Abstract
Purpose
DAPK1, a tumor suppressor, is a rate-limiting effector in an ER stress-dependent apoptotic pathway. Its expression is epigenetically suppressed in several tumors. A mechanistic basis for epigenetic/transcriptional repression of DAPK1 was investigated in certain forms of AML with poor prognosis, which lacked ER stress-induced apoptosis.
Experimental Design
Heterogeneous primary AMLs were screened to identify a subgroup with Flt3ITD in which repression of DAPK1, among NF-κB- and c- jun-responsive genes, was studied. RNAi knockdown studies were performed in Flt3ITD+ve cell line, MV-4-11, to establish genetic epistasis in the pathway Flt3ITD-TAK1-DAPK1 repression, and chromatin immunoprecipitations were performed to identify proximate effector proteins, including TAK1-activated p52NF-κB, at the DAPK1 locus.
Results
AMLs characterized by normal karyotype with Flt3ITD were found to have 10-100-fold lower DAPK1 transcripts normalized to the expression of c-jun, a transcriptional activator of DAPK1, as compared to a heterogeneous cytogenetic category. Meis1, a c-jun-responsive adverse AML prognostic gene signature was also measured as control. These Flt3ITD+ve AMLs over-express relB, a transcriptional repressor, which forms active heterodimers with p52NF-κB. Chromatin immunoprecipitation assays identified p52NF-κB binding to the DAPK1 promoter along with HDAC2 and HDAC6 in the Flt3ITD+ve human AML cell line MV-4-11. Knockdown of p52NF-κB or its upstream regulator, NIK, de-repressed DAPK1. DAPK1-repressed primary Flt3ITD+ve AMLs had selective nuclear activation of p52NF-κB.
Conclusions
Flt3ITD promotes a non-canonical pathway via TAK1 and p52NF-κB to suppress DAPK1 in association with HDACs, which explains DAPK1 repression in Flt3ITD+ve AML.
Keywords: DAPK1, NFκB, histone deacetylase, endoplasmic reticulum, Flt3
Introduction
Recent evidence suggests that attenuation of the unfolded protein response (UPR)- an ER-dependent stress response- may explain therapeutic failure of acute myeloid leukemia (AML) (1-3). Downstream transcriptional mediators of Flt3 internal tandem duplication (ITD) in AML may impose such status by regulating expression of effectors that control stress dependent apoptosis. For example, the ratio of ER levels of bcl-2 vs. DAPK1 as well as other effectors may determine this output at a defined setpoint (4-6).
DAPK1 is a calcium-calmodulin dependent serine-threonine protein kinase, which suppresses tumor cell survival and metastasis via autophagy and apoptosis. It plays a central role in ER stress-dependent apoptosis (7). DAPK1 expression is affected by a variety of oncogenic signals (7, 8). We previously demonstrated the existence of a Flt3/JNK1/ c-jun pathway in Flt3ITD+ve AML (9). c-jun is known to drive the expression of not only bcl-2, but also DAPK1 (10, 11). However, the latter circumstance would be antagonistic to the progression of poor-prognosis Flt3ITD+ve AML. On the other hand, NF-κB and CRE/c-jun regulatory sites co-exist on the promoters of certain tumor suppressor or cytokine genes including DAPK1, and the importance of NF-κB signaling in AML is known (12-15). We hypothesized that a resistance to apoptosis in certain AMLs occurs via severe repression of DAPK1, through recruitment of p52NF-κB to the putative NF-κB site at - 134bp of its promoter (12, 13). Indeed, expression of DAPK1 is lost in number of human cancers, including leukemias (16). Although epigenetic suppression of DAPK1 is well reported, the upstream mechanisms that contribute to tumor promotion via the recruitment of epigenetic apparatus to the DAPK1 promoter are not defined (16-19).
We hypothesized that tandem activation of both JNK1 and IKK/NF-κB may be involved in concerted regulation of anti-apoptotic as well as pro-apoptotic genes to achieve an anti-apoptotic/pro-apoptotic effector balance (e.g. bcl-2/DAPK1) to permit higher aggressiveness in Flt3ITD+ve AML (4, 6, 14, 20). This postulation was tested in context of prior functional and cohort analyses of AML blasts performed in our laboratory that linked Flt3 phosphorylation/activation to JNK1 phosphorylation (9); and in view of the known role for c-jun/AP-1 in DAPK1 and bcl-2 expression (10,11). We also inferred that a conserved dual activation mechanism for JNK1/c-jun and IKK/NF-κB may exist, which relies upon TAK1 (21), to promote the optimal anti-apoptotic/pro-apoptotic effector balance. This hypothesis was given emphasis by recognition that TAK1 is among the most highly expressed genes in a LSC (leukemic stem cell) signature of poor-risk AML in which DAPK1 repression co-exists (22,23).
Here, we show that TAK1 activated p52NF-κB, binds at the tandem NF-κB and CRE sites of DAPK1, and recruits certain transcriptional repressors, belonging to the histone deacetylase (HDAC) family (12). As p52NF-κB is a downstream target of Flt3 signaling, we hypothesized that interruption of this signaling arm of Flt3 would result in a derepression of DAPK1 to contribute toward enhanced apoptosis in Flt3 ITD+ve AMLs. Lastly, we suggest a therapeutic model for the rational combination of Flt3- and HDAC-inhibitors for suppressing AML growth.
Materials and Methods
Cell culture
The human leukemic cell lines, HL-60 and MV-4-11 (derived from a biphenotypic leukemia), were obtained from the American Type Culture Collection (ATCC, Manassas, VA) in McCoy's and RPMI media respectively, supplemented with 10% fetal calf serum. The MV-4-11 cell line overexpresses Flt3-ITD and harbors 11q23 translocation [t (4; 11)] involving the MLL gene (9). Blast cells from the bone marrow of patients with AML were obtained at the time of diagnosis, after informed consent. The buoyant fraction was isolated over Ficoll-Hypaque, and then washed with phosphate-buffered saline (PBS) before processing. The cohort of AMLs subjected to gene expression analysis demonstrated mean 88± 10% blasts (Supplemental Table 1). Cells were lysed and fractionated into Nuclear and Cytoplasmic fractions using the NE-PER Extraction Kit (Pierce Biotechnology, Rockford, IL). For Western blot analysis bone marrow samples with ≥70% blast cells in the purified aspirate were used.
Transfections and reporter assays
MV-4-11 cells were electroporated using the Amaxa system and then placed in McCoy's medium supplemented with 10% FBS. Cells were transfected with a luciferase reporter (13) driven by the DAPK1 promoter (1.2 κB) harboring either wildtype or mutated CRE site (−177 bp). Wildtype c-jun or vector control was co-transfected in some experiments with the DAPK1-reporter. A Renilla luciferase was co-transfected to normalize for variations in transfection efficiency.
Western blot analysis
Cytosolic or nuclear proteins were subjected to Western blotting with indicated antibodies as described (9). Densitometry was performed to quantify specific bands, and data were normalized to either cytoplasmic (GAPDH) or nuclear (Sp1) internal controls depending on the case.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed as described elsewhere (13). In brief, DNA: protein complexes were cross-linked by incubating 5 × 107 cells with 1% formaldehyde. After washing, cells were lysed and chromatin was sheared to yield DNA fragments (~ 800 bp) using a Branson Digital Sonifier. The lysate was then centrifuged, and soluble chromatin was immunoprecipitated with 5 μg of the indicated antibody. The protein-DNA complexes were collected after incubating with protein-A-magnetic beads, washed and reverse cross-linked by heating at 65°C overnight using 1% SDS and 0.1 M NaHCO3. DNA released from the complexes was purified using the QIAQuick spin kit (Qiagen, Inc) and colleted. The purified DNA samples from the input, immunoprecipitated with either a non-specific IgG (control) or specific IgG (experimental), were subjected to either PCR or qPCR analyses with DAPK1 promoter specific primers: 5’-AGTCCTCAGAAATCTC ATGCAAG-3’ and 5’-CATTAGAGTCCAAGACAGTA-3’. DNA extracted from soluble chromatin was used as an input control for each reaction. Each experiment was repeated at least 3 independent times with multiple samples (n =4) for ensuring the consistency of the results.
Real-time RT PCR analysis in a gene-set enrichment array panel
Total RNAs from blood/bone marrow specimens were isolated using the RNeasy Kit (Qiagen, MD) according to manufacturer's instructions. Total RNA (500 ng) from each specimen was converted to cDNA using the Superscript III First strand cDNA synthesis kit (Invitrogen, Carlsbad, CA). Relative gene expression was quantified using TaqMan Gene Expression Assays (Applied Biosystems, USA) and ABI PRISM 7900 real-time PCR machine. qRT-PCR for 31 AML associated genes and one housekeeping gene was performed at lowdensity array (LDA) format according to the manufacturer's protocol (TaqMan Gene Expression Micro Fluidic card, 4346799, Applied Biosystems). The amount of cDNA was 200 ng per card. 18S rRNA was chosen as an internal control. Relative expression was calculated using RQ manager Ver 1.2 (Applied Biosystems) using a one-patient volunteer sample (CBF+, very low c-jun and negative for Flt3ITD and Meis-1 expression) as a calibrator . Copy number or fold-change in expression was calculated using the 2-ΔΔCt method (24).
Statistical Analyses
Individual patient gene expression data within the defined cohorts was subjected to statistical analysis using Mann- Whitney/Wilcox test and a p value <0.05 was considered significant.
Results
Flt3ITD signaling is associated with loss of DAPK1 protein expression
Bcl-2 and DAPK1 are targets of transcriptionally-active p-jun, and are potentially relevant to the pathogenesis of Flt3ITD AML (10, 11). However, optimal progression of Flt3ITD-driven AML would be served by repression of DAPK1 with simultaneously overexpressed bcl-2, whose transcription is critically dependent upon a CRE/c-jun site (25).
We postulated that the poor prognosis of Flt3ITD AMLs may relate to a lack of DAPK1 expression, dependent on a transcriptional milieu affecting the tandem CRE and κB elements of its promoter, thus distinguishing DAPK1 regulation in Flt3ITD+ve vs. Flt3ITD-ve AML. Expression of DAPK1 in Flt3ITD+ve AML may be selectively blocked by a p52NF-κB/HDAC-associated complex at the κB/CRE site (12). Both NF-κB and c-jun can be generated by activation mechanisms (IKK and JNK1) regulated by the MAP3K, TAK1, respectively (21).
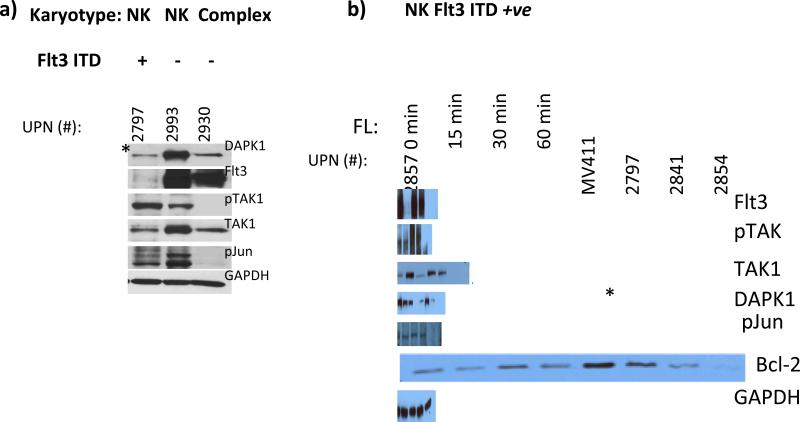
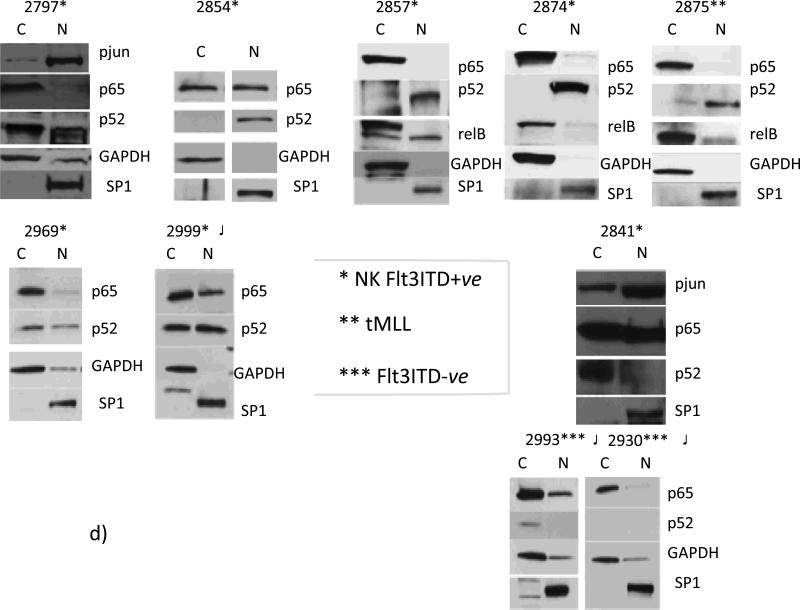
Moreover, analysis of AML blasts separated by their Flt3ITD status, in chronologic succession, showed an apparent relationship with expression of phospho-c-jun and TAK1 activation, via phosphorylation (Fig. 1a & b, and data not shown). Interestingly, in an Flt3ITD+ve sample (#2797) with very high pTAK1/p-jun activity, the lowest expression of DAPK1 was observed, when compared to 2 other AMLs lacking the Flt3ITD mutation (Fig. 1a). This is contrary to the expectation based on the known role for c-jun and its transcriptional partner canonical NF-κB on DAPK1 expression (refs. 11, 13, 15, and see below), and expression levels of p-jun were quite similar in Flt3ITD-ve AML #2993 (normal karyotype) with high DAPK1, compared with NKFlt3ITD+ve 2797 (Fig. 1a).
Fig. 1. Down regulation of DAPK1 expression in MV-4-11 cell line that bears Flt3ITD, and series of primary AML blasts with normal karyotypes and Flt3ITD, as compared to FltITD-ve AMLs.
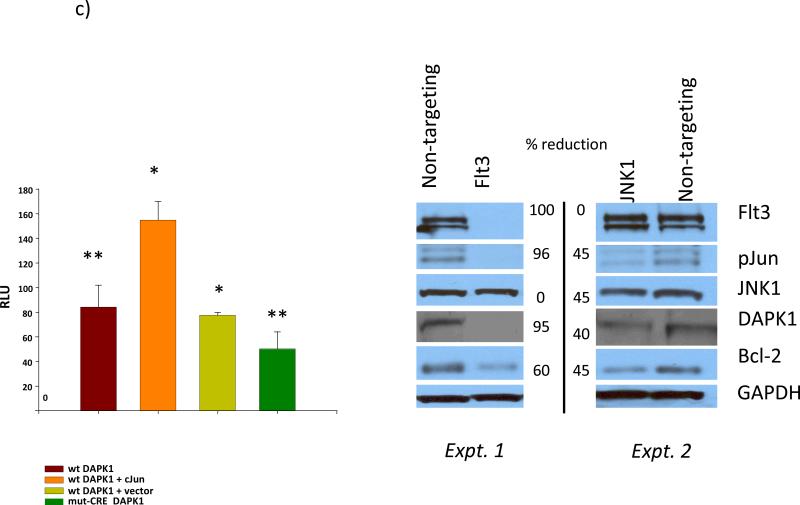
Western blot analyses were performed. A) Flt3ITD+ve AML 2797 and FltITD-ve AMLs-#2930 and 2993 demonstrated correlation of TAK1 phosphorylation levels with phosphorylation status of the secondary (TAK1-JNK1) downstream transcription factor target c-jun. Also, DAPK1 (a c-jun target gene) levels were analyzed in these samples: lower expression levels are found in the Flt3ITD+ve AML 2797 (denoted in two places by asterisk) (1.5-fold and >6-fold reduction) with strong TAK1 activation, when compared with FltITD-ve AMLs-#2930 and 2993 respectively. B) DAPK1 levels were quantified densitometrically in Flt3ITD+ AMLs and MV-4-11 cell line: for #2841, 3-to-4-fold higher compared with 2797 and 2854, respectively, as well as 1.5-fold higher vs. 2857). C) DAPK1 promoter activity is significantly augmented 2-fo l d b y c-jun (* p<0.004) in MV-4-11 cells. DAPK1 promoter vector with a mutated CRE site showed reduced activity (** p<0.03 ). Data represent mean ±SE replicate determinations, from 3 experiments. Results of Western blotting of MV-4-11 cells treated with RNAi for Flt3 or JNK1, compared to non-targeting control siRNA. Knockdown of Flt3 ITD reduces p-jun activation (by 96%) and DAPK1 and bcl-2 expression by 95% and 60%, respectively. RNAi-mediated knockdown of JNK1 (45%), reduces p-jun levels by 45%, and DAPK1 and bcl-2 levels by 40% and 45%, respectively,
To further explore the role of Flt3ITD/TAK1 in promoting cooperation between p-jun and NF-κB species, a series of AML blasts characterized by normal karyotype with Flt3-ITD (NK-Flt3ITD) was studied for c-jun phosphorylation and TAK1 activation. We found that, like the positive control cell line MV-4-11, the normal karyotype Flt3ITD+ve patient blasts express significant levels of Flt3 protein, active/total TAK1 and p-jun (Fig. 1). In one of the AML blasts (#2857), treatment with Flt3-ligand ex vivo led to down-regulation of Flt3, followed by transient upregulation of phospho-TAK1 and DAPK1 levels, which returned to baseline (Fig. 1b). Indeed, DAPK1-Luc reporter analyses in MV-4-11 revealed abundant activity, and exogenous c-jun could significantly augment luciferase - expression by 2-fold (p<0.004). However, a mutant promoter lacking the CRE site had blunted activity. RNAi with Flt3 and JNK1 demonstrated that interference with Flt3/JNK1 axis inhibited DAPK1 and bcl-2 expression (Fig. 1c).
On the other hand, Western blot analyses for DAPK1 expression in relation to p-jun, showed widely varied expression of DAPK1 between blast samples, contrary to expectation if only p-jun were the regulatory determinant (Fig. 1b). All Flt3ITD+ve samples except #2841 had an extremely low (#2857) or barely detectable DAPK1 expression (#2797, 2854), similar to that observed in MV-4-11- where DAPK1 was undetectable (Fig.1). This was despite high and largely invariant levels of p-jun (Fig. 1). Further, contrary to expectation was the occurrence of highest bcl-2 levels in #2797 primary AML blasts with lowest DAPK1, quite similar to the situation in MV-4-11 (Fig.1).
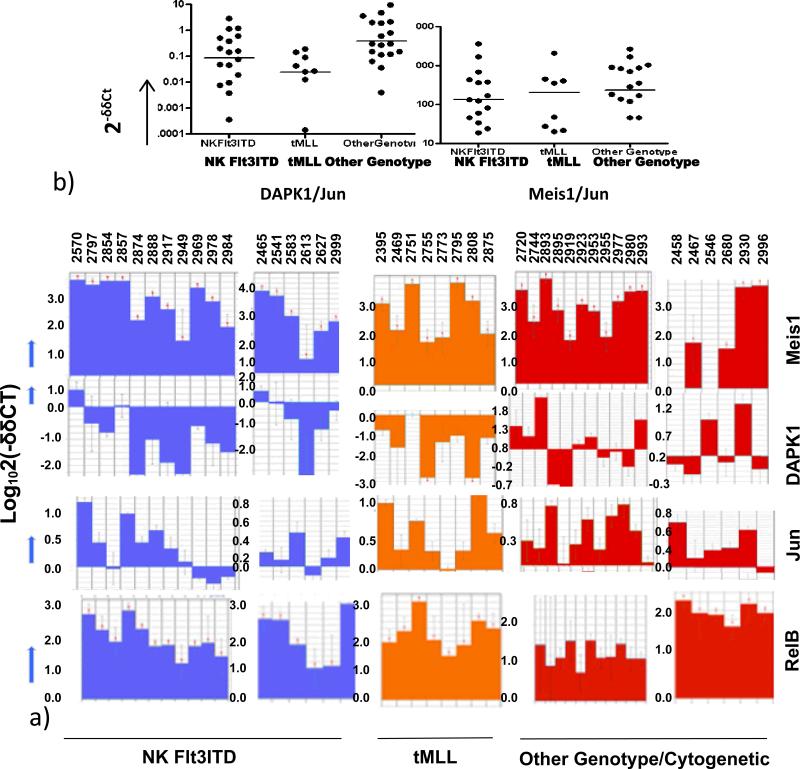
We next pursued the clinical/biological significance of these findings in MV-4-11 and in a larger series of NKFlt3ITD+ve samples. Genome-wide gene expression profiling coupled with real-time RT-PCR was performed using AML blasts from patients with defined cytogenetic and molecular Flt3ITD status from 3 distinct cohorts: 1) normal karyotype Flt3ITD; 2) tMLL-AML; 3) other genotypic/cytogenetic alterations, including those with monosomy 5/7 and a complex karyotype. These analyses included the cohort of blasts from the normal karyotype Flt3ITD AML patients (Fig. 1). We studied the expression of DAPK1, since it is responsive to c-jun/AP-1 and (p65) canonical NF-κB, to examine the basis for gene repression guided by p52NF-κB in the absence of p65NF-κB. In such circumstance, p52 NF-κB heterodimers with relB could recruit transcriptional co-repressors (12,14, 26, 27). In addition to c-jun and relB (26), expression of another c-jun target gene meis1 (11) was analyzed as a positive control in real-time RT-PCR.
Compared with CBF+ve AML or PML-RAR+ve AML (which express low c-jun and which fail to express hoxA9/meis1) (data not shown, and manuscript submitted), the NK Flt3ITD+ve samples had very high meis1 and c-jun expression (Fig. 2). However, there was a significant suppression of DAPK1 transcripts (Fig. 2). In most of the examples noted above, DAPK1 levels were 10 to 100-fold lower than the controls, when normalized to c-jun (Fig. 2a). In fact, a statistically significant difference in DAPK1/c-jun expression was noted when we compared the populations with NKFlt3ITD (or tMLL, as a positive-control for DAPK1 repression (see ref. 19)) to AML's having other cytogenetic/genotypic profiles, where differences in Meis1/c-jun levels were not significantly different (Fig. 2b; DAPK1/c- jun: Mann-Whitney/Wilcoxon one-sided test: NKFlt3ITD or tMLL vs other cytogenetic/genotype groups: p<0.041 or p<0.0006, respectively; Meis1/c-jun=N.S.). RelB, which is in part c-jun-responsive (26), was uniformly highly expressed (Fig. 2a).
Fig. 2. A dichotomy exists in Meis1, c-jun, and relB expression (which are known to be c-jun-dependent) vs. DAPK1, (the dually-responsive c-jun/NF κB-dependent gene) in patient groups with normal karyotype Flt3ITD or MLL translocation.
A). Expression levels of DAPK1, is significantly different in NK Flt3 ITD or MLL translocation AML's when compared with cases without Flt3ITD or nonrandom cytogenetic abnormalities. B) In normal karyotype Flt3ITD and the tMLL AML group the expression ratios of DAPK1 to c-jun were statistically less compared with other cytogenetic/genotype categories, but there was no difference in the ratios of Meis1 to jun.
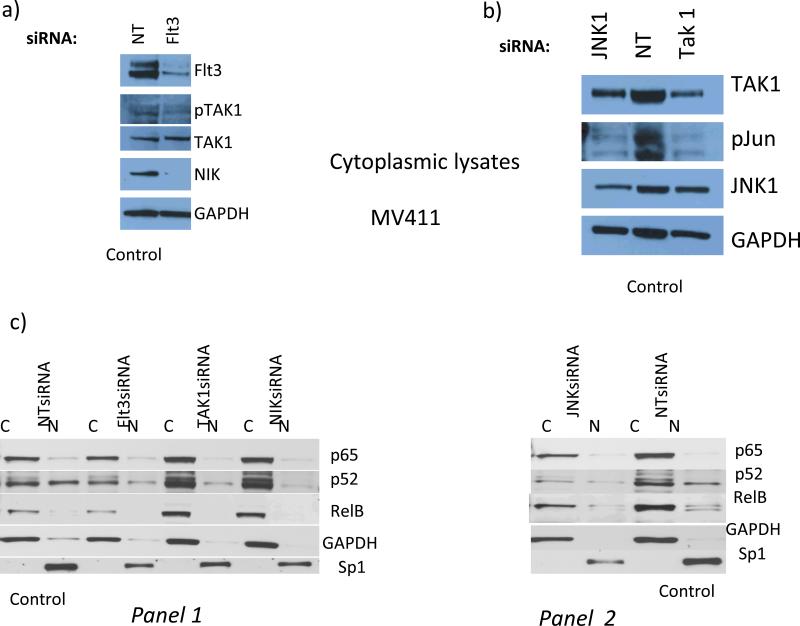
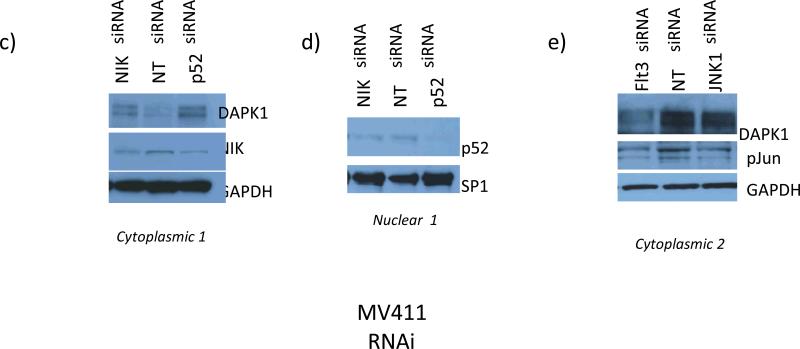
Therefore, Flt3ITD+ve MV-4-11 cell line was used as a model to further explore the relationship between Flt3, TAK1, and JNK1/phospho-c- jun vs. IKK/NF-κB (Fig. 3). We first knocked down Flt3 using RNAi. Flt3 knockdown (78%) resulted in a significant loss of TAK1 phosphorylation (53%) (Fig.3a). (We previously showed that Flt3 knockdown leads to a loss of JNK activity and c-jun phosphorylation (Fig. 1c and ref. 9)). Indeed, RNAi-mediated knockdown of either JNK1 or TAK1 also suppressed the expression of phosphorylated c-jun, by 100% or 80%, respectively (Fig. 3b). We also found that TAK1-inactivation led to essentially total loss/destabilization of NIK (Fig. 3a). NIK is a MAP3K required for non-canonical NF-κB activation but sensitive to TAK1 activation-dependent stabilization (ref. 28). Thus, loss of Flt3 would be expected to affect activation of either canonical NF-κB (solely involving TAK1) or non-canonical NF-κB (TAK1/NIK) (Fig. 3a, and see below).
Fig.3. In Flt3ITD+ve AMLs Flt3-to-TAK1 pathway is involved in establishing an effector apparatus involving p52NFκB for DAPK1 repression, whereas primary Flt3ITD-ve AMLs with high DAPK1 expression lack nuclear p52NFκB.
Flt3 knockdown blocked TAK1 activating phosphorylation (78% and 53% by densitometry, respectively) and led to NIK degradation, which inhibits the activation of non- canonical NFκB (panel a). Panels b and c : (b) JNK1 and TAK1 knockdown lead to diminution of c-jun phosphorylation by 100% and 80%, respectively. (c): control MV-4-11 cells (NT siRNA x2), nuclear fraction is characterized by dominant expression levels of p52NFκB and relB, but not p65NFκB. Flt3, TAK1, or NIK knock down lead to diminution of nuclear (N) p52NFκB levels (by 46%, 73%, or 76%, respectively) (panel 1). (Panel 2), JNK1 knockdown led to diminution of p52NFκB levels (by 76%). Flt3 knockdown also reduced overall p52NFκB levels (by 50%) (panel 1). D) Nuclear translocation of p52NFκB is dominant among NKFlt3ITD+ve AML with DAPK1 repression ((#2797, 49%; #2854, 100%; #2857, 70%; #2874, 100%; #2969, 35%; #2999, 50%). Note that ITD+ve sample #2999 was exposed on the same blot with ITD-ve #2993 and 2930, with no nuclear p52NFκB).
To study the activation status of NF-κB species, cellular lysates were fractionated into cytoplasmic and nuclear components because nuclear retention of these proteins determines their transcriptional activity. We and others have demonstrated that phosphorylated c-jun (at S63, S73) is always predominantly present in the nucleus as a heterodimer with other proteins (refs. 9, 29). Surprisingly, we found that in MV-4-11 cells, p65 NF-κB was largely absent in the nuclear fraction, though an abundant quantity of Sp1 (a constitutively nuclear transcription factor), but not GAPDH (a cytoplasmic marker), was found (Fig. 3c). Further, the RNAi-mediated knockdown of Flt3, TAK1, NIK, and JNK1, failed to increase nuclear level of p65-NF-κB (Fig. 3c).
In contrast, control MV-4-11 (Fig. 3c) had almost 50% fraction of p52NF-κB in the nucleus, and a lesser amount of relB, (a primary heterodimeric partner of p52NFκB) (Fig. 3c). Knockdown of either TAK1 or NIK strongly decreased nuclear p52NF-κB levels by 73% or 76%, respectively, with a corresponding rise in cytoplasmic levels (Fig. 3c panel 1, and see below). Knockdown of JNK1 or Flt3 appeared to diminish overall cytoplasmic and nuclear content of both relB and p52NF-κB by 76% and 50%, respectively (Fig. 3c panels 1,2). This is consistent with stability of this heterodimeric complex relying on partner relB, whose expression is induced by c-jun (Fig. 3c, ref. 26). Taken together, our data support the existence of an Flt3/p52NFκB pathway, which may negatively regulate DAPK1 expression (Figs. 1-3).
As 3/4 of primary Flt3ITD+ve AML cases presented in Fig. 1b were refractory to primary treatment (all except #2841) and had low DAPK1 expression, we hypothesized that this pathway might have a biologic/prognostic significance. Indeed, sample #2841 with high DAPK1 expression differed greatly from 7 other Flt3ITD+ve samples and a control tMLL AML because it had trivial amounts of nuclear p52NF-κB and abundant and predominantly nuclear p65NF-κB (Fig. 3d, and see below). These NKFlt3ITD+ve AMLs with little or no DAPK1 expression, as in the control tMLL AML, were distinguished by predominantly nuclear p52 NF-κB (mean % nuclear translocation p52NF-κB, 72.8+/− 9.9%; vs mean % nuclear translocation p65 NF-κB, 19.6 +/− 6.6%). In fact, qPCR analysis showed very low DAPK1 transcripts among samples with no nuclear p65, e.g. 2797, 2874, 2857 (compare Fig. 3d to Fig. 2a above). On the other hand, the higher and dominant content of nuclear p65 NF-κB in #2841 was more typical of the above-noted Flt3ITD-ve patients-where DAPK1 levels were high, and had little or no nuclear p52NF-κB (#2930, 2993 in Figs. 1, 2 and Fig. 3d). Also, dominant nuclear presence for p52NF-κB vs. p65 NF-κB in the different cohorts was confirmed by gel mobility shift assays in addition to immunoblotting (Supplemental Fig. 1). We have ascertained that the isolation procedure/washing had not created artifactual deficiency of nuclear p65NF- κB (upon any potential autocrine cytokine washout) by performing timed re-additions to cultured AML blasts of Flt3 ligand (ref. 30) (data not shown).
p52NF-κB and HDAC repress DAPK1 transcription in MV-4-11 and primary AML blasts with Flt3ITD or MLL translocation
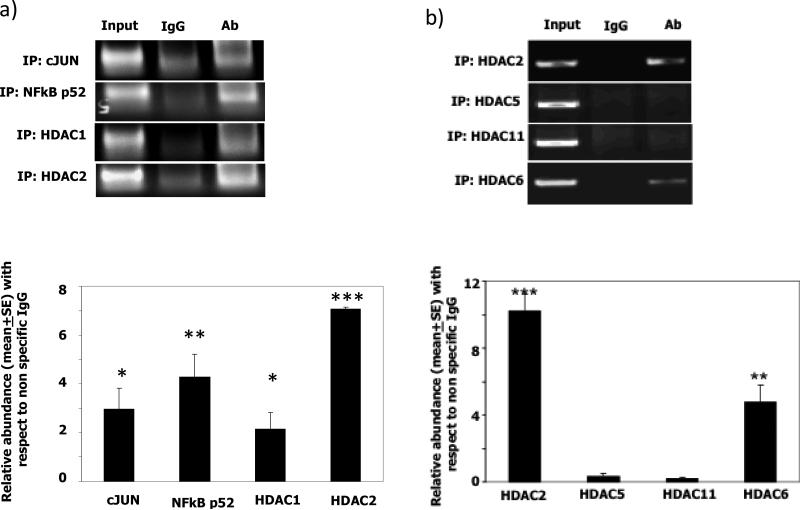
We next investigated a possible Flt3ITD-driven mechanism that linked p52NF-κB to DAPK1 repression in MV-4-11. Since histone deacetylases (HDAC) are known to cause gene repression, we next determined whether p52NF-κB, c-jun, and HDAC bound to the DAPK1 promoter. Chromatin immunoprecipitation assays revealed a strong binding of both p52NF-κB and HDAC2 to the DAPK1 promoter (Fig. 4). In contrast, a relatively weaker binding of c-jun and HDAC1 to DAPK1 promoter was observed, when compared with HDAC2 (Fig. 4a). In another series of ChIP experiments, we found that, among HDACs, strong binding of the HDAC2 (class I) to the promoter accompanied a corresponding absence of HDAC-5 (class II) and class IV HDAC11, which have been found to be predominantly localized to cytoplasm in certain hematopoietic cells and are expressed low in MV-4-11 (Fig. 4b). On the other hand, a moderate binding of the class II HDAC6 occurred. The latter has a nuclear localization signal and has been found to shuttle between nucleus (e.g. in association with c-jun/CREB/RUNX2/steroid receptors on chromatin) and cytoplasm for functions. In the cytoplasm, a dominant role for HDAC6 in regulating hsp90 chaperone function has been noted in AML cell lines as well (Fig. 4b, and refs. 31-36).
Fig. 4. ChIP analyses demonstrate that the tandem CRE and NF-κB sites of the proximal DAPK1 promoter are occupied by c-jun and to greater extent, by p52NFκBκB and HDAC2.
A& B) Typical PCR patterns obtained in ChIP assays with DAPK1-specific primers in MV-4-11 cells were shown. For input control reactions, one-fifth of the soluble chromatin used for the ChIP analysis was employed. In each case thirty cycles of PCR were performed. Non specific IgG, HDAC1, HDAC2, p52 NF-κB and c-jun, as well as HDAC5, HDAC11, and HDAC6 IgGs were used at 5 μg each/reaction. Real-time PCR analysis of DAPK1 promoter fragments recovered in ChIP assays performed with the indicated antibodies. Each bar represents the mean abundance of DAPK1 promoter fragments for specific antibody when compared with non-specific IgG. SE of 6 separate reactions from 2 independent experiments were shown. ‘***’ p-value, <0.001, ‘**’ p-value <0.01 and ‘*’ p-value <0.05. RNAi-mediated knockdown of p52NFκB or NIK (panels c&d) upregulated DAPK1 expression. Flt3 or JNK1 knockdown reduce phospho-c-jun and DAPK1 levels by 60%, 40%, respectively (panels c, d, e).
To provide additional evidence for a role of p52 NF-κB in repressing DAPK1, knockdown experiments in MV-4- 11 cells were undertaken (Fig. 4c-e). RNAi-mediated knockdown of p52NF-κB, or its upstream activator NIK, caused an increase in DAPK1 protein expression by 2-fold (Fig. 4c &d). As expected, knockdown of either JNK1 or Flt3 in MV-4-11 downregulated DAPK1 levels by 40% or 60%, respectively (Fig.4e) similar to the data shown in Fig 1c. Therefore, DAPK1 was derepressed following the knockdown of p52NF-κB when JNK1 arm of Flt3 signaling is sustained (Fig. 4). In addition, the identification of severe DAPK1 repression (Figs. 2, 3d) is consistent with the relationship between p52NF-κB and DAPK1 in MV-4-11.
Discussion
Genome-wide sequencing studies to identify gene mutations among the majority of solid tumors can be compared to similar screens performed with AML. Such comparison suggests that the mutational complexity of adult AML is modest and may involve 10-to-300-fold lower numbers of mutated genes (37-42). In the case of normal karyotype AMLs with Flt3ITD mutation, only 10-14 genes have been found mutated and the important founder mutations appear to involve not only the Flt3 and NPM1 genes, but also epigenetically-active enzymes (39-42). Perhaps because of the involvement of intrinsic epigenetic mechanisms in many examples of the heterogenous disease process, clinical use of single-agent Flt3-selective tyrosine kinase inhibitors has not demonstrated truly significant disease-free survival in most Flt3ITD+ve AML patients.
As the epigenetic signature of AMLs is robust, particularly in the setting of Flt3ITD mutation (41-43), we postulated that the unique signaling pathway used by Flt3ITD may contribute to those “stress-induced” steps involved in epigenetic reprogramming in Flt3ITD+ve AMLs (12, 17, 18). We further postulated TAK1, which is overexpressed in a poor-risk AML LSC signature (22), may contribute to p52NF-κB -induced epigenetic silencing, as similar to the ability for Evi-1 to attract a cis-localized epigenetic apparatus to distinct genes in an AML subgroup (44). Indeed, our data point to the existence of an instructive mechanism (as opposed to a “stochastic” origin) for transcriptional repression of certain tumor suppressor genes, as demonstrated here for DAPK1. DAPK1 is regulated by a binary cis-element consisting of CRE/jun and -κB sites (12, 13). These sites are subject to dominant-negative regulation by p52NF-κB and HDAC2/HDAC6 under the Flt3-/TAK1 pathway (Figs 3, 4, 5). Inhibition of p52NF-κB caused a derepression of DAPK1.
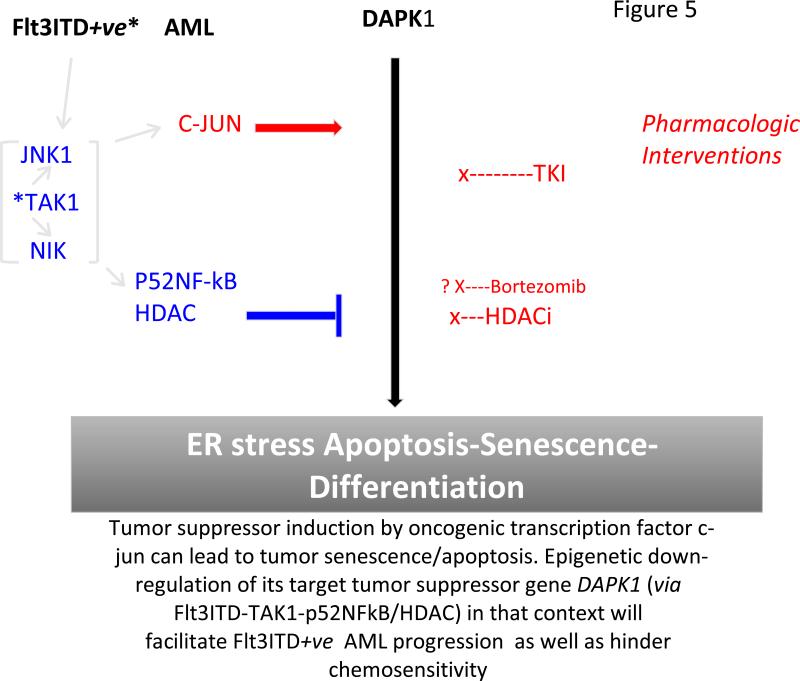
Fig.5. A model for the repression of DAPK1 by Flt3ITD-induced signals.
Whereas c-jun elicited by Flt3ITD/JNK1 drives DAPK1 transcription. This activity is blocked by p52NFκB which recruits HDAC2/HDAC6. Derepression of DAPK1 can be achieved by use of TKI/Flt3 inhibitor, especially in combination with HDAC inhibitor (eg. SAHA/Vorinostat) and/or Bortezomib as a proteasomal inhibitor of p52NF-κB
Very little is known about the role of p52NF-κB as a regulator of gene expression in AML (45). In one study, epigenetic repression of another tumor suppressor gene, DIF2 was indirectly linked to the activity of p52NF-κB. There has also been inadequate explanation for the severe reduction of DAPK1 expression in poor-risk AML. In fact, previous studies have identified much lower levels of DAPK1 promoter hypermethylation than would have been expected of the observed extent of transcriptional repression in AML (16, 41-43, 46, 47). This is in contrast to MLL-r infant ALL, where the DAPK1 repression was associated with its promoter hypermethylation (19). In this report we demonstrate that for the subset of AML's characterized by Flt3ITD, often with additional NPM1 and/or IDH1/2 or DNMT3 mutation, upstream signals may provide the stimulus for p52NF-κB, and its partner rel B, to attract HDACs and possibly other repressors to specific gene promoters to enforce a repression (12, 47).
The Flt3-/TAK1- dependent activation of IKK's/NF-κB and JNK1 that we identified in this study is pertinent to activation of these latter two crucial downstream tumor effectors. TAK1 may also bear upon other anti-apoptotic participants, including AMP kinase, which is activated by TAK1 for additional anti- apoptotic function in the TRAIL pathway (48, 49). Although TAK1 has more frequently been associated with the canonical NF-κB pathway rather than the NIK- dependent non-canonical pathway we identified, there is ample precedent that TAK1 can activate NIK, and that TAK1/TRAF6-ubiquitin conjugates activate IKKα as well as IKKβ (28, 50).
The data we report has also implications for the design of a mechanistically-driven therapeutic regimen-to inhibit Flt3ITD+ve AML. DAPK1 suppression in context of Flt3ITD activation of p52NF-κB implies that one criterion for selection of tyrosine kinase inhibitors, classified as Flt3 inhibitors, is in their ability to inhibit p52NF-κB activation in this subgroup of AMLs. In turn, HDAC inhibitors contribute independently toward DAPK1 derepression for its participation in ER stress apoptosis. Bortezomib is also known to inhibit steps required for proteasomal activation of p52NF-κB (51) This suggests a synergistic potential for combining these drug classes in “targeted” therapy of Flt3ITD+ve AML(Fig. 5). Indeed, we have both in vitro and in vivo evidence that such an approach may enforce apoptosis via enhanced ER stress following DAPK1 derepression in association with p52NF-κB depletion by the combination of a Flt3 inhibitor and an HDAC inhibitor in Flt3ITD+ve AML (H Sayar et al. in preparation).
Finally, the enzymatic activity of the co-oncogene PIN1(peptidyl prolylisomerase 1), required for tyrosine kinase-mediated JNK/c-jun signaling in breast cancer and AML, is abrogated by DAPK1-mediated phosphorylation on serine 71 of PIN1(52-54). Thus, the pathophysiologic significance of DAPK1 repression in these malignancies is further emphasized, along with the therapeutic strategy for combining TKI and HDAC inhibitor.
Supplementary Material
Translational Relevance.
Acute myeloid leukemia (AML) is a collection of complex and heterogeneous diseases, which can be grouped according to cytogenetic/genotypic features of the blast cells, and activated signaling pathways. These signaling pathways cooperate to promote blast cell survival and to prevent tumor suppressor induced senescence. We report here a tyrosine kinase (Flt3ITD)-initiated non-canonical NF-κB signaling pathway in a subset of AMLs, where p52NF-κB in association with certain histone deacetylases (HDACs) repress the tumor suppressor gene DAPK1. DAPK1 is an essential player in endoplasmic reticulum (ER)-stress induced apoptosis, and is implicated in poor outcome of AML by its repression. The mechanism for repression of DAPK1 by p52NF-κB and HDACs, influenced by Flt3ITD, was found to involve the MAP3K, TAK1. Because TAK1 is among the most highly expressed genes in a leukemic stem cell signature for poor-risk AML, these studies focus attention on the interface between signal transduction and epigenetic remodeling in AML.
Acknowledgements
This work was supported in part by Department of Veterans Affairs Merit Review Award (HSB), and stipend in memory of Dr. Gary D. Tollefson. DVK is supported by NIH grant CA78282.
Footnotes
Author Contributions
RS planned and performed experiments, analyzed results and wrote the manuscript; PG planned and performed experiments, analyzed results and wrote the manuscript; AWW planned and performed experiments, and analyzed results, SG planned and performed experiments; HS screened patients and edited the manuscript; AS analyzed results and edited the manuscript; CG analyzed results and reviewed the manuscript; LL analyzed results and reviewed the manuscript; AAC analyzed results and reviewed the manuscript; TA analyzed results and reviewed the manuscript; KJS screened patients and reviewed the manuscript; LDC screened patients and reviewed the manuscript; DVK planned experiments, analyzed results and wrote the manuscript; HSB designed the study, planned and performed experiments, analyzed results and wrote the manuscript.
References
- 1.Schardt JA, Weber D, Eyholzer M, Mueller BU, Pabst T. Activation of the unfolded protein response is associated with favorable prognosis in acute myeloid leukemia. Clin Cancer Res. 2009;15:3834–41. doi: 10.1158/1078-0432.CCR-08-2870. [DOI] [PubMed] [Google Scholar]
- 2.Choudhary C, Olsen JV, Brandts C, et al. Mislocalized activation of oncogenic RTKs switches downstream signaling outcomes. Mol Cell. 2009;36:326–39. doi: 10.1016/j.molcel.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 3.Rahmani M, Davis EM, Crabtree TR, et al. The kinase inhibitor sorafenib induces cell death through a process involving induction of endoplasmic reticulumstress. Mol Cell Biol. 2007;27:5499–513. doi: 10.1128/MCB.01080-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szegezdi E, Macdonald DC, Ní Chonghaile T, Gupta S, Samali A. Bcl-2 family on guard at the ER. Am J Physiol Cell Physiol. 2009;296:C941–53. doi: 10.1152/ajpcell.00612.2008. [DOI] [PubMed] [Google Scholar]
- 5.Chang NC, Nguyen M, Germain M, Shore GC. Antagonism of Beclin 1-dependent autophagy by BCL-2 at the endoplasmic reticulum requires NAF-1. EMBO J. 2010;29:606–18. doi: 10.1038/emboj.2009.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2008;7:1013–30. doi: 10.1038/nrd2755. Review. [DOI] [PubMed] [Google Scholar]
- 7.Bialik S, Kimchi A. The Death-Associated Protein Kinases: Structure, function, and Beyond. Ann Rev Biochem. 2006;75:189–210. doi: 10.1146/annurev.biochem.75.103004.142615. [DOI] [PubMed] [Google Scholar]
- 8.Martioriati A, Doumont G, Alcalay M, et al. dapk1, encoding an activator of p19arfp53-mediated apoptotic checkpoint, is a transcriptional target of p53. Oncogene. 2005;24:1461–66. doi: 10.1038/sj.onc.1208256. [DOI] [PubMed] [Google Scholar]
- 9.Hartman AD, Wilson-Weekes A, Suvannasankha A, et al. Constitutive c-jun N-terminal kinase activity in acute myeloid leukemia derives from Flt3 and affects survival and proliferation. Exp Hematol. 2006;34:1360–76. doi: 10.1016/j.exphem.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 10.Hess P, Pihan G, Sawyers CL, Flavell RA, Davis RJ. Survival signaling mediated by c-Jun NH(2)-terminal kinase in transformed B lymphoblasts. Nat Genet. 2002;32:201–5. doi: 10.1038/ng946. [DOI] [PubMed] [Google Scholar]
- 11.Hayakawa J, Mittal S, Wang Y, et al. Identification of promoters bound by c-Jun/ATF2 during rapid large-scale gene activation following genotoxic stress. Mol Cell. 2004;16:521–35. doi: 10.1016/j.molcel.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 12.Puto LA, Reed JC. Daxx represses RelB target promoters via DNA methyltransferase recruitment and DNA hypermethylation. Genes Dev. 2008;22:998–1010. doi: 10.1101/gad.1632208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gade P, Roy SK, Li H, Nallar SC, Kalvakolanu DV. Critical role for transcription factor C/EBP-beta in regulating the expression of death-associated protein kinase 1. Mol Cell Biol. 2008;28:2528–48. doi: 10.1128/MCB.00784-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang W, Ghisletti S, Perissi V, Rosenfeld MG, Glass CK. Transcriptional integration of TLR2 and TLR4 signaling at the NCoR derepression checkpoint. Mol Cell. 2009;35:48–57. doi: 10.1016/j.molcel.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guzman ML, Upchurch D, Grimes B, et al. Expression of tumor-suppressor genes interferon regulatory factor 1 and death-associated protein kinase in primitive acute myelogenous leukemia cells. Blood. 2001;97:2177–9. doi: 10.1182/blood.v97.7.2177. [DOI] [PubMed] [Google Scholar]
- 16.Fandy TE, Herman JG, Kerns P, et al. Early epigenetic changes and DNA damage do not predict clinical response in an overlapping schedule of 5-azacytidine and entinostat in patients with myeloid malignancies. Blood. 2009;114:2764–73. doi: 10.1182/blood-2009-02-203547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnstone SE, Baylin SB. Stress and the epigenetic landscape: a link to the pathobiology of human diseases? Nat Rev Genet. 2010;11:806–12. doi: 10.1038/nrg2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gazin C, Wajapeyee N, Gobeil S, Virbasius CM, Green MR. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449:1073–7. doi: 10.1038/nature06251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schafer E, Irizarry R, Negi S, et al. Promoter hypermethylation in MLL-r infant acute lymphoblastic leukemia: biology and therapeutic targeting. Blood. 2010;115:4798–4809. doi: 10.1182/blood-2009-09-243634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlenk RF, Döhner K, Krauter J, et al. German- Austrian Acute Myeloid Leukemia Study Group. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358:1909–18. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 21.Sanna MG, da Silva Correia J, Ducrey O, et al. IAP suppression of apoptosis involves distinct mechanisms: the TAK1/JNK1 signaling cascade and caspase inhibition. Mol Cell Biol. 2002;22:1754–66. doi: 10.1128/MCB.22.6.1754-1766.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eppert K, Takenaka K, Lechman ER, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011;17:1086–93. doi: 10.1038/nm.2415. [DOI] [PubMed] [Google Scholar]
- 23.Valk PJ, Verhaak RG, Beijen MA, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350:1617–28. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Wilson BE, Mochon E, Boxer LM. Induction of bcl-2 expression by phosphorylated CREB proteins during B cell activation and rescue from apoptosis. Mol Cell Biol. 1996;16:5546–56. doi: 10.1128/mcb.16.10.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mineva ND, Rothstein TL, Meyers JA, Lerner A, Sonenshein GE. CD40 ligand-mediated activation of the de novo RelB NF-kappaB synthesis pathway in transformed B cells promotes rescue from apoptosis. J Biol Chem. 2007;282:17475–85. doi: 10.1074/jbc.M607313200. [DOI] [PubMed] [Google Scholar]
- 27.Compagno M, Lim WK, Grunn A, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459(7247):717–21. 28. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninomiya-Tsuji J, Kishimoto K, Hiyama A, Inoue J, Cao Z, Matsumoto K. The kinase TAK1 can activate the NIK-I kappaB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature. 1999;398(6724):252–6. doi: 10.1038/18465. [DOI] [PubMed] [Google Scholar]
- 29.Liu H, Deng X, Shyu YJ, Li JJ, Taparowsky EJ, Hu CD. Mutual regulation of c-Jun and ATF2 by transcriptional activation and subcellular localization. EMBO J. 2006;25:1058–69. doi: 10.1038/sj.emboj.7601020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ludwig D, Hicklin D, Witte L, Li Y, Small D. FLT3 ligand causes autocrine signaling in acute myeloid leukemia cells. Blood. 2004;103(1):267–74. doi: 10.1182/blood-2003-06-1969. [DOI] [PubMed] [Google Scholar]
- 31.Keedy KS, Archin NM, Gates AT, Espeseth A, Hazuda DJ, Margolis DM. A limited group of class I histone deacetylases acts to repress human immunodeficiency virus type 1 expression. J Virol. 2009;83:4749–56. doi: 10.1128/JVI.02585-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bradbury CA, Khanim FL, Hayden R, et al. Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia. 2005;19:1751–9. doi: 10.1038/sj.leu.2403910. [DOI] [PubMed] [Google Scholar]
- 33.Xu X, Xie C, Edwards H, Zhou H, Buck SA, Ge Y. Inhibition of histone deacetylases 1 and 6 enhances cytarabine-induced apoptosis in pediatric acute myeloid leukemia cells. PLoS One. 2011;6:e17138. doi: 10.1371/journal.pone.0017138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Govindan MV. Recruitment of cAMP-response element-binding protein and histone deacetylase has opposite effects on glucocorticoid receptor gene transcription. J Biol Chem. 2010;285:4489–510. doi: 10.1074/jbc.M109.072728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Westendorf JJ, Zaidi SK, Cascino JE, et al. Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21(CIP1/WAF1) promoter. Mol Cell Biol. 2002;22:7982–92. doi: 10.1128/MCB.22.22.7982-7992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bali P, Pranpat M, Bradner J, et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005;280:26729–34. doi: 10.1074/jbc.C500186200. [DOI] [PubMed] [Google Scholar]
- 37.Pleasance ED, Cheetham RK, Stephens PJ, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463(7278):191–6. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pleasance ED, Stephens PJ, O'Meara S, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463(7278):184–90. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ley TJ, Mardis ER, Ding L, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456(7218):66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mardis ER, Ding L, Dooling DJ, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–66. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ley TJ, Ding L, McLellan MD, et al. DNMT3A mutations in acute myeloid leukemia. N Eng J Med. 2010;363:2424–33. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Figureroa ME, Abel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:533–67. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Figueroa ME, Lugthart S, Li Y, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17:13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lugthart S, Figueroa ME, Bindels E, et al. Aberrant DNA hypermethylation signature in acute myeloid leukemia directed by EVI1. Blood. 2011;117:234–41. doi: 10.1182/blood-2010-04-281337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laurenzana A, Petruccelli LA, Pettersson F, et al. Inhibition of DNA methyltransferase activates tumor necrosis factor alpha- induced monocytic differentiation in acute myeloid leukemia cells. Cancer Res. 2009;69:55–64. doi: 10.1158/0008-5472.CAN-08-0245. [DOI] [PubMed] [Google Scholar]
- 46.Claus R, Hackanson B, Poetsch AR, et al. Quantitative analyses of DAPK1 methylation in AML and MDS. Int J Cancer. 2011 doi: 10.1002/ijc.26429. published online Sep 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cedar H, Bergmann Y. Linking DNA methylation and histone modification : patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 48.Morioka S, Omori E, Kajino T, Kajino-Sakamoto R, Matsumoto K, Ninomiya-Tsuji J. TAK1 kinase determines TRAIL sensitivity by modulating reactive oxygen species and cIAP. Oncogene. 2009;28:2257–65. doi: 10.1038/onc.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Herrero-Martín G, Høyer-Hansen M, García-García C, et al. TAK1 activates AMPK-dependent cytoprotective autophagy in TRAIL-treated epithelial cells. EMBO J. 2009;28:677–85. doi: 10.1038/emboj.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xia ZP, Sun L, Chen X, et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461(7260):114–9. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-kappaB regulatory pathways. Annu Rev Biochem. 2009;78:769–96. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- 52.Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T, Petkova V, Lu KP. Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. EMBO J. 2001;20:3459–72. doi: 10.1093/emboj/20.13.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pulikkan JA, Dengler V, Peer Zada AA, et al. Elevated PIN1 expression by C/EBPalpha-p30 blocks C/EBPalpha-induced granulocytic differentiation through c-Jun in AML. Leukemia. 2010;24:914–23. doi: 10.1038/leu.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee TH, Chen CH, Suizu F, et al. Death-associated protein kinase 1 phosphorylates Pin1 and inhibits its prolyl isomerase activity and cellular function. Mol Cell. 2011;42:147–59. doi: 10.1016/j.molcel.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.