SUMMARY
Toxin-antitoxin (TA) systems are ubiquitous on bacterial chromosomes, yet the mechanisms regulating their activity, and the molecular targets of toxins, remain incompletely understood. Here, we identify SocAB, a new TA system in Caulobacter crescentus. Unlike canonical TA systems, the toxin SocB is unstable and constitutively degraded by the protease ClpXP; this degradation requires the antitoxin, SocA, as a proteolytic adaptor. We find that the toxin, SocB, blocks replication elongation through an interaction with the sliding clamp, driving replication fork collapse. Mutations that suppress SocB toxicity map to either the hydrophobic cleft on the clamp that binds DNA polymerase III or a clamp-binding motif in SocB. Our findings suggest that SocB disrupts replication by outcompeting other clamp-binding proteins. Collectively, our results expand the diversity of mechanisms employed by TA systems to regulate toxin activity and inhibit bacterial growth, and they suggest that inhibiting clamp function may be a generalizable antibacterial strategy.
INTRODUCTION
Toxin-antitoxin (TA) systems are genetic modules that are widely present on plasmids and bacterial chromosomes, with some species encoding more than 50 TA pairs (Pandey and Gerdes, 2005). Each TA system is typically comprised of a toxin and its cognate antitoxin that are encoded together in an operon; normally, both the toxin and antitoxin are synthesized and form a stable, non-toxic complex. However, under stressful conditions, the more labile antitoxin can be degraded, freeing the stable toxin to inhibit bacterial growth (Wang et al., 2011). TA systems are implicated in a number of important cellular processes, including plasmid stability, bacterial persistence, stress responses and resistance to phage (Gerdes and Maisonneuve, 2012; Yamaguchi and Inouye, 2011).
The toxins from characterized TA systems inhibit growth by targeting a relatively limited set of critical cellular functions such as translation, DNA replication, or cell wall growth. For instance, the toxin Doc inhibits translation by phosphorylating EF-Tu (Castro-Roa et al., 2013), whereas other toxins, such as MazF, block translation by cleaving mRNAs (Zhang et al., 2003). The toxins CcdB and ParE inhibit replication by binding DNA gyrase, which is required to relieve supercoiling that occurs ahead of the replication fork (Yuan et al., 2010). Additionally, the PezT toxin blocks cell wall growth by phosphorylating peptidoglycan precursors, thereby inhibiting the initial step in peptidoglycan synthesis (Mutschler et al., 2011). Strikingly, small-molecule antibiotics inhibit cellular proliferation by targeting a very similar set of processes, including translation (aminoglycosides), DNA replication (fluoroquinolones), and cell wall growth (β-lactams) (Walsh, 2003). The recent rise in antibiotic resistance (Bush et al., 2011), and the lack of diversity of current antibiotics (Coates et al., 2011), highlight the need to identify new targets within bacteria to inhibit their proliferation. The study of bacterial toxins may help elucidate such targets for the development of novel small-molecule therapeutics.
The DNA replication machinery, or replisome, could be a prime target, but is surprisingly not targeted by any known TA systems or antibiotics in clinical use (Robinson et al., 2012). Bacterial DNA replication is catalyzed by a multi-component complex known as DNA polymerase III (Pol III). The Pol III core (subunit composition αεθ) is weakly processive on its own and can only incorporate 1–10 nucleotides per binding event (Johnson and O'Donnell, 2005). To increase its processivity, Pol III core associates with the β sliding clamp, DnaN, a ring-shaped protein that encircles DNA topologically. Binding to DnaN increases the processivity of Pol III over three orders of magnitude (Johnson and O'Donnell, 2005; Maki and Kornberg, 1988). In E. coli, DnaN also associates with DNA Pols I, II, IV, and V (Indiani et al., 2005); the mismatch repair proteins MutS and MutL (Lopez de Saro et al., 2006); and the replication regulator Hda (Kurz et al., 2004). The interaction of these proteins with DnaN is required for a number of processes, including translesion synthesis (Lenne-Samuel et al., 2002) and mismatch repair (Lenhart et al., 2013). These clamp-binding proteins contain variants of a short peptide motif, QL[SD]LF, that mediates binding to a conserved hydrophobic cleft on DnaN (Dalrymple et al., 2001). DnaN thus forms a central hub for DNA replication and repair in bacteria.
Here, we identify an atypical TA system in Caulobacter crescentus, SocAB. We find that the toxin SocB is normally unstable and constitutively degraded by the protease ClpXP. In contrast to canonical TA systems, in which the antitoxin neutralizes its toxin by sequestration, we find that the antitoxin SocA neutralizes SocB by acting as an adaptor for the degradation of SocB by ClpXP. The requirement of ClpXP for SocB degradation explains for the first time why ClpXP is essential for viability in Caulobacter. Additionally, we provide evidence that SocB inhibits replication elongation through a direct interaction with DnaN. Mutations in DnaN or SocB that block their association occur in the hydrophobic cleft in DnaN or in a DnaN-binding motif in SocB, suggesting that SocB binds to DnaN in a similar manner as known clamp-binding proteins and competes for binding to DnaN during replication. In sum, our work elucidates novel mechanisms employed by TA systems to regulate toxin function and to inhibit cellular proliferation. Our results further suggest that protein interaction hubs such as DnaN may be ideal targets for the development of new protein- or small-molecule-based antimicrobials.
RESULTS
Mutations in the Toxin socB Can Bypass the Essentiality of clpXP
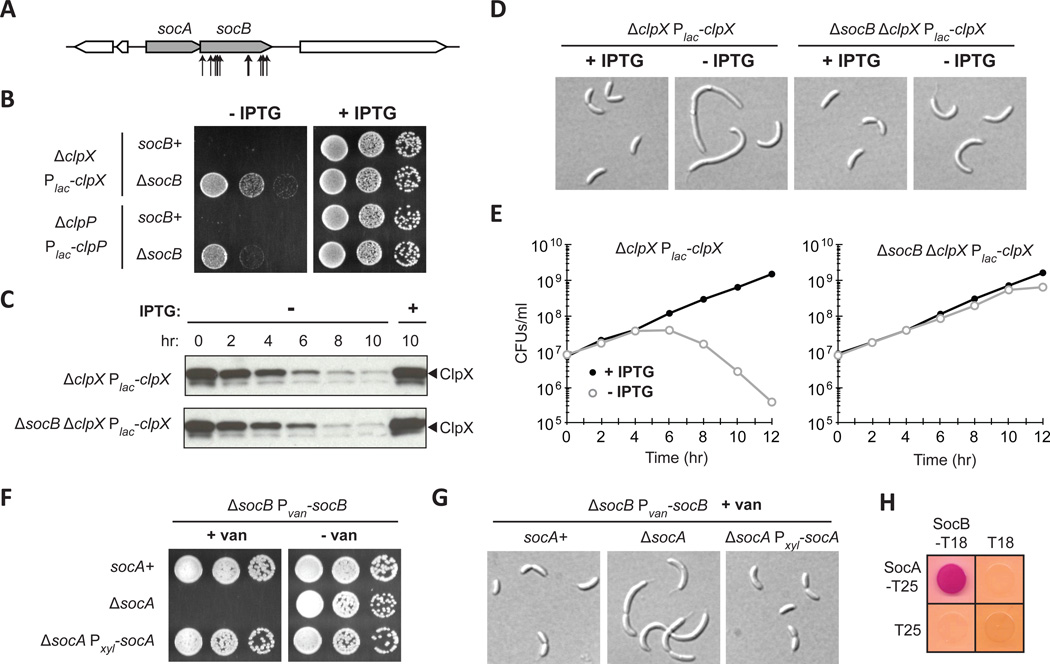
ClpXP is a widely conserved AAA+ protease that uses the power of ATP hydrolysis to unfold and proteolyze substrates within cells (Sauer and Baker, 2011). ClpXP is comprised of two proteins: the unfoldase ClpX, which recognizes and unfolds substrates, and the peptidase ClpP, which degrades the unfolded substrates that it receives from ClpX. Unlike most bacteria, clpX and clpP are essential for viability in Caulobacter (Jenal and Fuchs, 1998). To identify factors responsible for clpP essentiality, we selected for transposon insertions that allow cells to grow in the absence of clpP expression. We identified multiple transposon insertions in a hypothetical gene (CCNA_03629) that we named socB for suppressor of clpXP (Fig. 1A). A clean deletion of socB allowed cells to grow in the absence of either clpX or clpP expression (Fig. 1B); however, growth was slower on medium that repressed clpX or clpP, consistent with the fact that ClpXP degrades a range of cellular proteins (Bhat et al., 2013).
Fig. 1. Mutations in the Toxin socB Bypass ClpXP Essentiality.
(A) Schematic of transposon insertions in socB (CCNA_03629) that suppressed the essentiality of clpP.
(B) Growth of clpX and clpP depletion strains in socB+ and ΔsocB backgrounds. Five-fold serial dilutions of the indicated strains were spotted onto media ± IPTG.
(C) Kinetics of ClpX depletion. Indicated strains were shifted to media ± IPTG, and samples were subjected to immunoblotting.
(D) Morphology of cells following ClpX depletion in socB+ and ΔsocB backgrounds. Strains from (C) were imaged by DIC microscopy at 10 hr.
(E) Viability of cells following ClpX depletion in socB+ and ΔsocB backgrounds. Colony forming units (CFUs)/ml of the strains from (C) are shown; mean of two biological replicates.
(F) Growth of strains expressing socB in the socA+ or ΔsocA backgrounds. The indicated strains were five-fold serially diluted onto media that induces or represses socB.
(G) Morphology of strains from (F). The indicated strains were grown for 4 hr in socB inducing conditions and then imaged by DIC microscopy.
(H) Bacterial two-hybrid analysis of the interaction between SocA and SocB. T18/T25 were included as a negative control; red indicates a positive interaction. Cells were grown for 1 day at 30°C.
See also Fig. S1.
To better assess the ability of socB mutants to suppress clpX essentiality, we used a clpX depletion strain and performed a time-course experiment following the switch to non-inducing medium. Using immunoblotting, we found that ClpX gradually declined to almost undetectable levels after 10 hours (Fig. 1C) (Jenal and Fuchs, 1998). In socB+ cells, the depletion of ClpX coincided with an increase in cellular filamentation (Fig. 1D) and a more than 1000-fold decrease in colony-forming units (CFUs) (Fig. 1E). However, in cells lacking socB, we observed only intermediate filamentation and no drop in CFUs (Fig. 1D–E). These data support the conclusion that a socB deletion bypasses the essentiality of clpXP.
socB is present in an operon with an upstream gene, socA, that is predicted to be essential (Fig. 1A) (Christen et al., 2011). This observation raised the possibility that socAB may encode a TA system. To test this hypothesis, we placed the putative toxin socB under an inducible promoter and tested whether its expression was toxic to cells in the presence or absence of its putative antitoxin socA. We found that inducing socB in socA+ cells had no significant effect on cell viability or morphology (Fig. 1F–G). However, inducing socB in ΔsocA cells inhibited colony formation and led to cellular filamentation (Fig. 1F–G). These phenotypes could be rescued by expressing socA in trans from a plasmid behind a different inducible promoter. Furthermore, time-course experiments revealed an approximately 10-fold decrease in viability in cells expressing socB for 5 hours in the absence of socA (Fig. S1A–B). Collectively, our data indicate that socAB behaves genetically like other TA systems.
For type II TA systems, the antitoxin functions by forming a complex with its cognate toxin and neutralizing its activity (Yamaguchi et al., 2011). To test whether SocA and SocB directly interact, we used the bacterial two-hybrid system based on complementation of the T18 and T25 fragments of adenylate cyclase (Karimova et al., 1998). We fused socA and socB to the T25 and T18 fragments, respectively, and co-expressed the gene fusions in E. coli. We observed a strong interaction between SocA and SocB, indicating that they likely form a complex (Fig. 1H). Interestingly, socB expression did not kill E. coli, indicating that its toxicity may be phylogenetically restricted. Indeed, homologs of socAB were identified only in the α-proteobacteria (Fig. S1C).
SocA Promotes SocB Degradation by ClpXP
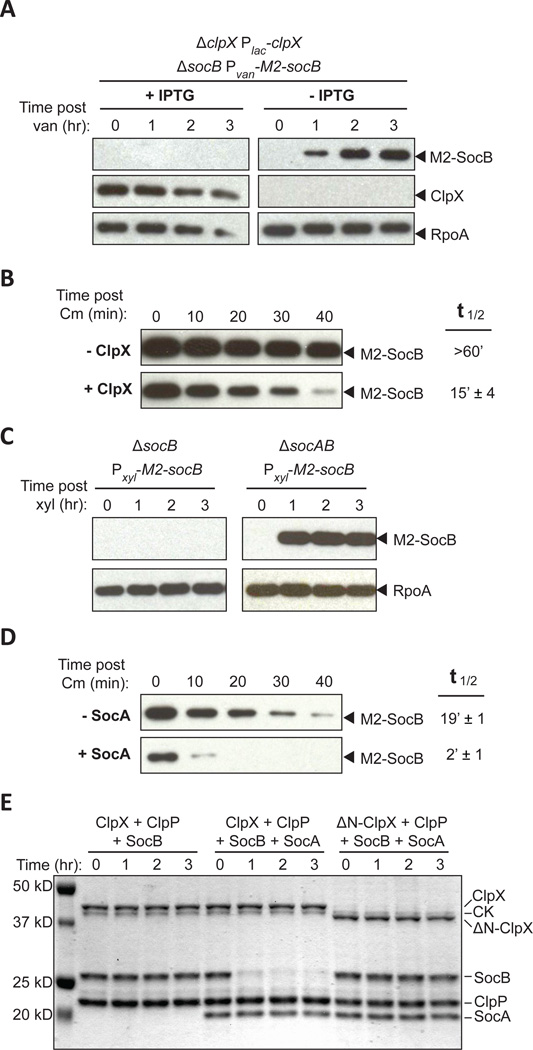
The observation that a deletion of socB can bypass the essentiality of ClpXP suggested that SocB may be a ClpXP substrate, and that accumulation of SocB in the absence of ClpXP inhibits growth. To test this possibility, we measured the accumulation of an M2-tagged variant of SocB in the presence or absence of ClpX. Whereas no M2-SocB was detected in the presence of ClpX, M2-SocB accumulation was observed when ClpX was first depleted for 12 hours (Fig. 2A). To confirm that the decreased abundance of SocB in the presence of ClpX results from a change in protein stability, we produced M2-SocB from a plasmid and measured its half-life in the presence or absence of ClpX using a chloramphenicol shut-off assay. We found that the presence of ClpX reduced the half-life of M2-SocB from >60 min to ~15 min, indicating that SocB is likely a ClpXP substrate (Fig. 2B, S2A).
Fig. 2. SocA Promotes SocB Degradation by ClpXP.
(A) Abundance of M2-SocB ± ClpX assessed by immunoblotting. The indicated strain was grown in clpX inducing or repressing conditions for 12 hr and M2-socB expression was induced at time zero. RpoA is a loading control.
(B) Stability of M2-SocB ± ClpX. clpX expression was repressed or induced for 12 hr, and then M2-socB expression was induced for 30 min prior to chloramphenicol (Cm) addition at time zero to shut off protein synthesis. Half-life ± S.E.M. quantified from three replicates (see Fig. S2A).
(C) Abundance of M2-SocB ± SocA assessed by immunoblotting. M2-socB expression was induced at time zero.
(D) Stability of M2-SocB ± SocA. M2-socB expression was induced for 2 hr, and then socA expression was induced for an additional 40 min prior to Cm addition at time zero. Half-life ± S.E.M. quantified from three replicates (see Fig. S2B).
(E) In vitro degradation of SocB by ClpXP ± SocA. Amounts were: 0.5 µM ClpX or ΔN-ClpX, 1 µM ClpP, 5 µM SocB, 5 µM SocA, 32 µg/ml creatine kinase (CK), 16 mM creatine phosphate, and 4 mM ATP. Reaction performed at 4°C.
See also Fig. S2.
Based on these results, we reasoned that SocB is normally present at low levels due to constitutive degradation by ClpXP. What role, then, does SocA play in the neutralization of SocB? Antitoxins of TA systems typically neutralize their cognate toxins by forming a stable complex (Yamaguchi et al., 2011). However, given that SocB is normally unstable, SocA may instead neutralize SocB by promoting its degradation. Indeed, we observed that M2-SocB accumulated in a strain lacking socA, but not in a socA+ strain (Fig. 2C). To test whether SocA affects the stability of SocB, we measured the half-life of M2-SocB with and without socA expression from a low-copy plasmid. We found that SocA reduced the half-life of M2-SocB from ~19 to ~2 min, indicating that SocA promotes the degradation of SocB (Fig. 2D, S2B). The half-life of M2-SocB in the presence of SocA (~2 min) was shorter than that measured above (~15 min), presumably due to differences in socA expression from a low-copy plasmid compared to its native chromosomal locus.
We hypothesized that SocA may be an adaptor for SocB degradation by ClpXP. Canonical adaptors, such as SspB, tether their substrates to the N-domain of ClpX. This tethering increases substrate concentration around the ClpX pore, which concomitantly increases the rate of substrate degradation (Dougan et al., 2003; Levchenko et al., 2000). To test whether SocA is a proteolytic adaptor, we purified SocA and SocB and performed an in vitro degradation reaction with ClpXP. When SocB was combined with ClpXP alone, no degradation was observed (Fig. 2E). However, when an equimolar amount of SocA was added to the reaction, we observed robust degradation of SocB, indicating that SocA promotes the degradation of SocB by ClpXP (Fig. 2E).
We performed the same reaction with a variant of ClpX that lacks the N-domain (residues 1–62) and, consequently, is catalytically active but deficient in adaptor-mediated degradation (Dougan et al., 2003). We observed no detectable degradation of SocB in the presence of SocA, ΔN-ClpX, and ClpP, indicating that the N-domain of ClpX is required for SocA to promote SocB proteolysis (Fig. 2E). Further, we observed a direct interaction between the ClpX N-domain and SocA (Fig. S2C), and found that mutations in the ClpX N-domain that abolish the interaction between ClpX and SocA prevent SocA from functioning as an antitoxin in vivo (Fig. S2D). Collectively, these data indicate that SocA is an adaptor for the degradation of SocB by ClpXP and they explain why clpXP and socA are essential for viability in Caulobacter.
Accumulation of SocB Blocks Replication Elongation
Why is the accumulation of SocB toxic to Caulobacter cells? To better study SocB function, we sought to develop a stabilized variant that is toxic even in the presence of SocA and ClpXP. Because ClpX often recognizes the free C-terminus of its substrates (Flynn et al., 2003), we tested the effect of appending an M2 tag to SocB. We found that SocB-M2 was stabilized over 40-fold relative to M2-SocB (Fig. S3A) and was toxic to socA+ clpXP+ cells (Fig. S3B). This stabilized variant was subsequently used to assess the effects of SocB on cellular physiology.
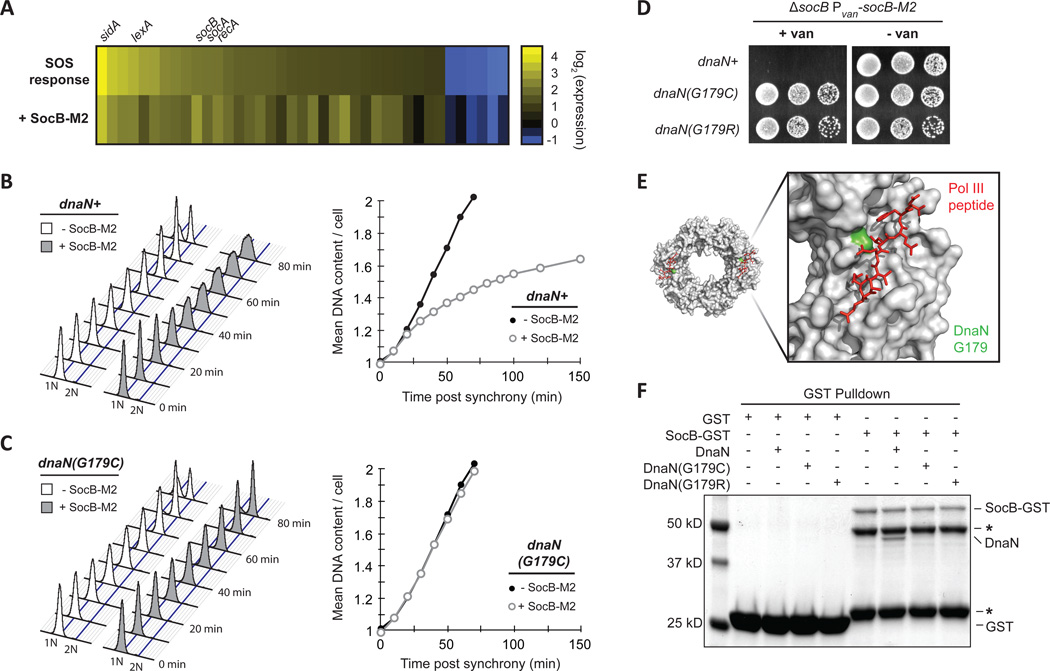
Toxins of TA systems target a diverse range of targets within bacterial cells (Yamaguchi et al., 2011). Two observations suggested that SocB inhibits DNA replication at the level of elongation. First, induction of socB-M2 caused cellular filamentation without chromosome accumulation, which may indicate that growth continues while replication elongation is blocked (Fig. S3C). Second, global expression profiling indicated that socB-M2 expression induced the SOS response (Fig. 3A). The SOS response is often induced in response to replication perturbations, such as DNA damage, that disrupt replication fork progression (Little and Mount, 1982). To directly test whether replication elongation is inhibited by SocB, we measured DNA content in synchronized populations of cells harboring an inducible copy of socB-M2. In non-inducing conditions, DNA content increased linearly as a function of time post-synchrony (Fig. 3B). In contrast, the induction of socB-M2 caused a decrease in the rate of replication elongation, and cells eventually arrested with a DNA content between 1N and 2N (Fig. 3B). As the completion of DNA replication is required for cell division, these cells also failed to divide (data not shown). These results indicate that SocB inhibits replication primarily at the level of elongation.
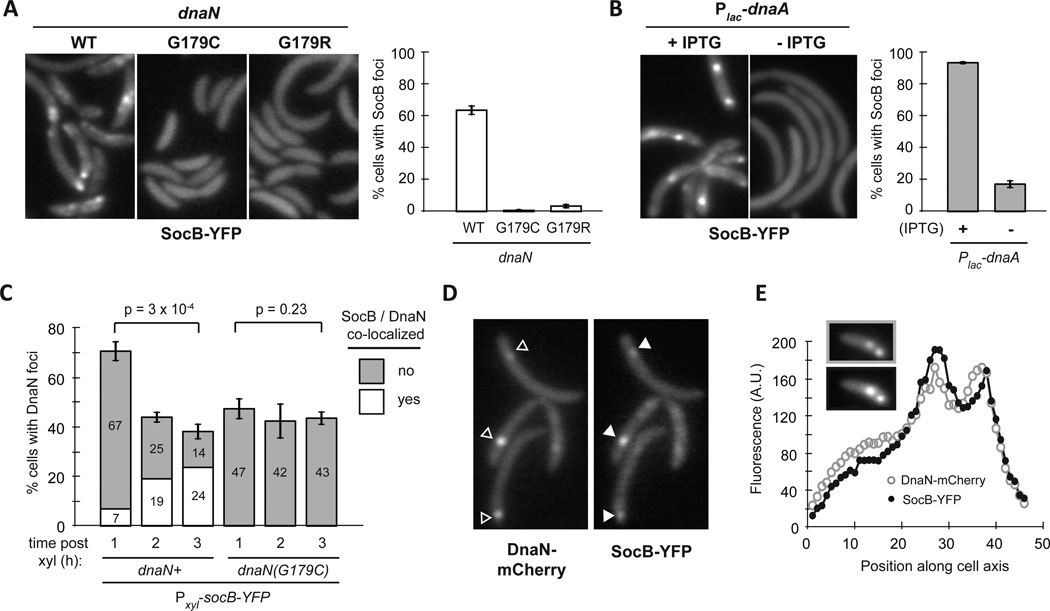
Fig. 3. SocB Blocks Replication Elongation Through an Interaction with DnaN.
(A) Analysis of gene expression changes following exposure to the DNA-damaging agent mitomycin C for 30 min (SOS response, top row) or following socB-M2 expression for 2 hr (+SocB-M2, bottom row). For each treatment, the genes induced or repressed more than 2-fold following mitomycin C treatment are shown.
(B) Flow cytometry of DNA content from synchronized cells grown ± socB-M2 expression. For the +SocB-M2 condition, socB-M2 was induced for 90 min prior to synchrony and release. Quantification of DNA content is shown on right.
(C) Same as (B), except performed with the dnaN(G179C) strain.
(D) Growth of indicated strains on socB-M2 inducing or repressing medium. Five-fold serial dilutions are shown.
(E) Structure of the E. coli sliding clamp in complex with a peptide derived from Pol III (PDB: 3D1F). Pol III peptide is in red, and the E. coli residue that corresponds to G179 in Caulobacter is colored in green.
(F) Interaction between SocB-GST and DnaN. For each condition, the indicated proteins were mixed with glutathione sepharose beads, washed, eluted, and then loaded on an SDS-PAGE gel. SocB-GST protein is unstable; asterisks indicate truncated SocB-GST products that retain GST tag.
See also Fig. S3.
SocB Blocks Replication Through an Interaction with DnaN
To identify the putative target of SocB, we screened for mutants that can tolerate high levels of socB-M2 expression. The first suppressors recovered were mutations in clpX that destabilized SocB-M2 (Fig. S3D). The largest group of these mutations clustered near the RKH loop, which protrudes from the ClpX pore and is important for the recognition of SsrA-tagged substrates (Farrell et al., 2007; Martin et al., 2008). Additional mutations were found at sites distant from the ClpX pore: Y76 and P326, for example, are proximal to the ATP-binding site and roughly 40 Å away from the nearest RKH loop. In each tested case, the mutations resulted in an approximate 10-fold reduction in SocB-M2 stability (Fig. S3E).
To identify suppressors outside of clpX, we continued our screen but focused on isolates that retained high levels of SocB-M2 and that did not harbor mutations in clpX. For two mutants, whole-genome sequencing revealed point mutations in dnaN, which encodes the β sliding clamp required for the processivity of DNA replication (Johnson and O'Donnell, 2005). Both mutations led to substitutions in glycine-179 of DnaN: G179C and G179R. We introduced these dnaN mutations into a clean genetic background and found that each was sufficient to bypass the replication block and growth inhibition normally observed following socB-M2 expression (Fig. 3C–D). Furthermore, these mutations were able to partially suppress the filamentation observed following SocB-M2 accumulation (Fig. S3F).
Glycine-179 resides within the hydrophobic groove on DnaN that is required for binding to Pol III and other replication proteins (Fig. 3E) (Georgescu et al., 2008). The identification of suppressor mutations affecting this residue of DnaN raised the possibility that SocB blocks replication through a direct interaction with DnaN. To test this possibility, we purified SocB-GST and DnaN and measured their binding by affinity chromatography in vitro. Production of SocB-GST is toxic to Caulobacter, indicating that this fusion protein is functional (Fig. S3G). We observed a strong interaction between DnaN and SocB-GST, but no interaction when DnaN was incubated with GST alone (Fig. 3F). Importantly, the suppressor mutations in DnaN, G179C and G179R, each disrupted the interaction with SocB-GST (Fig. 3F). We obtained similar results using a bacterial two-hybrid system, confirming that DnaN and SocB can directly interact (Fig. S3H).
The suppressor mutations isolated in DnaN reside within a hydrophobic groove required for its interaction with DnaN-binding proteins such as HdaA (Jonas et al., 2011), which must bind DnaA to regulate replication initiation (Kato and Katayama, 2001). To determine whether these mutations disrupt the interaction of DnaN with HdaA, we repeated our interaction assay and found that DnaN(G179C) retained the ability to interact with HdaA, whereas the G179R mutant did not (Fig. S3H). Consistent with these binding data, cells producing DnaN(G179C) appeared similar to wild type in the absence of socB expression, suggesting that the mutant version of DnaN supported wild-type like growth. In contrast, cells producing DnaN(G179R) were often filamentous (Fig. S3F), indicating that the interaction between DnaN and other proteins such as HdaA may be compromised. Taken together, our results support a model in which SocB blocks replication elongation through an interaction with DnaN, and that mutations in the hydrophobic cleft on DnaN can abrogate binding of SocB.
SocB Induces Loss of DnaN Replication Foci and Replication Fork Collapse
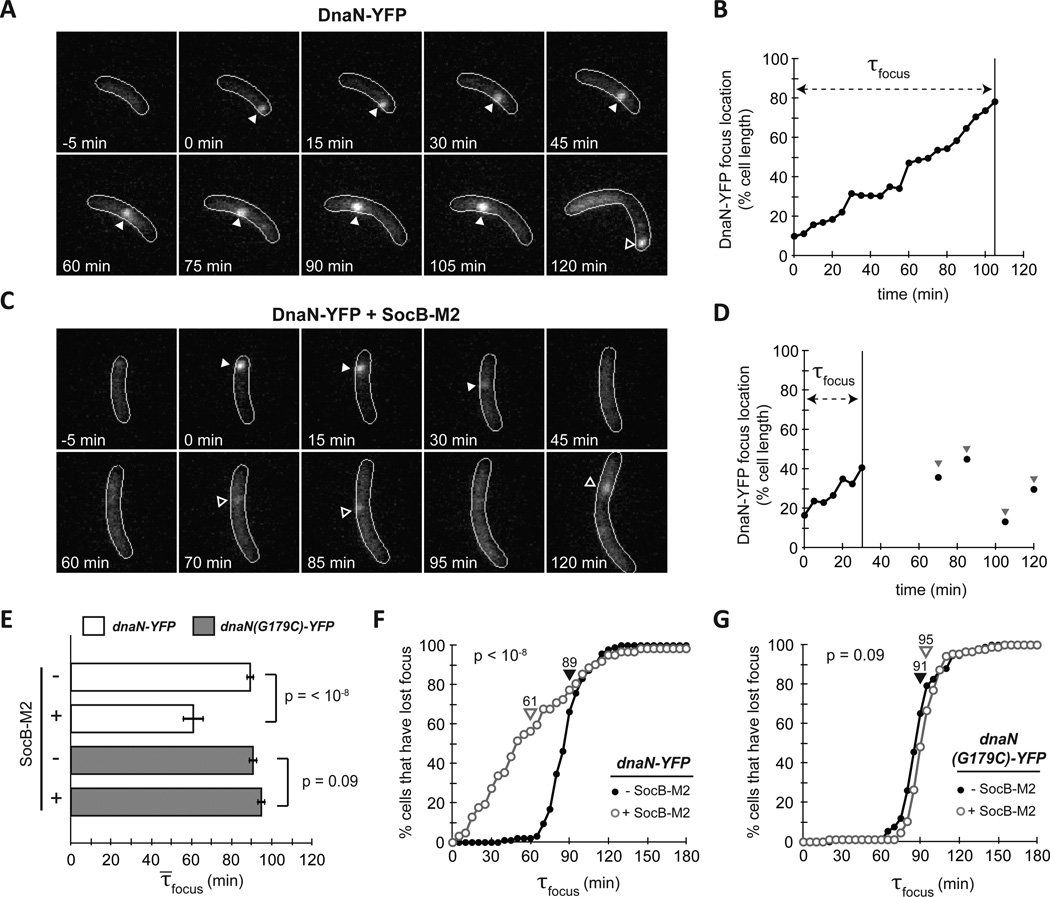
During DNA replication, DnaN accumulates dynamically behind the lagging strand polymerase as a result of discontinuous DNA replication; consequently, YFP-tagged DnaN typically forms a discrete focus within cells during replication (Su'etsugu and Errington, 2011). Additionally, in Caulobacter, DnaN translocates along the major axis of the cell during replication (Collier and Shapiro, 2009). To determine whether DnaN dynamics (and by proxy, ongoing replication) are affected by SocB, we imaged YFP-tagged DnaN in a strain harboring an inducible copy of socB-M2. In the absence of socB-M2 expression, G1-phased cells typically showed diffuse DnaN-YFP followed by formation of a single focus after the initiation of DNA replication (Fig. 4A). The focus moved along the major axis of the cell until its dispersal at the end of replication (Fig. 4B). In the presence of socB-M2 expression, we also observed formation of a single DnaN-YFP focus following initiation, but the focus often dispersed much earlier (Fig. 4C–D). In cells that lost their DnaN-YFP focus earlier, transient focus formation was sometimes observed following initial focus loss (Fig. 4C–D) although these foci often lasted only a single frame and appeared at varying points along the cell axis.
Fig. 4. SocB Induces Replication Fork Collapse.
(A) Fluorescence microscopy of strain bearing dnaN-YFP on a low-copy plasmid. dnaN-YFP was pre-induced for 2 h, and cells were synchronized and imaged every 5 min. Filled arrows indicates calculated position of DnaN-YFP focus; hollow arrows indicate a new round of replication following division.
(B) Calculated DnaN-YFP focus position of the cell from (A). τfocus is calculated as the time from focus formation to loss.
(C–D) Same as (A–B), except socB-M2 expression was induced for 90 min prior to synchrony and following release. Note the pre-mature focus loss between 30 and 45 min. Hollow arrows indicate transient DnaN-YFP focus formation events that may be attempts at replication restart.
(E) Average τ̄focus for dnaN-YFP strain (white bars) and dnaN(G179C)-YFP strain (grey bars) ± socB-M2 expression. Error bars indicate ± S.E.M; n≥60 cells per condition.
(F) Percentage of cells that have lost their DnaN-YFP focus as a function of time post initiation in the absence (filled circles) or presence (hollow circles) of socB-M2 expression. Average τ̄focus denoted by arrows. n≥60 cells per condition.
(G) Same as (F), but with dnaN(G179C)-YFP strain. n≥60 cells per condition.
To quantify these effects, we measured the time from focus formation to dispersal (τfocus), which reflects how long Pol III is engaged in replication before disengaging (either as a result of replication completion or pre-mature termination). We found that the average focus duration, τ̄focus, dropped from 89 to 61 minutes in the presence of socB-M2 expression (Fig. 4E, p<10−8). Multiple observations indicated that the decrease in τ̄focus represented fork collapse prior to the completion of replication. First, measurements of τfocus in the absence of socB-M2 expression indicated that replication takes, on average, 89 minutes to complete, and that no cells finish prior to 40 minutes post initiation (Fig. 4F); in the presence of socB-M2 expression, however, 34% of cells terminated replication prior to 40 minutes, indicating that these cells most likely suffered collapsed forks and harbored incompletely replicated chromosomes. Second, measurements of DNA content in the DnaN-YFP strain expressing socB-M2 indicated that these cells arrest with DNA content between 1N and 2N (data not shown). Third, transient DnaN-YFP focus formation following initial dispersal is suggestive of abortive attempts at replication re-start. These findings, in addition to the observed induction of the SOS response upon expression of socB-M2 (Fig. 3A), are consistent with a model in which SocB induces replication fork collapse.
To test whether the mutations identified in dnaN bypass replication fork collapse, we imaged strains expressing dnaN(G179C)-YFP in the presence or absence of socB-M2 expression and calculated τfocus. In contrast to cells expressing dnaN-YFP, we no longer observed a significant difference in τfocus following the production of SocB (p = 0.09) (Fig. 4E,G). Whereas 34% of dnaN-YFP cells expressing socB-M2 lost their foci within 40 minutes of replication initiation, only 1% of dnaN(G179C)-YFP cells did. These results indicate that mutations in DnaN that block binding to SocB also prevent replication fork collapse, further supporting the conclusion that SocB inhibits replication elongation through a direct interaction with DnaN.
SocB Co-Localizes with DnaN in a Replication-Dependent Manner
The interaction between SocB and DnaN suggested that SocB may localize to the replisome. To examine the subcellular localization of SocB, we integrated an inducible socB-YFP fusion on the chromosome and imaged cells by fluorescence microscopy. Expression of socB-YFP inhibited colony formation, indicating that this translational fusion is functional (Fig. S4A). After inducing socB-YFP for three hours, we observed the formation of SocB-YFP foci in a majority of cells (Fig. 5A). The formation of these foci was dependent on the ability of SocB to bind DnaN, as foci were rarely seen in cells producing DnaN(G179C) or DnaN(G179R) (Fig. 5A). SocB foci were also dependent on ongoing replication, as we saw a significant decrease in foci formation in cells depleted of DnaA, the replication initiator protein (Fig. 5B).
Fig. 5. SocB Forms Foci that Co-Localize with DnaN.
(A) Fluorescence microscopy of indicated strains at 3 hr post socB-YFP induction. Percentage of cells containing SocB-YFP foci is shown on the right. Errors bars indicate mean ± S.D. for three biological replicates (n>400 cells per replicate).
(B) Fluorescence microscopy of Plac-dnaA cells grown in the presence or absence of IPTG for 2 hr prior to socB-YFP induction for 3 h. Percentage of cells containing SocB-YFP foci calculated as in (A). Errors bars indicate mean ± S.D. for three biological replicates (n>400 cells per replicate).
(C) Percentage of cells with DnaN-mCherry foci as a function of time post socB-YFP induction. The percentage of total cells with co-localized (white) or not co-localized (grey) DnaN-mCherry and SocB-YFP foci is shown within each bar. Error bars indicate mean ± S.D. for three biological replicates (n>500 cells per replicate).
(D) Co-localization of DnaN-mCherry foci (hollow arrows) and SocB-YFP foci (filled arrows) after induction of socB-YFP for 3 h.
(E) Co-localization of multiple DnaN-mCherry and SocB-YFP foci in a single cell. Fluorescence profile for DnaN-mCherry (top inset, grey hollow circles) and SocB-YFP (bottom inset, black filled circles) is shown.
See also Fig. S4.
To test whether SocB and DnaN co-localize during replication, we fused dnaN to mCherry at the native chromosomal dnaN locus. We then integrated an inducible copy of socB-YFP on the chromosome and imaged cells by fluorescence microscopy at hour-long intervals post socB-YFP induction. Using an automated image analysis pipeline (Fig. S4B–C and Extended Experimental Procedures), we calculated the percentage of cells that have DnaN-mCherry foci, and of these cells, the percentage that also have co-localized SocB-YFP foci. As expected, we observed a decrease in the percentage of cells with DnaN-mCherry foci as a function of time post socB-YFP induction (Fig. 5C, p=3 × 10−4). The percentage of cells with DnaN-mCherry foci did not decrease to zero, presumably due to cells attempting to re-start replication following initial fork collapse (Fig. 4C). Although the percentage of cells with DnaN-mCherry foci decreased over time, the percentage of cells with co-localized DnaN-mCherry and SocB-YFP increased significantly (Fig. 5C). Most co-localization-positive cells had co-localized foci at a single point along the cell axis (Fig. 5D). In a minority of cells, multiple co-localized foci could be observed, which may occur when the left- and right-arm replisomes are no longer overlapping (Fig. 5E).
To test whether these localization effects were dependent on a direct interaction between SocB and DnaN, we repeated these microscopy experiments in a strain with dnaN(G179C)-mCherry integrated at its native chromosomal locus. In contrast to the dnaN-mCherry strain, we observed no significant decrease in the percentage of cells with DnaN(G179C)-mCherry foci as a function of time post socB-YFP induction (Fig. 5C, p=0.23). Further, fewer than 0.3% of cells exhibited co-localization between DnaN(G179C)-mCherry and SocB-YFP at any of the measured time points, in contrast to the 24% of cells that exhibit co-localization between DnaN-mCherry and SocB-YFP at three hours post induction (Fig. 5C). These results are consistent with a model in which SocB forms foci through its association with DnaN during active replication.
SocB Interacts with DnaN Through a DnaN-Binding Motif
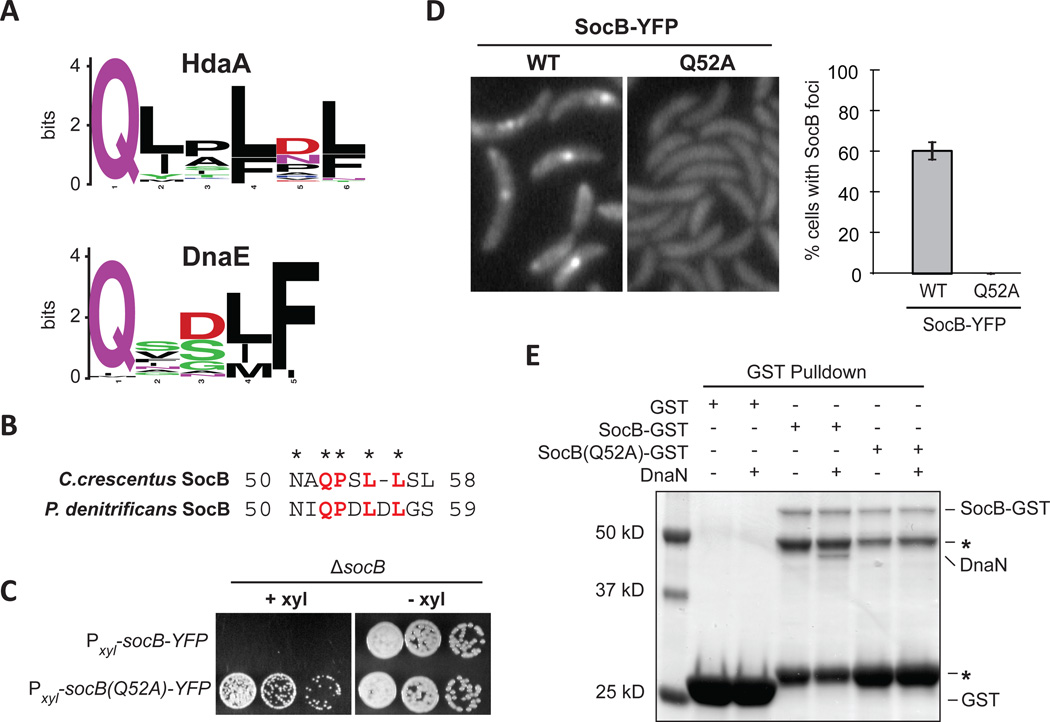
In γ-proteobacteria, DnaN-binding proteins such as Hda and DnaE often contain a shared motif (QL[SD]LF) for binding the β-sliding clamp. Examination of HdaA (the Caulobacter ortholog of Hda) and DnaE orthologs from α-proteobacteria revealed a similar, putative DnaN binding motif (Fig. 6A). SocB contained a short region similar to the DnaN-binding motif found in HdaA orthologs (Fig. 6B). To test whether this region is required for the interaction of SocB with DnaN, we generated a Q52A mutant of socB-YFP. Mutation of this conserved glutamine in HdaA is sufficient to abolish its interaction with DnaN (Jonas et al., 2011). We found that this mutation abolished the toxicity of socB-YFP expression (Fig. 6C), the formation of SocB-YFP foci (Fig. 6D), and also the interaction between SocB and DnaN (Fig. 6E, S5). Importantly, this mutation did not affect the ability of SocB to interact with SocA, indicating that SocB(Q52A) is likely properly folded (Fig. S5). These results support a model in which SocB inhibits replication by binding to DnaN using a motif similar to that of HdaA.
Fig. 6. SocB Interacts with DnaN Through a Conserved Motif.
(A) Sequence logo of the DnaN-binding motif in HdaA or DnaE from α-proteobacteria.
(B) Putative DnaN-binding motif in SocB from C. crescentus and P. denitrificans.
(C) Growth of indicated strains on media that induces (+xyl) or represses (−xyl) socB-YFP or socB(Q52A)-YFP expression. Five-fold serial dilutions are shown.
(D) Fluorescence microscopy of socB-YFP or socB(Q52A)-YFP 3 hr post induction. Percentage of cells with SocB-YFP foci ± S.D. for three biological replicates is shown on right (n>500 cells per replicate).
(E) Interaction between SocB-GST, SocB(Q52A)-GST, and DnaN. Performed as in Fig. 3H; as before, asterisk indicates SocB-GST N-terminal degradation products.
See also Fig. S5.
DISCUSSION
Essentiality of ClpXP and SocA Mechanism of Action
Unlike most bacteria, ClpXP is essential for the viability of Caulobacter cells (Jenal and Fuchs, 1998); however, the reason for this essentiality was not clear until now. Our work reveals that ClpXP is required for the degradation of a toxin, SocB, that is constitutively produced in cells (Fig. 7, left). In the absence of ClpXP or SocA, SocB accumulates, leading to the collapse of replication forks, induction of the SOS response, and eventual cell death (Fig. 7, right). Consequently, mutations in socB can bypass the essentiality of clpX or clpP (Fig. 1B). We note, however, that cells lacking socB and either clpX or clpP do not grow as rapidly as wild-type, likely due to defects in the turnover of other ClpXP substrates such as the cell cycle regulator CtrA (Bhat et al., 2013).
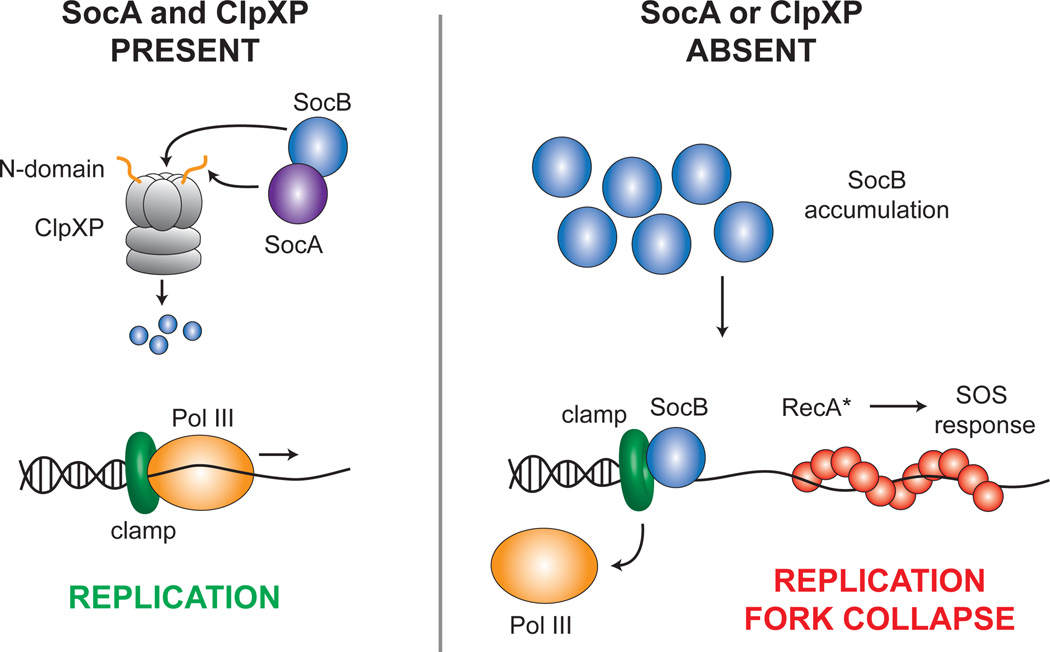
Fig. 7. Model for SocAB Function.
Under normal growth conditions, the toxin SocB is delivered to ClpXP for degradation by its antitoxin SocA. Pol III thus remains in association with the clamp, and replication proceeds normally (left). However, in the absence of either ClpXP or SocA, SocB accumulates and competes for binding to the clamp with Pol III and other replication factors. This competition eventually results in the collapse of replication forks and induction of the RecA-mediated SOS response (right).
The rapid turnover of SocB is unusual, given that, for most TA systems, the toxin is more stable than its cognate antitoxin. The reduced stability of SocB stems from the atypical mechanism of its antitoxin, SocA. Whereas most antitoxins inhibit their cognate toxins through sequestration, SocA is instead an adaptor for the degradation of SocB by ClpXP. SocA binds both SocB (Fig. 1H) and the N-domain of ClpX (Fig. S2C) and thereby promotes the degradation of SocB in vivo and in vitro (Fig. 2D–E). SocA binding to the N-domain appears to be essential for this activity, as SocA was unable to promote SocB proteolysis when the N-domain was truncated from ClpX (Fig. 2E). The observation that SocA decreases SocB stability does not rule out that SocA also blocks SocB toxicity through sequestration. However, multiple results argue against this role for SocA: (1) targeted mutations in the N-domain of ClpX that abolish SocA binding also prevent SocA from functioning as an antitoxin in vivo (Fig. S2C–D); and (2) production of the stable SocB-M2 variant in socA+ cells is lethal, even though SocB-M2 and SocA can still form a complex (Fig. S3B, data not shown). Thus, we propose that the antitoxin activity of SocA results principally from its ability to promote SocB proteolysis through ClpXP, which makes SocA necessary, but not sufficient, for counteracting SocB toxicity.
In many bacteria, ClpXP is not formally essential for viability, although it is often required for other critical processes. In E. coli, deletions of clpX or clpP have no significant effect on viability or growth rate (Schweder et al., 1996), but ClpXP contributes to the proteolysis of a range of cellular substrates (Flynn et al., 2003). In B. subtilis, clpX and clpP deletions are viable but have major defects in sporulation and competence (Nakano et al., 2001). These defects are mostly due to accumulation of a transcriptional regulator, Spx, that is normally degraded by ClpXP. ClpX or ClpP has, however, been found to be essential in several bacteria, including the pathogens Mycobacterium tuberculosis and Streptococcus pneumoniae (Piotrowski et al., 2009; Raju et al., 2012). The essentiality of clpX can be bypassed in Streptococcus by mutations in an uncharacterized gene, spr1630, suggesting that this gene product may accumulate in the absence of ClpXP and inhibit growth, similar to SocB. Characterization of the gene products that bypass ClpXP essentiality may reveal novel regulators of cell growth or physiology.
Mechanism of SocB Inhibition of Replication Elongation
Our results indicate that, upon accumulating, SocB blocks replication elongation and triggers an SOS response (Fig. 3). Multiple observations suggest that SocB mediates these effects through a direct interaction with DnaN: (1) SocB and DnaN interact in vitro and in a bacterial two-hybrid assay (Fig. 3F, S3H); (2) mutations in DnaN that abolish its interaction with SocB also bypass the SocB-induced replication block (Fig. 3C,F, S3H); (3) SocB and DnaN co-localize, and this co-localization is dependent on the ability of SocB and DnaN to interact (Fig. 5); and (4) SocB contains a DnaN-binding motif that is required for its interaction with DnaN and toxicity (Fig. 6). In sum, these results are consistent with a model in which SocB binds to DnaN, and that this association leads to catastrophic replication fork collapse (Fig. 4).
How, then, does the interaction between SocB and DnaN lead to the premature termination of replication? The simplest model is that SocB competes with Pol III for binding to DnaN, potentially disrupting both lagging and leading strand synthesis. During lagging strand replication, the polymerase extends discontinuously and presumably must associate with a new clamp to produce each Okazaki fragment (Johnson and O'Donnell, 2005). Disruption of the Pol III-DnaN interaction could rapidly inhibit production of new Okazaki fragments and consequently block synthesis of the lagging strand. In contrast, during leading strand replication, the polymerase can extend continuously and may not need to load new clamps following initiation. However, depending on the stability of the Pol III-DnaN interaction, SocB may still be able to compete for binding to DnaN on the leading strand if Pol III and DnaN ever transiently dissociate, such as upon encountering a DNA lesion. Additionally, blocking synthesis on the lagging strand would also indirectly inhibit synthesis on the leading strand, given that the leading and lagging strand polymerases are physically tethered by the γ clamp loader complex. Such stalling of replication could eventually lead the replisome to disassemble.
Notably, the effects of SocB on DNA replication are not immediate; replication first slows down approximately 120 min post-induction (Fig. 3B, note the 90 min pre-induction prior to time zero). This timing coincides with the first observable decrease in viability (Fig. S1B) and an increase in the co-localization of SocB with DnaN (Fig. 5C), but is later than may be expected if SocB immediately outcompetes Pol III for binding to DnaN. The delay may simply reflect a need for SocB to accumulate to sufficient levels before outcompeting Pol III and other DnaN-binding proteins. Alternatively, SocB may only disrupt new associations of DnaN and Pol III such that replication can initially continue following the accumulation of SocB by using DnaN molecules already in association with Pol III.
An additional question is why SocB, despite the presence of a DnaN-binding motif, is not toxic to E. coli which has a similar hydrophobic pocket on DnaN (Fig. 1H, data not shown). It is possible that that SocB makes contacts with DnaN at a secondary site that is less well conserved between bacteria. Interestingly, a co-crystal structure of DnaN and the little finger domain of DNA Pol IV, a DnaN-interacting protein, revealed the presence of a secondary binding interface outside of the hydrophobic pocket (Bunting et al., 2003). The secondary interface comprised over 70 percent of the buried surface area, indicating that it likely contributes significantly to the affinity of interaction. The requirement of a secondary binding interface may explain why appending a DnaN-binding motif to GFP only results in weak DnaN-dependent foci formation in B. subtilis (Su'etsugu and Errington, 2011).
Given the ability of SocB to inhibit replication progression, an important remaining question is when SocB normally accumulates in wild-type cells. Genetically, we revealed that a loss of clpX, clpP, or socA is sufficient to allow SocB accumulation (Fig. 1B, 2). Whether the stability or activity of ClpXP, or SocA, is ever modulated to induce SocB accumulation is not yet clear. Intriguingly, the socAB operon is induced by the DNA-damaging agent MMC (Fig. 3A), suggesting that it may play a regulatory role during the response to DNA damage. One attractive possibility is that sublethal amounts of SocB accumulate following DNA damage, or other stresses, to regulate the rate at which replication proceeds. Recent work has begun to reveal mechanisms by which cells regulate replication at the level of elongation. For example, the accumulation of (p)ppGpp during the stringent response in B. subtilis leads to an arrest of replication elongation through the inhibition of DNA primase (Wang et al., 2007). Although replication is often controlled at the level of initiation, mechanisms for modulating elongation may be widespread.
Protein Interaction Hubs and Antibiotic Targets
In addition to blocking replication elongation, the binding of SocB to DnaN may disrupt other replication-associated processes. In addition to Pol III, the clamp interacts with proteins required for DNA repair and the regulation of replication initiation, including Pol IV and Pol V (translesion synthesis), MutS and MutL (mismatch repair), and Hda (regulation of initiation). These proteins all bind to DnaN within the same hydrophobic cleft on its surface that is likely bound by SocB (Robinson et al., 2010), suggesting that SocB may disrupt several cellular processes required for growth and genome maintenance.
Interaction hubs such as DnaN may be ideal targets for new antibiotic development because they coordinate multiple cellular processes and may be unable to mutate to prevent small molecule binding without significantly compromising their native functions. The ability of a protein toxin like SocB to arrest DNA replication and kill cells suggests that the hydrophobic cleft on DnaN may be a prime target for small molecule inhibitors. In fact, the small molecule RU-7 was recently shown to bind within this hydrophobic cleft on the clamp and to prevent its association with Pol III in vitro (Georgescu et al., 2008). This interaction was specific for the bacterial sliding clamp, as the interaction between the eukaryotic PCNA clamp and Pol δ was unaffected. A separate screen for inhibitors of an in vitro bacterial replication system identified six compounds that share the same core structure as RU-7, suggesting that they also function through binding to the clamp (Dallmann et al., 2010). The recent rise in multi-drug resistant strains of various pathogenic bacteria highlights the continued need to develop novel antibiotics to combat these infections. The study of bacterially-encoded toxins and the mechanisms by which they inhibit cellular proliferation may prove valuable in the identification of promising new targets.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Media
Caulobacter strains used in this study are listed in Table S1. For details on strain construction and growth conditions, see Extended Experimental Procedures.
Suppressor Screening and Mapping
Suppressors of a clpP depletion strain were obtained by EZ-Tn5 mutagenesis, and transposon insertion sites in surviving colonies were identified by rescue cloning of circularized DNA in pir-116 E. coli. Suppressors of socB-M2 suppression were first screened for mutations in socB, clpX, and clpP; suppressors that contained no mutations in these genes were then subjected to whole genome sequencing to identify point mutations. For details, see Extended Experimental Procedures.
Protein Purification and Degradation Assays
His6-SocA and His6-SocB were purified by Ni-NTA affinity chromatography, and for His-SocA, by subsequent gel filtration chromatography (see Extended Experimental Procedures). ClpX, ΔN-ClpX, and ClpP were prepared as described previously (Chien et al., 2007). Degradation reactions were performed in PD-KCl-200 buffer at 4°C. Reaction conditions were as follows: 0.5 µM ClpX, 0.5 µM ΔN-ClpX, 1 µM ClpP, 5 µM SocB, 5 µM SocA, 32 µg/ml creatine kinase, 16 mM creatine phosphate, and 4 mM ATP.
Microscopy and Image Analysis
For details on image acquisition and processing, see Extended Experimental Procedures.
Microarrays
Expression data for cells exposed to the DNA-damaging agent mitomycin C was previously published (Modell et al., 2011). Expression profiling of cells expressing socB-M2 was performed as described previously (Gora et al., 2010).
Supplementary Material
HIGHLIGHTS.
SocAB is an atypical toxin-antitoxin system in Caulobacter crescentus
The antitoxin SocA is required for ClpXP-mediated degradation of the toxin SocB
In the absence of SocA or ClpXP, SocB accumulates and inhibits DNA replication
SocB inhibits replication by binding the β sliding clamp
ACKNOWLEDGEMENTS
We thank members of the Laub laboratory for discussions and comments on the manuscript. We thank Peter Chien for providing purified ClpX, ΔN-ClpX, and ClpP. M.T.L. is an Early Career Investigator at the Howard Hughes Medical Institute. This work was supported by an HHMI Summer Medical Fellowship to D.H., an NSF Graduate Research Fellowship to C.D.A., and an NIH grant (R01GM082899) to M.T.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
The expression data for cells producing SocB-M2 has been deposited in ArrayExpress with accession number E-MEXP-3990.
REFERENCES
- Bhat NH, Vass RH, Stoddard PR, Shin DK, Chien P. Identification of ClpP substrates in Caulobacter crescentus reveals a role for regulated proteolysis in bacterial development. Mol. Microbiol. 2013;88:1083–1092. doi: 10.1111/mmi.12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting KA, Roe SM, Pearl LH. Structural basis for recruitment of translesion DNA polymerase Pol IV/DinB to the beta-clamp. EMBO J. 2003;22:5883–5892. doi: 10.1093/emboj/cdg568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K, Courvalin P, Dantas G, Davies J, Eisenstein B, Huovinen P, Jacoby GA, Kishony R, Kreiswirth BN, Kutter E, et al. Tackling antibiotic resistance. Nat. Rev. Microbiol. 2011;9:894–896. doi: 10.1038/nrmicro2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Roa D, Garcia-Pino A, De Geiter S, van Nuland NAJ, Loris R, Zenkin N. The Fic protein Doc uses an inverted substrate to phosphorylate and inactivate EF-Tu. Nat. Chem. Biol. 2013 doi: 10.1038/nchembio.1364. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien P, Perchuk BS, Laub MT, Sauer RT, Baker TA. Direct and adaptor-mediated substrate recognition by an essential AAA+ protease. Proc. Natl. Acad. Sci. USA. 2007;104:6590–6595. doi: 10.1073/pnas.0701776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen B, Abeliuk E, Collier JM, Kalogeraki VS, Passarelli B, Coller JA, Fero MJ, McAdams HH, Shapiro L. The essential genome of a bacterium. Mol. Syst. Biol. 2011;7:528. doi: 10.1038/msb.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates AR, Halls G, Hu Y. Novel classes of antibiotics or more of the same? Br. J. Pharmacol. 2011;163:184–194. doi: 10.1111/j.1476-5381.2011.01250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier J, Shapiro L. Feedback control of DnaA-mediated replication initiation by replisome-associated HdaA protein in Caulobacter. J. Bacteriol. 2009;191:5706–5716. doi: 10.1128/JB.00525-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallmann HG, Fackelmayer OJ, Tomer G, Chen J, Wiktor-Becker A, Ferrara T, Pope C, Oliveira MT, Burgers PM, Kaguni LS, et al. Parallel multiplicative target screening against divergent bacterial replicases: identification of specific inhibitors with broad spectrum potential. Biochemistry. 2010;49:2551–2562. doi: 10.1021/bi9020764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalrymple BP, Kongsuwan K, Wijffels G, Dixon NE, Jennings PA. A universal protein-protein interaction motif in the eubacterial DNA replication and repair systems. Proc. Natl. Acad. Sci. USA. 2001;98:11627–11632. doi: 10.1073/pnas.191384398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan DA, Weber-Ban E, Bukau B. Targeted delivery of an ssrA-tagged substrate by the adaptor protein SspB to its cognate AAA+ protein ClpX. Mol. Cell. 2003;12:373–380. doi: 10.1016/j.molcel.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Farrell CM, Baker TA, Sauer RT. Altered specificity of a AAA+ protease. Mol. Cell. 2007;25:161–166. doi: 10.1016/j.molcel.2006.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA. Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell. 2003;11:671–683. doi: 10.1016/s1097-2765(03)00060-1. [DOI] [PubMed] [Google Scholar]
- Georgescu RE, Yurieva O, Kim SS, Kuriyan J, Kong XP, O'Donnell M. Structure of a small-molecule inhibitor of a DNA polymerase sliding clamp. Proc. Natl. Acad. Sci. USA. 2008;105:11116–11121. doi: 10.1073/pnas.0804754105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Maisonneuve E. Bacterial persistence and toxin-antitoxin loci. Annu. Rev. Microbiol. 2012;66:103–123. doi: 10.1146/annurev-micro-092611-150159. [DOI] [PubMed] [Google Scholar]
- Gora KG, Tsokos CG, Chen YE, Srinivasan BS, Perchuk BS, Laub MT. A cell-type-specific protein-protein interaction modulates transcriptional activity of a master regulator in Caulobacter crescentus. Mol. Cell. 2010;39:455–467. doi: 10.1016/j.molcel.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indiani C, McInerney P, Georgescu R, Goodman MF, O'Donnell M. A sliding-clamp toolbelt binds high- and low-fidelity DNA polymerases simultaneously. Mol. Cell. 2005;19:805–815. doi: 10.1016/j.molcel.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Jenal U, Fuchs T. An essential protease involved in bacterial cell-cycle control. EMBO J. 1998;17:5658–5669. doi: 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A, O'Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- Jonas K, Chen YE, Laub MT. Modularity of the bacterial cell cycle enables independent spatial and temporal control of DNA replication. Curr. Biol. 2011;21:1092–1101. doi: 10.1016/j.cub.2011.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J, Katayama T. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 2001;20:4253–4262. doi: 10.1093/emboj/20.15.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz M, Dalrymple B, Wijffels G, Kongsuwan K. Interaction of the sliding clamp beta-subunit and Hda, a DnaA-related protein. J. Bacteriol. 2004;186:3508–3515. doi: 10.1128/JB.186.11.3508-3515.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenhart JS, Sharma A, Hingorani MM, Simmons LA. DnaN clamp zones provide a platform for spatiotemporal coupling of mismatch detection to DNA replication. Mol. Microbiol. 2013;87:553–568. doi: 10.1111/mmi.12115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenne-Samuel N, Wagner J, Etienne H, Fuchs RP. The processivity factor beta controls DNA polymerase IV traffic during spontaneous mutagenesis and translesion synthesis in vivo. EMBO Rep. 2002;3:45–49. doi: 10.1093/embo-reports/kvf007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko I, Seidel M, Sauer RT, Baker TA. A specificity-enhancing factor for the ClpXP degradation machine. Science. 2000;289:2354–2356. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- Little JW, Mount DW. The SOS regulatory system of Escherichia coli . Cell. 1982;29:11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- Lopez de Saro FJ, Marinus MG, Modrich P, O'Donnell M. The beta sliding clamp binds to multiple sites within MutL and MutS. J. Biol. Chem. 2006;281:14340–14349. doi: 10.1074/jbc.M601264200. [DOI] [PubMed] [Google Scholar]
- Maki S, Kornberg A. DNA polymerase III holoenzyme of Escherichia coli II. A novel complex including the gamma subunit essential for processive synthesis. J. Biol. Chem. 1988;263:6555–6560. [PubMed] [Google Scholar]
- Martin A, Baker TA, Sauer RT. Diverse pore loops of the AAA+ ClpX machine mediate unassisted and adaptor-dependent recognition of ssrA-tagged substrates. Mol. Cell. 2008;29:441–450. doi: 10.1016/j.molcel.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modell JW, Hopkins AC, Laub MT. A DNA damage checkpoint in Caulobacter crescentus inhibits cell division through a direct interaction with FtsW. Genes Dev. 2011;25:1328–1343. doi: 10.1101/gad.2038911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutschler H, Gebhardt M, Shoeman RL, Meinhart A. A novel mechanism of programmed cell death in bacteria by toxin-antitoxin systems corrupts peptidoglycan synthesis. PLoS Biol. 2011;9:e1001033. doi: 10.1371/journal.pbio.1001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano MM, Hajarizadeh F, Zhu Y, Zuber P. Loss-of-function mutations in yjbD result in ClpX- and ClpP-independent competence development of Bacillus subtilis. Mol. Microbiol. 2001;42:383–394. doi: 10.1046/j.1365-2958.2001.02639.x. [DOI] [PubMed] [Google Scholar]
- Pandey DP, Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski A, Burghout P, Morrison DA. spr1630 is responsible for the lethality of clpX mutations in Streptococcus pneumoniae. J. Bacteriol. 2009;191:4888–4895. doi: 10.1128/JB.00285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju RM, Unnikrishnan M, Rubin DH, Krishnamoorthy V, Kandror O, Akopian TN, Goldberg AL, Rubin EJ. Mycobacterium tuberculosis ClpP1 and ClpP2 function together in protein degradation and are required for viability in vitro and during infection. PLoS Pathog. 2012;8:e1002511. doi: 10.1371/journal.ppat.1002511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A, Brzoska AJ, Turner KM, Withers R, Harry EJ, Lewis PJ, Dixon NE. Essential biological processes of an emerging pathogen: DNA replication, transcription, and cell division in Acinetobacter spp. Microbiol. Mol. Biol. Rev. 2010;74:273–297. doi: 10.1128/MMBR.00048-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A, Causer RJ, Dixon NE. Architecture and conservation of the bacterial DNA replication machinery, an underexploited drug target. Curr. Drug Targets. 2012;13:352–372. doi: 10.2174/138945012799424598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- Schweder T, Lee KH, Lomovskaya O, Matin A. Regulation of Escherichia coli starvation sigma factor (sigma s) by ClpXP protease. J. Bacteriol. 1996;178:470–476. doi: 10.1128/jb.178.2.470-476.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su'etsugu M, Errington J. The replicase sliding clamp dynamically accumulates behind progressing replication forks in Bacillus subtilis cells. Mol. Cell. 2011;41:720–732. doi: 10.1016/j.molcel.2011.02.024. [DOI] [PubMed] [Google Scholar]
- Walsh C. Antibiotics: actions, origins, resistance. Washington, D.C.: ASM Press; 2003. [Google Scholar]
- Wang JD, Sanders GM, Grossman AD. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell. 2007;128:865–875. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kim Y, Hong SH, Ma Q, Brown BL, Pu M, Tarone AM, Benedik MJ, Peti W, Page R, et al. Antitoxin MqsA helps mediate the bacterial general stress response. Nat. Chem. Biol. 2011;7:359–366. doi: 10.1038/nchembio.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Inouye M. Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat. Rev. Microbiol. 2011;9:779–790. doi: 10.1038/nrmicro2651. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y, Park JH, Inouye M. Toxin-antitoxin systems in bacteria and archaea. Annu. Rev. Genet. 2011;45:61–79. doi: 10.1146/annurev-genet-110410-132412. [DOI] [PubMed] [Google Scholar]
- Yuan J, Sterckx Y, Mitchenall LA, Maxwell A, Loris R, Waldor MK. Vibrio cholerae ParE2 poisons DNA gyrase via a mechanism distinct from other gyrase inhibitors. J. Biol. Chem. 2010;285:40397–40408. doi: 10.1074/jbc.M110.138776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang J, Hoeflich KP, Ikura M, Qing G, Inouye M. MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol. Cell. 2003;12:913–923. doi: 10.1016/s1097-2765(03)00402-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.