Abstract
Prodrugs are effective tools in overcoming drawbacks typically associated with drug formulation and delivery. Those employing esterase-triggered functional groups are frequently utilized to mask polar carboxylic acids and phenols, increasing drug-like properties such as lipophilicity. Herein we detail a comprehensive assessment for strategies that effectively release hydroxyl and phenolic moieties in the presence of an esterase. Matrix metalloproteinases (MMPs) serve as our proof-of-concept target. Three distinct ester-responsive protecting groups are incorporated into MMP proinhibitors containing hydroxyl moieties. Analytical evaluation of the proinhibitors demonstrates that the use of a benzyl ether group appended to the esterase trigger leads to considerably faster kinetics of conversion and enhanced aqueous stability when compared to more conventional approaches where the trigger is directly attached to the inhibitor. Biological assays confirm that all protecting groups effectively cleave in the presence of esterase to generate the active inhibitor.
Keywords: esterase, matrix metalloproteinases, metalloenzymes, prodrugs
Introduction
Prodrugs, or chemically modified versions of bioactive substances, represent a class of pharmaceuticals that are particularly useful in overcoming barriers to drug formulation.[1] These barriers often involve difficulties associated with drug delivery and poor pharmacokinetic properties including, but not limited to poor solubility, chemical instability, and inadequate oral absorption.[1] A stimulus-responsive promoiety can be appended to a drug to render it inactive until a chemical or enzymatic transformation event occurs, leading to metabolic conversion to the desired pharmacophore. The use of a prodrug strategy often improves the physiochemical and/or pharmacokinetic properties of a drug.[1b] The combined benefits associated with prodrugs mentioned above has made prodrugs increasingly popular, with ~10% of all drugs approved worldwide classified as prodrugs.[1a, 2]
The most common prodrug approach to deliver pharmacologically potent compounds is through esterase bioconversion.[1b]. The esterases involved in drug metabolism are mainly localized in the liver; among these are carboxyl- and butyrylcholinesterase, which can recognize acetate and phenylacetate groups as substrates.[3] Ester-based prodrugs have previously been shown to improve the properties of small-molecule drugs including solubility, stability and oral bioavailability.[3c] Prodrugs containing ester promoieties are generally easy to synthesize, further adding to the appeal of this approach. Esterase-activated prodrugs effectively mask polar moieties with a non-polar ester bond, often increasing lipophilicity, thus membrane permeability.[3c] The vast majority of ester prodrugs mask carboxylic acids, with fewer accounts documenting their use to release hydroxyl and phenolic moieties upon hydrolysis. In the latter cases, the hydroxyl moiety (hydroxyl or phenol) is directly esterified and esterase bioconversion leads to release of the drug.
Metalloenzyme inhibitors are a class of compounds that can greatly benefit from a prodrug approach. In fact, the most clinically successful metalloenzyme prodrugs involve alkyl and aryl ester modified carboxylates that target angiotensin-converting enzyme (ACE). In the case of enalapril (marketed as Vasotec, Merck), the ethyl ester prodrug is metabolically converted by esterases to a free carboxylic acid group that can bind to the catalytic Zn(II) ion and attenuate enzyme activity.[1b] Other reports of metalloenzyme prodrug development include matrix metalloproteinase (MMP) inhibitors.[4] MMPs are a family of >20 Zn(II)-dependent endopeptidases that are capable of degrading all components of the extracellular matrix. MMP expression and activity is a highly regulated process under normal physiological conditions.[5] Overexpression and misregulation of MMPs has implicated these proteases in a number of pathologies including arthritis and tumor cell metastasis.[6] Previous MMP broad-spectrum and selective inhibitors have been developed, but have seen limited clinical success due, in part, to undesired side effects from off-target inhibition and poor bioavailability.[7] Thus, MMP inhibitors (MMPi) stand to benefit from a prodrug strategy. For this reason, MMPs were chosen as our targets of interest for proof-of-concept studies regarding esterase activation of hydroxyl functionalities.
Recently, prodrug strategies utilizing glucose and hydrogen peroxide-responsive triggering groups were investigated.[8] In an attempt to expand the understanding and chemical tools available for prodrug design, three esterase-responsive strategies were examined, measuring both the aqueous stability and release kinetics for each. For this study, different promoieties were coupled to distinct metal-binding groups (MBGs) that serve as the core scaffold for metalloenzyme inhibitors. The three different approaches for release of the MBG were studied in the presence of an esterase to identify the best system for the development of potential prodrugs.
Results and Discussion
Assessment of Ester-Responsive Triggers
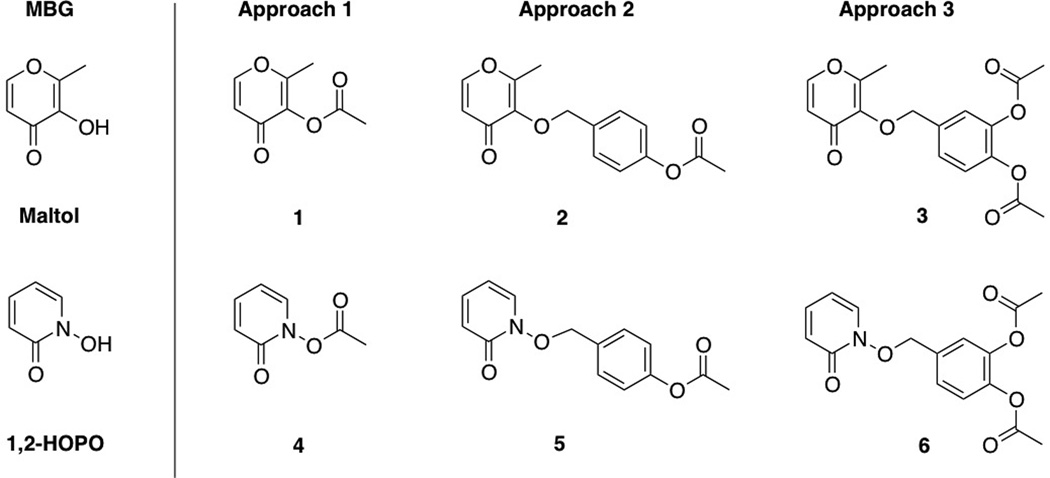
The approaches investigated here consist of the following ester-responsive promoieties, all of which are appended to the hydroxyl group of the MBG: (1) direct acetylation, (2) a benzyl ether protecting group containing an acetylated phenol, and (3) a doubly acetylated catechol-based linker (Scheme 1). Approach 1 is comparable to successful strategies widely reported for masking hydroxyl groups, where the direct appendage of the ester moiety inactivates the drug. Approaches 2 and 3 represent reaction-based strategies wherein the stimulus event (deacetylation) initiates an elimination reaction that leads to release of the inhibitor (Scheme 3). Previous studies indicate that the benzyl ether linkage is superior to the more prevalent carbonate linkage in prodrug design with respect to kinetics of release and stability, thus this linkage was incorporated into our prodrug study here.[9] Approach 2 has been previously reported to release phosphonates in the presence of esterase;[10] however, the utility of this design has not been thoroughly studied.
Scheme 1.
Model prodrugs appended with three distinct release strategies.
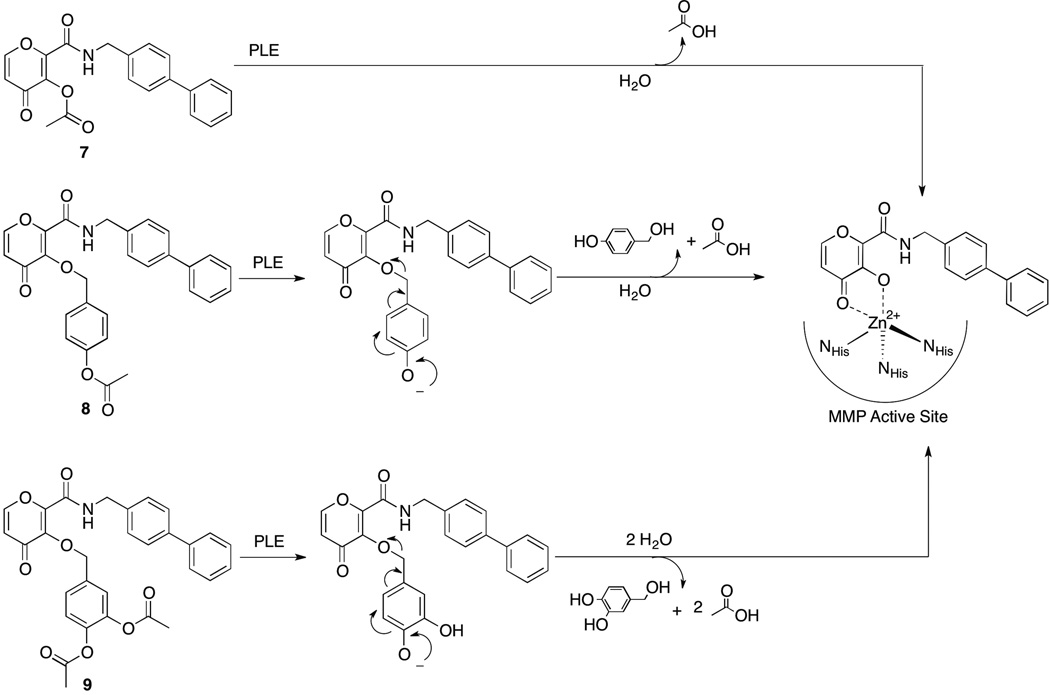
Scheme 3.
Deprotection mechanisms of proMMPi 7–9 by esterase to generate PY-2, a potent inhibitor of MMP-8 and MMP-12.
Synthesis of compounds 1–6 was straightforward and were relatively high yielding. The reactivity of 1–6 with an esterase was analysed via UV-Vis absorption spectroscopy. Upon the addition of porcine liver esterase (PLE), the absorbance of the reaction mixture was monitored. The emergence of a new spectrum with a λmax coinciding with that of the parent MBGs was observed, indicating complete conversion to the respective MBGs, maltol and 1-hydroxy-2-pyridinone (1,2-HOPO) (Figures S1–S6) was achieved. This trend was observed for 1–6, demonstrating that all three different protective approaches are effective for esterase-mediated conversion.
It is worth noting that approaches 2 and 3 generate side products that are released upon cleavage. That is, a deacetylation event by esterase at the para position leads to a spontaneous cascade reaction releasing a quinone-methide intermediate in both approaches. Quinone-methides are electrophilic Michael acceptors that react rapidly with water to generate 4-hydroxybenzyl alcohol (approach 2) and 3,4-dihydroxybenzyl alcohol (approach 3). This work did not study the effects these side products; however, 4-hydroxybenzyl alcohol is a known neuroprotective agent,[11] while 3,4-dihydroxybenzyl alcohol is found in virgin olive oil, suggesting an innocuous nature for each.[12]
Applying Strategies to Full-Length inhibitors
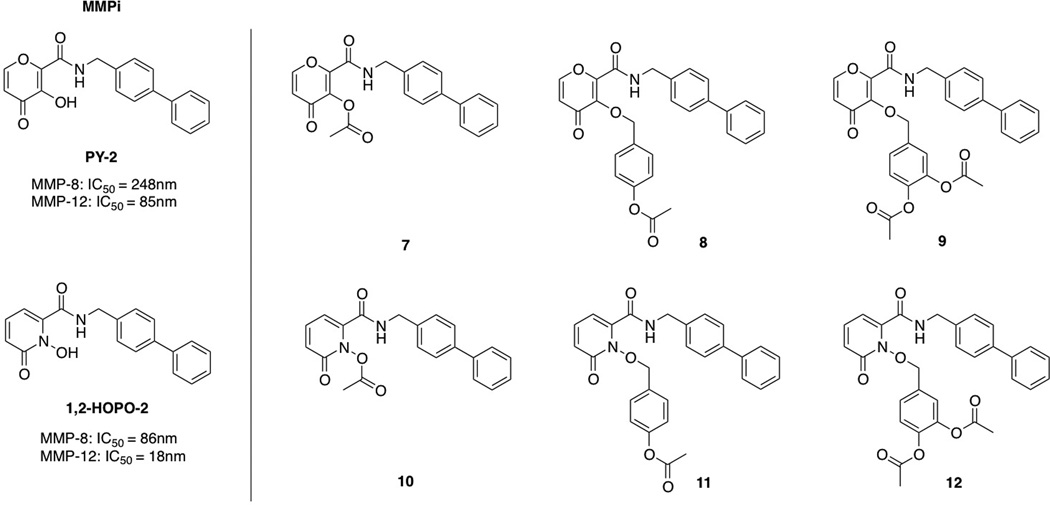
The successful conversion of the proMBGs to the parent chelators prompted the exploration of full-length matrix metalloproteinase proinhibitors (proMMPi). Previous studies in our laboratory have led to the discovery of specific inhibitors of MMP-8 and MMP-12 termed PY-2 and 1,2-HOPO-2 (Scheme 2). The biphenyl backbone of these MMPi selects against MMPs possessing shallow S1′ pockets, leading to semi-selective inhibition of deep-pocket MMPs with IC50 values in the low nanomolar range (Scheme 2).[13] The addition of an esterase-responsive protecting group to the two MMPi was performed in the same manner as 1–6.
Scheme 2.
Full-length proMMPi appended with the three protecting strategies.
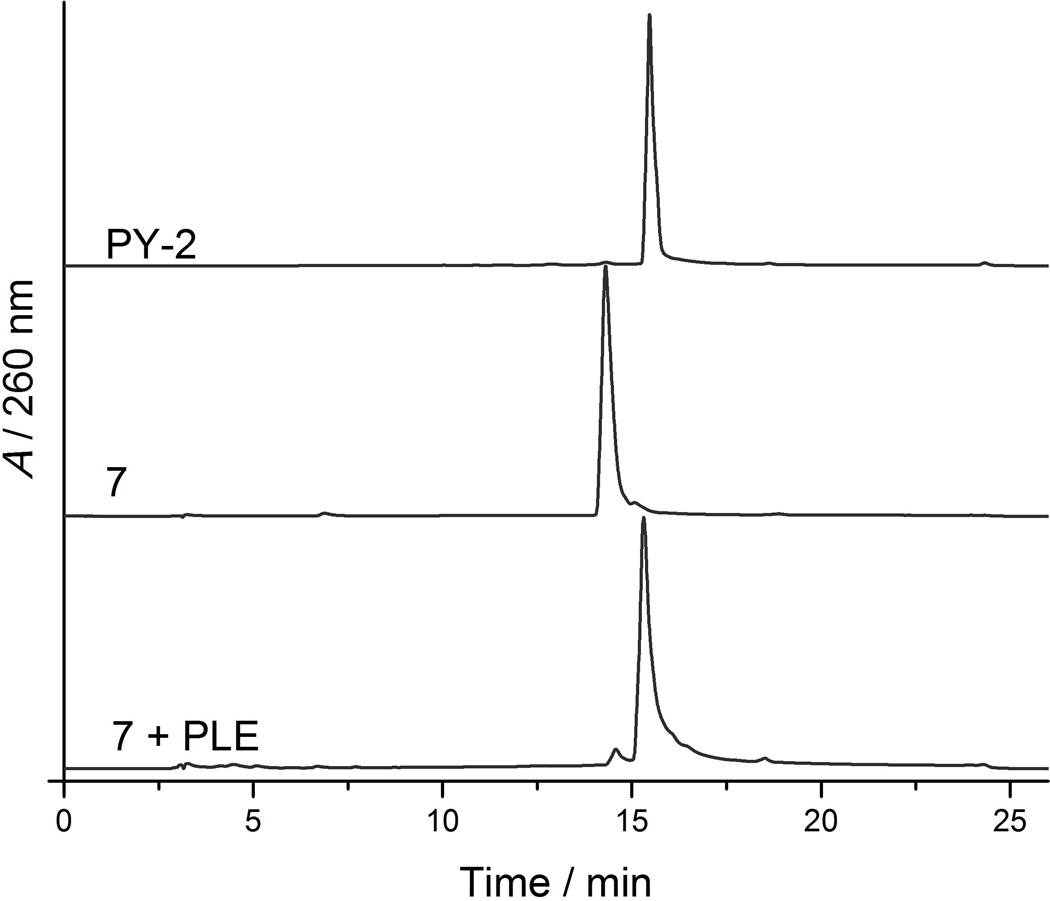
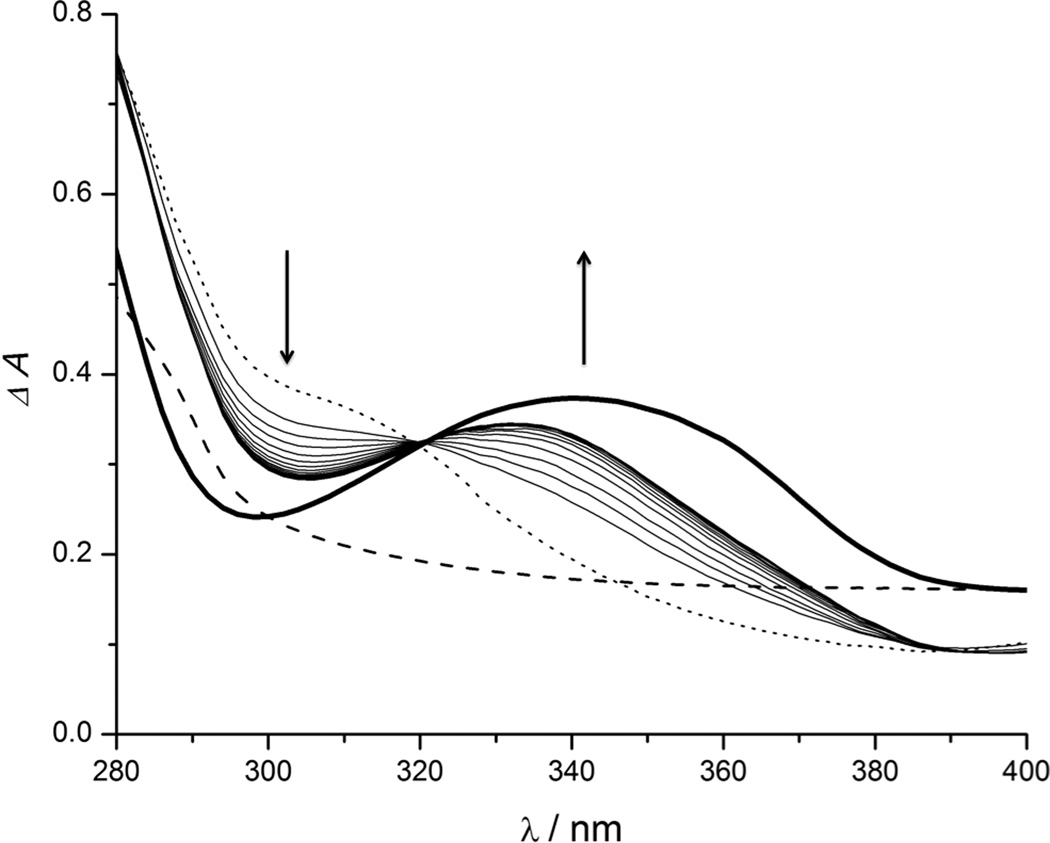
Conversion of proMMPi 7–9 was monitored via analytical HPLC, due to unclear spectral overlap between proinhibitors and parent inhibitors observed via UV-Vis spectroscopy. Treating compounds 7–9 with PLE produced HPLC traces corresponded to an authentic sample of PY-2 (Figure 1, Figures S8–S9), indicating successful prodrug release. proMMPi 10–12 were similarly converted by PLE as evidenced by UV-Vis absorption spectroscopy, where the emergence of spectral features matching that of the MMPi 1,2-HOPO-2 were clearly observed (Figure 2, Figure S7). The final absorbance spectrum shown in Figure 2 contains both 1,2-HOPO-2 and 4-hydroxybenzyl alcohol in a 1:1 ratio (vide supra), so that the resulting spectrum possesses features of both compounds. This side product was not detected via HPLC monitoring at 260 nm, further demonstrating the value of absorption spectroscopy for these studies. Nevertheless, both methods successfully show the responsiveness of the proMMPi to esterase with release of the parent inhibitors observed in every case. A summary of the deprotection mechanisms for each esterase-activated prodrug approach for PY-2 is summarized in Scheme 3.
Figure 1.
HPLC traces of PY-2 (top), 7 (middle) and 7 after treatment with PLE for 1 h (bottom). The retention time of 7 + PLE matches with that of an authentic sample of PY-2 (15.4 min), indicative of deprotection.
Figure 2.
Absorption spectra of 11 in the presence of PLE monitored every 30 sec for 8 min. The dotted line represents initial absorbance spectrum. The dashed line represents an authentic spectrum of 4-hydroxybenzyl alcohol, and the bold solid line depicts the absorption of an authentic sample of 1,2-HOPO-2. The arrows indicate spectral change over time.
Hydrolytic Stability Studies of Full-Length Proinhibitors
ProMMPi were evaluated for aqueous stability under simulated physiological conditions (50 mM HEPES, pH 7.4). An initial HPLC trace was obtained immediately after preparation in aqueous buffer and a second trace was collected after 24 h incubation at 37 °C. After 24 h, ~35% of 7 was hydrolyzed to PY-2, while 10 underwent rapid, complete hydrolysis to 1,2-HOPO-2 (data not shown). However, compounds 8, 9, 11, and 12 were all >90% stable to hydrolysis under these simulated physiological conditions for 24 h. These measurements clearly demonstrate the superior hydrolytic stability of the benzyl ether linkage (approach 2 or 3) over direct acetylation (approach 1) for these inhibitors.
Release Kinetics
To determine the sensitivity of these compounds to esterase in a quantitative fashion, pseudo first-order kinetic measurements were performed using UV-Vis absorption spectroscopy (Table 1). Pyrone-based proMBGs 1–3 and the full-length proMMPi 7–9 were evaluated to compare the three prodrug approaches. Compounds 2 and 3 displayed similar rate of conversion with kobs of 245±8 s−1 and 280±97 s−1, respectively. Surprisingly, these rates were >25× faster than that observed for the directly acetylated proMBG 1 (kobs of 9±0.4 s−1). A similar trend was observed for the proMMPi, where the directly acetylated compound 7 displayed slower kinetics than the proMMPi containing the acetylated trigger appended via either benzyl ether linker (8 and 9). Liberation of 8 and 9 were approximately 4× and 8× faster than that of 7, respectively. These values are consistent with previous reports showing that the rate of deprotection is enhanced with the presence of electron-donating substituents in the aromatic ring.[14] Overall, these findings highlight that the kinetic rates of release can be greatly attenuated by using different promoities.
Table 1.
Psuedo first-order rates of conversion in the presence of PLE
| Compound | kobs (s−1) | Compound | kobs (s−1) |
|---|---|---|---|
| 1 | 9.0 ± 0.4 | 7 | 160 ± 12 |
| 2 | 245 ± 8 | 8 | 742 ± 90 |
| 3 | 280 ± 97 | 9 | 1249 ± 60 |
Rate constant values were obtained by averaging three independent trials.
MMP Inhibition Studies
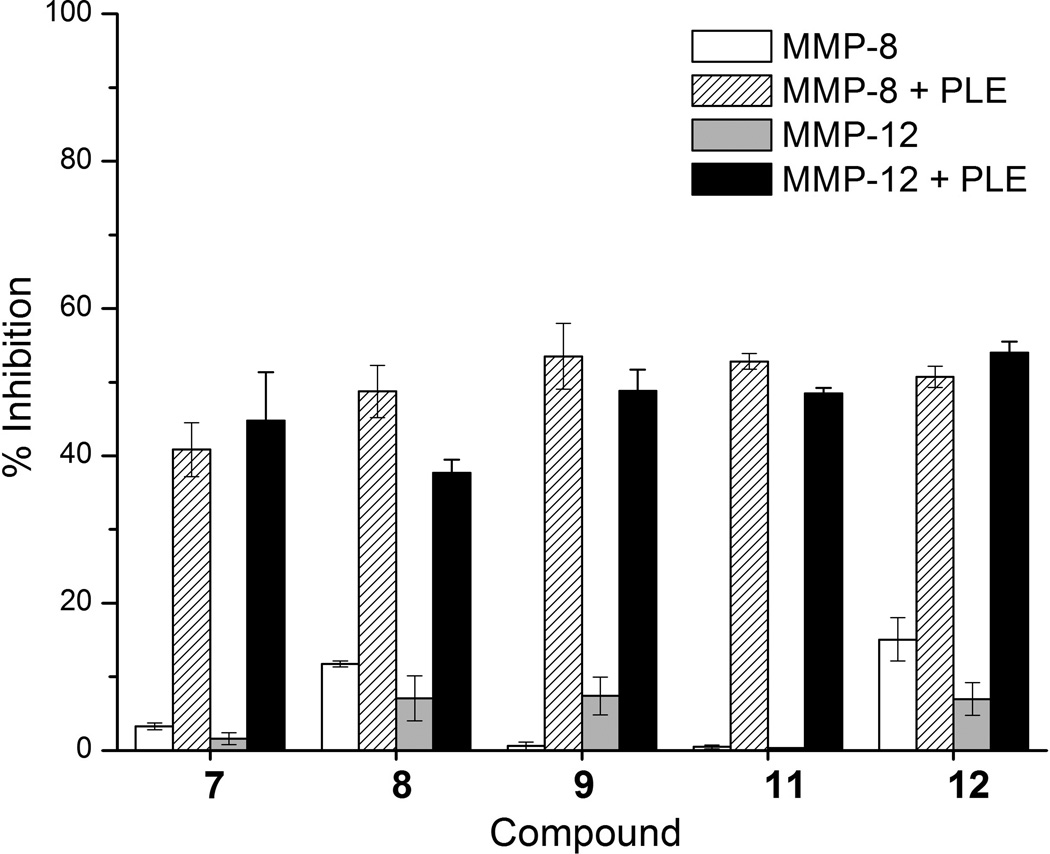
To determine the efficacy of these prodrug approaches, the ability of the proMMPi 7–12 to inhibit MMP-8 and MMP-12 in the absence and presence of esterase was performed. Compound 10 was excluded in these studies as it was found to be unstable in aqueous buffer upon preparation. MMP activity assays utilizing a cleavable fluorescent resonance energy transfer (FRET) substrate were employed (Figure 3).[15] Before treatment with PLE, compounds 7, 9, and 11 showed essentially no inhibition against MMP-8 and MMP-12. Compounds 8 and 12 showed minimal inhibition (<10%) of MMP-8 and 12. Upon the addition of PLE the percent inhibition increased to 40–50% inhibition for all compounds, indicative of activation to PY-2 and 1,2-HOPO-2. These biochemical assays demonstrate that esterase-responsive prodrugs are an effective class of proMMPi.
Figure 3.
Inhibition assays for compounds 7–9, 11 and 12 against MMP-8 (white) and MMP-12 (grey) and in the presence if PLE (striped and black, respectively.
Conclusion
We have demonstrated three different approaches to liberate phenol or hydroxyl moieties upon conversion by esterase, using MMP prodrugs as our proof-of-concept system. The benzyl ether linkage (approach 2 or 3) is superior to the conventional direct linkage of the acetate protecting group (approach 1) with respect to kinetics and aqueous stability. Testing of these compounds in a biochemical assay shows no inhibition by the proinhibitors against either MMP-8 or MMP-12. Upon treatment with esterase, the promoieties effectively cleave to generate the active MMPi, which inhibits the targets as expected. We hope that the superior reaction-based strategies presented here will serve as a platform for esterase-responsive prodrug design.
Experimental Section
Synthesis and Characterization
The detailed synthesis and characterization of compounds 1–12 are provided in the Supporting Information. All chemicals were purchased from commercial suppliers (Sigma-Aldrich, Acros Organics, TCI America) and were used without further purification. Chromatography was performed using a CombiFlash Rf-200 automated system from TeledyneISCO. NMR spectra were recorded on a Varian FT-400 MHz NMR instrument. Mass spectrometry was performed at the Molecular Mass Spectrometry Facility in the Department of Chemistry and Biochemisty at the University of California, San Diego.
UV-Vis Spectroscopy
Absorption spectra of compounds 1–6, 11, and 12 were collected on a Perkin-Elmer Lambda 25 UV-visible spectrophotometer. To a 1.0 mL solution at 0.05 mM concentration in HEPES buffer (50 mM, pH 7.5) was added PLE (3.57 U). Spectra were monitored over time at room temperature (Figures S1–S7).
Calculation of Kinetic Rate Constant
Pseudo-first order rate constants were calculated by monitoring the absorption spectra over time in the presence of PLE. To a 1.0 mL solution of 50 µM of each compound in HEPES buffer (50 mM, pH 7.5) was added PLE so that each sample contained 0.178 U of protein. Spectra were monitored over 10–20 min at room temperature with at least 100 spectra recorded for each sample. The change in absorption was monitored at 274 nm for the maltol series (1–3) and at 338 nm for the PY-2 series (7–9) we term Amax. The rate constant (kobs) was determined by monitoring the appearance of the absorption peak by plotting the linear slope of ln[(Amax - A)/(Amax)].
HPLC Analysis
Analytical HPLC was performed on a HP Series 1050 system equipped with a Vydac® C18 reverse phase column (218TP, 250×4.6 mm, 5 µm). Separation was achieved with a flow rate of 1 mL/min and the following solvents: solvent A is 5% MeOH and 0.1% formic acid in H2O and solvent B is 0.1% formic acid in MeOH. Starting with 95% A and 5% B, an isocratic gradient was run for 15 min to a final solvent mixture of 5% A and 95% B, which was held for 5 min before ramping back down to 95% A and 5% B in 2 min and holding for an additional 4 min. Compounds were prepared in HEPES buffer (50 mM, pH 7.5) at a concentration of 1 mM. Retention times of compounds PY-2, and 1,2-HOPO-2 were determined under identical HPLC conditions prior to evaluation of esterase cleavage of the protected compounds.
To determine the efficiency of esterase cleavage for the proMMPi, 1 mL samples of each compound were prepared at a concentration of 1 mM in HEPES buffer (50 mM, pH 7.5). To each sample was added 50 U of PLE and incubated at 25 °C for 1 h prior to analysis (Figures S8–S9).
To evaluate the hydrolytic stability of the proMMPi, a 1.0 mL sample of each compound was prepared at a concentration of 1 mM in HEPES buffer (50 mM, pH 7.4) and a trace was obtained immediately. This sample was incubated in the buffer solution for 24 h at 37° C before a second trace was obtained. The stability of each sample was determined based on the area under the curve (Figures S11–S14).
MMP Inhibition assays
Inhibition values of 7–9, and 11–12 were determined using a previously described commercially available fluroscent-based assay kit.[15] MMP activity was measured in 96-well plates using a Bio-Tek Flx800 fluorescent plate reader. The protected MMPi were dissolved in DMSO to a concentration of 1 mM and diluted in HEPES buffer (50 mM, pH 7.5) to a concentration of 50 µM. To each sample was added PLE such that 50 U of protein was present. This mixture was incubated for 1 h at room temperature. The esterase was removed via micro centrifugation using 10 kDa molecular weight cut-off filters. The filtered esterase-treated compounds were then added to appropriate wells at their respective IC50 values. Each well contained 20 µL of MMP-8 or MMP-12 (1.82 U/mL or 0.35U/mL, respectively), 60 µL MMP assay buffer (50 mM HEPES, 10mM CaCl2, 0.10% Brij-35, pH 7.5), and the esterase-treated MMPi (10 µL). After a 30 min incubation at 37 °C, a reaction was initiated with the addition of 10 µL (40 µM) of the fluorescent substrate (Mca-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2) where Mca = (7-methoxycoumarin-4-yl)-acetyl and Dpa = N-3-(2,4-dinitrophenyl)-L-α-β-diaminopropionyl)) and kinetic activity was monitored every 40 sec for 30 min with excitation and emission wavelengths at 335 nm and 405 nm, respectively. Enzymatic activity and thus inhibition was calculated with respect to the control experiment (no inhibitor present). Measurements were performed in duplicate in two independent experiments.
Supplementary Material
Acknowledgements
We thank Dr. Y. Su (UCSD MMSF) for performing mass spectrometry experiments. We acknowledge California Alliance for Minority Participation (C.P.). This work was funded by a grant from the National Institutes of Health through the National Institute of General Medical Sciences NIH (R01 GM098435).
References
- 1.a) Huttunen KM, Raunio H, Rautio J. Pharmacol. Rev. 2011;63:750–771. doi: 10.1124/pr.110.003459. [DOI] [PubMed] [Google Scholar]; b) Rautio J, Kumpulainen H, Heimbach T, Oliyai R, Oh D, Jarvinen T, Savolainen J. Nat. Rev. Drug Discov. 2008;7:255–270. doi: 10.1038/nrd2468. [DOI] [PubMed] [Google Scholar]
- 2.Stella VJ. J Pharm Sci-Us. 2010;99:4755–4765. doi: 10.1002/jps.22205. [DOI] [PubMed] [Google Scholar]
- 3.a) Liederer BM, Borchardt RT. J. Pharm. Sci. 2006;95:1177–1195. doi: 10.1002/jps.20542. [DOI] [PubMed] [Google Scholar]; b) Satoh T, Hosokawa M. Annu. Rev. Pharmacol. 1998;38:257–288. doi: 10.1146/annurev.pharmtox.38.1.257. [DOI] [PubMed] [Google Scholar]; c) Ettmayer P, Amidon GL, Clement B, Testa B. J. Med. Chem. 2004;47:2393–2404. doi: 10.1021/jm0303812. [DOI] [PubMed] [Google Scholar]
- 4.a) Failes TW, Cullinane C, Diakos CI, Yamamoto N, Lyons JG, Hambley TW. Chem. Eur. J. 2007;13:2974–2982. doi: 10.1002/chem.200601137. [DOI] [PubMed] [Google Scholar]; b) Failes TW, Hambley TW. Dalton Trans. 2006:1895–1901. doi: 10.1039/b512322d. [DOI] [PubMed] [Google Scholar]; c) Failes TW, Hambley TW. J. Inorg. Biochem. 2007;101:396–403. doi: 10.1016/j.jinorgbio.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 5.a) Hu JL, Van den Steen PE, Sang QXA, Opdenakker G. Nat. Rev. Drug Discov. 2007;6:480–498. doi: 10.1038/nrd2308. [DOI] [PubMed] [Google Scholar]; b) Whittaker M, Floyd CD, Brown P, Gearing AJH. Chem. Rev. 1999;99:2735–2776. doi: 10.1021/cr0100345. [DOI] [PubMed] [Google Scholar]
- 6.a) Egeblad M, Werb Z. Nat. Rev. Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]; b) Overall CM, Kleifeld O. Br. J. Cancer. 2006;94:941–946. doi: 10.1038/sj.bjc.6603043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.a) Coussens LM, Fingleton B, Matrisian LM. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]; b) Overall CM, Lopez-Otin C. Nat. Rev. Cancer. 2002;2:657–672. doi: 10.1038/nrc884. [DOI] [PubMed] [Google Scholar]
- 8.a) Daniel KB, Jourden JLM, Negoescu KE, Cohen SM. J. Biol. Inorg. Chem. 2011;16:313–323. doi: 10.1007/s00775-010-0727-x. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Major Jourden JL, Cohen SM. Chem. Commun. 2010;46:1241–1243. doi: 10.1039/b923302d. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Major Jourden JL, Cohen SM. Angew. Chem. Int. Ed. 2010;49:6795–6797. doi: 10.1002/anie.201003819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Major Jourden JL, Daniel KB, Cohen SM. Chem. Commun. 2011;47:7968–7970. doi: 10.1039/c1cc12526e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schultz C. Bioorg. Med. Chem. 2003;11:885–898. doi: 10.1016/s0968-0896(02)00552-7. [DOI] [PubMed] [Google Scholar]
- 11.Yu SS, Zhao J, Zhen WP, Zhao Y. Brain. Res. 2010;1308:167–175. doi: 10.1016/j.brainres.2009.10.037. [DOI] [PubMed] [Google Scholar]
- 12.Saitta M, Salvo F, Di Bella G, Dugo G, La Torre GL. Food Chem. 2009;112:525–532. [Google Scholar]
- 13.Agrawal A, Romero-Perez D, Jacobsen JA, Villarreal FJ, Cohen SM. ChemMedChem. 2008;3:812–820. doi: 10.1002/cmdc.200700290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) Hay MP, Sykes BM, Denny WA, O'Connor CJ. J. Chem. Soc. Perkin Trans. 1999;1:2759–2770. [Google Scholar]; b) Sykes BM, Hay MP, Bohinc-Herceg D, Helsby NA, O'Connor CJ, Denny WA. J. Chem. Soc. Perkin Trans. 2000;1:1601–1608. [Google Scholar]
- 15.Knight CG, Willenbrock F, Murphy G. FEBS Lett. 1992;296:263–266. doi: 10.1016/0014-5793(92)80300-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.