Abstract
The Chromosome-centric Human Proteome Project (C-HPP) was recently initiated as an international collaborative effort. Our team adopted chromosome 9 (Chr 9) and performed a bioinformatics and proteogenomic analysis to catalog Chr 9-encoded proteins from normal tissues, lung cancer cell lines and lung cancer tissues. Approximately 74.7% of the Chr 9 genes of the human genome were identified, which included approximately 28% of missing proteins (46 of 162) on Chr 9 compared with the list of missing proteins from the neXtProt master table (2013-09). In addition, we performed a comparative proteomics analysis between normal lung and lung cancer tissues. Based on the data analysis, 15 proteins from Chr 9 were detected only in lung cancer tissues. Finally, we conducted a proteogenomic analysis to discover Chr 9-residing single nucleotide polymorphisms (SNP) and mutations described in the COSMIC cancer mutation database. We identified 21 SNPs and 4 mutations containing peptides on Chr 9 from normal human cells/tissues and lung cancer cell lines, respectively. In summary, this study provides valuable information of the human proteome for the scientific community as part of C-HPP. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium with the data set identifier PXD.
Keywords: C-HPP, missing proteins, lung cancer, biomarker
INTRODUCTION
The Chromosome-centric Human Proteome Project (C-HPP) was initiated to annotate and characterize all proteins encoded by each human chromosome1. Approximately 30% of all chromosome-encoded proteins currently lack of any experimental proof at the protein level2. The primary goal of this project was to identify and characterize chromosome-encoded proteins that exist based on genetic evidence but currently lack mass spectrometry (MS) evidence or antibody (Ab) detection, termed “missing proteins”. The C-HPP initiative defined missing proteins as those missing from the three mass spectrometry databases (neXtProt, GPMdb and PeptideAtlas) and an antibody-based database (Human Protein Atlas)1. Twenty-four groups worldwide have joined the C-HPP initiative to share the tasks including the annotation of identified protein-coding genes and new protein isoforms, including new types of variants such as SNPs3. Human Chr 9, our focus in this study, is known to have approximately 141 million base pairs, representing approximately 4.5% of the total DNA. At the time of the preparation of this manuscript, the number of protein-coding genes on Chr 9 was 821 based on the C-HPP Master Table (Ensembl v72, 2013-09) at the Yokohama HUPO meeting.
Mutations of genes on Chr 9 have been associated with various types of cancer, including lung cancers4–6. Our previous studies have focused on lung cancer biomarkers, but there is still a lack of specific proteins or their variants, in particular, as biomarkers for lung cancer. Reports of chromosomal genetic changes serve as useful information to identify specific variant forms of proteins during carcinogenesis. Specific chromosomal aberrations have been associated with particular cancer types. The partial loss of the Chr 9p arm has been reported as one of the most frequent genetic aberrations in lung cancer patients, and lost chromosomal segments have proven to harbor candidate tumor suppressor genes7, 8. Several well-characterized tumor suppressor genes, such as CDKN2A, MTAP and P16β, have been mapped to 9p, and the DMRT1, DMRT3, and DOCK8 genes, located on 9p24, and TSC1 gene, mapped to 9q, are often involved in amplifications, translocations or deletions in lung cancers9–11. Some of the proteins encoded by genes on Chr 9 with a high frequency of genetic alterations, such as single nucleotide polymorphism (SNPs) and alternative splice variants, can be identified by proteogenomic approaches and provide useful information for diagnoses and mechanistic studies.
In this study, we performed Chr 9-based data analysis to catalog Chr 9-encoded missing proteins and to identify Chr 9-based lung cancer-specific proteins, SNPs and mutations. To do so, we collected high quality mass spectrometry datasets using tandem mass spectral data that were acquired on a high resolution mass analyzer in the high-high mode with the HCD fragmentation method. Our bioinformatics analysis catalogued a number of proteins encoded by 614 genes on Chr 9 that included 46 missing proteins. In addition, we performed lung cancer biomarker discovery from the proteins on Chr 9 identified by mass spectrometry-based proteomics on human lung cancer tissues. We also performed an extensive analysis to identify peptide evidence of single nucleotide polymorphisms (SNPs) and mutations from the normal lung and lung cancer cell lines/tissues datasets. Our results will provide information about tissue-/cell line-specific Chr 9-encoded proteins.
EXPERIMENTAL METHODS
Chromosome 9 based protein identification
The publicly available high quality proteomics datasets were collected (PMID: 23933261, PMID: 22278370), using both MS and MS/MS scans that were acquired on an Orbitrap mass analyzer at high-high mode. Briefly, each sample was prepared for 24 fractions (high pH RPLC or SDS-PAGE) that were analyzed on either LTQ-Orbitrap Velos or LTQ-Orbitrap Elite (Thermo Fisher, San Jose, CA) with Agilent’s 1200 nano-LC system to a trap column (2 cm × 75 μm, C18 material 5 μm, 120 Å) and an analytical column (10 cm × 75 μm, C18 material 5 μm, 120 Å). Peptide samples were loaded onto trap column in 3% solvent B (90% acetonitrile in 0.1% formic acid) and washed for 5 minutes. Peptides were eluted using a gradient of 3–35% solvent B for 60 minutes at a constant flow rate of 0.4 μl/min. All tandem spectra were generated by the Higher-energy collision dissociation (HCD) using 40% normalized collision energy. We reanalyzed these datasets (more than 2,000 raw files) using our pipeline with the Proteome Discoverer Platform and performed searches against the human RefSeq Protein Database (version 50) using the SEQUEST and Mascot database search algorithms. A list of non-redundant peptides was collected, and the resulting peptide sequences were searched against the human genome database. After identifying the proteins, spectral counts were normalized by total spectral counts for each MS data. These sums were then scaled so that they were equivalent. Next, the peptides uniquely mapped to human Chr 9 were selected for further analysis in this study.
Missing proteins on human Chr 9
In this study, we integrated the following databases to develop a list of missing proteins on human Chr 9. We used C-HPP wiki website databases (SIB Swiss Institute of Bioinformatics, rel. 2013-09-26), which comprised database information from Ensembl (version 72), neXtProt (rel. 2013-09, 3,844 missing genes), GPMdb (green, rel. 2013-08), PeptideAtlas (rel. 2013-08), and Human Protein Atlas (rel. 2012-12).
Protein identification of lung cancer tissue lysates
Lung cancer tissues and adjacent normal lung tissues (1 mg each of tissues pooled from 5 patients) were lysed and suspended in RIPA buffer (Thermo, USA) containing protease inhibitors (1:200) (Roche, Germany). The samples were sonicated, vortexed on ice and centrifuged at 14,000 rpm at 4 °C for 10 min to collect the protein supernatants, followed by the evaporation of water using a speed vacuum. Tryptic digestion was conducted based on the filter-aided sample preparation protocol (FASP)12. Briefly, 1 mg of the protein sample was solubilized with 8 M urea in a 10 kDa cut-off Amicon spin column (Millipore, MA, USA). The proteins were reduced with 10 mM dithiothreitol (DTT) at 60 °C for 30 min and then alkylated with 10 mM iodoacetamide (IAA) in the dark at room temperature for 30 min. Following alkylation, the samples were washed with 8 M urea and then with ammonium bicarbonate solution; the samples were finally subjected to tryptic digestion overnight. The peptides were eluted from the spin column by centrifugation at 3,000 rpm. The OFFGEL electrophoresis fractionation (Agilent, 12 fractions) and tandem mass spectrometric analysis of normal lung and lung cancer tissue lysate samples were performed. The LTQ-Orbitrap mass spectrometry was used for acquiring the mass spectra to identify proteins. Briefly, Nano high-performance liquid chromatography (nano-HPLC) analysis was performed using an Easy n-LC system (Thermo Fisher). The capillary column used (150 × 0.075 mm) for LC-MS/MS analysis was obtained from Phoenix S&T (Chester, PA, USA), and the slurry was packed in-house using a 5-μm, 100-Å pore size Magic C18 stationary phase resin (Michrom Bio Resources, Auburn, CA, USA). The mobile phase A for LC separation was 0.1% formic acid in deionized water, and the mobile phase B was 0.1% formic acid in acetonitrile. The chromatography gradient was designed for a linear increase from 0 to 8% B in 5 min, 5 to 25% B in 100 min, 25 to 45% B in 10 min, and 45 to 60% B in 10 min. Orbitrap full MS scans were acquired from m/z 350 to 1500 at a resolution of 15 000 (at m/z 400). The minimum threshold was set to 100 000 ion counts. Parent ions were fragmented using the LTQ (isolation width of 2 m/z units) with a maximum injection time of 100 ms combined with an AGC value of 1 ×104 using three fragmentation modes such as collision-induced dissociation (CID) alone, the reagent ion source emission current, reagent ion electron energy, and reagent ion source chemical ionization pressure were set to 35 mA, 70 V, and 26 psi, respectively. The tandem mass spectra were extracted, and searches were conducted against the UniProt database (rel. 2012-06, 86,875 entries) using MASCOT software (version 2.2.04). The search parameters used were the same as those previously reported13.
Customization of peptides database for SNPs and mutations
The dbSNP (version 138) and COSMIC (version 66) (Forbes et. al. 2011, PMID: 20952405, Sherry et. al, 2001 PMID: 11125122) databases were downloaded from their respective FTP servers. Every genomic coordinate was searched against the hg19 human reference genome for confirmation. A total of 3,052,321 non-synonymous SNPs and 1,100,191 mutations were used to create the databases. The non-synonymous SNPs and mutations were selected and incorporated one at a time into the protein sequences. The altered protein sequences were trypsinized in silico to create all possible fully tryptic peptides with lengths between 6 and 30 amino acids. All non-synonymous SNPs and mutations were considered if they caused alterations of fully tryptic peptide sequences that were observable by mass spectrometry. When a fully tryptic peptide was altered compared with the annotated protein database, it was deposited to a custom peptide database in Fasta format. This annotation tool was developed using the Java 2 Platform, Standard Edition. Finally, SNP-specific and mutation-specific peptide databases were built for the proteogenomic analysis. The SNP database comprised a total of 1,746,675 fully tryptic peptide sequences, and the COSMIC database contained 426,869 peptide sequences. Mapping of SNPs found in the protein coding regions of the gene (cSNPs) was automated by a tool developed in-house using shell scripts.
Identification of SNPs and mutation on Chr 9
All the unassigned tandem mass spectra were collected. In short, a tandem spectrum was set aside when it was not matched to any peptide at a 1% false discovery rate (FDR) in the SEQUEST and MASCOT searches. All unassigned spectra were then merged into peaklist files that were used to search for SNPs and mutations by SEQUEST. At a 1% FDR, peptides with SNPs or mutations derived from Chr 9 were collected for further analysis.
RESULTS AND DICUSSION
Catalog of Chr 9 proteome
The overall workflow is illustrated in Figure 1. Chr 9-encoded proteins were cataloged by using publicly available datasets. We analyzed Chr 9-encoded proteins using large human proteome profiling data of normal human samples (18 adult tissues, 6 primary hematopoietic cells and 6 fetal tissues) from Pandey lab (Supplementary Table 1). Based on these normal proteome datasets, the protein products of a total of 614 Chr 9 genes were detected (including peptides mapped to multiple protein entries), which were identified from 11,065 peptides. These identified proteins covered approximately 74.7% of the Chr 9 protein-coding genes compared with the neXtProt database. We also focused on Chr 9-centric analysis using lung cancer datasets, which resulted in the identification of a number of proteins derived from a total of 6,824 protein-coding genes, including 255 proteins located on Chr 9.
Figure 1.
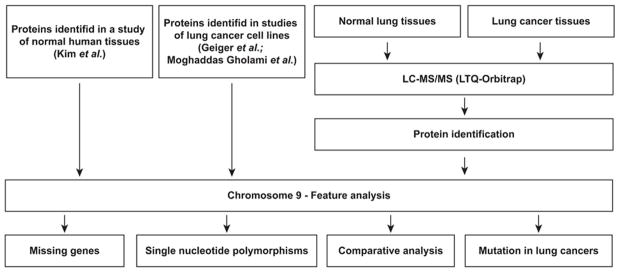
Schematic diagram of the overall workflow for the analysis of human chromosome 9-encoded missing proteins, single nucleotide polymorphisms, lung cancer-selective comparative analysis and mutational analysis.
The list of missing proteins on Chr 9 was generated by in silico database analysis, as described in the Experimental Methods. A total of 162 missing proteins that are awaiting validation are listed as Chr 9-encoded proteins, which have no proteomics evidence and have only protein level existence as neXtProt uncertain proteins. The entire chromosome 9-encoded missing protein data lists identified from normal human samples as well as normal lung and lung cancer cell lines and tissues were found from the missing protein list by manually searching by gene symbols and descriptions. When using all three datasets, missing proteins from 46 genes were detected, including Ankyrin repeat domain 20 families and olfactory receptor families (Table 1 and Supplementary Table 2). Interestingly, we have found FAM157B for Chr 9 in the adult colon, OR1J1 for Chr 9 in the fetal liver, and OR1L1 in the placenta. Some of the missing proteins were detected with a high number of unique peptides. For example, Actin-like 7B (ACTL7B) and Calicin (CCIN) were detected with more than 10 unique peptides. The missing protein list was compared with the GPMDB and HPA databases (Supplementary Table 2). Most of the missing proteins had no or low levels detected by MS or antibody, except ACTL7B (green level in GPMdb and high level in HPA).
Table 1.
The list of newly detected missing proteins on chromosome 9. The detailed information on the peptide sequences is available in Supplementary Table 2.
| NCBI Accession No. | Gene Symbol | Description |
|---|---|---|
| NP_006677.1 | ACTL7B | actin-like 7B |
| NP_671728.2 | ANKRD18A | ankyrin repeat domain 18A |
| NP_115626.2 | ANKRD20A1 | ankyrin repeat domain 20 family, member A1 |
| NP_001012421.1 | ANKRD20A2 | ankyrin repeat domain 20 family, member A2 |
| NP_001012419.1 | ANKRD20A3 | ankyrin repeat domain 20 family, member A3 |
| NP_001092275.1 | ANKRD20A4 | ankyrin repeat domain 20 family, member A4 |
| NP_054729.3 | ASTN2 | astrotactin 2 |
| NP_001012520.2 | C9orf117 | chromosome 9 open reading frame 117 |
| NP_997394.1 | C9orf139 | chromosome 9 open reading frame 139 |
| NP_955382.3 | C9orf50 | chromosome 9 open reading frame 50 |
| NP_001122090.1 | C9orf57 | chromosome 9 open reading frame 57 |
| NP_689782.2 | C9orf66 | chromosome 9 open reading frame 66 |
| NP_001074020.1 | C9orf84 | chromosome 9 open reading frame 84 |
| NP_775821.2 | CCDC171 | coiled-coil domain containing 171 |
| NP_005884.2 | CCIN | calicin |
| NP_001124337.1 | DMRT2 | doublesex and mab-3 related transcription factor 2 |
| NP_001138721.1 | FAM157B | family with sequence similarity 157, member B |
| NP_001001710.1 | FAM166A | family with sequence similarity 166, member A |
| NP_001157782.1 | FAM166B | family with sequence similarity 166, member B |
| NP_001092749.1 | FOXD4L2 | forkhead box D4-like 2 |
| NP_954586.4 | FOXD4L3 | forkhead box D4-like 3 |
| NP_954714.2 | FOXD4L4 | forkhead box D4-like 4 |
| NP_001119806.1 | FOXD4L5 | forkhead box D4-like 5 |
| NP_001078945.1 | FOXD4L6 | forkhead box D4-like 6 |
| NP_002164.1 | IFNA16 | interferon, alpha 16 |
| NP_009110.2 | INSL6 | insulin-like 6 |
| NP_001017969.2 | KIAA2026 | Uncharacterized protein KIAA2026 |
| NP_997393.3 | LCNL1 | lipocalin-like 1 |
| NP_055396.1 | OBP2B | odorant binding protein 2B |
| NP_001001919.1 | OR13C4 | olfactory receptor, family 13, subfamily C, member 4 |
| NP_001004483.1 | OR13C8 | olfactory receptor, family 13, subfamily C, member 8 |
| NP_001004450.1 | OR1B1 | olfactory receptor, family 1, subfamily B, member 1 |
| NP_001004451.1 | OR1J1 | olfactory receptor, family 1, subfamily J, member 1 |
| NP_001005236.3 | OR1L1 | olfactory receptor, family 1, subfamily L, member 1 |
| NP_001005234.1 | OR1L3 | olfactory receptor, family 1, subfamily L, member 3 |
| NP_001005235.1 | OR1L4 | olfactory receptor, family 1, subfamily L, member 4 |
| NP_001004453.2 | OR1L6 | olfactory receptor, family 1, subfamily L, member 6 |
| NP_001001923.1 | OR5C1 | olfactory receptor, family 5, subfamily C, member 1 |
| NP_001128691.1 | PIP5KL1 | phosphatidylinositol-4-phosphate 5-kinase-like 1 |
| NP_659488.2 | RNF183 | ring finger protein 183 |
| NP_001034484.3 | SPATA6L | spermatogenesis associated 6-like |
| NP_001007472.2 | TRPM3 | transient receptor potential cation channel, subfamily M, member 3 |
| NP_001012361.1 | WDR31 | WD repeat domain 31 |
| NP_001038941.1 | WDR38 | WD repeat domain 38 |
| NP_003399.1 | ZFP37 | ZFP37 zinc finger protein |
Tissue-wise expression pattern of the Chr 9 proteome
Chr 9-encoded proteins in the human normal proteome dataset are presented in Table 2 and Supplementary Table 1. Among all the samples, the highest number of Chr 9-encoded proteins was identified in the adult testis (372), and the smallest number was identified in platelets (174). We observed four Chr 9-encoded proteins that were detected most abundantly in all sample types: Heat shock 70 kDa protein 5 (HSPA5), Spectrin alpha non-erythrocytic 1 (SPTAN1), Talin 1 (TLN1), and Valosin-containing protein (VCP). HSPA5, a glucose-related heat shock protein, plays a role in facilitating the assembly of multimeric protein complexes inside the endoplasmic reticulum14. SPTAN1, similar to erythrocyte spectrin, is involved in secretion and also interacts with calmodulin in the calcium-dependent movement of the cytoskeleton at the membrane15. TLN1, a cell junction protein, is involved in connections of major cytoskeletal structures to the plasma membrane16. VCP is necessary for the fragmentation of Golgi stacks during mitosis and is involved in the formation of the transitional ER17. Moreover, tissue or blood cell line selective abundant proteins (dominantly detected by more than 95% in one type of tissue or cell) were revealed in our large data set analysis, including Paired box 5 (PAX5) in B cells, Calicin (CCIN) in the adult testis, and Src homology 2 domain containing transforming protein 3 (SHC3) in the adult frontal cortex. PAX5 is known to be involved in B-cell differentiation, neural development and spermatogenesis18. CCIN is a cytoskeletal element involved in spermiogenic differentiation19. SHC3 is a signaling adaptor that couples activated growth factor receptors to signaling pathways in neurons and is highly expressed in the cerebral cortex and frontal lobes20. Our data indicate that the multi-tissue proteome analysis strongly confirms the previously known protein-tissue relationship and biology.
Table 2.
The number of chromosome 9-encoded proteins identified and the sum of normalized spectra counts in the normal human samples.
| Samples | Protein numbers identified | Sum of normalized specrta count |
|---|---|---|
| Adult testis | 372 | 28439 |
| Adult ovary | 353 | 27908 |
| Adult retina | 346 | 29355 |
| Adult pancreas | 336 | 26923 |
| Fetal heart | 333 | 27438 |
| CD8+ T cells | 328 | 30468 |
| Fetal testis | 323 | 24545 |
| Fetal ovary | 317 | 35325 |
| Fetal liver | 315 | 28612 |
| Fetal brain | 305 | 33194 |
| Adult prostate | 301 | 21395 |
| Adult frontal cortex | 296 | 42961 |
| Adult liver | 293 | 32734 |
| B cells | 293 | 22692 |
| Adult adrenal gland | 291 | 34811 |
| Fetal gut | 288 | 23047 |
| Adult colon | 285 | 21742 |
| CD4+ T cells | 273 | 18485 |
| NK cells | 269 | 23882 |
| Adult rectum | 265 | 24815 |
| Adult spinal cord | 258 | 21308 |
| Adult urinary bladder | 257 | 18523 |
| Adult gallbladder | 249 | 23021 |
| Monocytes | 247 | 16888 |
| Adult heart | 230 | 15517 |
| Adult kidney | 228 | 28469 |
| Adult lung | 216 | 22252 |
| Placenta | 214 | 21349 |
| Adult esophagus | 181 | 16445 |
| Platelets | 174 | 34265 |
For the identification of the tissue or blood cell line-selective abundant Chr 9 protein lists, we compared the data for 30 different samples. A list of the top 10 abundantly detected Chr 9 proteins in each of the 30 different samples is presented in Supplementary Table 3. Spectrin alpha non-erythrocytic 1 (SPTAN1) was the most abundantly expressed Chr 9 protein in the 5 fetal tissues, except the fetal liver (heat shock 70 kDa protein 5, HSPA5). SPTAN1 was the most abundantly expressed Chr 9 protein in the 10 adult tissues, followed by talin 1 (TLN1) (5 out of 18). TLN1 was the most abundant Chr 9 protein in 5 out of 6 immune cell lines analyzed. The cell-/tissue-specific protein lists are provided in Table 3 with normalized spectra counts. With the detection sensitivity of mass spectrometry used in this study, a total of 122 proteins were identified as cell-/tissue-specific Chr 9 proteins (i.e., detected in only one sample and not in any others). Among 108 cell-/tissue-specific Chr 9-encoded proteins from the normal human samples, the adult retina showed the highest number of proteins (12); conversely, no tissue-specific protein was detected in the adult lung, adult rectum, and adult prostate. Most of the tissue-specific Chr 9-encoded proteins were identified with normalized spectra counts of 2.2 to 10. Notably, the proteins detected with lower spectra counts would not show high selectivity for the tissue and might be detected in other tissues using more sensitive analytical approaches. However, some of the specific proteins were identified as being highly specific, including Chr 9 open reading frame 169 in adult esophagus and CD 72 molecule in B cells. These results show that important tissue-selective Chr 9 encoded proteins are differentially expressed in not only fetal and adult tissues but also immune cells.
Table 3.
The list of cell-/tissue-specific proteins detected in normal human samples.
| Tissue | Gene | Sum of normalized spectra count | NCBI accession ID | Protein existence | Proteomics | Antibody | Description |
|---|---|---|---|---|---|---|---|
| Fetal heart | AGTPBP1 | 4.8 | NP_056054.2 | protein level | yes | no | ATP/GTP binding protein 1 |
| C9orf66 | 4.7 | NP_689782.2 | transcript level | no | yes | chromosome 9 open reading frame 66 | |
| IFNA14 | 4.7 | NP_002163.2 | protein level | no | no | interferon, alpha 14 | |
| LCN6 | 2.9 | NP_945184.1 | protein level | no | no | lipocalin 6 | |
| ZDHHC21 | 2.9 | NP_848661.1 | protein level | yes | no | zinc finger, DHHC-type containing 21 | |
| CBWD1 | 2.8 | NP_001138827.1 | protein level | yes | no | COBW domain containing 1 | |
| *Common Peptide A | 2.8 | - | - | - | - | - | |
| ZNF510 | 2.8 | NP_055745.1 | transcript level | yes | yes | zinc finger protein 510 | |
| Fetal liver | FKTN | 10.6 | NP_001073270.1 | protein level | yes | yes | fukutin |
| OR5C1 | 5.5 | NP_001001923.1 | transcript level | no | no | olfactory receptor, family 5, subfamily C, member 1 | |
| USP20 | 5.5 | NP_006667.3 | protein level | yes | yes | ubiquitin specific peptidase 20 | |
| OR1J1 | 2.7 | NP_001004451.1 | transcript level | no | no | olfactory receptor, family 1, subfamily J, member 1 | |
| IFNA21 | 2.3 | NP_002166.2 | protein level | no | no | interferon, alpha 21 | |
| IFNK | 2.3 | NP_064509.2 | protein level | no | no | interferon, kappa | |
| SLC35D2 | 2.3 | NP_008932.2 | protein level | yes | no | solute carrier family 35, member D2 | |
| Fetal gut | LCN15 | 2.2 | NP_976222.1 | protein level | yes | no | lipocalin 15 |
| *Common Peptide B | 2.2 | - | - | - | - | - | |
| OBP2A | 2.2 | NP_055397.1 | protein level | no | yes | odorant binding protein 2A | |
| Fetal ovary | C9orf85 | 4.8 | NP_872311.2 | protein level | yes | no | chromosome 9 open reading frame 85 |
| GKAP1 | 3.6 | NP_001129425.1 | protein level | yes | yes | G kinase anchoring protein 1 | |
| NTMT1 | 3.6 | NP_054783.2 | protein level | yes | yes | N-terminal Xaa-Pro-Lys N-methyltransferase 1 | |
| Fetal testis | SPATA6L | 7.9 | NP_001034484.3 | transcript level | no | yes | spermatogenesis associated 6-like |
| *Common Peptide C | 2.4 | - | - | - | - | - | |
| Fetal brain | NOTCH1 | 2.8 | NP_060087.3 | protein level | yes | yes | notch 1 |
| Adult frontal cortex | LRSAM1 | 6.6 | NP_001005374.1 | protein level | yes | yes | leucine rich repeat and sterile alpha motif containing 1 |
| NTRK2 | 6.3 | NP_001018074.1 | protein level | yes | yes | neurotrophic tyrosine kinase, receptor, type 2 | |
| PIP5K1B | 5.2 | NP_003549.1 | protein level | yes | yes | phosphatidylinositol-4-phosphate 5-kinase, type I, beta | |
| *Common Peptide D | 4.1 | - | - | - | - | - | |
| INSL4 | 3 | NP_002186.1 | protein level | no | no | insulin-like 4 (placenta) | |
| CAMSAP1 | 2.8 | NP_056262.3 | protein level | yes | yes | calmodulin regulated spectrin-associated protein 1 | |
| PRUNE2 | 2.6 | NP_056040.2 | protein level | yes | yes | prune homolog 2 (Drosophila) | |
| *Common Peptide E | 2.6 | - | - | - | - | - | |
| GPR21 | 2.6 | NP_005285.1 | transcript level | yes | yes | G protein-coupled receptor 21 | |
| LRRC8A | 2.6 | NP_001120717.1 | protein level | yes | yes | leucine rich repeat containing 8 family, member A | |
| SLC24A2 | 2.6 | NP_001180217.1 | protein level | yes | no | solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 | |
| Adult spinal cord | CNTLN | 3.3 | NP_001107867.1 | protein level | yes | yes | centlein, centrosomal protein |
| FANCC | 2.4 | NP_000127.2 | protein level | yes | yes | Fanconi anemia, complementation group C | |
| ZBTB34 | 2.4 | NP_001092740.1 | protein level | yes | yes | zinc finger and BTB domain containing 34 | |
| Adult retina | PDCL | 10.1 | NP_005379.3 | protein level | yes | yes | phosducin-like |
| FIBCD1 | 7.4 | NP_116232.3 | protein level | yes | no | fibrinogen C domain containing 1 | |
| KIAA1958 | 5.9 | NP_597722.1 | protein level | yes | yes | KIAA1958 | |
| ADAMTS13 | 5.5 | NP_620595.1 | protein level | yes | yes | ADAM metallopeptidase with thrombospondin type 1 motif, 13 | |
| OLFM1 | 3.4 | NP_055094.1 | protein level | yes | no | olfactomedin 1 | |
| ZMYND19 | 3.4 | NP_612471.1 | protein level | yes | yes | zinc finger, MYND-type containing 19 | |
| LHX3 | 2.5 | NP_055379.1 | protein level | no | no | LIM homeobox 3 | |
| LHX2 | 2.5 | NP_004780.3 | transcript level | yes | no | LIM homeobox 2 | |
| MPDZ | 2.5 | NP_003820.2 | protein level | yes | yes | multiple PDZ domain protein | |
| RASEF | 2.5 | NP_689786.2 | protein level | yes | yes | RAS and EF-hand domain containing | |
| TMEM215 | 2.5 | NP_997723.2 | transcript level | yes | yes | transmembrane protein 215 | |
| TRPM3 | 2.5 | NP_079247.5 | transcript level | no | no | transient receptor potential cation channel, subfamily M, member 3 | |
| Adult heart | INPP5E | 4.7 | NP_063945.2 | protein level | yes | no | inositol polyphosphate-5-phosphatase, 72 kDa |
| WDR31 | 4.7 | NP_001012361.1 | transcript level | no | yes | WD repeat domain 31 | |
| TRUB2 | 3.8 | NP_056494.1 | protein level | yes | yes | TruB pseudouridine (psi) synthase homolog 2 (E. coli) | |
| CREB3 | 3.3 | NP_006359.3 | protein level | yes | no | cAMP responsive element binding protein 3 | |
| MUSK | 3.3 | NP_005583.1 | protein level | no | no | muscle, skeletal, receptor tyrosine kinase | |
| Adult liver | ENTPD8 | 2.8 | NP_001028285.1 | protein level | yes | yes | ectonucleoside triphosphate diphosphohydrolase 8 |
| RGP1 | 2.8 | NP_001073965.2 | protein level | yes | yes | RGP1 retrograde golgi transport homolog (S. cerevisiae) | |
| CNTRL | 2.8 | NP_008949.4 | protein level | yes | yes | centriolin | |
| Adult ovary | BNC2 | 2.6 | NP_060107.3 | protein level | yes | yes | basonuclin 2 |
| CCL21 | 2.6 | NP_002980.1 | protein level | yes | yes | chemokine (C-C motif) ligand 21 | |
| KLF9 | 2.6 | NP_001197.1 | protein level | yes | yes | Kruppel-like factor 9 | |
| NR5A1 | 2.6 | NP_004950.2 | protein level | no | no | nuclear receptor subfamily 5, group A, member 1 | |
| PBX3 | 2.6 | NP_001128250.1 | protein level | yes | no | pre-B-cell leukemia homeobox 3 | |
| ZNF658 | 2.6 | NP_149350.3 | transcript level | yes | yes | zinc finger protein 658 | |
| Adult testis | DCAF12 | 10.8 | NP_056212.1 | protein level | yes | yes | DDB1 and CUL4 associated factor 12 |
| C9orf91 | 5.4 | NP_694590.2 | transcript level | yes | yes | chromosome 9 open reading frame 91 | |
| AK8 | 2.7 | NP_689785.1 | protein level | no | yes | adenylate kinase 8 | |
| C9orf139 | 2.7 | NP_997394.1 | transcript level | no | yes | chromosome 9 open reading frame 139 | |
| ZBTB5 | 2.7 | NP_055687.1 | protein level | yes | yes | zinc finger and BTB domain containing 5 | |
| Adult lung | - | - | - | - | - | - | - |
| Adult adrenal gland | ABCA1 | 7.2 | NP_005493.2 | protein level | yes | no | ATP-binding cassette, sub-family A (ABC1), member 1 |
| WDR38 | 7.2 | NP_001038941.1 | transcript level | no | yes | WD repeat domain 38 | |
| CDKN2A | 2.8 | NP_001182061.1 | protein level | yes | yes | cyclin-dependent kinase inhibitor 2A | |
| Adult gallbladder | ASS1 | 8.7 | NP_446464.1 | protein level | yes | yes | argininosuccinate synthase 1 |
| ACO1 | 2.9 | NP_002188.1 | protein level | yes | yes | aconitase 1, soluble | |
| ARID3C | 2.9 | NP_001017363.1 | transcript level | yes | no | AT rich interactive domain 3C (BRIGHT-like) | |
| LCNL1 | 2.9 | NP_997393.3 | transcript level | no | no | lipocalin-like 1 | |
| Adult pancreas | MSMP | 6.1 | NP_001037729.1 | protein level | yes | yes | microseminoprotein, prostate associated |
| ADAMTSL2 | 2.9 | NP_055509.2 | protein level | yes | yes | ADAMTS-like 2 | |
| C9orf50 | 2.9 | NP_955382.3 | transcript level | no | yes | chromosome 9 open reading frame 50 | |
| LURAP1L | 2.9 | NP_981948.1 | transcript level | yes | yes | leucine rich adaptor protein 1-like | |
| Adult kidney | BARHL1 | 4.4 | NP_064448.1 | transcript level | yes | yes | BarH-like homeobox 1 |
| C9orf135 | 4.2 | NP_001010940.1 | protein level | yes | yes | chromosome 9 open reading frame 135 | |
| Adult esophagus | C9orf169 | 17.8 | NP_945352.2 | protein level | yes | yes | chromosome 9 open reading frame 169 |
| IFNA5 | 5.4 | NP_002160.1 | protein level | no | no | interferon, alpha 5 | |
| Adult colon | GALNT12 | 3.1 | NP_078918.3 | protein level | yes | yes | N-acetylgalactosaminyltransferase 12 |
| ABO | 2.6 | NP_065202.2 | protein level | yes | no | alpha 1–3-N-acetylgalactosaminyltransferase | |
| FAM157B | 2.6 | NP_001138721.1 | homology | no | no | family with sequence similarity 157, member B | |
| Adult rectum | - | - | - | - | - | - | - |
| Adult urinary bladder | GOLGA2 | 6.9 | NP_004477.3 | protein level | yes | yes | golgin A2 |
| OR1K1 | 2.5 | NP_543135.1 | protein level | yes | no | olfactory receptor, family 1, subfamily K, member 1 | |
| Adult prostate | - | - | - | - | - | - | - |
| Placenta | OR1L1 | 5.2 | NP_001005236.3 | transcript level | no | no | olfactory receptor, family 1, subfamily L, member 1 |
| *Common Peptide F | 3.4 | - | - | - | - | - | |
| B cells | CD72 | 42.9 | NP_001773.1 | protein level | yes | yes | CD72 molecule |
| ZNF79 | 9.5 | NP_009066.2 | protein level | yes | yes | zinc finger protein 79 | |
| NAIF1 | 7.8 | NP_931045.1 | protein level | yes | no | nuclear apoptosis inducing factor 1 | |
| CDK20 | 2.6 | NP_848519.1 | protein level | yes | yes | cyclin-dependent kinase 20 | |
| ZBTB6 | 2.4 | NP_006617.1 | protein level | yes | yes | zinc finger and BTB domain containing 6 | |
| CD4+ T cells | MGC50722 | 12.2 | NP_976223.1 | - | - | - | uncharacterized MGC50722 |
| SPIN1 | 3.3 | NP_006708.2 | protein level | yes | yes | spindlin 1 | |
| CD8+ T cells | *Common Peptide G | 5.3 | - | - | - | - | - |
| IFNA16 | 2.6 | NP_002164.1 | transcript level | no | no | interferon, alpha 16 | |
| NK cells | LOC100287368 | 2.4 | XP_002342958.2 | - | - | - | protein FAM27D1-like |
| Monocytes | *Common Peptide H | 14.7 | - | - | - | - | - |
| KLF4 | 6 | NP_004226.3 | protein level | yes | yes | Kruppel-like factor 4 (gut) | |
| CBWD6 | 5.6 | NP_001078926.1 | protein level | yes | no | COBW domain containing 6 | |
| Platelets | CDC14B | 2.2 | NP_001070649.1 | protein level | yes | yes | cell division cycle 14B |
Eight peptides are shared between more than two Chr 9 proteins. Protein existence, Proteomics, and Antibody column information were used by the C-HPP wiki website neXtProt database. (*Common Peptide A~H): *Common Peptide A - ZNF658;ZNF883 (NP_149350.3;NP_001094808.1); *Common Peptide B - LOC100132859;LOC100505781 (XP_003960500.1;XP_003119217.1); *Common Peptide C - FOXD4;FOXD4L5;FOXD4L4;FOXD4L2;FOXD4L6;FOXD4L3 (NP_997188.2;NP_001119806.1;NP_954714.2;NP_001092749.1;NP_001078945.1;NP_954586.4); *Common Peptide D - GNAQ;GNA14 (NP_002063.2;NP_004288.1); *Common Peptide E - CNTNAP3B;CNTNAP3 (NP_001188309.1;NP_387504.2); *Common Peptide F - OR1L4;OR1L6 (NP_001005235.1;NP_001004453.2); *Common Peptide G - ZNF782;ZNF79 (NP_001001662.1;NP_009066.2); *Common Peptide H - FCN2;FCN1 (NP_004099.2;NP_001994.2).
Cancer-specific Chr 9 encoded proteins in lung cancer tissues
We also performed proteomic analysis and comparative data analysis using lung cancer tissues along with the adjacent normal lung tissue (Supplementary Table 4). The Chr 9-encoded proteins matched data analysis revealed that 38 and 47 proteins were allocated to Chr 9-encoded proteins in normal lung and lung cancer tissues, respectively. The portion of Chr 9 proteins was 5.3% (38 out of 723) in normal lung tissue and 4.8% (47 out of 987) in lung cancer tissue.
Compared with normal lung tissue, 15 lung cancer-selective Chr 9 proteins were identified (Table 4). Based on a literature search, we present herein the significance of those proteins with respect to lung cancer and their speculated functions in lung cancer. The UV excision repair protein RAD23 homolog B (RAD23B) was identified as a lung cancer-selective protein and is known to be involved in nucleotide excision repair. It is also capable of binding polyubiquitinated substrates and delivers ubiquitinated proteins to the proteasome21. Nucleotide excision repair proteins play a key role in reversing DNA damage from exposure to environmental carcinogens. RAD23B variants have been reported to be associated with primary lung cancer risk in different ethnic groups22. 40S ribosomal protein S6 (RPS6) is the major substrate of protein kinase in eukaryote ribosomes23. RPS6 plays a role in AKT signaling in drug resistance. Strong activation was observed in the drug-resistant clones for several key AKT substrates, including RPS6 in a non-small cell lung cancer cell line24. SET is a proto-oncogene involved in apoptosis, transcription, nucleosome assembly and histone chaperoning. The protein encoded by this gene inhibits the acetylation of nucleosomes, especially histone H4, by histone acetylases25. The nuclear exit of SET correlates with cell spreading, and SET cooperates with Rac1 to stimulate cell migration26. The acidic leucine-rich nuclear phosphoprotein 32B (ANP32B) works as a cell cycle progression factor and a cell survival factor, with functions that are largely unknown27. ANP32B is known to contribute to the retinoic acid-induced differentiation of leukemic cells28. Far upstream element-binding protein 3 (FUBP3) is a single strand DNA binding protein that recognizes the far upstream element (FUSE; originally identified upstream of the c-myc promoter)29. FUBP 3, as an activator of c-myc, has an important function in the high proliferation rate of renal cell carcinoma30. Perilipin 2 (PLIN2) is involved in the formation of lipid droplets for the development and maintenance of adipose tissues31. PLIN2 protein levels were increased in the plasma samples from colorectal cancer patients32. Actin-related protein 2/3 (Arp2/3) complex subunit 5-link protein (ARPC5L) is a component of the Arp2/3 complex, which is involved in the regulation of actin polymerization and the branched actin network. Actin is rapidly formed in response to specific cellular signals that converge on the Arp2/3 complex to regulate its assembly33. The increased expression of ARPC5 is known to reduce the levels of tumor suppressor miR-133a in lung squamous cell carcinoma34. Histidine triad nucleotide-binding protein 2 (HINT2) is a nuclear-encoded mitochondrial hydrolase. HINT2 overexpression sensitizes hepatocellular carcinoma to apoptosis35. HINT2 has been demonstrated to have low expression in endometrial cancer cells compared with control cells36. Sialic acid synthase (NANS) produces N-acetylneuraminic acid. Glycan structures of glycoprotein change during carcinogenesis and affect various biological behaviors of tumor cells. Typical glycan changes have also been reported to occur at the level of fucose and sialic acid in cancer cells37. Constitutive coactivator of PPAR-gamma-like protein 1 (FAM120A) was found in an mRNA-protein complex and might participate in mRNA transport throughout the cytoplasm38. FAM120A is a critical component of the oxidative stress-induced survival signaling pathway and is highly expressed in gastric scirrhous carcinoma compared with normal gastric mucosa39. Dipeptidyl peptidase 2 (DPP2/DPP7) is a serine protease that prevents spontaneous cell cycle progression in quiescent cells40. DPP2 inhibition tends to induce apoptosis of 60% of chronic lymphocytic leukemia cells41, suggesting its oncogenic activity. The list of lung cancer–selective Chr 9 proteins shows that most of these proteins are not well known as being associated with lung cancer; however, their functions are considered to be related to lung cancer. The selective biomarkers described here need further verification.
Table 4.
The list of lung cancer tissue-selective proteins on chromosome 9 compared with adjacent normal lung tissues.
| NCBI accession ID | Gene symbol | Description |
|---|---|---|
| NP_055037.1 | NDUFA8 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8 |
| NP_001231653.1 | RAD23B | UV excision repair protein RAD23 homolog B |
| NP_001001.2 | RPS6 | 40S ribosomal protein S6 |
| NP_001116293.1 | SET | Protein SET |
| NP_001139580.1 | PTGR1 | Prostaglandin reductase 1 |
| NP_056073.1 | FKBP15 | FK506-binding protein 15 |
| NP_006392.1 | ANP32B | Acidic leucine-rich nuclear phosphoprotein 32 family member B |
| NP_003925.1 | FUBP3 | Far upstream element-binding protein 3 |
| NP_001113.2 | PLIN2 | Perilipin-2 |
| NP_112240.1 | ARPC5L | Actin-related protein 2/3 complex subunit 5-like protein |
| NP_115982.1 | HINT2 | Histidine triad nucleotide-binding protein 2, mitochondrial |
| NP_061819.2 | NANS | Sialic acid synthase |
| NP_064530.1 | SH3GLB2 | Endophilin-B2 |
| NP_055427.2 | FAM120A | Constitutive coactivator of PPAR-gamma-like protein 1 |
| NP_037511.2 | DPP7 | Dipeptidyl peptidase 2 |
Identification of SNP- and mutation-containing peptides derived from Chr 9-encoded proteins
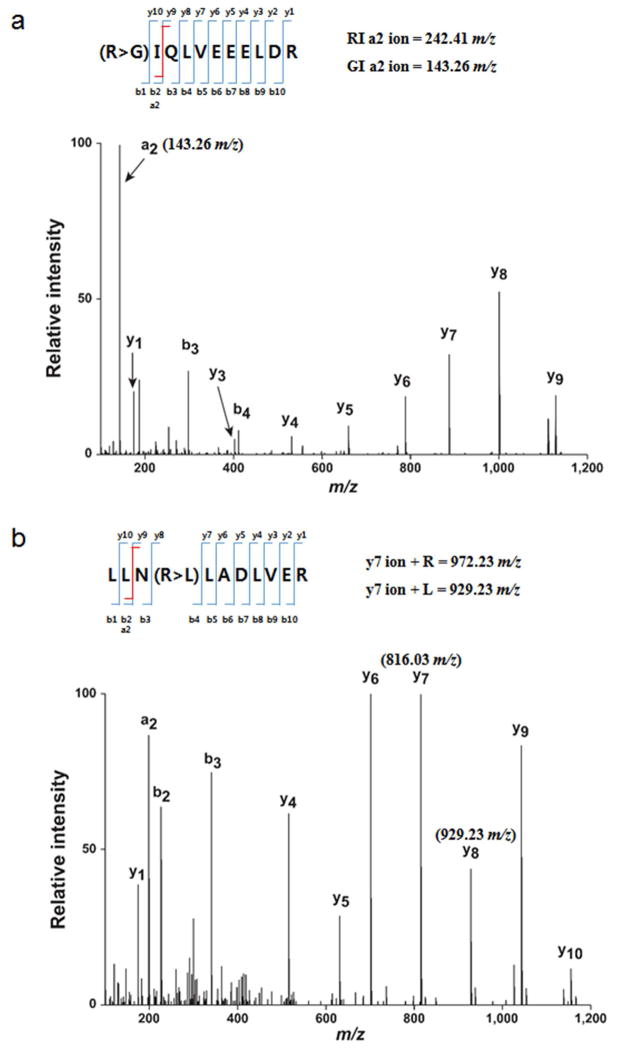
We performed a proteogenomic analysis by searching against two customized peptides sequence databases: the SNP-specific and mutation-specific databases. To decrease the false discovery rate, we first generated peaklist files that contained only tandem mass spectra that did not match any peptides from the initial protein database searches. Unmatched spectra from two available lung cancer cell line datasets (A549 and NCI-H460) and our adult lung tissue dataset were used to search against the two databases through the Proteome Discover Platform (version 1.4) using SEQUEST. We identified 249 SNPs on Chr 9 genes from normal human cells/tissues, with 21 SNP-containing peptides being mapped to 19 genes that were identified from normal adult lung tissue. For example, we observed a peptide (GIQLVEEELDR) that differed by one amino acid from the sequence of the tropomyosin beta chain (TPM2) protein in the databases (glycine instead of arginine at position 91), likely due to a known SNP in the gene (rs104894127) (Figure 2a). In addition, we observed four peptides from the two lung cancer cell lines that contained amino acid substitutions, which could be explained on the basis of mutations in the COSMIC database. For example, a peptide derived from the gene ALDH1B1 (aldehyde dehydrogenase X, mitochondrial) (LLNRLADLVER) was identified from a mutant-specific database search that contained a single amino acid substitution (L107R) from the database sequence (Figure 2b). These results show that peptides with genetic alterations can be identified by such a proteogenomic analysis.
Figure 2.
Two representative mass spectrometry peak profiles that show the identification of two mutations in peptide sequences. (a) A peptide (GIQLVEEELDR) in tropomyosin beta chain (TPM2) differed by one amino acid identified in normal adult lung tissue from the known sequence of the TPM2 protein in databases. Arginine at position 91 was changed to glycine, likely due to a known SNP in the gene (rs104894127). (b) Based on mutations in the COSMIC database, a peptide (LLNRLADLVER) derived from the gene aldehyde dehydrogenase X, mitochondrial (ALDH1B1) was identified by a mutant-specific database search. A single amino acid substitution, arginine at position 107 to leucine, was identified from the database sequence.
CONCLUSIONS
In this study, we have participated in the first year of C-HPP project implementation to annotate proteins encoded by genes located on Chr 9 using proteomics approaches. We have analyzed data pertaining to Chr 9-encoded proteins from a panel of normal human samples and lung cancer cell lines/tissues. We have obtained data regarding the distribution of proteins in normal human tissues that are encoded by genes located on Chr 9; this information could be further mined for the study of tissue-selective biomarkers. Proteomics and data comparison analyses for the human lung cancer tissues and adjacent normal lung tissues provided 15 lung cancer-specific Chr 9-encoded proteins, most of which are functionally associated with cancer. We have also described 46 Chr 9-encoded proteins that have not been detected previously and account for ~28% of the missing proteins located on Chr 9. These results demonstrate how a global proteomic analysis of a large number of samples using high-resolution MS instruments can increase the possibility of detecting missing proteins on each chromosome. In addition, we performed preliminary analyses to find peptide-based evidence of SNPs and mutations in the Chr 9-encoded proteins from the normal lung and lung cancer cell line/tissue datasets.
In the future, we will seek more Chr 9 protein information with the MS data, such as splicing variant proteins, expressed pseudogenes and post-translationally modified proteins. To detect additional missing proteins encoded by Chr 9, a deeper analysis of the proteins from multiple tissues after subcellular fractionation or the enrichment of certain classes of proteins should be performed. These data will also assist in the analysis of genomics data generated by next generation sequencing. Another strategy is to analyze the quantitative levels of missing proteins and lung cancer-associated Chr 9-encoded proteins in various samples using targeted MRM proteomics approaches.
Supplementary Material
Total chromosome 9-encoded protein list from normal human samples
The list of newly detected missing proteins on chromosome 9.
The list of detected top 10 abundance chromosome 9-encoded proteins among the normal human sample data.
The lists in the identification of chromosome 9 proteins from normal lung and lung cancer tissues.
Acknowledgments
We thank Gyoung-Beom Heo for his assistance in the data arrangement of the Excel files. We acknowledge Srikanth S. Manda for assistance with the proteogenomic analysis. This research was supported by the 21C Frontier Follow-On Support Research and Development Program (No. 2012M3C5A1053342), Proteogenomics Research Program grants through the National Research Foundation of Korea (NRF) grant and Converging Research Center Program (2012K001536) funded by Ministry of Science, ICT & Future Planning (MSIP). This work was supported by NCI’s Clinical Proteomic Tumor Analysis Consortium initiative (U24CA160036).
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supplementary Tables 1–4, as described in the main text, are provided as MS Excel files (*.xlsx). Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org. The table descriptions are as follows.
References
- 1.Marko-Varga G, Omenn GS, Paik YK, Hancock WS. A first step toward completion of a genome-wide characterization of the human proteome. J Proteome Res. 2013;12(1):1–5. doi: 10.1021/pr301183a. [DOI] [PubMed] [Google Scholar]
- 2.Legrain P, Aebersold R, Archakov A, Bairoch A, Bala K, Beretta L, Bergeron J, Borchers CH, Corthals GL, Costello CE, Deutsch EW, Domon B, Hancock W, He F, Hochstrasser D, Marko-Varga G, Salekdeh GH, Sechi S, Snyder M, Srivastava S, Uhlen M, Wu CH, Yamamoto T, Paik YK, Omenn GS. The human proteome project: current state and future direction. Mol Cell Proteomics. 2011;10(7):M111 009993. doi: 10.1074/mcp.M111.009993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paik YK, Jeong SK, Omenn GS, Uhlen M, Hanash S, Cho SY, Lee HJ, Na K, Choi EY, Yan F, Zhang F, Zhang Y, Snyder M, Cheng Y, Chen R, Marko-Varga G, Deutsch EW, Kim H, Kwon JY, Aebersold R, Bairoch A, Taylor AD, Kim KY, Lee EY, Hochstrasser D, Legrain P, Hancock WS. The Chromosome-Centric Human Proteome Project for cataloging proteins encoded in the genome. Nat Biotechnol. 2012;30(3):221–3. doi: 10.1038/nbt.2152. [DOI] [PubMed] [Google Scholar]
- 4.Aravidis C, Panani AD, Kosmaidou Z, Thomakos N, Rodolakis A, Antsaklis A. Detection of numerical abnormalities of chromosome 9 and p16/CDKN2A gene alterations in ovarian cancer with fish analysis. Anticancer Res. 2012;32(12):5309–13. [PubMed] [Google Scholar]
- 5.Dagher J, Dugay F, Verhoest G, Cabillic F, Jaillard S, Henry C, Arlot-Bonnemains Y, Bensalah K, Oger E, Vigneau C, Rioux-Leclercq N, Belaud-Rotureau MA. Histologic prognostic factors associated with chromosomal imbalances in a contemporary series of 89 clear cell renal cell carcinomas. Hum Pathol. 2013;44(10):2106–15. doi: 10.1016/j.humpath.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 6.Narayanan V, Pollyea DA, Gutman JA, Jimeno A. Ponatinib for the treatment of chronic myeloid leukemia and Philadelphia chromosome-positive acute lymphoblastic leukemia. Drugs Today (Barc) 2013;49(4):261–9. doi: 10.1358/dot.2013.49.4.1950147. [DOI] [PubMed] [Google Scholar]
- 7.Merlo A, Gabrielson E, Askin F, Sidransky D. Frequent loss of chromosome 9 in human primary non-small cell lung cancer. Cancer Res. 1994;54(3):640–2. [PubMed] [Google Scholar]
- 8.Zhu Y, Spitz MR, Strom S, Tomlinson GE, Amos CI, Minna JD, Wu X. A case-control analysis of lymphocytic chromosome 9 aberrations in lung cancer. Int J Cancer. 2002;102(5):536–40. doi: 10.1002/ijc.10762. [DOI] [PubMed] [Google Scholar]
- 9.Kang JU, Koo SH, Kwon KC, Park JW. Frequent silence of chromosome 9p, homozygous DOCK8, DMRT1 and DMRT3 deletion at 9p24. 3 in squamous cell carcinoma of the lung. Int J Oncol. 2010;37(2):327–35. [PubMed] [Google Scholar]
- 10.Panani AD, Maliaga K, Babanaraki A, Bellenis I. Numerical abnormalities of chromosome 9 and p16CDKN2A gene deletion detected by FISH in non-small cell lung cancer. Anticancer Res. 2009;29(11):4483–7. [PubMed] [Google Scholar]
- 11.Shibukawa K, Miyokawa N, Tokusashi Y, Sasaki T, Osanai S, Ohsaki Y. High incidence of chromosomal abnormalities at 1p36 and 9p21 in early-stage central type squamous cell carcinoma and squamous dysplasia of bronchus detected by autofluorescence bronchoscopy. Oncol Rep. 2009;22(1):81–7. doi: 10.3892/or_00000409. [DOI] [PubMed] [Google Scholar]
- 12.Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6(5):359–62. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 13.Jeong SK, Lee HJ, Na K, Cho JY, Lee MJ, Kwon JY, Kim H, Park YM, Yoo JS, Hancock WS, Paik YK. GenomewidePDB, a proteomic database exploring the comprehensive protein parts list and transcriptome landscape in human chromosomes. J Proteome Res. 2013;12(1):106–11. doi: 10.1021/pr3009447. [DOI] [PubMed] [Google Scholar]
- 14.Dana RC, Welch WJ, Deftos LJ. Heat shock proteins bind calcitonin. Endocrinology. 1990;126(1):672–4. doi: 10.1210/endo-126-1-672. [DOI] [PubMed] [Google Scholar]
- 15.Holaska JM, Kowalski AK, Wilson KL. Emerin caps the pointed end of actin filaments: evidence for an actin cortical network at the nuclear inner membrane. PLoS Biol. 2004;2(9):E231. doi: 10.1371/journal.pbio.0020231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo G, Herrera AH, Horowits R. Molecular interactions of N-RAP, a nebulin-related protein of striated muscle myotendon junctions and intercalated disks. Biochemistry. 1999;38(19):6135–43. doi: 10.1021/bi982395t. [DOI] [PubMed] [Google Scholar]
- 17.Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429(6994):841–7. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- 18.Adams B, Dorfler P, Aguzzi A, Kozmik Z, Urbanek P, Maurer-Fogy I, Busslinger M. Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis. Genes Dev. 1992;6(9):1589–607. doi: 10.1101/gad.6.9.1589. [DOI] [PubMed] [Google Scholar]
- 19.von Bulow M, Heid H, Hess H, Franke WW. Molecular nature of calicin, a major basic protein of the mammalian sperm head cytoskeleton. Exp Cell Res. 1995;219(2):407–13. doi: 10.1006/excr.1995.1246. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura T, Sanokawa R, Sasaki Y, Ayusawa D, Oishi M, Mori N. N-Shc: a neural-specific adapter molecule that mediates signaling from neurotrophin/Trk to Ras/MAPK pathway. Oncogene. 1996;13(6):1111–21. [PubMed] [Google Scholar]
- 21.Sugasawa K, Ng JM, Masutani C, Maekawa T, Uchida A, van der Spek PJ, Eker AP, Rademakers S, Visser C, Aboussekhra A, Wood RD, Hanaoka F, Bootsma D, Hoeijmakers JH. Two human homologs of Rad23 are functionally interchangeable in complex formation and stimulation of XPC repair activity. Mol Cell Biol. 1997;17(12):6924–31. doi: 10.1128/mcb.17.12.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang JS, Wrensch MR, Hansen HM, Sison JD, Aldrich MC, Quesenberry CP, Jr, Seldin MF, Kelsey KT, Kittles RA, Silva G, Wiencke JK. Nucleotide excision repair genes and risk of lung cancer among San Francisco Bay Area Latinos and African Americans. Int J Cancer. 2008;123(9):2095–104. doi: 10.1002/ijc.23801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roux PP, Shahbazian D, Vu H, Holz MK, Cohen MS, Taunton J, Sonenberg N, Blenis J. RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J Biol Chem. 2007;282(19):14056–64. doi: 10.1074/jbc.M700906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iida M, Brand TM, Campbell DA, Starr MM, Luthar N, Traynor AM, Wheeler DL. Targeting AKT with the allosteric AKT inhibitor MK-2206 in non-small cell lung cancer cells with acquired resistance to cetuximab. Cancer Biol Ther. 14(6):481–91. doi: 10.4161/cbt.24342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.ten Klooster JP, Leeuwen I, Scheres N, Anthony EC, Hordijk PL. Rac1-induced cell migration requires membrane recruitment of the nuclear oncogene SET. Embo J. 2007;26(2):336–45. doi: 10.1038/sj.emboj.7601518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam BD, Anthony EC, Hordijk PL. Cytoplasmic targeting of the proto-oncogene SET promotes cell spreading and migration. FEBS Lett. 587(2):111–9. doi: 10.1016/j.febslet.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 27.Tochio N, Umehara T, Munemasa Y, Suzuki T, Sato S, Tsuda K, Koshiba S, Kigawa T, Nagai R, Yokoyama S. Solution structure of histone chaperone ANP32B: interaction with core histones H3-H4 through its acidic concave domain. J Mol Biol. 401(1):97–114. doi: 10.1016/j.jmb.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Yu Y, Shen SM, Zhang FF, Wu ZX, Han B, Wang LS. Acidic leucine-rich nuclear phosphoprotein 32 family member B (ANP32B) contributes to retinoic acid-induced differentiation of leukemic cells. Biochem Biophys Res Commun. 423(4):721–5. doi: 10.1016/j.bbrc.2012.06.025. [DOI] [PubMed] [Google Scholar]
- 29.Davis-Smyth T, Duncan RC, Zheng T, Michelotti G, Levens D. The far upstream element-binding proteins comprise an ancient family of single-strand DNA-binding transactivators. J Biol Chem. 1996;271(49):31679–87. doi: 10.1074/jbc.271.49.31679. [DOI] [PubMed] [Google Scholar]
- 30.Weber A, Kristiansen I, Johannsen M, Oelrich B, Scholmann K, Gunia S, May M, Meyer HA, Behnke S, Moch H, Kristiansen G. The FUSE binding proteins FBP1 and FBP3 are potential c-myc regulators in renal, but not in prostate and bladder cancer. BMC Cancer. 2008;8:369. doi: 10.1186/1471-2407-8-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heid HW, Moll R, Schwetlick I, Rackwitz HR, Keenan TW. Adipophilin is a specific marker of lipid accumulation in diverse cell types and diseases. Cell Tissue Res. 1998;294(2):309–21. doi: 10.1007/s004410051181. [DOI] [PubMed] [Google Scholar]
- 32.Matsubara J, Honda K, Ono M, Sekine S, Tanaka Y, Kobayashi M, Jung G, Sakuma T, Nakamori S, Sata N, Nagai H, Ioka T, Okusaka T, Kosuge T, Tsuchida A, Shimahara M, Yasunami Y, Chiba T, Yamada T. Identification of adipophilin as a potential plasma biomarker for colorectal cancer using label-free quantitative mass spectrometry and protein microarray. Cancer Epidemiol Biomarkers Prev. 20(10):2195–203. doi: 10.1158/1055-9965.EPI-11-0400. [DOI] [PubMed] [Google Scholar]
- 33.Nurnberg A, Kitzing T, Grosse R. Nucleating actin for invasion. Nat Rev Cancer. 11(3):177–87. doi: 10.1038/nrc3003. [DOI] [PubMed] [Google Scholar]
- 34.Moriya Y, Nohata N, Kinoshita T, Mutallip M, Okamoto T, Yoshida S, Suzuki M, Yoshino I, Seki N. Tumor suppressive microRNA-133a regulates novel molecular networks in lung squamous cell carcinoma. J Hum Genet. 57(1):38–45. doi: 10.1038/jhg.2011.126. [DOI] [PubMed] [Google Scholar]
- 35.Martin J, Magnino F, Schmidt K, Piguet AC, Lee JS, Semela D, St-Pierre MV, Ziemiecki A, Cassio D, Brenner C, Thorgeirsson SS, Dufour JF. Hint2, a mitochondrial apoptotic sensitizer down-regulated in hepatocellular carcinoma. Gastroenterology. 2006;130(7):2179–88. doi: 10.1053/j.gastro.2006.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee LR, Teng PN, Nguyen H, Hood BL, Kavandi L, Wang G, Turbov JM, Thaete LG, Hamilton CA, Maxwell GL, Rodriguez GC, Conrads TP, Syed V. Progesterone enhances calcitriol antitumor activity by upregulating vitamin d receptor expression and promoting apoptosis in endometrial cancer cells. Cancer Prev Res (Phila) 6(7):731–43. doi: 10.1158/1940-6207.CAPR-12-0493. [DOI] [PubMed] [Google Scholar]
- 37.Hakomori S. Glycosylation defining cancer malignancy: new wine in an old bottle. Proc Natl Acad Sci U S A. 2002;99(16):10231–3. doi: 10.1073/pnas.172380699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobayashi Y, Suzuki K, Kobayashi H, Ohashi S, Koike K, Macchi P, Kiebler M, Anzai K. C9orf10 protein, a novel protein component of Puralpha-containing mRNA-protein particles (Puralpha-mRNPs): characterization of developmental and regional expressions in the mouse brain. J Histochem Cytochem. 2008;56(8):723–31. doi: 10.1369/jhc.2008.950733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanaka M, Sasaki K, Kamata R, Hoshino Y, Yanagihara K, Sakai R. A novel RNA-binding protein, Ossa/C9orf10, regulates activity of Src kinases to protect cells from oxidative stress-induced apoptosis. Mol Cell Biol. 2009;29(2):402–13. doi: 10.1128/MCB.01035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Underwood R, Chiravuri M, Lee H, Schmitz T, Kabcenell AK, Yardley K, Huber BT. Sequence, purification, and cloning of an intracellular serine protease, quiescent cell proline dipeptidase. J Biol Chem. 1999;274(48):34053–8. doi: 10.1074/jbc.274.48.34053. [DOI] [PubMed] [Google Scholar]
- 41.Danilov AV, Danilova OV, Brown JR, Rabinowitz A, Klein AK, Huber BT. Dipeptidyl peptidase 2 apoptosis assay determines the B-cell activation stage and predicts prognosis in chronic lymphocytic leukemia. Exp Hematol. 38(12):1167–77. doi: 10.1016/j.exphem.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Total chromosome 9-encoded protein list from normal human samples
The list of newly detected missing proteins on chromosome 9.
The list of detected top 10 abundance chromosome 9-encoded proteins among the normal human sample data.
The lists in the identification of chromosome 9 proteins from normal lung and lung cancer tissues.