Abstract
For the analysis of rare-variant data in population-based designs, we propose a method to detect study subjects that may create population substructure in the study sample. Our approach is computationally fast and simple, permitting applications to whole-genome sequencing studies. The method does not require the variants to be in linkage equilibrium and can be applied to all the genetic loci that are available in the study. For both rare and common variants, we assess the performance of our approach by its application to the 1000 Genome Project data, and in simulation studies. The results are compared to the commonly used outlier detection algorithm based on principal component analysis (PCA). The statistical power of both approaches to detect outliers are comparable in most of the scenarios, but the power of PCA to detect outliers is lower than the novel approach in the presence of linkage disequilibrium and for subpopulations that are genetically similar. The data analysis and the simulation studies suggest that the number of false-positive results appears to be different for the two approaches. Our approach maintains the type I error rate while the outlier detection approach based on PCA does not. Taking additionally into account the minimal computational requirements of our approach and the ability to incorporate all the marker information, the proposed method will have important application in sequencing studies and genome-wide association studies.
Keywords: population substructure, outlier detection, GWAS, sequence data
Introduction
Genetic association analysis has proven to be a powerful statistical tool for the identification of disease loci in the human genome [Consortium, 2007; McCarthy et al., 2008; Stranger et al., 2011]. Population-based association analysis is straight-forward and computationally fast, even at a whole-genome level. One of the main caveats of population-based association analysis, however, is that it can be susceptible to bias due to genetic confounding, i.e., population substructure.
This issue has been the focus of statistical research for some time. In designs of unrelated individuals, most genetic association tests take the form of a score test in which the numerator sums the contributions of the study subjects to the statistics and the denominator calculates the variance of the statistic, assuming independence of the study subjects. In the presence of mating among relatives or population substructure, the genotypes of the study subjects are no longer independent, leading to a potentially biased estimate for the variance of the test statistic. This can cause the test statistic to become anticonservative. Genomic control approach adjusts for the bias in the variance of the test statistic by estimating a variance inflation factor at a set of reference loci and scaling the variance of the test statistic accordingly [Devlin and Roeder, 1999; Reich and Goldstein, 2001]. With the arrival of Genome-wide genetic (GWAS) data, principal component analysis (PCA) gained popularity [Patterson et al., 2006; Price et al., 2006]. They infer population substructure and admixture based on the PCA of the variance-covariance matrix of genotyped markers [McVean, 2009;Novembre and Stephens, 2008]. Then, the principal components (PCs) are either used to identify genetically homogeneous subpopulations in the study [Luca et al., 2008] or to adjust the association for genetic confounding [Price et al., 2006].
For the association analysis of rare variants, the application of such approaches to avoid bias due to population substructure and admixture can be problematic. In PCA approach, the estimation of the variance/covariance matrix can become unstable for genetic loci with low minor allele frequencies, making the results of this approach less reliable. For example, the investigators usually select markers with allele frequencies greater than 10% before applying PCA [He et al., 2011; Sladek et al., 2007]. An alternative that could be considered here is to assess population substructure for loci with common alleles and apply the PC results to the rare-variant analysis, assuming that the population substructures for rare and common variants are the same. The transferability of population substructure between common and rare genetic loci is a hypothesis which has not been assessed thoroughly based on real data so far. The general applicability of this concept seems to be problematic in light of the age of the different variant types, i.e., common variants are genetically much older than rare variants [Mathieson and McVean, 2012]. Although rare-variant approaches rely mostly on permutation tests for the assessment of the significance, the concept of genomic control generally can be modified and applied to rare-variant analysis. However, it can give a reduced power [Price et al., 2010] and cannot be utilized to identify homogenous subpopulations.
Here, we propose a simple, computationally fast approach that allows the identification of genetic outliers to obtain a genetically homogeneous subpopulation in studies with sequence data, minimizing the impact of population substructure on rare-variants analysis. The approach is able to utilize the information on all available genetic loci and does not require the selection of a subset of markers that are in linkage equilibrium (LE). The test statistic is computed for each individual based on all the rare-variant information available. The power and the type I error of the approach are examined in simulation studies and by the applications to the 1000 Genome Project data. We compare the performance of our approach with the outlier detection algorithm based on PCA.
Methods
Introducing Test Statistics T1 and T2
Suppose in a genetic association study of unrelated individuals, genotype data are available at m bi-allelic loci for all the study subjects. We denote the number of the minor alleles at the ith marker locus by Xi for one subject. We define the genetic residual byΔ Xi = Xi − E (Xi) where E (Xi) is the expected number of the minor alleles at the ith locus in the study population. The genetic residual can be considered as the genetic deviation of the subject at ith locus from the study population. We define two genome-wide scores that measure the distance between a particular individual and the population across the genome. The scores are given by
and
Based on the scores, we can construct the score tests T1 and T2 which are given by
and
The first score aggregates the residuals over all the marker loci for one subject. If, for the study population and the population where the outliers are from, there is preferentially a one-direction difference in the minor allele frequency (MAF), i.e., most of the markers have smaller MAF in one population than in the other population, then the test score S1 will be more powerful in detecting the population outliers. This situation can occur due to the founder effects in one subpopulation [Reich et al., 2001; Roy-Gagnon et al., 2011], long-range haplotypes [Price et al., 2008], etc. However, if the differences in minor allele frequencies between two subpopulations do not follow this patterns, test statistic S2 is generally better suited to identify genetically different subjects. In the supplementary Note III, we provide the theoretically justification for that. We will further outline these features of the score statistics S1 and S2 in the simulation section of this paper.
Under the assumption of Hardy-Weinberg equilibrium (HWE), the expected marker score can be calculated based on the minor allele frequency, i.e., E (Xi) = 2pi, where p i is the true minor allele frequency at ith marker locus. For large datasets, we can estimate the allele frequency p i by the observed frequency of the minor allele in the actual data and the asymptotic distribution stays the same. Alternatively, the allele frequencies can be obtained from the corresponding reference populations. Assuming the absence of LD between the loci, the mean and variance of S1 and S2 can be derived analytically based on the allele frequencies, as shown in supplementary Note I. Then the test statistics are given by:
| (1) |
| (2) |
Then, under the null hypothesis of no population substructure, both test statistics T1 and T2 follow a χ2 distribution with one degree of freedom asymptotically.
Adjusting T1 and T2 in the Presence of LD
For sequence data, the no LD assumption is not reasonable unless only a subset of loci that are in LE is selected. In the presence of LD, both standardized scores have to be adjusted accordingly. Since the variances of S1 and S2 do not depend on the actual genotype of the study subject and are constant across the subjects, ideally, we would need to adjust T1 by
This adjustment is only reasonable when the effect of LD is a linear inflation of T1, which is the case here. This is due to the fact that the denominator of the test statistic is the same across all the subjects as it aggregates over all subjects and does not depend on the each subject’s genotype. Thus, we can use one value to correct for the inflation under LD. To estimate the inflation factor, since the calculation of the correlations of the residuals across the genome requires a great amount of computation time, the genomic inflation factor for each test statistic can be estimated based on the distribution of the test statistic across the study subjects. For test statistic T1, we estimate the genomic inflation λ1 by
| (3) |
where 0.455 is the 50th percentile of a distribution. Similarly for T2, we estimate the genomic inflation factor λ2 by
| (4) |
In the presence of LD, we can adjust T1 using the subject inflation factor λ1 by
| (5) |
The adjusted test statistic T2 is derived in the same way. Under the null-hypothesis that the subject is from the study population, the test statistics T1 and T2 have an asymptotic χ2-distribution with one degree of freedom.
The Optimal Test and Its Asymptotic Distribution
Since, prior to the calculation of the test statistic, we do not have any knowledge whether test statistic T1 or T2 is more suitable for the analyzed study subject, we define the genome-wide test statistic to detect genetic outliers in rare-variant data as:
| (6) |
We already know that assuming no LD between the markers, and under the null hypothesis that the subject under study is from the given population, the standardized test statistics T1 and T2 follow a distribution asymptotically. To derive the asymptotic distribution of Topt, we need to incorporate the correlation between the test statistics T1 and T2. In the absence of LD between the genetic loci, an estimator of the correlation between R1 and R2 based on the allele frequencies of the loci can be easily derived (supplementary Note II). As an alternative approach or in the presence of LD, the correlation between R1 and R2 can also be estimated by the empirical correlation between the statistics R1 and R2 in the study (supplementary Note II). Given the estimate for the correlation/covariance of R1 and R2, the asymptotic distribution of Topt can be obtained under the null hypothesis, by simulating from a bivariate normal distribution with the estimated correlation. In supplementary Note II, we outline the derivation of the asymptotic distribution for Topt in more details.
Results
We examined the performance of the proposed test statistic Topt by its applications to the third version of 1000 Genome Project data, and in simulation studies with sequencing and GWAS data. In all applications and simulation scenarios, the approach was compared to the outlier detection algorithm based on PCA. For this comparison, we selected the smartpca implementation of PCA in the package EIGENSOFT version 3.0 [Price et al., 2006].
Applications to 1000 Genome Project Data
The 1000 Genome Project [The 1000 Genomes Project Consortium, 2010] data provide a unique framework to validate our approach based on real data. We applied the novel test to the third release of the variant call set based on both low coverage and exome whole-genome sequence data from the 1000 Genome Project [The 1000 Genomes Project Consortium, 2010]. The release contains the genotype calls of 1,092 samples from 14 different populations. We combined three pairs of populations to investigate the power, type I error, and family-wise error rate (FWER) of the test. The three pairs are Han Chinese in Beijing, China (CHB) and Japanese in Tokyo, Japan (JPT) (Fst = 0.007; supplementary information of Altshuler et al. [2010]), Tuscany in Italy (TSI) and Finnish from Finland (FIN; Fst = 0.020) [Nelis et al., 2009], Yoruba in Ibadan, Nigeria (YRI) and Luhya in Webuye, Kenya (LWK) (Fst = 0.008; supplementary information of Altshuler et al. [2010]). Since these populations can be considered to be genetically homogeneous they are an ideal validation tool for methodology to detect population substructure. The general idea is to create datasets that consist of one population, and include one additional subject that is not part of the population.
We focused only on the single-nucleotide polymorphisms (SNPs) calls, thus any information on the short Indels or large deletions was ignored. Quality control process has been implemented (autosomal SNPs with call rate > 98%, HWE P-value > 0.000001 that are not in the long-range LD regions [Price et al., 2008], and unrelated subjects with call rate > 98%), we are left with approximately 11M variants for the combined datasets CHB and JPT, and FIN and TSI, and with approximately 19M variants for the combined datasets LWK and YRI. To apply PCA, the three combined datasets, CHB and JPT, FIN and TSI, and LWK and YRI, have been pruned to include SNPs with MAF > 10% and with pairwise r2 < 0.05 in each 50 SNPs window with a step size of five SNPs. This pruned dataset for CHB and JPT includes about 92K SNPs, similar for FIN and TSI. The pruned dataset for LWK and YRI includes about 150K SNPs. To compare with the new test, PCA was also applied to the variants with MAF≤ 5% without any LD pruning. There are about 6M–7M such variants for the combined datasets of CHB and JPT, and of FIN and TSI, and there are about 13M such variants (MAF ≤ 5%) for the combined datasets of LWK and YRI.
In each application/replicate, we assess whether the two methods correctly identify the subject that is not part of the population as an outlier. A subject is rejected as an outlier if its test statistic Topt is greater than the value corresponding to the significance level 0.05/n where n is the number of subjects in the dataset. The type I error is the average percentage of incorrectly rejected subjects among the combined datasets for each scenario. The family-wise error rate (FWER) is the percentage of times that there is at least one incorrectly rejected subjects in the datasets. The methods were applied first to all the common SNPs, which, for PCA, are the pruned SNPs with minor allele frequency >10% and which, for our approach, are all the available SNPs, including rare SNPs and SNPs in the long-range LD regions [Price et al., 2008]. Then, we applied the two approaches to the rare SNPs (minor allele frequency < 5%).
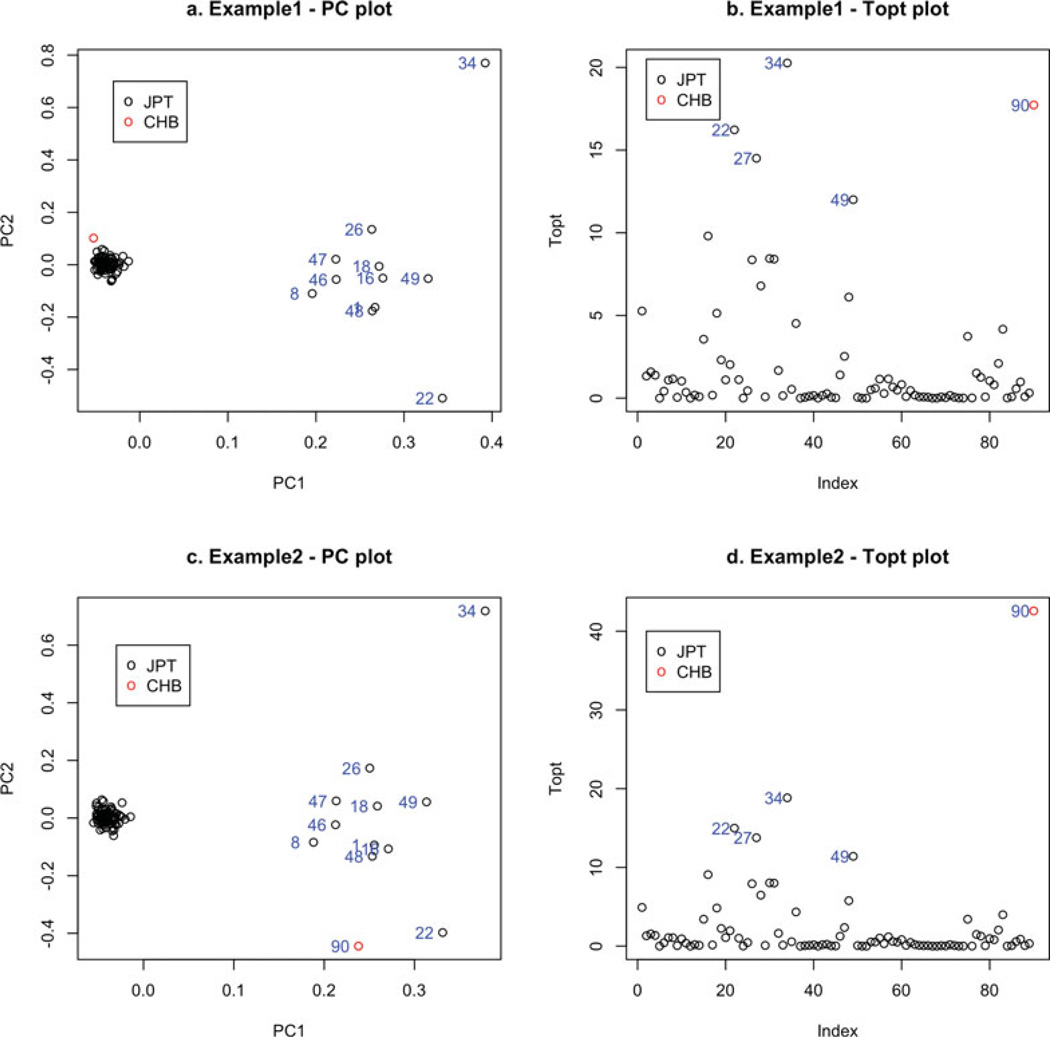
The power, type I error, and FWER estimates are shown in Table 1. For this table, we used the default values recommended in the package, i.e., 10 for the number of PCs used for determining outliers, and 6 for the number of standard deviations of which the subject must deviate in any of the top 10 PCs to be removed as an outlier. We also used the default maximum number of outlier removal iterations, which is 5 in the process. The table shows that PCA cannot detect the outlier using the pruned SNP set with MAF > 10% due to the small number of SNPs included in the pruned data and the closeness of the two populations. PCA has a good power to detect the outlier using SNPs with MAF ≤ 5%. However, the outlier detection algorithm based on PCA does not control for the type I error or the FWER, which would result in the unnecessary removal of samples. The new statistic Topt has a good power to detect the outliers, especially for the more distant pairs, TSI and FIN, and LWK and YRI. The type I error and the FWER are mostly controlled well except the case where the JPT population is combined with one CHB subject. In this scenario, there are three JPT subjects that are detected as outliers in most of the combined datasets due to the small genetic difference between JPT and CHB (Fst = 0.007; supplementary information of Altshuler et al. [2010]), and two examples are shown in Figure 1. We can see that in the examples, Topt is able to detect the outlier, but it also detected some CHB subjects as the outliers, whereas the outlier detection algorithm based on PCA applying to SNPs with MAF < 5% could not identify the CHB outlier, but rejects the JPT subject at index 34 (NA18978).
Table 1.
The estimated family-wise error rate (FWER), the average type I error (TI) and the power of Topt and the outlier detection process based on PCA when they were applied to the combined 1,000 genome datasets
| Estimates | Pop Outlier |
CHB JPT |
JPT CHB |
TSI FIN |
FIN TSI |
LWK YRI |
YRI LWK |
|---|---|---|---|---|---|---|---|
| PCA (MAF > 10%) | FWER | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 | 0.00 |
| TI | 0.00 | 0.00 | 0.00 | 0.00 | 0.0125 | 0.00 | |
| POWER | 0.00 | 0.00 | 0.151 | 0.00 | 0.0349 | 0.00 | |
| PCA (MAF < 5%) | FWER | 1.00 | 1.00 | 0.151 | 0.990 | 1.00 | 1.00 |
| TI | 0.144 | 0.0935 | 0.00351 | 0.0765 | 0.0489 | 0.0234 | |
| POWER | 0.843 | 0.443 | 0.957 | 0.888 | 0.570 | 1.00 | |
| Topt (MAF < 5%) | FWER | 0.00 | 1.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| TI | 0.00 | 0.0415 | 0.00 | 0.00 | 0.00 | 0.00 | |
| POWER | 0.146 | 0.495 | 1.00 | 1.00 | 0.988 | 1.00 | |
| Topt (all SNPs) | FWER | 0.0225 | 1.00 | 0.882 | 0.00 | 0.00 | 0.00 |
| TI | 0.00 | 0.0365 | 0.000869 | 0.00 | 0.00 | 0.00 | |
| POWER | 0.0562 | 0.0928 | 0.720 | 0.969 | 0.00 | 0.861 |
Figure 1.
PC plots and Topt plots for two randomly selected datasets with JPT subjects and one CHB subject. The outlier (one CHB subject) in both examples has an index of 90. Only SNPs with MAF < 5% were considered. The indices of some of the subjects are shown on the left of their points in the plots.
Note that there are a few surprises here. One is that the asymmetry in the power for the dataset of LWK with one YRI sample as the outlier and the dataset of YRI with one LWK as the outlier. This can be explained by the larger genetic variation of the LWK population than the YRI population. However, we observe that, using SNPs with MAF ≤ 5%, we have a much better power to detect the YRI outliers included in the LWK samples than using all the SNPs. This may be due to the fact that a lot of variants that contribute in distinguishing the two populations are rare since the separation of the two populations are relatively recent. Adding more common variants only adds noise to the difference and it overwhelms the information given by the rare variants. This suggests the importance of using SNPs with small MAF to distinguish genetically close populations. This is also true for the other combined datasets.
In practice, researchers would examine the PC plots manually to determine the outliers rather than using the default parameters and thresholds. However, due to the large number of simulated datasets we have, we are unable to do that for each combined datasets. To maximize the performance of the outlier detection algorithm based on PCA, we examined the effects of different parameters used in the outlier removal process based on PCA. Table 2 shows the effects of including different numbers of PCs on the power, type I error, and FWER to detect the outlier. The number of PCs to be used can be determined using the Tracy-Widom statistic in practice [Patterson et al., 2006]. There is one outlier included in each combined dataset and only SNPs with MAF < 5% are considered. We observe a decrease in the type I error rate as the number of PCs decreases, but it is still above the significance level (0.05/n where n is the number of subjects in the dataset). Also, we observe a decrease in the power to detect outliers and a constant FWER of 1.0 as the number of PCs decreases in most of the cases. Similarly, as the number of standard deviations used to determine the outlier decreases, the power increases but the type I error rate also increases. We also changed the number of iterations to 1 rather than using the default value 5, and we observe lower type I error in most of the cases, but the power is lower too (supplementary Table S1). Therefore, changing the parameters used in the outlier detection process based on PCA does not improve its overall performance in terms of both FWER and power comparing to Topt as Topt performs better in both aspects.
Table 2.
The estimated family-wise error rate (FWER), the average type I error (TI), and the power of the outlier detection process based on PCA when different numbers of principal components are used to determine the outliers. There is one outlier included in each combined dataset and only SNPs with MAF < 5% are used
| Pop | Outlier | Number of PC | 2 | 4 | 10 | 20 |
|---|---|---|---|---|---|---|
| CHB | JPT | FWER | 1.00 | 1.00 | 1.00 | 1.00 |
| TI | 0.010 | 0.064 | 0.144 | 0.147 | ||
| Power | 0.169 | 0.427 | 0.843 | 0.888 | ||
| JPT | CHB | FWER | 1.00 | 1.00 | 1.00 | 1.00 |
| TI | 0.044 | 0.057 | 0.0935 | 0.120 | ||
| Power | 0.289 | 0.340 | 0.443 | 0.567 | ||
| TSI | FIN | FWER | 0.118 | 0.118 | 0.151 | 0.151 |
| TI | 0.0024 | 0.00318 | 0.00351 | 0.00351 | ||
| Power | 0.785 | 0.946 | 0.957 | 0.957 | ||
| FIN | TSI | FWER | 0.418 | 0.929 | 0.990 | 1.00 |
| TI | 0.0045 | 0.0106 | 0.0765 | 0.0856 | ||
| Power | 0.398 | 0.827 | 0.888 | 0.929 | ||
| LWK | YRI | FWER | 1.00 | 1.00 | 1.00 | 1.00 |
| TI | 0.038 | 0.0489 | 0.0489 | 0.0511 | ||
| Power | 0.00 | 0.00 | 0.570 | 0.733 | ||
| YRI | LWK | WER | 1.00 | 1.00 | 1.00 | 1.00 |
| TI | 0.0233 | 0.0233 | 0.0234 | 0.0234 | ||
| Power | 1.00 | 1.00 | 1.00 | 1.00 |
We further investigated the performance of the test by introducing more than one outliers into the dataset. We randomly selected 5 or 10 outliers from the outlier population and they were combined with the corresponding study population to assess the performance of the approaches. There are 1,000 such randomly generated datasets in all the scenarios except for evaluating the performance of PCA on variants with MAF ≤ 5%. We observe that even with more outliers included in the dataset, our method performs generally better than PCA, especially in the combined datasets of TSI and FIN, and LWK and YRI. PCA continues to have a large type I error rate and FWER in all the scenarios. We also observe again that the performance of the novel test is much better using the SNPs with MAF≤ 5%, than using all the SNPs. The results are shown in supplementary Tables S2 and S3.
Note that as the proportion of outliers included in the dataset continues to increase, the power of our test decreases. This is because that as more outliers are included in the dataset, the estimated MAF and the expected number of alleles obtained from the data are biased toward the outlier population. Then the test statistics would be biased and would not follow the same distribution as under the null hypothesis Thus, the novel method is mainly used to detect a relatively small set of outliers. The effect of the proportion of the outliers included in the dataset on the test statistics also depends on the genetic distance of the populations.
Simulations
In addition to the analyses on the 1000 Genome Project data, we performed simulation studies under the alternative hypothesis to examine whether the proposed test Topt has sufficient power to detect outliers. We assessed the power of the approach to detect genetic outliers based on both rare variants and common variants. In our simulations, the Balding-Nichols model [Balding and Nichols, 1995] was applied to generate the allele frequencies of the two subpopulations: , where pi1, pi2 are the allele frequencies of marker locus i for the two subpopulations; F is the Fst, the genetic distance between the two subpopulations [Holsinger and Weir, 2009]; and the parameter pi is the background allele frequency for marker locus i. In the simulation studies for the rare variants, the background allele frequencies pi were generated from Wright’s distribution [Wright, 1949] using Metropolis-Hastings algorithm: f (p) = cpβs −1(1 − p)βn−1eσ(1−p), where the scaled mutation rates are elected to be βs = 0.001, βn = βs/3, the selection rate σ = 12, and c is a normalizing constant. Wright’s distribution is expected to simulate the MAF spectrum of the human genome, where most of the MAFs generated are smaller than 5% [Pritchard, 2001]. To generate common variant data, we generated the background allele frequencies pi from the Uniform distribution Unif (0, 0.5).
Under the models defined by these parameters, we draw two sets of allele frequencies for the two subpopulations in each trial. In analogy to the 1000 Genome Project analysis, one study subject was generated from the first subpopulation, while the remaining study subjects in the dataset were generated from the second subpopulation.
Power
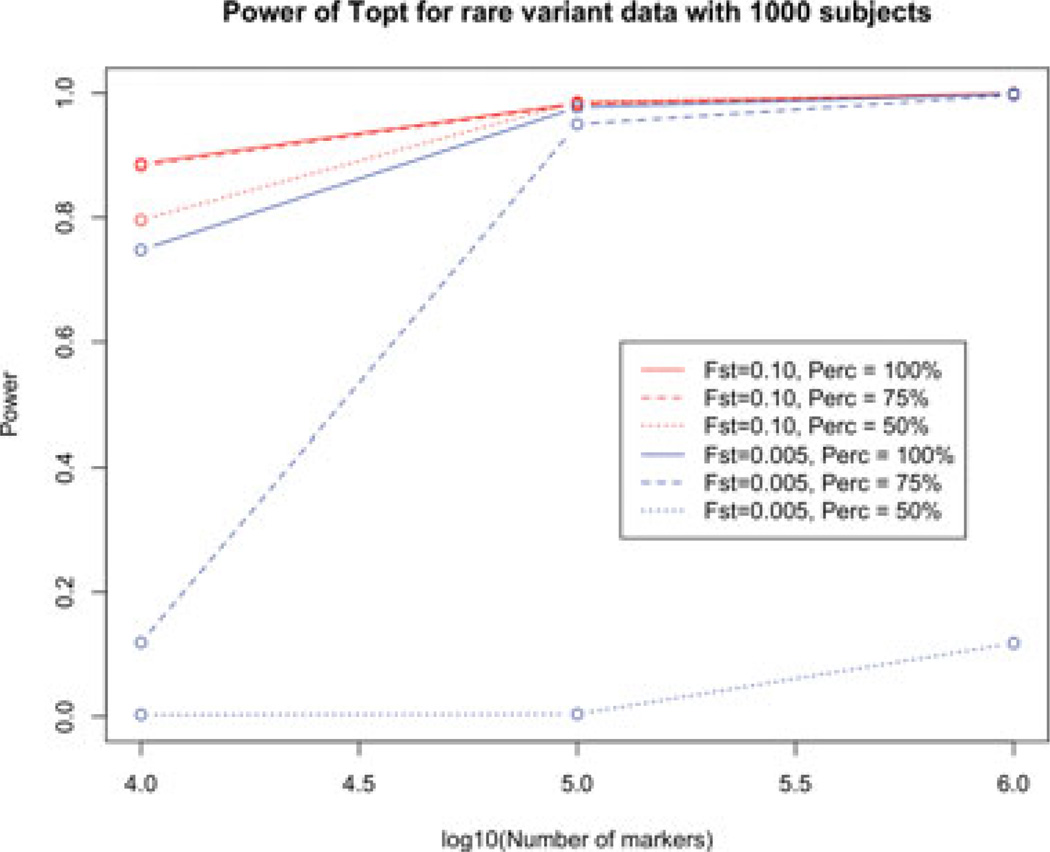
Based on 1,000 replicates, we estimated the power of the test under each scenario for both common variant and rare-variant data. The power is estimated by the percentage of trials in which the outlier is detected using Topt, where the test statistic is adjusted for multiple comparisons, i.e., study subjects, using the Bonferroni correction. The results are shown in Table 3 and supplementary Table S4. Table 3 suggests that the test Topt has sufficient power in most of the scenarios for rare-variant data, especially for datasets with 1 million markers, as shown in Figure 2. This is expected since, as the number of markers increases, there is more information about the genetic structure of the population, and it is easier for the test to capture any small difference between the outlier and the rest of the subjects in the dataset. The genetic distance of the two subpopulations, Fst, is varied over a wide range, and we observe a decrease in the power of the test as Fst decreases, as we would expect. Also, we find that the percentage of markers with smaller MAF in the first population than in the second population also influences the power of the test, especially when the two populations are genetically close to each other. However, after assessing the power for the percentage between 50% and 75% (data not shown), we found that, as long as the percentage is above 50%, i.e., there is LD in the sample, we have a good power to detect the outlier under the scenarios we considered.
Table 3.
Power of Topt for rare-variant data
| Fst | Percentage | 500 × 10k | 1,000 × 10k | 500 × 100k | 1,000 × 100k | 500 × 1M | 1,000 × 1M |
|---|---|---|---|---|---|---|---|
| 0.20 | 100 | 0.898 | 0.905 | 0.991 | 0.990 | 0.998 | 1.000 |
| 75 | 0.879 | 0.867 | 0.990 | 0.995 | 0.997 | 0.998 | |
| 50 | 0.914 | 0.870 | 0.988 | 0.986 | 1.000 | 1.000 | |
| 0.15 | 100 | 0.904 | 0.900 | 0.983 | 0.987 | 1.000 | 0.998 |
| 75 | 0.854 | 0.857 | 0.989 | 0.954 | 0.999 | 0.998 | |
| 50 | 0.880 | 0.856 | 0.987 | 0.987 | 1.000 | 1.000 | |
| 0.10 | 100 | 0.894 | 0.887 | 0.988 | 0.984 | 1.000 | 1.000 |
| 75 | 0.859 | 0.833 | 0.981 | 0.982 | 0.998 | 0.999 | |
| 50 | 0.835 | 0.796 | 0.981 | 0.986 | 1.000 | 0.999 | |
| 0.05 | 100 | 0.878 | 0.875 | 0.980 | 0.983 | 0.997 | 0.996 |
| 75 | 0.807 | 0.777 | 0.973 | 0.974 | 0.998 | 0.997 | |
| 50 | 0.388 | 0.354 | 0.967 | 0.970 | 0.998 | 0.996 | |
| 0.01 | 100 | 0.828 | 0.825 | 0.973 | 0.979 | 0.999 | 0.999 |
| 75 | 0.453 | 0.401 | 0.968 | 0.963 | 0.995 | 0.997 | |
| 50 | 0.003 | 0.006 | 0.031 | 0.025 | 0.863 | 0.839 | |
| 0.005 | 100 | 0.757 | 0.748 | 0.987 | 0.977 | 0.997 | 0.999 |
| 75 | 0.183 | 0.119 | 0.947 | 0.950 | 0.993 | 0.997 | |
| 50 | 0.005 | 0.003 | 0.002 | 0.004 | 0.149 | 0.118 |
The number of subjects in the datasets is either 500 or 1,000. The number of SNPs included is 10,000, 100,000, or 1 million. The first column refers to the genetic distance of the two subpopulations in the dataset. The second column shows the percentage of the markers with a smaller MAF in the first subpopulation than in the second subpopulation.
Figure 2.
The power of Topt for rare-variant data with 1,000 subjects at F st = 0.1 and 0.005 with the three percentage levels as a function of the number of SNPs included in the dataset, which are 10k, 100k, and 1 million.
For common variant data, the power of the test becomes very small when the percentage of markers with smaller MAF in one population than in the other population is approximately 50%, even for large number of SNPs. However, as the percentage increases, the power increases rapidly. This is due to the small power offered by the score S2 since S1 does not have much power under the 50% scenario. It is important to note that, in a real dataset, we would not expect the percentage to be exactly 50% if all available genetic loci are included in the calculation of the test statistic and there is LD between the loci.
We also compared our approach and the PCA approach for rare-variant data, as shown in Table 4. We observe that both the proposed test statistic and the outlier detection algorithm based on PCA have sufficient statistical power in most scenarios. However, when there is a systematic difference in allele frequencies between the two populations, the PCA approach does not perform well. In practice, this effect on PCA can be minimized by the removal of long-range LD regions and LD-pruning for common variant analysis, but would be unavoidable for sequence data. Note that there are only 10k SNPs included in the simulation due to the computational cost of PCA, and the power of Topt increases dramatically as the number of SNPs included increases, whereas the results of the PCA analysis would be biased since LD would be introduced as the number of SNPs included increases.
Table 4.
Power of Topt and the outlier detection process based on PCA for rare-variant data
| 500 subjects × 10k | 1,000 subjects × 10k | ||||
|---|---|---|---|---|---|
| Fst | Percentage | PCA | Topt | PCA | Topt |
| 0.2 | 100 | 0.05 | 0.93 | 0.00 | 0.90 |
| 75 | 0.90 | 0.84 | 0.90 | 0.87 | |
| 50 | 0.93 | 0.87 | 0.93 | 0.87 | |
| 0.15 | 100 | 0.06 | 0.92 | 0.05 | 0.87 |
| 75 | 0.86 | 0.88 | 0.90 | 0.79 | |
| 50 | 0.94 | 0.90 | 0.96 | 0.92 | |
| 0.10 | 100 | 0.03 | 0.96 | 0.05 | 0.88 |
| 75 | 0.65 | 0.84 | 0.67 | 0.81 | |
| 50 | 0.92 | 0.80 | 0.94 | 0.86 | |
| 0.05 | 100 | 0.03 | 0.89 | 0.02 | 0.88 |
| 75 | 0.20 | 0.80 | 0.38 | 0.77 | |
| 50 | 0.90 | 0.33 | 0.94 | 0.28 | |
| 0.01 | 100 | 0.04 | 0.84 | 0.04 | 0.78 |
| 75 | 0.05 | 0.37 | 0.04 | 0.37 | |
| 50 | 0.37 | 0.00 | 0.48 | 0.00 | |
| 0.005 | 100 | 0.00 | 0.73 | 0.00 | 0.73 |
| 75 | 0.03 | 0.15 | 0.07 | 0.12 | |
| 50 | 0.10 | 0.00 | 0.19 | 0.01 | |
The ancestral MAFs are generated from Wright’Õs distribution for the datasets. The number of SNPs included is 10,000.
Type I Error
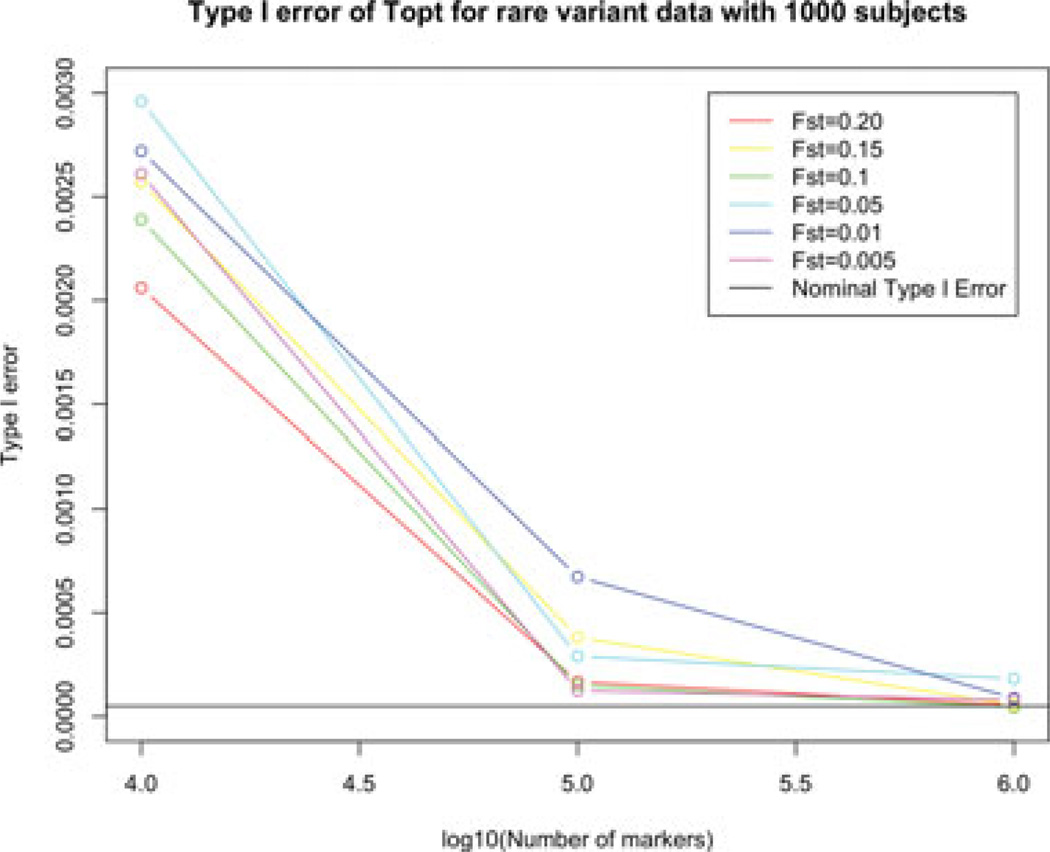
Using the same set of simulated data, the type I error is estimated as the average percentage of subjects who are incorrectly rejected. Part of the results for the rare variants is shown in Figure 3, and the complete results are in supplementary Table S5. In the scenarios considered, the nominal type I error is 0.05/n, where n is the number of subjects included in the dataset, to maintain the FWER at 0.05 level. Thus, the nominal type I error is 0.0001 for 500 subjects and 0.00005 for 1,000 subjects. From the results, we observe that for rare SNPs, the type I error for 10,000 SNPs is inflated, but for datasets with a large number of SNPs, the type I error rate is acceptable. For common variants, the type I error is well maintained in all the scenarios (data not shown).
Figure 3.
The type I error rate of Topt for rare-variant data with 1,000 subjects at six different Fst values with the percentage level 75% as a function of the number of SNPs included in the dataset, which are 10k, 100k, and 1 million.
The same pattern is observed for the FWER as for the type I error rate. FWER is estimated as the number of trials among 1,000 trials such that at least one subject is wrongly rejected. For common variant data, the FWER is well below 0.05 for all the scenarios. For rare-variant data, we do see an inflation in the FWER as the type I error rate when the number of SNPs included in the dataset is small. However, in real dataset, e.g., whole-exome sequencing data, GWAS, etc., we expect that a sufficient number of loci is available to guarantee that the FWER is maintained.
As a last comparison, we assessed the performance of both approaches under the null hypothesis. As shown in Table 5, for rare variants generated from Wright’s distribution, PCA has much larger FWER compared to Topt. In almost all the trails, PCA rejected at least one subject incorrectly, whereas Topt has been shown above that the FWER is acceptable when the number of SNPs included is sufficiently large. For common variants, the FWER of both approaches is well maintained with 500 or 1,000 subjects included in the datasets.
Table 5.
FWER of Topt and the outlier detection process based on PCA for rare-variant data
| 500 × 10k | 1,000 × 10k | 500 × 100k | 1,000 × 100k | |||||
|---|---|---|---|---|---|---|---|---|
| FWER dist | PCA | Topt | PCA | Topt | PCA | Topt | PCA | Topt |
| Wright | 0.912 | 0.106 | 0.978 | 0.123 | 0.928 | 0.066 | 0.989 | 0.060 |
Discussion
The large-scale applications of next-generation sequencing technology to association studies require the development of robust and powerful analysis approaches. While substantial progress has been made in terms of the development of association tests for rare variants [Ionita-Laza et al., 2011; Li and Leal, 2008; Madsen and Browning, 2009; Mukhopadhyay et al., 2010; Neale et al., 2011], there is yet no standard statistical approach that addresses the issues of population substructure for sequence data.
Recently, a permutation procedure is proposed by Epstein et al. [2012] to address the problem in association tests of rare variations. It provides the option for the rare-variant association tests that cannot correct for confounding to adjust for covariates. However, it is subject to the same problem as other rare-variant association tests that may be adjusted for ancestry due the fact that the ancestry covariates obtained using PCA may not be accurate as the type I error of the association tests after adjusting for ancestry using PCA has been shown to be still inflated under certain scenarios [Mathieson and McVean, 2012]. Here, we try to approach the problem from a different direction, by obtaining a homogeneous subpopulation to remove confounding and avoid the hassle of estimating the ancestry covariates for rare variants. In this communication, we proposed a method that can detect study subject that introduce population substructure in the sample, potentially confounding the association analysis. Our approach is computationally fast and simple, i.e., the method is computed based on all available genetic loci, making LD estimation and pruning unnecessary. This is especially useful for data from Exome chips or disease-specific fine-mapping chips, in which case the pruning of the SNPs would not be desirable, and thus PCA would give biased results. The approach works well for both rare and common variants. We illustrated this by the applications to the 1000 Genome Project data, and in our simulation studies.
While these are the advantages over the standard PCA analysis for rare-variant data, our approach assumes that most of the subjects in the dataset are from the same population and there is only a small proportion of outliers included. Also, our method does not assess the pairwise similarity of study subjects, e.g., PC plots. This restricts our approach to the role of an outlier detection tool. Unlike PCs, an integration of the test statistic into a regression model as an adjustment for population substructure is problematic for this reason. Additional research on this topic is required.
Supplementary Material
Acknowledgments
We would like to acknowledge the generous support from the Department of Biostatistics, Harvard School of Public Health. Also, we thank A. L. Price for his generous and helpful comments on the topic. The project described was supported by award numbers (R01MH081862, R01MH087590) from the National Institute of Mental Health and award numbers (U01HL089856, U01HL089897) from the National Heart, Lung, and Blood Institute. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Mental Health or the National Heart, Lung, and Blood Institute.
Footnotes
Supporting Information is available in the online issue at wileyonlinelibrary.com.
References
- Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Bonnen PE, de Bakker PI, Deloukas P, Gabriel SB, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467(7311):52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balding DJ, Nichols RA. A method for quantifying differentiation between populations at multi-allelic loci and its implications for investigating identity and paternity. Genetica. 1995;96(1–2):3–12. doi: 10.1007/BF01441146. [DOI] [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55(4):997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Epstein MP, Duncan R, Jiang Y, Conneely KN, Allen AS, Satten GA. A permutation procedure to correct for confounders in case-control studies, including tests of rare variation. Am J Hum Genet. 2012;91:215–223. doi: 10.1016/j.ajhg.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Zhang X, Ding L, Baye TM, Kurowski BG, Martin LJ. Effect of population stratification analysis on false-positive rates for common and rare variants. BMC Proc. 2011;5(Suppl 9):S116. doi: 10.1186/1753-6561-5-S9-S116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsinger KE, Weir BS. Genetics in geographically structured populations: defining, estimating and interpreting F(ST) Nat Rev Genet. 2009;10(9):639–650. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionita-Laza I, Buxbaum JD, Laird NM, Lange C. A new testing strategy to identify rare variants with either risk or protective effect on disease. PLoS Genet. 2011;7(2):e1001289. doi: 10.1371/journal.pgen.1001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83(3):311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca D, Ringquist S, Klei L, Lee AB, Gieger C, Wichmann HE, Schreiber S, Krawczak M, Lu Y, Styche A, et al. On the use of general control samples for genome-wide association studies: genetic matching highlights causal variants. Am J Hum Genet. 2008;82(2):453–463. doi: 10.1016/j.ajhg.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5(2):e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson I, McVean G. Differential confounding of rare and common variants in spatially structured populations. Nat Genet. 2012;44(3):243–246. doi: 10.1038/ng.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9(5):356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- McVean G. A genealogical interpretation of principal components analysis. PLoS Genet. 2009;5(10):e1000686. doi: 10.1371/journal.pgen.1000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay I, Feingold E, Weeks DE, Thalamuthu A. Association tests using kernel-based measures of multi-locus genotype similarity between individuals. Genet Epidemiol. 2010;34(3):213–221. doi: 10.1002/gepi.20451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Rivas MA, Voight BF, Altshuler D, Devlin B, Orho-Melander M, Kathiresan S, Purcell SM, Roeder K, Daly MJ. Testing for an unusual distribution of rare variants. PLoS Genet. 2011;7(3):e1001322. doi: 10.1371/journal.pgen.1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelis M, Esko T, MŠgi R, Zimprich F, Zimprich A, Toncheva D, Karachanak S, Piskackova T, Balascak I, Peltonen L, et al. Genetic structure of Europeans: a view from the NorthD-East. PLoS ONE. 2009;4(5):e5472. doi: 10.1371/journal.pone.0005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J, Stephens M. Interpreting principal component analyses of spatial population genetic variation. Nat Genet. 2008;40(5):646–649. doi: 10.1038/ng.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2(12):e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Price AL, Weale ME, Patterson N, Myers SR, Need AC, Shianna KV, Ge D, Rotter JI, Torres E, Taylor KD, et al. Long-range LD can confound genome scans in admixed populations. Am J Hum Genet. 2008;83(1):132–135. doi: 10.1016/j.ajhg.2008.06.005. author reply 135-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Zaitlen NA, Reich D. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 2010;11:459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK. Are rare variants responsible for susceptibility to complex diseases? Am J Hum Genet. 2001;69(1):124–137. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, Lavery T, Kouyoumjian R, Farhadian SF, Ward R, et al. Linkage disequilibrium in the human genome. Nature. 2001;411(6834):199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- Reich DE, Goldstein DB. Detecting association in a case-control study while correcting for population stratification. Genet Epidemiol. 2001;20(1):4–16. doi: 10.1002/1098-2272(200101)20:1<4::AID-GEPI2>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Roy-Gagnon MH, Moreau C, Bherer C, St-Onge P, Sinnett D, Laprise C, Vezina H, Labuda D. Genomic and genealogical investigation of the French Canadian founder population structure. Hum Genet. 2011;129(5):521–531. doi: 10.1007/s00439-010-0945-x. [DOI] [PubMed] [Google Scholar]
- Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445(7130):881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- Stranger BE, Stahl EA, Raj T. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics. 2011;187(2):367–383. doi: 10.1534/genetics.110.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium. Amap of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Adaptation and selection. In: Jepson G, Simpson G, Mayr E, editors. Genetics, Paleontology and Evolution. Princeton: Princeton University Press; 1949. pp. 365–389. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.