Summary
Genome sequencing projects are discovering millions of genetic variants in humans, and interpretation of their functional effects is essential for understanding the genetic basis of variation in human traits. Here we report sequencing and deep analysis of mRNA and miRNA from lymphoblastoid cell lines of 462 individuals from the 1000 Genomes Project – the first uniformly processed RNA-seq data from multiple human populations with high-quality genome sequences. We discovered extremely widespread genetic variation affecting regulation of the majority of genes, with transcript structure and expression level variation being equally common but genetically largely independent. Our characterization of causal regulatory variation sheds light on cellular mechanisms of regulatory and loss-of-function variation, and allowed us to infer putative causal variants for dozens of disease-associated loci. Altogether, this study provides a deep understanding of the cellular mechanisms of transcriptome variation and of the landscape of functional variants in the human genome.
Introduction and data set
Interpreting functional consequences of millions of discovered genetic variants is one of the biggest challenges in human genomics1. While genome-wide association studies have linked genetic loci to various human phenotypes and the functional annotation of the genome is improving,2,3, we still have limited understanding of the underlying causal variants and biological mechanisms. One approach to address this challenge has been to analyze variants affecting cellular phenotypes, such as gene expression,4–8 known to affect many human diseases and traits.9,10
In this study, we characterize functional variation in human genomes by RNA-sequencing hundreds of samples from the 1000 Genomes project1, the most important reference data set of human genetic variation, thus creating the biggest RNA sequencing data set of multiple human populations to date. We not only catalogue novel loci with regulatory variation but also, for the first time, discover and characterize molecular properties of causal functional variants.
We performed mRNA and small RNA sequencing on lymphoblastoid cell line (LCL) samples from 5 populations: the CEPH (CEU), Finns (FIN), British (GBR), Toscani (TSI) and Yoruba (YRI). After quality control, we had 462 and 452 individuals (89–95 per population) with mRNA and miRNA data, respectively (Fig. S1–11, Table S1). Of these, 421 are in the 1000 Genomes Phase 1 dataset1, and the remaining were imputed from SNP array data (Fig. S3, Table S2). RNA-seq was performed in seven laboratories, and the smaller amount of variation between laboratories than individuals demonstrated that RNA sequencing is a mature technology ready for distributed data production (MW p < 2.2 × 10−16 for mRNA, p = 1.34 × 10−10 for miRNA; Fig. 1a, S11;11). To discover genetic regulatory variants, we mapped cis-QTLs to transcriptome traits of protein-coding and miRNA genes separately in the European (EUR) and Yoruba (YRI) populations (Fig. S12, Table S3, Table 1). The RNA-seq read, quantification, genotype and QTL data are available open-access (see Data Access section).
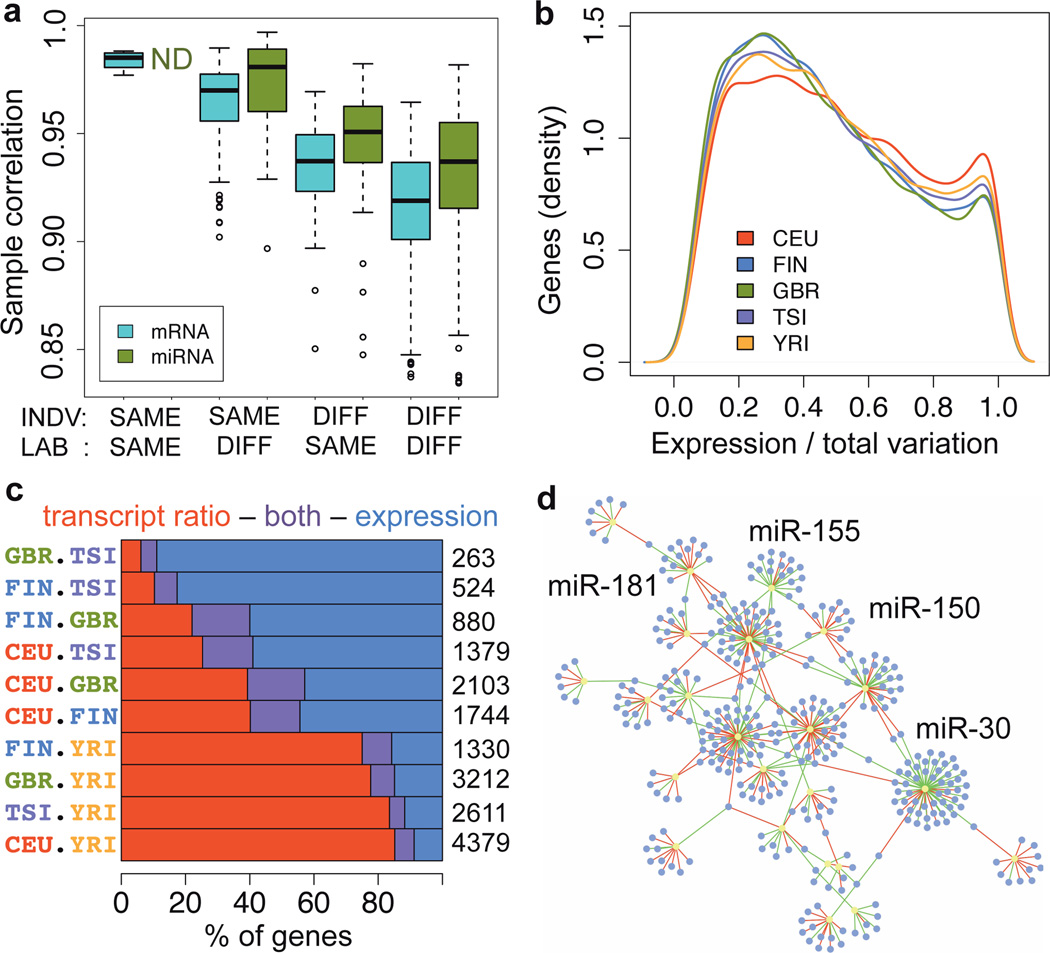
Figure 1. Transcriptome variation.
a) Spearman rank correlation of replicate samples, based on mRNA exon and miRNA quantifications of 5 individuals sequenced 8 and 7 times for mRNA and miRNA, respectively, and separated by the individual or the sequencing lab being the same or different. The quantifications have been normalized only for the total number of mapped reads (see Fig. S11 for correlations after normalization). b) The proportion of expression level variation (as opposed to splicing) of the total transcription variation between individuals in each population, measured per gene. c) Proportion of genes with differential expression levels and/or transcript usage between population pairs, out of the total listed on the right-hand side. d) Network of significant miRNA families (P<0.001; yellow) and their significantly associated mRNA targets (P<0.05; purple). The edges display negative (green) and positive (red) associations.
Table 1.
Numbers of transcriptome features with a QTL (FDR 5%)
| Total | EUR (n=373) | YRI (n=89) | Union | |
|---|---|---|---|---|
| exon eQTL | 12981 genes | 7390 | 2369 | 7825 |
| gene eQTL | 13703 genes | 3259 | 501 | 3773 |
| transcript ratio QTL | 7855 genes | 620 | 83 | 639 |
| mirQTL | 644 miRNAs | 57 | 15 | 60 |
| Transcribed repeat eQTL | 43875 repeats | 5763 | 1055 | 6069 |
Transcriptome variation in populations
This first uniformly processed RNA-seq data set from multiple human populations allowed high-resolution analysis of transcriptome variation. Individual and population differences in transcription can manifest in (1) overall expression levels, and (2) relative abundance of transcripts from the same gene (transcript ratios). Deconvolution of the relative contribution of these12 indicates that this ratio is characteristic for each gene with transcript ratio being on average more dominant (Fig. 1b, Fig.S13, S14). Population differences explain a small but significant proportion of 3% of total variation (MW p < 2.2 × 10−16). In addition to this genome-wide perspective to population variation, we identified 263–4379 genes with differential expression and/or transcript ratios between population pairs (PGF, JM, MGP, MB, TL, TW, MRF, A Guin, MAR, TGC, PR, ETD, RG, MS, submitted). Interestingly, continental differences between YRI-EUR population pairs have much higher contribution of genes with different transcript usage than European population pairs (75–85% versus 6–40%; Fig. 1c, Fig. S14). This has not been observed before in humans, but it is consistent with splicing patterns capturing phylogenetic differences between species better than expression levels13,14.
We quantify a total of 644 autosomal miRNAs in >50% individuals of which 60 have significant cis-mirQTLs for miRNA expression (Fig. S15, Table 1), showing that genetic effects on miRNA expression are much more widespread than the previously identified loci15. To complement previous studies of miRNA function in cell perturbation experiments, we analyzed miRNA-mRNA interaction in our steady-state population sample. Of 100 miRNA families, 32 correlated with the expression of predicted target exons in a highly connected network (P<0.001, Fig. 1d, Table S4), including miRNA families with important immunological or lymphocyte functions, such as miR-150, miR-155, miR-181, and miR-14616. Interestingly, 45% of the associations were positive – consistent with previous results15 – even though based on knockout experiments miRNAs mostly downregulate genes. Analyzing the direction of causality, cis-mirQTLs had small trans-eQTL effects to predicted targets only when effects were negative (pi1 = 0.11 versus pi1 = 0, Fig.S16), suggesting that miRNAs indeed downregulate their targets. Positive correlations may be driven by other effects, which is supported by overrepresentation of transcription factors in the network (29%, Fisher p= 2.1 × 10−7 for negative targets and 26% p=4.0 × 10−4 for positive targets). This suggests feedback loops of both mRNA and miRNA genes affecting the expression of each other, and supports the idea that under steady-state conditions miRNAs confer robustness to expression programs17. Altogether, these results highlight the added insight into the role of miRNAs in regulatory networks from analysis of population variation.
Genetic effects on the transcriptome
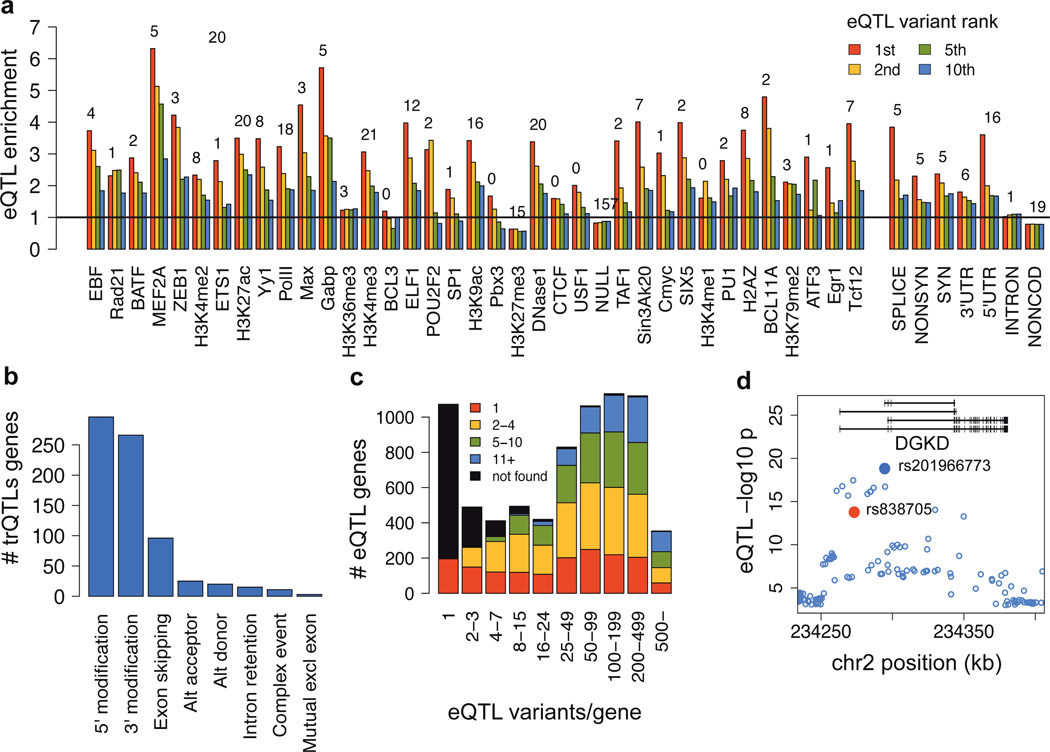
Expression QTL (eQTL) analysis of protein-coding and lincRNA genes uncovered extremely widespread regulatory variation, with 3,773 genes having a classical eQTL for gene expression levels (Table 1). While the potential of RNA-seq to discover other transcriptome traits such as splicing variation is widely known7,8,18–20, a comprehensive analysis has been lacking. To this end, we first mapped eQTLs for exon quantifications that can capture both gene expression and splicing variation, discovering as many as 7,825 genes with an eQTL, referred to as eQTLs in this paper unless otherwise specified. Regressing out the most significantly associated variant from the EUR eQTL analysis showed that as many as 34% of the genes have a second, independent eQTL for any of their exons (of which 7% for the exon of the first association). Thus, there is substantial allelic heterogeneity for regulatory effects on a single gene and independence of exons of the same gene (Fig.S17), To investigate genetic effects specifically on splicing, we discovered 639 genes with transcript ratio QTLs (trQTLs) affecting the ratio of each transcript to the gene total – the largest number of genetic effects on transcript structure identified to date. The lower number relative to gene eQTLs is likely caused by higher noise in model-based transcript quantifications than in gene counts. To characterize the relationship of genetic variants affecting expression versus splicing, we regressed out the best trQTL variant from the gene eQTL analysis in 279 genes with both types of QTLs. The results showed that the causal variants are independent in ≥57% of these genes (Fig. S18), suggesting that transcriptional activity and transcript usage are usually controlled by different regulatory elements of the genome.
The transcript differences driven by trQTLs involve exon skipping only in 15% of genes, with as much as 48% and 43% varying in 5’ and 3’ ends, respectively (in EUR; categories not mutually exclusive; Fig. 2b). To further analyze transcript modifications through unannotated transcript elements, we mapped cis-eQTL for expressed retrotransposon-derived elements (repeat elements) outside genes, known to be an important source for evolution of new transcripts.21 We detected widespread sharing between the 5,763 cis-eQTLs discovered for repeat elements (Fig. S19, Table 1) and nearby exon eQTLs: of the best repeat eQTLs variants in EUR, 49% were significant and 6% the top eQTLs variants for exons of a nearby gene (3.8× and 26× enrichment; Fisher p<2.2 × 10−16). This suggests that retrotransposon-derived elements can share regulatory elements with nearby genes. These results provide the first genome-wide characterization of genetic effects on transcript structure through annotated and unannotated 3’ and 5’ changes, which may predominate exon skipping that previous studies have focused on19. This opens new perspectives for understanding their cellular and high-level effects, as end modifications will rarely change protein structure but may affect post-transcriptional regulation.
Figure 2. Transcriptome QTLs.
a) Enrichment of EUR exon eQTLs in functional annotations for the 1st, 2nd, 5th and 10th best associating eQTL variant per gene, relative to a matched null set of variants denoted by the horizontal line. The numbers are −log10 p-values of a Fisher test between the best eQTL and the null. b) Classification of changes caused by transcript ratio QTLs. c) The rank of the best Omni2.5M SNP among the significant EUR eQTL variants per gene. d) DGKD gene locus where an intronic SNP rs838705 is associated to calcium levels (red), and the top eQTL variant 21 kb downstream (blue) is a very likely causal variant, close the TSS of two transcripts in the MEF2A,C binding region.
Altogether, we present the largest and the most diverse catalog of cis-regulatory variants discovered in a single tissue to date. The majority of the analyzed genes – 8,329 out of 13,970 – have one or several QTLs for different transcript traits, a resolution enabled by in-depth analysis of high-quality transcriptome and genome sequencing data. These results highlight both allelic heterogeneity of regulatory variants and phenotypic heterogeneity of diverse transcriptome traits of individual genes.
Properties of regulatory variants
To understand how eQTLs affect gene expression, we compared the properties of the top (most significant) eQTL variant per gene to a null of non-eQTL variants (matched for distance from TSS and minor allele frequency). The best eQTL variant may not always be the causal variant due to noise in genotype and phenotype data, and to estimate our ability to pinpoint causal variants, we contrasted the properties of the 1st eQTL to the 2nd, 5th and 10th best eQTL variants (Fig. 2a).
First, comparing the eQTL with the best p-value to the matched null showed an enrichment of indels among top eQTLs (13% = 1.22× enrichment; Fisher p = 1.9 × 10−3 in EUR; Fig.S20), suggesting that indels are more likely to have functional effects than SNPs. eQTLs are highly enriched in several noncoding elements from the Ensembl Regulatory Build, such as many transcription factor peaks (median enrichment 3.3×, median p = 0.009 in EUR; Fig. 2a, S21), DNase1 hypersensitive sites (3.4×, p = 1.00 × 10−20), as well as in chromatin states of active promoters (3.5×, p = 1.08 × 10−36) and strong enhancers (median 2.4×, median p = 1.14 × 10−5). Within genes, splice-site (3.8×, p = 1.65 × 10−5) and nonsynonymous (2.3×, p = 4.84 × 10−6) enrichments point to putative regulatory functions of coding variants. Transcript ratio QTLs are overrepresented in splice sites (6.8×, p = 2.44 × 10−7), as expected, but also for example in 3’UTRs (2.5×, p = 1.83 × 10−6; Fig. S22) and promoters (2.4×, p = 5.79 × 10−6). Altogether, the higher resolution of annotations and eQTLs relative to previous studies22,23 provides important insight into the role of individual transcription factors and other regulatory elements mediating genetic regulatory effects.
Functional enrichment typically decreases rapidly from the best eQTL variant towards lower ranks. To estimate how often the first variant is likely to be the causal regulatory variant, we calculated the annotation enrichment of the best eQTL variants relative to the null for (1) all eQTL loci, and (2) loci where the best eQTL variant is very likely causal due to having a log10 p-value >1.5 higher than the second variant (Fig.S23). The ratio of the enrichments (1) and (2) yields an approximation of the best variant being causal in 55% of EUR and 74% of YRI eQTLs, with more conservative estimates being 34% and 41%, respectively (Fig.S23). Thus, we have reasonable power to pinpoint causal regulatory variants from unbiased p-value distributions alone without annotation priors23. This is enabled by not relying on SNP array data22: in 81% of the cases the best variant is not on the Omni 2.5M chip (Fig.2c, Fig.S25). Validating the putative causal effects, we observed that the best eQTL variants in CTCF peaks showed more allele-specific binding compared to matched null variants (p = 2.0 × 10−3, Fig.S24) in CTCF ChIP-seq data from 6 individuals24, and the best eQTLs were enriched in DNase1 hypersensitivity QTLs25 (3.3×, p = 2.51 × 10−6 in EUR, 7.9×, p < 2.2 × 10−16 in YRI). In conclusion, we not only identify broad eQTL loci but also substantially increase our confidence to pinpoint individual causal variants and their functional mechanisms.
Of the 6,473 variants in the GWAS catalog26, 16% are eQTLs and 1.8% are trQTLs in EUR or YRI, but a high overlap is observed also by chance for a frequency-matched GWAS null (11% and 0.84%, respectively). The modest (albeit significant: eQTL chi2 p < 2.2 × 10−16; trQTL p = 7.2 × 10−9) enrichment9,10 is due to eQTLs being very ubiquitous, and consequently, a GWAS variant being an eQTL does not mean that the regulatory change is necessarily driving the disease association. Our data offers a unique opportunity to address the key question of whether the causal eQTL variant is also causal for the disease. The enrichment of GWAS SNPs in the top eQTL ranks (p=1.18 × 10−7; Fig. S26) is a genome-wide signal of shared causality. To further characterize individual loci, we selected 78 eQTL regions that are likely causal signals for 91 GWAS SNPs (estimated by the RTC method),6,9, and in these loci our best eQTL variant is the putative disease-causing variant (Fig.S27, Table S5). Figure 2d shows an example of the DGKD gene where an intronic SNP rs838705 is associated to calcium levels27, and 21 kb downstream the top eQTL – a 2bp insertion – is the likely causal variant affecting calcium levels. Thus, the integration of genome sequencing and cellular phenotype data helps not only to understand causal genes and biological processes but also to pinpoint putative causal genetic variants underlying GWAS associations.
Allelic and oss-of-function effects
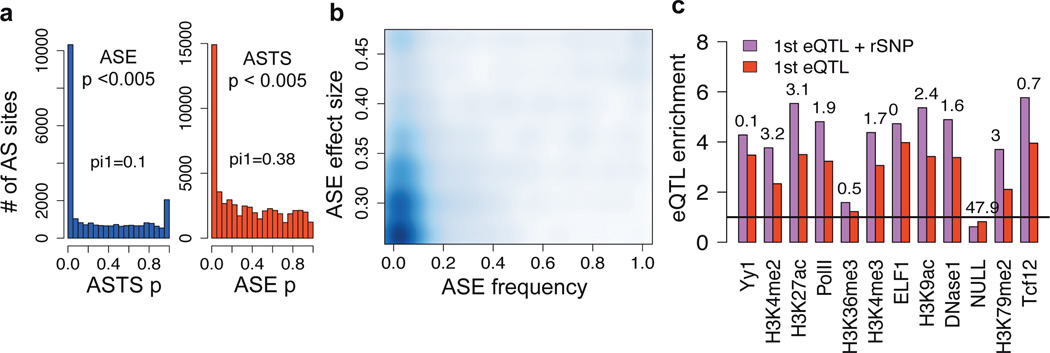
Transcript differences between the two haplotypes of an individual allow quantification of regulatory variation even when eQTLs cannot be detected e.g. due to low allele frequency. We analyzed both allele-specific expression (ASE) and allele-specific transcript structure (ASTS), a novel approach based on exonic distribution of reads (Fig.S2, S28–33). This first genome-wide quantification of allelic effects on transcript structure shows that it is almost equally common as ASE, with significant (p < 0.005) ASE and ASTS in a median of 6.5% and 5.6% sites (out of 8,420 and 2,135) per individual, respectively. Furthermore, the substantial overlap of ASE and ASTS signals (Fig.3a) suggests that ASE is actually often driven by transcript structure variation. The low population frequency of the vast majority of ASE (Fig.3b) and ASTS (Fig.S30) events points to widespread rare regulatory variation that is undetectable in eQTL analysis.
Figure 3. Allele-specific effects on expression and transcript structure.
a) Sharing of allele-specific expression (ASE) and transcript structure (ASTS) signals: the distribution of ASTS p-value of the sites with significant (p<0.005) ASE in the same individual, and vice versa. The ASE p-values are calculated from sites sampled to exactly 30 reads. The numbers denote the pi1 statistic measuring the enrichment of low p-values. b) Frequency of significant ASE event in the population (x-axis) and their effect size (|0.5 – REF/TOTAL|), calculated per ASE SNP. Only ASE SNPs with >=20 heterozygote individuals with >=30 reads were included, and the estimates were corrected for coverage bias and false positives by sampling and permutations. c) Enrichment of variants in regulatory annotations relative to a matched null distribution for the most significant eQTL variants, and for the subset of these that are also rSNPs. Categories with highest amount of data are shown (see Fig. S36 for all categories, see also Fig. 2a).
An important caveat in ASE analysis has been the possibility that it can be driven by purely epigenetic effects rather than cis-regulatory genetic variants. We investigated this by a novel approach to quantify concordance between ASE and putative regulatory variants (prSNPs), where heterozygotes but not homozygotes for a true rSNP should have differential expression of the two haplotypes, i.e. allelic imbalance in an aseSNP (Fig. S2, S34). We calculated concordance of allelic ratios of 5,479 aseSNPs and genotypes of all SNPs +/− 100kb from TSS, with an empirical p-value from 100–1000 permutations. Assigning the prSNPs with empirical p-value <0.01 to p<0.001 as likely rSNPs yielded a total of 224,640 rSNPs (7.4% of tested, Table S6) that clustered close to TSS as expected for regulatory variants5 and replicate the majority of eQTL signals (Fig. S35). Nearly all aseSNPs (95%) had more observed rSNPs than expected; thus ASE appears to be nearly always genetic rather than driven by genotype-independent allelic epigenetic effects. rSNP signals are widespread and robust also outside eQTL genes (Table S6, Fig.S35), indicating potential to capture novel effects. Variants that are both eQTLs and rSNPs show higher enrichment in functional annotations (Fig. S3c, S36), suggesting that integrated analysis may improve resolution to find causal regulatory variants. Altogether, we show evidence that ASE effects are mostly rare and nearly always genetic, and ASE-based analyses may complement eQTL analysis in identification of especially low-frequency regulatory variants in future studies.
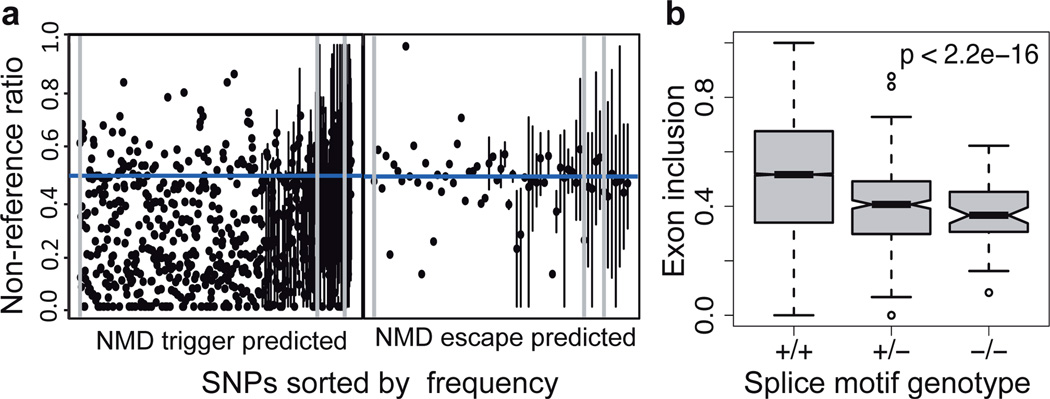
While QTL and prSNP analyses aim at identifying previously unknown regulatory variants, we can also quantify functional effects of predicted loss-of-function variants.28 Our RNA-seq data captures 839 premature stop codon and 849 splice-site variants, with the much higher number than in previous studies enabling proper quantification of their transcriptome effects. As expected, premature stop variants often show loss of the variant allele (Fig, S37) indicating nonsense-mediated decay29 as in previous studies28,30. Variants close to the end of the transcript appear to escape NMD as predicted29. However, of the variants predicted to trigger NMD, in 68% (54% of rare variants MAF<1%) the ASE results do not support this (Fig. 4), suggesting currently unknown mechanisms of NMD escape.
Figure 4. Transcriptome effects of loss-of-function variants.
A) Nonsense-mediated decay due to premature stop codon variants was measured using allele-specific expression. The distribution of non-reference allele ratios (on the y-axis) for premature stop variants sorted on the x-axis according to derived allele frequency, split to sites predicted to trigger and escape NMD. The dots denote the median across individuals, and the vertical lines show the range of ratios for variants carried by several individuals. The grey vertical lines denote derived allele frequencies of 0, 0.001 and 0.01. B) Exon inclusion scores for variable exons for individuals that carry 0, 1 or 2 copies of variants that destroy a splice motif, with p-value from Mann-Whitney test.
Finally, we modeled how genetic variants affect splicing affinity in the entire splicing motif rather than only the canonical splice site, which is the first comprehensive set of such predictions genome-wide (PGF et al., submitted). Nonreference alleles have a lower splicing affinity on average (p<2.2 × 10−16, Fig. S38). For the 10% of these variants predicted to destroy the motif, individuals carrying two motif-destroying alleles have 29% lower median inclusion rates of the affected exon (p<2.2 × 10−16, Fig.4c), indicating that our RNA-seq data is consistent with predictions of splicing effects.
Conclusions
By integrated analysis of RNA and DNA sequencing data we were able to obtain a unique view to variation of the transcriptome and its genetic causes, moving beyond eQTL catalogs to a high-resolution view of genetic regulatory variants. We deconvoluted the effect of gene expression and transcript structure in population differences of the transcriptome, in QTLs, and in allele-specific effects, and show that these two dimensions of transcript variation appear equally common but largely independent. Genetic regulatory variation is the rule rather than the exception in the genome with widespread allelic heterogeneity, and is the major determinant of allelic expression. For the first time, we were able to predict large numbers of causal regulatory variants, and thus provide a detailed view into cellular mechanisms of regulatory and loss-of-function variation, which is essential for future functional prediction of variants discovered in personal genomes.
A subset of this functional variation at the cellular level will also have effects on higher-level traits. We demonstrate how eQTL data can be used to pinpoint putative causal GWAS variants of individual loci, which is important as a new paradigm of how integration of cellular phenotypes and genome sequencing data can uncover causal variants and biological mechanisms underlying diseases. The landscape of regulatory variation in this study adds a functional dimension to the 1000 Genomes data, which is used in effectively all disease studies, and together they form an important joint reference data set of variation and function of the human genome. Ultimately, this study illustrates the power of combining genome sequence analysis with a high-depth functional readout such as the transcriptome.
Methods
Total RNA was extracted from EBV transformed lymphoblastoid cell line pellets by the TRIzol reagent (Ambion), and mRNA and small RNA sequencing of 465 unique individuals was performed on the Illumina HiSeq2000 platform, with paired-end 75bp mRNA-seq and single-end 36bp small RNA-seq. Five samples were sequenced in replicate in each of the seven sequencing laboratories. The mRNA and small RNA reads were mapped with GEM31 and miraligner32, respectively, with an average of 48.9M mRNA-seq reads and 1.2M miRNA reads per sample after QC. Numerous transcript features were quantified using Gencode v1233 and miRBase v1834 annotations: protein-coding and lincRNA genes (16,084 detected in >50% of samples), transcripts (67,603; with FluxCapacitor7), exons (146,498), annotated splice junctions (129,805; analyzed in detail in Ferreira et al. submitted), transcribed repetitive elements (47,409), and mature miRNAs (715). Data quality was assessed by sample correlations and read and gene count distributions, and technical variation was removed by PEER normalization35 for the QTL and miRNA-mRNA correlation analyses11. The samples clustered uniformly both before and after normalization. The genotype data was obtained from 1000 Genomes Phase 1 data set for 421 samples (80× average exome and 5× whole genome read depth), and the remaining 41 samples were imputed from Omni 2.5M SNP array data. Furthermore, we did functional reannotation for all the 1000 Genomes variants using Gencode v12. QTL mapping was done with linear regression, using genetic variants with >5% frequency in 1MB window and normalized quantifications transformed to standard normal. Permutations were used to adjust FDR to 5%. Full details are provided in Supplementary Methods.
Supplementary Material
Acknowledgements
We would like to thank Emilie Falconnet, Luciana Romano, Alexandra Planchon, Deborah Bielsen, Alisa Yurovsky, Alfonso Buil, Julies Bryois, Alexandra Nica, Ivan Topolsky, Nicolo Fusi, Sebastian Waszak, Carlos Bustamante, Johan Rung, Nikolay Kolesnikov, Asier Roa, Eugene Bragin, Simon Brent, Justo Gonzalez, Marta Morell, Anna Puig, Emilio Palumbo, Marina Ventayol Garcia, Jeroen F.J. Laros, Julie Blanc, Rahnehild Birkelund, Gloria Plaja, Matt Ingham, Jordi Camps, Monica Bayes, Lidia Agueda, Anais Gouin, Marie-Laure Yaspo, Elisabeth Graf, Anett Walther, Carola Fischer, Sandy Loesecke, Bianca Schmick, Daniela Balzereit, Simon Dökel, Matthias Linser, Alexander Kovacsovics, Melanie Friskovec, Catharina von der Lancken, Melanie Schlapkohl, Anita Dietsch, Markus Schilhabel, the SNP&SEQ Technology Platform in Uppsala, Sascha Sauer, the Vital-IT high-performance computing center of the SIB Swiss Institute of Bioinformatics, Bernadette Goldstein and others at the Coriell Institute, and James Cooper, Edward Burnett, Karen Ball and others at the European Collection of Cell Cultures (ECACC) and the 1000 Genomes Consortium.
This project was funded by the European Commission 7th Framework Program (FP7) (261123; GEUVADIS); the Swiss National Science Foundation (130326, 130342), the Louis Jeantet Foundation, andERC(260927) (E.T.D.); NIH-NIMH(MH090941) (E.T.D.,M.I.M., R.G.); Spanish Plan Nacional SAF2008-00357 (NOVADIS), the Generalitat de Catalunya AGAUR 2009 SGR-1502, and the Instituto de Salud Carlos III (FIS/FEDER PI11/00733) (X.E.); Spanish Plan Nacional (BIO2011-26205) and ERC (294653) (R.G.); ESGI, READNA (FP7 Health-F4-2008-201418), Spanish Ministry of Economy and Competitiveness (MINECO) and the Generalitat de Catalunya (I.G.G.); DFG Cluster of Excellence Inflammation at Interfaces, the INTERREG4A project HIT-ID, and the BMBF IHEC project DEEP SP 2.3 (P.Ro.); German Centre for Cardiovascular Research (DZHK) and the GermanMinistry of Education and Research (01GR0802, 01GM0867, 01GR0804, 16EX1020C) (T.M.); EurocanPlatform (FP7 260791), ENGAGE and CAGEKID (241669) (A.B.); FP7/2007-2013, ENGAGE project, HEALTH-F4-2007- 201413, and the Centre for Medical Systems Biology within the framework of The Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific and Research (NWO) (P.AC.H & G.-J.v.O.); The Swedish Research Council (C0524801, A028001) and the Knut and Alice Wallenberg Foundation (2011.0073) (A.-C.S.); The Swiss National Science Foundation (127375, 144082) and ERC (249968) (S.E.A.); Instituto de Salud Carlos III (FIS/FEDER PS09/02368) (A.C.); German Federal Ministry of Education and Research (01GS08201) (R.S.); Max Planck Society (H.L.); Wellcome Trust (WT085532) and the European Molecular Biology Laboratory (P.F.); ENGAGE, Wellcome Trust (081917, 090367, 090532, 098381), and Medical Research Council UK (G0601261) (M.I.M.); Wellcome Trust Centre for Human Genetics (090532/Z/09/ Z, 075491/Z/04/B), Wellcome Trust (098381, 090367, 076113, 083270), the WTCCC2 project (085475/B/08/Z, 085475/Z/08/Z), Royal Society Wolfson Merit Award, Wellcome Trust Senior Investigator Award (095552/Z/11/Z) (P.D.); EMBO long-term fellowship EMBO-ALTF 2010-337 (H.K.); NIH-NIGMS (R01 GM104371) (D.G.M.); Marie Curie FP7 fellowship (O.S.); Scholarship by the Clarendon Fund of the University of Oxford, and the Nuffield Department of Medicine (M.A.R.); EMBO long-term fellowship ALTF225-2011 (M.R.F.); Emil Aaltonen Foundation and Academy of Finland fellowships (T.L.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions
Designed the study: TL, TGi, SBM, PACH, EL, HL, SS, RS, AC, SEA, RH, ACS, GJvO, AB, TM, Pro, RG, IGG, XE, ETD
Coordinated the project: TL, TGi, GB, XE, ETD
Participated in data production: TL, TGi, IPa, MSu, EL, SB, MG, VA, KK, DE, PR, OK
Analyzed the data: TL, MSa, MRF, PACH, JM, MAR, MGo, NK, TGr, PGF, MB, TW, LG, MvI, JA, PRi, IPu, DE, AT, MSu, DGM, ML, EL, HPJB, IPa, TS, OK, HO, HK, SB, MGu, KK, VA, OS, MP, PD, MIM, PF, TMS
Drafted the paper: TL, ETD
Principal Investigators of the Geuvadis Consortium: See Supplementary Note
Data access
The Geuvadis RNA-sequencing data, genotype data, variant annotations, splice scores, quantifications, and QTL results are freely and openly available with no restrictions. The main portal for accessing the data is EBI ArrayExpress (accessions E-GEUV-1, E-GEUV-2, E-GEUV-3; see the data access schema in Fig. S39). For visualization of the results we created the Geuvadis Data Browser (www.ebi.ac.uk/Tools/geuvadis-das) where quantifications and QTLs can be viewed, searched, and downloaded (Fig. S40). The project webpage www.geuvadis.org provides full documentation and links to all files, and the analysis group wiki is open to the public in geuvadiswiki.crg.es.
The authors declare no competing financial interests
References
- 1.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. doi:nature11632 [pii] 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. doi:nature05911 [pii] 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. doi:nature11247 [pii] 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emilsson V, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. doi:nature06758 [pii] 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 5.Stranger BE, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. doi:ng2142 [pii] 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grundberg E, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat Genet. 2012;44:1084–1089. doi: 10.1038/ng.2394. doi:ng.2394 [pii] 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montgomery SB, et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–777. doi: 10.1038/nature08903. doi:nature08903 [pii] 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pickrell JK, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. doi:nature08872 [pii] 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nica AC, et al. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 2010;6:e1000895. doi: 10.1371/journal.pgen.1000895. doi:10.1371/journal.pgen.1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicolae DL, et al. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. doi:10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.'t Hoen PA, Friedländer MR, Almlöf J, Sammeth M, Pulyakhina I, Anvar SY, Laros JF, Buermans HP, Karlberg O, Brännvall M, The GEUVADIS Consortium. den Dunnen JT, van Ommen GJ, Gut IG, Guigó R, Estivill X, Syvänen AC, Dermitzakis ET, Lappalainen T. Reproducibility of high-throughput mRNA and small RNA sequencing across laboratories. Nat Biotechnol. 2013;31:1015–1022. doi: 10.1038/nbt.2702. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez-Porta M, Calvo M, Sammeth M, Guigo R. Estimation of alternative splicing variability in human populations. Genome Res. 2012;22:528–538. doi: 10.1101/gr.121947.111. doi:gr.121947.111 [pii] 10.1101/gr.121947.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2013;338:1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barbosa-Morais NL, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338:1587–1593. doi: 10.1126/science.1230612. doi:338/6114/1587 [pii] 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- 15.Parts L, et al. Extent, causes, and consequences of small RNA expression variation in human adipose tissue. PLoS Genet. 2012;8:e1002704. doi: 10.1371/journal.pgen.1002704. doi:10.1371/journal.pgen.1002704 PGENETICS-D-12-00282 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136:26–36. doi: 10.1016/j.cell.2008.12.027. doi:S0092-8674(08)01633-4 [pii] 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 17.Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. doi:S0092-8674(12)00464-3 [pii] 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pickrell JK, Pai AA, Gilad Y, Pritchard JK. Noisy splicing drives mRNA isoform diversity in human cells. PLoS Genet. 2010;6:e1001236. doi: 10.1371/journal.pgen.1001236. doi:10.1371/journal.pgen.1001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee Y, et al. Variants affecting exon skipping contribute to complex traits. PLoS Genet. 2012;8:e1002998. doi: 10.1371/journal.pgen.1002998. doi:10.1371/journal.pgen.1002998 PGENETICS-D-11-02434 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Djebali S, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. doi:nature11233 [pii] 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. doi:nrg2640 [pii] 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Veyrieras JB, et al. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS Genet. 2008;4:e1000214. doi: 10.1371/journal.pgen.1000214. doi:10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaffney DJ, et al. Dissecting the regulatory architecture of gene expression QTLs. Genome Biol. 2012;13:R7. doi: 10.1186/gb-2012-13-1-r7. doi:gb-2012-13-1-r7 [pii] 10.1186/gb-2012-13-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDaniell R, et al. Heritable individual-specific and allele-specific chromatin signatures in humans. Science. 2010;328:235–239. doi: 10.1126/science.1184655. doi:science.1184655 [pii] 10.1126/science.1184655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Degner JF, et al. DNase I sensitivity QTLs are a major determinant of human expression variation. Nature. 2012;482:390–394. doi: 10.1038/nature10808. doi:nature10808 [pii] 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hindorff LA, Junkins HA, Hall PN, Mehta JP, Manolio TA. A Catalog of Published Genome-Wide Association Studies. 2010 www.genome.gov/gwastudies.
- 27.O'Seaghdha CM, et al. Common variants in the calcium-sensing receptor gene are associated with total serum calcium levels. Hum Mol Genet. 2010;19:4296–4303. doi: 10.1093/hmg/ddq342. doi:ddq342 [pii] 10.1093/hmg/ddq342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.MacArthur DG, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. doi:335/6070/823 [pii] 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. doi:S0968-0004(98)01208-0 [pii] [DOI] [PubMed] [Google Scholar]
- 30.Montgomery SB, Lappalainen T, Gutierrez-Arcelus M, Dermitzakis ET. Rare and common regulatory variation in population-scale sequenced human genomes. PLoS Genet. 2011;7:e1002144. doi: 10.1371/journal.pgen.1002144. doi:10.1371/journal.pgen.1002144 PGENETICS-D-10-00589 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marco-Sola S, Sammeth M, Guigo R, Ribeca P. The GEM mapper: fast, accurate and versatile alignment by filtration. Nat Methods. 2012 doi: 10.1038/nmeth.2221. doi:nmeth.2221 [pii] 10.1038/nmeth.2221. [DOI] [PubMed] [Google Scholar]
- 32.Pantano L, Estivill X, Marti E. SeqBuster, a bioinformatic tool for the processing and analysis of small RNAs datasets, reveals ubiquitous miRNA modifications in human embryonic cells. Nucleic Acids Res. 2010;38:e34. doi: 10.1093/nar/gkp1127. doi:gkp1127 [pii] 10.1093/nar/gkp1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. doi:22/9/1760 [pii] 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. doi:gkq1027 [pii] 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stegle O, Parts L, Durbin R, Winn J. A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput Biol. 2010;3:e1000770. doi: 10.1371/journal.pcbi.1000770. doi:10.1371/journal.pcbi.1000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.