Abstract
The bloodmeal hosts used by Culex tarsalis Coquillett collected along the Salton Sea in Coachella Valley, CA, during 1998–2002 were identified using sequences of the cytochrome c oxidase I gene identified from Barcode of Life database. Overall, 265 (83.3%) of 318 bloodmeals were identified, of which 76.6% fed on birds, 18.1% on mammals, and 5.3% on reptiles. Forty-seven different hosts were identified, none of which comprised >12.5% of the total. Although Cx. tarsalis exhibits specific host-seeking flight patterns, bloodmeals seemed to be acquired opportunistically, thereby limiting potential arbovirus transmission efficiency in species-rich environments.
Keywords: bloodmeal host, Culex tarsalis, Salton Sea
The frequency of bloodmeal acquisition from competent hosts is a key factor in understanding the vectorial capacity of Culex mosquitoes for the North American encephalitides, including West Nile virus (WNV). The distribution of bloodmeals among competent and noncompetent hosts in rural areas with high host diversity seems to create a dilution effect that limits virus amplification to spill-over levels where tangential transmission to humans and equines leads to outbreaks (Swaddle and Calos 2008). This dilution effect may be compensated by the vector preferentially selecting hosts that are moderately or highly competent, such as the apparent preference by Culex pipiens Linnaeus for American Robins in the eastern United States (Kilpatrick et al. 2006).
The Salton Sea, associated wetlands, and upland agricultural habitats in southeastern California are exploited by a remarkably diverse avifauna as well as abundant mosquito populations dominated by the encephalitis vector, Culex tarsalis Coquillett. Warm winters and hot summers combine to create ideal conditions for the enzootic maintenance and amplification of arboviruses, including, historically, western equine encephalomyelitis and St. Louis encephalitis (SLEV) viruses (Reisen et al. 2002) and, recently, WNV (Reisen et al. 2008). However, few human or equine cases have been reported from this area of California (http://www.cdc.gov/ncidod/dvbid/westnile/USGS_frame.html), supporting the notion that high avian diversity may dampen transmission efficiency in rural areas (Ezenwa et al. 2006, Loss et al. 2009). Possible mechanisms leading to these patterns relate to mosquito host-seeking and opportunistic blood-feeding behaviors that may divert bloodmeals to low and moderate virus competent avian and mammalian hosts, including humans and equines. This “dilution concept” (Swaddle and Calos 2008) is similar in outcome to the “zooprophylaxis” effect of domestic mammals described previously for encephalitides (Hess and Hayes 1970).
The host selection patterns of mosquitoes at the Salton Sea initially were investigated using serological techniques (Tempelis 1975). The initial survey of females collected by traps at farmsteads in Imperial Valley found that Cx. tarsalis fed equally on mammals (horses, cattle) and birds (chickens, passerines) (Gunstream et al. 1971). Subsequently, an attempt was made to collect replete resting females from natural settings, but except for rabbits, bloodmeals were again taken mostly from passerine birds (Lothrop et al. 1997), which could not be identified to species using serological methods. Recently, we used molecular methods based on sequencing of the cytochrome c oxidase I (COI) gene to specifically identify host species (Thiemann et al. 2012a) using the Barcode of Life system (Ratnasingham and Hebert 2007). This system was used to identify the bloodmeal hosts of Cx. tarsalis and Culex quinquefasciatus Say collected throughout the Coachella Valley (Thiemann et al. 2012b). This survey found that Cx. tarsalis females fed on a wide variety of avian and mammalian hosts, with 36 host species identified from 69 bloodmeals collected during 2007–2009. Interestingly, few bloodmeals were detected from rabbits.
The current survey reports on the identity of bloodmeal hosts of Cx. tarsalis females collected during 1998–2002. Between this period and the previous 2007–2009 survey (Thiemann et al. 2012b), the Salton Sea was intentionally lowered (Fig. 1), markedly altering shoreline ecology and eliminating much of the seminatural wetlands historically exploited by a variety of wetland bird species. Our current survey compares bloodmeal host diversity during these two sampling periods and extends our knowledge of Cx. tarsalis host selection at wetlands and surrounding agricultural habitats.
Fig. 1.
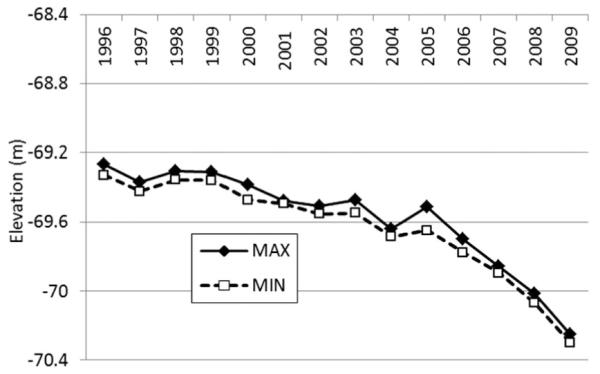
Elevation of the Salton Sea in Coachella Valley, CA. Plotted are the mean annual maximum and minimum elevation for the years 1996–2009.
Materials and Methods
Salton Sea
Blood-fed Cx. tarsalis females were collected along the north, western, and southeastern shores of the Salton Sea within the Coachella Valley of Riverside County, CA. The Sea lies within the Salton Sink of the Colorado Desert of Southeastern California; it was ≈70 m below sea level and covered an area of ≈1,300 km2 at the time of this study. The Sea was created in 1905 by flooding of the Colorado River into Imperial Valley, an event prevented thereafter by the building of the Hoover Dam. As the Sea has no outflow, it has become progressively saline over time, with a current salinity of ≈44 g/L (http://ceimperial.ucanr.edu/Custom_Program275/Salton_Sea_and_Salinity/). High salinity/alkalinity, hot summer temperatures, and algal blooms, possibly created by fertilizer run-off, have grossly simplified the fauna of the sea; however, artesian upwelling and irrigation canals have facilitated extensive upland agriculture, lowland aquaculture, and wetlands managed for migratory waterfowl. In combination, these diverse habitats and the Pacific flyway have created one of the most diverse avifauna found in North America, with more than 400 bird species recorded from the Sea and its environs. In 2003/2004, the California State Legislature directed the California Resources Agency to prepare a plan for the restoration of the Salton Sea, which included reducing the available surface area from 945 to 381 km2, thereby the lowering of the elevation of the Sea (http://saltonsea.ca.gov/media/ppr_summary.pdf). To pay for this restoration, starting in 2004, Colorado River water was diverted from the Sea, with a target elevation of −71 m below sea level.
Mosquito Collection
During 1998–2002, partial and replete blood-fed females were retained from samples taken during a variety of ecological studies. Replete flying females were collected by nonbaited suction traps deployed along vegetative ecotones to study mosquito flight paths (Lothrop and Reisen 2001). Partially blood-fed females were collected host-seeking at dry ice-baited Centers for Disease Control and Prevention (CDC) style traps (Newhouse et al. 1966). Resting females were aspirated from walk-in red boxes (Meyer 1987) or plastic garbage cans (Lothrop et al. 2007) as well as from vegetation during attempts to monitor the abundance of the resting population. The 2007–2009 survey used similar sampling methods at generally the same habitats sampled during 1998–2002, although specific sites varied because of changes in the landscape. Mosquitoes were anesthetized with triethylamine, identified and sorted by feeding status under a dissecting scope, blood-fed Cx. tarsalis females placed into individual gelatin capsules, and then stored in labeled cryovials at −80°C until tested during 2012.
Bloodmeal Identification
DNA was extracted from blood in mosquito abdomens and identified using procedures described previously (Thiemann et al. 2012a). This system was validated initially using blood samples from more than 120 avian species collected from California as well as selected domestic mammals, including cow, horse, goat, and dog. Briefly, DNA was extracted using the DNAeasy 96 Blood and Tissue Kit (Qiagen, Valencia, CA) using the Animal Tissue protocol. A nested polymerase chain reaction was used to first amplify the mitochondrial gene COI and then the 658-bp “barcoding region of the COI gene” (Ivanova et al. 2006, Cooper et al. 2007). Amplicons were purified and then capillary array sequenced by the College of Agricultural and Environmental Sciences Genomics Facility at the University of California Davis. Sequences were identified using the Barcode of Life Data Systems (BOLD) Identification Engine (Ratnasingham and Hebert 2007).
Bird Relative Abundance
The frequency of occurrence of avian species throughout the year during the two sampling periods 1998–2002 and 2007–2009 was downloaded from eBird (Sullivan et al. 2009), a project developed by Cornell Laboratory of Ornithology and the National Audubon Society to monitor avian occurrence from citizen-based reporting. We considered abundance as the proportion of citizen-submitted checklists that reported a species from the Salton Sea reporting area of California during each of the two sampling periods.
Results
In total, 265 (83.3%) bloodmeals were identified from the 318 Cx. tarsalis samples tested (Table 1). Of the 53 not identified to species, 45 showed no DNA amplification likely because of sample degradation and/or low blood volumes in mosquitoes collected from CO2 traps, whereas five produced good sequences that were visible as DNA on gels, but could not be adequately matched using the BOLD search engine and database. A similar matching problem also was encountered for a few samples collected during the 2007–2009 period (Thiemann et al. 2012b). Three samples produced a mixed sequence probably related to successive partial bloodmeals taken from more than one host species.
Table 1.
Identification of bloodmeal hosts of Culex tarsalis collected during 1998–2002 near the Salton Sea, Coachella Valley, Riverside County, CA
| Common name | Species | Collection method |
Total | Percent | ||
|---|---|---|---|---|---|---|
| Suction trap | CO2 trap | Resting | ||||
| Mourning Dove | Zenaida macroura | 20 | 3 | 10 | 33 | 12.5% |
| Black-Crowned Night Heron | Nycticorax nycticorax | 4 | 25 | 2 | 31 | 11.7% |
| Chicken | Gallus gallus | 21 | 2 | 23 | 8.7% | |
| Greater Roadrunner | Geococcyx californianus | 7 | 15 | 22 | 8.3% | |
| White-Winged Dove | Zenaida asiatica | 4 | 2 | 12 | 18 | 6.8% |
| Green Heron | Butorides virescens | 5 | 6 | 3 | 14 | 5.3% |
| House Sparrow | Passer domesticus | 1 | 5 | 6 | 2.3% | |
| Gambel’s Quail | Callipepla gambelii | 4 | 1 | 5 | 1.9% | |
| Common Ground Dove | Columbina passerina | 4 | 4 | 1.5% | ||
| Least Bittern | Ixobrychus exilis | 1 | 3 | 4 | 1.5% | |
| Northern Mockingbird | Mimus polyglottos | 3 | 1 | 4 | 1.5% | |
| Bullock’s Oriole | Icterus bullockii | 2 | 1 | 3 | 1.1% | |
| House Finch | Carpodacus mexicanus | 3 | 3 | 1.1% | ||
| American Kestrel | Falco sparverius | 1 | 1 | 2 | 0.8% | |
| Black Necked stilt | Himantopus mexicanus | 1 | 1 | 2 | 0.8% | |
| Black-Tailed Gnatcatcher | Polioptila melanura | 2 | 2 | 0.8% | ||
| Brown-Headed Cowbird | Molothrus ater | 2 | 2 | 0.8% | ||
| Cliff Swallow | Petrochelidon pyrrhonota | 1 | 1 | 2 | 0.8% | |
| Cooper’s Hawk | Accipiter cooperii | 2 | 2 | 0.8% | ||
| European Starling | Sturnus vulgaris | 2 | 2 | 0.8% | ||
| Hooded Oriole | Icterus cucullatus | 2 | 2 | 0.8% | ||
| Spotted Dove | Streptopelia chinensis | 2 | 2 | 0.8% | ||
| American Coot | Fulica americana | 1 | 1 | 0.4% | ||
| Brewer’s Blackbird | Euphagus cyanocephalus | 1 | 1 | 0.4% | ||
| Bronzed Cowbird | Molothrus aeneus | 1 | 1 | 0.4% | ||
| Cinnamon Teal | Anas cyanoptera | 1 | 1 | 0.4% | ||
| Hermit Thrush | Catharus guttatus | 1 | 1 | 0.4% | ||
| Lesser nighthawk | Chordeiles acutipennis | 1 | 1 | 0.4% | ||
| Lincoln’s Sparrow | Melospiza lincolnii | 1 | 1 | 0.4% | ||
| MacGillivray’s Warbler | Oporornis tolmiei | 1 | 1 | 0.4% | ||
| Mallard | Anas platyrhynchos | 1 | 1 | 0.4% | ||
| Song Sparrow | Melospiza melodia | 1 | 1 | 0.4% | ||
| Swainson’s Thrush | Catharus ustulatus | 1 | 1 | 0.4% | ||
| Hermit Thrush | Catharus guttatus | 1 | 1 | 0.4% | ||
| Virginia Rail | Rallus limicola | 1 | 1 | 0.4% | ||
| Western Tanager | Piranga ludoviciana | 1 | 1 | 0.4% | ||
| Yellow-Rumped Warbler | Dendroica coronata | 1 | 1 | 0.4% | ||
| Bird total | 44 | 107 | 52 | 203 | 76.6% | |
| Percent of total birds | 21.7% | 52.7% | 25.6% | |||
| Rat | Rattus rattus | 15 | 15 | 5.7% | ||
| Dog | Canis lupus | 1 | 5 | 4 | 10 | 3.8% |
| Horse | Equus caballus | 8 | 8 | 3.0% | ||
| Desert Cottontail | Sylvilagus audubonii | 5 | 1 | 6 | 2.3% | |
| Human | Homo sapiens | 1 | 2 | 3 | 1.1% | |
| Cat | Felis catus | 2 | 2 | 0.8% | ||
| Coyote | Canis latrans | 2 | 2 | 0.8% | ||
| Raccoon | Procyon lotor | 2 | 2 | 0.8% | ||
| Mammal total | 2 | 31 | 15 | 48 | 18.1% | |
| Percent of total mammals | 4.2% | 64.6% | 31.3% | |||
| Long-Tailed Brush Lizard | Urosaurus graciosus | 10 | 2 | 2 | 14 | 5.3% |
| Reptile total | 10 | 2 | 2 | 14 | 5.3% | |
| Percent of total reptiles | 71.4% | 14.3% | 14.3% | |||
| Total identified | 56 | 140 | 69 | 265 | 83.3% | |
| No match | Good sequence; no match | 2 | 2 | 1 | 5 | |
| Mixed | mixed sequence, no result | 1 | 1 | 1 | 3 | |
| Negative | No amplification | 9 | 31 | 5 | 45 | |
| Total unidentified or negative | 12 | 34 | 7 | 53 | 16.7% | |
| Total specimens tested | 68 | 174 | 76 | 318 | ||
Overall, Cx. tarsalis ingested blood from 47 different hosts. Most females (76.6% of identified bloodmeals) fed on avian hosts, but these included 37 different species, none of which comprised >12.5% of the total (Table 1). Cx. tarsalis fed most frequently on relatively large birds, including Mourning Dove, Black-Crowned Night Heron, and chicken, and there did not seem to be a preference for specific taxa or birds with a particular night-roosting behavior. Noticeably absent were bloodmeals from corvids, which are relatively rare in Coachella Valley, and abundant gulls, shore birds, and pelicans that roost on sand spits. Previously, we reported that few Cx. tarsalis were collected host-seeking over open habitats such as sand spits as well as at snags over water in the Sea, although both habitats often supported large numbers of night-roosting birds (Lothrop and Reisen 2001). Surprisingly few bloodmeals were taken from Gambel’s Quail that were abundant within desert brush and were frequently infected with arboviruses (Reisen et al. 2006). It also was interesting to note variation in host selection among collection methods; for example, Cx. tarsalis females that fed on Mourning Dove, White-Winged Dove, and Greater Roadrunner were collected in suction traps and resting, but rarely were taken partially fed in CO2 traps. Conversely, females that fed on Black-Crowned Night Heron and chicken were collected most frequently in CO2 traps, perhaps indicating that they were frequently disturbed while feeding, took a partial meal, and were seeking to complete the feeding process when collected. These results also were affected by the juxtaposition of sampling methods to night-roosting locations.
Mammalian hosts comprised 18.1% of the total bloodmeals identified (Table 1). Among the mammalian hosts, a species in the genus Rattus and domestic Dog were the most common hosts; only three females fed on humans. Rattus were identified in BOLD as Rattus tanezumi, but likely are Rattus rattus Lineage II (Conroy et al. 2013), as concluded previously (Thiemann et al. 2012b). In Coachella Valley, R. rattus frequently live in trees where they forage at night for fruit, thereby exposing themselves to Cx. tarsalis host-seeking within the canopy. Interestingly, all rat feeds were identified from females collected host-seeking at CO2 traps. Similar to the 2007–2009 period (Thiemann et al. 2012b), few females fed on rabbits, whereas our earlier survey using serological methods (Lothrop et al. 1997) identified lagomorphs as a relatively frequent host.
Identification of the reptile hosts was difficult. A single reptile species, Long-tailed Brush Lizard, comprised 5.3% of the total bloodmeals identified. Five bloodmeals listed as “good sequence, no match” most likely also were reptiles based on search results, but remain unidentified. Previous bloodmeal surveys indicated that Cx. tarsalis rarely feed on poikilotherms (Reisen and Reeves 1990), but were attracted to reptile hosts exposed in lard can traps (Dow et al. 1957).
Discussion
Despite ecological changes at the Salton Sea, utilization of different sampling sites, and more intensive mosquito control to combat the WNV outbreak, the diversity of bloodmeal hosts identified during 2007–2009 (0.38 host species/bloodmeal tested, 36 species, n = 94) was greater than that observed during 1998–2002 (0.17 host species/bloodmeal, 47 species, n = 265), with 20 of the same host species identified during both surveys. Mourning Dove and chicken were among the most frequently selected hosts in both surveys; however, most bloodmeals seemed to be acquired because of flight path encounters rather than strong selection and/or questing for specific taxa. Similar high host diversity also was described for Cx. tarsalis collected at wetland habitats near Davis in the Sacramento Valley (Thiemann et al. 2012b).
Using data from the Salton Sea area downloaded from eBird, there did not appear to be marked changes in overall diversity (number of species reported) or the frequency of reporting of species representative of different habitats, including herons that roost and nest at snags and trees, shorebirds that roost along the margin of the Sea, and passerines that nest in reed beds (Table 2). Three of three passerine species from reed habitats showed slight declines, but so did two control species that exploit upland habitats.
Table 2.
Frequency of representative species appearing on checklists per year reported to eBird during the 1998–2002 and 2007–2009 periods
| Species | 1998–2002 | 2007–2009 |
|---|---|---|
| Herons | ||
| Least Bitterna | 0.05 | 0.01 |
| Great Blue Heronb | 0.71 | 0.76 |
| Black-Crowned Night-Herona | 0.43 | 0.38 |
| Green Herona | 0.39 | 0.27 |
| Shore birds | ||
| Black-Necked Stilta | 0.74 | 0.79 |
| American Avocet | 0.67 | 0.73 |
| Spotted Sandpiper | 0.26 | 0.25 |
| Reed beds | ||
| Red-Winged Blackbird | 0.59 | 0.44 |
| Yellow-Headed Blackbird | 0.16 | 0.14 |
| Great-Tailed Grackleb | 0.61 | 0.53 |
| Controls | ||
| House Fincha | 0.41 | 0.37 |
| Brewer’s Blackbirda | 0.32 | 0.21 |
| For all species | ||
| No reports | 148 | 397 |
| No total species/taxa | 269 | 294 |
| No species reports | 3,667 | 5,093 |
Reports per week for the sampling period were summed and standardized by the number of total reports.
Fed on during both surveys.
Fed on only during 2007–2009 survey.
During 2003–2006, 7,900 sera were taken from 91 species of birds collected by mist netting or grain-baited traps at many of the same habitats used for mosquito collection near the Salton Sea, and sera were tested for antibodies against SLEV and WNV, of which 5.7% were positive (Reisen et al. 2008). Columbiforms (Common Ground Dove, 29.8%; Rock Pigeon, 39%; White-winged Dove, 42.1%; Mourning Dove, 4%), Gambel’s Quail (10.3%), and Least Bittern (61%) were frequently positive, whereas species such as Black-Crowned Night Heron and Greater Roadrunner were collected too infrequently for comparison. Interestingly, quail were frequently seropositive, but comprised a relatively infrequent blood source in the current and previous surveys. With the exception of the Least Bittern, whose host competency is unknown, most frequently seropositive hosts were considered as low competent and probably dead-end hosts for SLEV and WNV (Reisen et al. 2003, 2005), unless infected when nestlings (Mahmood et al. 2004).
In summary, Cx. tarsalis seemed to feed opportunistically on a wide variety of hosts encountered while host-seeking along flight paths (Lothrop and Reisen 2001), and did not focus on specific taxa or birds exhibiting specific nesting/roosting patterns. Therefore, the distribution of competent hosts along flight paths seemed to determine where infectious bloodmeals were acquired and where transmission would occur most efficiently. In Coachella Valley, the wide diversity of avian hosts fed on by Cx. tarsalis, many of which have unknown or low competence for WNV, most likely dampened the efficiency of WNV enzootic transmission at locations near the Salton Sea, precluding effective amplification, and perhaps, in part, limiting the number of human cases reported from this region.
Acknowledgments
W.K.R. acknowledges support from the Research and Policy for Infectious Disease Dynamics (RAPIDD) program of the Science & Technology Directorate, Department of Homeland Security and Fogarty International Center, National Institutes of Health. This research was supported, in part, by funds from the Coachella Valley Mosquito and Vector Control District and Research Grant AI55607 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
References Cited
- Conroy CJ, Rowe KC, Rowe KMC, Kamath PL, Aplin KP, Hui LT, James DK, Moritz C, Patton JL. Cryptic genetic diversity in Rattus of the San Francisco Bay region, California. Biol. Invasions. 2013;15:741–758. [Google Scholar]
- Cooper JK, Sykes G, King S, Cottrill K, Ivanova NV, Hanner R, Ikonomi P. Species identification in cell culture: a two-pronged molecular approach. In Vitro Cell. Dev. Biol. Anim. 2007;43:344–351. doi: 10.1007/s11626-007-9060-2. [DOI] [PubMed] [Google Scholar]
- Dow RP, Reeves WC, Bellamy RE. Field tests of avian host preference of Culex tarsalis Coq. Am. J. Trop. Med. Hyg. 1957;6:294–303. doi: 10.4269/ajtmh.1957.6.294. [DOI] [PubMed] [Google Scholar]
- Ezenwa VO, Godsey MS, King RJ, Guptill SC. Avian diversity and West Nile virus: testing associations between biodiversity and infectious disease risk. Proc. Biol. Sci. 2006;273:109–117. doi: 10.1098/rspb.2005.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunstream SE, Chew RM, Hagstrum DW, Tempelis CH. Feeding patterns of six species of mosquitoes in arid southeastern California. Mosq. News. 1971;31:99–101. [Google Scholar]
- Hess AD, Hayes RO. Relative potentials of domestic animals for zooprophylaxis against mosquito vectors of encephalitis. Am. J. Trop. Med. Hyg. 1970;19:327–334. doi: 10.4269/ajtmh.1970.19.327. [DOI] [PubMed] [Google Scholar]
- Ivanova NV, deWaard JR, Hebert PD. An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol. Ecol. Notes. 2006;6:998–1002. [Google Scholar]
- Kilpatrick AM, Daszak P, Jones MJ, Marra PP, Kramer LD. Host heterogeneity dominates West Nile virus transmission. Proc. R. Soc. Lond. B Biol. Sci. 2006;273:2327–2333. doi: 10.1098/rspb.2006.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loss SR, Hamer GL, Walker ED, Ruiz MO, Goldberg TL, Kitron UD, Brawn JD. Avian host community structure and prevalence of West Nile virus in Chicago, Illinois. Oecologia. 2009;159:415–424. doi: 10.1007/s00442-008-1224-6. [DOI] [PubMed] [Google Scholar]
- Lothrop HD, Reisen WK. Landscape affects the host-seeking patterns of Culex tarsalis (Diptera: Culicidae) in the Coachella Valley of California. J. Med. Entomol. 2001;38:325–332. doi: 10.1603/0022-2585-38.2.325. [DOI] [PubMed] [Google Scholar]
- Lothrop HD, Tempelis CH, Reisen WK. Host selection by Culex tarsalis around the margin of the Salton Sea in the Coachella Valley of California. Proc. Mosq. Vector Control Assoc. Calif. 1997;65:28–29. [Google Scholar]
- Lothrop HD, Lothrop B, Palmer M, Wheeler SS, Gutierrez A, Miller P, Gomsi DE, Reisen WK. Evaluation of Pyrenone aerial ULV applications for adult Culex tarsalis control in the desert environments of the Coachella Valley of California. J. Am. Mosq. Control Assoc. 2007;23:405–419. doi: 10.2987/5623.1. [DOI] [PubMed] [Google Scholar]
- Mahmood F, Chiles RE, Fang Y, Barker CM, Reisen WK. Role of nestling mourning doves and house finches as amplifying hosts of St. Louis encephalitis virus. J. Med. Entomol. 2004;41:965–972. doi: 10.1603/0022-2585-41.5.965. [DOI] [PubMed] [Google Scholar]
- Meyer RP. The “walk-in” type red box for sampling resting adult mosquitoes. Proc. N. J. Mosq. Control Assoc. 1987;72:104–105. [Google Scholar]
- Newhouse VF, Chamberlain RW, Johnston JG, Jr., Sudia WD. Use of dry ice to increase mosquito catches of the CDC miniature light trap. Mosq. News. 1966;26:30–35. [Google Scholar]
- Ratnasingham S, Hebert PDN. BOLD: the Barcode of Life Data System. Mol. Ecol. Notes. 2007;7:355–364. doi: 10.1111/j.1471-8286.2007.01678.x. ( www.barcodinglife.org) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisen WK, Reeves WC. Bionomics and ecology of Culex tarsalis and other potential mosquito vector species. In: Reeves WC, editor. Epidemiology and Control of Mosquito-Borne Arboviruses in California, 1943–1987. California Mosquito and Vector Control Association; Sacramento, CA: 1990. pp. 254–329. [Google Scholar]
- Reisen WK, Lothrop HD, Chiles RE, Cusack R, Green EG, Fang Y, Kensington M. Persistence and amplification of St. Louis encephalitis virus in the Coachella Valley of California, 2000–2001. J. Med. Entomol. 2002;39:793–805. doi: 10.1603/0022-2585-39.5.793. [DOI] [PubMed] [Google Scholar]
- Reisen WK, Chiles RE, Martinez VM, Fang Y, Green EN. Experimental infection of California birds with western equine encephalomyelitis and St. Louis encephalitis viruses. J. Med. Entomol. 2003;40:968–982. doi: 10.1603/0022-2585-40.6.968. [DOI] [PubMed] [Google Scholar]
- Reisen WK, Fang Y, Martinez VM. Avian host and mosquito (Diptera: Culicidae) vector competence determine the efficiency of West Nile and St. Louis encephalitis virus transmission. J. Med. Entomol. 2005;42:367–375. doi: 10.1093/jmedent/42.3.367. [DOI] [PubMed] [Google Scholar]
- Reisen WK, Martinez VM, Fang Y, Garcia S, Ashtari S, Wheeler SS, Carroll BD. Role of California (Callipepla californica) and Gambel’s (Callipepla gambelii) quail in the ecology of mosquito-borne encephalitis viruses in California, USA. J. Wildl. Dis. 2006;6:248–260. doi: 10.1089/vbz.2006.6.248. [DOI] [PubMed] [Google Scholar]
- Reisen WK, Lothrop HD, Wheeler SS, Kennsington M, Gutierrez A, Fang Y, Garcia S, Lothrop B. Persistent West Nile virus transmission and the apparent displacement St. Louis encephalitis virus in southeastern California, 2003–2006. J. Med. Entomol. 2008;45:494–508. doi: 10.1603/0022-2585(2008)45[494:pwnvta]2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BL, Wood CL, Iliff M, Bonney RE, Fink D, Kelling S. eBird: a citizen-based bird obervation network in the biological sciences. Biol. Conserv. 2009;142:2282–2292. [Google Scholar]
- Swaddle JP, Calos SE. Increased avian diversity is associated with lower incidence of human West Nile infection: observation of the dilution effect. PLoS ONE. 2008;3:e2488. doi: 10.1371/journal.pone.0002488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempelis CH. Host-feeding patterns of mosquitoes, with a review of advances in analysis of blood meals by serology. J. Med. Entomol. 1975;11:635–653. doi: 10.1093/jmedent/11.6.635. [DOI] [PubMed] [Google Scholar]
- Thiemann TC, Brault AC, Ernest HB, Reisen WK. Development of a high-throughput microsphere-based molecular assay to identify 15 common bloodmeal hosts of Culex mosquitoes. Mol. Ecol. Resour. 2012a;12:238–246. doi: 10.1111/j.1755-0998.2011.03093.x. (doi:10.1111/j.1755-0998.2011.03093.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiemann TC, Lemenager DA, Kluh S, Carroll BD, Lothrop HD, Reisen WK. Spatial variation in host feeding patterns of Culex tarsalis and the Culex pipiens complex (Diptera: Culicidae) in California. J. Med. Entomol. 2012b;49:903–916. doi: 10.1603/me11272. [DOI] [PMC free article] [PubMed] [Google Scholar]


