The ability to sensitively detect biomolecules and monitor complex biological processes has enabled many important fundamental and technological advances in the life sciences.[1] These include novel and powerful ways of diagnosing disease, pharmaceutical screening procedures, and a variety of biosafety applications.[2–4] Over the past couple of decades, a number of different detection methods have been developed for high-sensitivity biomolecule detection.[5–19] Many of these rely on nanostructures with novel signal transduction or signal amplification modes. Most can be categorized either as spectroscopic[5–11] or electrical approaches[12–17] and have strengths and weaknesses depending upon the analyte, environment, and intended uses.[1–3] With researchers gaining increasing control over the architectural parameters of nanostructures, one can begin to envision systems with multiple layers of complexity and corresponding functional capabilities. Electrical approaches are attractive because in principle they can be miniaturized in the form of relatively simple and even portable devices.[13,14,17] Spectroscopic approaches often rely on an additional labeling material and have the virtue of offering greater amounts of chemical information about the analytes being probed.[6,8] The combination of these two assay formats within the context of a single nanostructure can provide built-in internal assay controls[20] and a way to independently measure multiple steps in a complex biological process. Herein, we describe how one can use segmented nanostructures[21–25] prepared by on-wire lithography (OWL)[23,24] to create a system with two orthogonally functional components: one that behaves as a diode-like detection system with electrical readout, and the other that acts as a Raman enhancing device that allows one to spectroscopically probe different but related reactions that occur on the same device (Figure 1A).
Figure 1.
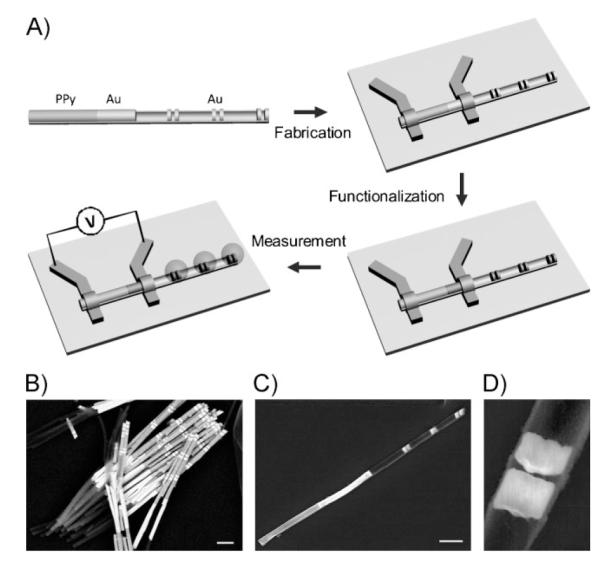
A) Schematic of the synthesis, device fabrication, surface functionalization, and measurement of hybrid PPy-rod/Au-rod/Au-disk-pair nanostructures. The Au–PPy nanorod portion is contacted with electrodes to form the electrical sensing elements, and the Au disks functionas SERS hotspots. The surface functionalization takes place over the entire exposed Au surface (disks and rods). B) An SEM image of assynthesized Au–Ni–Au–PPy nanorods. The Ni portions are subsequently removed by chemical etching to form the multigap structures (C). Scale bar = 1 μm. C) An SEM image of one PPy-rod/Au-rod/Au-disk-pair nanostructure made by OWL with a SiO2 backing layer. The structures consist of a PPy–Au rod segment (the PPy portion has a lower contrast than the Au portion) and three pairs of Au disks. Scale bar = 1 μm. D) Zoomed-in SEM image of a single pair of Au disks (disk thickness ≈120 nm, disk separation ≈30 nm).
Developed in our lab, the OWL method can be used to fabricate 1D wire structures with precise control over the compositional blocks that comprise such wires and the location and size of disks and gaps within such structures.[23] We have used this capability to create: i) platforms for studying molecular-transport junctions;[26,27] ii) electrical nanotraps that can be used to localize and spectroscopically detect small amounts of charged materials;[28] and iii) libraries of disk-like nanostructures that can be used for encoding purposes[29] and to probe the relationship between nanostructure architectures and the well-known surface-enhanced Raman scattering (SERS) phenomenon.[24,29,30] Herein, we report a new hybrid nanostructure, which consists of a combination of a polypyrrole (PPy) nanorod, an Au nanorod, and three Au nanodisk pair segments, for parallel electrical and spectroscopic detection of biomolecules. Briefly, multisegmented nanorods (Figure 1B) composed of Au, Ni, and PPy segments were synthesized by electrochemical deposition using porous alumina membranes as templates. Afterwards, the hybrid PPy-rod/Au-rod/Au-disk-pair nanostructures were fabricated by OWL,[23] during which a thin layer (≈50-nm-thick) of silicon dioxide was deposited onto one side of the entire nanostructure by plasma-enhanced chemical vapor deposition (PECVD), followed by the etching of the sacrificial Ni segments to form the gaps between gold disks (Figure 1C and D). The thickness of the 360-nm-diameter disks is ≈120 nm and the gap size is ≈30 nm, which for this sample yields an optimal SERS response under 632.8-nm laser excitation.[24] The distance between adjacent disk pairs is set at ≈1 μm to avoid interference from different nanogap-generated Raman hotspots within one structure.
After the OWL process, electron-beam lithography was used to define electrode contacts at both the Au and PPy segments of the nanostructure. The metal contacts were subsequently passivated by depositing a thin layer of silicon oxide by PECVD to reduce the leakage current through the electrolyte solution. Since PPy is a hole-carrying conducting polymer, the intrarod junction between the PPy and Au segments behaves as a diode-like Schottky barrier. Indeed, its conductivity can be modulated by the Schottky barrier height, that is, the energy difference between the valence band of PPy (5.0 eV)[31] and the Fermi level of Au (5.5 eV).[32] During a biodetection assay, charged biomolecules will bind to Au segments (previously modified with thiolated receptors) and subsequently modulate the Fermi level of the Au rod and thus the Schottky barrier height between Au and PPy. These binding events will lead to a change in the electrical conductance of the PPy–Au nanodiode. A similar mechanism has previously been reported in carbon-nanotube-based electrical sensing.[13,14] In addition, the Au disk pairs on the same nanostructures are also functionalized with surface receptors via the same thiol-on-Au reaction. When biomolecules bound at these sites are subsequently sandwiched with a second Raman dye-labeled target-selective receptor, they can be spectroscopically characterized because of the strong Raman signal-enhancing properties of the disk pairs.[29]
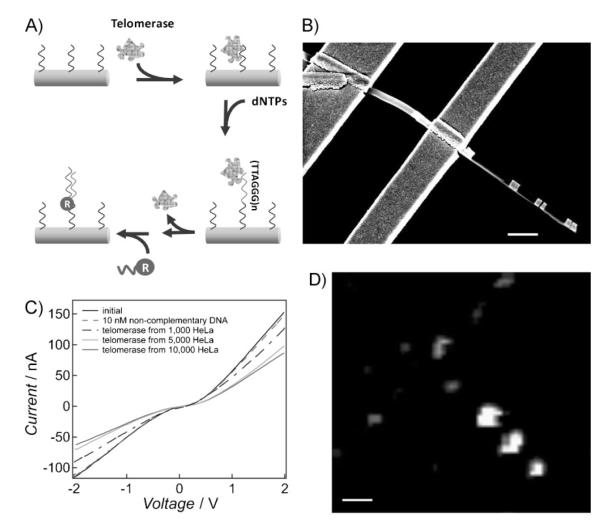
To evaluate the potential of these novel hybrid nanostructures for parallel electrical and optical detection of biological analytes, we investigated a nucleic acid based marker for telomerase activity. Telomerase is a reverse transcriptase enzyme that catalyzes the addition of the telomeric repeat sequence TTAGGG onto the 3′-end of the human chromosome.[33–35] Our experimental design allows for electrical measurement of the telomerase binding on its oligonucleotide substrate, followed by Raman readout of oligonucleotide elongation catalyzed by the telomerase enzyme (Figure 2A). In a typical assay, the Au surface of the hybrid nanostructures (both nanorods and nanodisks) is first functionalized with oligonucleotide strands (sequence: 5′ HS T5 AAT CCG TCG AGC AGA GTT 3), which serve as telomerase binding substrates. Different sample solutions were delivered to the device surface by a custom-made microfluidic channel. The electrical conductivity of Au–PPy rod segment was cyclically measured by applying aDC bias between −2and 2V (Figure 2B and C). The telomerase reagents (extracted from HeLa cells) bind to the immobilized nucleic acids and subsequently elongate the template strands by adding TTAGGG repeat units in the presence of deoxyribonucleotide triphosphate (dNTP) monomers. After removing the telomerase with sodium dodecyl sulfate (SDS), a solution containing Cy5-labeled oligonucleotides (reporter strands) complementary to the telomerase-elongated sequence is added to the solution, which results in hybridization between the two sequences. Confocal Raman spectroscopy allows one to identify the Cy5-labeled oligonucleotides in and near the gaps formed by the Au disk pairs.
Figure 2.
A) Schematic of telomerase binding and elongation on the Au nanostructure (both Au nanorod and Au disk pair) surface. The oligonucleotide sequence serving as the telomerase binding substrate is 5′ HS T5 AAT CCG TCG AGC AGA GTT 3′. B) An SEM image of a representative device for both electrical and optical detection (C and D). Scale bar = 1 μm. C) Current versus voltage plots of the Au–PPy nanorod segment with binding of telomerase extracted from solutions of HeLa cells at different concentrations. D) As canning Raman microscope image of the same device in (C) showing the Cy5 signal upon binding of the oligonucleotide reporter strands. Scale bar = 1 μm.
In a control experiment, when a non-complementary DNA sequence was delivered onto the device surface, it did not bind to the telomerase capture strands that were used to pre-functionalize the Au nanorod surface. Therefore, the conductivity of the Au–PPy rod remained unchanged. When solutions of telomerase samples extracted from different numbers of HeLa cells (1 000, 5 000, and 10 000 HeLa cells in 200 μL solution, Chemicon International Inc.) were delivered onto the device surface, a decrease in conductivity was measured, and the signal measured was dependent on added telomerase (Figure 2C). Since the telomerase is positively charged in the buffer solution,[36] when it binds to the Au portion of the nanorods, the amount of positive charge on the device surface increases. This results in an increase in the energy barrier height between Au and PPy segments, and thus a decrease in the electrical conductance. Electrical measurements on over 15 different devices with similar Au–PPy structures show that the electrical conductivity change for 1 000 HeLa cells was 14%±5% of the original conductivity of the devices. With the limit of detection of this assay at 1 000 HeLa cells, the sensitivity of this proof-of-concept system is moderate. The relatively high salt concentration of the buffer solution, which is required for efficient hybridization (≈50 mM phosphate buffer), screens the charge around the nanorod sensing element and decreases the magnitude of signal change upon target binding. This screening effect can be reduced by measuring the conductivity change in the dried state,[37] although this will inevitably cause the loss of enzymatic activity of telomerase for subsequent reactions. Alternatively, it may be possible to use capture elements made of peptide nucleic acids (PNAs) or other designed oligonucleotides, which are less affected by salt.[38]
After hybridization of the reporter strands to the telomerase-elongated sequence on the Au surface, a strong SERS signal from the Au disk pairs can be measured (Figure 2D). These data show that the nanogaps exhibit an intense Raman signal (integrated intensity from Raman spectrum at the band of 1 170–1 210 cm−1), which appears as a bright spot against a dark, smooth background (the higher the intensity, the higher the counts). Little Raman signal is observed from the flat portion of the chip surface, while a very low Raman signal may also be seen from some portions of the titanium electrode surface due to nonspecific binding. These complementary electrical and optical measurements demonstrate that the binding and elongation steps of telomerase can be rigorously identified by this hybrid nanostructure device.
In conclusion, we developed a new platform composed of both electrically and optically sensitive elements for the detection of telomerase binding and enzymatic activity. The diode junction between the PPy and Au rod segments is used to record the binding step of telomerase to its oligonucleotide substrate, while the Au disk pairs are used to monitor the elongation of the oligonucleotide strands through telomerase enzymatic activity. This system can be further optimized for greater sensitivity by using different surface receptors and sensor materials such as inorganic semiconductors, and may open up a host of new opportunities for studying many biological reactions in the life sciences.
Experimental Section
Materials synthesis and device fabrication
Multisegmented nanorods were electrically deposited using an anodic aluminum oxide (AAO) porous membrane (Whatman, Anodisc 0.02-μm pores, 47-mm outer diameter) as a template.[23,29] Commercially available Orotemp 24RTU and Nickel Sulfamate SEMI Bright RTU electroplating solutions (Technic. Inc.) were used for electrochemical deposition of Au and Ni, respectively. PPy was deposited from an aqueous solution of pyrrole (340-μL pyrrole and 0.212-g LiClO4 in 20-mL H2O). Different segments of the nanorods (Au, Ni, and PPy) were electrochemically deposited at a constant potential (Au: −900 mV, Ni: −850 mV, PPy: 750 mV vs. Ag/AgCl), and the lengths of each nanorod segment were tailored by varying the amount of charge passing through the electrodes. After electrodeposition and dissolving the AAO template, a layer of SiO2 (≈50-nm-thick) was deposited onto one side of these multisegmented nanorods by PECVD. Afterwards, the Ni segments were chemically dissolved with 6 m HCl to form the multigap structures. These nanorods were then dispersed onto a silicon substrate with 600-nm-thick thermal oxide. The electrodes connecting to individual nanorods were defined by electron-beam lithography (Model: Quanta-600, FEI Inc.) followed by evaporation of 300 nm of titanium. A layer of SiO2 (≈100-nm-thick) was deposited on top of the titanium surface by PECVD before lifting off the electron-beam resist. Scanning electron microscopy (SEM) images were recorded using a SEM (Model S4800, Hitachi Inc.).
Device functionalization
The nanorod devices were plasma-cleaned (30 W, 30 s) and incubated overnight with the oligonucleotide strands (100 μm; sequence: 5′ HS T5 AAT CCG TCG AGC AGA GTT 3′) serving as telomerase binding substrates in 10 mm phosphate buffer (with 0.3 m NaCl, 0.1% Tween-20, pH 7.4).
Telomerase extraction
The telomerase was extracted by the CHAPS method.[33] HeLa cell pellets (1 million cells, Chemicon International Inc.) were first suspended in 200-μL 1×CHAPS lysis buffer (Chemicon International Inc.) and incubated in an ice bath for 30 min. Finally, the mixture was spun at 12 000×g for 20 min at 4 °C and the supernatant was transferred to separate tubes and stored at −80 °C. The telomerase extracts are stable at −80 °C for over one year.[33]
Telomerase activity measurement
Telomerase samples at different concentrations were incubated with oligonucleotide-functionalized multisegmented nanorod devices for 1 h. The devices then were rinsed with deionized water to remove unbound telomerase. Electrical measurements (current vs. voltage) were carried out before and after telomerase incubation, respectively, in a two-probe configuration. A 16-bit digital acquisition board (DAQ, National Instruments, Austin, TX) was used as the DC voltage source and a current preamplifier (Model 1211, DL Instrument, Ithaca, NY) was used to measure the current. The device was incubated with a solution containing 1 nm of oligonucleotide reporting strands (sequence: 5′ CCC TAA CCC TAA Cy5 3′) for 30 min, rinsed, and characterized by confocal Raman microscopy (Model CRM200, WITec Inc.) equipped with a piezoscanner and 100×microscope objective (NA=0.90, Nikon Inc.). [29] A He–Ne laser (632.8 nm) was used to excite the Cy5 dye. Each pixel in the images was constructed by integrating the Raman intensity of the main band from 1 170–1 210 cm−1.[6]
Footnotes
C.A.M. acknowledges support from AFOSR, NSF-MRSEC, and DARPA. C.A.M. is also grateful for an NIH Director’s Pioneer Award and an NSSEF Fellowship from the Department of Defense.
References
- [1].Rosi NL, Mirkin CA. Chem. Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- [2].Etzioni R, Urban N, Ramsey S, Mclntosh M, Schwartz S, Reid B, Radich J, Anderson G, Hartwell L. Nat. Rev. Cancer. 2003;3:243–252. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- [3].Ferrari M. Nat. Rev. Cancer. 2005;5:161–171. doi: 10.1038/nrc1566. [DOI] [PubMed] [Google Scholar]
- [4].Zheng G, Daniel WL, Mirkin CA. J. Am. Chem. Soc. 2008;130:9644–9645. doi: 10.1021/ja803035p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Barnes WL, Dereux A, Ebbesen TW. Nature. 2003;424:824–830. doi: 10.1038/nature01937. [DOI] [PubMed] [Google Scholar]
- [6].Cao YC, Jin RC, Nam JM, Thaxton CS, Mirkin CA. J. Am. Chem. Soc. 2003;125:14676–14677. doi: 10.1021/ja0366235. [DOI] [PubMed] [Google Scholar]
- [7].Cao YC, Jin RC, Mirkin CA. Science. 2002;297:1536–1540. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- [8].Kneipp K, Kneipp H, Itzkan I, Dasari RR, Feld MS. Chem. Rev. 1999;99:2957–2975. doi: 10.1021/cr980133r. [DOI] [PubMed] [Google Scholar]
- [9].Stoermer RL, Cederquist KB, McFarland SK, Sha MY, Penn SG, Keating CD. J. Am. Chem. Soc. 2006;128:16892–16903. doi: 10.1021/ja0658261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tang J, Signarvic RS, DeGrado WF, Gai F. Biochemistry. 2007;46:13856–13863. doi: 10.1021/bi7018404. [DOI] [PubMed] [Google Scholar]
- [11].Weiss S. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- [12].Beer PD, Gale PA. Angew. Chem. Int. Ed. 2001;40:486–516. [PubMed] [Google Scholar]
- [13].Chen RJ, Choi HC, Bangsaruntip S, Yenilmez E, Tang X, Wang Q, Chang Y, Dai H. J. Am. Chem. Soc. 2004;126:1563–1568. doi: 10.1021/ja038702m. [DOI] [PubMed] [Google Scholar]
- [14].Heller I, Janssens AM, Mannik J, Minot ED, Lemay SG, Dekker C. Nano Lett. 2008;8:591–595. doi: 10.1021/nl072996i. [DOI] [PubMed] [Google Scholar]
- [15].Skladal P. Electroanalysis. 1997;9:737–745. [Google Scholar]
- [16].Wang J. Talanta. 2002;56:223–231. doi: 10.1016/s0039-9140(01)00592-6. [DOI] [PubMed] [Google Scholar]
- [17].Zheng G, Patolsky F, Cui Y, Wang WU, Lieber CM. Nat. Biotechnol. 2005;23:1294–1301. doi: 10.1038/nbt1138. [DOI] [PubMed] [Google Scholar]
- [18].Jersch J, Maletzky T, Fuchs H. Rev. Sci. Instrum. 2006;77:083701. [Google Scholar]
- [19].Wu GH, Datar RH, Hansen KM, Thundat T, Cote RJ, Majumdar A. Nat. Biotechnol. 2001;19:856–860. doi: 10.1038/nbt0901-856. [DOI] [PubMed] [Google Scholar]
- [20].Patolsky F, Zheng G, Hayden O, Lakadamyali M, Zhuang X, Lieber CM. Proc. Natl. Acad. Sci. USA. 2004;101:14017–14022. doi: 10.1073/pnas.0406159101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Martin CR. Science. 1994;266:1961–1966. doi: 10.1126/science.266.5193.1961. [DOI] [PubMed] [Google Scholar]
- [22].Martin BR, Dermody DJ, Reiss BD, Fang MM, Lyon LA, Natan MJ, Mallouk TE. Adv. Mater. 1999;11:1021–1025. [Google Scholar]
- [23].Qin L, Park S, Huang L, Mirkin CA. Science. 2005;309:113–115. doi: 10.1126/science.1112666. [DOI] [PubMed] [Google Scholar]
- [24].Qin L, Zou SL, Xue C, Atkinson A, Schatz SC, Mirkin CA. Proc. Natl. Acad. Sci. USA. 2006;103:13300–13303. doi: 10.1073/pnas.0605889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Liu S, Tok JBH, Bao ZN. Nano Lett. 2005;5:1071–1076. doi: 10.1021/nl050581w. [DOI] [PubMed] [Google Scholar]
- [26].Chen X, Jeon YM, Jang JW, Qin L, Huo F, Wei W, Mirkin CA. J. Am. Chem. Soc. 2008;130:8166–8167. doi: 10.1021/ja800338w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chen X, Braunschweig AB, Wiester MJ, Yeganeh S, Ratner MA, Mirkin CA. Angew. Chem. Int. Ed. 2009;48:5178–5181. doi: 10.1002/anie.200806028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Zheng G, Qin L, Mirkin CA. Angew. Chem. Int. Ed. 2008;47:1938–1941. doi: 10.1002/anie.200705312. [DOI] [PubMed] [Google Scholar]
- [29].Qin L, Banholzer MJ, Millstone JE, Mirkin CA. Nano Lett. 2007;7:3849–3853. doi: 10.1021/nl072606s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kelly KL, Coronado E, Zhao LL, Schatz GC. J. Phys. Chem. B. 2003;107:668–677. [Google Scholar]
- [31].Somani PR, Radhakrishnan S. Chem. Phys. Lett. 2003;379:401–405. [Google Scholar]
- [32].Hache F, Ricard D, Flytzanis C, Kreibig U. Appl. Phys. A: Mater. 1988;47:347–357. [Google Scholar]
- [33].Kim NW, Piatyszek MA, Prowse KR, Harkey CB, West MD, Ho PLC, Coviello GM, Wright WE, Weinrich SL, Shay JW. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- [34].Morin GB. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- [35].Weizmann Y, Patolsky F, Lioubashevski O, Willner I. J. Am. Chem. Soc. 2004;126:1073–1080. doi: 10.1021/ja038257v. [DOI] [PubMed] [Google Scholar]
- [36].Bryan TM, Sperger JM, Chapman KB, Cech TR. Proc. Natl. Acad. Sci. USA. 1998;95:8479–8484. doi: 10.1073/pnas.95.15.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wang X, Ozkan CS. Nano Lett. 2008;8:398–404. doi: 10.1021/nl071180e. [DOI] [PubMed] [Google Scholar]
- [38].Weiler J, Gausepohl H, Hauser N, Jensen ON, Hoheisel JD. Nucleic Acids Res. 1997;25:2792–2799. doi: 10.1093/nar/25.14.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]