oligotrophs; fast and slow growers; and extremophiles and mesophiles (see Table 4 and S2). The definition of microbial weeds is qualitatively distinct from that of other microbial groups (see Table S2): only weeds are characterized by their ability to dominate communities and the associated/underlying biology (Tables 2) and it is only weeds that are linked via their ecology and evolution with open habitats (Tables 1, S1, and S2). The biology and ecology of microbial weed species cuts across established groupings of microorganisms (i.e. the former does not represent a parallel category; see Table S2). Some microbes have a considerable number of weed traits but only rarely emerge as dominant members of communities (e.g. Pseudoxylaria spp., E. coli and M. smegmatis; see Tables S1 and5 and 6). Such species serve as a reminder that weeds and non-weeds represent two extremes of an otherwise continuous scale. An insightful study of Termitomyces ecology, for example, discusses the biology of Pseudoxylaria; a fungus that is present in Termitomyces habitats but is typically inactive and/or present at negligible cell densities (Visser et al., 2011). Pseudoxylaria could be ecologically disadvantaged if present in greater numbers as termites, which can detect VOCs produced during interactions between Pseudoxylaria and Termitomyces, remove the unwanted fungus from their Termitomyces cultures. The extant Pseudoxylaria strains thereby appear to have been selected for/have evolved a low competitive ability (Visser et al., 2011). Nevertheless Pseudoxylaria can occasionally emerge to dominate the substrate (Visser et al., 2011), a fertile open habitat consisting of decomposing plant material (Table S1). Non-weed species make up a loose affiliation of species that are only linked by their poor ability to dominate communities in open habitats and may not share any specific metabolic or phenotypic traits (Table S2). We propose that this ‘dustbin’ group includes Synodropsis spp., Polypaecilum pisce, Metschnikowia orientalis, Caulobacter crescentus and Salmonella species (Table S2). Several ecophysiological classification systems that are based on microbial growth-phenotype and/or substrate utilization (Table S2). Microbial generalists can occupy a wide range of ecological niches, and typically achieve this via the ability to utilize a wide range of nutrients and substrates (Table S2). Conversely microbial specialists, such as S. cerevisiae, H. werneckii and many basidiomycetes have specific nutritional and/or physicochemical requirements (de Fine Licht et al., 2005; Kashangura et al., 2006). Obligate extremophiles commonly occupy, and are adapted to grow optimally in, niches that are hostile to the majority of microorganisms (Table S2). There have been relatively few studies (and is consequently inadequate terminology) to properly describe microbes that are extremo-intolerant such as xero-intolerant species (see Table 4), with the exception of mesophilic species that function optimally at non-extreme temperatures (though the term mesophiles can be used in a general sense to refer to non-extremophiles: Table S2; Gostinčar et al., 2010). Non-extremophilic microbes grow optimally in physicochemical conditions which favour metabolic activity and growth of the majority of microbial species, and this grouping includes both weed and non-weed species (Table S2). While fast-growing microbes have short doubling times, the vast majority of these species do not have weed-like capabilities and are unable to out-grow their competitors (Tables 4 and S2). It may be that relatively few oligotrophic microbes or K-strategists are weed species, although some mycobacteria do have a weed phenotype (Tables S1,5 and 6). Pseudomonas putida is generally considered to be a model r-strategist (e.g. Margesin et al., 2003) yet P. putida has an extraordinary competitive ability (see Tables S1 and6), and may not be the best example of an r-strategist (the latter are characterized by a weak competitive ability; Table S2). By contrast P. putida appears to be an exceptional example of a microbial weed both in terms of its cellular biology and its ability to dominate diverse types of open habitat.
Concluding remarks
It is irrefutable that a small minority of microbial species recurrently appear as the dominant members of microbial communities; and we suggest here that these microbes have shared phenotypic traits. This implies that open-habitat ecology has impacted the evolutionary trajectories of these microbes, and that the activities of weeds determine microbial diversity in many localities within the biosphere. Whereas microbial weed species are not always useful to humankind (see Table 1), they make up a substantial portion of the world's biotechnology sector. Saccharomyces cerevisiae supports global industries in fermented products, biofuel, biocides (ethanol is also an important disinfectant; McDonnell and Russell, 1999), and other white biotechnologies; P. anomala produces a substantial catalogue of biotechnologically useful substances and is employed in production of foodstuffs (see Walker, 2011); lactic acid bacteria form the basis of food- and drinks-production processes where the antimicrobial volatiles and organic acids that they secrete act as flavour compounds and natural preservatives, and are used as probiotics, and for the production of fine chemicals (Rose, 1982; Steinkraus, 1983; Molly et al., 1996; Nomura et al., 1998; Tiwari et al., 2009; Randazzo et al., 2010; Bron et al., 2012); P. putida is used for the commercial production of bulk and fine chemicals, industrial biocatalysis, and as the basis of many other industrial and environmental biotechnologies (Kruijt et al., 2009; Puchałka et al., 2008; Hassan et al., 2011; Poblete-Castro et al., 2012); a number of weed species, including Aspergillus spp., P. putida and Clostridium spp. play primary roles in the treatment or decomposition of plant matter, oil and hydrocarbons, xenobiotics in polluted soils, sludge and slurry treatments, and other organic wastes (Tables S2 and4; Narihiro and Sekiguchi, 2007; Timmis, 2009); and Aspergillus species are used for production of foods (Rose, 1982; Tamang, 2010), fine chemicals (e.g. citric acid, secondary metabolites) and extracellular enzymes (e.g. amylase, cellulase, pectinase and protease), for industrial biocatalysis, and have potential for bioremediation and biosorption applications (Ward et al., 2006). It is the metabolic activities of substances that suppress potential competitors which make S. cerevisiae, Rhodotorula spp., P. anomala, P. putida, lactic acid bacteria and other weed species invaluable as agents of biological control, for preventing spoilage of drinks, foodstuffs and animal feeds (see Calvente et al., 1999; Kruijt et al., 2009; Hassan et al., 2011; Olstorpe and Passoth, 2011; Walker, 2011).
Microbial weeds are also used as model systems for research, yet key aspects of their biology remain enigmatic. Many plant weeds have exceptional reproduction and dispersal systems (Table 1) and it remains unclear whether microbial weed species produce more propagules, have superior dispersal mechanisms, or evolved propagules with exceptional longevity, and/or the ability to germinate over a wider range of conditions than their non-weed comparators. Could it be that, like plant weeds, the competitive ability of microbial weeds is enhanced by an abnormally high phenotypic plasticity and intra-specific variability (see Table 1)? Out of the world's plant flora it has been estimated that only several hundred plant species are weeds (Hill, 1977); what percentage of microbes might exhibit weed phenotypes and behaviour? Is there evidence preserved in rocks, sediments, peat, ice, or the hypersaline fluid inclusions of salt crystals that can shed light on the evolutionary trajectories of microbial weed species? Is there an ideal genome size for prokaryotic and eukaryotic weed species? What are the minimum/essential characteristics required to enable a microorganism to dominate a specific habitat; can we design and manufacture a microbial weed via a synthetic biology approach? Will hitherto uninvestigated microbial weeds prove to be rich sources of antimicrobial compounds for novel biotechnological and clinical applications? Can microbial weed ecology shed light on wound microbiology, lead to the prevention of the microbial colonization of prosthetic devices, or minimize the emergence of opportunistic pathogens? Could knowledge-based interventions in weed ecology enhance our ability to manipulate and manage microbes at various levels of food-production and food-spoilage, or lead to better management of environments that harbor potentially dangerous species?
Crop plants such as sunflowers, potatoes, wheat and barley are known to have weedy ancestors (Anderson, 1952) and it is thought that the evolution of both plant weeds and crop plants has been driven by anthropogenically opened habitats. We believe that open (microbial) habitats also select for the phenotypes, and therefore act as a potent driving force that determines the evolutionary trajectories, of microbial weeds and their communities: open-habitat ecology may therefore be worthy of recognition as a distinct scientific field that spans all organismal biology including microbiology. There is a maxim that everything is everywhere but that the environment selects; this implies that microbial species are everywhere on Earth but that the local environment determines which are able to grow (Baas Becking, 1934; de Wit and Bouvier, 2006). While not necessarily untrue, this idea implies a level of passivity on the part of the microbial cell. Yet microorganisms are inherently variable and dynamic systems with considerable genetic dexterity and phenotypic plasticity (able to both respond and adapt). Baas Becking (1934) acknowledged that cells are able to modify their extracellular environment, but microbes are also proactive in their manipulation of each other. It may be that microbial weeds are the most accomplished examples of this.
Acknowledgments
We are most grateful to T.A. Hill (University of London, UK) for his inspirational lectures on the biology of plant weeds; and for scientific enjoyable and fruitful discussions with E. J. Carpenter (San Francisco State University, USA), L.R. Cooke (Agri-Food and Biosciences Institute, Belfast); B.T.M. Dentinger (Royal Botanic Gardens, Kew, UK), N. Gunde-Cimerman (University of Ljubljana, Slovenia), S. Iwaguchi (Nara Women's University, Japan), M.-A. Lachance (University of Western Ontario, Canada), K.D. Farnsworth, I.R. Grant, J.D.R. Houghton, C.J. Law, A.L. McCann, J.W. McGrath, J. Megaw, J. P. Quinn, and J.T. Russell (Queen's University Belfast), J.L. Green, and A.M. Womack (University of Oregon, USA), T.J. McGenity (University of Essex, UK), T. Mock (University of East Anglia), A. Oren (The Hebrew University of Jerusalem, Israel), I.S. Pretorius (University of South Australia, Australia), R. Santos (Laboratório de Análises of the Instituto Superior Técnico, Lisbon, Portugal), R.S. Singhal (Institute of Chemical Technology, Mumbai, India), G.G. Stewart (Heriot-Watt University, UK), J.P. Tamang (Sikkim University, India), K.N. Timmis (Helmholtz Centre for Infection Research, Braunschweig, Germany), M.A. Voytek (NASA Headquaters, USA).
Conflict of interest
None declared.
Supporting information
Additional Supporting Information may be found in the online version of this article:
Table S1. Examples of open habitats in which specific microbes attain dominance.
Table S2. Biology of microbial weeds compared with that of other ecophysiological groups.
References
- Albergaria H, Francisco D, Gori K, Arneborg N, Gírio F. Saccharomyces cerevisiae CCMI 885 secretes peptides that inhibit the growth of some non-Saccharomyces wine-related strains. Appl Microbiol Biotechnol. 2010;86:965–972. doi: 10.1007/s00253-009-2409-6. [DOI] [PubMed] [Google Scholar]
- Albertyn J, Hohmann S, Thevelein JM, Prior B. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high osmolarity glycerol response pathway. Mol Cell Biol. 1994;14:4135–4144. doi: 10.1128/mcb.14.6.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsaadawi IS, Salih NMM. Allelopathic potential of Cyperus rotundus L. I. Interference with crops. Allelopathy J. 2009;23:297–303. [Google Scholar]
- Al-Tahhan RA, Sandrin TR, Bodour AA, Maier RM. Rhamnolipid-induced removal of lipopolysaccharide from Pseudomonas aeruginosa: effect on cell surface properties and interaction with hydrophobic substrates. Appl Environ Microbiol. 2000;66:3263–3268. doi: 10.1128/aem.66.8.3262-3268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altermann E, Russell WM, Azcarate-Peril MA, Barrangou R, Buck BL, McAuliffe O, et al. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc Natl Acad Sci USA. 2005;102:3906–3912. doi: 10.1073/pnas.0409188102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Ordóñez A, Begley M, Prieto M, Messens W, López M, Bernardo A, et al. Salmonella spp. survival strategies within the host gastrointestinal tract. Microbiology. 2011;157:3268–3281. doi: 10.1099/mic.0.050351-0. [DOI] [PubMed] [Google Scholar]
- Andersen D, Renshaw JC, Wiebe MG. Rhodotorulic acid production by Rhodotorula mucilaginosa. Mycol Res. 2003;107:949–956. doi: 10.1017/s0953756203008220. [DOI] [PubMed] [Google Scholar]
- Anderson E. Plants, Man and Life. Boston, MA, USA: Little, Brown and Company; 1952. [Google Scholar]
- Anderson RC, Flythe MD, Krueger NA, Callaway TR, Edrington TS, Harvey RB, et al. Decreased competiveness of the foodborne pathogen Campylobacter jejuni during co-culture with the hyper-ammonia producing anaerobe Clostridium aminophilum. Folia Microbiol. 2010;55:309–311. doi: 10.1007/s12223-010-0046-1. [DOI] [PubMed] [Google Scholar]
- Anderssen EL, Diep DB, Nes IF, Eijsink VGH, Nissen-Meyer J. Antagonistic activity of Lactobacillus plantarum C11: two new two-peptide bacteriocins, plantaricins EF and JK, and the induction factor plantaricin A. Appl Environ Microbiol. 1998;64:2269–2272. doi: 10.1128/aem.64.6.2269-2272.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antón J, Oren A, Benlloch S, Rodríguez-Valera F, Amann R, Rosselló-Mora R. Salinibacter ruber gen. nov., sp. nov., a novel, extremely halophilic member of the Bacteria from saltern crystallizer ponds. Int J Syst Evol Microbiol. 2002;52:485–491. doi: 10.1099/00207713-52-2-485. [DOI] [PubMed] [Google Scholar]
- Aranda JS, Salgado E, Taillandier P. Trehalose accumulation in Saccharomyces cerevisiae cells: experimental data and structured modeling. Biochem Eng J. 2004;17:129–140. [Google Scholar]
- Arsène F, Tomoyasua T, Bukau B. The heat shock response of Escherichia coli. Int J Food Microbiol. 2000;55:3–9. doi: 10.1016/s0168-1605(00)00206-3. [DOI] [PubMed] [Google Scholar]
- Aziz S, Memon HR, Shah FA, Rajoka MI, Soomro SA. Production of ethanol by indigenous wild and mutant strain of thermotolerant Kluyveromyces marxianus under optimized fermentation conditions. Pak J Anal Environ Chem. 2009;10:25–33. [Google Scholar]
- Baas Becking LGM. Geobiologie of inleiding tot de milieukunde. The Hague, the Netherlands: W.P. Van Stockum 0026; Zoon; 1934. (in Dutch) [Google Scholar]
- Baati H, Guermazi S, Amdouni R, Gharsallah N, Sghir A, Ammar E. Prokaryotic diversity of a Tunisian multipond solar saltern. Extremophiles. 2008;12:505–518. doi: 10.1007/s00792-008-0154-x. [DOI] [PubMed] [Google Scholar]
- Back JF, Oakenfull D, Smith MB. Increased thermal-stability of proteins in the presence of sugars and polyols. Biochemistry. 1979;18:5191–5196. doi: 10.1021/bi00590a025. [DOI] [PubMed] [Google Scholar]
- Baker HG. Characteristics and modes of origin of weeds. In: Baker HG, Stebbins GL, editors. Genetics of Colonizing Species. New York, USA: Academic Press; 1965. pp. 147–168. [Google Scholar]
- Bakker EG, Montgomery B, Nguyen T, Eide K, Chang J, Mockler TC, et al. Strong population structure characterizes weediness gene evolution in the invasive grass species Brachypodium distachyon. Mol Ecol. 2009;18:2588–2601. doi: 10.1111/j.1365-294X.2009.04225.x. [DOI] [PubMed] [Google Scholar]
- Balashov SP, Imasheva ES, Boichenko VA, Anton J, Wang JM, Lanyi JK. Xanthorhodopsin: a proton pump with a light-harvesting carotenoid antenna. Science. 2005;309:2061–2064. doi: 10.1126/science.1118046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballerstedt H, Volkers RJM, Mars AE, Hallsworth JE, Martins dos Santos VA, Puchałka J, et al. Genomotyping of Pseudomonas putida strains using P. putida KT2440-based high-density DNA microarrays: implications for transcriptomics studies. Appl Microbiol Biotechnol. 2007;75:1133–1142. doi: 10.1007/s00253-007-0914-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardavid RE, Oren A. Sensitivity of Haloquadratum and Salinibacter to antibiotics and other inhibitors: implications for the assessment of the contribution of Archaea and Bacteria to heterotrophic activities in hypersaline environments. FEMS Microbiol Ecol. 2008;63:309–315. doi: 10.1111/j.1574-6941.2007.00433.x. [DOI] [PubMed] [Google Scholar]
- Bardavid RE, Oren A. The amino acid composition of proteins from anaerobic halophilic bacteria of the order Halanaerobiales. Extremophiles. 2012;16:567–572. doi: 10.1007/s00792-012-0455-y. [DOI] [PubMed] [Google Scholar]
- Bardavid RE, Khristo P, Oren A. Interrelationships between Dunaliella and halophilic prokaryotes in saltern crystallizer ponds. Extremophiles. 2008;12:5–14. doi: 10.1007/s00792-006-0053-y. [DOI] [PubMed] [Google Scholar]
- Behrends V, Williams KJ, Jenkins VA, Robertson BD, Bundy JG. Free glucosylglycerate is a novel marker of nitrogen stress in Mycobacterium smegmatis. J Proteome Res. 2012;11:3888–3896. doi: 10.1021/pr300371b. [DOI] [PubMed] [Google Scholar]
- Bell ANW, Magill E, Hallsworth JE, Timson DJ. Effects of alcohols and compatible solutes on the activity of β-galactosidase. Appl Biochem Biotechnol. 2013 doi: 10.1007/s12010-012-0003-3. doi: 10.1007/s12010-012-0003-3. [DOI] [PubMed] [Google Scholar]
- Bellinger BJ, Van Mooy BAS. Nonphosphorus lipids in Periphyton reflect available nutrients in the Florida Everglades, USA. J Phycol. 2012;48:303–311. doi: 10.1111/j.1529-8817.2012.01125.x. [DOI] [PubMed] [Google Scholar]
- Benning C. Biosynthesis and function of the sulfolipid sulfoquinovosyl diacylglycerol. Annu Rev Plant Phys. 1998;49:53–75. doi: 10.1146/annurev.arplant.49.1.53. [DOI] [PubMed] [Google Scholar]
- Bental M, Pick U, Avron M, Degani H. Polyphosphate metabolism in the alga Dunaliella salina studied by P-31-NMR. Biochim Biophys Acta. 1991;1092:21–28. doi: 10.1016/0167-4889(91)90173-u. [DOI] [PubMed] [Google Scholar]
- Berdjeb L, Pollet T, Domaizon I, Jacquet S. Effect of grazers and viruses on bacterial community structure and production in two contrasting trophic lakes. BMC Microbiol. 2011;11 doi: 10.1186/1471-2180-11-88. Article number 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal P, Seguar A, Ramos J-L. Compensatory role of the cistrans-isomerase and cardiolipin synthase in the membrane fluidity of Pseudomonas putida DOT-T1E. Environ Microbiol. 2007;9:1658–1664. doi: 10.1111/j.1462-2920.2007.01283.x. [DOI] [PubMed] [Google Scholar]
- Berrah G, Konetzka WA. Selective and reversible inhibition of the synthesis of bacterial deoxyribonucleic acid by phenethyl alcohol. J Bacteriol. 1962;83:738–744. doi: 10.1128/jb.83.4.738-744.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry C, Fernando WGD, Loewen PC, de Kievit TR. Lipopeptides are essential for Pseudomonas sp. DF41 biocontrol of Sclerotinia sclerotiorum. Biol Control. 2010;55:211–218. [Google Scholar]
- Bhaganna P, Volkers RJM, Bell ANW, Kluge K, Timson DJ, McGrath JW, et al. Hydrophobic substances induce water stress in microbial cells. Microb Biotechnol. 2010;3:701–716. doi: 10.1111/j.1751-7915.2010.00203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolhuis H, Palm P, Wende A, Falb M, Rampp M, Rodriguez-Valera F, et al. The genome of the square archaeon Haloquadratum walsbyi: life at the limits of water activity. BMC Genomics. 2006;7:169. doi: 10.1186/1471-2164-7-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet MP, Poulin M. Numerical modelling of the planktonic succession in a nutrient-rich reservoir: environmental and physiological factors leading to Microcystis aeruginosa dominance. Ecol Modell. 2002;156:93–112. [Google Scholar]
- Borowitzka LJ. The microflora. Adaptation to life in extremely saline lakes. Hydrobiologia. 1981;81:33–46. [Google Scholar]
- Boutibonnes P, Auffray Y, Malherbe C, Kogbo W, Marais C. Antibacterial and genotoxic properties of 33 mycotoxins. Mycopathologia. 1984;87:43–49. doi: 10.1007/BF00436626. [DOI] [PubMed] [Google Scholar]
- Boynton ZL, Bennett GN, Rudolph FB. Cloning, sequencing, and expression of genes encoding phosphotransacetylase and acetate kinase from Clostridium acetobutylicum ATCC 824. Appl Environ Microbiol. 1996;62:2758–2766. doi: 10.1128/aem.62.8.2758-2766.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britigan BE, Miller RA, Hassett DJ, Pfaller MA, McCormick ML, Rasmussen GT. Antioxidant enzyme expression in clinical isolates of Pseudomonas aeruginosa: Identification of an atypical form of manganese superoxide dismutase. Infect Immun. 2001;69:7396–7401. doi: 10.1128/IAI.69.12.7396-7401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bron PA, Van Baarlen P, Kleerebezem M. Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat Rev Microbiol. 2012;10:66-U90. doi: 10.1038/nrmicro2690. [DOI] [PubMed] [Google Scholar]
- Brown AD. Microbial Water Stress Physiology – Principles and Perspectives. Chichester, UK: Wiley; 1990. [Google Scholar]
- Buzzini P, Gasparetti C, Turchetti B, Cramarossa MR, Vaughan-Martini A, Martini A, et al. Production of volatile organic compounds (VOCs) by yeasts isolated from the ascocarps of black (Tuber melanosporum Vitt.) and white (Tuber magnatum Pico) truffles. Arch Microbiol. 2005;184:187–193. doi: 10.1007/s00203-005-0043-y. [DOI] [PubMed] [Google Scholar]
- Cabrol L, Malhautier L, Poly F, Lepeuple A-S, Fanlo J-L. Bacterial dynamics in steady-state biofilters: beyond functional stability. FEMS Microbiol Ecol. 2012;79:260–271. doi: 10.1111/j.1574-6941.2011.01213.x. [DOI] [PubMed] [Google Scholar]
- Calvente V, Benuzzi D, de Tosetti MIS. Antagonistic action of siderophores from Rhodotorula glutinis upon the postharvest pathogen Penicillium expansum. Int Biodeterior Biodegradation. 1999;43:167–172. [Google Scholar]
- Capone KA, Dowd SE, Stamatas GN, Nikolovski J. Diversity of the human skin microbiome early in life. J Invest Dermatol. 2011;131:2026–2032. doi: 10.1038/jid.2011.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadei MA, Mañas P, Niven G, Needs E, Mackey BM. Role of membrane fluidity in pressure resistance of Escherichia coli NCTC 8164. Appl Environ Microbiol. 2002;68:5965–5972. doi: 10.1128/AEM.68.12.5965-5972.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrol CD, Koretsky AP, Domach MM. NMR-observed phosphate trafficking and polyphosphate dynamics in wild-type and vph1-1 mutant Saccharomyces cerevisiae in response to stresses. Biotechnol Prog. 1999;15:65–73. doi: 10.1021/bp9800743. [DOI] [PubMed] [Google Scholar]
- Cerdeño-Tárraga AM, Patrick S, Crossman LC, Blakely G, Abratt V, Lennard N, et al. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science. 2005;307:1463–1465. doi: 10.1126/science.1107008. [DOI] [PubMed] [Google Scholar]
- Chakrabarty AM, Roy SC. Nature of endogenous reserve material in a strain of Pseudomonas fluorescens-putida intermediate. Biochem J. 1964;92:105–112. doi: 10.1042/bj0920105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W-S, van de Mortel M, Nielsen L, de Guzman GN, Li X, Halverson LJ. Alginate production by Pseudomonas putida creates a hydrated microenvironment and contributes to biofilm architecture and stress tolerance under water-limiting conditions. J Bacteriol. 2007;189:8290–8299. doi: 10.1128/JB.00727-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Bradford WD, Seidel CW, Li R. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature. 2012;482:246–250. doi: 10.1038/nature10795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Jiang J-G. Osmotic responses of Dunaliella to the changes of salinity. J Cell Physiol. 2009;219:251–258. doi: 10.1002/jcp.21715. [DOI] [PubMed] [Google Scholar]
- Chen J, Lee SM, Mao Y. Protective effect of exopolysaccharide colanic acid of Escherichia coli O157:H7 to osmotic and oxidative stress. Int J Food Microbiol. 2004;93:281–286. doi: 10.1016/j.ijfoodmicro.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Chen M, Tang HY, Ma HZ, Holland TC, Ng KYS, Salley SO. Effect of nutrients on growth and lipid accumulation in the green algae Dunaliella tertiolecta. Bioresour Technol. 2011;102:1649–1655. doi: 10.1016/j.biortech.2010.09.062. [DOI] [PubMed] [Google Scholar]
- Chin JP, Megaw J, Magill CL, Nowotarski K, Williams JP, Bhaganna P, et al. Solutes determine the temperature windows for microbial survival and growth. Proc Natl Acad Sci USA. 2010;107:7835–7840. doi: 10.1073/pnas.1000557107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirife J, Favetto G, Fontán CF. Microbial growth at reduced water activities: some physicochemical properties of compatible solutes. J Appl Bacteriol. 1984;56:259–268. doi: 10.1111/j.1365-2672.1984.tb01346.x. [DOI] [PubMed] [Google Scholar]
- Chiruvolu V, Eskridge K, Cregg J, Meagher M. Effects of glycerol concentration and pH on growth of recombinant Pichia pastoris yeast. Appl Biochem Biotechnol. 1998;75:163–173. [Google Scholar]
- Chivian D, Brodie EL, Alm EJ, Culley DE, Dehal PS, DeSantis TZ, et al. Environmental genomics reveals a single-species ecosystem deep within Earth. Science. 2008;322:275–278. doi: 10.1126/science.1155495. [DOI] [PubMed] [Google Scholar]
- Chou CH. Role of allelopathy in sustainable agriculture: use of allelochemicals as naturally occurring bio-agrochemicals. Allelopathy J. 2010;25:3–16. [Google Scholar]
- Coleman AW, Heywood P. Structure of the chloroplast and its DNA in chloromonadophycean algae. J Cell Sci. 1981;49:401–409. doi: 10.1242/jcs.49.1.401. [DOI] [PubMed] [Google Scholar]
- Collado-Fabbri S, Vaulot D, Ulloa O. Structure and seasonal dynamics of the eukaryotic picophytoplankton community in a wind-driven coastal upwelling ecosystem. Limnol Oceanogr. 2011;56:2334–2346. [Google Scholar]
- Colomb A, Yassaa N, Williams J, Peeken I, Lochte K. Screening volatile organic compounds (VOCs) emissions from five marine phytoplankton species by head space gas chromatography/mass spectrometry (HS-GC/MS) J Environ Monit. 2008;10:325–330. doi: 10.1039/b715312k. [DOI] [PubMed] [Google Scholar]
- Corcelli A, Lattanzio VMT, Mascolo G, Babudri F, Oren A, Kates M. Novel sulfonolipid in the extremely halophilic bacterium Salinibacter ruber. Appl Environ Microbiol. 2004;70:6678–6685. doi: 10.1128/AEM.70.11.6678-6685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcuff R, Mercier J, Tweddell R, Arul J. ) Effect of water activity on the production of volatile organic compounds by Muscodor albus and their effect on three pathogens in stored potato. Fungal Biol. 2011;115:220–227. doi: 10.1016/j.funbio.2010.12.005. [DOI] [PubMed] [Google Scholar]
- Cordero OX, Wildschutte H, Kirkup B, Proehl S, Ngo L, Hussain F, et al. Ecological populations of bacteria act as socially cohesive units of antibiotic production and resistance. Science. 2012;337:1228–1231. doi: 10.1126/science.1219385. [DOI] [PubMed] [Google Scholar]
- Costa V, Amorim MA, Reis E, Quintanilha A, Moradas-Ferreira P. Mitochondrial superoxide dismutase is essential for ethanol tolerance of Saccharomyces cerevisiae in the post-diauxic phase. Microbiology. 1997;43:1649–1656. doi: 10.1099/00221287-143-5-1649. [DOI] [PubMed] [Google Scholar]
- Couturier E, Rocha EPC. Replication-associated gene dosage effects shape the genomes of fast-growing bacteria but only for transcription and translation genes. Mol Microbiol. 2006;59:1506–1518. doi: 10.1111/j.1365-2958.2006.05046.x. [DOI] [PubMed] [Google Scholar]
- Cox RA. Quantitative relationships for specific growth rates and macromolecular compositions of Mycobacterium tuberculosisStreptomyces coelicolor A3(2) and Escherichia coli B/r: an integrative theoretical approach. Microbiology. 2004;150:1413–1426. doi: 10.1099/mic.0.26560-0. [DOI] [PubMed] [Google Scholar]
- Cramer HH. Plant Protection and World Crop Production. Pflanzenschutz: Nachrichten Bayer; 1967. [Google Scholar]
- Cray JA, Russell JT, Timson DJ, Singhal RS, Hallsworth JE. A universal measure of chaotropicity and kosmotropicity. Environ Microbiol. 2013;15:287–296. doi: 10.1111/1462-2920.12018. [DOI] [PubMed] [Google Scholar]
- Crowe JH, Crowe LM, Chapman D. Preservation of membranes in anhydrobiotic organisms – the role of trehalose. Science. 1984;223:701–703. doi: 10.1126/science.223.4637.701. [DOI] [PubMed] [Google Scholar]
- D'Amore T, Panchal CJ, Russell I, Stewart GG. A study of ethanol tolerance in yeast. Crit Rev Biotechnol. 1990;9:287–304. doi: 10.3109/07388558909036740. [DOI] [PubMed] [Google Scholar]
- D'Souza-Ault MR, Smith LT, Smith GM. Roles of N-acetylglutaminylglutamine amide and glycine betaine in adaptation of Pseudomonas aeruginosa to osmotic stress. Appl Environ Microbiol. 1993;59:473–478. doi: 10.1128/aem.59.2.473-478.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffonchio D, Borin S, Brusa T, Brusetti L, van der Wielen PWJJ, Bolhuis H, et al. Stratified prokaryote network in the oxic–anoxic transition of a deep sea halocline. Nature. 2006;440:203–207. doi: 10.1038/nature04418. [DOI] [PubMed] [Google Scholar]
- Daniels C, Godoy P, Duque E, Molina-Henares MA, de la Torre J, del Arco JM, et al. Global regulation of food supply by Pseudomonas putida DOT-T1E. J Bacteriol. 2010;192:2169–2181. doi: 10.1128/JB.01129-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Fine Licht HH, Andersen A, Aanen DK. Termitomyces sp. associated with the termite Macrotermes natalensis has a heterothallic mating system and multinucleate cells. Mycol Res. 2005;109:314–318. doi: 10.1017/s0953756204001844. [DOI] [PubMed] [Google Scholar]
- Desai JD, Banat IM. Microbial production of surfactants and their commercial potential. Microbiol Mol Biol Rev. 1997;61:47–64. doi: 10.1128/mmbr.61.1.47-64.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar R, Sägesser R, Weikert C, Yuan J, Wagner A. Adaptation of Saccharomyces cerevisiae to saline stress through laboratory evolution. J Evol Biol. 2011;24:1135–1153. doi: 10.1111/j.1420-9101.2011.02249.x. [DOI] [PubMed] [Google Scholar]
- Díaz Muñoz G, Montalvo-Rodríguez R. Halophilic black yeast Hortaea werneckii in the Cabo Rojo solar salterns: Its first record for this extreme environment in Puerto Rico. Caribb J Sci. 2005;41:360–365. [Google Scholar]
- Djelal H, Amrane A, Lahrer F, Martin G. Effect of medium osmolarity on the bioproduction of glycerol and ethanol by Hansenula anomala growing on glucose and ammonium. Appl Microbiol Biotechnol. 2005;69:341–349. doi: 10.1007/s00253-005-1987-1. [DOI] [PubMed] [Google Scholar]
- Domínguez JM, Gong CS, Tsao GT. Production of xylitol from d-xylose by Debaryomyces hansenii. Appl Biochem Biotechnol. 1997;63-65:117–127. doi: 10.1007/978-1-4612-2312-2_12. [DOI] [PubMed] [Google Scholar]
- Domínguez-Cuevas P, González-Pastor J-E, Marqués S, Ramos J-L, de Lorenzo V. Transcriptional tradeoff between metabolic and stress-response programs in Pseudomonas putida KT2440 cells exposed to toluene. J Biol Chem. 2006;281:11981–11991. doi: 10.1074/jbc.M509848200. [DOI] [PubMed] [Google Scholar]
- Duda VI, Danilevich VN, Suzina NE, Shorokhova AP, Dmitriev VV, Mokhova ON, et al. Changes in the fine structure of microbial cells induced by chaotropic salts. Microbiology (Russia) 2004;73:341–349. [PubMed] [Google Scholar]
- Duiff BJ, Meijer JW, Bakker PAHM, Schippers B. Siderophore-mediated competition for iron and induced resistance in the suppression of Fusarium wilt of carnation by fluorescent Pseudomonas spp. Neth J Plant Pathol. 1993;99:277–289. [Google Scholar]
- Egan MW, Spratt DA, Ng YL, Lam JM, Moles DR, Gulabivala K. Prevalence of yeasts in saliva and root canals of teeth associated with apical periodontitis. Int Endod J. 2002;35:321–329. doi: 10.1046/j.1365-2591.2002.00478.x. [DOI] [PubMed] [Google Scholar]
- El Ghachi M, Bouhss A, Blanot D, Mengin-Lecreulx D. The bacA gene of Escherichia coli encodes an undecaprenyl pyrophosphate phosphatase activity. J Biol Chem. 2004;279:30106–30113. doi: 10.1074/jbc.M401701200. [DOI] [PubMed] [Google Scholar]
- Enya J, Shinohara H, Yoshida S, Negishi TTH, Suyama K, Tsushima S. Culturable leaf-associated bacteria on tomato plants and their potential as biological control agents. Microb Ecol. 2007;53:524–536. doi: 10.1007/s00248-006-9085-1. [DOI] [PubMed] [Google Scholar]
- Escapa IF, García JL, Bühler B, Blank LM, Prieto MA. The polyhydroxyalkanoate metabolism controls carbon and energy spillage in Pseudomonas putida. Environ Microbiol. 2012;14:1049–1063. doi: 10.1111/j.1462-2920.2011.02684.x. [DOI] [PubMed] [Google Scholar]
- Eshkol N, Sendovski M, Bahalul M, Katz-Ezov T, Kashi Y, Fishman A. Production of 2-phenylethanol from L-phenylalanine by a stress tolerant Saccharomyces cerevisiae strain. J Appl Microbiol. 2009;106:534–542. doi: 10.1111/j.1365-2672.2008.04023.x. [DOI] [PubMed] [Google Scholar]
- Ezeji T, Milne C, Price N, Blaschek H. Achievements and perspectives to overcome the poor solvent resistance in acetone and butanol-producing microorganisms. Appl Microbiol Biotechnol. 2010;85:1697–1712. doi: 10.1007/s00253-009-2390-0. [DOI] [PubMed] [Google Scholar]
- Farkas I, Hardy TA, DePaoli-Roach AA, Roach PJ. Isolation of the GSY1 gene encoding yeast glycogen synthase and evidence for the existence of a second gene. J Biol Chem. 1990;265:20879–20886. [PubMed] [Google Scholar]
- Favero MS, Carson LA, Bond WW, Petersen NJ. Pseudomonas aeruginosa – growth in distilled water from hospitals. Science. 1971;173:836–838. doi: 10.1126/science.173.3999.836. [DOI] [PubMed] [Google Scholar]
- Ferrer M, Chernikova TN, Yakimov MM, Golyshin PN, Timmis KN. Chaperonins govern growth of Escherichia coli at low temperatures. Nat Biotechnol. 2003;21:1266–1267. doi: 10.1038/nbt1103-1266. [DOI] [PubMed] [Google Scholar]
- Fialho MB, Ferreira LFR, Monteiro RTR, Pascholati SF. Antimicrobial volatile organic compounds affect morphogenesis-related enzymes in Guignardia citricarpa, causal agent of citrus black spot. Biocontrol Sci Technol. 2011;21:797–807. [Google Scholar]
- Fillet S, Daniels C, Pini C, Krell T, Duque E, Bernal P, et al. Transcriptional control of the main aromatic hydrocarbon efflux pump in Pseudomonas. Environ Microbiol Rep. 2012;4:158–167. doi: 10.1111/j.1758-2229.2011.00255.x. [DOI] [PubMed] [Google Scholar]
- Fleming A. On the antibacterial action of cultures of a Penicillium, with special reference to their use in the isolation of B. influenza. Exp Pathol. 1929;10:226–236. [Google Scholar]
- Flipphi M, Sun JB, Robellet X, Karaffa L, Fekete E, Zeng AP, et al. Biodiversity and evolution of primary carbon metabolism in Aspergillus nidulans and other Aspergillus spp. Fungal Genet Biol. 2009;46:S19–S44. doi: 10.1016/j.fgb.2008.07.018. [DOI] [PubMed] [Google Scholar]
- Freitas JS, Silva EM, Leal J, Gras DE, Martinez-Rossi NM, dos Santos LD, et al. Transcription of the Hsp30, Hsp70, and Hsp90 heat shock protein genes is modulated by the PalA protein in response to acid pH-sensing in the fungus Aspergillus nidulans. Cell Stress Chaperones. 2011;16:565–572. doi: 10.1007/s12192-011-0267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajic O, Buist G, Kojic M, Topisirovic L, Kuipers OP, Kok J. Novel mechanism of bacteriocin secretion and immunity carried out by Lactococcal multidrug resistance proteins. J Biol Chem. 2003;278:34291–34298. doi: 10.1074/jbc.M211100200. [DOI] [PubMed] [Google Scholar]
- Garvie EI. Bacterial lactate dehydrogenases. Microbiol Rev. 1980;44:106–139. doi: 10.1128/mr.44.1.106-139.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George HA, Johnson JL, Moore WEC, Holdeman LV, Chen JS. Acetone, isopropanol, and butanol production by Clostridium beijerinckii (syn. Clostridium butylicum) and Clostridium aurantibutyricum. Appl Environ Microbiol. 1983;45:1160–1163. doi: 10.1128/aem.45.3.1160-1163.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubev VI, Churkina LG. Intrageneric killing patterns of Rhodotorula mucilaginosa mycocins. Izv Akad Nauk Ser Biol. 1993;(4):550–557. [Google Scholar]
- Golubev VI, Churkina LG, Seregina SA. Intergeneric spectra of action of the Rhodotorula mucilaginosa mycocins. Izv Akad Nauk Ser Biol. 1996;(5):523–529. [Google Scholar]
- Görner W, Durchschlag E, Martinez-Pastor MT, Estruch F, Ammerer G, Hamilton B, et al. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostinčar C, Grube M, de Hoog S, Zalar P, Gunde-Cimerman N. Extremotolerance in fungi: evolution on the edge. FEMS Microbiol Ecol. 2010;71:2–11. doi: 10.1111/j.1574-6941.2009.00794.x. [DOI] [PubMed] [Google Scholar]
- Gostinčar C, Gunde-Cimerman N, Turk M. Genetic resources of extremotolerant fungi: a method for identification of genes conferring stress tolerance. Bioresour Technol. 2012;111:360–367. doi: 10.1016/j.biortech.2012.02.039. [DOI] [PubMed] [Google Scholar]
- Goyal R, Das S, Arora A, Aggarwal A. Rhodotorula mucilaginosa as a cause of persistent femoral nonunion. J Postgrad Med. 2008;54:25–27. doi: 10.4103/0022-3859.39186. [DOI] [PubMed] [Google Scholar]
- Grant WD. Life at low water activity. Philos Trans R Soc Lond B Biol Sci. 2004;359:1249–1266. doi: 10.1098/rstb.2004.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillot A, Gitton C, Anglade P, Mistou M-Y. Proteomic analysis of Lactococcus lactis, a lactic acid bacterium. Proteomics. 2003;3:337–354. doi: 10.1002/pmic.200390047. [DOI] [PubMed] [Google Scholar]
- Guillou V, Plourde-Owobi L, Parrou JL, Goma G, François J. Role of reserve carbohydrates in the growth dynamics of Saccharomyces cerevisiae. FEMS Yeast Res. 2004;4:773–787. doi: 10.1016/j.femsyr.2004.05.005. [DOI] [PubMed] [Google Scholar]
- Gulati A, Sharma N, Vyas P, Sood S, Rahi P, Pathania V, et al. Organic acid production and plant growth promotion as a function of phosphate solubilization by Acinetobacter rhizosphaerae strain BIHB 723 isolated from the cold deserts of the trans-Himalayas. Arch Microbiol. 2010;192:975–983. doi: 10.1007/s00203-010-0615-3. [DOI] [PubMed] [Google Scholar]
- Gunde-Cimerman N, Zalar P, de Hoog S, Plemenitas A. Hypersaline waters in salterns – natural ecological niches for halophilic black yeasts. FEMS Microbiol Ecol. 2000;32:235–240. doi: 10.1111/j.1574-6941.2000.tb00716.x. [DOI] [PubMed] [Google Scholar]
- Hallsworth JE. Ethanol-induced water stress in yeast. J Ferment Bioeng. 1998;85:125–137. [Google Scholar]
- Hallsworth JE, Magan N. Effect of carbohydrate type and concentration on polyols and trehalose in conidia of three entomopathogenic fungi. Microbiology. 1994a;140:2705–2713. [Google Scholar]
- Hallsworth JE, Magan N. Improved biological control by changing polyols/trehalose in conidia of entomopathogens. In: McKim FM, editor. Brighton Crop Protection Council – Pests And Diseases 1994. Farnham, Surrey, UK: British Crop Protection Council; 1994b. pp. 1091–1096. [Google Scholar]
- Hallsworth JE, Magan N. Effects of KCl concentration on accumulation of acyclic sugar alcohols and trehalose in conidia of three entomopathogenic fungi. Lett Appl Microbiol. 1994c;18:8–11. [Google Scholar]
- Hallsworth JE, Magan N. Manipulation of intracellular glycerol and erythritol enhances germination of conidia at low water availability. Microbiology. 1995;141:1109–1115. doi: 10.1099/13500872-141-5-1109. [DOI] [PubMed] [Google Scholar]
- Hallsworth JE, Nomura Y. A simple method to determine the water activity of ethanol-containing samples. Biotechnol Bioeng. 1999;62:242–245. doi: 10.1002/(sici)1097-0290(19990120)62:2<242::aid-bit15>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Hallsworth JE, Nomura Y, Iwahara M. Ethanol-induced water stress and fungal growth. J Ferment Bioeng. 1998;86:451–456. [Google Scholar]
- Hallsworth JE, Heim S, Timmis KN. Chaotropic solutes cause water stress in Pseudomonas putida. Environ Microbiol. 2003a;5:1270–1280. doi: 10.1111/j.1462-2920.2003.00478.x. [DOI] [PubMed] [Google Scholar]
- Hallsworth JE, Prior BA, Nomura Y, Iwahara M, Timmis KN. Compatible solutes protect against chaotrope (ethanol)-induced, nonosmotic water stress. Appl Environ Microbiol. 2003b;69:7032–7034. doi: 10.1128/AEM.69.12.7032-7034.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallsworth JE, Yakimov MM, Golyshin PN, Gillion JLM, D'Auria G, Alves FDL, et al. Limits of life in MgCl2-containing environments: chaotropicity defines the window. Environ Microbiol. 2007;9:801–813. doi: 10.1111/j.1462-2920.2006.01212.x. [DOI] [PubMed] [Google Scholar]
- Hartmans S, Smits JP, van der Werf MJ, Volkering F, de Bont JAM. Metabolism of styrene oxide and 2-phenylethanol in the styrene-degrading Xanthobacter strain 124X. Appl Environ Microbiol. 1989;55:2850–2855. doi: 10.1128/aem.55.11.2850-2855.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan MN, Afghan S, Hafeez FY. Biological control of red rot in sugarcane by native pyoluteorin-producing Pseudomonas putida strain NH-50 under field conditions and its potential modes of action. Pest Manag Sci. 2011;67:1147–1154. doi: 10.1002/ps.2165. [DOI] [PubMed] [Google Scholar]
- Hellström AM, Almgren A, Carlsson N-G, Svanberg U, Andlid TA. Degradation of phytate by Pichia kudriavzevii TY13 and Hanseniaspora guilliermondii TY14 in Tanzanian togwa. Int J Food Microbiol. 2012;153:73–77. doi: 10.1016/j.ijfoodmicro.2011.10.018. [DOI] [PubMed] [Google Scholar]
- Hentzer M, Teitzel GM, Balzer GJ, Heydorn A, Molin S, Givskov M, et al. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J Bacteriol. 2001;183:5395–5401. doi: 10.1128/JB.183.18.5395-5401.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero M, Ibáñez E, Cifuentes A, Reglero G, Santoyo S. Dunaliella salina microalga pressurized liquid extracts as potential antimicrobials. J Food Prot. 2006;69:2471–2477. doi: 10.4315/0362-028x-69.10.2471. [DOI] [PubMed] [Google Scholar]
- Hill TA. The Biology of Weeds. London, UK: E. Arnold; 1977. [Google Scholar]
- Hirasawa T, Yoshikawa K, Nakakura Y, Nagahisa K, Furusawa C, Katakura Y, et al. Identification of target genes conferring ethanol stress tolerance to Saccharomyces cerevisiae based on DNA microarray data analysis. J Bacteriol. 2007;131:34–44. doi: 10.1016/j.jbiotec.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Hirsch P. Microbial life at extremely low nutrient levels. Adv Space Res. 1986;6:287–298. doi: 10.1016/0273-1177(86)90097-9. [DOI] [PubMed] [Google Scholar]
- Hocking AD. Responses of xerophilic fungi to changes in water activity. In: Jennings DH, editor. Stress Tolerance of Fungi. New York, USA: Marcel Decker; 1993. pp. 233–256. [Google Scholar]
- Hoffland E, Kuyper TW, Wallander H, Plassard C, Gorbushina AA, Haselwandter K, et al. The role of fungi in weathering. Front Ecol Environ. 2004;2:258–264. [Google Scholar]
- Hong J, Tamaki H, Akiba S, Yamamoto K, Kumagai H. Cloning of a gene encoding a highly stable endo-β-1,4-glucanase from Aspergillus niger and its expression in yeast. J Biosci Bioeng. 2001;92:434–441. doi: 10.1263/jbb.92.434. [DOI] [PubMed] [Google Scholar]
- Horinouchi T, Tamaoka K, Furusawa C, Ono N, Suzuki S, Hirasawa T, et al. Transcriptome analysis of parallel-evolved Escherichia coli strains under ethanol stress. BMC Genomics. 2010;11:579. doi: 10.1186/1471-2164-11-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter PJ, Hand P, Pink D, Whipps JM, Bending GD. Both leaf properties and microbe-microbe interactions influence within-species variation in bacterial population diversity and structure in the lettuce (Lactuca species) phyllosphere. Appl Environ Microbiol. 2010;76:8117–8125. doi: 10.1128/AEM.01321-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inderjit, Evans H, Crocoll C, Bajpai D, Kaur R, Feng YL, et al. Volatile chemicals from leaf litter are associated with invasiveness of a Neotropical weed in Asia. Ecology. 2011;92:316–324. doi: 10.1890/10-0400.1. [DOI] [PubMed] [Google Scholar]
- Ingram LO, Buttke TM. Effects of alcohols on microorganisms. Adv Microb Physiol. 1984;25:253–300. doi: 10.1016/s0065-2911(08)60294-5. [DOI] [PubMed] [Google Scholar]
- Izgü F, Altinbay D, Sertkaya A. Enzymic activity of the K5-type yeast killer toxin and its characterization. Biosci Biotechnol Biochem. 2005;69:2200–2206. doi: 10.1271/bbb.69.2200. [DOI] [PubMed] [Google Scholar]
- Jahid IK, Silva AJ, Benitez JA. Polyphosphate stores enhance the ability of Vibrio cholerae to overcome environmental stresses in a low-phosphate environment. Appl Environ Microbiol. 2006;72:7043–7049. doi: 10.1128/AEM.00924-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang M-H, Ha K, Joo G-J, Takamura N. Toxin production of cyanobacteria is increased by exposure to zooplankton. Freshw Biol. 2003;48:1540–1550. [Google Scholar]
- Jiménez JI, Miñambres B, García JL, Díaz E. Genomic analysis of the aromatic catabolic pathways from Pseudomonas putida KT2440. Environ Microbiol. 2002;4:824–841. doi: 10.1046/j.1462-2920.2002.00370.x. [DOI] [PubMed] [Google Scholar]
- Jorge RMF, Livingston AG. A novel method for characterisation of microbial growth kinetics on volatile organic compounds. Appl Microbiol Biotechnol. 1999;52:174–178. [Google Scholar]
- Jüttner F. Evidence that polyunsaturated aldehydes of diatoms are repellents for pelagic crustacean grazers. Aquat Ecol. 2005;39:271–282. [Google Scholar]
- Jüttner F, Watson SB, von Elert E, Koster O. β-Cyclocitral, a grazer defence signal unique to the cyanobacterium Microcystis. J Chem Ecol. 2010;36:1387–1397. doi: 10.1007/s10886-010-9877-0. [DOI] [PubMed] [Google Scholar]
- Kachalkin AV, Yurkov AM. Yeast communities in Sphagnum phyllosphere along the temperature-moisture ecocline in the boreal forest-swamp ecosystem and description of Candida sphagnicola sp. nov. Antonie Van Leeuwenhoek. 2012;102:29–43. doi: 10.1007/s10482-012-9710-6. [DOI] [PubMed] [Google Scholar]
- Kadouri D, Jurkevitch E, Okon Y, Castro-Sowinski S. Ecological and agricultural significance of bacterial polyhydroxyalkanoates. Crit Rev Microbiol. 2005;31:55–67. doi: 10.1080/10408410590899228. [DOI] [PubMed] [Google Scholar]
- Kaino T, Takagi H. Gene expression profiles and intracellular contents of stress protectants in Saccharomyces cerevisiae under ethanol and sorbitol stresses. Appl Microbiol Biotechnol. 2008;79:273–283. doi: 10.1007/s00253-008-1431-4. [DOI] [PubMed] [Google Scholar]
- Kanekar SP, Kanekar PP, Sarnaik SS, Gujrathi NP, Shede PN, Kedargol MR, Reardon KF. Bioremediation of nitroexplosive wastewater by an yeast isolate Pichia sydowiorum MCM Y-3 in fixed film bioreactor. J Ind Microbiol Biotechnol. 2009;36:253–260. doi: 10.1007/s10295-008-0493-8. [DOI] [PubMed] [Google Scholar]
- Karlin S, Mrázek J, Campbell A, Kaiser D. Characterizations of highly expressed genes of four fast-growing bacteria. J Bacteriol. 2001;183:5025–5040. doi: 10.1128/JB.183.17.5025-5040.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashangura C, Hallsworth JE, Mswaka AY. Phenotypic diversity amongst strains of Pleurotus sajor-caju: implications for cultivation in arid environments. Mycol Res. 2006;110:312–317. doi: 10.1016/j.mycres.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Kazda J, Falkinham JO., III . Mycobacteria in sphagnum, peats and potting soils. In: Kazda J, Pavlik I, Falkinham JO III, Hruska K, editors. The Ecology of Mycobacteria: Impact on Animal's and Human's Health. Heidelberg: Springer; 2009. pp. 89–95. [Google Scholar]
- Kets EPW, Galinski EA, de Wit M, de Bont JAM, Heipieper HJ. Mannitol, a novel bacterial compatible solute in Pseudomonas putida S12. J Bacteriol. 1996;178:6665–6670. doi: 10.1128/jb.178.23.6665-6670.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khemakhem H, Elloumi J, Moussa M, Aleya L, Ayadi H. The concept of ecological succession applied to phytoplankton over four consecutive years in five ponds featuring a salinity gradient. Estuar Coast Shelf Sci. 2010;88:33–44. [Google Scholar]
- Kiran GS, Hema TA, Gandhimathi R, Selvin J, Thomas TA, Rajeetha Ravji T, et al. Optimization and production of a biosurfactant from the sponge-associated marine fungus Aspergillus ustus MSF3. Colloids Surf B Biointerfaces. 2009;73:250–256. doi: 10.1016/j.colsurfb.2009.05.025. [DOI] [PubMed] [Google Scholar]
- Klappenbach JA, Dunbar JM, Schmidt TM. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol. 2000;66:1328–1333. doi: 10.1128/aem.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Klei IJ, Harder W, Veenhuis M. Biosynthesis and assembly of alcohol oxidase, a peroxisomal matrix protein in methylotrophic yeasts: a review. Yeast. 1991;7:195–209. doi: 10.1002/yea.320070302. [DOI] [PubMed] [Google Scholar]
- Klinke T, Kneist S, de Soet JJ, Kuhlisch E, Mauersberger S, Förster A, et al. Acid production by oral strains of Candida albicans and Lactobacilli. Caries Res. 2009;43:83–91. doi: 10.1159/000204911. [DOI] [PubMed] [Google Scholar]
- Koch AL. Oligotrophs versus copiotrophs. Bioessays. 2001;23:657–661. doi: 10.1002/bies.1091. [DOI] [PubMed] [Google Scholar]
- Kolaczkowski M, van der Rest M, Cybularz-Kolaczkowska A, Soumillion J-P, Konings WN, Goffeau A. Anticancer drugs, ionophoric peptides, and steroids as substrates of the yeast multidrug transporter Pdr5p. J Biol Chem. 1996;271:31543–31548. doi: 10.1074/jbc.271.49.31543. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Meinke D. The development of Arabidopsis as a model plant. Plant J. 2010;61:909–921. doi: 10.1111/j.1365-313X.2009.04086.x. [DOI] [PubMed] [Google Scholar]
- Kotyk A, Lapathitis G, Krenková S. Glucose- and K+-induced acidification in different yeast species. Folia Microbiol (Praha) 1999;44:295–298. doi: 10.1007/BF02818550. [DOI] [PubMed] [Google Scholar]
- Krallish I, Gonta S, Savenkova L, Bergauer P, Margesin R. Phenol degradation by immobilized cold-adapted yeast strains of Cryptococcus terreus and Rhodotorula creatinivora. Extremophiles. 2006;10:441–449. doi: 10.1007/s00792-006-0517-0. [DOI] [PubMed] [Google Scholar]
- Krappmann S, Braus GH. Nitrogen metabolism of Aspergillus and its role in pathogenicity. Med Mycol. 2005;43:S31–S40. doi: 10.1080/13693780400024271. (Suppl. 1): [DOI] [PubMed] [Google Scholar]
- Kruijt M, Tran H, Raaijmakers JM. Functional, genetic and chemical characterization of biosurfactants produced by plant growth-promoting Pseudomonas putida 267. J Appl Microbiol. 2009;107:546–556. doi: 10.1111/j.1365-2672.2009.04244.x. [DOI] [PubMed] [Google Scholar]
- Kushner DJ, editor. Microbial Life in Extreme Environments. London, UK: Academic Press; 1978. Life at high salt and solute concentrations: halophilic bacteria; pp. 318–368. [Google Scholar]
- Lahav R, Fareleira P, Nejidat A, Abeliovich A. The identification and characterization of osmotolerant yeast isolates from chemical wastewater evaporation ponds. Microb Ecol. 2002;43:388–396. doi: 10.1007/s00248-002-2001-4. [DOI] [PubMed] [Google Scholar]
- Lahav R, Nejidat A, Abeliovich A. Alterations in protein synthesis and levels of heat shock 70 proteins in response to salt stress of the halotolerant yeast Rhodotorula mucilaginosa. Antonie Van Leeuwenhoek. 2004;85:259–269. doi: 10.1023/B:ANTO.0000020361.81006.2b. [DOI] [PubMed] [Google Scholar]
- Lambou K, Lamarre C, Beau R, Dufour N, Latge JP. Functional analysis of the superoxide dismutase family in Aspergillus fumigatus. Mol Microbiol. 2010;75:910–923. doi: 10.1111/j.1365-2958.2009.07024.x. [DOI] [PubMed] [Google Scholar]
- Langaee TY, Gagnon L, Huletsky A. Inactivation of the ampD Gene in Pseudomonas aeruginosa leads to moderate-basal-level and hyperinducible AmpC β-lactamase expression. Antimicrob Agents Chemother. 2000;44:583–589. doi: 10.1128/aac.44.3.583-589.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bihan T, Martin SF, Chirnside ES, van Ooijen G, Barrios-LLerena ME, O'Neill JS, et al. Shotgun proteomic analysis of the unicellular alga Ostreococcus tauri. J Proteomics. 2011;74:2060–2070. doi: 10.1016/j.jprot.2011.05.028. [DOI] [PubMed] [Google Scholar]
- Leão PN, Ramos V, Vale M, Machado JP, Vasconcelos VM. Microbial community changes elicited by exposure to cyanobacterial allelochemicals. Microb Ecol. 2012;63:85–95. doi: 10.1007/s00248-011-9939-z. [DOI] [PubMed] [Google Scholar]
- Lee GD, Shin SY, Maeng C-Y, Jin ZZ, Kim KL, Hahm K-S. Isolation and characterization of a novel antifungal peptide from Aspergillus niger. Biochem Biophys Res Commun. 1999;263:646–651. doi: 10.1006/bbrc.1999.1428. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Choi YR, Lee SY, Park JT, Shim JH, Park KH, et al. Screening wild yeast strains for alcohol fermentation from various Fruits. Mycobiology. 2011;39:33–39. doi: 10.4489/MYCO.2011.39.1.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong SL, Pettersson OV, Rice T, Hocking AD, Schnürer J. The extreme xerophilic mould Xeromyces bisporus – growth and competition at various water activities. Int J Food Microbiol. 2011;145:57–63. doi: 10.1016/j.ijfoodmicro.2010.11.025. [DOI] [PubMed] [Google Scholar]
- Lesuisse E, Blaiseau PL, Dancis A, Camadro JM. Siderophore uptake and use by the yeast Saccharomyces cerevisiae. Microbiology. 2001;147:289–298. doi: 10.1099/00221287-147-2-289. [DOI] [PubMed] [Google Scholar]
- Levine DW, Cooney CL. Isolation and characterization of a thermotolerant methanol-utilizing yeast. Appl Microbiol. 1973;26:982–990. doi: 10.1128/am.26.6.982-990.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman LM, Sozzani R, Benfey PN. Integrative systems biology: an attempt to describe a simple weed. Curr Opin Plant Biol. 2012;15:162–167. doi: 10.1016/j.pbi.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libkind D, Sampaio JP. Rhodotorula. In: Liu D, editor. Molecular Detection of Foodborne Pathogens. Florida, USA: CRC Press; 2010. pp. 603–618. [Google Scholar]
- Lifshits M, Carmeli S. Metabolites of Microcystis aeruginosa bloom material from Lake Kinneret, Israel. J Nat Prod. 2012;75:209–219. doi: 10.1021/np200909x. [DOI] [PubMed] [Google Scholar]
- Lillie SH, Pringle JR. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J Bacteriol. 1980;143:1384–1394. doi: 10.1128/jb.143.3.1384-1394.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisichkina GA, Bab'eva IP, Dlu S. Alkalitolerant yeasts from natural biotopes. Mikrobiologia. 2003;72:695–698. [PubMed] [Google Scholar]
- Liu I-C, Whang L-M, Ren W-J, Lin P-Y. The effect of pH on the production of biohydrogen by clostridia: thermodynamic and metabolic considerations. Int J Hydrogen Energy. 2011;36:439–449. [Google Scholar]
- Liu J, Nikaido H. A mutant of Mycobacterium smegmatis defective in the biosynthesis of mycolic acids accumulates meromycolates. Proc Natl Acad Sci USA. 1999;96:4011–4016. doi: 10.1073/pnas.96.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Y-C, Lu W-C, Chen C-Y, Chen W-M, Chang J-S. Characterization and high-level production of xylanase from an indigenous cellulolytic bacterium Acinetobacter junii F6-02 from southern Taiwan soil. Biochem Eng J. 2010;53:77–84. [Google Scholar]
- Lo Nostro P, Ninham BW, Lo Nostro A, Pesavento G, Fratoni L, Baglioni P. Specific ion effects on the growth rates of Staphylococcus aureus and Pseudomonas aeruginosa. Phys Biol. 2005;2:1–7. doi: 10.1088/1478-3967/2/1/001. [DOI] [PubMed] [Google Scholar]
- Loaces I, Ferrando L, Scavino AF. Dynamics, diversity and function of endophytic siderophore-producing bacteria in rice. Microb Ecol. 2011;61:606–618. doi: 10.1007/s00248-010-9780-9. [DOI] [PubMed] [Google Scholar]
- Lobasso S, Lopalco P, Vitale R, Saponetti MS, Capitanio G, Mangini V, et al. The light-activated proton pump Bop I of the archaeon Haloquadratum walsbyi. Photochem Photobiol. 2012;88:690–700. doi: 10.1111/j.1751-1097.2012.01089.x. [DOI] [PubMed] [Google Scholar]
- Long HC. Weeds of Grass Land. London, UK: H.M.S.O; 1932. [Google Scholar]
- Louvel B, Cébron A, Leyval C. Root exudates affect phenanthrene biodegradation, bacterial community and functional gene expression in sand microcosms. Int Biodeter Biodegr. 2011;65:947–953. [Google Scholar]
- Lowe DP, Arendt EK. The use and effects of lactic acid bacteria in malting and brewing with their relationships to antifungal activity, mycotoxins and gushing: a review. J Inst Brew. 2004;110:163–180. [Google Scholar]
- Ma M, Liu LZ. Quantitative transcription dynamic analysis reveals candidate genes and key regulators for ethanol tolerance in Saccharomyces cerevisiae. BMC Microbiol. 2010;10:169. doi: 10.1186/1471-2180-10-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur JV. Microbial Ecology: An Evolutionary Approach. Oxford, UK: Elsevier; 2006. [Google Scholar]
- McCammick EM, Gomase VS, Timson DJ, McGenity TJ, Hallsworth JE. Water-hydrophobic compound interactions with the microbial cell. In: Timmis KN, editor. Handbook of Hydrocarbon and Lipid Microbiology – Hydrocarbons, Oils and Lipids: Diversity, Properties and Formation. Vol. 2. New York, USA: Springer; 2009. pp. 1451–1466. [Google Scholar]
- McDonald IR, Murrell JC. The methanol dehydrogenase structural gene mxaF and its use as a functional gene probe for methanotrophs and methylotrophs. Appl Environ Microbiol. 1997;63:3218–3224. doi: 10.1128/aem.63.8.3218-3224.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell G, Russell AD. Antiseptics and disinfectants: activity, action, and resistance. Clin Microbiol Rev. 1999;12:147–179. doi: 10.1128/cmr.12.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack RN, Simberloff D, Lonsdale WM, Evans H, Clout M, Bazzaz F. Biotic invasions: causes, epidemiology, global consequences, and control. Ecol Appl. 2000;10:689–710. [Google Scholar]
- McNerney R, Mallard K, Okolo PI, Turner C. Production of volatile organic compounds by mycobacteria. FEMS Microbiol Lett. 2012;328:150–156. doi: 10.1111/j.1574-6968.2011.02493.x. [DOI] [PubMed] [Google Scholar]
- Magan N, Hope R, Cairns V, Aldred D. Post-harvest fungal ecology: impact of fungal growth and mycotoxin accumulation in stored grain. Eur J Plant Pathol. 2003;109:723–730. [Google Scholar]
- Mansure JJC, Panek AD, Crowe LM, Crowe JH. Trehalose inhibits ethanol effects on intact yeast cells and liposomes. Biochim Biophys Acta. 1994;1191:309–316. doi: 10.1016/0005-2736(94)90181-3. [DOI] [PubMed] [Google Scholar]
- Margesin R, Labbe D, Schinner F, Greer CW, Whyte LG. Characterization of hydrocarbon-degrading microbial populations in contaminated and pristine alpine soils. Appl Environ Microbiol. 2003;69:3085–3092. doi: 10.1128/AEM.69.6.3085-3092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz C, Bergfeld T, Rice SA, Kjelleberg S. Microcolonies, quorum sensing and cytotoxicity determine the survival of Pseudomonas aeruginosa biofilms exposed to protozoan grazing. Environ Microbiol. 2004;6:218–226. doi: 10.1111/j.1462-2920.2004.00556.x. [DOI] [PubMed] [Google Scholar]
- Mavrodi DV, Blankenfeldt W, Thomashow LS. Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu Rev Phytopathol. 2006;44:417–445. doi: 10.1146/annurev.phyto.44.013106.145710. [DOI] [PubMed] [Google Scholar]
- Meagher MM, Tao BY, Chow JM, Reilly PJ. Kinetics and subsite mapping of a d-xylobiose- and d-xylose-producing Aspergillus niger endo-(1→4)-β-d-xylanase. Carbohydr Res. 1988;173:273–283. doi: 10.1016/s0008-6215(00)90823-1. [DOI] [PubMed] [Google Scholar]
- Meksawat S, Pornprom T. Allelopathic effect of itchgrass (Rottboellia cochinchinensis) on seed germination and plant growth. Weed Biol Manag. 2010;10:16–24. [Google Scholar]
- Middelhoven WJ, de Waard MA, Mulder EG. The ferrous ion as the cofactor of arginase in vivo. II. Experiments on the replacement of ferrous ions in native yeast arginase by other cations in vivo. Biochim Biophys Acta. 1969;191:122–129. doi: 10.1016/0005-2744(69)90321-0. [DOI] [PubMed] [Google Scholar]
- Minerdi D, Bossi S, Gullino ML, Garibaldi A. Volatile organic compounds: a potential direct long-distance mechanism for antagonistic action of Fusarium oxysporum strain MSA 35. Environ Microbiol. 2009;11:844–854. doi: 10.1111/j.1462-2920.2008.01805.x. [DOI] [PubMed] [Google Scholar]
- Minnikin DE, Abdolrahimzadeh H, Baddiley J. Replacement of acidic phosphates by acidic glycolipids in Pseudomonas diminuta. Nature. 1974;249:268–269. doi: 10.1038/249268a0. [DOI] [PubMed] [Google Scholar]
- Mlouka A, Comte K, Castets AM, Bouchier C, de Marsac N. The gas vesicle gene cluster from Microcystis aeruginosa and DNA rearrangements that lead to loss of cell buoyancy. J Bacteriol. 2004;186:2355–2365. doi: 10.1128/JB.186.8.2355-2365.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moliné M, Flores MR, Libkind D, Diéguez MDC, Farías ME, van Broock M. Photoprotection by carotenoid pigments in the yeast Rhodotorula mucilaginosa: the role of torularhodin. Photochem Photobiol Sci. 2010;9:1145–1151. doi: 10.1039/c0pp00009d. [DOI] [PubMed] [Google Scholar]
- Molly K, DeSmet I, Nollet L, Vande Woestyne M, Verstraete W. Effect of lactobacilli on the ecology of the gastro-intestinal microbiota cultured in the SHIME reactor. Microb Ecol Health. 1996;9:79–89. [Google Scholar]
- Mongodin EF, Nelson KE, Daugherty S, DeBoy RT, Wister J, Khouri H, et al. The genome of Salinibacter ruber: convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc Natl Acad Sci USA. 2005;102:18147–18152. doi: 10.1073/pnas.0509073102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales P, Fernández-García E, Nuñez M. Volatile compounds produced in cheese by Pseudomonas strains of dairy origin belonging to six different species. J Agric Food Chem. 2005;53:6835–6843. doi: 10.1021/jf050717b. [DOI] [PubMed] [Google Scholar]
- Morohoshi T, Oseki K, Ikeda T. Exopolysaccharide production is influenced by sugars, N-acylhomoserine, lactone, and transcriptional regulators RcsA and RcsB, but does not affect pathogenicity in the plant pathogen Pantoea ananatis. Biosci Biotechnol Biochem. 2011;75:997–999. doi: 10.1271/bbb.100888. [DOI] [PubMed] [Google Scholar]
- Moskvina E, Schüller C, Maurer CTC, Mager WH, Ruis H. A search in the genome of Saccharomyces cerevisiae for genes regulated via stress response elements. Yeast. 1998;14:1041–1050. doi: 10.1002/(SICI)1097-0061(199808)14:11<1041::AID-YEA296>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Muñoz J, Mudge SM, Sandoval A. Effects of ionic strength on the production of short chain volatile hydrocarbons by Dunaliella salina (Teodoresco) Chemosphere. 2004;54:1267–1271. doi: 10.1016/j.chemosphere.2003.09.019. [DOI] [PubMed] [Google Scholar]
- Nair S, Finkel SE. Dps protects cells against multiple stresses during stationary phase. J Bacteriol. 2004;186:4192–4198. doi: 10.1128/JB.186.13.4192-4198.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narihiro T, Sekiguchi Y. Microbial communities in anaerobic digestion processes for waste and wastewater treatment: a microbiological update. Curr Opin Biotechnol. 2007;18:273–278. doi: 10.1016/j.copbio.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Naumov GI, Naumova ES, Schnürer J. Genetic characterization of the nonconventional yeast Hansenula anomala. Res Microbiol. 2001;152:551–562. doi: 10.1016/s0923-2508(01)01229-3. [DOI] [PubMed] [Google Scholar]
- Newton RJ, Kent AD, Triplett EW, McMahon KD. Microbial community dynamics in a humic lake: differential persistence of common freshwater phylotypes. Environ Microbiol. 2006;8:956–970. doi: 10.1111/j.1462-2920.2005.00979.x. [DOI] [PubMed] [Google Scholar]
- Nishino K, Yamaguchi A. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J Bacteriol. 2001;183:5803–5812. doi: 10.1128/JB.183.20.5803-5812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisiotou AA, Spiropoulos AE, Nychas GE. Yeast community structures and dynamics in healthy and Botrytis-affected grape must fermentations. Appl Environ Microbiol. 2007;73:6705–6713. doi: 10.1128/AEM.01279-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisiotou AA, Chorianopoulos N, Nychas G-JE, Panagou EZ. Yeast heterogeneity during spontaneous fermentation of black Conservolea olives in different brine solutions. J Appl Microbiol. 2010;108:396–405. doi: 10.1111/j.1365-2672.2009.04424.x. [DOI] [PubMed] [Google Scholar]
- Nitschke M, Costa SGVAO, Contiero J. Rhamnolipids and PHAs: recent reports on Pseudomonas-derived molecules of increasing industrial interest. Process Biochem. 2011;46:621–630. [Google Scholar]
- Nomura Y, Hallsworth JE, Iwahara M, Tanaka T, Ishizaki A. Rapid and effective production of L-lactate from xylose using electrodialysis culture-associated product separation. World J Microb Biot. 1998;14:911–916. [Google Scholar]
- Nordström K, Dasgupta S. Copy-number control of the Escherichia coli chromosome: a plasmidologist's view. EMBO Rep. 2006;7:484–489. doi: 10.1038/sj.embor.7400681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak-Thompson B, Chaney N, Wing JS, Gould SJ, Loper JE. Characterization of the pyoluteorin biosynthetic gene cluster of Pseudomonas fluorescens Pf-5. J Bacteriol. 1999;181:2166–2174. doi: 10.1128/jb.181.7.2166-2174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes FMN, Veloso MCC, Pereira PAP, de Andrade JB. Gas-phase ozonolysis of the monoterpenoids (S)-(+)-carvone, (R)-(−)-carvone, (−)-carveol, geraniol and citral. Atmos Environ. 2005;39:7715–7730. [Google Scholar]
- Oatham MP. Water relations of Zygophyllum hamienseHeliotropium kotschyi and Panicum turgidum and their response to rangeland management techniques in Abu Dhabi. J Arid Environ. 1997;35:95–110. [Google Scholar]
- Ogino A, Koshikawa H, Nakahara T, Uchiyama H. Succession of microbial communities during a biostimulation process as evaluated by DGGE and clone library analyses. J Appl Microbiol. 2001;91:625–635. doi: 10.1046/j.1365-2672.2001.01424.x. [DOI] [PubMed] [Google Scholar]
- Olstorpe M, Passoth M. Pichia anomala in grain preservation. Antonie Van Leeuwenhoek. 2011;99:57–62. doi: 10.1007/s10482-010-9497-2. [DOI] [PubMed] [Google Scholar]
- Oren A. Bioenergetic aspects of halophilism. Microbiol Mol Biol Rev. 1999;63:334–338. doi: 10.1128/mmbr.63.2.334-348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A, Heldal M, Norland S, Galinski EA. Intracellular ion and organic solute concentrations of the extremely halophilic bacterium Salinibacter ruber. Extremophiles. 2002;6:491–498. doi: 10.1007/s00792-002-0286-3. [DOI] [PubMed] [Google Scholar]
- Ozaki K, Ohta A, Iwata C, Horikawa A, Tsuji K, Ito E, et al. Lysis of cyanobacteria with volatile organic compounds. Chemosphere. 2008;71:1531–1538. doi: 10.1016/j.chemosphere.2007.11.052. [DOI] [PubMed] [Google Scholar]
- Pacheco CC, Passos JF, Moradas-Ferreira P, De Marco P. Strain PM2, a novel methylotrophic fluorescent Pseudomonas sp. FEMS Microbiol Lett. 2003;227:279–285. doi: 10.1016/S0378-1097(03)00692-X. [DOI] [PubMed] [Google Scholar]
- Pan F, Yang Q, Zhang Y, Zhang S, Yang M. Biodegradation of polycyclic aromatic hydrocarbons by Pichia anomala. Biotechnol Lett. 2004;26:803–806. doi: 10.1023/b:bile.0000025882.33234.91. [DOI] [PubMed] [Google Scholar]
- Park W, Jeon CO, Cadillo H, DeRito C, Madsen EL. Survival of naphthalene-degrading Pseudomonas putida NCIB 9816-4 in naphthalene-amended soils: toxicity of naphthalene and its metabolites. Appl Microbiol Biotechnol. 2004;64:429–435. doi: 10.1007/s00253-003-1420-6. [DOI] [PubMed] [Google Scholar]
- Park HC, Bae YU, Cho SD, Kim SA, Moon JY, Ha KC, et al. Toluene-induced accumulation of trehalose by Pseudomonas sp. BCNU 106 through expression of otsA and otsB homologues. Lett Appl Microbiol. 2007;1:50–55. doi: 10.1111/j.1472-765X.2006.02036.x. [DOI] [PubMed] [Google Scholar]
- Parkinson N, Bryant R, Bew J, Elphinstone J. Rapid phylogenetic identification of members of the Pseudomonas syringae species complex using the rpoD locus. Plant Pathol. 2011;60:338–344. [Google Scholar]
- Parret AHA, Schoofs G, Proost P, Mot DM. Plant lectin-like bacteriocin from a rhizosphere-colonizing Pseudomonas isolate. J Bacteriol. 2003;185:897–908. doi: 10.1128/JB.185.3.897-908.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrou JL, Teste MA, François J. Effects of various types of stress on the metabolism of reserve carbohydrates in Saccharomyces cerevisiae: genetic evidence for a stress-induced recycling of glycogen and trehalose. Microbiology. 1997;143:1891–1900. doi: 10.1099/00221287-143-6-1891. [DOI] [PubMed] [Google Scholar]
- Passoth V, Fredlund E, Druvefors U, Schnürer J. Biotechnology, physiology and genetics of the yeast Pichia anomala. FEMS Yeast Res. 2006;6:3–13. doi: 10.1111/j.1567-1364.2005.00004.x. [DOI] [PubMed] [Google Scholar]
- Paul D, Pandey G, Meier C, van der Meer JR, Jain RK. Bacterial community structure of a pesticide-contaminated site and assessment of changes induced in community structure during bioremediation. FEMS Microbiol Ecol. 2006;57:116–127. doi: 10.1111/j.1574-6941.2006.00103.x. [DOI] [PubMed] [Google Scholar]
- Paz Y, Shimoni E, Weiss M, Pick U. Effects of iron deficiency on iron binding and internalization into acidic vacuoles in Dunaliella salina. Plant Physiol. 2007;144:1407–1415. doi: 10.1104/pp.107.100644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecoraro V, Zerulla K, Lange C, Soppa J. Quantification of ploidy in proteobacteria revealed the existence of monoploid, (mero-)oligoploid and polyploid species. PLoS ONE. 2011;6:e16392. doi: 10.1371/journal.pone.0016392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltomaa E, Ojala A. Size-related photosynthesis of algae in a strongly stratified humic lake. J Plankton Res. 2010;32:341–355. [Google Scholar]
- Perniola R, Faneschi ML, Manso E, Pizzolante M, Rizzo A, Sticchi Damiani A, et al. Rhodotorula mucilaginosa outbreak in neonatal intensive care unit: microbiological features, clinical presentation, and analysis of related variables. Eur J Clin Microbiol Infect Dis. 2006;25:193–196. doi: 10.1007/s10096-006-0114-2. [DOI] [PubMed] [Google Scholar]
- Pfeiffer T, Schuster S, Bonhoeffer S. Cooperation and competition in the evolution of ATP-producing pathways. Science. 2001;292:504–507. doi: 10.1126/science.1058079. [DOI] [PubMed] [Google Scholar]
- de Pina CG, Hogg TA. Microbial and chemical changes during the spontaneous ensilage of grape pomace. J Appl Microbiol. 1999;86:777–784. [Google Scholar]
- Piškur J, Rozpędowska E, Polakova S, Merico A, Compagno C. How did Saccharomyces evolve to become a good brewer? Trends Genet. 2006;22:183–186. doi: 10.1016/j.tig.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Pitt JI. Xerophilic fungi and the spoilage of foods of plant origin. In: Duckworth RB, editor. Water Relations of Foods. 1st edn. New York, USA: Academic Press; 1975. pp. 273–307. [Google Scholar]
- Pitt JI, Christian JHB. Water relations of xerophilic fungi isolated from prunes. Appl Microbiol. 1968;16:1853–1858. doi: 10.1128/am.16.12.1853-1858.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt JI, Hocking AD. Influence of solute and hydrogen-ion concentration on water relations of some xerophilic fungi. J Gen Microbiol. 1977;101:35–40. doi: 10.1099/00221287-101-1-35. [DOI] [PubMed] [Google Scholar]
- Poblete-Castro I, Becker J, Dohnt J, dos Santos VM, Wittman C. Industrial biotechnology of Pseudomonas putida and other related species. Appl Microbiol Biotechnol. 2012;93:2279–2290. doi: 10.1007/s00253-012-3928-0. [DOI] [PubMed] [Google Scholar]
- Pollegioni L, Piubelli L, Sacchi S, Pilone MS, Molla G. Physiological functions of d-amino acid oxidases: from yeast to humans. Cell Mol Life Sci. 2007;64:1373–1394. doi: 10.1007/s00018-007-6558-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praphailong W, Fleet GH. The effect of pH, sodium chloride, sucrose, sorbate and benzoate on the growth of food spoilage yeasts. Food Microbiol. 1997;14:459–468. [Google Scholar]
- Pretorius IS. Tailoring wine yeast for the new millennium: novel approaches to the ancient art of winemaking. Yeast. 2000;16:675–729. doi: 10.1002/1097-0061(20000615)16:8<675::AID-YEA585>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Puchałka J, Oberhardt MA, Godinho M, Bielecka A, Regenhardt D, Timmis KN, et al. Genome-scale reconstruction and analysis of the Pseudomonas putida KT2440 metabolic network facilitates applications in biotechnology. PLoS Comput Biol. 2008;4:e1000210. doi: 10.1371/journal.pcbi.1000210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purvis JE, Yomano LP, Ingram LO. Enhanced trehalose production improves growth of Escherichia coli under osmotic stress. Appl Environ Microbiol. 2005;71:3761–3769. doi: 10.1128/AEM.71.7.3761-3769.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragaert P, Devlieghere F, Loos S, Dewulf J, van Langenhove H, Foubert I, et al. Role of yeast proliferation in the quality degradation of strawberries during refrigerated storage. Int J Food Microbiol. 2006;108:42–50. doi: 10.1016/j.ijfoodmicro.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Rainieri S, Zambonelli C, Hallsworth JE, Pulvirenti A, Giudici P. Saccharomyces uvarum, a distinct group within Saccharomyces sensu stricto. FEMS Microbiol Lett. 1999;177:177–185. doi: 10.1111/j.1574-6968.1999.tb13729.x. [DOI] [PubMed] [Google Scholar]
- Rajala T, Peltoniemi M, Hantula J, Mäkipää R, Pennanen T. RNA reveals a succession of active fungi during the decay of Norway spruce logs. Fungal Ecol. 2011;4:437–448. [Google Scholar]
- Ralphs MH. Ecological relationships between poisonous plants and rangeland condition: a review. J Range Manag. 2002;55:285–290. [Google Scholar]
- Ramos JL, Duque E, Gallegos M-T, Godoy P, Ramos-González MI, Rojas A, et al. Mechanisms of solvent tolerance in Gram-negative bacteria. Annu Rev Microbiol. 2002;56:743–768. doi: 10.1146/annurev.micro.56.012302.161038. [DOI] [PubMed] [Google Scholar]
- Randazzo CL, Pitino I, Ribbera A, Caggia C. Pecorino Crotonese cheese: study of bacterial population and flavour compounds. Food Microbiol. 2010;27:363–374. doi: 10.1016/j.fm.2009.11.010. [DOI] [PubMed] [Google Scholar]
- Rao D, Webb JS, Kjelleberg S. Competitive interactions in mixed-species biofilms containing the marine bacterium Pseudoalteromonas tunicate. Appl Environ Microbiol. 2005;71:1729–1736. doi: 10.1128/AEM.71.4.1729-1736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raspor P, Milek DM, Polanc J, Možina SS, Čadež N. Yeasts isolated from three varieties of grapes cultivated in different locations of the Dolenjska vine-growing region, Slovenia. Int J Food Microbiol. 2006;109:97–102. doi: 10.1016/j.ijfoodmicro.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Reis RS, Pereira AG, Neves BC, Freire DMG. Gene regulation of rhamnolipid production in Pseudomonas aeruginosa – a review. Bioresour Technol. 2011;102:6377–6384. doi: 10.1016/j.biortech.2011.03.074. [DOI] [PubMed] [Google Scholar]
- Rengefors K, Pålsson C, Hansson L-A, Heiberg L. Cell lysis of competitors and osmotrophy enhance growth of the bloom-forming alga Gonyostomum semen. Aquat Microb Ecol. 2008;51:87–96. [Google Scholar]
- Renouf V, Claisse O, Lonvaud-Funel A. Understanding the microbial ecosystem on the grape berry surface through numeration and identification of yeast and bacteria. Aust J Grape Wine Res. 2005;11:316–327. [Google Scholar]
- Renshaw JC, Robson GD, Trinci APJ, Wiebe MG, Livens FR, Collison D, et al. Fungal siderophores: structures, functions and applications. Mycol Res. 2002;106:1123–1142. [Google Scholar]
- Restaino L, Bills S, Tscherneff K, Lenovich LM. Growth-characteristics of Saccharomyces rouxii isolated from chocolate syrup. Appl Environ Microbiol. 1983;45:1614–1621. doi: 10.1128/aem.45.5.1614-1621.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynisson E, Lauzon HL, Magnússon H, Jónsdóttir R, Ólafsdóttir G, Marteinsson V, et al. Bacterial composition and succession during storage of North-Atlantic cod (Gadus morhua) at superchilled temperatures. BMC Microbiol. 2009;9:250. doi: 10.1186/1471-2180-9-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Peña JM, García R, Nombela C, Arroyo J. The high-osmolarity glycerol (HOG) and cell wall integrity (CWI) signalling pathways interplay: a yeast dialogue between MAPK routes. Yeast. 2010;27:495–502. doi: 10.1002/yea.1792. [DOI] [PubMed] [Google Scholar]
- Roels JA. Energetics and Kinetics in Biotechnology. Amsterdam, the Netherlands: Elsevier University Press; 1983. [Google Scholar]
- Rohlfs M, Albert M, Keller NP, Kempken F. Secondary chemicals protect mould from fungivory. Biol Lett. 2007;3:523–525. doi: 10.1098/rsbl.2007.0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röling WFM, Milner MG, Jones DM, Fratepietro F, Swannell RPJ, Head IM. Bacterial community dynamics and hydrocarbon degradation during a field-scale evaluation of bioremediation on a mudflat beach contaminated with buried oil. Appl Environ Microbiol. 2004;70:2603–2613. doi: 10.1128/AEM.70.5.2603-2613.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa CA, Novak FR, Almeida JAG, Mendonça-Hagler LC, Hagler AN. Yeasts from human milk collected in Rio de Janeiro, Brazil. Rev Microbiol. 1990;21:361–363. [Google Scholar]
- Rose AH. Fermented Foods. New York, USA: Academic Press; 1982. [Google Scholar]
- Sadeghi AM, Starr JL, Teasdale JR, Rosecrance RC, Rowland RA. Real-time soil profile water content as influenced by weed-corn competition. Soil Sci. 2007;172:759–769. [Google Scholar]
- Salisbury E. Weeds and Aliens. London, UK: Collins; 1961. [Google Scholar]
- Salonen K, Rosenberg M. Advantages from diel vertical migration can explain the dominance of Gonyostomum semen (Raphidophyceae) in a small, steeply-stratified humic lake. J Plankton Res. 2000;22:1841–1853. [Google Scholar]
- Salvadó Z, Arroyo-López FN, Guillamón JM, Salazar G, Querol A, Barrio E. Temperature adaptation markedly determines evolution within the genus Saccharomyces. Appl Environ Microbiol. 2011;77:2292–2302. doi: 10.1128/AEM.01861-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Santos Sant'Ana G, Paes L, Vieira Paiva AF, Fietto LG, Totola AH, Trópia MJM, et al. Protective effect of ions against cell death induced by acid stress in Saccharomyces. FEMS Yeast Res. 2009;9:701–712. doi: 10.1111/j.1567-1364.2009.00523.x. [DOI] [PubMed] [Google Scholar]
- Santos PM, Benndorf D, Sá-Correia I. Insights into Pseudomonas putida KT2440 response to phenol- induced stress by quantitative proteomics. Proteomics. 2004;4:2640–2652. doi: 10.1002/pmic.200300793. [DOI] [PubMed] [Google Scholar]
- Saponetti MS, Bobba F, Salerno G, Scarfato A, Corcelli A, Cucolo A. Morphological and structural aspects of the extremely halophilic archaeon Haloquadratum walsbyi. PLoS ONE. 2011;6:e18653. doi: 10.1371/journal.pone.0018653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saum SH, Pfeiffer F, Palm P, Rampp M, Schuster SC, Müller V, et al. Chloride and organic osmolytes: a hybrid strategy to cope with elevated salinities by the moderately halophilic, chloride-dependent bacterium Halobacillus halophilus. Environ Microbiol. 2012 doi: 10.1111/j.1462-2920.2012.02770.x. doi: 10.1111/j.1462-2920.2012.02770.x. [DOI] [PubMed] [Google Scholar]
- Schnitzler A, Bailey J. Genetic polymorphism and phenotypic plasticity: two advantages for the dispersal of Japanese knotweeds. Rev Ecol. 2008;63:209–217. [Google Scholar]
- Segura A, Godoy P, van Dillewijn P, Hurtado A, Arroyo A, Santacruz S, Ramos JL. Proteomic analysis reveals the participation of energy- and stress- related proteins in the response of Pseudomonas putida DOT-T1E to toluene. J Bacteriol. 2005;187:5937–5945. doi: 10.1128/JB.187.17.5937-5945.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selezska K, Kazmiercszak M, Müsken M, Garbe J, Schobert M, Häussler S, et al. Pseudomonas aeruginosa population structure revisited under environmental focus: impact of water quality and phage pressure. Environ Microbiol. 2012;14:1952–1967. doi: 10.1111/j.1462-2920.2012.02719.x. [DOI] [PubMed] [Google Scholar]
- Senthilkumar K, Udaiyan K, Manian S. Successional pattern of mycoflora associated with litter degradation in a Cymbopogon caesius-dominated tropical grassland. Trop Grasslands. 1993;27:121–127. [Google Scholar]
- Seward R, Willetts JC, Dinsdale MG, Lloyd D. The effects of ethanol, hexan-1-ol, and 2-phenylethanol on cider yeast growth, viability, and energy status. J Inst Brew. 1996;102:439–443. [Google Scholar]
- Shi XL, Yang LY, Niu XJ, Xiao L, Kong ZM, Qin BQ, et al. Intracellular phosphorus metabolism of Microcystis aeruginosa under various redox potential in darkness. Microbiol Res. 2003;158:345–352. doi: 10.1078/0944-5013-00214. [DOI] [PubMed] [Google Scholar]
- Shukurov RR, Voblikova VD, Nikonorova AK, Komakhin RA, Komakhina VV, E-gorov TA, et al. Transformation of tobacco and Arabidopsis plants with Stellaria media genes encoding novel hevein-like peptides increases their resistance to fungal pathogens. Trans Res. 2012;21:313–325. doi: 10.1007/s11248-011-9534-6. [DOI] [PubMed] [Google Scholar]
- Sicard D, Legras J-L. Bread, beer and wine: yeast domestication in the Saccharomyces sensu stricto complex. C R Biol. 2011;334:229–236. doi: 10.1016/j.crvi.2010.12.016. [DOI] [PubMed] [Google Scholar]
- Siegel SM, Speitel TW. Performance of fungi in low temperature and hypersaline environments. Life Sci Space Res. 1976;14:351–354. [PubMed] [Google Scholar]
- Sikkema J, de Bont JAM, Poolman B. Mechanisms of membrane toxicity of hydrocarbons. Microbiol Rev. 1995;59:201–222. doi: 10.1128/mr.59.2.201-222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetta MP, Verga R, Fretta A, Hanozet GM. Induction of d-amino-acid oxidase by d-alanine in Rhodotorula gracilis grown in defined medium. J Gen Microbiol. 1989;135:593–600. [Google Scholar]
- Sinha M, Srinivasa H, Macaden R. Antibiotic resistance profile 0026; extended spectrum β-lactamase (ESBL) production in Acinetobacter species. Indian J Med Res. 2007;126:63–67. [PubMed] [Google Scholar]
- Snow D. The germination of mould spores at controlled humidities. Ann Appl Biol. 1949;36:1–13. doi: 10.1111/j.1744-7348.1949.tb06395.x. [DOI] [PubMed] [Google Scholar]
- Steinkraus KH, editor. Handbook of Indigenous Fermented Foods. New York, USA: Marcel Dekker; 1983. [Google Scholar]
- Stinson MW, Hayden C. Secretion of phospholipase-c by Pseudomonas aeruginosa. Infect Immun. 1979;25:558–564. doi: 10.1128/iai.25.2.558-564.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoynova-Bakalova E, Toncheva-Panova T. Subcellular adaptation to salinity and irradiance in Dunaliella salina. Biol Plant. 2003;47:233–236. [Google Scholar]
- Stratford M. Food and beverage spoilage yeasts. In: Querol A, Fleet GH, editors. The Yeast Handbook. Berlin, Heidelberg: Springer-Verlag; 2006. pp. 335–379. [Google Scholar]
- Straus E, Wu N. Radioimmunoassay of tuberculoprotein derived from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1980;77:4301–4304. doi: 10.1073/pnas.77.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki C, Ando Y, Machida S. Interaction of SMKT, a killer toxin produced by Pichia farinosa, with the yeast cell membranes. Yeast. 2001;18:1471–1478. doi: 10.1002/yea.791. [DOI] [PubMed] [Google Scholar]
- Syn CKC, Magnuson JK, Kingsley MT, Swarup S. Characterization of Pseudomonas putida genes responsive to nutrient limitation. Microbiology. 2004;150:1661–1669. doi: 10.1099/mic.0.26657-0. [DOI] [PubMed] [Google Scholar]
- Szopinska A, Morsomme P. Quantitative proteomic approaches and their application in the study of yeast responses. OMICS. 2010;14:639–649. doi: 10.1089/omi.2010.0045. [DOI] [PubMed] [Google Scholar]
- Tamang JP. Himalayan Fermented Foods: Microbiology, Nutrition, and Ethnic Values. Boca Raton, USA: CRC Press; 2010. [Google Scholar]
- Tark M, Tover A, Tarassova K, Tegova R, Kivi G, Hõrak R, et al. A DNA polymerase V homologue encoded by TOL plasmid pWW0 confers evolutionary fitness on Pseudomonas putida under conditions of environmental stress. J Bacteriol. 2005;187:5203–5213. doi: 10.1128/JB.187.15.5203-5213.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therkildsen MS, Isaksen MF, Lomstein BA. Urea production by the marine bacteria Delaya venusta and Pseudomonas stutzeri grown in a minimal medium. Aquat Microb Ecol. 1997;13:213–217. [Google Scholar]
- Thomas KC, Hynes SH, Ingledew WM. Excretion of proline by Saccharomyces cerevisiae during fermentation of arginine-supplemented high gravity wheat mash. J Ind Microbiol. 1993;12:93–98. [Google Scholar]
- Thomas R, Grimsley N, Escande ML, Subirana L, Derelle E, Moreau H. Acquisition and maintenance of resistance to viruses in eukaryotic phytoplankton populations. Environ Microbiol. 2011;13:1412–1420. doi: 10.1111/j.1462-2920.2011.02441.x. [DOI] [PubMed] [Google Scholar]
- Thurston MI, Pallett DW, Cortina-Borja M, Edwards ML, Raybould AF, Cooper JI. The incidence of viruses in wild Brassica nigra in Dorset (UK) Ann Appl Biol. 2001;139:277–284. [Google Scholar]
- Timberlake WE, Frizzell MA, Richards KD, Gardner RC. A new yeast genetic resource for analysis and breeding. Yeast. 2011;28:63–80. doi: 10.1002/yea.1821. [DOI] [PubMed] [Google Scholar]
- Timmis KN. Pseudomonas putida: a cosmopolitan opportunist par excellence. Environ Microbiol. 2002;4:779–781. doi: 10.1046/j.1462-2920.2002.00365.x. [DOI] [PubMed] [Google Scholar]
- Timmis KN, editor. Handbook of Hydrocarbon and Lipid Microbiology – Hydrocarbons, Oils and Lipids: Diversity, Properties and Formation. Vol. 2. New York, USA: Springer; 2009. [Google Scholar]
- Tiwari BK, Valdramidis VP, O'Donnell CP, Muthukumarappan K, Bourke P, Cullen PJ. Application of natural antimicrobials for food preservation. J Agric Food Chem. 2009;57:5987–6000. doi: 10.1021/jf900668n. [DOI] [PubMed] [Google Scholar]
- Tobin MB, Peery RB, Skatrud PL. Genes encoding multiple drug resistance-like proteins in Aspergillus fumigatus and Aspergillus flavus. Gene. 1997;200:11–23. doi: 10.1016/s0378-1119(97)00281-3. [DOI] [PubMed] [Google Scholar]
- Tobin KM, McGrath JW, Mullan A, Quinn JP, O'Connor KE. Polyphosphate accumulation by Pseudomonas putida CA-3 and other medium-chain-length polyhydroxyalkanoate-accumulating bacteria under aerobic growth conditions. Appl Environ Microbiol. 2007;73:1383–1387. doi: 10.1128/AEM.02007-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PP, Lucero CA, Falkinham JO. Health impacts of environmental mycobacteria. Clin Microbiol Rev. 2004;17:98–106. doi: 10.1128/CMR.17.1.98-106.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh H, Kayingo G, van der Merwe MJ, Kilian SG, Hallsworth JE, Hohmann S, et al. Implications of FPS1 deletion and membrane ergosterol content for glycerol efflux from Saccharomyces cerevisiae. FEMS Yeast Res. 2001;1:205–211. doi: 10.1111/j.1567-1364.2001.tb00035.x. [DOI] [PubMed] [Google Scholar]
- Trigal C, Goedkoop W, Johnson RK. Changes in phytoplankton, benthic invertebrate and fish assemblages of boreal lakes following invasion by Gonyostomum semen. Freshw Biol. 2011;56:1937–1948. [Google Scholar]
- Trindade RC, Resende MA, Silva CM, Rosa CA. Yeasts associated with fresh and frozen pulps of Brazilian tropical fruits. Syst Appl Microbiol. 2002;25:294–300. doi: 10.1078/0723-2020-00089. [DOI] [PubMed] [Google Scholar]
- Tsirogianni E, Aivaliotis M, Papasotiriou DG, Karas M, Tsiotis G. Identification of inducible protein complexes in the phenol degrader Pseudomonas sp. strain phDV1 by blue native gel electrophoresis and mass spectrometry. Amino Acids. 2006;30:63–72. doi: 10.1007/s00726-005-0219-4. [DOI] [PubMed] [Google Scholar]
- Tsubo M, Nishihara E, Nakamatsu K, Cheng YX, Shinoda M. Plant volatiles inhibit restoration of plant species communities in dry grassland. Basic Appl Ecol. 2012;13:76–84. [Google Scholar]
- Turk M, Plemenitaš A, Gunde-Cimerman N. Extremophilic yeasts: plasma-membrane fluidity as determinant of stress tolerance. Fungal Biol. 2011;115:950–958. doi: 10.1016/j.funbio.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Vallentyne JR. Environmental biophysics and microbial ubiquity. Ann N Y Acad Sci. 1963;108:342–352. doi: 10.1111/j.1749-6632.1963.tb13387.x. [DOI] [PubMed] [Google Scholar]
- Vasileva-Tonkova E, Gesheva V. Biosurfactant production by Antarctic facultative anaerobe Pantoea sp. during growth on hydrocarbons. Curr Microbiol. 2007;54:136–141. doi: 10.1007/s00284-006-0345-6. [DOI] [PubMed] [Google Scholar]
- Visser AA, Kooij PW, Debets AJM, Kuyper TW, Aanen DK. Pseudoxylaria as stowaway of the fungus-growing termite nest: interaction asymmetry between PseudoxylariaTermitomyces and free-living relatives. Fungal Ecol. 2011;4:322–332. [Google Scholar]
- Vital MJS, Abranches J, Hagler AN, Mendonça-Hagler LC. Mycocinogenic yeasts isolated from Amazon soils of the Maracá Ecological Station, Roraima-Brazil. Braz J Microbiol. 2002;33:230–235. [Google Scholar]
- Volkers RJM, de Jong AL, Hulst AG, van Baar BLM, de Bont JAM, Wery J. Chemostat-based proteomic analysis of toluene-affected Pseudomonas putida S12. Environ Microbiol. 2006;8:1674–1679. doi: 10.1111/j.1462-2920.2006.01056.x. [DOI] [PubMed] [Google Scholar]
- Volkers RJM, Ballerstedt H, Ruijssenaars HJ, de Bont JA, de Winde JH, Wery J. TrgI, toluene repressed gene I, a novel gene involved in toluene-tolerance in Pseudomonas putida S12. Extremophiles. 2009;13:283–297. doi: 10.1007/s00792-008-0216-0. [DOI] [PubMed] [Google Scholar]
- Volkers RJM, Ballerstedt H, de Winde JH, Ruijssenaars HJ. Isolation and genetic characterization of an improved benzene-tolerant mutant of Pseudomonas putida S12. Environ Microbiol Rep. 2010;2:456–460. doi: 10.1111/j.1758-2229.2010.00172.x. [DOI] [PubMed] [Google Scholar]
- Walker GM. Pichia anomala: cell physiology and biotechnology relative to other yeasts. Antonie Van Leeuwenhoek. 2011;99:25–34. doi: 10.1007/s10482-010-9491-8. [DOI] [PubMed] [Google Scholar]
- Wang R-L, Staehelin C, Peng S-L, Wang W-T, Xie X-M, Lu H-N. Responses of Mikania micrantha, an invasive weed to elevated CO2: induction of β-caryophyllene synthase, changes in emission capability and allelopathic potential of β-caryophyllene. J Chem Ecol. 2010;36:1076–1082. doi: 10.1007/s10886-010-9843-x. [DOI] [PubMed] [Google Scholar]
- Ward OP, Qin WM, Dhanjoon J, Ye J, Singh A. Physiology and biotechnology of Aspergillus. Adv Appl Microbiol. 2006;58:1–75. [PubMed] [Google Scholar]
- Watanabe A, Yano K, Ikebukuro K, Karube I. Cyanide hydrolysis in a cyanide-degrading bacterium, Pseudomonas stutzeri AK61, by cyanidase. Microbiology. 1998;144:1677–1682. doi: 10.1099/00221287-144-6-1677. [DOI] [PubMed] [Google Scholar]
- Wecke T, Bauer T, Harth H, Mäder U, Mascher T. The rhamnolipid stress response of Bacillus subtilis. FEMS Microbiol Lett. 2011;323:113–123. doi: 10.1111/j.1574-6968.2011.02367.x. [DOI] [PubMed] [Google Scholar]
- Wendell DL, Bisson LF. Expression of high-affinity glucose transport protein Hxt2p of Saccharomyces cerevisiae is both repressed and induced by glucose and appears to be regulated post-translationally. J Bacteriol. 1994;176:3730–3737. doi: 10.1128/jb.176.12.3730-3737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westenberg B, Boller T, Wiemken A. Lack of arginine- and polyphosphate-storage pools in a vacuole-deficient mutant (endl) of Saccharomyces cerevisiae. FEBS Lett. 1989;254:133–136. [Google Scholar]
- Wheeler KA, Hocking AD. Interactions among xerophilic fungi associated with dried salted fish. J Appl Bacteriol. 1993;74:164–169. doi: 10.1111/j.1365-2672.1993.tb03010.x. [DOI] [PubMed] [Google Scholar]
- Wiesenborn DP, Rudolph FB, Papoutsakis ET. Phosphotransbutyrylase from Clostridium acetobutylicum ATCC-824 and its role in acidogenesis. Appl Environ Microbiol. 1989;55:317–322. doi: 10.1128/aem.55.2.317-322.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JP, Hallsworth JE. Limits of life in hostile environments: no barriers to biosphere function? Environ Microbiol. 2009;11:3292–3308. doi: 10.1111/j.1462-2920.2009.02079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit R, Bouvier T. ‘Everything is everywhere, but, the environment selects’; what did Baas Becking and Beijerinck really say? Environ Microbiol. 2006;8:755–758. doi: 10.1111/j.1462-2920.2006.01017.x. [DOI] [PubMed] [Google Scholar]
- Wu SJ, Zhang HX, Yu X, Chen JM. Identification and cloning of a gene encoding dichloromethane dehalogenase from a methylotrophic bacterium, Bacillus circulans WZ-12 CCTCC M 207006. Bioprocess Biosyst Eng. 2009;32:845–852. doi: 10.1007/s00449-009-0311-3. [DOI] [PubMed] [Google Scholar]
- Wu T-H, Liao M-H, Kuo W-Y, Huang C-H, Hsieh H-L, Jinn T-L. Characterization of copper/zinc and manganese superoxide dismutase in green bamboo (Bambusa oldhamii): cloning, expression and regulation. Plant Physiol Biochem. 2011;49:195–200. doi: 10.1016/j.plaphy.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Xing W, Huang W-M, Li D-H, Liu Y-D. Effects of iron on growth, pigment content, photosystem II efficiency, and siderophores production of Microcystis aeruginosa and Microcystis wesenbergii. Curr Microbiol. 2007;55:94–98. doi: 10.1007/s00284-006-0470-2. [DOI] [PubMed] [Google Scholar]
- Xuan TD, Chung MI, Khanh TD, Tawata S. Identification of phytotoxic substances from early growth of barnyard grass (Echinochloa crusgalli) root exudates. J Chem Ecol. 2006;32:895–906. doi: 10.1007/s10886-006-9035-x. [DOI] [PubMed] [Google Scholar]
- Yalcin SK, Ozbas ZY. Effects of pH and temperature on growth and glycerol production kinetics of two indigenous wine strains of Saccharomyces cerevisiae from Turkey. Braz J Microbiol. 2008;39:325–332. doi: 10.1590/S1517-838220080002000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancey PH. Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J Exp Biol. 2005;208:2819–2830. doi: 10.1242/jeb.01730. [DOI] [PubMed] [Google Scholar]
- Yang Z, Kong F. Formation of large colonies: a defense mechanism of Microcystis aeruginosa under continuous grazing pressure by flagellate Ochromonas sp. J Limnol. 2012;71:61–66. [Google Scholar]
- Zhu H-Y, Xu H, Dai X-Y, Zhang Y, Ying H-J, Ouyang P-K. Production of d-arabitol by a newly isolated Kodamaea ohmeri. Bioproc Biosyst Eng. 2010;33:565–571. doi: 10.1007/s00449-009-0378-x. [DOI] [PubMed] [Google Scholar]
- Zuroff TR, Curtis WR. Developing symbiotic consortia for lignocellulosic biofuel production. Appl Microbiol Biotechnol. 2012;93:1423–1435. doi: 10.1007/s00253-011-3762-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Examples of open habitats in which specific microbes attain dominance.
Table S2. Biology of microbial weeds compared with that of other ecophysiological groups.