Abstract
Costello syndrome is linked to activating mutations of a residue in the p-loop or the NKCD/SAK motifs of Harvey Ras (HRas). More than 10 HRas mutants that induce Costello syndrome have been identified; G12S HRas is the most prevalent of these. However, certain HRas p-loop mutations also are linked to cancer formation that are exemplified with G12V HRas. Despite these relations, specific links between types of HRas mutations and diseases evade definition because some Costello syndrome HRas p-loop mutations, such as G12S HRas, also often cause cancer. This study established novel kinetic parameter-based equations that estimate the value of the cellular fractions of the GTP-bound active form of HRas mutant proteins. Such calculations differentiate between two basic kinetic mechanisms that populate the GTP-bound form of Ras in cells. (i) The increase in GTP-bound Ras by the HRas mutation-mediated perturbation of the intrinsic kinetic characteristics of Ras. This generates a broad spectrum of the population of the GTP-bound form of HRas that typically causes Costello syndrome. The upper end of this spectrum of HRas mutants, as exemplified by G12S HRas, can also cause cancer. (ii) The increase in GTP-bound Ras because the HRas mutations perturb the p120GAP action on Ras. This causes production of a significantly high population of the only GTP-bound form of HRas linked merely to cancer formation. The HRas mutant G12V belongs to this category.
Keywords: Costello Syndrome, Ras, Cancer, HRas G12S
Ras family proteins such as Harvey Ras (HRas), Neuroblastoma Ras (NRas), and Kirsten Ras (KRas) each generate distinct signals despite their interactions with a common set of regulators.1 These Ras proteins function by cycling between inactive GDP-bound and active GTP-bound states, and various regulators control this GDP/GTP cycling.2 These regulators include guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs).3 Ras GEFs belong to the positive Ras regulators. Several isoforms of Ras GEFs, including Son of Sevenless (SOS), Ras guanine-nucleotide-release factor (RasGRF), and Ras guanyl nucleotide-releasing protein (RasGRP) have been identified.4–6 These GEFs contain a common catalytic core domain Cdc25 and facilitate the intrinsically slow rate of guanine nucleotide exchange (GNE) of Ras-bound GDP/GTP with cellular free GDP/GTP.7 Because the cellular concentration of GTP is ~10-fold higher than the concentration of GDP,8 GEF-mediated Ras GNE populates Ras in their biologically active GTP-bound states. In turn, activated Ras proteins interact with a variety of downstream effector proteins that modulate numerous cellular signaling processes such as cellular proliferation and gene expression.9 Ras GAPs are the negative Ras regulators. They consist of p120GAP, neurofibromin1 (NF1), and the GAP1 family.10 All of these Ras GAPs share a RasGAP catalytic core domain.11 GAPs increase the intrinsically slow rate of GTP hydrolysis for most Ras family proteins to permit conversion of the bound GTP into GDP, thereby terminating Ras downstream signaling.12
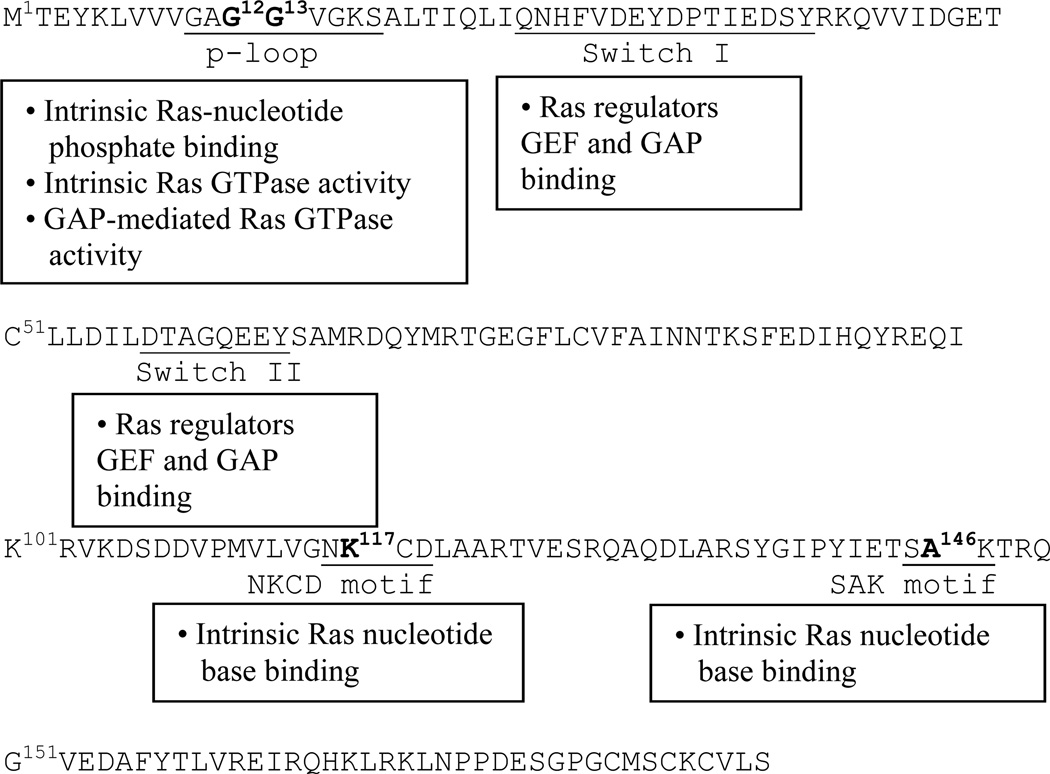
Several Ras motifs (Fig. 1) — the phosphate-binding loop (P-loop; Gly10-Ser17), Switch I (Gln25-Tyr40), Switch II (Asp57- Tyr64), the nucleotide base-binding NKCD (Asn116-Asp119), and SAK (Ser145-Lys147) motifs (HRas numbering) — are known to be involved in these binding interactions with GDP and GTP. Many of these Ras motif residues are also directly or indirectly involved in the catalytic functions of GEFs and GAPs (Fig. 1).7, 11
Figure 1. Sequence of wt HRas.
The p-loop, Switch I, Switch II, NKCD, and SAK motifs of wt HRas are underlined. The HRas residues that are known to mutate to produce Costello syndrome-relevant HRas mutants are shown in boldface. These include Gly12 and Gly13 in the p-loop, Lys117 in the NKCD, and Ala146 in the SAK motif. The boxes depict the contributions of these motifs in the binding and/or catalytic function of Ras with nucleotide and/or regulators.
Costello syndrome is a genetic disorder that affects, but is not limited to, the skin and joints; it also often causes heart abnormalities.13 Other characteristics are postnatal growth delays, mental retardation, and facial dysmorphism.13 Mutations in the HRAS gene at codons Gly12 and Gly13 in the p-loop as well as at Lys117 and Ala146 in the NKCD and SAK motifs cause the syndrome.14 The Costello syndrome-relevant HRas mutants for the p-loop Gly12 residue include G12A, G12S, G12C, G12D, and G12E (Table 1). Notably, the well-known oncogenic HRas p-loop mutant G12V15 also causes Costello syndrome (Table 1). Other HRas p-loop mutants that induce Costello syndrome include G13S, G13C, and G13D (Table 1). Symptoms of Costello syndrome also are linked to an unusual HRas NKCD-motif mutant K117R and to the HRas SAK-motif mutants A146T and A146V (Table 1). Of all these HRas mutants, G12S is the most prevalent mutation found in Costello syndrome patients, occurring five times more often than other mutations combined (Table 1). When it occurs in NRas and KRas, the G12S mutation often causes certain cancers.16–18 Because of its weak oncogenicity,15, 19 there has been speculation that G12S HRas is present to serve as a partially active form of HRas to upregulate HRas-dependent cellular signaling.13 The second most prevalent HRas mutant found in Costello Syndrome patients is G12A (Table 1); however, this mutant, unlike G12S, is not commonly found in cancers. Nonetheless, unlike other common HRas p-loop mutants, such as G12V, the biochemical properties of the listed Costello syndrome-relevant HRas mutants (Table 1) have not been fully investigated either with or without their regulators. Accordingly, no definite link has been established between these HRas mutations and the cellular populations of the GTP-bound form of these HRas mutants. This study used the kinetic parameters of Ras and its regulators to formulate novel equations that allow one to estimate the theoretical populations of the GTP-bound Ras in cells. Equipped with these novel equations, this study estimated the values of the theoretical populations of the GTP-bound form of these HRas mutants. The theoretical values were further evaluated with the directly measured cellular populations of the GTP-bound form of selected HRas mutants. These biochemical approaches reveal the kinetic mechanisms by which these HRas mutations contribute to Costello syndrome.
Table 1. Frequency of HRas mutations in Costello syndrome.
Previous reports of the relative occurrence in Costello syndrome of p-loop and NKCD/SAK HRas mutants are summarized.
| Aoki et al36 |
Gripp et al37 |
Estep et al38 |
Kerr et al39 |
Sol-Church et al40 |
Zampino et al41 |
Van der Brugt et al42 |
Gripp et al43 |
Burkitt-Wright et al44 |
Niihori et al45 |
Digilio et al46 |
Tidyman et al47 |
Gripp et al48 |
Kuniba et al31 |
Piccione et al49 |
Schulz et al50 |
Denayer et al51 |
Sinico et al52 |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p-loop mutants | G12A | 2 | 2 | 2 | 3 | 2 | 1 | 3 | 3 | ||||||||||
| G12C | 2 | 1 | |||||||||||||||||
| G12D | 1 | 1 | |||||||||||||||||
| G12E | 1 | ||||||||||||||||||
| G12S | 7 | 30 | 30 | 30 | 39 | 8 | 16 | 3 | 4 | 23 | |||||||||
| G12V | 1 | 1 | 4 | ||||||||||||||||
| G13C | 1 | 1 | 1 | 12 | 1 | 1 | |||||||||||||
| G13D | 2 | 1 | 1 | ||||||||||||||||
| G13S | 1 | ||||||||||||||||||
| G13Va | |||||||||||||||||||
| NKCD/SAK mutants | K117R | 1 | 1 | ||||||||||||||||
| A146T | 1 | ||||||||||||||||||
| A146V | 1 | ||||||||||||||||||
| Totals | 12 | 33 | 33b | 37 | 42 | 9 | 4c | 2c | 4 | 21 | 4 | 4 | 12 | 1 | 1 | 31c | 1 | 1 | |
For comparison with Tables 2, 3, and 4 (see below), G13V HRas also is listed. However, the G13V HRas mutant has been linked to cancer, but not to Costello syndrome.53
In this study it was noted that 20 of the 33 patients also participated in the investigation done by Gripp et al37
In these studies, the total number of Costello syndrome patients were noted, but only the relevant HRas mutants were specified.
Materials and Methods
Protein preparations
All protein constructs, including wild type (wt) HRas, p120GAP, and Cdc25, were derived from humans. A full-length wt HRas (1–189) and HRas mutant proteins including G12V, G12A, G12S, G12C, G12D, G12E, G13V, G13S, G13C, G13D, K117R, A146T, and A146V were expressed in and purified from E. coli, as described in the previous study.20
A full-length p120GAP (1–1047) was overexpressed in insect Sf9 cells by using the pIEx vector (Novagen) and then purified using a sequence of columns, including the Sephadex G-150 gel filtration and FPLC Mono-Q columns, as described in the previous study.21 However, because of the high cellular expression of p120GAP, the fast-flow S-Sepharose column step was omitted. The Ras SOS1 catalytic core domain Cdc25 (564–1049) was expressed in and purified from E. coli, as described in the previous study.22
Kinetic assay conditions
All kinetic analyses were performed using proteins more than 95% pure, as judged by SDS-PAGE. The buffer used for all assays consisted of 50 mM NaCl, 10 mM MgCl2, 100 µM diethylenetriaminepentaacetic acid, and 100 mM TrisHCl (pH 7.4).
Estimation of kinetic parameters of Ras GNE in the presence and absence of Cdc25
Kinetic parameters of the intrinsic Ras GTP and GDP binding interactions were determined, respectively, with minor modifications, according to the previously established method.23 The values of k−1 — the intrinsic rate constant of GTP dissociation from Ras protein — were determined by using the nonhydrolyzable radioactive GTP analog [35S]GTPγS (~400 cpm/pmol) paired with GDP. Ras (1 µM) loaded with [35S]GTPγS was placed in an assay buffer containing GDP (1 µM). Aliquots of the assay mixture were withdrawn at specific intervals over a period of 20,000 s, and spotted onto nitrocellulose filters. The nitrocellulose filters were washed three times with the assay buffer, and the filter-bound radioactivity was measured using a scintillation counter (Beckman LS 6000). These data were fit to single exponential decays that give values of k−1. The values of k−2 — the intrinsic rate constant of GDP dissociation from Ras proteins — also were measured as the measurement of k−1, as described above, except that the radioactive [8-3H]GDP (~200 cpm/pmol) paired with GTP were used instead of the [35S]GTPγS paired with GDP.
Kinetic parameters of the Cdc25-mediated Ras-bound GTP and GDP exchange with GDP and GTP were measured, respectively, as described in the previous study.22 The values of k+3 — the exchange rate constant of the Ras-bound GTP with GDP in the presence of Cdc25 — were determined using the 2'(3')-O-(N-methylanthraniloyl) 5'-guanylyl-imidodiphosphate (mantGppNHp; a nonhydrolyzable GTP analog) paired with GDP. In brief, the reaction was initiated by the addition of the various concentrations of the mantGppNHp-loaded Ras (0–500 µM) into an assay buffer containing GDP (5 mM) and Cdc25 (500 nM). Dissociation of mantGppNHp from Ras was monitored over a period of 20,000 s by using a Fluorescence Spectrometer (LS 55, PerkinElmer). These data were fit to a single exponential decay to determine the apparent dissociation rates of mantGppNHp from Ras in the presence of Cdc25. Once determined, these rates were then replotted against the concentrations of the mantGppNHp-loaded Ras used. These rates were then fit to a hyperbola to determine the maximal velocity (Vmax) and the Michaelis–Menten constant (KM) of the Cdc25-mediated dissociation of mantGppNHp from Ras. According to the Theorell-Chance type mechanism,24 the dissociation of GTP from Ras couples with the association of GEF, which is then immediately displaced with GDP (if only GDP is present). Hence, the dissociation rate of mantGppNHp from Ras represents the exchange rate of mantGppNHp with GDP. Therefore, from the perspective of the Theorell-Chance type of mechanism, the values of Vmax of the dissociation of the Ras-bound mantGppNHp from Ras per the total Cdc25 enzyme (Eo) are, respectively, equivalent to k+3. With one exception, the methods and analyses used for the determination of the value of k+3 also were applied to determination of the values of k+4 — the exchange rate constant of the Ras-bound GDP with GTP in the presence of Cdc25. The exception was the use of the 2'(3')-O-(N-methylanthraniloyl) guanosine diphosphate (mantGDP) paired with GTP instead of the mantGppNHp paired with GDP.
Estimation of kinetic parameters of Ras GTP hydrolysis in the presence and absence of p120GAP
The intrinsic kinetic parameters of Ras GTPase activity were measured, with minor modifications, as described in the previous study.25 The values of k+6 — the rate constant for the intrinsic Ras GTPase activities — were determined by using [γ-32P]GTP (~500 cpm/pmol). As-purified Ras (1 µM) was added to the assay buffer containing [γ-32P]GTP (50 µM). Aliquots were taken from the assay solution at specific intervals over a period of 20,000 sec and spotted onto nitrocellulose filters. The radioactivity of filtrants that contain only free [γ-32P] was determined by using a scintillation counter (Beckman LS 6000). The values of k+6 were then determined by the fit of these data to a function of exponential decay.
It is of interest that, unlike the GEF-mediated enzymatic process, the GAP-mediated enzymatic process does not follow the Theorell-Chance type of mechanism.24 This is because the formative process of the Ras•GTP•GAP ternary complex intermediate is not so transient.11 Nonetheless, within this study, the parameters associated with the p120GAP-mediated Ras GTP hydrolysis were calculated based on the values of the KM and Vmax of p120GAP for Ras. In brief, the values of k+8 — the rate constant for the hydrolysis of GTP of the Ras•GTP•p120GAP ternary complex to produce Ras•GDP — were estimated by dividing the total Ras concentration (Eo) into Vmax; this yields Vmax/Eo, which is equivalent to kcat, which is the same as k+8. The values of KM and kcat of p120GAP for Ras were measured by using radioactive [γ-32P]GTP (~500 cpm/pmol) as described in the previous study.26 As-purified Ras (1 µM) was added to the assay buffer containing [γ-32P]GTP (50 µM) and various concentrations of p120GAP (0–35 µM). Aliquots of the assay mixture were drawn at specific intervals over a period of 20,000 sec and spotted onto nitrocellulose filters. The radioactivity of only the free [32P]-containing liquid filtrants was determined with a scintillation counter (Beckman LS 6000). The apparent rates of Ras GTPase activity in the presence of p120GAP were then determined by the fit of these data to a function of a single exponential decay. The plots of apparent rates against the concentration of p120GAP that were thus determined were fit to a hyperbola to obtain the values of Vmax and KM of p120GAP for Ras GTP hydrolysis.
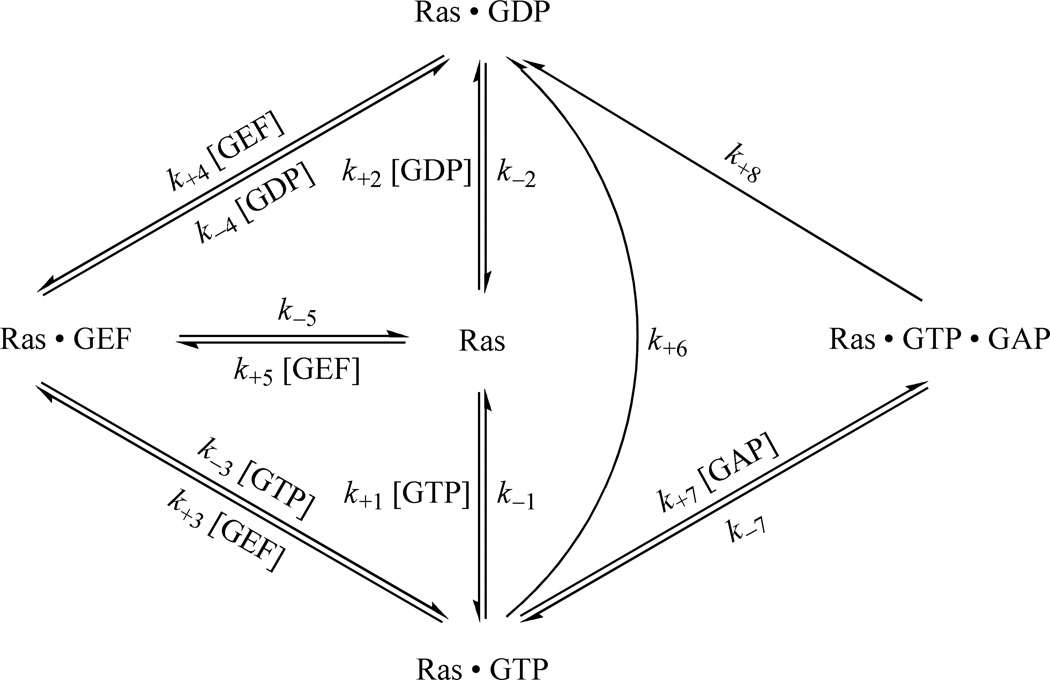
Comprehensive kinetic scheme of Ras activity regulation in cells
Scheme 1 shows a comprehensive web of kinetic paths that describe the regulation of the binding of Ras with its ligands, GTP and GDP. Scheme 1 comprehensively encompasses all of the known cellular kinetic paths that modulate the Ras binding states with GTP and GDP in the presence and absence of Ras regulators, including GEF and GAP. The kinetic parameters denoted in Scheme 1 stand for the kinetic rate constants associated with the processes of the reactions that occur through the kinetic paths. The kinetic parameters shown in Scheme 1 also represent the steps of the reaction processes that occur through the given kinetic paths. According to Scheme 1, three essential kinetic processes in effect determine the featured state of the Ras binding with GTP and GDP. These three are the intrinsic Ras GNE and GTP hydrolysis, the GEF-mediated Ras GNE, and the GAP-mediated Ras GTP hydrolysis in conjunction with the term of concentrations of GTP and GDP ([GTP] and [GDP], respectively). The kinetic steps of k+1, k−1, k+2, and k−2 are involved in the intrinsic Ras GNE. The kinetic steps of k+3, k−3, k+4, k−4, k+5, and k−5 in combination with the concentration of GEF ([GEF]) are implicated in the GEF-mediated Ras GNE. The kinetic step of k+6 is engaged in the intrinsic Ras GTP hydrolysis. The kinetic steps of k+7, k−7, and k+8 in combination with the concentration of GAP ([GAP]) are linked to the process of the GAP-mediated Ras GTP hydrolysis.
Scheme 1.
Calculation of the theoretical cellular population of the GTP-bound Ras
The result of the regulation of Ras binding interactions with GTP and GDP through these paths (Scheme 1) determines the overall comprehensive cellular fraction of the GTP-bound Ras over the GTP- and GDP-bound Ras (the comprehensive of Ras. The value of the comprehensive fRas•GTP of Ras is of particular interest because it refers to the overall cellular population of the biologically active form of Ras.
Equation 1 defines the value of the comprehensive fRas•GTP of Ras that enables calculation of the theoretical comprehensive cellular population of the GTP-bound form of Ras of interest by using the intrinsic kinetic parameters of Ras as well as the kinetic parameters of GEF and GAP with Ras shown in Scheme 1 (see Supporting Information for the derivation).
| Equation 1 |
Equation 2 expresses the intrinsic cellular fraction of the GTP-bound Ras over the GTP-and GDP-bound Ras (the intrinsic f Ras•GTP) of Ras in terms of the kinetic parameters of intrinsic Ras GNE and GTP hydrolysis shown in Scheme 1 (see Supporting Information for the derivation).
| Equation 2 |
Equation 3 denotes the cellular fraction of the GTP-bound Ras over the GTP- and GDP-bound Ras (the GEF-mediated f Ras•GTP) of Ras in the presence of GEF in cells shown in Scheme 1 (see Supporting Information for the derivation).
| Equation 3 |
Equation 4 expresses the cellular fraction of the GTP-bound Ras over the GTP- and GDP-bound Ras (the GAP-mediated f Ras•GTP) of Ras in the presence of GAP in cells shown in Scheme 1 (see Supporting Information for the derivation).
| Equation 4 |
Quantification of the Ras-bound GTP in cells
Unstimulated NIH 3T3 cells were stably transfected with various HRAS constructs (#1–189) using a mammalian expression vector pCAGGS with FuGene-6-reagent (Roche). Cells were cultured for four days in a complete RPMI 1640 medium, and washed with phosphate-free Eagle's minimum essential medium. Resuspended cells were then incubated for 5 hr with a serum- and phosphate-free RPMI 1640 medium containing ~50 µCi of 32P/mL. Cells were washed twice with ice-cold PBS (Sigma) and then scraped into an ice-cold homogenization buffer containing 500 mM NaCl, 10 mM MgCl2, 5 mM DTT, 1 mM EDTA, 0.1% Triton X-100, 0.005% SDS, protease inhibitors (0.5 mM phenylmethylsulfonyl fluoride, 1 µg/mL peptstain A, 1 µg/mL aproptinin, and 1 µg/mL leupeptin), and 10 mM TrisHCl (pH 7.4). Cells in the ice-cold homogenization buffer were sonificated 4 × 5 sec (40 watts) on ice followed by centrifugation (100,000 × g) at 4 °C for 20 min. The supernatant of the cell lysate extract was nutated with Amine Immobilization Resin (Thermo Scientific) coupled with a pan Ras F132 antibody (Santa Cruz Biotechnology) at 4 °C for 5 hr. Resin was collected by centrifugation (10,000 × g) for 30 min; the proteins bound to the resin were washed with a buffer containing 500 mM NaCl, 10 mM MgCl2, 5 mM DTT, 1 mM EDTA, 0.1% Triton X-100, and 10 mM TrisHCl (pH 7.4). Nucleotides were eluted from the resin-bound proteins by treatment with a buffer containing 500 mM NaCl, 10 mM MgCl2, 5 mM DTT, 2.5% SDS, and 10 mM TrisHCl (pH 6.8) at 4 °C for 1 hr. The nucleotides were then analyzed by thin-layer chromatography and autoradiographed using a densitometry (Bio-Rad GS-670) as described in the previous study27 to determine the actual cellular fraction of the GTP-bound HRas proteins in the presence of both GEFs and GAPs (the cellular fRas•GTP).
Results
The course of this study involved preparation of wt HRas and multitudes of p-loop and NKCD/SAK HRas mutants listed in Table 1. Cdc25 and p120GAP were prepared, respectively, as GEF and GAP. The key kinetic parameters (depicted in Scheme 1) of these Ras proteins with and without Cdc25 and/or p120GAP were then determined (Figs. 2 and 3). The kinetic values are summarized in Tables 2 and 3. Using Equations 1–4 in conjunction with these values of the kinetic parameters, the intrinsic, GEF- and GAP-mediated and comprehensive values of the fRas•GTP of these Ras proteins were assessed (Table 4) and analyzed in detail.
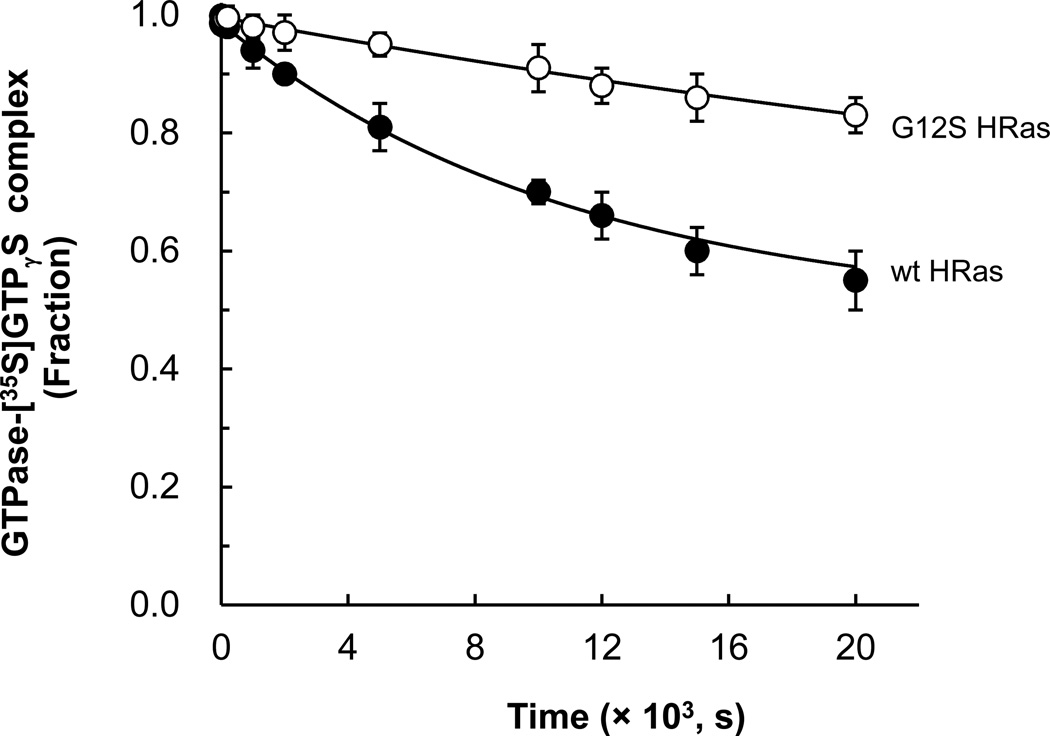
Figure 2. Estimation of the kinetic constants of the intrinsic GTP dissociations from wt and G12S HRas.
Measurements of the rate constants for the Ras GTP dissociation using [35S]GTPγS are described in Materials and Methods. Radioactivity values determined for the Ras-bound [35S]GTPγS at various time points were fractionated against the initial radioactivity value of the Ras-bound [35S]GTPγS (time = 0 s); the fractionated radioactivity values were plotted against time. Mean values with the SD of each data point of the plots as derived from three separate independent experiments are shown. Table 2 summarizes the k−1 values that were determined by the fit of these plotted values of wt and G12S HRas to a single exponential decay with regression values (r2) of > 0.9595.
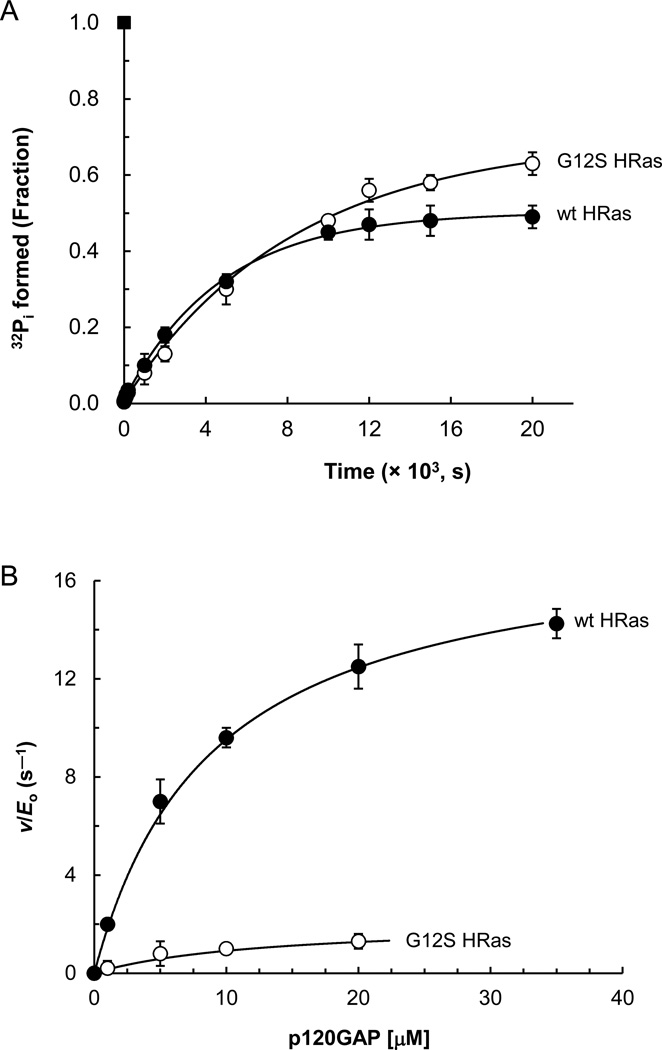
Figure 3. Determination of the kinetic constants for the intrinsic and p120GAP-mediated activities of wt and G12S HRas.
Kinetic constants of the phosphatase activity of Ras with and without p120GAP were assessed by using [γ-32P]GTP as described in Materials and Methods. A. All apparent intrinsic values determined for radioactivity were fractionated against the radioactivity value of [γ-32P]GTP that was initially added to the assay mixture and then plotted against time. All plot values represent mean values with the SD from three separate independent experiments. The estimated values of k+6 as determined by the fit of these plot data to a single exponential function with r2 > 0.9650 are summarized in Table 3. B. Ras GTPase activity assays in the presence of various concentrations of p120GAP were monitored over a period as described in Materials and Methods. The radioactivity values were plotted against time and fit to a single exponential function with r2 >0.9965 to determine apparent rates of Ras GTPase activities in the presence of various concentrations of p120GAP. The apparent rates with the SD of Ras GTPase activities were then plotted against the concentration of p120GAP. All apparent values shown are mean values with the SD from three separate independent experiments. The plots were determined to fit to a hyperbola with r2 > 0.9695 to give the Vmax and KM values of various Ras proteins coupled with p120GAP for the GTP hydrolysis. The Vmax values were converted into kcat(Vmax/Eo) values as described in Materials and Methods. The estimated kcat and KM values are summarized in Table 3.
Table 2. Kinetic parameters for the intrinsic and GEF-mediated GDP and GTP dissociation from wt HRas and its mutants.
The kinetic values with standard deviations (SD) of the intrinsic GTP dissociation from wt HRas and G12S were taken from Fig. 2. The kinetic values with the SD of intrinsic GTP dissociation from all other listed HRas mutants were obtained as described in Fig. 2. In addition, the kinetic values of intrinsic GDP dissociation with the SD from these Ras proteins also were obtained as described in Fig. 2, except that [3H]GDP, instead of [γ-32P]GTP, was used. The kcat (Vmax/Eo) values of the Cdc25-mediated nucleotide dissociation from these Ras proteins were obtained by using a saturation kinetic analysis essentially as described in Materials and Methods. A fixed concentration of Ras (1 µM) loaded with either mantGDP or mantGppNHp and variable concentrations of Cdc25 (0–900 µM) was used in this analysis. The values of KM also were obtained from this saturation kinetic analysis. However, because all determined KM values of these HRas mutants were indistinguishable from those of wt HRas (KM = ~340 µM), a listing of the detailed KM values of these HRas mutants is omitted. The SD of the estimated values were within 10% of the values shown. The values of "a, b, c, d, and e" were taken from the references “28, 23, 54, 21, and 51,” respectively.
| Intrinsic rate constants for Ras nucleotide dissociation (10−4 s−1) |
Kinetic parameters associated with Cdc25 (103 s−1 M−1) |
|||||
|---|---|---|---|---|---|---|
| GTP dissociation (k−1) |
GDP dissociation (k−2) |
GEF-mediated GTP dissociation (k+3) |
GEF-mediated GDP dissociation (k+4) |
|||
| HRas | wt HRas | 0.9 | 1.2 | 7.2 | 7.8 | |
| p-loop mutants | G12A | 0.9 | 0.5 | 7.5 | 7.3 | |
| G12C | 0.6 | 2.2 | 7.4 | 8.2 | ||
| G12D | 9.5 (89c) | 1.6 (1.4c) | 7.6 | 7.0 | ||
| G12E | 4.9 | 1.5 | 8.0 | 7.5 | ||
| G12S | 0.2 | 4.8 | 7.0 | 8.0 | ||
| G12V | 0.9 (0.78a) | 0.2 (0.38b) | 7.5 | 7.1 | ||
| G13C | 0.8 | 2.5 | 7.7 | 8.2 | ||
| G13D | 6.3 | 1.6 | 7.4 | 7.0 | ||
| G13S | 0.7 (0.83d) | 3.6 (3.83d) | 8.2 | 8.8 | ||
| G13V | 1.2 (5d) | 3.4 (105d) | 7.4 | 7.9 | ||
| NKCD/SAK mutants | K117R | 11.1 | 13.0 (32e) | 8.6 | 9.0 | |
| A146T | 8.1 | 9.7 | 8.1 | 8.2 | ||
| A146V | 9.3 | 13.5 | 8.4 | 8.8 | ||
Table 3.
Kinetic constants for the intrinsic and p120GAP-mediated GTPase activity of wt HRas and its mutants.
| Intrinsic Ras GTP hydrolysis rate (k+6) (10−4 s−1) |
Kinetic parameters associated with p120GAP |
|||||
|---|---|---|---|---|---|---|
|
kcat (k+8) (s−1) |
KM(µM) |
kcat/KM (× 106 s−1 M−1) |
||||
| HRas | wt HRas | 2.1 | 18.0 | 8.9 | 2.02 | |
| p-loop mutants | G12A | 0.5 | 0.5 | 46.8 | 0.01 | |
| G12C | 1.8 | 3.2 | 12.3 | 0.26 | ||
| G12D | 1.4 (1.7a) | 0.5 | 4.8 | 0.10 | ||
| G12E | 1.6 | 0.9 | 4.7 | 0.20 | ||
| G12S | 1.2 | 2.1 | 15.8 | 0.13 | ||
| G12V | 0.05 (< 0.03a; 0.33b) | 0.01 | < 100 | ~0 | ||
| G13C | 1.1 | 1.9 | 26.6 | 0.03 | ||
| G13D | 1.9 | 1.3 | 5.2 | 0.25 | ||
| G13S | 3.2 (5.33d) | 1.5 (0.5d) | 19.6 (21.6d) | 0.01 (0.023d) | ||
| G13V | 2.0 (2.17d) | 0.1 | 87.0 | <0.01 | ||
| NKCD/SAK mutants | K117R | 1.9 | 16.2 | 11.9 | 1.36 | |
| A146T | 1.9 | 12.5 | 10.7 | 1.05 | ||
| A146V | 1.8 (same as wte) | 10.7 | 13.8 | 0.78 | ||
The kinetic data of the intrinsic and p120GAP-mediated GTPase activities of wt HRas and G12S were taken from Fig. 2. Data for other HRas mutants were also collected as described in the legends of Figs. 2 and 3. The standard errors of the values were determined to be less than 10% of the values shown. The values of "a, b, c, d, and e" were taken from the references "55, 28, 33, 21, and 56, respectively.
Table 4. Theoretically estimated comprehensive f Ras•GTP values of wt HRas and its mutants.
The various fRas•GTP values of wt HRas and its mutants were estimated by using various kinetic parameters (Tables 2 and 3) and Equations 1–4 as described in the Supporting Information. The data in this table reflects: (i) the aintrinsic fRas•GTP, and the bGEF-mediated fRas•GTP (I, minimally active 5 nM Cdc25; II, highly active 5 nM Cdc25; and III, highly active 0.6 µM Cdc25); (ii) the cGAP-mediated fRas•GTP (10 nM p120GAP), and (iii) the dcomprehensive fRas•GTP (I, minimally active 5 nM Cdc25 with 10 nM p120GAP; II, highly active 5 nM Cdc25 with 10 nM p120GAP; and III, highly active 0.6 µM Cdc25 with 10 nM p120GAP).
| f Ras•GTP values (fraction) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Intrinsica | GEF-mediatedb | GAP-mediatedc | Comprehensived | |||||||
| I | II | III | I | II | III | |||||
| HRas | wt HRas | 0.33 | 0.39 | 0.89 | 0.90 | 0.01 | 0.01 | 0.44 | 0.89 | |
| p-loop mutants | G12A | 0.43 | 0.55 | 0.89 | 0.89 | 0.19 | 0.28 | 0.88 | 0.89 | |
| G12C | 0.51 | 0.55 | 0.90 | 0.90 | 0.07 | 0.08 | 0.80 | 0.90 | ||
| G12D | 0.37 | 0.42 | 0.89 | 0.90 | 0.10 | 0.12 | 0.84 | 0.90 | ||
| G12E | 0.39 | 0.44 | 0.88 | 0.89 | 0.06 | 0.07 | 0.80 | 0.89 | ||
| G12S | 0.78 | 0.79 | 0.90 | 0.91 | 0.23 | 0.24 | 0.85 | 0.91 | ||
| G12V | 0.55 | 0.74 | 0.90 | 0.90 | 0.46 | 0.67 | 0.89 | 0.90 | ||
| G13S | 0.50 | 0.52 | 0.89 | 0.89 | 0.23 | 0.25 | 0.85 | 0.89 | ||
| G13C | 0.65 | 0.68 | 0.88 | 0.90 | 0.21 | 0.24 | 0.86 | 0.90 | ||
| G13D | 0.36 | 0.40 | 0.89 | 0.90 | 0.05 | 0.06 | 0.78 | 0.90 | ||
| G13V | 0.59 | 0.61 | 0.89 | 0.90 | 0.54 | 0.57 | 0.89 | 0.90 | ||
| NKCD/SAK mutants | K117R | 0.79 | 0.79 | 0.89 | 0.90 | 0.08 | 0.08 | 0.57 | 0.89 | |
| A146T | 0.76 | 0.76 | 0.89 | 0.90 | 0.07 | 0.07 | 0.58 | 0.89 | ||
| A146V | 0.81 | 0.82 | 0.90 | 0.90 | 0.13 | 0.07 | 0.58 | 0.89 | ||
Calculation of the intrinsic fRas•GTP value of Ras
The value of the intrinsic fRas•GTP of Ras denotes the cellular population of the active GTP-bound form of Ras in the absence of any Ras regulators. A number of the intrinsic fRas•GTP values of Ras proteins (Table 4) were reached by determination of several kinetic parameters for subsequent calculation in Equation 2. These kinetic parameters include (i) the value of the intrinsic GNE of Ras proteins (k−1 and k−2) (Table 2) and (ii) the value of the intrinsic GTP hydrolysis of Ras proteins (k+6) (Table 3). This calculation used the previously reported average values of [GTP] (~300 µM) and [GDP] (~40 µM) in human cells.8
Analysis of the intrinsic fRas•GTP value of wt HRas
The intrinsic fRas•GTP value of wt HRas was calculated to be 0.33 (Table 4). The values of k−1, k−2, and k+6 of wt HRas (Tables 2 and 3) that determine the intrinsic fRas•GTP value of wt HRas did not deviate significantly from those of the previously reported values.21, 26, 28, 29
Analysis of the intrinsic fRas•GTP values of HRas mutants
All of the intrinsic fRas•GTP values of the listed HRas mutants exceeded the intrinsic fRas•GTP value of wt HRas (Table 4).
The HRas p-loop mutants G12A and G12V both have smaller k−2 values than wt HRas but have similar k−1 values (Table 2). Although smaller k−2 values in Equation 2 would suggest a decrease in intrinsic fRas•GTP values, an increase in intrinsic fRas•GTP values (Table 4) is present due to the k+6 values in Equation 2 that are much smaller than those of wt HRas (Table 3).
Compared with the values of wt HRas, the values of HRas p-loop mutants G12C, G13S, and G13C have a smaller k−1 and a larger k−2 (Table 2). Nevertheless, these HRas mutants have slightly decreased k+6 values compared with the k+6 value of wt HRas (Table 3). These combinations of k values in Equation 2 favor a higher value of intrinsic fRas•GTP than is found in wt HRas. Another HRas p-loop mutant, G12S, is intriguing, because it has the same trait ― a smaller k−1 and a larger k−2 value than that of wt HRas ― but to a larger extent than the HRas mutants G12C, G13S, and G13C (Table 2). G12S HRas also has a slightly increased k+6 value compared with the k+6 value of wt HRas (Table 3). The combination of these unique k values in Equation 2 is reflected in the intrinsic fRas•GTP value, because G12S has the largest intrinsic fRas•GTP value, 0.78, among these mutants (Table 4).
HRas p-loop mutants G12D, G13D, and G12E exhibit much larger k−1 values than wt HRas does, but display only marginally increased k−2 and marginally decreased k+6 values compared with those that wt HRas exhibits for the same elements (Tables 2 and 3). However, the intrinsic fRas•GTP values of these HRas mutants increased slightly in comparison with these values of wt HRas. This is the opposite of the decrease that had been expected (Table 4). This demonstrates again that the k−1 value contributes little to the denominator in Equation 2 compared with the contribution of k−2 and k+6.
The k−1 and k−2 values of G13V HRas are much higher than these same values of wt HRas (Table 2), but the k+6 value of G13V HRas is similar to that of wt HRas (Table 3). The higher k−1 and k−2 but invariable k+6 values indicate the faster Ras GNE. Besides, when the k+6 value is unchanged, the higher k−1 and k−2 values in Equation 2 produce a larger intrinsic fRas•GTP value of Ras. Hence, the larger intrinsic fRas•GTP value of G13V HRas than that of wt HRas (Table 4) reflects that the GNE of G13V HRas is faster than that of wt HRas. This is highlighted again in the case of the NKCD/SAK HRas mutants K117R, A146T, and A146V in which the significant change lies in their increased k−1 and k−2 values, but not in their k+6 value, in comparison with wt HRas (Tables 2 and 3). This causes the intrinsic fRas•GTP values of the NKCD/SAK HRas mutants to be nearly twice that of wt HRas (Table 4).
Calculation of the GEF-mediated fRas•GTP value of Ras
The cellular effects of the action of the positive Ras regulator GEFs on Ras proteins were assessed by calculation of the theoretical value of the GEF-mediated fRas•GTP of Ras (Equation 3). The value of the GEF-mediated fRas•GTP of Ras represents the cellular populations of the active GTP-bound form of Ras in the presence of GEFs. The factors that determine the GEF-mediated fRas•GTP of Ras are not only GEF-mediated Ras GNE but also intrinsic Ras GNE and GTP hydrolysis. Accordingly, Equation 3 consists of the GEF-relevant kinetic terms in addition to the intrinsic kinetic terms that determine the intrinsic fRas•GTP value. In Equation 3, the GEF-relevant kinetic terms are defined as k+3 and k+4 in combination with [GEF] (i.e., k+3·[GEF] and k+4·[GEF]).
[GEF] in Equation 3 stands for the total cellular concentration of GEFs, such as SOS, RasGRF, and RasGRP. The concentration of the dominantly expressed GEF is vastly different from cell to cell. For example, it has been shown that SOS was dominant in HEK-293T cells at a concentration of 5 nM.30 In Jurkat T cells, however, the SOS concentration was reported as 0.6 µM.31 For the purposes of this study, these two reported dominant cellular levels of SOS were essentially used as [GEF] in Equation 3. Although these two reported cases of variations in the levels of SOS expression do not necessarily represent its range of concentration in various cells, they certainly cover a cellularly relevant range of its concentrations from the scope of nM to µM.
Within this study, the values of the kinetic terms associated with GEF (i.e., k+3, and k+4) in Equation 3 were determined by using the Cdc25 from SOS. This Cdc25 retains the basal activity of SOS, and thus these estimated unmodified k+3 and k+4 values of Cdc25 represent the kinetic parameters of the minimally active SOS. SOS activity in cytoplasm is reported to be increased up to 500 times by formation of a complex of SOS with Ras on a plasma membrane.32 Hence, these values of k+3 and k+4 multiplied by 500 correspond to the kinetic parameters of the highly active SOS.
Taking into account variations in the levels of cellular expression and activities of SOS permits consideration of three different sets of the values of the GEF-relevant kinetic terms. These are represented by: (I) the minimally active 5 nM SOS (designating the action of 5 nM SOS with the original k+3 and k+4 values); (II) the highly active 5 nM SOS (representing the action of 5 nM SOS with k+3 and k+4 values multiplied by 500); and (III) the highly active 0.6 µM SOS (denoting the action of 0.6 µM SOS with k+3 and k+4 values multiplied by 500). Note that the case of the minimally active 0.6 µM SOS is unlisted. This is because the values of the case (II) "5 nM SOS with k+3 and k+4 values multiplied by 500" are equal to the values of the case "0.6 µM SOS with the original k+3 and k+4 values." Accordingly, we assessed three different values of the GEF-mediated fRas•GTP of Ras: (i) the GEF-mediated fRas•GTP of Ras with the minimally active 5 nM SOS (termed the GEF-mediated fRas•GTP (I) of Ras); (ii) the GEF-mediated fRas•GTP of Ras with the highly active 5 nM SOS (termed the GEF-mediated fRas•GTP (II) of Ras); and (iii) the GEF-mediated fRas•GTP of Ras with the highly active 0.6 µM SOS (termed the GEF-mediated fRas•GTP (III) of Ras).
Analysis of the GEF-mediated fRas•GTP values of wt HRas
All of the values of the GEF-mediated fRas•GTP (I), (II), and (III) of wt HRas, 0.39, 0.89, and 0.90, respectively, were larger than the value of 0.33 of the intrinsic fRas•GTP of wt HRas (Table 4). This is not too surprising, because the comparison of Equations 2 and 3 predicts that, in general, the presence of the GEF-relevant terms ― k+3·[GEF] and k+4·[GEF] ― in Equation 3 produces a GEF-mediated fRas•GTP (I) of Ras that is at least similar to or larger than the intrinsic fRas•GTP value of Ras. Also, Equation 3 predicts that a larger value of the GEF-relevant terms produces a larger value of the GEF-mediated fRas•GTP of wt HRas. Hence, it stands to reason that the GEF-mediated fRas•GTP (III) of wt HRas has a larger value than the GEF-mediated fRas•GTP (II) of wt HRas. The same is true for the larger value of the GEF-mediated fRas•GTP (II) of wt HRas in comparison with the value of the GEF-mediated fRas•GTP (I) of wt HRas.
Analysis of the GEF-mediated fRas•GTP values of HRas mutants
Similar to the values associated with wt HRas, the values of the GEF-mediated fRas•GTP (I) of the listed HRas p-loop mutants were larger than the values of the intrinsic fRas•GTP of the listed HRas p-loop mutants (Table 4). This is, as with wt Ras, because of the presence of the GEF-relevant terms of these p-loop HRas mutants in Equation 3. Intriguingly, among these listed HRas p-loop mutants, G12A and G12V show the largest increases in the GEF-mediated fRas•GTP (I) values in comparison with intrinsic fRas•GTP values (Table 4). Nevertheless, such value increases exceed expectations. One key contributor to such a value increase, in addition to the values of the GEF-relevant terms associated with G12A and G12V HRas, is the particularly small value of the k−2 of G12A and G12V HRas (Table 2). The presence of a smaller k−2 value in Equation 3 allows for the GEF-relevant terms to weigh more significantly in the determination of the value of the GEF-mediated fRas•GTP (I) of Ras. The opposite is true of the NKCD/SAK HRas mutants K117R, A146T, and A146V. Because these HRas mutants have a much larger k−2 value (Table 2), the GEF-relevant terms in Equation 3 have relatively little effect on the determination of the value of the GEF-mediated fRas•GTP (I) of Ras (Table 4).
Unlike the case of the GEF-mediated fRas•GTP (I) of Ras, the values of the GEF-mediated fRas•GTP (II) and (III) of all of the listed HRas mutants almost uniformly parallel the values of the GEF-mediated fRas•GTP (II) and (III) of wt HRas. These similarities occur because the values of the GEF-relevant terms of these HRas mutants associated with the highly active 5 nM and 0.6 µM SOS were significantly higher than the values of the combined intrinsic kinetic parameters of these HRas mutants in Equation 3. As a result, the values of the GEF-mediated fRas•GTP (II) and (III) of these HRas mutants reached their maximum.
Calculation of the GAP-mediated fRas•GTP value of Ras
The cellular consequences of the actions of the negative Ras regulator GAP on Ras were assessed by calculating the theoretical values of the GAP-mediated fRas•GTP of Ras (Equation 4). The value of the GAP-mediated fRas•GTP of Ras represents the cellular populations of the active GTP-bound Ras in the presence of GAPs.
Equation 4 contains not only the intrinsic kinetic terms that determine the intrinsic fRas•GTP value but also the GAP-relevant kinetic terms; these include k+7, k−7, and k+8 in conjunction with [GAP] (i.e., k+7·k+8/(k−7 + k+8)·[GAP]). Intriguingly, the GAP-relevant kinetic terms, k+7·k+8/(k−7 + k+8), in the denominator of Equation 4 are equivalent to the catalytic efficiency, kcat/KM, of GAP on Ras. Hence, with [GAP], the factors that determine the value of the GAP-mediated fRas•GTP of Ras that is of interest are those that govern the catalytic efficiency of GAP on Ras.
[GAP] in Equation 4 signifies the total concentration of the various types of GAPs in cells. Among these GAPs, p120GAP and NF1 are often dominantly and simultaneously expressed in cells.33 The kinetic values of kcat and KM of GAP for Ras (Table 3) were determined by using p120GAP with Ras. Therefore, the GAP-relevant kinetic values listed in Table 3 can only be used to assess the catalytic action of the p120GAP, but not NF1, on Ras. The concentration of p120GAP across various mammalian cells, including NIH 3T3 cells, was estimated to be ~10 nM.33 Yet, unlike with GEFs (i.e., 0.6 µM of SOS), the cellular concentration of p120GAP in the range of µM has not been reported. Taking these factors into account, only one value of the GAP-relevant kinetic terms ― the estimated values of kcat and KM in combination with 10 nM p120GAP ― is used within this study for calculation of the values of the GAP-mediated fRas•GTP of Ras.
Analysis of the GAP-mediated fRas•GTP value of wt HRas
The estimated GAP-mediated fRas•GTP value of wt HRas approached 0.01, a value much less than the intrinsic fRas•GTP value of 0.33 of wt HRas (Table 4). This large difference is due to the magnitude of the value of the GAP-relevant terms ― (k+7·k+8/(k−7 + k+8)·[GAP], which is equivalent to kcat/KM·[GAP] ― added to the denominator in Equation 4.
Analysis of the GAP-mediated fRas•GTP values of HRas mutants
All of the GAP-mediated fRas•GTP values of these HRas mutants exceeded the GAP-mediated fRas•GTP value of wt HRas. HRas mutants G12A, G12C, G12D, G12E, G12S, G13C, G13D, and G13S have GAP-mediated fRas•GTP values that lie between the two extremes of wt HRas and G12V mutant and also between the two extremes of wt HRas and G13V HRas mutant (see below) (Table 4). Compared with the values of wt HRas, the values of the GAP-relevant kinetic terms of these mutants are smaller, thus closer in value to the intrinsic kinetic terms. The decreased catalytic efficiency of p120GAP on these p-loop HRas mutants allows the intrinsic kinetic terms in Equation 4 to contribute more to the GAP-mediated fRas•GTP value compared with what occurs with wt HRas. This contribution by the intrinsic kinetic terms is responsible for the partially active states of these p-loop HRas mutants.
Other p-loop HRas mutants such as G12V and G13V have the largest GAP-mediated fRas•GTP values. They also have GAP-mediated fRas•GTP values that changed the least from their intrinsic fRas•GTP value. This is because of the miniscule catalytic efficiency of p120GAP on these HRas mutants (Table 3).
Unlike what happens with the p-loop HRas mutants, the catalytic efficiency of p120GAP on the NKCD/SAK HRas mutants K117R, A146T, and A146V does not significantly differ from wt HRas (Table 3). However the GAP-mediated fRas•GTP values of these NKCD/SAK HRas mutants are significantly higher than in wt HRas (Table 4). The increase in the GAP-mediated fRas•GTP value is because of the sufficiently large intrinsic kinetic values of these NKCD/SAK HRas mutants that counteract the values of the GAP-relevant kinetic terms in Equation 4 that are associated with these NKCD/SAK HRas mutants.
Calculation of the comprehensive fRas•GTP value of Ras
The effects of the simultaneous actions of both GEF and GAP on Ras were analyzed by calculation of the theoretical value of the comprehensive fRas•GTP of Ras (Equation 1). The comprehensive fRas•GTP of Ras represents the populations of the active GTP-bound Ras in the presence of both GEF and GAP in cells.
Within this study, three different values of the GEF-relevant kinetic terms were used to calculate the GEF-mediated fRas•GTP values of Ras (see above). For calculation of the GAP-mediated fRas•GTP values of Ras, only one value of the GAP-relevant kinetic terms was used (see above). Three distinct combinational value sets of the GEF/GAP-relevant kinetic terms are possible. These are represented by: (I) the minimally active 5 nM SOS/10 nM p120GAP; (II) the highly active 5 nM SOS/10 nM p120GAP; and (III) the highly active 0.6 µM SOS/10 nM p120GAP. Accordingly, three different sets of the comprehensive fRas•GTP values of Ras were calculated: the comprehensive fRas•GTP of Ras with the minimally active 5 nM SOS/10 nM p120GAP (termed the comprehensive fRas•GTP (I) of Ras); the comprehensive fRas•GTP of Ras with the highly active 5 nM SOS/10 nM p120GAP (termed the comprehensive fRas•GTP (II) of Ras); and the comprehensive fRas•GTP of Ras with the highly active 0.6 µM SOS/10 nM p120GAP (termed the comprehensive fRas•GTP (III) of Ras). These values of the comprehensive fRas•GTP (I), (II), and (III) of the Ras of interest, including wt, G12V, and G12S HRas, were further used to analyze the corresponding actual values of the fraction of the GTP-bound form (the cellular fRas•GTP) of Ras measured in NIH 3T3 cells.
Analysis of the comprehensive fRas•GTP values of wt HRas
The values of the comprehensive fRas•GTP (I), (II), and (III) of wt HRas are, respectively, 0.01, 0.44, and 0.89 (Table 4). The results suggest that the comprehensive fRas•GTP value of wt HRas depends largely on the combinational actions of SOS with p120GAP.
The cellular fRas•GTP value of the transfected wt HRas in unstimulated NIH 3T3 cells was estimated to be 0.12 (Fig. 4). Notably, a previously measured cellular fRas•GTP value of the transfected wt HRas in unstimulated NIH 3T3 cells was reported to be 0.025.34 One possible reason for such different values would be the different quantification methods used for the GTP-and GDP-bound Ras isolated from cells. The difference in cell culture conditions may also affect the ratio of the GTP-bound Ras over the GDP-bound Ras. Regardless of the value differences, those of the estimated cellular fRas•GTP values of the transfected and endogenous wt HRas in unstimulated NIH 3T3 cells fall between the values of the comprehensive fRas•GTP (I) and (II) of wt HRas (Table 4). This is not unexpected because the cellular expression and activity of GEFs and GAPs in unstimulated NIH 3T3 cells would not duplicate the value sets in Equation 1 for the calculations of any of these comprehensive fRas•GTP (I), (II), and (III) values of wt HRas. By setting the values of the GEF- and GAP-relevant kinetic terms for the calculation of the comprehensive fRas•GTP (I), (II), and (III) values of wt HRas as default values, a comparison of these comprehensive fRas•GTP values of wt HRas with the cellular fRas•GTP values of wt HRas in unstimulated NIH 3T3 cells may predict the status of SOS in unstimulated NIH 3T3 cells. For example, the estimated value of the cellular fRas•GTP of the transfected wt HRas in unstimulated NIH 3T3 cells (Fig. 4) can be obtained by a value combination of ~80% of the minimally active 5 nM SOS with ~20% of the highly active 5 nM SOS plus 10 nM p120GAP in Equation 1. Nevertheless, direct measurements of the cellular expression levels and activities of GEFs and GAPs are necessary. Such measurements increase the likelihood of obtaining meaningful estimated percentages for the cellular expression levels and activities of SOS and p120GAP associated with the comprehensive fRas•GTP value of wt HRas.
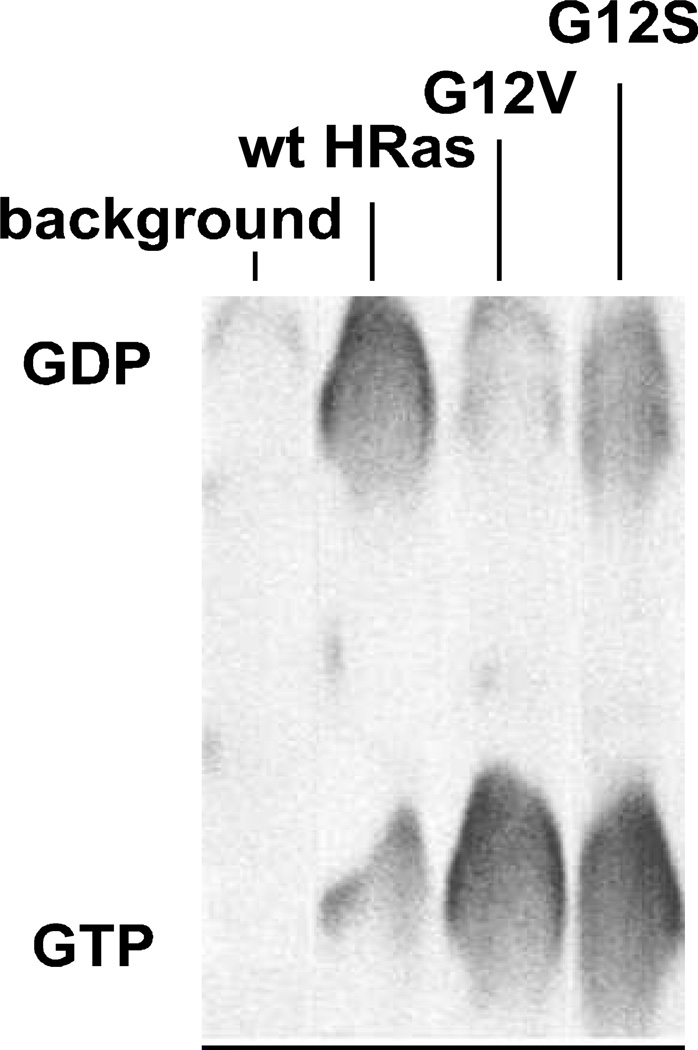
Figure 4. Determination of the fractions of the HRas-bound GTP in cells.
GTP fractions bound to various Ras expressed in unstimulated NIH 3T3 cells were determined as described in Materials and Methods. Western blot analysis showed that all transfected Ras proteins were evenly expressed. The fraction values of the Ras-bound GTP using densitometry estimation of GTP and GDP concentrations were calculated to be: background (transfection of the mammalian expression vector without ras gene), not determined; wt HRas, 0.12; G12V HRas, 0.81; and G12S HRas, 0.62. The presented values are the averages of the values of the independent triplicate measurements using separate cell culture samples, and the SD are less than 10% of the GTP fraction values that were indicated.
Note that the previous study also showed that the fraction of the cellular fRas•GTP value of the transfected wt HRas in unstimulated NIH 3T3 cells does not significantly differ from the cellular fRas•GTP value of the endogenous wt HRas in unstimulated NIH 3T3 cells.34 This lack of a significant difference was found despite the expression level of the transfected wt HRas being much higher than that of the endogenous wt HRas in unstimulated NIH 3T3 cells. Hence, the feature analysis of the cellular fRas•GTP value of the transfected wt HRas in unstimulated NIH 3T3 cells that was performed by using the comprehensive fRas•GTP (I), (II), and (III) values of wt HRas (see above) may reflect the cellular traits of the endogenous wt HRas. This endogenous wt HRas is associated with the cellular expressions and activities of SOS and p120GAP in unstimulated NIH 3T3 cells.
Analysis of the comprehensive fRas•GTP values of HRas mutants
To one degree or another, all of the values of the comprehensive fRas•GTP (I) of these HRas mutants exceeded the values of the comprehensive fRas•GTP (I) of wt HRas.
According to the main kinetic factor that changes the value of the comprehensive fRas•GTP (I) of Ras, the values of the comprehensive fRas•GTP (I) of the p-loop and NKCD/SAK HRas mutants can be divided into two types for purposes of comparison with the values of the comprehensive fRas•GTP (I) of wt HRas. First, the values of the GAP-relevant kinetic terms of HRas mutants in Equation 1 are almost 0; as a consequence, the outcome of the value of the comprehensive fRas•GTP (I) of HRas mutants is significantly large. Mechanically, such small values of the GAP-relevant kinetic terms of HRas mutants are rooted in the almost total impairment of the catalytic action of p120GAP on HRas mutants. The two p-loop HRas mutants G12V and G13V possess this kinetic characteristic. Second, the higher values of the intrinsic kinetic terms of HRas mutants and/or the lower values of the GAP-relevant kinetic terms for HRas mutants, when compared with the values associated with wt HRas in Equation 1, yield larger values of the comprehensive fRas•GTP (I) of HRas mutants than the value of the comprehensive fRas•GTP (I) of wt HRas. In all of these cases, these altered values are because of the perturbed intrinsic kinetic processes of HRas mutants and/or GAP catalytic efficiencies on HRas mutants. With the exceptions of G12V and G13V HRas, all of the examined HRas mutants have this kinetic trait.
Similar to the case of the comprehensive fRas•GTP (I) of these HRas mutants, all of the values of the comprehensive fRas•GTP (II) of HRas mutants were higher under the same conditions than the values of the comprehensive fRas•GTP of wt HRas (Table 4). Based upon the main kinetic factor that contributes to altering the value of the comprehensive fRas•GTP (II) of Ras, the values of the comprehensive fRas•GTP (II) of the p-loop and NKCD/SAK HRas mutants fall into two categories when compared with the value of the comprehensive fRas•GTP (II) of wt HRas. First, the higher values of the intrinsic kinetic terms of these HRas mutants — in conjunction with the comparatively lower values of the catalytic efficiencies of p120GAP on these HRas mutants than those of wt HRas — mainly contribute to give higher values for the comprehensive fRas•GTP (II) of HRas mutants than for the comprehensive fRas•GTP (II) of wt HRas. The perturbed intrinsic processes of HRas mutants and GAP catalytic efficiencies on HRas mutants are, respectively, responsible for such value changes in the intrinsic kinetic terms and/or the GAP-relevant kinetic terms of HRas mutants. All p-loop HRas mutants, but not these NKCD/SAK HRas mutants, have this kinetic trait. Second, only a significantly high value of the intrinsic kinetic terms of HRas mutants, compared with those of wt HRas, contributes to the generation of higher values of the comprehensive fRas•GTP (II) of HRas mutants than of the comparative values of wt HRas. The significantly higher values of the intrinsic kinetic terms of HRas mutants, compared with those of wt HRas, is because of the perturbed intrinsic processes of HRas mutants. In this case, no causation can be attributed to the change in the values of the catalytic efficiencies of p120GAP of HRas mutants. The NKCD/SAK HRas mutants, including K117R, A146T, and A146V HRas, have this kinetic feature.
All values of the comprehensive fRas•GTP (III) of these HRas mutants are almost uniformly similar to the value of the comprehensive fRas•GTP (III) of wt HRas, which is extremely high (i.e., ≥ 0.87) (Table 4). This similarity is because the value of the GEF-relevant kinetic terms (the value associated with the highly active 0.6 µM SOS) overwhelms all other kinetic parameters associated with the value of the GAP-relevant kinetic terms (the value associated with 10 nM p120GAP) and the intrinsic kinetic parameter values of Ras in Equation 1. In addition, as discussed elsewhere, this highly active 0.6 µM SOS/10 nM p120GAP condition can also generate an extremely high value for the comprehensive fRas•GTP of wt HRas. The highly active 0.6 µM SOS/10 nM p120GAP generates significantly higher values for the comprehensive fRas•GTP (III) of Ras than it does for any examined HRas proteins. Because of these high values, it is certain that a cellular condition with the highly active 0.6 µM SOS/10 nM p120GAP is sufficient to produce uncontrolled Ras-dependent cellular signaling events without regard to the values of any other examined HRas mutants or wt HRas.
G12S HRas is one of the most predominant forms found in Costello syndrome (Table 1).35 Hence, the value of the cellular fRas•GTP of G12S HRas in unstimulated NIH 3T3 cells was determined as a way to use various combinations of activities and expression levels of SOS with p120GAP to evaluate the various comprehensive fRas•GTP values of G12S HRas. Because of the important role of G12V HRas in cancer formation, the cellular fRas•GTP value of G12V HRas in unstimulated NIH 3T3 cells also was determined to be a way to use various combinations of activities and expression levels of SOS with p120GAP to evaluate these various comprehensive fRas•GTP (I), (II), (III) values of G12V HRas. The cellular fRas•GTP value of G12V HRas in unstimulated NIH 3T3 cells also serves as a positive control for the analysis of the cellular fRas•GTP value of G12S HRas in unstimulated NIH 3T3 cells. The value of the cellular fRas•GTP of wt HRas in unstimulated NIH 3T3 cells serves as a negative control for this analysis. This cellular fRas•GTP value of G12V and G12S HRas in unstimulated NIH 3T3 cells was determined to be 0.80 and 0.63, respectively, which is ~8.0- and 6.3-fold higher than the cellular fRas•GTP value of wt HRas in unstimulated NIH 3T3 cells (Fig. 4).
Discussion
This study established novel kinetic parameter-based calculations of the values of intrinsic, GEF- and GAP-mediated, and comprehensive fRas•GTP of Ras proteins that represent the cellular content of the GTP-bound form of Ras in the presence and absence of GEF and/or GAP. The kinetic characterizations linked with the calculations of the population of the GTP-bound form of Ras first provide an overall picture of the inherited causality between Ras mutations and changes in the cellular population of the GTP-bound Ras. These linkages explain the biochemical roles of these HRas mutants in various diseases. These roles include diseases such as Costello syndrome and certain cancers.
Depending on the cellular activity and expression of GEFs in combination with GAPs, three sets of values of the comprehensive fRas•GTP of Ras ― the comprehensive fRas•GTP (I), (II), and (III) of these HRas ― were calculated. Comparison of these calculated values with the values of the cellular fRas•GTP of selected HRas proteins in the unstimulated NIH 3T3 cells suggests that the comprehensive fRas•GTP (I) values of HRas mainly represents the actual cellular f Ras•GTP values of HRas in the unstimulated NIH 3T3 cells. This recognition takes into account the component of the comprehensive fRas•GTP (II) of HRas. Intriguingly, although there are no clear-cut dividing lines, the spectrum of the calculated values of the comprehensive fRas•GTP (I) of HRas mutants can be classified into three groups. The first group encompasses the values of 0.57–0.67 and is associated with G12V and G13V HRas mutants. The G12V and G13V HRas mutations are the only ones linked specifically and exclusively to cancer formation. The second group spans 0.24–0.28 and is linked to G12A, G12S, G13S, and G13C HRas mutations. These HRas mutations are mainly linked to development of Costello syndrome, but they are also often linked to cancers. Finally, the third group has a range of 0.06–0.12 and is associated with all other listed HRas mutants. This group includes G12C, G12D, G12E, G13D, K117R, A146T, and A146V HRas mutations that are only linked to development of Costello syndrome. These groups and their links to cancer and/or Costello syndrome suggest that the high end of the spectrum of values of the comprehensive fRas•GTP (I) of HRas mutants is certainly linked to cancer formation, but the low end of this spectrum is only associated with the development of Costello syndrome. Values in the midrange of this spectrum are linked with the development of both of Costello syndrome and cancer. Accordingly, it is possible to postulate that the values of the comprehensive fRas•GTP (I) of HRas mutants can be used to gauge whether Ras mutants cause development of diseases such as cancers and/or Costello syndrome. For example, if a comprehensive fRas•GTP (I) value of a certain HRas mutant is 0.25, this HRas mutation is likely to cause development of Costello syndrome and/or cancers. However, if the same value of a certain HRas mutant is 0.10, this HRas mutation is likely to lead only to development of Costello syndrome. The cellular fRas•GTP values of HRas proteins from unstimulated NIH 3T3 cells were used as references for the analyses of the comprehensive fRas•GTP (I) values of HRas proteins. Therefore, the analytical results discussed above cannot be applied immediately to diseases associated with Ras mutations in other cells. Use of our results to gauge other diseases must await further evaluation of the comprehensive fRas•GTP (I) of HRas by using the cellular fRas•GTP values of HRas from various other cells.
As was discussed in the Results section, the reason for such a high spectrum of values for the comprehensive fRas•GTP (I) of HRas mutants ― G12V and G13V HRas ― is because the catalytic action of p120GAP on these HRas mutants is impaired. However, the middle and the lower end of this spectrum — which includes all of the HRas mutants of this study except G12V and G13V HRas ―reflects the perturbation of the intrinsic kinetic parameters of HRas mutants in combination with the partial perturbation of these mutants by the catalytic action of p120GAP. Accounting for the linkages between certain groups of the comprehensive fRas•GTP (I) values of these HRas mutants and certain types of diseases (see above), the features of the mechanical perturbation of these HRas mutants can be further linked to the type of the diseases that they are associated with. The severe impairment of the catalytic action of p120GAP on HRas mutants results in the high-end values of the comprehensive fRas•GTP (I) of Ras. These values at the upper end of the spectrum cause development of cancers. The perturbation of the intrinsic kinetic parameters of HRas mutants in combination with the partial perturbation of HRas mutants by the catalytic action of p120GAP leads to values in the middle or low ranges of the spectrum. Values in these ranges are sufficient for development of cancers and/or the Costello syndrome.
Earlier, the main factor in increases in the cellular population of the GTP-bound Ras was thought to be caused by the impaired catalytic action of GAP on HRas, with HRas mutations as the culprits in the impairment. This outlook remains consistent with the values at the upper end of the spectrum of values of the comprehensive fRas•GTP (I) of the tumorigenic G12V and G13V HRas mutants. However, this study is the first to show that perturbation that these HRas mutations cause in the intrinsic kinetic properties of Ras also plays a key role in increases in the cellular population of the GTP-bound HRas. This notion is supported by the middle and low end spectrum of values of the comprehensive fRas•GTP (I) of HRas proteins. Values in these ranges of the spectrum include all of the listed HRas mutants, except G12V and G13V HRas. One of the best examples of this is the value of the comprehensive fRas•GTP (I) of the G12S HRas mutation. This occurrence is intriguing because G12S HRas is the most prevalent form of HRas mutant found in patients with Costello syndrome.
The value of the comprehensive fRas•GTP (III) of Ras represents a case in which the population of the GTP-bound form of Ras exists under conditions of extreme SOS expression and activity in cells. Regardless of the features of the HRas mutants, the values of the comprehensive fRas•GTP (III) of these HRas mutants are significantly high. Intriguingly, the development of at least one case of Noonan syndrome has been linked to high cellular fRas•GTP values of wt KRas and wt NRas.13 The high values encountered in this case are suspected to be the result of upregulation of SOS that was caused by mutations of the sos1 gene.13 This SOS upregulation-dependent development of Noonan syndrome can be explained by an incident in the calculation of the value of the comprehensive fRas•GTP value (III) of wt HRas that dovetails with high SOS activity and expression.
Because other GAPs, such as NF1, have not been included in these analyses, an assessment of the effect of p120GAP on the cellular population of the GTP-bound Ras through the action of these HRas mutations should not be overinterpreted. Moreover, no exploration has been undertaken of the possibility of a change in the p120GAP expression-dependent cellular population of the GTP-bound form of Ras that is associated with these HRas mutations. This limitation reflects the lack of evidence of higher or lower cellular expression of p120GAP other than as 10 nM p120GAP. Future studies are expected to examine the possibility that the various cellular expressions of p120GAP as well as NF1 modulate the cellular population of these HRas mutations in the GTP-bound form of Ras.
Supplementary Material
Acknowledgments
Funding Source Statement: This work was supported by NIH grant 1R15AI096146-01A1 to J.H.
Abbreviations and Textual Footnotes
- GAP
GAP protein concentration
- GEF
GEF protein concentration
- Cdc25
Ras SOS1 catalytic core domain
- GAPs
GTPase-activating proteins
- GEFs
guanine nucleotide exchange factors
- GNE
guanine nucleotide exchange
- HRas
Harvey Ras
- KRas
Kirsten Ras
- mantGDP
2'(3')-O-(N-methylanthraniloyl) guanosine diphosphate
- mantGppNHp
the 2'(3')-O-(N-methylanthraniloyl) 5'-guanylyl-imidodiphosphate
- NF1
neurofibromin1
- NRas
Neuroblastoma Ras
- RasGRF
Ras guanine-nucleotide-release factor
- RasGRP
Ras guanyl nucleotide-releasing protein
- SOS
Son of Sevenless
- the comprehensive fRas•GTP (I) of Ras
the comprehensive fRas•GTP of Ras with the minimally active 5 nM SOS/5 nM p120GAP
- the comprehensive fRas•GTP (II) of Ras
the comprehensive fRas•GTP of Ras with the highly active 5 nM SOS/5 nM p120GAP
- the comprehensive fRas•GTP (III) of Ras
the comprehensive fRas•GTP of Ras with the highly active 0.6 µM SOS/5 nM p120GAP
- the comprehensive fRas•GTP
the overall comprehensive cellular fraction of the GTP-bound Ras over the GTP- and GDP-bound Ras in the presence of GEF and GAP
- the GAP-mediated fRas•GTP
the cellular fraction of the GTP-bound Ras over the GTP- and GDP-bound Ras in the presence of GAP
- the GEF-mediated fRas•GTP (I) of Ras
the GEF-mediated fRas•GTP of Ras with the minimally active 5 nM SOS
- the GEF-mediated fRas•GTP (II) of Ras
the GEF-mediated fRas•GTP of Ras with the highly active 5 nM SOS
- the GEF-mediated fRas•GTP (III) of Ras
the GEF-mediated fRas•GTP of Ras with the highly active 0.6 µM SOS
- the GEF-mediated fRas•GTP
the cellular fraction of the GTP-bound Ras over the GTP- and GDP-bound Ras in the presence of GEF
- the intrinsic fRas•GTP
the intrinsic cellular fraction of the GTP-bound Ras over the GTP- and GDP-bound Ras
- wild type
wt
Footnotes
Supporting Information
The derivation of Equations 1–4 is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Oxford G, Theodorescu D. Ras superfamily monomeric G proteins in carcinoma cell motility. Cancer Lett. 2003;189:117–128. doi: 10.1016/s0304-3835(02)00510-4. [DOI] [PubMed] [Google Scholar]
- 2.Marshall CB, Meiri D, Smith MJ, Mazhab-Jafari MT, Gasmi-Seabrook GMC, Rottapel R, Stambolic V, Ikura M. Probing the GTPase cycle with realtime NMR: GAP and GEF activities in cell extracts. Methods. 2012;57:473–485. doi: 10.1016/j.ymeth.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 3.Geyer M, Wittinghofer A. GEFs, GAPs, GDIs and effectors: taking a closer (3D) look at the regulation of Ras-related GTP-binding proteins. Curr. Opin. Struct. Biol. 1997;7:786–792. doi: 10.1016/s0959-440x(97)80147-9. [DOI] [PubMed] [Google Scholar]
- 4.Bonfini L, Karlovich CA, Dasgupta C, Banerjee U. The Son of sevenless gene product: a putative activator of Ras. Science. 1992;255:603–606. doi: 10.1126/science.1736363. [DOI] [PubMed] [Google Scholar]
- 5.Ebinu JO, Bottorff DA, Chan EY, Stang SL, Dunn RJ, Stone JC. RasGRP, a Ras guanyl nucleotide- releasing protein with calcium- and diacylglycerol-binding motifs. Science. 1998;280:1082–1086. doi: 10.1126/science.280.5366.1082. [DOI] [PubMed] [Google Scholar]
- 6.Bottorff D, Ebinu J, Stone JC. RasGRP, a Ras activator: mouse and human cDNA sequences and chromosomal positions. Mamm. Genome. 1999;10:358–361. doi: 10.1007/s003359901001. [DOI] [PubMed] [Google Scholar]
- 7.Boriack-Sjodin PA, Margarit SM, Bar-Sagi D, Kuriyan J. The structural basis of the activation of Ras by Sos. Nature. 1998;394:337–343. doi: 10.1038/28548. [DOI] [PubMed] [Google Scholar]
- 8.Traut TW. Physiological concentrations of purines and pyrimidines. Mol. Cell. Biochem. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 9.Sprang S. GEFs: master regulators of G-protein activation. Trends Biochem. Sci. 2001;26:266–267. doi: 10.1016/s0968-0004(01)01818-7. [DOI] [PubMed] [Google Scholar]
- 10.Grewal T, Koese M, Tebar F, Enrich C. Differential Regulation of RasGAPs in Cancer. Genes Cancer. 2011;2:288–297. doi: 10.1177/1947601911407330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, Schmitz F, Wittinghofer A. The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 12.Boguski MS, McCormick F. Proteins regulating Ras and its relatives. Nature. 1993;366:643–654. doi: 10.1038/366643a0. [DOI] [PubMed] [Google Scholar]
- 13.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat. Rev. Cancer. 2007;7:295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 14.Roberts PJ, Der CJ. Targeting the Raf-MEK-ERK mitogen-activated protein kinase cascade for the treatment of cancer. Oncogene. 2007;26:3291–3310. doi: 10.1038/sj.onc.1210422. [DOI] [PubMed] [Google Scholar]
- 15.Fasano O, Aldrich T, Tamanoi F, Taparowsky E, Furth M, Wigler M. Analysis of the transforming potential of the human H-ras gene by random mutagenesis. Proc. Natl. Acad. Sci. U. S. A. 1984;81:4008–4012. doi: 10.1073/pnas.81.13.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuda K, Shimada A, Yoshida N, Ogawa A, Watanabe A, Yajima S, Iizuka S, Koike K, Yanai F, Kawasaki K, Yanagimachi M, Kikuchi A, Ohtsuka Y, Hidaka E, Yamauchi K, Tanaka M, Yanagisawa R, Nakazawa Y, Shiohara M, Manabe A, Kojima S. Spontaneous improvement of hematologic abnormalities in patients having juvenile myelomonocytic leukemia with specific RAS mutations. Blood. 2007;109:5477–5480. doi: 10.1182/blood-2006-09-046649. [DOI] [PubMed] [Google Scholar]
- 17.Jakubauskas A, Griskevicius L. KRas and BRaf mutational status analysis from formalin-fixed, paraffin-embedded tissues using multiplex polymerase chain reaction-based assay. Arch. Pathol. Lab. Med. 2010;134:620–624. doi: 10.5858/134.4.620. [DOI] [PubMed] [Google Scholar]
- 18.Sahu RP, Batra S, Kandala PK, Brown TL, Srivastava SK. The role of K-ras gene mutation in TRAIL-induced apoptosis in pancreatic and lung cancer cell lines. Cancer Chemother. Pharmacol. 2011;67:481–487. doi: 10.1007/s00280-010-1463-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seeburg PH, Colby WW, Capon DJ, Goeddel DV, Levinson AD. Biological properties of human c-Ha-ras1 genes mutated at codon 12. Nature. 1984;312:71–75. doi: 10.1038/312071a0. [DOI] [PubMed] [Google Scholar]
- 20.Hall A, Self AJ. The effect of Mg2+ on the guanine nucleotide exchange rate of p21N-ras. J. Biol. Chem. 1986;261:10963–10965. [PubMed] [Google Scholar]
- 21.Gideon P, John J, Frech M, Lautwein A, Clark R, Scheffler JE, Wittinghofer A. Mutational and kinetic analyses of the GTPase-activating protein (GAP)-p21 interaction: the C-terminal domain of GAP is not sufficient for full activity. Mol. Cell. Biol. 1992;12:2050–2056. doi: 10.1128/mcb.12.5.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenzen C, Cool RH, Prinz H, Kuhlmann J, Wittinghofer A. Kinetic analysis by fluorescence of the interaction between Ras and the catalytic domain of the guanine nucleotide exchange factor Cdc25Mm . Biochemistry. 1998;37:7420–7430. doi: 10.1021/bi972621j. [DOI] [PubMed] [Google Scholar]
- 23.Cotton FA, Wilkinson G. Advanced Inorganic Chemistry. 5th. New York: John Wiley & Sons; 1988. [Google Scholar]
- 24.Heo J, Campbell SL. Superoxide Anion Radical Modulates the Activity of Ras and Ras-related GTPases by a Radical-based Mechanism Similar to that of Nitric Oxide. J. Biol. Chem. 2005;280:12438–12445. doi: 10.1074/jbc.M414282200. [DOI] [PubMed] [Google Scholar]
- 25.Leupold CM, Goody RS, Wittinghofer A. Stereochemistry of the elongation factor Tu X GTP complex. Eur. J. Biochem. 1983;135:237–241. doi: 10.1111/j.1432-1033.1983.tb07643.x. [DOI] [PubMed] [Google Scholar]
- 26.Eccleston JF, Moore KJ, Morgan L, Skinner RH, Lowe PN. Kinetics of interaction between normal and proline 12 Ras and the GTPase-activating proteins, p120-GAP and neurofibromin. The significance of the intrinsic GTPase rate in determining the transforming ability of ras. J. Biol. Chem. 1993;268:27012–27019. [PubMed] [Google Scholar]
- 27.Downward J, Graves JD, Warne PH, Rayter S, Cantrell DA. Stimulation of p21ras upon T-cell activation. Nature. 1990;346:719–723. doi: 10.1038/346719a0. [DOI] [PubMed] [Google Scholar]
- 28.Krengel U, Schlichting I, Scherer A, Schumann R, Frech M, John J, Kabsch W, Pai EF, Wittinghofer A. Three-dimensional structures of H-ras p21 mutants: molecular basis for their inability to function as signal switch molecules. Cell. 1990;62:539–548. doi: 10.1016/0092-8674(90)90018-a. [DOI] [PubMed] [Google Scholar]
- 29.John J, Frech M, Wittinghofer A. Biochemical properties of Ha-ras encoded p21 mutants and mechanism of the autophosphorylation reaction. J. Biol. Chem. 1988;263:11792–11799. [PubMed] [Google Scholar]
- 30.Maurer T, Garrenton LS, Oh A, Pitts K, Anderson DJ, Skelton NJ, Fauber BP, Pan B, Malek S, Stokoe D, Ludlam MJ, Bowman KK, Wu J, Giannetti AM, Starovasnik MA, Mellman I, Jackson PK, Rudolph J, Wang W, Fang G. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc. Natl. Acad. Sci. U. S. A. 2012;109:5299–5304. doi: 10.1073/pnas.1116510109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuniba H, Pooh RK, Sasaki K, Shimokawa O, Harada N, Kondoh T, Egashira M, Moriuchi H, Yoshiura K, Niikawa N. Prenatal diagnosis of Costello syndrome using 3D ultrasonography amniocentesis confirmation of the rare HRAS mutation G12D. Am. J. Med. Genet. A. 2009;149A:785–787. doi: 10.1002/ajmg.a.32335. [DOI] [PubMed] [Google Scholar]
- 32.Gureasko J, Galush WJ, Boykevisch S, Sondermann H, Bar-Sagi D, Groves JT, Kuriyan J. Membrane-dependent signal integration by the Ras activator Son of sevenless. Nat. Struct. Mol. Biol. 2008;15:452–461. doi: 10.1038/nsmb.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bollag G, McCormick F. Differential regulation of rasGAP and neurofibromatosis gene product activities. Nature. 1991;351:576–579. doi: 10.1038/351576a0. [DOI] [PubMed] [Google Scholar]
- 34.Scheele JS, Rhee JM, Boss GR. Determination of absolute amounts of GDP and GTP bound to Ras in mammalian cells: comparison of parental and Ras-overproducing NIH 3T3 fibroblasts. Proc. Natl. Acad. Sci. U. S. A. 1995;92:1097–1100. doi: 10.1073/pnas.92.4.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gelb BD, Tartaglia M. Noonan syndrome and related disorders: dysregulated RAS-mitogen activated protein kinase signal transduction. Hum. Mol. Genet. 2006;15(Spec No 2):R220–R226. doi: 10.1093/hmg/ddl197. [DOI] [PubMed] [Google Scholar]
- 36.Aoki Y, Niihori T, Kawame H, Kurosawa K, Ohashi H, Tanaka Y, Filocamo M, Kato K, Suzuki Y, Kure S, Matsubara Y. Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat. Genet. 2005;37:1038–1040. doi: 10.1038/ng1641. [DOI] [PubMed] [Google Scholar]
- 37.Gripp KW, Lin AE, Stabley DL, Nicholson L, Scott CI, Jr., Doyle D, Aoki Y, Matsubara Y, Zackai EH, Lapunzina P, Gonzalez-Meneses A, Holbrook J, Agresta CA, Gonzalez IL, Sol-Church K. HRAS mutation analysis in Costello syndrome: genotype and phenotype correlation. Am. J. Med. Genet. A. 2006;140:1–7. doi: 10.1002/ajmg.a.31047. [DOI] [PubMed] [Google Scholar]
- 38.Estep AL, Tidyman WE, Teitell MA, Cotter PD, Rauen KA. HRAS mutations in Costello syndrome: detection of constitutional activating mutations in codon 12 and 13 and loss of wild-type allele in malignancy. Am. J. Med. Genet. A. 2006;140:8–16. doi: 10.1002/ajmg.a.31078. [DOI] [PubMed] [Google Scholar]
- 39.Kerr B, Delrue MA, Sigaudy S, Perveen R, Marche M, Burgelin I, Stef M, Tang B, Eden OB, O'Sullivan J, De Sandre-Giovannoli A, Reardon W, Brewer C, Bennett C, Quarell O, M'Cann E, Donnai D, Stewart F, Hennekam R, Cave H, Verloes A, Philip N, Lacombe D, Levy N, Arveiler B, Black G. Genotype-phenotype correlation in Costello syndrome: HRAS mutation analysis in 43 cases. J. Med. Genet. 2006;43:401–405. doi: 10.1136/jmg.2005.040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sol-Church K, Stabley DL, Nicholson L, Gonzalez IL, Gripp KW. Paternal bias in parental origin of HRAS mutations in Costello syndrome. Hum. Mutat. 2006;27:736–741. doi: 10.1002/humu.20381. [DOI] [PubMed] [Google Scholar]
- 41.Zampino G, Pantaleoni F, Carta C, Cobellis G, Vasta I, Neri C, Pogna EA, De Feo E, Delogu A, Sarkozy A, Atzeri F, Selicorni A, Rauen KA, Cytrynbaum CS, Weksberg R, Dallapiccola B, Ballabio A, Gelb BD, Neri G, Tartaglia M. Diversity, parental germline origin, and phenotypic spectrum of de novo HRAS missense changes in Costello syndrome. Hum. Mutat. 2007;28:265–272. doi: 10.1002/humu.20431. [DOI] [PubMed] [Google Scholar]
- 42.van der Burgt I, Kupsky W, Stassou S, Nadroo A, Barroso C, Diem A, Kratz CP, Dvorsky R, Ahmadian MR, Zenker M. Myopathy caused by HRAS germline mutations: implications for disturbed myogenic differentiation in the presence of constitutive HRas activation. J. Med. Genet. 2007;44:459–462. doi: 10.1136/jmg.2007.049270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gripp KW, Innes AM, Axelrad ME, Gillan TL, Parboosingh JS, Davies C, Leonard NJ, Lapointe M, Doyle D, Catalano S, Nicholson L, Stabley DL, Sol-Church K. Costello syndrome associated with novel germline HRAS mutations: an attenuated phenotype? Am. J. Med. Genet. A. 2008;146A:683–690. doi: 10.1002/ajmg.a.32227. [DOI] [PubMed] [Google Scholar]
- 44.Burkitt-Wright EM, Bradley L, Shorto J, McConnell VP, Gannon C, Firth HV, Park SM, D'Amore A, Munyard PF, Turnpenny PD, Charlton A, Wilson M, Kerr B. Neonatal lethal Costello syndrome and unusual dinucleotide deletion/insertion mutations in HRAS predicting p.Gly12Val. Am. J. Med. Genet. A. 2012;158A:1102–1110. doi: 10.1002/ajmg.a.35296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niihori T, Aoki Y, Okamoto N, Kurosawa K, Ohashi H, Mizuno S, Kawame H, Inazawa J, Ohura T, Arai H, Nabatame S, Kikuchi K, Kuroki Y, Miura M, Tanaka T, Ohtake A, Omori I, Ihara K, Mabe H, Watanabe K, Niijima S, Okano E, Numabe H, Matsubara Y. HRAS mutants identified in Costello syndrome patients can induce cellular senescence: possible implications for the pathogenesis of Costello syndrome. J. Hum. Genet. 2011;56:707–715. doi: 10.1038/jhg.2011.85. [DOI] [PubMed] [Google Scholar]
- 46.Digilio MC, Lepri F, Baban A, Dentici ML, Versacci P, Capolino R, Ferese R, De Luca A, Tartaglia M, Marino B, Dallapiccola B. RASopathies: Clinical Diagnosis in the First Year of Life. Mol. Syndromol. 2011;1:282–289. doi: 10.1159/000331266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tidyman WE, Lee HS, Rauen KA. Skeletal muscle pathology in Costello and cardio-facio-cutaneous syndromes: developmental consequences of germline Ras/MAPK activation on myogenesis. Am. J. Med. Genet. C. Semin. Med. Genet. 2011;157:104–114. doi: 10.1002/ajmg.c.30298. [DOI] [PubMed] [Google Scholar]
- 48.Gripp KW, Hopkins E, Sol-Church K, Stabley DL, Axelrad ME, Doyle D, Dobyns WB, Hudson C, Johnson J, Tenconi R, Graham GE, Sousa AB, Heller R, Piccione M, Corsello G, Herman GE, Tartaglia M, Lin AE. Phenotypic Analysis of Individuals With Costello Syndrome due to HRAS p.G13C. Am. J. Med. Genet. A. 2011;155A:706–716. doi: 10.1002/ajmg.a.33884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Piccione M, Piro E, Pomponi MG, Matina F, Pietrobono R, Candela E, Gabriele B, Neri G, Corsello G. A Premature Infant With Costello Syndrome Due to a Rare G13C HRAS Mutation. Am. J. Med. Genet. A. 2009;149A:487–489. doi: 10.1002/ajmg.a.32674. [DOI] [PubMed] [Google Scholar]
- 50.Schulz AL, Albrecht B, Arici C, van der Burgt I, Buske A, Gillessen-Kaesbach G, Heller R, Horn D, Hubner CA, Korenke GC, Konig R, Kress W, Kruger G, Meinecke P, Mucke J, Plecko B, Rossier E, Schinzel A, Schulze A, Seemanova E, Seidel H, Spranger S, Tuysuz B, Uhrig S, Wieczorek D, Kutsche K, Zenker M. Mutation and phenotypic spectrum in patients with cardio-facio-cutaneous and Costello syndrome. Clin. Genet. 2008;73:62–70. doi: 10.1111/j.1399-0004.2007.00931.x. [DOI] [PubMed] [Google Scholar]
- 51.Denayer E, Parret A, Chmara M, Schubbert S, Vogels A, Devriendt K, Frijns JP, Rybin V, de Ravel TJ, Shannon K, Cools J, Scheffzek K, Legius E. Mutation analysis in Costello syndrome: functional and structural characterization of the HRAS p.Lys117Arg mutation. Hum. Mutat. 2008;29:232–239. doi: 10.1002/humu.20616. [DOI] [PubMed] [Google Scholar]
- 52.Sinico M, Bassez G, Touboul C, Cave H, Vergnaud A, Zirah C, Fleury-Feith J, Gettler S, Vojtek AM, Chevalier N, Amram D, Alsamad IA, Haddad B, Encha-Razavi F. Excess of neuromuscular spindles in a fetus with Costello syndrome: a clinicopathological report. Pediatr. Dev. Pathol. 2011;14:218–223. doi: 10.2350/09-06-0664-CR.1. [DOI] [PubMed] [Google Scholar]
- 53.Piispanen AE, Bonnefoi O, Carden S, Deveau A, Bassilana M, Hogan DA. Roles of Ras1 membrane localization during Candida albicans hyphal growth and farnesol response. Eukaryot. Cell. 2011;10:1473–1484. doi: 10.1128/EC.05153-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Franken SM, Scheidig AJ, Krengel U, Rensland H, Lautwein A, Geyer M, Scheffzek K, Goody RS, Kalbitzer HR, Pai EF, et al. Three-dimensional structures and properties of a transforming and a nontransforming glycine-12 mutant of p21H–ras. Biochemistry. 1993;32:8411–8420. doi: 10.1021/bi00084a005. [DOI] [PubMed] [Google Scholar]
- 55.Gibbs JB, Schaber MD, Allard WJ, Sigal IS, Scolnick EM. Purification of ras GTPase activating protein from bovine brain. Proc. Nat. Acad. Sci. U. S. A. 1988;85:5026–5030. doi: 10.1073/pnas.85.14.5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feig LA, Cooper GM. Relationship among guanine nucleotide exchange, GTP hydrolysis, and transforming potential of mutated ras proteins. Mol. Cell Biol. 1988;8:2472–2478. doi: 10.1128/mcb.8.6.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.