Summary
Escherichia coli-based bioreporters for arsenic detection are typically based on the natural feedback loop that controls ars operon transcription. Feedback loops are known to show a wide range linear response to the detriment of the overall amplification of the incoming signal. While being a favourable feature in controlling arsenic detoxification for the cell, a feedback loop is not necessarily the most optimal for obtaining highest sensitivity and response in a designed cellular reporter for arsenic detection. Here we systematically explore the effects of uncoupling the topology of arsenic sensing circuitry on the developed reporter signal as a function of arsenite concentration input. A model was developed to describe relative ArsR and GFP levels in feedback and uncoupled circuitry, which was used to explore new ArsR-based synthetic circuits. The expression of arsR was then placed under the control of a series of constitutive promoters, which differed in promoter strength, and which could be further modulated by TetR repression. Expression of the reporter gene was maintained under the ArsR-controlled Pars promoter. ArsR expression in the systems was measured by using ArsR–mCherry fusion proteins. We find that stronger constitutive ArsR production decreases arsenite-dependent EGFP output from Pars and vice versa. This leads to a tunable series of arsenite-dependent EGFP outputs in a variety of systematically characterized circuitries. The higher expression levels and sensitivities of the response curves in the uncoupled circuits may be useful for improving field-test assays using arsenic bioreporters.
Introduction
Bacterial bioreporters are genetically modified strains that express a reporter protein, typically a spectroscopically or electrochemically active protein, in response to a specific unique or group of related target chemicals (van der Meer and Belkin, 2010). Bioreporter assays can be a useful complement for analysis of toxic compounds in, e.g. water (Tecon et al., 2010) or soil samples (Paton et al., 2009), air (de las Heras and de Lorenzo, 2011), food-stuffs (Baumann and van der Meer, 2007), urine (Lewis et al., 1984) or blood serum (Turner et al., 2007). In certain cases where chemical analyses are too expensive or logistically difficult to perform, bioreporter assays can present an appropriate quantitative substitution. As an example, Siegfried and colleagues (2012) and Trang and colleagues (2005) successfully demonstrated large-scale and quantitative use of an Escherichia coli-based bioreporter assay for arsenic in drinking water from local wells in villages in Bangladesh and Vietnam respectively.
The central element in bioreporter strains is a genetic circuit formed by the gene for a ‘sensor/transducer’ protein (e.g. a transcription regulator) and a ‘switch’ (the DNA region to which the transcription regulator binds), which controls the promoter driving expression of the reporter gene (Daunert et al., 2000). The DNA ‘parts’ for the genetic circuit are commonly mined from natural systems and placed in a different host cell context. Genetic circuits for arsenic detection (Ramanathan et al., 1997; Tauriainen et al., 1997; Stocker et al., 2003) are typically based on the bacterial arsenic defence system, like, for instance, encoded by the arsRDABC operon on E. coli plasmid R773 (Hedges and Baumberg, 1973). This system is homeostatically regulated by the ArsR and ArsD trans-acting repressors at the level of ars expression (Wu and Rosen, 1993; Bruhn et al., 1996; Chen and Rosen, 1997). Both ArsR and ArsD are 13 kDa protomers and form homodimers (Wu and Rosen, 1993; Rosen, 1995), but they share no sequence similarity. ArsR is an AsIII/SbIII-responsive repressor with high affinity for its DNA operator (named ArsR binding site or ABS), which is positioned upstream of the ars promoter (Fig. 1A) (Wu and Rosen, 1993; Rosen, 1995). ArsR binds the ABS in absence of arsenite and is thought to hinder RNA polymerase from starting transcription, thereby controlling the background expression of the ars operon, including of the arsR gene itself. Binding of arsenite or antimonite to ArsR decreases its affinity for the ABS (Wu and Rosen, 1991), and unleashes ars transcription. Expression of the ars operon is thus controlled via a feedback loop, since arsR is the first gene to be transcribed after derepression. ArsD is a metallochaperone that increases cellular resistance by delivering arsenite to the ArsA subunit of the extrusion system (Lin et al., 2006). It also controls the maximal level of expression of the ars operon by binding with a two orders of magnitude lower affinity than ArsR to the ABS, eventually turning ars expression off (Chen and Rosen, 1997). Escherichia coli additionally has a chromosomally encoded ars operon, which is formed by the arsRBC genes (Diorio et al., 1995; Chen and Rosen, 1997). ArsRR773 and ArsRK12 share 74% amino acid similarity (Fig. S1). The arsK12 operon lacks arsD and arsA, an ATPase that forms a complex with the arsenite-specific membrane channel ArsB to produce the active arsenite extrusion complex (Zhou et al., 2000).
Figure 1.
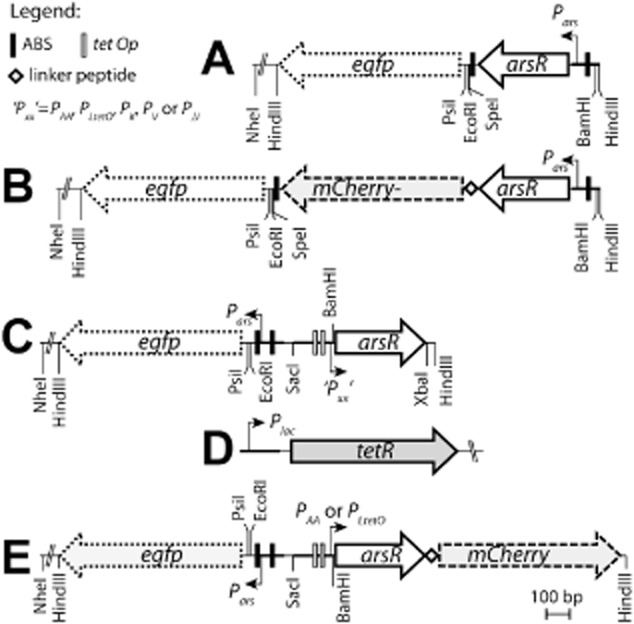
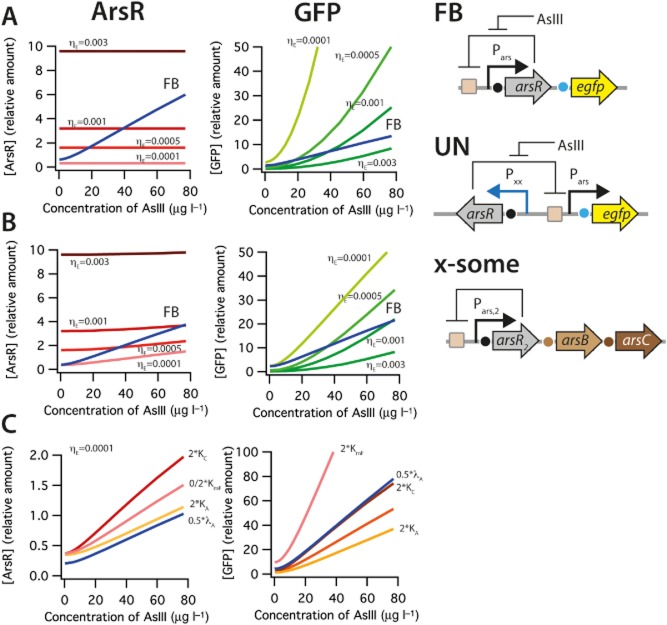
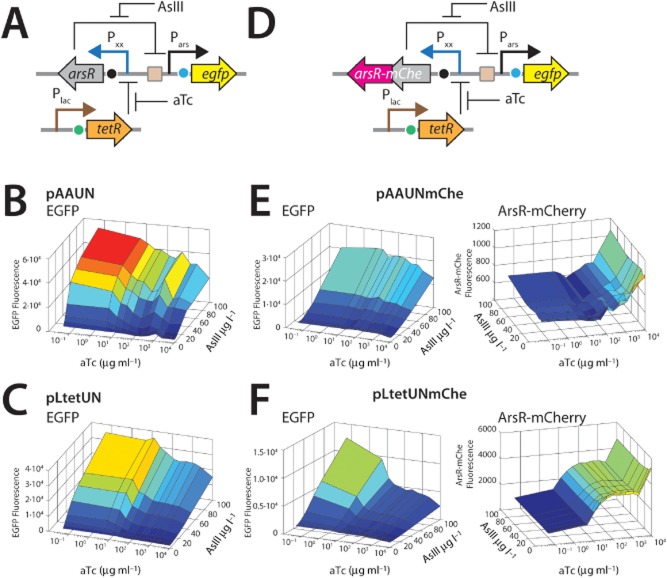
Schematic organization of the ArsR-controlled genetic circuits assembled on plasmids in E. coli.A. Elements building the feedback arsR-egfp construct.B. As (A), but with the arsR–mCherry fusion gene.C. The uncoupled arsenic bioreporter circuits.D. The tetR gene under control of the lac promoter. E. Uncoupled circuit with the arsR–mCherry fusion gene. The position of the binding site for ArsR on the DNA is depicted by dark vertical bars (ABS); those for TetR by grey vertical bars. Positions of restriction sites relevant for cloning are indicated. Outline of (C) indicative for plasmids pAAUN, pLtetOUN, pIIUN, pKUN, pVUN and pJJUN. Those in (E) for pAAUNmChe and pLtetOUNmChe.
Most arsenic bioreporters except one (Tani et al., 2009) have been designed to have the reporter gene downstream of arsR under ArsR-feedback control of Pars (Ramanathan et al., 1997; Tauriainen et al., 1997; Stocker et al., 2003). When such reporter cells encounter arsenite, this will bind to the ArsR-dimer, causing it to dissociate from its binding site and unleashing further expression of itself and of the reporter gene. The increase in reporter protein expression and activity is approximately linear in the range between 5 and 80 μg of arsenite per litre (Stocker et al., 2003; Baumann and van der Meer, 2007), and can be used to quantify arsenite concentrations in unknown samples. However, since the feedback loop is essentially a bit leaky to allow formation of ArsR that needs to repress the system, background reporter gene expression in the absence of arsenite may be disturbingly high (Stocker et al., 2003). In a conceptually very different reporter circuit configuration, expression of arsR is uncoupled from its feedback loop, whereas the reporter gene expression is maintained under ArsR control via the Pars promoter and the ABS (Fig. 1C). In this case an arsenite-independent promoter controls the expression of arsR such that ArsR levels are sufficient to repress the background expression of the reporter gene from the Pars promoter are constitutively produced.
The objectives of the current work were to systematically explore the effects of arsenite concentration-dependent reporter gene expression in the uncoupled circuitry mode. A mechanistic model was developed for ArsR repression of Pars based on mass action kinetics, analogous to a model for LacI repression of Plac (Lee and Bailey, 1984) to predict the effects of feedback and uncoupled circuitry on ArsR and EGFP expression. The model was tested experimentally by varying ArsR concentrations over a wide range using two promoters with different maximal strength that were placed under control of TetR and could be derepressed by addition of anhydrotetracycline (aTc). In order to estimate relative changes in intracellular ArsR concentrations we used additional gene circuitry with arsR–mCherry fusions instead of arsR (Fig. 1B and E). Since pre-induction with aTc is not practical in field assays, we then replaced TetR-regulatable expression by a set of constitutive promoters with different (published) strengths (Alper et al., 2005) (Fig. S2), and tested the EGFP output as a function of arsenite concentrations in E. coli strains with or without chromosomal arsRBC gene cassette. We find that uncoupling can have important gain on reporter output and can result in modulatable maximum reporter levels.
Results
Uncoupling arsR expression is predicted to produce tunable reporter signal development
The behaviour of the ArsR-Pars feedback (FB) system can be predicted using a mechanistic model based on mass action binding equilibria between ArsR and its DNA binding sites, ArsR and arsenite, and RNA polymerase and the arsR promoter, analogous to a model described for LacI control of the lac promoter (Lee and Bailey, 1984) (Supporting information). The predicted relative concentrations of ArsR and EGFP produced under steady-state conditions as a function of exposure to arsenite both increase over the range of 0–80 μg of AsIII per litre (Fig. 2A, FB), for a situation with arsR present only on a plasmid in the cell. Note that we consider here only the typical measurement range of arsenite concentrations for the arsenic bioreporter. The model in Supporting information (SI) File 1 allows interested readers to test other concentration ranges. In case of an additional chromosomal arsR copy, the arsenite-dependent production of ArsR would be slightly lower and that of EGFP slightly higher (Fig. 2B, FB). We next examined the model prediction for the case where expression of ArsR is ‘uncoupled’ from its feedback control, whereas that of EGFP is maintained under arsenite-dependent ArsR/Pars control. In this scenario arsR transcription can be varied by using different strength promoters, or giving different transcription efficiencies (ηE in the model). Accordingly, the model predicts that by varying the promoter strength for arsR expression across a 30-fold range (ηE in Fig. 2A and B) one could achieve ArsR levels in the cell that are constantly lower (ηE = 0.0001) or higher (ηE = 0.003) than in the feedback system. Interestingly, maintaining constant ArsR production at different levels is predicted to result in largely different response curves of the EGFP signal produced from Pars. Higher ArsR levels (e.g. ηE = 0.003) will lead to less steep EGFP response curves as a function of arsenite exposure, whereas lower levels (ηE = 0.0001) are predicted to lead to steeper response curves (Fig. 2A). Noteworthy, predictions suggest that maintaining a chromosomal arsR copy would result in slightly lower EGFP outputs for the case of the uncoupled gene circuitry. It is important to further note that the model is not a data ‘fitting’ but a mechanistic model, allowing to systematically explore variations in underlying parameters. As an example, the model predicts the reporter output to be relatively sensitive to changes in the equilibrium binding constant of ArsR with AsIII (KC, Fig. 2C).
Figure 2.
A and B. Predictions of ArsR and EGFP mass action equilibrium concentrations as a function of arsenite (AsIII) exposure for the original plasmid feedback circuit (FB), for the plasmid uncoupled circuit (UN), in absence (A) or presence (B) of an extra chromosomal arsR (x-some). Parameters: ηE, transcription efficiency of the constitutive promoter for arsR (Pxx).C. Sensitivity analysis on case (B) with ηE = 0.0001 varying KmF, EGFP translation efficiency; KC, equilibrium constant of ArsR for AsIII; KA, equilibrium constant of ArsR with its DNA binding site; λA, number of ArsR protein per arsR mRNA. For details of model assumptions, see Supporting information and SI File 1.
Tunable uncoupling effects on EGFP expression
To experimentally explore and verify the predicted effects of uncoupling the ArsR-Pars feedback loop on reporter gene induction, we constructed a series of new topologies in which arsR expression is controlled from a defined promoter, whereas ArsR still controls the expression of the reporter gene (egfp) via Pars (Fig. 2, UN). Since the native Pars expression feedback loop has a relatively high background expression, we used a variant in which a second ArsR binding site is inserted downstream of arsR in the feedback circuit, which reduces background expression in the absence of arsenite (Stocker et al., 2003). This secondary ArsR binding site is maintained in the uncoupled versions (Fig. 1). Furthermore, to experimentally create the condition of having only a plasmid-located arsR gene circuit we deleted the chromosomal arsRBC cassette in E. coli MG1655. Tunable expression of arsR was achieved by using two constitutive promoters (PLtetO and PAA) that have additional TetR recognition sites within their promoters (Figs S2 and S3). Expression of arsR can then be brought under control of TetR by including a Plac-expressed tetR gene on a secondary plasmid (Figs 1 and 3A). The output of the PLtetO and PAA promoters was systematically increased by pre-incubation with defined aTc concentrations for 2 h, after which the cells were exposed to arsenite to follow reporter induction from Pars. Increasing the aTc concentration will on average lead to more derepression of TetR control on ArsR, as a result of which more ArsR is produced that can repress the Pars promoter. The consequence of this is a less steep EGFP reporter curve (Fig. 3B and C). In the absence of aTc repression by TetR is maximal, causing minimal ArsR production and highest arsenite-dependent EGFP expression. At the highest aTc concentration ArsR levels were maximal and arsenite-dependent production of EGFP was minimal, which is conform the model predictions. One can observe that the PLtetO promoter is indeed stronger than PAA since the EGFP response curve is lower at the highest aTc concentrations. Interestingly, both model predictions and experimental data confirm that even the strongest promoter for arsR expression will not completely abolish arsenite-dependent expression from Pars (Figs 2 and 3).
Figure 3.
Systematic effects of varying ArsR production on the arsenite-dependent EGFP synthesis from Pars in E. coli MG1655 ΔRBC.A–C. (A) Relevant uncoupled circuitry design with Pxx being either PAA (B) or PLtetO (C).D–F. Relevant uncoupled circuitry design for the arsR–mCherry fusion variant circuitry with Pxx being either PAA (E) or PLtetO (F). Notice log scale for aTc addition and different scale for EGFP or ArsR–mCherry fluorescence between panels. Fluorescence measured by flow cytometry on cells pre-induced for 2 h with aTc, and subsequently 3 h with arsenite.
To demonstrate that indeed higher ArsR levels are responsible for this behaviour we produced variant reporter circuits in which the arsR gene is fused via a short linker to mCherry, which leads to an ArsR–mCherry fusion protein (Fig. 3D). Comparatively, the circuits with the ArsR–mCherry fusion protein produced only half the EGFP reporter output as those with ArsR (Fig. 3E and F). This indicated that ArsR–mCherry is still functional, but the model predicts that it must have a stronger binding constant to the ArsR binding site, since EGFP production is lower than for the ArsR system at the same arsenite concentration (Fig. 2C, stronger binding constant would be equivalent to changing the value for KA. Compare EGFP responses for KA and 2×KA). As expected from the model predictions the amount of ArsR–mCherry protein, taken as the intensity of mCherry fluorescence, increased with increasing aTc concentration in the pre-incubation step, was independent of the arsenite concentration, and was higher for the PLtetO -driven than the PAA-driven system (Fig. 3E and F).
Uncoupling effects in modular strains with different constitutive arsR control
Because pre-induction with aTc is not a practical solution for a bioassay we tested the same circuits in a background without tetR but varying only the promoter strength for arsR. Indeed, we observed that the levels of ArsR-mCherry were independent of the arsenite concentration in the strains with the uncoupled circuits (Fig. 4B), whereas those in the strain with the feedback circuit increased with increasing arsenite concentration. As expected from the model the circuit with the stronger promoter for arsR–mCherry (PLtetO) produced more ArsR–mCherry but less EGFP output than the circuit with the weaker PAA promoter (Fig. 4A). In contrast, but also according to model predictions, the background EGFP expression in absence of arsenite was higher in the uncoupled circuit with the weaker promoter.
Figure 4.
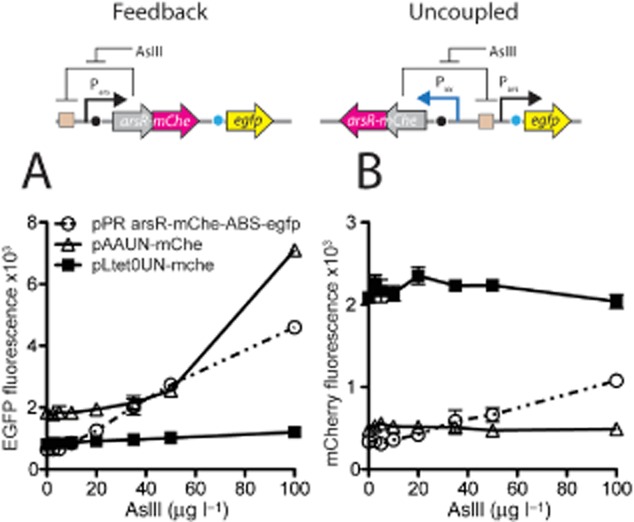
Effects of uncoupled versus feedback circuit in the absence of TetR control in E. coli MG1655 ΔRBC.A. EGFP fluorescence as a function of arsenite exposure, measured 180 min after induction using flow cytometry.B. mCherry fluorescence from ArsR–mCherry as a function of arsenite exposure, in the same cells as in (A).
Because these circuits were tested with the ArsR–mCherry variant, which had a stronger repression effect than native ArsR, we finally replaced the native arsR gene back instead of arsR–mCherry under control of variant constitutive promoters with different (published) strengths, from the weakest PII to the strongest PLtetO (Alper et al., 2005) (Table 1, Fig. S2). Results showed a range of EGFP outputs with increasing fluorescence for the same arsenite exposure concentration at weaker promoter strengths for arsR expression. The weakest promoter for arsR in the construct pIIUN resulted in up to fivefold higher EGFP fluorescence than in the original feedback construct pPR-ArsR-ABS at the same arsenite concentration (Fig. 5B). Interestingly, and stronger than expected from the model, the EGFP output of the same circuit in E. coli without the chromosomal arsRBC cassette (MG1655ΔRBC) was more than twice as strong as in wild-type E. coli MG1655 (Fig. 5A). The reason for this may be that because the arsR chromosomal copy is not completely identical to the plasmid arsR copy, their mutual repression is different than the model assumes for reasons of simplicity. Incidentally, measuring the induction from the same reporter circuits by fluorometry produces approximately similar response curves (Fig. S4). Kinetic profiles of reporter gene induction under the used assay conditions all show a typical 40 min lag during which hardly any increase of reporter signal is observed (Fig. S5).
Table 1.
Used strains and plasmids in this study
| Strain number | Host strain | Relevant genotype | Plasmid | Reference |
|---|---|---|---|---|
| 1598 | Escherichia coli DH5α | KmR | pPR-arsR-ABS-egfp | Stocker et al. (2003) |
| 3391 | E. coli MG1655 ΔRBC | Deletion of arsRBC, KmR | pAAUN | This study |
| 3316 | E. coli MG1655 ΔRBC | Deletion of arsRBC, KmR | pPR-arsR-ABS-egfp | This study |
| 3304 | E. coli MG1655 ΔRBC | Deletion of arsRBC | – | This study |
| 3328 | E. coli MG1655 | Wild-type, KmR | pPR-arsR-ABS-egfp | This study |
| 3307 | E. coli MG1655 | KmR | pAAUN | This study |
| 3612 | E. coli MG1655 | KmR | pJJUN | This study |
| 3633 | E. coli MG1655 | KmR, ArsR–mCherry fusion | pJJUN-mChe | This study |
| 3636 | E. coli MG1655 | KmR | pKUN | This study |
| 3614 | E. coli MG1655 | KmR | pLtetOUN | This study |
| 3634 | E. coli MG1655 | KmR, ArsR–mCherry fusion | pLtetOUN-mChe | This study |
| 3652 | E. coli MG1655 ΔRBC | Deletion of arsRBC, KmR | pLtetOUN | This study |
| 3653 | E. coli DH5α | KmR | pLtetOUN | This study |
| 3660 | E. coli MG1655 ΔRBC | Deletion of arsRBC, KmR, ArsR–mCherry fusion | pLtetOUN-mCherry | This study |
| 3665 | E. coli DH5α | KmR | pAAUN | This study |
| 3668 | E. coli DH5α | KmR, ArsR–mCherry fusion | pAAUN-mChe | This study |
| 3670 | E. coli MG1655 | KmR | pVUN | This study |
| 3792 | E. coli DH5α | KmR, ArsR–mCherry fusion | pPR-arsR-mChe-ABS-egfp | This study |
| 3795 | E. coli MG1655 ΔRBC | Deletion of arsRBC, KmR, ArsR–mCherry fusion | pPR-arsR-mChe-ABS-egfp - | This study |
| 4210 | E. coli MG1655 ΔRBC | Deletion of arsRBC, KmR, ApR, ArsR–mCherry fusion, Plac driven TetR expression | pLtetOUN-mCherry/pGem-TetR | This study |
| 4222 | E. coli MG1655 ΔRBC | Deletion of arsRBC, KmR, ApR, ArsR–mCherry fusion, Plac driven TetR expression | pAAOUN-mCherry/pGem-TetR | This study |
ApR, ampicillin resistance; KmR, kanamycin resistance.
Figure 5.
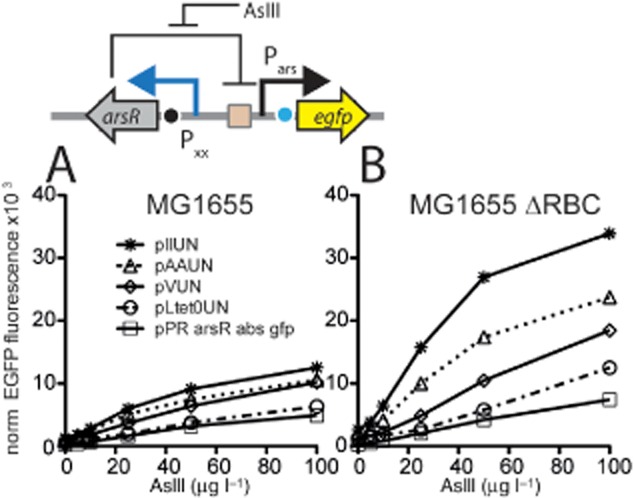
Arsenite-dependent EGFP fluorescence in cultures of E. coli MG1655 (A), and E. coli MG1655 ΔRBC (without the chromosomal arsRBC cassette; B) carrying the original feedback circuit (pPR-arsR-ABS) or four uncoupled arsR reporter circuits with different promoter strengths driving arsR expression (pAAUN, pLtetOUN, pIIUN, pVUN). Fluorescence measured by flow cytometry after 180 min induction time. Data symbols represent the average from independent biological triplicates. Whiskers, SD (when not visible lay within the symbol size).
Cell to cell variation in reporter expression in feedback versus uncoupled circuits
EGFP expression heterogeneity among individual cells (expressed as the mean SD from the FC FITC channel distributions) was significantly smaller (∼ 1.4-fold, when expressed as ratio between the SD normalized as percentage of the respective means) for cells in the presence of arsenite than without, but only in the case of the feedback circuit (Fig. 6, pPR-arsR-mChe-ABS-egfp; Table 2). Single-cell EGFP and mCherry fluorescence correlated positively for the feedback but not for the uncoupled circuit (Table 2). Normalized SD of both EGFP and mCherry among individual cells were lower for the stronger promoter in the uncoupled circuit (pLtetOUN, Fig. 6).
Figure 6.
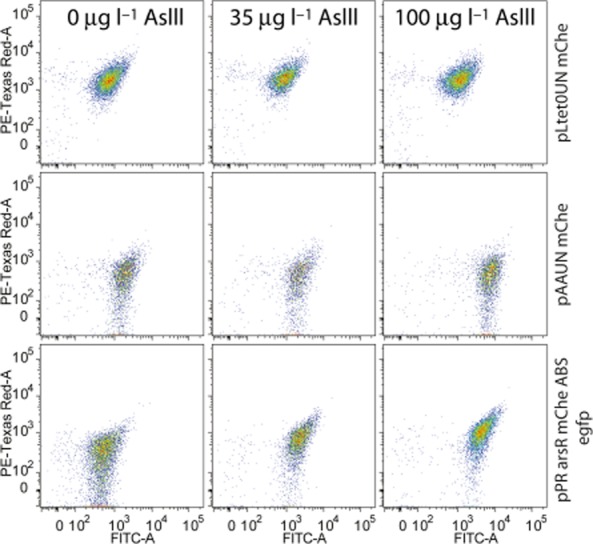
FC analysis of single-cell heterogeneity of ArsR–mCherry and EGFP expression in E. coli MG1655 ΔRBC coupled and uncoupled bioreporters after 3 h exposure to arsenite at the indicated concentrations. Dot plots show EGFP (FITC-A channel) versus mCherry fluorescence (PE-Texas Red-A channel) of single cells on a 10log-scale for ∼ 2000 events per sample.
Table 2.
Analysis of reporter protein variation in feedback versus uncoupled ArsR-controlled circuits in E. coli MG1655 ΔRBC
| Circuit | Arsenite concentration (μg l−1) | Average EGFPa | Average SDb | SD per cent of averagec | Average mCherrya | Average SD mCherryb | SD per cent of averagec | Pearson correlation factor (r)d |
|---|---|---|---|---|---|---|---|---|
| pPR-arsR-mChe-ABS | 0 | 605 ± 18 | 504 ± 136 | 83 | 345 ± 18 | 345 ± 18 | 98 | 0.5216 |
| 2.5 | 646 ± 33 | 477 ± 66 | 74 | 339 ± 26 | 364 ± 11 | 107 | 0.4221 | |
| 5 | 655 ± 33 | 471 ± 19 | 72 | 311 ± 9 | 375 ± 35 | 121 | 0.6183 | |
| 10 | 843 ± 65 | 536 ± 13 | 64 | 357 ± 17 | 381 ± 18 | 107 | 0.5805 | |
| 20 | 1253 ± 50 | 733 ± 7 | 58 | 419 ± 8 | 398 ± 18 | 95 | 0.6181 | |
| 35 | 2028 ± 257 | 1169 ± 102 | 58 | 584 ± 136 | 495 ± 85 | 85 | 0.7143 | |
| 50 | 2733 ± 123 | 1504 ± 98 | 55 | 661 ± 83 | 543 ± 73 | 82 | 0.7075 | |
| 100 | 4601 ± 47 | 2295 ± 90 | 50 | 1077 ± 22 | 708 ± 63 | 66 | 0.748 | |
| pAAUN-mChe | 0 | 1836 ± 24 | 1095 ± 50 | 60 | 483 ± 12 | 485 ± 29 | 100 | 0.4986 |
| 2.5 | 1779 ± 113 | 1207 ± 13 | 68 | 523 ± 9 | 558 ± 22 | 107 | 0.5924 | |
| 5 | 1836 ± 208 | 1243 ± 117 | 68 | 554 ± 44 | 569 ± 51 | 103 | 0.5777 | |
| 10 | 1835 ± 133 | 1305 ± 32 | 71 | 518 ± 3 | 533 ± 26 | 103 | 0.5352 | |
| 20 | 1942 ± 172 | 1396 ± 48 | 72 | 515 ± 26 | 557 ± 48 | 108 | 0.6204 | |
| 35 | 2146 ± 231 | 1516 ± 233 | 71 | 507 ± 56 | 549 ± 30 | 108 | 0.5626 | |
| 50 | 2549 ± 199 | 1753 ± 61 | 69 | 470 ± 7 | 501 ± 31 | 107 | 0.5063 | |
| 100 | 7101 ± 129 | 3712 ± 85 | 52 | 489 ± 12 | 500 ± 56 | 102 | 0.3341 | |
| pLtet0UN-mChe | 0 | 799 ± 14 | 424 ± 12 | 53 | 2084 ± 44 | 1320 ± 83 | 63 | 0.6042 |
| 2.5 | 851 ± 17 | 463 ± 12 | 54 | 2261 ± 102 | 1446 ± 151 | 64 | 0.5457 | |
| 5 | 845 ± 25 | 457 ± 15 | 54 | 2165 ± 137 | 1862 ± 746 | 86 | 0.5623 | |
| 10 | 868 ± 7 | 479 ± 9 | 55 | 2136 ± 92 | 1343 ± 33 | 63 | 0.5299 | |
| 20 | 912 ± 8 | 500 ± 24 | 55 | 2351 ± 107 | 1653 ± 182 | 70 | 0.5866 | |
| 35 | 956 ± 6 | 511 ± 6 | 53 | 2231 ± 67 | 1361 ± 53 | 61 | 0.5414 | |
| 50 | 1016 ± 25 | 550 ± 36 | 54 | 2234 ± 65 | 1310 ± 56 | 59 | 0.4568 | |
| 100 | 1199 ± 17 | 638 ± 13 | 53 | 2037 ± 81 | 1504 ± 455 | 74 | 0.497 |
Averages from three independent replicates ± one calculated standard deviation (SD) on the average. Signals averaged from 10 000 events per replicate.
Average of SD calculated from 10 000 events per replicate ± one calculated SD on the average. This average is an indication for the variation of reporter expression among single cells in the population.
Percentage of the average SD of the total average EGFP or mCherry.
Correlation between EGFP and mCherry signals of each single cell.
Discussion
We focused in this work on a systematic analysis of the effects on reporter gene expression from the ars promoter when decoupling synthesis of ArsR itself from its regular feedback loop. Controlling the expression of the circuit regulator by synthetic constitutive rather than cognate promoters has been shown previously to improve reporter output (Wu et al., 2009) but this has not been tested very systematically. As a first research question we examined whether the level of constitutive expression of ArsR would influence the reporter output from Pars. A mechanistic model was developed for ArsR-dependent EGFP expression from Pars, which non-intuitively predicted that constitutive promoters with a 30-fold different ‘strength’ would largely change the output of the circuitry in response to arsenite (Fig. 2). Experimental verification using TetR-aTc modulatable expression of arsR confirmed the model predictions to a large extent, except for details in the background expression level in absence of arsenite. Although the TetR-aTc system can be used for stepwise modulation of ArsR production, the pre-incubation with aTc is not very practical in a field assay. We then therefore replaced the TetR-aTc by a subset of constitutive promoters of different strength, which had been derived from PLtetO (Lutz and Bujard, 1997) by random mutagenesis (Alper et al., 2005) (Table 1, Fig. S2). Based upon the relative amount of mRNA produced from the specific promoter compared with the amount produced from PLtetO (= 1), Alper and colleagues (2005) ranked the promoters as 0.57 for PV, 0.30 for PK, 0.24 for PAA, 0.16 for PJJ and 0.06 for PII. By deducing from the fluorescence light intensity of ArsR–mCherry fusion proteins in single cells (FC) we could confirm that PLtetO was the stronger promoter than PAA (Figs 3 and 4). This experiment showed directly that higher ArsR–mCherry production leads to a reduction of the formation of EGFP from Pars as a function of arsenite exposure (Fig. 4).
Second, the model also predicted that different strength constitutive promoters for arsR expression would influence the shape of the arsenite-dependent response curve of the reporter protein (Fig. 2). Experiments with all six promoters confirmed that the amount of EGFP reporter signal produced from Pars as a function of arsenite exposure can be tuned by the strength of the promoter controlling the transcription of arsR (Fig. 5). Interestingly, these results also demonstrated that even the strong PLtetO promoter is insufficient to produce ArsR to such a level as to completely repress Pars in presence of arsenite (Figs 2 and 5). In contrast, the ArsR–mCherry fusion protein produced from PLtetO (Fig. 4A) was sufficient to completely repress Pars, which suggest that although ArsR–mCherry is functional and responsive to arsenite, its stability or DNA binding properties are enhanced and, consequently, its repression of the ars promoter is more severe.
Single-cell analysis of the reporter responses in the feedback (pPR-ArsR-mChe-ABS-egfp) and uncoupled system (pAAUN-mChe, pLTet0UN-mChe) showed that the cells with the feedback system tend to have larger variation in EGFP and mCherry produced from Pars at low arsenite concentrations, which successively become smaller at higher arsenite concentrations (Table 2). EGFP and ArsR–mCherry fluorescence in those cells correlate positively at higher arsenite exposures, meaning that cells which accidentally have higher ArsR–mCherry levels also (have) produce(d) more EGFP. This is conform the model predictions in Fig. 2 but counterintuitive for the supposed negative feedback exerted by ArsR, which would dictate that (temporarily) higher intracellular ArsR concentrations would tend to suppress the Pars promoter. In that case, there would not be a correlation between ArsR–mCherry and EGFP levels in the same cell. While keeping in mind that ArsR–mCherry does not behave exactly as ArsR itself our observations thus suggests either that there are oscillations in Pars expression at single-cell level which we cannot detect because of using stable EGFP, or that a part of the produced ArsR–mCherry is not engaged in binding its promoter (e.g. by being permanently bound to arsenite). Modelling and experimental measures of GFP output from an engineered lacI-based negative feedback circuit showed that single feedback circuits can indeed produce reporter oscillations, although not as pronounced as typical oscillatory double loop genetic circuits (Stricker et al., 2008). This may be further explored for the ArsR-controlled circuits by expanding the mechanistic model presented here to a stochastic single-cell model. The EGFP reporter variation per cell in the uncoupled circuits depends on the strength of the promoter used to produce ArsR–mCherry, and diminishes at higher ArsR–mCherry levels. However, in contrast to the feedback circuit, variation in EGFP expression for the uncoupled circuits across single cells in a population does not diminish at higher arsenite concentrations (Table 2). Also, there is a poorer correlation for the uncoupled circuits between the ArsR–mCherry level in single cells and their EGFP level (Table 2), meaning that cells can have considerable variation in ArsR–mCherry but still produce the same amount of EGFP from Pars. Understanding and controlling single-cell variation in reporter gene expression may be useful for more assays that capture the responses of relatively few cells such as, e.g. in microfluidics systems (Buffi et al., 2011).
In summary, the results of the presented work show how the Pars-arsR feedback loop can be uncoupled to produce a tunable expression system with the advantage of increasing the linear operational range or intensity of the response. The higher reporter outputs may be useful for improving the detection range in, e.g. field test assays focusing on measuring arsenic in potable water sources (Trang et al., 2005; Siegfried et al., 2012). Better understanding of the ArsR-feedback circuit may also provide alternative models for genetic circuitry, which typically concentrate on a limited number of inducible or repressible systems with little relevance for environmentally useful bioreporters.
Experimental procedures
Strains and culture conditions
All strains, plasmids and relevant characteristics are listed in Table 1. Escherichia coli strains were generally cultured on LB medium (Sambrook and Russell, 2001) at 37°C with inclusion of the appropriate antibiotics to maintain the plasmid reporter constructs, as indicated in Table 1.
Design of the arsenic reporter circuits
In the new ars reporter circuits the expression of arsR is uncoupled from its own natural Pars promoter, whereas the reporter gene remains under ArsR-repressible Pars control (Fig. 1). A synthetic DNA fragment was produced (DNA2.0, Menlo Park, CA, USA) containing arsR positioned under the control of the weak PAA constitutive promoter described by Alper and colleagues (2005), fused to a divergently oriented Pars promoter and a second ArsR binding site (ABS, Fig. 1). This 688 bp fragment (ABS_Pars_PAA_arsR) further contains specific unique restriction sites by which each individual element is interchangeable (Fig. 1). The fragment was cloned in front of the egfp reporter gene of pPROBE-tagless (Miller et al., 2000) using EcoRI and XbaI digestion. After ligation and transformation into E. coli DH5α this resulted in plasmid pAAUN. pLtetOUN, pVUN, pIIUN, pJJUN and pKUN derive from pAAUN by substituting PAA with the resynthesized PLtetO, PV, PII, PJJ or PK promoter fragments (Alper et al., 2005) (DNA2.0) via cloning in the unique SacI and BamHI sites. The integrity of the new assemblies on both plasmids was verified by DNA sequencing. The relevant part of the DNA sequence characteristic for this new family of constructs with all the different promoter regions is presented in Fig. S2 (Supporting information). An arsR–mCherry fusion was created by using a previously developed plasmid encoding a variant mCherry with a 15-amino-acid linker at its N-terminal end (Miyazaki et al., 2012). Plasmids pAAUN-mChe and pLtetOUN-mChe resulted from cloning the linker-mCherry fragment in plasmids pAAUN or pLtetOUN using the HindIII site. Proper insertions were validated by DNA sequencing (Fig. 1C and D). All the primers used for sequence verification are listed in Table S1 of Supporting information. An equivalent variant of pPR-ArsR-ABS was constructed by resynthesizing an arsR–mCherry gene fragment (DNA2.0) with the appropriate restriction sites (BamHI and SpeI) and replacing the arsR-gene in pPR-ArsR-ABS with the arsR–mCherry fusion (Fig. 1B). TetR was amplified from pME6012 (Heeb et al., 2000) in the PCR with specific primers (Table S1), and cloned in pGEM-T-Easy (Promega) downstream of Plac to produce pGem-TetR. The correct direction of the insertion was determined by sequencing on the resulting plasmids. Plasmid pGem-TetR was then introduced into E. coli MG1655 ΔRBC carrying either pAAUN or pLtet0UN.
Construction of chromosomal ars gene deletion
In order to test the influence of the native chromosomal ars operon on the functioning of the arsenic reporter constructs, we deleted the complete (ΔarsRBC) ars operon of E. coli MG1655. This was accomplished using a modification of the I-SceI recombination–digestion system (Martinez-Garcia and de Lorenzo, 2011). This system is composed of a suicide plasmid pJP5603-IsceIv2, containing a kanamycin resistance cassette and a site for the intron-specific restriction enzyme I-SceI, on each side of which two regions identical to the areas flanking the chromosomal fragment to be deleted can be cloned. For the complete ars operon deletion this consisted of fragments upstream of arsR and downstream of arsC (pJP5603-SceIv2ExtRC). Up- and downstream fragments were amplified by PCR using primers listed in Table S1, then cloned into pGEM-T-Easy and verified for correctness by DNA sequencing. Subsequently, they were retrieved by restriction digestion and cloned into pJP5603-ISceIv2. Appropriate purified pJP5603-derived plasmids were transformed into E. coli MG1655 and single recombinants were selected for kanamycin resistance. Recombination was verified by PCR amplification and when correct, those strains were subsequently transformed with the second plasmid pSW(ISceI), which carries an ampicillin resistance and bears the gene for I-SceI under the control of the Pm m-toluate-inducible promoter (Martinez-Garcia and de Lorenzo, 2011). Transformants were selected by ampicillin resistance and then induced for production of I-SceI by adding m-toluate at 15 mM. Ampicillin-resistant but kanamycin-sensitive colonies were subsequently screened by PCR for the absence of the targeted chromosomal region or for reversion to wild-type (Table S1). In case of correct deletions the strains were grown in multiple batch passages on LB medium without ampicillin until they were cured from the pSW(ISceI) plasmid.
Bioreporter cultivation
Starting from a single colony, the bioreporter strain was grown for 16 h at 37°C in LB medium in the presence of 50 μg ml−1 kanamycin to select for the presence of the pPROBE-based reporter plasmid and, when required, 100 μg ml−1 ampicillin to select for pGem-TetR, with 160 r.p.m. agitation of the culture flask. The bacterial culture was then 100-fold diluted into fresh LB medium plus kanamycin and incubated for 2 h at 160 r.p.m. agitation until the culture turbidity at 600 nm had reached between 0.3 and 0.4 for the flow cytometry (FC) assay, and between 0.4 and 0.7 for the fluorimeter assay (representative for mid-exponential-phase cells). When pre-incubation with anhydrotetracycline (aTc) was required the bacterial culture was 50-fold diluted in 5 ml of LB media supplemented with kanamycin, ampicillin and 50 μl of stock solutions of aTc ranging between 0 and 1 mg per millilitre, prepared by dissolution and successive serial dilutions in HPLC degree ethanol of pure anhydrotetracycline (IBA, Göettingen).
Cells from 10 ml of culture, or 5 ml in case of aTc pre-incubation, were harvested by centrifugation at 4000 g for 5 min and at room temperature. The cell pellet was resuspended into 30°C preheated MOPS medium to a final optical density at 600 nm of 0.4 for the fluorimeter assay and 0.2 for the FC assay [MOPS medium contains 10% (v/v) of MOPS buffer, 2 mM MgCl2, 0.1 mM CaCl2, 2 g of glucose per litre, and is set at pH 7.0]. MOPS buffer itself was prepared by dissolving, per litre: 5 g of NaCl, 10 g of NH4Cl, 98.4 g of 3-([N-morpholino]propanesulfonic acid, sodium salt), 0.59 g of Na2HPO4·2H2O and 0.45 g of KH2PO4.
Bioreporter assay preparation and readout
Both fluorimeter and FC bioreporter assays were prepared in triplicates in 96-well microplates (Greiner μCLEAR-BLACK). An aliquot of 180 μl of bioreporter suspension was mixed with 20 μl of aqueous solution containing between 0 and 1000 μg of arsenite (AsIII) per litre, prepared by serial dilution of a 0.05 M solution of NaAsO2 (Merck) in arsenic-free tap water. Bioreporter assays for fluorometry were incubated at 30°C and were mixed at 500 r.p.m. for 30 s every 10 min using a multiplate reader (FLUOstar Omega, BMG LABTECH), after which EGFP fluorescence (at 480 nm excitation and 520 nm collection) and culture turbidity (at 600 nm) were measured automatically. Reported EGFP and mCherry fluorescence values from fluorometry were normalized for culture turbidity (NFU). Bioreporter assays measured by FC were incubated at 30°C and were mixed at 500 r.p.m. for 3 h in a 96-well thermostated shaker (THERMOstar-BMG Labtech). After incubation 5 μl of all samples were diluted twice in 195 μl of distilled water, and 3 μl volume of each triplicate was aspired and analysed on a Becton Dickinson LSR-Fortessa (BD Biosciences, Erembodegem, Belgium). mCherry fluorescence of individual cells was collected in the ‘Texas-Red’ channel (610/20 nm), whereas EGFP fluorescence was registered in the ‘FITC’ channel (530/30 nm). FC fluorescence values were reported as such and not further normalized.
Modelling the ArsR-Pars system in the feedback and uncoupled configurations
A mechanistic model was developed for ArsR-mediated control of the Pars promoter using equilibrium binding affinities, in analogy of a LacI-Plac model developed by Lee and Bailey (Lee and Bailey, 1984). This model can be solved algebraically under equilibrium conditions and allows to express formation of ArsR and EGFP as a function of arsenite concentration. Essentially four configurations were modelled: (1) arsR and egfp under control of Pars (Feedback), but only plasmid copies, (2) as (1), but including a chromosomal copy of arsR and Pars, (3) arsR expression under control of a constitutive promoter with defined strength, egfp expression under control of Pars (uncoupled), but only plasmid copies, and (4) as (3), but including a chromosomal copy of arsR and Pars. Details of the model descriptions, mathematical functions and parameters are presented in Supporting information. An Excel version of the model is included as SI File 1, by which interested readers can vary model parameters or arsenite concentration ranges.
Conflict of interest
None declared.
Supporting information
Additional Supporting Information may be found in the online version of this article:
Mathematical model for ArsR circuits.
Table S1. List of all the primers used in the present work showing sequence, length and melting temperature (Tm).
Fig. S1. Nucleotide alignment of the arsRR73 and the chromosomal arsRK12 genes.
Fig. S2. Relevant part of the DNA sequence of the different promoters used for uncoupled expression of arsRR773.
Fig. S3. Relevant construction details of the feedback (A) and uncoupled (B) circuits. Sequences show part of the arsR gene, the various promoters, the ArsR Binding Sites (ABS) and the start of the egfp reporter gene.
Fig. S4. Arsenite-dependent EGFP fluorescence in cultures of E. coli MG1655 with different uncoupled arsR reporter circuits (pAAUN, pLtetOUN, pJJUN, pVUN, pKUN) compared with the feedback-controlled arsR-egfp circuit on pPR-arsR-ABS-egfp. NFU, culture density normalized fluorescence after 120 min induction time using fluorimeter measurements. Data symbols represent the average from independent biological triplicates. Whiskers, SD (when not visible lay within the symbol size).
Fig. S5. Time response kinetics of the EGFP fluorescence signal in E. coli MG1655 carrying the different feedback and uncoupled bioreporter circuits, at different arsenite concentrations between 0 and 20 μg l−1 and measured in fluorimetry. NFU, culture density normalized fluorescence. Data points show triplicate averages ± one SD.
SI File 1. Excel version of the ArsR-Pars models in feedback or uncoupled modes.
References
- Alper H, Fischer C, Nevoigt E, Stephanopoulos G. Tuning genetic control through promoter engineering. Proc Natl Acad Sci USA. 2005;102:12678–12683. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann B, van der Meer JR. Analysis of bioavailable arsenic in rice with whole cell living bioreporter bacteria. J Agric Food Chem. 2007;55:2115–2120. doi: 10.1021/jf0631676. [DOI] [PubMed] [Google Scholar]
- Bruhn DF, Li J, Silver S, Roberto F, Rosen BP. The arsenical resistance operon of IncN plasmid R46. FEMS Microbiol Lett. 1996;139:149–153. doi: 10.1111/j.1574-6968.1996.tb08195.x. [DOI] [PubMed] [Google Scholar]
- Buffi N, Merulla D, Beutier J, Barbaud F, Beggah S, van Lintel H, et al. Development of a microfluidics biosensor for agarose-bead immobilized Escherichia coli bioreporter cells for arsenite detection in aqueous samples. Lab Chip. 2011;11:2369–2377. doi: 10.1039/c1lc20274j. [DOI] [PubMed] [Google Scholar]
- Chen Y, Rosen BP. Metalloregulatory properties of the ArsD repressor. J Biol Chem. 1997;272:14257–14262. doi: 10.1074/jbc.272.22.14257. [DOI] [PubMed] [Google Scholar]
- Daunert S, Barrett G, Feliciano JS, Shetty RS, Shrestha S, Smith-Spencer W. Genetically engineered whole-cell sensing systems: coupling biological recognition with reporter genes. Chem Rev. 2000;100:2705–2738. doi: 10.1021/cr990115p. [DOI] [PubMed] [Google Scholar]
- Diorio C, Cai J, Marmor J, Shinder R, DuBow MS. An Escherichia coli chromosomal ars operon homolog is functional in arsenic detoxification and is conserved in gram-negative bacteria. J Bacteriol. 1995;177:2050–2056. doi: 10.1128/jb.177.8.2050-2056.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges RW, Baumberg S. Resistance to arsenic compounds conferred by a plasmid transmissible between strains of Escherichia coli. J Bacteriol. 1973;115:459–460. doi: 10.1128/jb.115.1.459-460.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeb S, Itoh Y, Nishijyo T, Schnider U, Keel C, Wade J, Haas D. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in gram-negative, plant-associated bacteria. Mol Plant Microbe Interact. 2000;13:232–237. doi: 10.1094/MPMI.2000.13.2.232. [DOI] [PubMed] [Google Scholar]
- de las Heras A, de Lorenzo V. In situ detection of aromatic compounds with biosensor Pseudomonas putida cells preserved and delivered to soil in water-soluble gelatin capsules. Anal Bioanal Chem. 2011;400:1093–1104. doi: 10.1007/s00216-010-4558-y. [DOI] [PubMed] [Google Scholar]
- Lee SB, Bailey JE. Genetically structured models for lac promoter-operator function in the Escherichia coli chromosome and in multicopy plasmids: Lac operator function. Biotechnol Bioeng. 1984;26:1372–1382. doi: 10.1002/bit.260261115. [DOI] [PubMed] [Google Scholar]
- Lewis C, Beggah S, Pook C, Guitart C, Redshaw C, van der Meer JR, et al. Novel use of a whole cell E. coli bioreporter as a urinary exposure biomarker. Environ Sci Technol. 2009;43:423–428. doi: 10.1021/es801325u. [DOI] [PubMed] [Google Scholar]
- Lin YF, Walmsley AR, Rosen BP. An arsenic metallochaperone for an arsenic detoxification pump. Proc Natl Acad Sci USA. 2006;103:15617–15622. doi: 10.1073/pnas.0603974103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia E, de Lorenzo V. Engineering multiple genomic deletions in Gram-negative bacteria: analysis of the multi-resistant antibiotic profile of Pseudomonas putida KT2440. Environ Microbiol. 2011;13:2702–2716. doi: 10.1111/j.1462-2920.2011.02538.x. [DOI] [PubMed] [Google Scholar]
- van der Meer JR, Belkin S. Where microbiology meets microengineering: design and applications of reporter bacteria. Nat Rev Microbiol. 2010;8:511–522. doi: 10.1038/nrmicro2392. [DOI] [PubMed] [Google Scholar]
- Miller WG, Leveau JH, Lindow SE. Improved gfp and inaZ broad-host-range promoter-probe vectors. Mol Plant Microbe Interact. 2000;13:1243–1250. doi: 10.1094/MPMI.2000.13.11.1243. [DOI] [PubMed] [Google Scholar]
- Miyazaki R, Minoia M, Pradervand N, Sulser S, Reinhard F, van der Meer JR. Cellular variability of RpoS expression underlies subpopulation activation of an integrative and conjugative element. PLoS Genet. 2012;8:e1002818. doi: 10.1371/journal.pgen.1002818. and. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton GI, Reid BJ, Semple KT. Application of a luminescence-based biosensor for assessing naphthalene biodegradation in soils from a manufactured gas plant. Environ Pollut. 2009;157:1643–1648. doi: 10.1016/j.envpol.2008.12.020. [DOI] [PubMed] [Google Scholar]
- Ramanathan S, Shi W, Rosen BP, Daunert S. Sensing antimonite and arsenite at the subattomole level with genetically engineered bioluminescent bacteria. Anal Chem. 1997;69:3380–3384. doi: 10.1021/ac970111p. [DOI] [PubMed] [Google Scholar]
- Rosen BP. Resistance mechanisms to arsenicals and antimonials. J Basic Clin Physiol Pharmacol. 1995;6:251–263. doi: 10.1515/jbcpp.1995.6.3-4.251. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, USA: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Siegfried K, Endes C, Bhuiyan AF, Kuppardt A, Mattusch J, van der Meer JR, et al. Field testing of arsenic in groundwater samples of Bangladesh using a test kit based on lyophilized bioreporter bacteria. Environ Sci Technol. 2012;46:3281–3287. doi: 10.1021/es203511k. [DOI] [PubMed] [Google Scholar]
- Stocker J, Balluch D, Gsell M, Harms H, Feliciano JS, Daunert S, et al. Development of a set of simple bacterial biosensors for quantitative and rapid field measurements of arsenite and arsenate in potable water. Environ Sci Technol. 2003;37:4743–4750. doi: 10.1021/es034258b. [DOI] [PubMed] [Google Scholar]
- Stricker J, Cookson S, Bennett MR, Mather WH, Tsimring LS, Hasty J. A fast, robust and tunable synthetic gene oscillator. Nature. 2008;456:516–519. doi: 10.1038/nature07389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani C, Inoue K, Tani Y, Harun-ur-Rashid M, Azuma N, Ueda S, et al. Sensitive fluorescent microplate bioassay using recombinant Escherichia coli with multiple promoter-reporter units in tandem for detection of arsenic. J Biosci Bioeng. 2009;108:414–420. doi: 10.1016/j.jbiosc.2009.05.014. [DOI] [PubMed] [Google Scholar]
- Tauriainen S, Karp M, Chang W, Virta M. Recombinant luminescent bacteria for measuring bioavailable arsenite and antimonite. Appl Environ Microbiol. 1997;63:4456–4461. doi: 10.1128/aem.63.11.4456-4461.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tecon R, Beggah S, Czechowska K, Sentchilo V, Chronopoulou PM, McGenity TJ, van der Meer JR. Development of a multistrain bacterial bioreporter platform for the monitoring of hydrocarbon contaminants in marine environments. Environ Sci Technol. 2010;144:1049–1055. doi: 10.1021/es902849w. and. [DOI] [PubMed] [Google Scholar]
- Trang PT, Berg M, Viet PH, Van Mui N, van der Meer JR. Bacterial bioassay for rapid and accurate analysis of arsenic in highly variable groundwater samples. Environ Sci Technol. 2005;39:7625–7630. doi: 10.1021/es050992e. and. [DOI] [PubMed] [Google Scholar]
- Turner K, Xu S, Pasini P, Deo S, Bachas L, Daunert S. Hydroxylated polychlorinated biphenyl detection based on a genetically engineered bioluminescent whole-cell sensing system. Anal Chem. 2007;79:5740–5745. doi: 10.1021/ac0705162. [DOI] [PubMed] [Google Scholar]
- Wu CH, Le D, Mulchandani A, Chen W. Optimization of a whole-cell cadmium sensor with a toggle gene circuit. Biotechnol Prog. 2009;25:898–903. doi: 10.1002/btpr.203. [DOI] [PubMed] [Google Scholar]
- Wu J, Rosen BP. The ArsR protein is a trans-acting regulatory protein. Mol Microbiol. 1991;5:1331–1336. doi: 10.1111/j.1365-2958.1991.tb00779.x. [DOI] [PubMed] [Google Scholar]
- Wu J, Rosen BP. Metalloregulated expression of the ars operon. J Biol Chem. 1993;268:52–58. [PubMed] [Google Scholar]
- Zhou T, Radaev S, Rosen BP, Gatti DL. Structure of the ArsA ATPase: the catalytic subunit of a heavy metal resistance pump. EMBO J. 2000;19:4838–4845. doi: 10.1093/emboj/19.17.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of all the primers used in the present work showing sequence, length and melting temperature (Tm).
Fig. S1. Nucleotide alignment of the arsRR73 and the chromosomal arsRK12 genes.
Fig. S2. Relevant part of the DNA sequence of the different promoters used for uncoupled expression of arsRR773.
Fig. S3. Relevant construction details of the feedback (A) and uncoupled (B) circuits. Sequences show part of the arsR gene, the various promoters, the ArsR Binding Sites (ABS) and the start of the egfp reporter gene.
Fig. S4. Arsenite-dependent EGFP fluorescence in cultures of E. coli MG1655 with different uncoupled arsR reporter circuits (pAAUN, pLtetOUN, pJJUN, pVUN, pKUN) compared with the feedback-controlled arsR-egfp circuit on pPR-arsR-ABS-egfp. NFU, culture density normalized fluorescence after 120 min induction time using fluorimeter measurements. Data symbols represent the average from independent biological triplicates. Whiskers, SD (when not visible lay within the symbol size).
Fig. S5. Time response kinetics of the EGFP fluorescence signal in E. coli MG1655 carrying the different feedback and uncoupled bioreporter circuits, at different arsenite concentrations between 0 and 20 μg l−1 and measured in fluorimetry. NFU, culture density normalized fluorescence. Data points show triplicate averages ± one SD.
SI File 1. Excel version of the ArsR-Pars models in feedback or uncoupled modes.