Summary
The members of the Lactobacillus genus are widely used in the food and feed industry and show a remarkable ecological adaptability. Several Lactobacillus strains have been marketed as probiotics as they possess health-promoting properties for the host. In the present study, we used two complementary next-generation sequencing technologies to deduce the genome sequences of two Lactobacillus casei strains LcA and LcY, which were isolated from the products Actimel and Yakult, commercialized as probiotics. The LcA and LcY draft genomes have, respectively, an estimated size of 3067 and 3082 Mb and a G+C content of 46.3%. Both strains are close to identical to each other and differ by no more than minor chromosomal re-arrangements, substitutions, insertions and deletions, as evident from the verified presence of one insertion-deletion (InDel) and only 29 single-nucleotide polymorphisms (SNPs). In terms of coding capacity, LcA and LcY are predicted to encode a comparable exoproteome, indicating that LcA and LcY are likely to establish similar interactions with human intestinal cells. Moreover, both L. casei LcA and LcY harboured a 59.6 kb plasmid that shared high similarities with plasmids found in other L. casei strains, such as W56 and BD-II. Further analysis revealed that the L. casei plasmids constitute a good evolution marker within the L. casei species. The plasmids of the LcA and LcY strains are almost identical, as testified by the presence of only three verified SNPs, and share a 3.5 kb region encoding a remnant of a lactose PTS system that is absent from the plasmids of W56 and BD-II but conserved in another smaller L. casei plasmid (pLC2W). Our observations imply that the results obtained in animal and human experiments performed with the Actimel and Yakult strains can be compared with each other as these strains share a very recent common ancestor.
Funding Information The present work was supported by the Center of Excellence in Microbial Food Safety Research (Academy of Finland, Grant 141140), Grant ERC 250172 – Microbes Inside from the European Research Council and Grants 137389 and 141130 from the Academy of Finland. F.P.D. was funded by a postdoctoral research fellowship (Academy of Finland, Grant 252123).
Introduction
Lactobacilli belong to the lactic acid bacteria (LAB) group and constitute a very large genus of Gram-positive, non-sporulating bacteria that show remarkable ecological adaptability and phylogenetic diversity (Axelsson, 2004). Commonly found in human body cavities, i.e. vagina, intestinal tract and oral cavity, they also naturally persist in a broad range of food environments, such as fermented milk, meat and plant (Kandler and Weiss, 1986). A number of lactobacilli species have been extensively implemented in various industrial processes, as starter or adjunct cultures (Stiles and Holzapfel, 1997). More recently, some strains of the species Lactobacillus casei, L. paracasei, L. reuteri, L. rhamnosus, L. acidophilus, L. plantarum and L. johnsonii have also been increasingly employed in food products marketed as probiotics (de Vos, 2011). The health benefits associated with the consumption of products containing some of these commercialized Lactobacillus strains have been demonstrated by a number of human clinical studies on patients with diverse disorders (Kalliomaki et al., 2003; Saxelin et al., 2005; Sykora et al., 2005; Almeida et al., 2012; Hajela et al., 2012). However, the large diversity of lactobacilli also suggested that probiotic functions and health-promoting properties are specific to each strain, justifying the need to examine probiotic lactobacilli strains individually on a genomic and molecular basis (Ventura et al., 2009). In this regard, the L. casei group is of particular interest, as it constitutes an important reservoir of probiotic-marketed strains. Although subject to recurrent and controversial changes over its taxonomy and nomenclature, the L. casei group currently consists of three species: L. casei, L. paracasei and L. rhamnosus (Collins et al., 1989; Klein et al., 1998; Dellaglio et al., 2002; Dobson et al., 2004; Sato et al., 2012). The L. casei group includes some well-documented strains, such as L. rhamnosus strain GG (Kankainen et al., 2009; Lebeer et al., 2012) or L. casei strain BL23 (Maze et al., 2010). Although not known to be commercialized, L. casei strain BL23 possesses potential probiotic functions which relate to its anti-inflammatory properties in an animal model (Foligne et al., 2007; Rochat et al., 2007) and its ability to bind extracellular matrix in vitro (Munoz-Provencio et al., 2010). A majority of L. casei strains are also resistant to stresses, such as exposure to acid and bile salts present in the gastrointestinal tract, a trait characteristic of probiotic strains (Broadbent et al., 2010; Alcántara and Zúñiga, 2012; Hamon et al., 2012).
Peculiarly, the genomes of some probiotic L. casei strains widely used in probiotic products have not been examined. Up to date, only six complete L. casei genomes have been sequenced: L. casei ATCC 334 (Makarova et al., 2006), L. casei BL23 (Maze et al., 2010), L. casei Zhang (Zhang et al., 2007), L. casei LCW2 (Chen et al., 2011), L. casei W56 (Hochwind et al., 2012) and L. casei BD-II (Ai et al., 2011) and 12 additional L. casei genomes have been partially sequenced (NCBI database, as on 1 March 2013). Recent comparative genomic analyses of the L. casei species provided important and valuable insights into the species diversity in regards to its ecological versatility and genome evolution (Cai et al., 2009; Broadbent et al., 2012). Broadbent and colleagues also demonstrated that the presence of other bacterial species residing in L. casei ecological habitats clearly impact the L. casei pan-genome, indicating that the Distributed Genome Hypothesis can be applied to non-pathogenic bacterial species, such as L. casei. This also illustrated the significant role of horizontal gene transfer events in lifestyle adaptation (Broadbent et al., 2012). Moreover, the significant genome decay observed in some L. casei strains adapted to the dairy environment, is believed to contribute to the diversity of the species (Broadbent et al., 2012).
Recently, we addressed the genome stability of Lactobacillus strains, used in industrial processes (Douillard et al., 2013). By genomic re-sequencing of product isolates of strain L. rhamnosus GG, we determined the stability of this widely used probiotic strain (Douillard et al., 2013). Identical probiotic functions were detected in strains isolated from products from bona fide producers, testifying for the stability of L. rhamnosus GG, which has a well-characterized 3.01 Mb genome (Kankainen et al., 2009). However, a different situation may be encountered when these or other strains are not handled appropriately by the manufacturer. This seems to be the case for some unspecified products with L. rhamnosus strains, as reported recently, although post-sampling events cannot be excluded (Sybesma et al., 2013). Specifically, a deletion was identified in a region encoding the genes for the production of extracellular pili that bind to human intestinal mucus (Kankainen et al., 2009; Reunanen et al., 2012; Sybesma et al., 2013). In contrast, we showed that L. casei strains isolated from two globally marketed products, Actimel and Yakult, contained pili genes that were highly conserved in sequence to that of L. rhamnosus GG but did not express these pili under the tested conditions (Douillard et al., 2013). This was explained by the absence of an IS-mediated promoter that drives constitutive pili expression in L. rhamnosus GG (Douillard et al., 2013). The present study expands on this by providing a further detailed comparative genomic characterization of these two L. casei strains that derive from strains branded as L. casei defensis and L. casei Shirota. It shows that these strains are highly similar though having a reportedly different history. Among others this is illustrated by the presence of 29 confirmed SNPs and 1 verified InDel, an identical predicted exoproteome and a ∼59 kb plasmid that is nearly identical in both strains.
Results and discussion
Genomic features of L. casei strain LcY isolated from the Yakult product
The probiotic-marketed product Yakult is commercialized by Yakult Honsha (Japan) and contains L. casei strain Shirota that has previously been subject to multiple studies with regards to its potential health-promoting properties in humans and animals (Rochat et al., 2007; Matsumoto et al., 2010; Naito et al., 2011; Nanno et al., 2011; Almeida et al., 2012). In the present study, we sequenced the genome of L. casei strain LcY that had been isolated from the Yakult product (Douillard et al., 2013) and is assumed to represent L. casei strain Shirota. Genomic DNA from L. casei LcY was isolated and sequenced by a combination of two next-generation sequencing platforms 454 GS-FLX+ (Life Sciences) and SOLiD (Life Technologies). The contig order and orientation were determined using L. casei BL23, W56, LC2W and BD-II as reference genomes (Maze et al., 2010; Ai et al., 2011; Chen et al., 2011; Hochwind et al., 2012), allowing us to assemble the de novo contigs into one single scaffold (Table 1). The thus constructed L. casei LcY draft genome consists of one circular chromosome with an estimated size of 3 082 048 bp and an overall GC content of 46.33%. It also harbours an estimated 59.6 kb plasmid designated pYAK that shows high sequence identity with plasmids pW56 and pBD-II respectively present in L. casei strains W56 and BD-II (Ai et al., 2011; Hochwind et al., 2012). Initial automated annotation of the L. casei LcY genome (and LcA; see below) was performed using RAST (Aziz et al., 2008). However, due to the very high sequence identity and synteny between some L. casei strains (Figs 1 and 2), we re-annotated both genomes using the Rapid Annotation Transfer Tool (RATT) (Aziz et al., 2008), as we observed that this tool provided more accurate gene annotations (data not shown). A total of 3119 coding DNA sequences (CDS) were predicted and annotated using RATT (Otto et al., 2011), including 3044 proteins (Table 1). Five rRNA operons, containing 5S, 16S and 23S rRNA genes, are scattered throughout the genome of LcY. The first three rRNA operons are located in the leading strand and the other two on the lagging strand. Forty-nine tRNAs were located in the vicinity of the rRNA genes, whereas the other 11 tRNAs were uniformly distributed on the chromosome. Interestingly, three unique tRNAs species were identified, e.g. tRNAs for cysteine, histidine and tryptophane.
Table 1.
General genomic features of L. casei strains LcA and LcY isolated from Actimel and Yakult probiotic-marketed products
| Genetic features | L. casei LcY | L. casei LcA | L. casei BL23 |
|---|---|---|---|
| Origin | Yakult | Actimel | N/A |
| Number of scaffolds | 1 | 1 | 1 |
| Estimated chromosome size | 3 082 048 | 3 066 955 | 3 079 196 |
| GC content (%) | 46.33 | 46.34 | 46.34 |
| Coding DNA sequences (CDS) | 3 119 | 3 109 | 3 119 |
| Protein coding sequences | 3 044 | 3 031 | 3 044 |
| RNA genes | 75 | 78 | 75 |
| rRNA genes | 15 | 16 | 15 |
| tRNA genes | 60 | 62 | 60 |
| Plasmids | 1 | 1 | 0 |
| CRISPR locus | 1 | 1 | 1 |
Data shown in the table have been estimated based on draft genome assembly and RATT transfer gene annotation. Features relative to the plasmids pACT and pYAK respectively present in strains LcA and LcY have not been incorporated in the table.
N/A, not available.
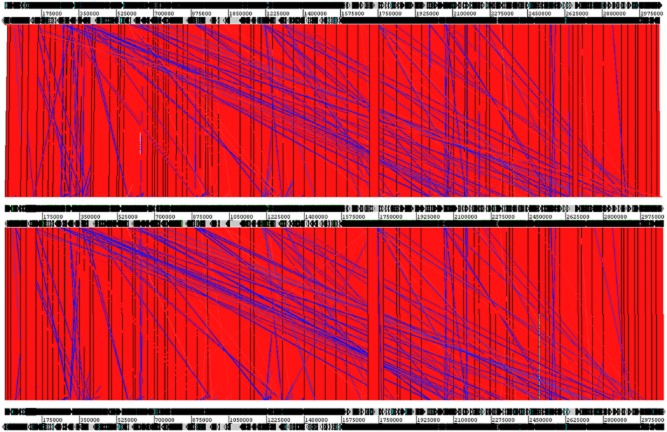
Figure 1.
Comparison of the genomes of L. casei LcA, LcY and BL23 using ACT (Artemis Comparison Tool). LcY, BL23 and LcA are respectively shown from top to bottom in the figure. Bars designate conserved chromosomal regions between LcA, LcY and BL23 (BlastN hits) that have the same orientation (red) or have been inverted (blue).
Figure 2.
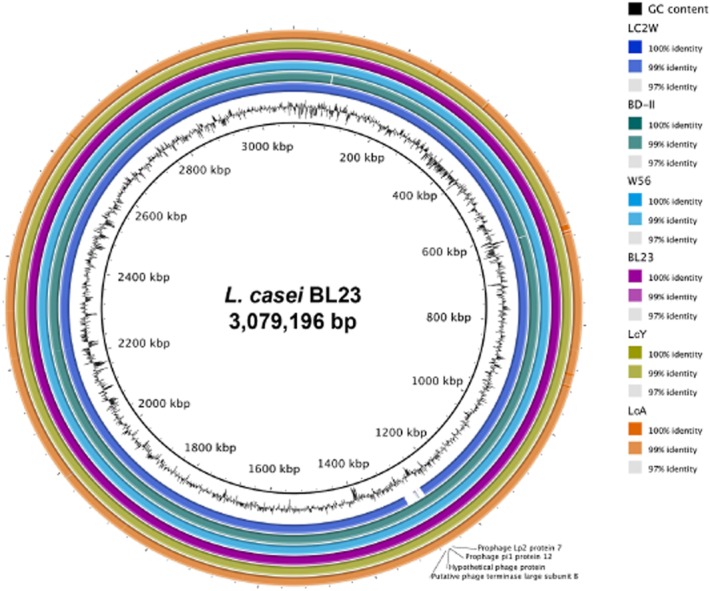
BLAST Ring Image Generator (BRIG) analysis of L. casei strains LcA, LcY, BD-II, W56, LC2W and BL23. Lactobacillus casei strain BL23 was used as the reference genome. Colours indicate the percentage of sequence identity.
Genomic features of L. casei strain LcA isolated from the Actimel product
The probiotic-marketed drinkable yogurt Actimel is produced by Danone and contains L. casei strain DN-114001, branded as L. casei defensis. Lactobacillus casei strain DN-114001 has been studied for its potential probiotic properties in various animal and human studies (Marcos et al., 2004; Sykora et al., 2005; Pawlowska et al., 2007; Guillemard et al., 2010). We previously isolated and characterized the L. casei strain LcA present in the Actimel product (Douillard et al., 2013). We assume that L. casei LcA is virtually identical to the seed culture of L. casei strain DN-114001. Using two next-generation sequencing platforms, we determined the genome sequence of L. casei strain LcA similarly as described above (Table 1). The thus obtained draft genome of L. casei LcA consists of one scaffold with an estimated size of 3 066 955 bp and a 59.6 kb plasmid (pACT). The overall G+C content of LcA chromosome is 46.34%. A total of 3109 CDS were predicted and annotated using RATT (Otto et al., 2011), including 3031 proteins (Table 1). The total number of CDS for both strains LcA and LcY is comparable to BL23 (3119 CDS) and in line with previous CDS range prediction in lactobacilli (2700–3700) (Makarova et al., 2006). Five rRNA operons, containing 5S, 16S and 23S rRNA genes, and 62 tRNA genes are present the genome of L. casei LcA and show a similar distribution as in L. casei LcY. Three tRNAs species were unique in LcA, e.g. tRNAs for cysteine, histidine and tryptophane and tyrosine. To reduce the analysis load, we did not address the exact number of pseudogenes and IS elements in this study.
Genetic relatedness and comparison with other members of the L. casei species
In terms of genomic features, there were clear similarities between the two strains, i.e. genome size, GC% content and CDS number (Table 1). We first compared L. casei LcA and LcY to L. casei BL23 as the latter strain also has been found to have potential probiotic properties, although it is not commercialized. Genomic alignments of L. casei strains LcA, LcY and BL23 further showed a high degree of synteny with large identical chromosomal blocks, indicating that genome differences are mostly due to minor genetic recombination events (Fig. 1). Further comparative genomic analysis of the genomes of L. casei LcA, LcY and other L. casei strains revealed their close genetic relatedness (Fig. 2), which is in line with a previous comparative genomic study on the L. casei species (Broadbent et al., 2012). In the species L. casei, six distinct sublineages were defined and one of them consists of three closely related strains BL23, BD-II and LC2W (Broadbent et al., 2012). Our data indicate that L. casei strains LcA, LcY and the recently sequenced strain W56 also belong to that very same cluster, whereas other sequenced L. casei strains, such as strain Zhang and ATCC334 distinctly evolved from the common ancestor. Lactobacillus casei strains LcA and LcY are highly similar to each other, as shown by synteny plot (Fig. 3A). However, some small differences were observed between L. casei LcA and LcY genomes. When comparing the number of shared genes between L. casei LcA and LcY, it is estimated that 3031 genes were present in both strains, which represent 99.6% of the gene content of strain BL23 (Maze et al., 2010). All genes of strain BL23 were found in L. casei LcY, whereas 13 genes present in strain BL23 were missing in LcA based on RATT analysis. These 13 genes that were predicted to encode putative proteins and transposases are likely missed due the present state of the genomes, since repetitive elements is one of the most common cause for assembly gaps (Kingsford et al., 2010). These results also suggest that L. casei LcA, LcY and BL23 have a virtually similar coding capacity. We then used both SOLiD and 454 data to look at the presence of any SNPs and InDels between the two L. casei draft genomes. SOLiD reads of LcA were mapped to the sequence of LcY and vice versa. Identical SNPs and InDels identified in both mappings were then manually inspected. We were able then to confidently identify the presence of one InDel and a total of 26 SNPs (Table 2 and Fig. 3B). The InDel found in LcA strain is located in the gene LCACT_2629 (gltB), encoding a NADPH-dependent glutamate synthase and it results in the truncation of the protein. Four of the SNPs are located in intercistronic regions, while the remaining ones did not generate any stop codons and only gave rise to some missense mutations, testifying for the similarity of the strains (Table 2). This is confirmed by the high degree of metabolic similarity that we described previously and indicates that the small differences in efficiency of sugar utilization are likely to be explained by experimental variations (Douillard et al., 2013).
Figure 3.
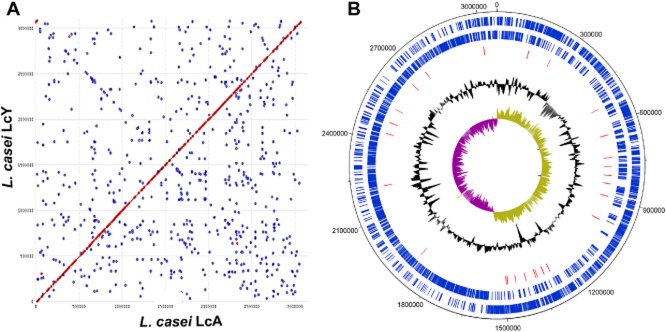
Comparison of the genomes of L. casei strains LcA and LcY.A. Synteny plot shows the comparison at the nucleotide level of the genome of L. casei LcY (vertical axis) with the genome of L. casei LcA (horizontal axis). Red dots indicate homologous chromosomal regions between genomes with the same orientation. Blue dots indicate homologous chromosomal regions which are in the opposite direction.B. Genome circle of L. casei LcA using DNAPlotter (Carver et al., 2009). Blue bars indicate CDS and red bars indicate SNPs and InDels identified when compared with L. casei LcY.
Table 2.
List of unequivocal SNPS and InDels identified and verified between LcA and LcY genomes, chromosome and plasmid included
| LcY coordinate | Base | Base | LcA coordinate | AA change LcY/LcA | Locus tag and predicted gene product |
|---|---|---|---|---|---|
| Chromosome | |||||
| 137132 | T | G | 136805 | N14H | LCACT_0132/LCY_0132, putative protein |
| 236020 | A | T | 235693 | N340I | LCACT_0235/LCY_0235, inulosucrase (fragment) |
| 530929 | G | A | 527365 | Q82Q | LCACT_0522/LCY_0523, ABC transporter |
| 649781 | C | A | 641841 | N/A | Intercistronic region |
| 649813 | T | G | 641373 | N/A | Intercistronic region |
| 744458 | A | C | 736004 | Y135D | LCACT_0725/LCY_0737, tagatose 1,6-diphosphate aldolase |
| 780579 | T | C | 772125 | I39M | LCACT_0759/LCY_0771, putative protein |
| 817971 | G | T | 809517 | R140R | LCACT_0800/LCY_0812, HAD-superfamily hydrolase |
| 852271 | A | G | 843663 | E13E | LCACT_0832/LCY_0844, thioredoxin |
| 904463 | A | G | 895886 | G26G | LCACT_0880/LCY_0892, putative protein |
| 1010123 | G | A | 996844 | R136K | LCACT_0988/LCY_1000, ABC transporter |
| 1314821 | A | G | 1300766 | K76K | LCACT_1311/LCY_1323, beta-lactamase family protein |
| 1342691 | A | T | 1328636 | I5F | LCACT_1341/LCY_1353, cysteine desulfurase |
| 1372643 | T | G | 1358501 | N/A | Intercistronic region |
| 1379614 | A | G | 1365472 | F97L | LCACT_1383/LCY_1395, ABC transporter related |
| 1423524 | C | T | 1409467 | P107L | LCACT_1434/LCY_1446, methyltransferase |
| 1467351 | A | G | 1453376 | T219A | LCACT_1475/LCY_1487, acetyltransferase |
| 1502680 | C | G | 1488607 | N/A | Intercistronic region |
| 1508926 | G | A | 1494853 | D338N | LCACT_1510/LCY_1522, acyltransferase |
| 1901798 | C | A | 1886618 | L313I | LCACT_1895/LCY_1907, sensor protein |
| 2231304 | G | A | 2217739 | G199D | LCACT_2201/LCY_2214, putative protein |
| 2447647 | T | C | 2433375 | N64S | LCACT_2405/LCY_2418, oxidoreductase |
| 2494439 | G | T | 2480167 | P166Q | LCACT_2449/LCY_2462, uridine phosphorylase |
| 2554046 | G | T | 2539774 | G76V | LCACT_2511/LCY_2524, transcriptional regulator |
| 2754290 | T | G | 2740097 | Q51P | LCACT_2701/LCY_2714, glutathione reductase |
| 3025404 | C | T | 3011097 | R383Q | LCACT_2974/LCY_2987, malate oxidoreductase |
| 2678962 | A | . | 2664876 | Frameshift | LCACT_2629/LCY_2642, NADPH-dependent glutamate synthase (truncation in LCACT_2629) |
| Plasmid | |||||
| 168 | C | A | 168 | N/A | Intercistronic region |
| 27281 | T | C | 27281 | Y691Y | pACT_0028/pYAK_0028, hypothetical protein |
| 37993 | G | A | 37993 | N/A | Intercistronic region |
The SNPs and InDels were identified based on reciproqual SOLiD mapping to each draft genome sequence of L. casei strains LcA and LcY.
N/A, not available.
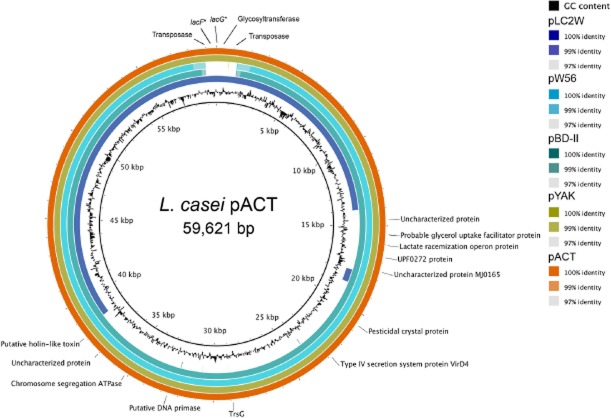
Plasmids pACT and pYAK
Genome sequencing of both L. casei strains LcA and LcY also revealed the presence of one 59.6 kb plasmid, respectively, named pACT and pYAK. PCR amplification was used to confirm the presence of the plasmid in LcA and LcY by joining all the contigs into a single circular molecule. Our data suggest that plasmids pACT and pYAK are nearly identical in terms of size and gene content and highly similar to two previously reported plasmids found in L. casei strains W56 and BD-II (Ai et al., 2011; Hochwind et al., 2012). The plasmid draft sequences were carefully compared using a similar approach as mentioned above. Three SNPs were detected and confirmed (Table 2). pACT and pYAK share all sequences present in the 38 kb plasmid pLC2W found in L. casei strain LC2W (Ai et al., 2011), indicating that a major deletion occurred in this plasmid (Fig. 4). Remarkably, this shared 38 kb region includes a 3.5 kb fragment not present in plasmids pW56 and pBD-II (Fig. 4). This fragment contains a remnant of lactose PTS system. No significant homologies between pACT, pYAK and plasmids from L. casei strains Zhang and ATCC334 were found. In spite of the recent sequencing of strains W56, BD-II and LC2W, their respective plasmids remained poorly annotated and comprehended. Similarly, plasmids pYAK and pACT are also predicted to encode mostly hypothetical proteins, transposases and a restriction–modification enzyme system. We did not observed any unique functional traits that are observed in LAB and in Lactobacilli, such a lactose hydrolysis (Cai et al., 2009) or copper resistance (Barré et al., 2007). To further explore the properties of these plasmids, comparative functional analysis of these strains and their plasmid-cured derivatives would possibly reveal some features of interest. It is noteworthy that a higher diversity was observed within the L. casei plasmids pACT, pYAK, pW56, pBD-II and pLC2W than within the chromosomes of the same L. casei strains, suggesting that plasmids evolves faster than the genomes and are suitable markers to differentiate L. casei strains (Fig. 4). Similar observations have recently been reported for Burkholderia and Vibrio (Cooper et al., 2010).
Figure 4.
BLAST Ring Image Generator (BRIG) analysis of the plasmids present in L. casei strains LcA, LcY, W56, BD-II and Lc2W. Plasmid of the L. casei strain LcA was used as the plasmid backbone. Colours indicate the percentage of sequence identity. Genes present in the variable regions are also indicated on the figure. Genes marked with an asterisk are altered.
Environmental traits
Chromosomal sequences that are known to be highly variable can be found in the CRISPR (clustered regularly interspaced short palindromic repeats) loci that are found in almost half of the prokaryotic genomes (Horvath and Barrangou, 2010). The CRISPRs and their associated cas-genes from a prokaryotic immune system and play an important role in regulating horizontal gene transfers. It has been well documented that some bacteria acquired a CRISPR locus as it provides protection and immunization against plasmids, phages and other mobile elements (Horvath and Barrangou, 2010; Marraffini and Sontheimer, 2010; Makarova et al., 2011). We identified the presence of a Type II-A/LsaI1 CRISPR-cas system in the genome of both L. casei LcA and LcY, consisting of four cas genes and an array of 21 spacers interspaced by a 36 bp direct repeat. Identical CRISPR locus could also be found in BL23, BD-II and LC2W (Ai et al., 2011; Hochwind et al., 2012; Broadbent et al., 2012). Another relevant property for industrial strains marketed as probiotics is their ability to endure a number of stresses that are typically encountered during production and also in the gastrointestinal tract. In the genomes of both L. casei LcA and LcY, we identified genes encoding heat (i.e. GroEL and GroES), cold (i.e. CspA and CspG) and alkaline shock proteins, SOS-response proteins and Clp proteases (ClpE, ClpX, ClpL and ClpP).
Prediction of the exoproteome, cell-surface proteins and other relevant functions
Proteins expressed on the bacterial cell surface play essential roles in bacteria–host cross-talk, virulence, colonization, immunogenicity and have been proposed to constitute probiotic traits. Using the SignalP prediction model, a total of 162 proteins were predicted to have a peptide signal in both L. casei LcA and LcY, indicating that they may be translocated across the cellular membrane and constitute the exoproteome. LcA and LcY strains possess a comparable set of secreted proteins. A total of 47% of these were predicted to be cell wall associated but have not been annotated, underlining the fact that a number of interaction players remain to be uncovered. Recent studies performed in L. casei identified new candidate proteins that could contribute to the bacteria–host interaction. For example, a gene encoding a fibronectin/fibrinogen-binding protein FbpA in BL23 was found to be involved in the attachment of the bacterial cells with the intestinal epithelium (Munoz-Provencio et al., 2010). Interestingly, an FbpA homologue was also encoded in both LcA and LcY genomes (LCACT_1599 and LCY_1611 respectively). Two other proteins of interest found in LcA and LcY are p40 (LCACT_0271; LCY_0271) and p75 (LCACT_0023; LCY_0023), and are identical to the two major secreted proteins in L. rhamnosus GG (Claes et al., 2012). These proteins have attracted considerable attention as they were found to display anti-apoptotic and cellular protective properties on the intestinal epithelium in L. rhamnosus GG (Yan et al., 2007) and L. casei BL23 (Bauerl et al., 2010). Other predicted secreted proteins (23%) were mostly associated with ABC transporters and phosphotransferase systems essential in the cell metabolism.
We further examined the genes encoding LPxTG or LPxTG-like proteins, as they typically have a pivotal role in virulence, colonization or persistence in ecological niches. Cell-wall-associated LPxTG proteins typically possess a C-terminal sorting signal domain consisting of a highly conserved LPxTG motif flanked by a chain of hydrophobic and positively charged amino acidic residues (Navarre and Schneewind, 1999; Mazmanian et al., 2001). The LPxTG motif is specifically recognized by membrane-associated sortases that cleave the LPXTG motif between the threonine (T) residue and the glycine (G) residue and then covalently anchors the proteins to the peptidoglycan layer (Schneewind and Missiakas, 2012). Although LPxTG proteins are well documented in Gram-positive pathogens, their roles in commensal intestinal lactobacilli only begin to be comprehended. In strains LcA and LcY, these include enzymes, adhesive glycoproteins and pilin-specific proteins (Table 3). Notably, six of them were encoding pili subunits and were clustered in two distinct pili operons, spaFED and spaCBA, termed after their homologous genes in L. rhamnosus GG (Kankainen et al., 2009). Both operons showed a similar genetic order where the three pilin-specific genes were associated with a pilin-specific sortase. Similarly, the strain LcA has these two pili gene clusters (Table 3). The genetic organization of the pili operon is widespread within the L. casei species, as previously noted (Broadbent et al., 2012). Despite the presence of pili operons, a recent study demonstrated that the strains LcA, LcY and BL23 did not express mucus-binding pili in vitro (Douillard et al., 2013). In L. rhamnosus GG, the orthologous pili gene cluster spaCBA-srtC is expressed and confers strong mucus-binding abilities to the host (Kankainen et al., 2009; Lebeer et al., 2012; Reunanen et al., 2012). We showed that the insertion of an IS element upstream the spaC adhesion pilin gene constituted a functional promoter in L. rhamnosus GG, whereas in the L. casei strains it is absent, explaining the lack of pili production under conditions that resemble industrial and intestinal conditions (Douillard et al., 2013).
Table 3.
List of predicted proteins in L. casei strains LcA and LcY harbouring a LPxTG or LPxTG-like motif
| Gene name | Predicted gene product | Gene name |
|---|---|---|
| Strain LcA | Strain LcY | |
| LCACT_0280 | β-N-acetylglucosaminidase precursor | LCY_0280 |
| LCACT_0513 | Pilus-specific adhesion protein (SpaC) | LCY_0514 |
| LCACT_0514 | Pilus-specific minor-backbone protein (SpaB) | LCY_0515 |
| LCACT_0515 | Pilus-specific major-backbone protein (SpaA) | LCY_0516 |
| LCACT_0525 | Cell envelope-associated proteinase | LCY_0526 |
| LCACT_0526 | Cell envelope-associated proteinase (PrtR) | LCY_0527 |
| LCACT_0651 | Hypothetical protein | LCY_0663 |
| LCACT_1204 | Hypothetical protein | LCY_1216 |
| LCACT_2049 | Hypothetical protein | LCY_2061 |
| LCACT_2373 | Cell-wall-associated serine proteinase | LCY_2386 |
| LCACT_2425 | Mucus-binding factor (MBF) | LCY_2438 |
| LCACT_2463 | Pilus-specific adhesion protein (SpaF) | LCY_2476 |
| LCACT_2462 | Pilus-specific minor backbone protein (SpaE) | LCY_2475 |
| LCACT_2461 | Pilus-specific major-backbone protein (SpaD) | LCY_2474 |
| LCACT_2524 | Outer membrane protein | LCY_2537 |
Reports on the function of other LPxTG proteins in lactobacilli are still sparse, in spite of the fact that several species contain strains that are marketed as probiotics, i.e. L. plantarum, L. rhamnosus, L. salivarius. Notably, two housekeeping sortases genes were identified in L. casei LcA and LcY, similar as in strain BL23 (Munoz-Provencio et al., 2012). In the latter strain, it was found that the housekeeping sortases SrtA are required for the cell wall anchoring of N-acetylglucosaminidases and proteinases (Munoz-Provencio et al., 2012). In addition to the pilin-specific proteins mentioned above, L. casei LcA and LcY are predicted to encode nine LPxTG and LPxTG-like proteins that are covalently coupled to the bacterial cell wall. It is noteworthy that the LPxTG motif is highly conserved in all these proteins along with the presence of hydrophobic tail essential for their retention on the cell surface prior to their sortase-mediated cell wall anchoring (Perry et al., 2002; Hendrickx et al., 2011). Most of these have no known function, but it is reasonable to think that these surface-anchored proteins may be involved in the intestinal colonization and lifestyle of L. casei. Interestingly, the genomes of both L. casei LcA and LcY also are predicted to encode the Mucus-Binding Factor (MBF). The MBF protein is well conserved in L. casei strains but also in the L. rhamnosus strains, where it has an orthologue that was shown to be involved in adhesion mechanisms in the intestinal tract (von Ossowski et al., 2011). However, the MBF does not seem to play a significant role in L. casei LcA and LcY, as both strains did not show any mucus-binding ability in vitro (Douillard et al., 2013).
Conclusions
Lactobacillus casei strains LcA and LcY are genomically closely related to a number of other L. casei strains, including the well-studied L. casei BL23. Comparative analysis of the L. casei plasmids revealed a greater diversity than found at the chromosomal level, indicating that, in spite of their extrachromosomal nature, plasmids are a good indicator of the evolution of L. casei species and further substantiates the high identity between LcA, LcY and other L. casei strains. Lactobacillus casei LcA and LcY contain a comparable set of genes encoding secreted proteins, underlining that they may display similar probiotic functions to the host. The limited number of SNPs and InDels detected in both chromosome and plasmid (in total 29 SNPs and one InDel) indicates that the strains LcA and LcY are very closely related. We also cannot exclude the possibility that some of this limited amount of diversity was generated during the strain isolation procedure. The genomic sequences reported in the present study offer a functional basis to facilitate the understanding of the well-known L. casei strains LcA and LcY. Our observations also imply that the results obtained in animal and human experimental studies performed with the L. casei strains used in Actimel and Yakult products may be compared with each other as these two strains share a very recent common ancestor.
Experimental procedures
Bacterial strains, growth conditions and DNA isolation
The two L. casei strains LcA and LcY were isolated from the products Yakult (Yakult Honsha, Japan) and Actimel (Danone, France), as previously described (Douillard et al., 2013). Bacterial strains were cultured in MRS broth at 37°C in anaerobic conditions overnight and 2 ml of cell suspension was used for genomic DNA isolation using Wizard Genomic DNA Purification Kit (Promega, WI, USA) as per the manufacturer's instructions.
Genome sequencing, assembly, annotation and analysis
The genomes of L. casei strains LcA and LcY were sequenced using two next-generation sequencing platforms (454 GS-FLX+, Life Sciences and SOLiD, Life Technologies) and assembled de novo at the Institute of Biotechnology (Helsinki, Finland). The combination of both platforms allowed a much greater sequencing depth and accuracy in contrast to a unique sequencing technology approach, as previously done for L. casei strains LcA and LcY (Douillard et al., 2013). The 454 coverage on basis of numbers of reads in contigs is averaging 14X for both LcA and LcY genomes and the SOLiD coverage estimated from mappings is of ∼88X for LcA and ∼107X for LcY. Using progressive Mauve genome alignment package (Darling et al., 2010), the draft contigs were ordered and oriented. The published genomes of L. casei BL23, W56, BD-II and LC2W were used as references, improving the draft genome assemblies. Automatic genome annotation was conducted using RAST pipelines (Aziz et al., 2008) and subsequently RATT (Otto et al., 2011). PCR amplification from total DNA was performed to confirm contig order and circularity of the plasmids identified in both L. casei strains LcA and LcY. Pseudogenes and transposases were not addressed in the present study. Signal sequences were predicted with SignalP v4.0 (Petersen et al., 2011) and LPxTG proteins were identified using CW-PRED (Litou et al., 2008). BLAST Ring maps were generated using BRIG (Alikhan et al., 2011). SNPs and InDels analysis of both draft genomes was performed as follows. The LcA SOLiD reads were mapped to the LcY draft genome sequence using SHRiMP2, and BAM files of the mapping result were generated using SAMtools (Li et al., 2009; Rumble et al., 2009). Similarly, the LcY SOLiD reads were mapped to the LcA draft genome sequence. SNPs and InDels were detected with the MUMmer software package using nucmer parameters ‘maxmatch’ and ‘-c 100’, and show-snps command was used with parameter ‘-C’ (Kurtz et al., 2004). Only unequivocal SNPs and InDels detected in both mappings were reported. Read mappings and annotations were inspected with Artemis (Carver et al., 2012).
Sequence data deposition
The Whole Genome Shotgun projects for L. casei strains LcA and LcY have been deposited at DDBJ/EMBL/GenBank under the accession AQPP00000000 and ARNV00000000, respectively. The versions of LcA and LcY draft genomes described in this paper are the first versions, AQPP01000000 and ARNV01000000, respectively. L. casei strains LcA and LcY are available from Prof. W. M. de Vos, Department of Veterinary Biosciences, University of Helsinki, Helsinki, Finland. Draft genome and plasmid annotations presented in this study are accessible in the Supporting Information.
Acknowledgments
We thank Pia Laine and Okeke Godfrey Uche for technical assistance in genome sequencing.
Conflict of interest
The authors declare no competing interests.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's web-site:
Draft genome annotation of LcA chromosome.
Draft genome annotation of pACT plasmid.
Draft genome annotation of LcY chromosome.
Draft genome annotation of pYAK plasmid.
References
- Ai L, Chen C, Zhou F, Wang L, Zhang H, Chen W, Guo B. Complete genome sequence of the probiotic strain Lactobacillus casei BD-II. J Bacteriol. 2011;193:3160–3161. doi: 10.1128/JB.00421-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcántara C, Zúñiga M. Proteomic and transcriptomic analysis of the response to bile stress of Lactobacillus casei BL23. Microbiology. 2012;158:1206–1218. doi: 10.1099/mic.0.055657-0. [DOI] [PubMed] [Google Scholar]
- Alikhan N-F, Petty N, Ben Zakour N, Beatson S. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida CC, Lorena SL, Pavan CR, Akasaka HM, Mesquita MA. Beneficial effects of long-term consumption of a probiotic combination of Lactobacillus casei Shirota and Bifidobacterium breve Yakult may persist after suspension of therapy in lactose-intolerant patients. Nutr Clin Pract. 2012;27:247–251. doi: 10.1177/0884533612440289. [DOI] [PubMed] [Google Scholar]
- Axelsson L. Lactic Acid Bacteria Microbiological and Functional Aspects. New York, USA: Marcel Dekker; 2004. [Google Scholar]
- Aziz R, Bartels D, Best A, DeJongh M, Disz T, Edwards R, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barré O, Mourlane F, Solioz M. Copper induction of lactate oxidase of Lactococcus lactis: a novel metal stress response. J Bacteriol. 2007;189:5947–5954. doi: 10.1128/JB.00576-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauerl C, Perez-Martinez G, Yan F, Polk DB, Monedero V. Functional analysis of the p40 and p75 proteins from Lactobacillus casei BL23. J Mol Microbiol Biotechnol. 2010;19:231–241. doi: 10.1159/000322233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadbent JR, Larsen RL, Deibel V, Steele JL. Physiological and transcriptional response of Lactobacillus casei ATCC 334 to acid stress. J Bacteriol. 2010;192:2445–2458. doi: 10.1128/JB.01618-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadbent JR, Neeno-Eckwall E, Stahl B, Tandee K, Cai H, Morovic W, et al. Analysis of the Lactobacillus casei supragenome and its influence in species evolution and lifestyle adaptation. BMC Genomics. 2012;13:533. doi: 10.1186/1471-2164-13-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Thompson R, Budinich M, Broadbent JR, Steele JL. Genome sequence and comparative genome analysis of Lactobacillus casei: insights into their niche-associated evolution. Genome Biol Evol. 2009;1:239–2357. doi: 10.1093/gbe/evp019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 2012;28:464–469. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ai LZ, Zhou FF, Wang L, Zhang H, Chen W, Guo BH. Complete genome sequence of the probiotic bacterium Lactobacillus casei LC2W. J Bacteriol. 2011;193:3419–3420. doi: 10.1128/JB.05017-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claes IJJ, Schoofs G, Regulski K, Courtin P, Chapot-Chartier M, Rolain T, et al. Genetic and biochemical characterization of the cell wall hydrolase activity of the major secreted protein of Lactobacillus rhamnosus GG. PLoS ONE. 2012;7:e31588. doi: 10.1371/journal.pone.0031588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MD, Phillips BA, Zanoni P. Deoxyribonucleic acid homology studies of Lactobacillus caseiLactobacillus paracasei sp. nov., subsp. paracasei and subsp. tolerans, and Lactobacillus rhamnosus sp. nov., comb. nov. Int J Syst Bacteriol. 1989;39:105–108. [Google Scholar]
- Cooper VS, Vohr SH, Wrocklage SC, Hatcher PJ. Why genes evolve faster on secondary chromosomes in bacteria. PLoS Comput Biol. 2010;6:e1000732. doi: 10.1371/journal.pcbi.1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaglio F, Felis GE, Torriani S. The status of the species Lactobacillus casei (Orla-Jensen 1916) Hansen and Lessel 1971 and Lactobacillus paracasei Collins et al. 1989. Request for an opinion. Int J Syst Evol Microbiol. 2002;52:285–287. doi: 10.1099/00207713-52-1-285. [DOI] [PubMed] [Google Scholar]
- Dobson CM, Chaban B, Deneer H, Ziola B. Lactobacillus caseiLactobacillus rhamnosus, and Lactobacillus zeae isolates identified by sequence signature and immunoblot phenotype. Can J Microbiol. 2004;50:482–488. doi: 10.1139/w04-044. [DOI] [PubMed] [Google Scholar]
- Douillard FP, Ribbera A, Järvinen HM, Kant R, Pietilä TE, Randazzo C, et al. Comparative genomic and functional analysis of Lactobacillus casei and Lactobacillus rhamnosus strains marketed as probiotics. Appl Environ Microbiol. 2013;79:1923–1933. doi: 10.1128/AEM.03467-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foligne B, Nutten S, Grangette C, Dennin V, Goudercourt D, Poiret S, et al. Correlation between in vitro and in vivo immunomodulatory properties of lactic acid bacteria. World J Gastroenterol. 2007;13:236–243. doi: 10.3748/wjg.v13.i2.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemard E, Tondu F, Lacoin F, Schrezenmeir J. Consumption of a fermented dairy product containing the probiotic Lactobacillus casei DN-114001 reduces the duration of respiratory infections in the elderly in a randomised controlled trial. Br J Nutr. 2010;103:58–68. doi: 10.1017/S0007114509991395. [DOI] [PubMed] [Google Scholar]
- Hajela N, Nair GB, Abraham P, Ganguly NK. Health impact of probiotics – vision and opportunities. Gut Pathog. 2012;4:1757–4749. doi: 10.1186/1757-4749-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon E, Horvatovich P, Bisch M, Bringel F, Marchioni E, Aoude-Werner D, Ennahar S. Investigation of biomarkers of bile tolerance in Lactobacillus casei using comparative proteomics. J Proteome Res. 2012;11:109–118. doi: 10.1021/pr200828t. [DOI] [PubMed] [Google Scholar]
- Hendrickx APA, Budzik JM, Oh S-Y, Schneewind O. Architects at the bacterial surface – sortases and the assembly of pili with isopeptide bonds. Nat Rev Microbiol. 2011;9:166–176. doi: 10.1038/nrmicro2520. [DOI] [PubMed] [Google Scholar]
- Hochwind K, Weinmaier T, Schmid M, Hemert van S, Hartmann A, Rattei T, Rothballer M. Draft genome sequence of Lactobacillus casei W56. J Bacteriol. 2012;194:6638. doi: 10.1128/JB.01386-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- Kalliomaki M, Salminen S, Poussa T, Arvilommi H, Isolauri E. Probiotics and prevention of atopic disease: 4-year follow-up of a randomised placebo-controlled trial. Lancet. 2003;361:1869–1871. doi: 10.1016/S0140-6736(03)13490-3. [DOI] [PubMed] [Google Scholar]
- Kandler O, Weiss N. Bergey's Manual of Systematic Bacteriology. Baltimore, MD, USA: Williams and Wilkins; 1986. pp. 1063–1065. [Google Scholar]
- Kankainen M, Paulin L, Tynkkynen S, Ossowski von I, Reunanen J, Partanen P, et al. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc Natl Acad Sci USA. 2009;106:17193–17198. doi: 10.1073/pnas.0908876106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsford C, Schatz M, Pop M. Assembly complexity of prokaryotic genomes using short reads. BMC Bioinformatics. 2010;11:21. doi: 10.1186/1471-2105-11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein G, Pack A, Bonaparte C, Reuter G. Taxonomy and physiology of probiotic lactic acid bacteria. Int J Food Microbiol. 1998;41:103–125. doi: 10.1016/s0168-1605(98)00049-x. [DOI] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher A, Smoot M, Shumway M, Antonescu C, Salzberg S. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeer S, Claes I, Tytgat HL, Verhoeven TL, Marien E, Ossowski von I, et al. Functional analysis of Lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl Environ Microbiol. 2012;78:185–193. doi: 10.1128/AEM.06192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litou ZI, Bagos PG, Tsirigos KD, Liakopoulos TD, Hamodrakas SJ. Prediction of cell wall sorting signals in gram-positive bacteria with a hidden markov model: application to complete genomes. J Bioinform Comput Biol. 2008;6:387–401. doi: 10.1142/s0219720008003382. [DOI] [PubMed] [Google Scholar]
- Makarova K, Slesarev A, Wolf Y, Sorokin A, Mirkin B, Koonin E, et al. Comparative genomics of the lactic acid bacteria. Proc Natl Acad Sci USA. 2006;103:15611–15616. doi: 10.1073/pnas.0607117103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR–Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos A, Warnberg J, Nova E, Gomez S, Alvarez A, Alvarez R, et al. The effect of milk fermented by yogurt cultures plus Lactobacillus casei DN-114001 on the immune response of subjects under academic examination stress. Eur J Nutr. 2004;43:381–389. doi: 10.1007/s00394-004-0517-8. [DOI] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Takada T, Shimizu K, Moriyama K, Kawakami K, Hirano K, et al. Effects of a probiotic fermented milk beverage containing Lactobacillus casei strain Shirota on defecation frequency, intestinal microbiota, and the intestinal environment of healthy individuals with soft stools. J Biosci Bioeng. 2010;110:547–552. doi: 10.1016/j.jbiosc.2010.05.016. [DOI] [PubMed] [Google Scholar]
- Maze A, Boel G, Zuniga M, Bourand A, Loux V, Yebra MJ, et al. Complete genome sequence of the probiotic Lactobacillus casei strain BL23. J Bacteriol. 2010;192:2647–2648. doi: 10.1128/JB.00076-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Ton-That H, Schneewind O. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol Microbiol. 2001;40:1049–1057. doi: 10.1046/j.1365-2958.2001.02411.x. [DOI] [PubMed] [Google Scholar]
- Munoz-Provencio D, Perez-Martinez G, Monedero V. Characterization of a fibronectin-binding protein from Lactobacillus casei BL23. J Appl Microbiol. 2010;108:1050–1059. doi: 10.1111/j.1365-2672.2009.04508.x. [DOI] [PubMed] [Google Scholar]
- Munoz-Provencio D, Rodriguez-Diaz J, Collado MC, Langella P, Bermudez-Humaran LG, Monedero V. Functional analysis of the Lactobacillus casei BL23 sortases. Appl Environ Microbiol. 2012;78:8684–8693. doi: 10.1128/AEM.02287-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito E, Yoshida Y, Makino K, Kounoshi Y, Kunihiro S, Takahashi R, et al. Beneficial effect of oral administration of Lactobacillus casei strain Shirota on insulin resistance in diet-induced obesity mice. J Appl Microbiol. 2011;110:650–657. doi: 10.1111/j.1365-2672.2010.04922.x. [DOI] [PubMed] [Google Scholar]
- Nanno M, Kato I, Kobayashi T, Shida K. Biological effects of probiotics: what impact does Lactobacillus casei Shirota have on us? Int J Immunopathol Pharmacol. 2011;24:45S–50S. [PubMed] [Google Scholar]
- Navarre WW, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski von I, Satokari R, Reunanen J, Lebeer S, De Keersmaecker SCJ, Vanderleyden J, et al. Functional characterization of a mucus-specific LPXTG surface adhesin from probiotic Lactobacillus rhamnosus GG. Appl Environ Microbiol. 2011;77:4465–4472. doi: 10.1128/AEM.02497-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto TD, Dillon GP, Degrave WS, Berriman M. RATT: Rapid Annotation Transfer Tool. Nucleic Acids Res. 2011;39:8. doi: 10.1093/nar/gkq1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowska J, Klewicka E, Czubkowski P, Motyl I, Jankowska I, Libudzisz Z, et al. Effect of Lactobacillus casei DN-114001 application on the activity of fecal enzymes in children after liver transplantation. Transplant Proc. 2007;39:3219–3221. doi: 10.1016/j.transproceed.2007.03.101. [DOI] [PubMed] [Google Scholar]
- Perry AM, Ton-That H, Mazmanian SK, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus: III. Lipid II is an in vivo peptidoglycan substrate for sortase-catalyzed surface protein anchoring. J Biol Chem. 2002;277:16241–16248. doi: 10.1074/jbc.M109194200. [DOI] [PubMed] [Google Scholar]
- Petersen TN, Brunak S, Heijne von G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Reunanen J, Ossowski von I, Hendrickx AP, Palva A, Vos de WM. Characterization of the SpaCBA pilus fibers in the probiotic Lactobacillus rhamnosus GG. Appl Environ Microbiol. 2012;78:2337–2344. doi: 10.1128/AEM.07047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochat T, Bermudez-Humaran L, Gratadoux J-J, Fourage C, Hoebler C, Corthier G, Langella P. Anti-inflammatory effects of Lactobacillus casei BL23 producing or not a manganese-dependant catalase on DSS-induced colitis in mice. Microb Cell Fact. 2007;6:22. doi: 10.1186/1475-2859-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumble SM, Lacroute P, Dalca AV, Fiume M, Sidow A, Brudno M. SHRiMP: accurate mapping of short color-space reads. PLoS Comput Biol. 2009;5:e1000386. doi: 10.1371/journal.pcbi.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Torimura M, Kitahara M, Ohkuma M, Hotta Y, Tamura H. Characterization of the Lactobacillus casei group based on the profiling of ribosomal proteins coded in S10-spc-alpha operons as observed by MALDI-TOF MS. Syst Appl Microbiol. 2012;35:447–454. doi: 10.1016/j.syapm.2012.08.008. [DOI] [PubMed] [Google Scholar]
- Saxelin M, Tynkkynen S, Mattila-Sandholm T, Vos de WM. Probiotic and other functional microbes: from markets to mechanisms. Curr Opin Biotechnol. 2005;16:204–211. doi: 10.1016/j.copbio.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Schneewind O, Missiakas DM. Protein secretion and surface display in Gram-positive bacteria. Philos Trans R Soc Lond B Biol Sci. 2012;367:1123–1139. doi: 10.1098/rstb.2011.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles ME, Holzapfel WH. Lactic acid bacteria of foods and their current taxonomy. Int J Food Microbiol. 1997;36:1–29. doi: 10.1016/s0168-1605(96)01233-0. [DOI] [PubMed] [Google Scholar]
- Sybesma W, Molenaar D, IJcken van W, Venema K, Kort R. Genome instability in Lactobacillus rhamnosus GG. Appl Environ Microbiol. 2013;79:2233–2239. doi: 10.1128/AEM.03566-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykora J, Valeckova K, Amlerova J, Siala K, Dedek P, Watkins S, et al. Effects of a specially designed fermented milk product containing probiotic Lactobacillus casei DN-114 001 and the eradication of H. pylori in children: a prospective randomized double-blind study. J Clin Gastroenterol. 2005;39:692–698. doi: 10.1097/01.mcg.0000173855.77191.44. [DOI] [PubMed] [Google Scholar]
- Ventura M, O'Flaherty S, Claesson MJ, Turroni F, Klaenhammer TR, Sinderen van D, O'Toole PW. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat Rev Microbiol. 2009;7:61–71. doi: 10.1038/nrmicro2047. [DOI] [PubMed] [Google Scholar]
- Vos de WM. Systems solutions by lactic acid bacteria: from paradigms to practice. Microb Cell Fact. 2011;10:S2. doi: 10.1186/1475-2859-10-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F, Cao H, Cover TL, Whitehead R, Washington MK, Polk DB. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology. 132:562–575. doi: 10.1053/j.gastro.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WY, Yu DL, Sun ZH, Wu RN, Chen X, Chen W, et al. Complete genome sequence of Lactobacillus casei Zhang, a new probiotic strain isolated from traditional homemade koumiss in Inner Mongolia, China. J Bacteriol. 2007;192:5268–5269. doi: 10.1128/JB.00802-10. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Draft genome annotation of LcA chromosome.
Draft genome annotation of pACT plasmid.
Draft genome annotation of LcY chromosome.
Draft genome annotation of pYAK plasmid.