Abstract
Studying genetics of development with small model organisms such as zebrafish Danio Rerio, the fruit fly Drosophila Melanogaster, and the soil-dwelling nematode Caenorhabditis elegans, provide unique opportunities for understanding related processes and diseases in human as well as potential drug screens. There have been sweeping developments of microfabrication and microfluidic technologies for manipulating and imaging small objects, which allow high-throughput quantitative biological studies. Here, we review recent progress on these microfluidic tools and project future directions in these fields.
Keywords: Developmental biology, Microfluidics, Model organisms, Zebrafish, Danio rerio, Nematode, Caenorhabditis elegans, Fruit fly, Drosophila melanogaster
1) Introduction
Development is a complex process that is in part determined by an intricate network of genes and their interactions with the environment. Model organisms have been used extensively in understanding how development and growth are controlled, and in some instances for understanding diseased processes in human. These model organisms, which have been chosen because of their intrinsic characteristics such as small size, optical transparency, ease of genetics and relatively short life span, have been extensively studied over a few decades of research [1–4]. Worms, flies, and fish have been widely used for the developmental studies due to their sequenced genomes, ease of introducing transgenes, and well-characterized anatomical information. For example, Zebrafish, Danio rerio, has been applied for human disease models as well as drug safety tests [5–7], and the nematode, Caenorhabditis elegans, has also been a powerful tool for the pharmaceutical industry as well as for fundamental studies of development and neurobiology [8, 9], due to the highly conserved molecular and genetic pathways between mammals and the model organisms. The fruit fly, Drosophila melanogaster, has also been frequently used as a model organism for studying human diseases and therapeutic discovery [10, 11]. Despite much progress, the traditional strategies to handle these organisms are labor intensive and time-consuming, which limit the throughput of the analysis and ultimately the speed of discovery. The macro-scale tools do not seem appropriate or adequate for handling these objects; further, obtaining high-resolution spatiotemporal information during the developments of the organisms in the precisely controlled dynamic microenvironments is almost impossible with the current conventional techniques. To overcome these limitations of the traditional manipulation tools and provide new paradigms in the developmental studies of small model organisms, a variety of microfabrication and microfluidic technologies has been utilized for the phenotyping and screening of the organisms or their embryos.
In this Review, we will discuss the contribution and potential contributions of microfluidic tools in genetics and developmental studies of small model organisms. We will also take a look at some prospects for future research directions.
2) Nematodes
The nematode, C. elegans, is one of the most widely studied model organisms to address fundamental questions in developmental biology, neurobiology, and behavior. C. elegans is a small, transparent, free-living round worm, whose size is about 1mm in length as an adult. The nematodes are easy to maintain in the laboratory as they feed on bacteria and have a relatively short life cycle – with a whole life span being 2–3 weeks and a reproductive cycle of ~3 days. Due to those characteristic advantages, the developmental cell lineage and the neural wiring diagram of the C. elegans have been completely mapped [12, 13]. The C. elegans genome has long been sequenced [14], and genetic/genomic manipulation such as knockouts, knockdown via RNAi, and making transgenic strains are routinely available.
Imaging is a required technology for phenotypic analysis of multicellular model organisms, especially for the genetic and developmental biology studies. Microfluidic tools have played a role mainly in optical imaging and manipulation of the nematodes. Immobilization is often required for high-resolution imaging [15–17] or the surgery via laser ablation techniques [18, 19]. In the traditional protocols, the worms are fully immobilized on an agarose pad with anesthetics, such as sodium azide. Partial immobilization for calcium imaging is usually achieved using a cyanoacrylate glue. The gluing method always requires manual handling processes, which are laborious and time-consuming.
Hulme et al. developed a passive immobilization method using tapered microchannel geometry, which physically confines the nematodes (Figure 1A) [20]. The chip has 128 parallel microchannels, whose width gradually narrows, and worms in suspension are injected into each channel by the pressure difference across each loading channel. Because loading is pressure-driven, the 128 channels essentially operate independently, resulting in the parallel immobilization of a large population of worms. Allen et al. applied this immobilization scheme based on tapered microchannels for laser ablation of single synapses in neurons in vivo[21]. Using this tool, they investigated the synaptic plasticity of the HSNL motor neuron in the nematode at the L4 development stage compared to that in the nematode at young adult stage.
Figure 1. Microfluidic tools for immobilization and confinement of nematodes.
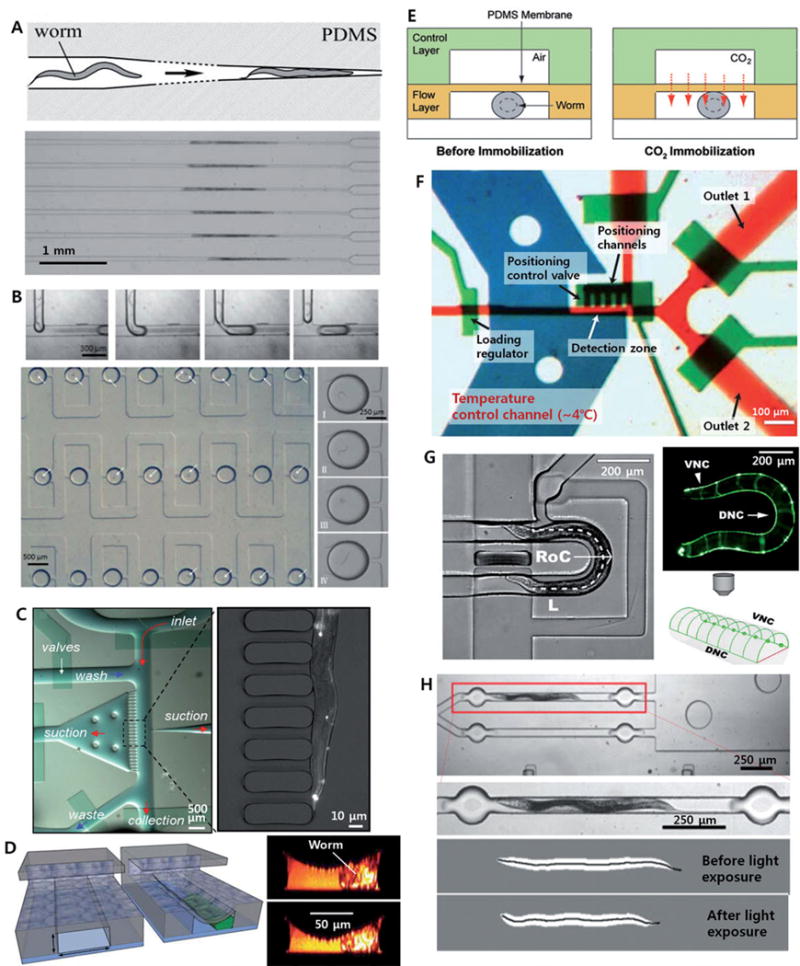
(A) Passive immobilization using a tapered microchannel ([20]–Reproduced by permission of The Royal Society of Chemistry); (B) microdroplet generation and trapping array ([27]–Reproduced by permission of The Royal Society of Chemistry); (C) suction-induced immobilization (Copyright 2007 National Academy of Sciences, USA [29]); (D) compressive immobilization (Reprinted by permission from Macmillan Publishers Ltd: [30] copyright 2008); (E) CO2 immobilization ([35]–Reproduced by permission of The Royal Society of Chemistry); (F) cooling method for fully immobilization [36]; (G) passive lateral orientation [40]; (H) photoactivation of motor neurons (Reprinted from [51] with permission from Elsevier) of nematodes inside microfluidic devices.
This tapered microchannel geometry has also been applied for imaging during the longitudinal experiments to observe whole lifespans of nematodes [22]. The modified device has 16 culture chambers each connected with a tapered microchannel to enable temporary immobilization of the nematode for imaging. Screw valves embedded in the PDMS device were also utilized for independent control of flows in each culture chamber [23]. This microfluidic platform, which enables long-term cultivation of the nematodes in independently controlled microenvironments, could be very useful for the longitudinal studies of nematodes such as development and aging [24, 25].
Droplet-based microfluidic platforms, in which nematode eggs or larvae were encapsulated into segmented droplets, have also been developed for the developmental studies and screenings of nematodes. Clausell-Tormos et al. demonstrated the encapsulation of nematode eggs into M9 media droplets containing Escherichia coli in a gas-permeable PTFE tubing and observed their growth for several days [26]. Shi et al. have developed a microfluidic device, in which all the processes from the encapsulation of single nematode into individual droplet to trapping and incubation of the droplets (Figure 1B) [27]. They applied a conventional T-junction droplet generator to encapsulate nematode larvae at L1 stage into microdroplets, whose volume is around 28 nL, and observed behavioral changes upon application of a neurotoxin, 1-methyl-4-phenylpyridinium (MPP+). They also combined their droplet microfluidic platform with the tapered microchannel array to allow fluorescence imaging on the neurons [28]. The device trapped 80 droplets containing individual nematodes for the behavioral assays. Then the nematodes were pinned into the tapered microchannel similar to Hulmes et al. They applied the system to examine the mobility defects, dopaminergic (DAergic) neuron degeneration, and increased oxidative stress of the nematodes in response to neurotoxin, 6-hydroxydopamine.
These passive platforms for trapping and immobilization of individual worms would allow simple fluidic manipulation, which is advantageous for the integration of the system. The drawback may be that the number of animals that can be simultaneously processed on a chip is restricted by the number of trapping positions in the parallel immobilization schemes. High-throughput sorting of specific individuals is also difficult for the array-based, enclosed system.
Alternatively, some active microfluidic platforms, in which external forces were applied to freely control the transport and immobilization of individual nematodes, have also been developed. Rohde et al. captured a nematode via suction in a microchannel (Figure 1C) [29]. PDMS membrane microvalves were integrated into the chip through a multi-layer soft lithography. Guo et al. applied the PDMS membrane valves to directly trap and immobilize a nematode in microchannel (See Figure 1D) [30]. With this platform, they investigated the nerve regeneration after a laser nanosurgery of the nematodes grown in a variety of culture conditions. Zeng et al. also reported the PDMS membrane-based device, which provides more stable immobilization of the worm via combination with micropillar array for suction-induced fixation [31]. Gilleland et al. demonstrated high-resolution confocal or two-photon imaging of nematodes as well as the nanosurgery [32] while Samara et al. suggested more advanced format of the microfluidic device for the membrane-based immobilization, which allows capture of a single worm via an aspiration channel, followed by cleaning of the channels and its immobilization [33]. Based on the nanosurgery system, they demonstrated in vivo screens for identifying compounds which affect neurite regeneration of C. elegans PLM neurons. The membrane-based immobilization technique has also been applied to screen for the effect of neurotoxin MPP onto the mobility defects and DAergic neuron degeneration in nematodes [34]. This microfluidic array system allowed both behavioral assays and fluorescence imaging of multiple nematodes in response to neurotoxin. Although they had already demonstrated the behavioral assays of nematodes for the same target, MPP, by encapsulating them into microdroplets [27], this integration would allow characterization of the behavioral and morphometric phenotypes. They also performed more studies based on the droplet microfluidic platform combined with the tapered microchannel method as an alternative approach [28].
There are also several other methods to immobilize C. elegans by controlling the microenvironments the animals are in, particularly using CO2[35] and cooling [36]. In the former case, CO2 gas was injected into the channel above a chamber that a nematode was located (See Figure 1E) [35]. The CO2molecules were transported to the trapped nematode through the thin gas-permeable PDMS membrane, which can also be used for the compressive immobilization at high pressure. The authors compared the mobility recovery of nematodes after the CO2 immobilization with that after the compressive immobilization. While the nematodes immobilized for 1 h by the deflected PDMS membrane did not show any locomotion activity, the nematodes immobilized by CO2were completely recovered after the 2.5 hours long-term immobilization. Although the effect of CO2 immobilization on the physiological changes in the nematodes is still largely unknown, this CO2 method may allow long-term fluorescent imaging with reduced photobleaching due to the low-oxygen condition.
In using transient cooling to immobilize worms, pre-cooled liquid was injected into the channel above a chamber that a nematode was confined. A thin membrane separates the cooling channel and the worm channel, and the worm is immobilized by being transiently cooled down (to ~4°C)for a few seconds [36]. When the worms are fully immobilized, z-stack images can be obtained. With a widely available epifluorescence microscope, Chung et al. demonstrated the ability to analyze the gene expression pattern in 3D; worms can be sorted in high-throughput based on the expression of reporter fluorescence proteins in vivo (Figure 1F). The cooling scheme has also been applied in a multiwell plate format, which allows time-lapse imaging of nematodes for the longitudinal studies as well as reduces complexity than the microvalve-based microfluidic devices [37]. The latter paper applied a temperature control element composed of thermoelectrically cooled aluminum pins, which can be inserted into each well containing a nematode and individually controlled. These cooling methods would be the only way to fully immobilize animals as well as provide the highest robustness among the variety of immobilization schemes. However, cooling is inappropriate for live imaging applications where the specimen needs to be observed at physiological conditions.
To overcome the difficulty for live imaging (at physiological conditions), Krajniak et al. recently developeda microfluidic device to fully immobilize the nematode at physiological temperature based on a gel whose phase change is temperature-sensitive (~2°C change) [38]. Slight increase in temperature in a separate channel is used to heat up the chambers containing the nematodes and the gel, amphiphilic block copolymer PF128, and the gelation result in the immobilization of the nematodes. The repetitive short-term immobilizations of the nematodes during longitudinal studies were possible by exchanging culture media with the PF127 solution because the sol-gel transition of PF127 is reversible. Despite a slight temperature shift and a relatively high complexity to operate, this method is the only way to fully immobilize the nematodes at physiological conditions up to now.
Besides the immobilization of specimen, it is sometimes important to control the orientation of the nematodes to observe certain structures. Although there have been abundant microfluidic tools for nematode research, none of them have been available for controlling the orientation of nematodes. Although anterior and posterior sides of the nematode have been distinguished via an image processing in a microfluidic device [39], there was no method to bias the orientation either ventral/dorsally or anterior/posterior using microfluidic devices. Recently, Cáceres et al. have developed an integrated microfluidic platform for passive orientation of nematodes into lateral body positions via curved microfluidic channel (See Figure 1G) [40]. In the curved channel geometry, more than 80% of the nematodes were oriented in to lateral body positions with the radii of curvature for the channel geometries being 125 to 145 μm. Based on this simple technology, they characterized the neural morphologies and found six mutants that have neuronal development or neurodegenerative defects, and sorted them in a high-throughput manner. The advantage of this approach is the simplicity; the passive operation scheme is compatible with the integrated microfluidic device for automatic sorting, where laborious fixation processes are avoided. Such this system, where active flow control and passive animal orientation exhibit are combined with the computer-assisted phenotyping based on the advanced image processing and hardware control [41], it could potentially present ultra-high performance system for large-scale phenotyping and morphological studies of the nematode development.
Another set of experiments that microfluidics enables is in optogenetic manipulations of neurons or muscles in worms. Optogenetics is the combination of genetic and optics to precisely control well-defined events within complex biological systems [42]. has emerged as one of the promising technologies for studying neural circuits in the model organisms. Some light-activated channels such as channelrhodopsin-2 [43] and halorhodopsin [44] have been actively applied for optical excitement and inhibition of neurons, respectively, in both nematodes [45–47] and zebrafish [48, 49]. Real-time optical control of specific targets in freely moving C. elegans has also been demonstrated on the basis of computer-assisted tracking of the motion of the nematodes and a digital micromirror device providing high spatial resolution [50]. A microfluidic platform combined with the optogenetic tools has also been reported [51]. In this study, the whole system was automatically operated by a computer and image processing was also applied for high-throughput, quantitative analysis of the contraction and relaxation of the muscles of C. elegans under photoactivation of motor neurons (See Figure 1H). In addition, trapping and parallel analysis of multiple nematodes inside microfluidic channels allowed high-throughput assays. As like this example, the combination of microfluidics, optogenetics and image processing would open a new way for small animal studies.
The microfabrication technologies can be applied not only for the applications based on the fluidic controls at microscale, but also for decorating microenvironments where the nematodes live. Lockery et al. fabricated a microchip consists of an agar microposts array that mimics natural soil environment [52]. Based on the soft-lithographic fabrication processes, they could easily modify the size and the gap between the posts. The artificial dirt chip provided more complex and realistic surface than abare agar surface, which has been used in traditional. This soil-like microchip could be utilized for the swimming motion of C. elegans [53]. Three-dimensional activity of the nematodes such as nictation has also been studied based on the artificial dirt chip in combination with the optogenetic tools [54]. The artificial soil device might potentially allow more reliable studies based on more natural-like environments in combination with chemical modification or fluidic controls.
Another way to study C. elegans, centrifugal microfluidic platforms, in which a centrifugal force due to the rotation of disc-shaped device is applied for the fluid transport without any external pumps or connectors, have also been applied to automatically cultivate nematodes under hypergravity conditions (~ 100G) [55] and to investigate its effect on neuronal function, muscular structure, and metabolic profiles in the nematodes [56]. This motor-based system might be a unique tool for applying the hypergravity on the whole animal via centrifugation. Although we cannot assert this platform to be suitable for longitudinal cultivation and high-throughput analysis of nematodes, further studies based on the tools of this sort might provide extensive perspectives on the developmental studies of the model organisms.
There also has been studied on electric field-induced behavior of worms in microfluidic environment [57, 58]. The electrotactic response was analyzed as a function of worm developmental stages and genetic background. The results suggest that longer animals, which might experience more potential drop across the body, were more sensitive to the electric field than shorter ones [57]. However, in the adult stage, the average locomotion decreased as the animals got older [58]. The differences in the electrotactic movements of worms may be another aspect of sensing and behavior that can be used to sort animals.
3) Fruit flies
Fruit fly, D. melanogaster, is an insect that has been used as a model organism in studies of genetics and developmental biology for over a hundred years [59]. It is one of the most long-established model organisms due to its short life cycle (~2 weeks), small and mapped genome, simple diet, and many well-established protocols for biochemical and genetic analysis.
Due to their inherent characteristic flying in the air, in the traditional protocols for the fruit fly research, an anesthetic mixture is generally used for reducing their movements to observe or sort them, which were bred in a plastic vial containing some media in the bottom. In this context, it is almost impossible to apply the microfluidic tools for the fruit flies at the adult stage. Therefore, most of the microfluidic studies on the fruit flies have targeted their embryos.
Embryogenesis in the fruit fries has been extensively studied as a favorite model system for geneticists and developmental biologists. For example, the first study about the effect of mutations on the differentiation and cellular functions in a complex organism was carried out on the basis of D. melanogaster embryos [60]. Discoveries of genes regulate the pattern formation during the fruit fly embryogenesis have also attracted much attention from many scientists. For example, perturbation in morphogens such as Bicoid protein via genetic or environmental variations induced the changes in embryo pattering such as posterior or anterior shift of anterior anlagen [61, 62]. On 2005, Lucchetta et al. applied a laminar flow in a simple Y-shaped microfluidic channel to regulate the local temperature of the fruit fly embryo in a spatial and temporal manner (See Figure 2A) [63]. This is also the first application of fluid flows at micro-scale, which is in almost all cases laminar, [64, 65] to the embryonic study of fruit fly. Anterior and posterior parts of the embryo were exposed to different temperatures formed by laminar streams of different temperatures, resulting in a step-like temperature profile across the channel [66]. The temperature profile around the embryo could be dynamically controlled based on the precise flow control as well as varied with the position of the embryo.
Figure 2. Microfluidic tools for studies of fruit fly embryos.
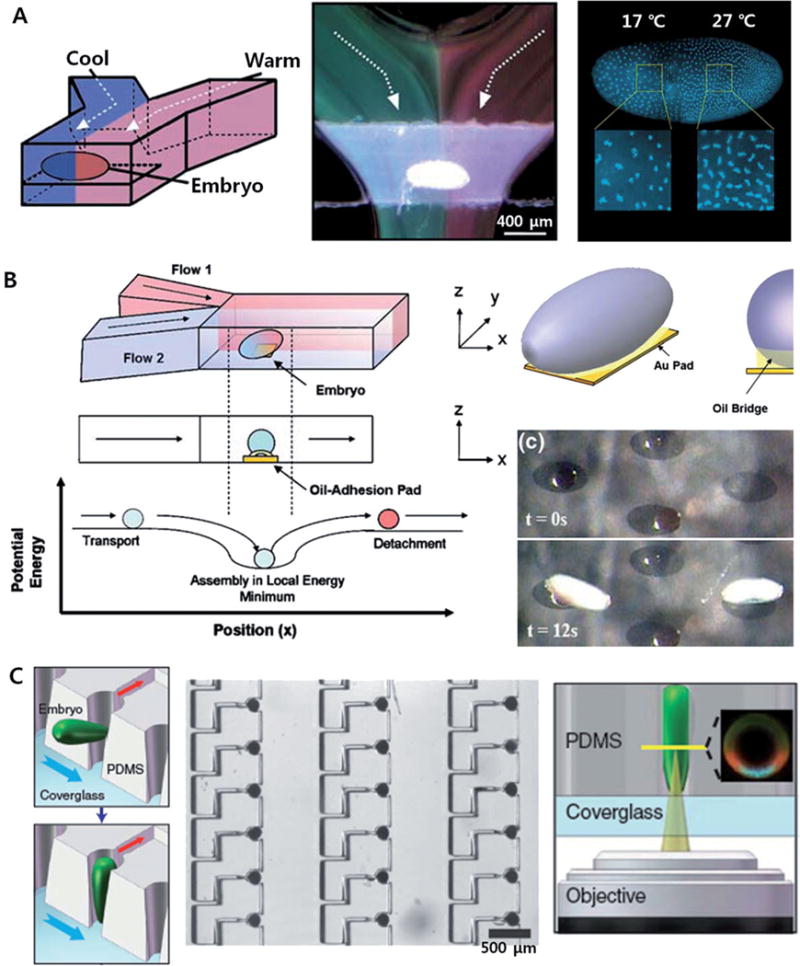
(A) Spatiotemporal control of local temperature in a fruit fly embryo with laminar flow (Reprinted by permission from Macmillan Publishers Ltd: [63] copyright 2005); (B) immobilization of embryos with oil-adhesion pads (Reproduced with permission from Springer Science and Business Media [67]); and (C) large-scale positioning and vertical orientation of fruit fly embryos [71].
Moreover, Dagani et al. have reported a method for the positioning of a fruit fly embryo in the middle of microfluidic channel via capillary force-induced self-assembly in an oil-adhesion pad (See Figure 2B) [67]. This microfluidic assembly method has also been applied to immobilize, position, and align the fruit fly embryos in 2D arrays [68]. The high-throughput process of multiple embryos could be embodied by using the advanced microinjection technologies being developed, such as a microfabricated injection needle arrays [69] or a micromachined piezoelectric vibratory microinjector [70].
The large-scale patterning of embryos would also be very useful tool for the high-throughput studies of the morphogenesis in the embryos. For the positioning and orientation of embryos, Chung et al. have developed a microfluidic embryo-trap array (Figure 2C) [71]. Different from the previously reported methods, Chung et al. demonstrated vertical orientation of embryos, in which their dorsoventral plane is in horizontal position, only with a simple microstructure leading the embryo to be oriented by a flow. The array-type device would allow high-throughput quantitative research of pattern formation in the early embryos of fruit fly, as well as new comprehensive studies of the dorsoventral system in the fruit fly embryo through statistical analysis of the images of the vertically oriented embryos.
4) Zebrafish
Zebrafish as a model organism has the advantage of a large number of offspring; many protocols for generating transgenic models have been developed. In addition, the transparency of embryos and larvae allow real-time imaging of internal processes and tracking gene activities. These advantages allow the zebrafish to become one of the most prominent genetic model organisms not only for aquaculture fish species [72] but also for human diseases such as diabetes [73], Parkinson’s disease [74], amyothrophic lateral sclerosis [75], and many other genetic diseases [76, 77].
For the studies of zebrafish genetics and development, conventional multi-well plates and standard liquid handling techniques have been applied to the embryonic cultures and screenings. In the traditional protocols, the embryos collected after spawning processes are manually sorted to remove dead or unfertilized ones. The selected healthy embryos are transferred into the multiwell plates for chemical treatments, following by the screenings based on optical observation [78]. Although this conventional approach enables the screening of zebrafish or their embryos based on their behavioral and morphological phenotypes, there are still some limitations in the manipulation, stimulation, and observation of the targets with high spatiotemporal resolution. For example, a laborious and time-consuming manual handling operation limits the throughput as well as the uniformity and the accuracy in treatment might be reduced in a loosely controlled environment. Delivery of target species to localized positions of the model organisms has also been unavailable. To deal with those limitations of the conventional techniques, several approaches on the basis of novel microfluidic and optical technologies have been developed.
Most of the microfluidic devices developed for the zebrafish research have been utilized mainly for the embryonic culture and development. Funfak et al. have first developed a microfluidic platform to observe the developmental processes of D. rerio embryos in microfluidic segments and to test the effect of some chemicals on them (See Figure 3A) [79]. The microfluidic segments were generated by alternate aspiration of perfluoromethyldecalin solution as an immiscible carrier phase and sample solution containing eggs using a syringe pump. They observed the embryonic development inside the segments of sample solution, in Teflon tubes more than 75 h with a total volume of ~6 μL. They also examined the effect of sodium dodecyl sulfate and CuCl2 on the survival and physiological development of the embryos inside the microfluidic segments. Although this segmented microfluidic system allows a simple method to form a variety of chemical environments for the embryonic developments, it is very difficult for the system to precisely control the number of embryos in each segment and to dynamically modulate the chemical microenvironments; it also requires manual steps for dipping the tubes into different solutions during loading.
Figure 3. Microfluidic tools for studies of zebrafish embryos.
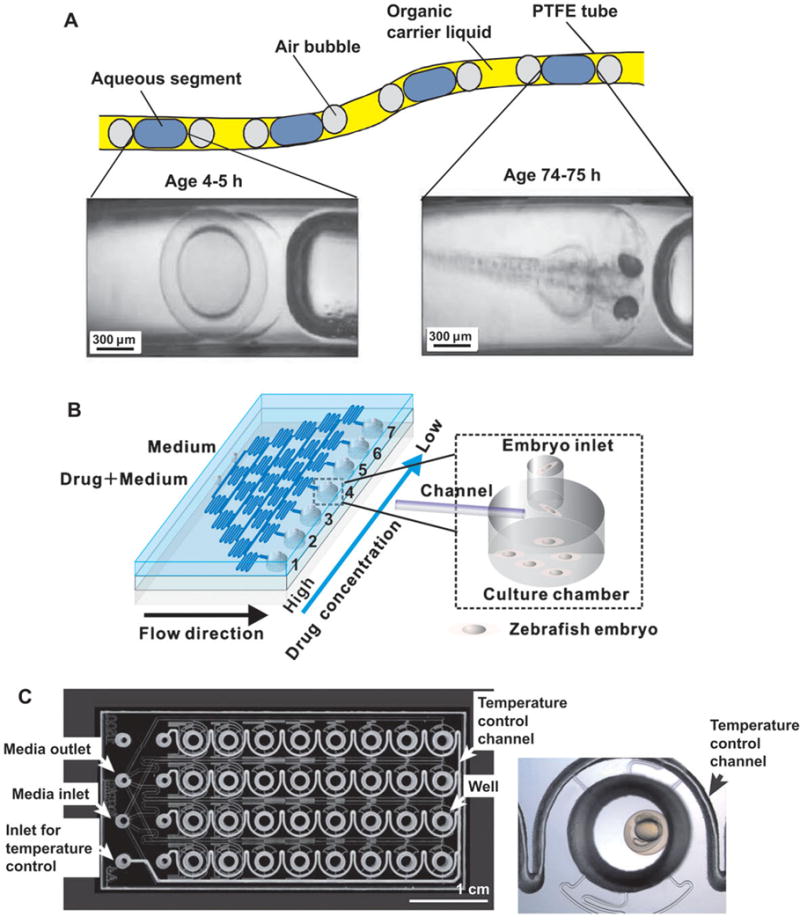
(A) Microfluidic segment system ([79]–Reproduced by permission of The Royal Society of Chemistry); (B) microfluidic array with drug concentration gradient generator (Reprinted with permission from [80]. Copyright 2011, American Institute of Physics); (C) microfluidic flow-through system ([85]–Reproduced by permission of The Royal Society of Chemistry).
To be able to screen for drug effects on zebrafish embryos, Yang et al. developed a microfluidic array system (See Figure 3B) [80]. This chip generates different dilutions of drugs and was used to evaluate the dynamic developmental toxicity and teratogenicity of clinical drugs. They applied a conventional microfluidic gradient generator [81, 82] to spontaneously form seven different concentrations of anti-proliferating drugs such as doxorubicin and 5-fluorouracil; each microchannel branch was connected to the culture chambers containing multiple zebrafish eggs at various developmental stages. This microfluidic array system could facilitate embryonic development assays based on a continuous perfusion of chemical solutions at different concentrations of interest. Choudhury et al. have recently applied the same strategy to study the effect of a drug, valproic acid on the development of zebrafish embryo [83]. They applied the silicon-glass chip (which could be fabricated more accurately) to quantify the effect of valproic acid in the range of 0.01 to 0.05 mM on the development of tail and eye from live imaging of embryos for 3 days.
Shen et al. uses a microfluidic approach for delivering growth factors or cytokines to distinct regions of an embryo. The device immobilizes the embryos in a funnel-shaped hole connected to a microfluidic channel, through which the stimulation factors are transported by a gravity-driven flows [84]. Here the embryo itself separates the fluid in the microchannel from the embryo media in the Petri dish, provided that the embryo seals the funnel-shaped hole. Shen et al. applied this system for testing the effect of a pro-inflammatory cytokine on the zebrafish embryonic developments.
Although the systems developed by Yang et al. and Shen et al. could be powerful tools for the studies of the embryonic developments under controlled microenvironments, those open systems may have undesired effects such as evaporation and contamination of the media; imaging is also an issue because of a distortion due to the varying depth and the curved meniscus of the buffer.
Wielhouwer et al. have developed a microfluidic flow-through system towards parallel and cost-effective experiments based on zebrafish embryos (Figure 3C) [85]. In this system, the embryos can be continuously accessed and they are isolated in parallel chamber arrays to prevent the cross-contamination. The temperature of each chamber is also controllable via water flowing through heating channels on the chip. They investigated the survival rate and the phenotypes of the embryos against the flow rates and the ethanol treatment in the microdevice and compared with those of the embryos cultured in the conventional 96-well plates. Despite some minor defects in the phenotypes of the embryos cultured in the microfluidic device, there was no increase in malformation during their organogenesis inside the chip. Although this microfluidic flow-through system can be simply treated as a miniaturized system of the flow-through multiwell plate [86], the completely enclosed polydimethylsiloxane (PDMS) microfluidic device [85] are advantageous in as the ability to allow high-quality imaging of the embryos, precisely controllable flow rate, rapid buffer exchanges, and significantly reduced volume of the buffers and the reagents.
Recently, a microfluidic device for trapping single zebrafish embryos in an array has also been reported [87]. They applied a simple hydrodynamic scheme very similar to that described in the previous work for trapping and orientation of fruit fly embryos [71]. The embryos immobilized in each trap were cultured up to 72 h under a variety of perfusion conditions. The effect of an anti-angiogenic compound on the formation of intersegmental vessels during the zebrafish embryonic development was also investigated on the basis of fluorescence imaging of the single embryo array in a microfluidic chamber.
In addition to culturing, perfusing, and imaging, microdevices have also been applied for the transfection of the embryos. The introduction of target materials such as drugs or transgenes into cells or embryos is critical in modern biotechnology. Huang et al. have developed a microchip for the transfection of zebrafish embryos via electroporation (See Figure 4A) [88]. They successfully delivered trypan blue dyes, water-soluble quantum dots, and transgenes encoding green fluorescence protein (GFP) using the micro-fabricated device. The device has 20 cavities containing 10 wells for one embryo each and enables a batch process. Two parallel plate electrodes located at the top and the bottom sides of the cavities were utilized to apply electrical pulses. A digital microfluidic platform for manipulating zebrafish embryo has also been developed [89]. By using an electrowetting-based droplet manipulation, a droplet containing a zebrafish embryo was transported and mixed with a digestive reagent to allow dechorionation. This is a step that facilitates transfection and accelerates growth by removing the chorion of an embryo. Noori et al. have developed a capillary microinjection system based on a PDMS microfluidic channels with electroosmotic dosage control (See Figure 4B) [90]. The device has two collinear channels into which a glass capillary for embryo trapping via suction and a tapered microinjection needle are embedded. A precise insertion of the injection needle into the trapped embryo was controlled via compliant deformation of the PDMS substrate on two separated glass substrates – one fixed and the other moving. They could deliver the target using an electroosmosis, which can provide accurate flow control during injections. These microfluidic systems for microinjection and electroporation of the embryos would potentially be useful platform for the development of advanced embryonic research tools. The range of applications is potentially enormous, including drug development, in vitro fertilization, and genetic engineering, if further systematic approaches for optimizing the processes and increasing the reproducibility of these systems are performed.
Figure 4. Microfluidic tools for embryo transfection.
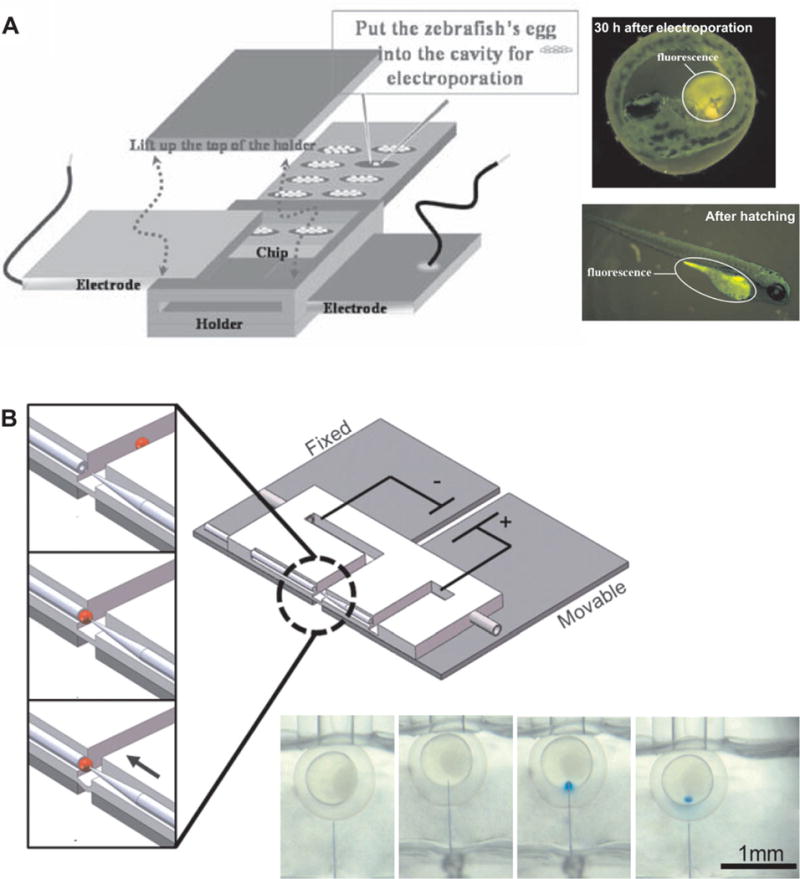
(A) Electroporation in microchamber array (Reproduced with permission from Springer Science and Business Media [88]); and (B) microinjection with electroosmotic flow control ([90]–Reproduced by permission of The Royal Society of Chemistry).
Despite elegant advantages of the microfluidic embryonic culture technologies, microchannels and culture chambers can restrict the growth of zebrafish larvae whose body length is 3–4 mm. This therefore is a disadvantage for the long-term studies using zebrafish larvae [91]. Pardo-Martin et al. have constructed a high-throughput platform for zebrafish larvae screenings based on a conventional multiwell plates and a capillary tube [92]. Here they controlled the flows with pinch valves and syringe pumps for transporting the larvae, and applied two optical systems: a simple setup composed of a light emitting device and a photodiode for discriminate fluorescent larvae from non-fluorecent ones, air bubbles and debris; and a high-speed confocal microscopy with an electron-multiplying charge-coupled device camera for high-speed wide-field fluorescence imaging of the fluorescent larvae. They could control the orientation of the larvae trapped in the capillary via high-precision stepping motors to rotate the capillary. They recently reported a modified system to achieve higher throughput of this automated screening technology by processing multiple fishes simultaneously [93]. These automated screening technologies allow high-throughput analysis of vertebrate model organisms based on high-quality images, which are obtainable without any manual handling procedures.
The throughput of the fully automated screening systems, in which all the handling processes were automated, is limited by the speed of image acquisition. In this context, several high-speed and high-quality imaging technologies have also been developed. For example, Huisken et al. have developed a special microscopic system to generate multidimensional high-resolution images of samples up to a few millimeters in size using a two-dimensional illumination of rotating agarose cylinder containing the sample [94]. Similarly, Keller et al. have developed a fluorescence microscopy based on a scanning light sheet for high spatiotemporal resolution imaging of embryogenesis in zebrafish and fly [95, 96]. They could simultaneously track the behavior of all cells in entire embryos during their developmental process inside a rotating agarose gel. However, these advanced imaging technologies are current not compatible with microfluidics. There is still plenty of room for development of advanced imaging tools combined with microfluidic devices for better controlled microenvironment as well as image processing technologies toward more extensive studies of small animal development.
5) Future Perspectives
As discussed in this paper, the microfluidic technologies have so far provided clear benefits for studying genetics and developmental biology based on small model organisms such as nematodes, fruit flies, and zebrafish. The advantages of these and other microfluidic tools can be further enhanced when used in combination with other advanced technologies. In particular, automated systems for high-speed image acquisition and processing may potentially be one of the most powerful tools to transform phenotyping and screening of small model organisms and their embryos. For example, in the developmental studies of the multicellular model organisms, wide-field and laser-scanning microscopic systems have frequently been required for obtaining high-quality images for accurate analyses and extensive studies [97–99]. However, there is typically a trade-off between image quality and acquisition speed in those imaging systems, limiting the high-throughput analysis based on high-quality images in spite of precise and rapid manipulation of the targets in microfluidic devices.
Post-processing of the acquired images is also a crucial part. For example, more advanced computational methods allowed high-throughput studies on gene expression, cell patterning, and neuronal morphology in development based on the 3D reconstructed image data [100–103]. Image processing tools have also been helpful for automated and quantitative analysis of movements in the animals such as swimming behavior of nematodes [104, 105] and eye movements in zebrafish [106]. Combination of microfluidic tools and the advanced image processing techniques would accelerate the development of automated platforms for high-throughput developmental studies on the model organisms.
Although small organisms such as nematodes, fruit flies, and zebrafish have some limitations as animal models of human development and diseases, they are informative models for many of these processes because many biological mechanisms are well conserved throughout evolution, and the advantages in both the genetics and the technological development make them attractive models for studying the basic biology and using them as drug screening platforms. These model systems are also much less expensive than mammalian models such as rodents and primates. Microfluidic platforms could dramatically enhance our ability to perform genetic and genomic research based on the small animal models by providing accurate sample handling and automated parallel processing. For example, large-scale culture of a number of animals and embryos in precisely controllable microenvironments would be possible using the microfluidic platforms. Phenotypic changes of the model animals by a variety of environmental cues, such as biochemical and chemical perturbations, physical stresses, and temperature change, during development could be studied in a high-throughput manner. Those platforms could also be applied for screening drugs or genes based on the morphological and behavioral changes of the wild-type or mutant animal although initial drug screens using these small animals would have to be validated by high-model organisms, e.g. mammalian systems, to move closer to translational or clinical research. It is more likely that novel scientific questions could be answered using innovative tools, and new lines of research can inspire new tool development. For instance, microfluidic tools designed for embryonic studies of C. elegans or studies of D. melanogaster larvae or adults have not been tried or not very commonly in use. We expect that the continuous technological innovation in microfluidics for the small model organism and the research that can benefit from them will lead to a new era of biology.
Acknowledgments
HL acknowledges the US National Science Foundation and the US National Institutes of Health for support.
Abbreviations
- PDMS
polydimethylsiloxane
- DAergic
dopaminergic
- MPP+
1-methyl-4-phenylpyridinium
Footnotes
The authors declare no conflict of interest.
References
- 1.Tissenbaum HA, Guarente L. Model organisms as a guide to mammalian aging. Dev Cell. 2002;2:9–19. doi: 10.1016/s1534-5807(01)00098-3. [DOI] [PubMed] [Google Scholar]
- 2.Jenner RA, Wills MA. The choice of model organisms in evo-devo. Nat Rev Genet. 2007;8:311–314. doi: 10.1038/nrg2062. [DOI] [PubMed] [Google Scholar]
- 3.Narashimhan MG. Model organisms in biology: Scientific other uses. J Biosci. 1999;24:141–142. [Google Scholar]
- 4.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berghmans S, Butler P, Goldsmith P, Waldron G, et al. Zebrafish based assays for the assessment of cardiac, visual and gut function — potential safety screens for early drug discovery. J Pharmacol Toxicol Methods. 2008;58:59–68. doi: 10.1016/j.vascn.2008.05.130. [DOI] [PubMed] [Google Scholar]
- 6.Brittijn SA, Duivesteijn SJ, Belmamoune M, Bertens LF, et al. Zebrafish development and regeneration: New tools for biomedical research. Int J Dev Biol. 2009;53:835–850. doi: 10.1387/ijdb.082615sb. [DOI] [PubMed] [Google Scholar]
- 7.Meeker ND, Trede NS. Immunology and zebrafish: Spawning new models of human disease. Dev Comp Immunol. 2008;32:745–757. doi: 10.1016/j.dci.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 8.Kaletta T, Hengartner MO. Finding function in novel targets: C. elegans as a model organism. Nat Rev Drug Discov. 2006;5:387–399. doi: 10.1038/nrd2031. [DOI] [PubMed] [Google Scholar]
- 9.Leung MCK, Williams PL, Benedetto A, Au C, et al. Caenorhabditis elegans: An emerging model in biomedical and environmental toxicology. Toxicol Sci. 2008;106:5–28. doi: 10.1093/toxsci/kfn121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pandey UB, Nichols CD. Human disease models in Drosophila melanogaster and the role of the fly in therapeutic drug discovery. Phamacol Rev. 2011;63:411–436. doi: 10.1124/pr.110.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Kane CJ. Drosophila as a model organism for the study of neuropsychiatric disorders. In: Hagan JJ, editor. Molecular and Functional Models in Neuropsychiatry. Springer-Verlag; Berlin, Heidelberg: 2011. pp. 37–60. [DOI] [PubMed] [Google Scholar]
- 12.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematodes. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 13.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode. Philos Trans R Soc Lond Ser B. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 14.C. elegans Sequencing Consortium, Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 15.Hilliard MA, Apicella AJ, Kerr R, Suzuki H, et al. In vivo imaging of C. elegans ASH neurons: Cellular response and adaptation to chemical repellents. EMBO J. 2005;24:63–72. doi: 10.1038/sj.emboj.7600493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerr R, Lev-Ram V, Baird G, Vincent P, et al. Optical imaging of calcium transients in neurons and pharyngeal muscle of C. elegans. Neuron. 2000;26:583–594. doi: 10.1016/s0896-6273(00)81196-4. [DOI] [PubMed] [Google Scholar]
- 17.Avery L, Shtonda BB. Food transport in the C. elegans pharynx. J Exp Biol. 2003;206:2441–2457. doi: 10.1242/jeb.00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yanik MF, Cinar H, Cinar HN, Chisholm AD, et al. Neurosurgery: Functional regeneration after laser axotomy. Nature. 2004;432:822. doi: 10.1038/432822a. [DOI] [PubMed] [Google Scholar]
- 19.Chung SH, Clark DA, Gabel CV, Mazur E, et al. The role of the AFD neuron in C. elegans thermotaxis analyzed using femtosecond laser ablation. BMC Neurosci. 2006;7:30. doi: 10.1186/1471-2202-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hulme SE, Shevkoplyas SS, Apfeld J, Fontana W, et al. A microfabricated array of clamps for immobilizing and imaging C. elegans. Lab Chip. 2007;7:1515–1523. doi: 10.1039/b707861g. [DOI] [PubMed] [Google Scholar]
- 21.Allen PB, Sgro AE, Chao DL, Doepker BE, et al. Single-synapse ablation and long-term imaging in live C. elegans. J Neurosci Methods. 2008;173:20–26. doi: 10.1016/j.jneumeth.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hulme SE, Shevkoplyas SS, McGuigan AP, Apfeld J, et al. Lifespan-on-a-chip: Microfluidic chambers for performing lifelong observation of C. elegans. Lab Chip. 2010;10:589–597. doi: 10.1039/b919265d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weibel DB, Kruithof M, Potenta S, Sia SK, et al. Torque-actuated valves for microfluidics. Anal Chem. 2005;77:4726–4733. doi: 10.1021/ac048303p. [DOI] [PubMed] [Google Scholar]
- 24.Golden TR, Melov S. W. The C. elegans Research Community. WormBook; 2006. Gene expression changes associated with aging in C. elegans (January 2, 2007) http://www.wormbook.org. (Ed.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antebi A. Genetics of aging in Caenorhabditis elegans. PLoS Genet. 2007;3:e129. doi: 10.1371/journal.pgen.0030129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clausell-Tormos J, Lieber D, Baret JC, El-Harrak A, et al. Droplet-based microfluidic platforms for the encapsulation and screening of mammalian cells and multicellular organism. Chem Biol. 2008;15:427–437. doi: 10.1016/j.chembiol.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 27.Shi W, Qin J, Ye N, Lin B. Droplet-based microfluidic system for individual Caenorhabditis elegans assay. Lab Chip. 2008;8:1432–1435. doi: 10.1039/b808753a. [DOI] [PubMed] [Google Scholar]
- 28.Shi W, Wen H, Lu Y, Shi Y, et al. Droplet microfluidics for characterizing the neurotoxin-induced responses in individual Caenorhabditis elegans. Lab Chip. 2010;10:2855–2863. doi: 10.1039/c0lc00256a. [DOI] [PubMed] [Google Scholar]
- 29.Rohde CB, Zeng F, Gonzalez-Rubio R, Angel M, et al. Microfluidic system for on-chip high-throughput whole-animal sorting and screening at subcellular resolution. Proceed Nat Acad Sci. 2007;104:13891–13895. doi: 10.1073/pnas.0706513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo SX, Bourgeois F, Chokshi T, Durr NJ, et al. Femtosecond laser nanoaxotomy lab-on-achip for in vivo nerve regeneration studies. Nat Methods. 2008;5:531–533. doi: 10.1038/nmeth.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeng F, Rohde CB, Yanik MF. Sub-cellular precision on-chip small-animal immobilization, multi-photon imaging and femtosecond-laser manipulation. Lab Chip. 2008;8:653–656. doi: 10.1039/b804808h. [DOI] [PubMed] [Google Scholar]
- 32.Gilleland CL, Rohde CB, Zeng F, Yanik MF. Microfluidic immobilization of physiologically active Caenorhabditis elegans. Nat Protoc. 2010;5:1888–1902. doi: 10.1038/nprot.2010.143. [DOI] [PubMed] [Google Scholar]
- 33.Samara C, Rohde CB, Gilleland CL, Norton S, et al. Large-scale in vivo femtosecond laser neurosurgery screen reveals small-molecule enhancer of regeneration. Proceed Nat Acad Sci. 2010;107:18342–18347. doi: 10.1073/pnas.1005372107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma H, Jiang L, Shi W, Qin J, et al. A programmable microvalve-based microfluidic array for characterization of neurotoxin-induced responses of individual C. elegans. Biomicrofluidics. 2009;3:044114. doi: 10.1063/1.3274313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chokshi TV, Ben-Yakar A, Chronis N. CO2 and compressive immobilization of C. elegans on-chip. Lab Chip. 2009;9:151–157. doi: 10.1039/b807345g. [DOI] [PubMed] [Google Scholar]
- 36.Chung K, Crane MM, Lu H. Automated on-chip rapid microscopy, phenotyping and sorting of C. elegans. Nat Methods. 2008;5:637–643. doi: 10.1038/nmeth.1227. [DOI] [PubMed] [Google Scholar]
- 37.Rohde CB, Yanik MF. Subcellular in vivo time-lapse imaging and optical manipulation of Caenorhabditis elegans in standard multiwell plates. Nat Communications. 2011;2:1–7. doi: 10.1038/ncomms1266. [DOI] [PubMed] [Google Scholar]
- 38.Krajniak J, Lu H. Long-term high-resolution imaging and culture of C. elegans in chip-gel hybrid microfluidic device for developmental studies. Lab Chip. 2010;10:1862–1868. doi: 10.1039/c001986k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chokshi TV, Bazopoulou D, Chronis N. An automated microfluidic platform for calcium imaging of chemosensory neurons in Caenorhabditis elegans. Lab Chip. 2010;10:2758–2763. doi: 10.1039/c004658b. [DOI] [PubMed] [Google Scholar]
- 40.Cáceres I d C, Valmas N, Hilliard MA, Lu H. Laterally orienting C. elegans using geometry at microscale for high-throughput visual screens in neurodegeneration and neuronal development studies. PLoS ONE. 2012;7:e35037. doi: 10.1371/journal.pone.0035037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crane MM, Chung K, Lu H. Computer-enhanced high-throughput genetic screens of C. elegans in a microfluidic system. Lab Chip. 2009;9:38–40. doi: 10.1039/b813730g. [DOI] [PubMed] [Google Scholar]
- 42.Deisseroth K. Optogenetics. Nat Methods. 2011;8:26–29. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagel G, Szellas T, Huhn W, Kateriya S, et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proceed Nat Acad Sci. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kolbe M, Besir H, Essen LO, Oesterhelt D. Structure of the light-driven chloride pump halorhodopsin at 1.8 Å resolution. Science. 2000;288:1390–1396. doi: 10.1126/science.288.5470.1390. [DOI] [PubMed] [Google Scholar]
- 45.Zhang F, Wang LP, Brauner M, Liewald JF, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 46.Guo ZV, Hart AC, Ramanathan S. Optical interrogation of neural circuits in Caenorhabditis elegans. Nat Methods. 2009;6:891–896. doi: 10.1038/nmeth.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stirman JN, Crane MM, Husson SJ, Wabnig S, et al. Real-time multimodal optical control of neurons and muscles in freely behaving Caenorhabditis elegans. Nat Methods. 2011;8:153–158. doi: 10.1038/nmeth.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wyart C, Bene FD, Warp E, Scott EK, et al. Optogenetic dissection of a behavioural module in the vertebrate spinal cord. Nature. 2009;461:407–410. doi: 10.1038/nature08323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schoonheim PJ, Arrenberg AB, Bene FD, Baier H. Optogenetic localization and genetic perturbation of saccade-generating neurons in zebrafish. J Neurosci. 2010;30:7111–7120. doi: 10.1523/JNEUROSCI.5193-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leifer AM, Fang-Yen C, Gershow M, Alkema MJ, et al. Optogenetic manipulation of neural activity in freely moving Caenorhabditis elegans. Nat Methods. 2011;8:147–152. doi: 10.1038/nmeth.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stirman JN, Brauner M, Gottschalk A, Lu H. High-throughput study of synaptic transmission at the neuromuscular junction enabled by optogenetics and microfluidics. J Neurosci Methods. 2010;191:90–93. doi: 10.1016/j.jneumeth.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lockery SR, Lawton KJ, Doll JC, Faumont S, et al. Artificial dirt: microfluidic substrates for nematode neurobiology and behavior. J Neurophysiol. 2008;99:3136–3143. doi: 10.1152/jn.91327.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park S, Hwang H, Nam SW, Martinez F, et al. Enhanced Caenorhabditis elegans locomotion in a structured microfluidic environment. PLoS ONE. 2008;3:e2550. doi: 10.1371/journal.pone.0002550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee H, Choi M-k, Lee D, Kim H-s, et al. Nictation, a dispersal behavior of the nematode Caenorhabditis elegans, is regulated by IL2 neurons. Nat Neurosci. 2012;15:107–112. doi: 10.1038/nn.2975. [DOI] [PubMed] [Google Scholar]
- 55.Kim N, Dempsey CM, Zoval JV, Sze JY, et al. Automated microfluidic compact disc (CD) cultivation system of Caenorhabditis elegans. Sens Actuators B. 2007;122:511–518. [Google Scholar]
- 56.Kim N, Dempsey CM, Kuan CJ, Zovel JV, et al. Gravity force transduced by the MEC-4/MEC-10 DEG/ENaC channel modulates DAF-16/FoxO activity in Caenorhabditis elegans. Genetics. 2007;177:835–845. doi: 10.1534/genetics.107.076901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rezai P, Siddiqui A, Selvaganapathy PR, Gupta BP. Electrotaxis of Caenorhabditis elegans in a microfluidic environment. Lab Chip. 2010;10:220–226. doi: 10.1039/b917486a. [DOI] [PubMed] [Google Scholar]
- 58.Manière X, Lebois F, Matic I, Ladoux B, et al. Running Worms: C. elegans Self-Sorting by Electrotaxis. PLoS ONE. 2011;6:e16637. doi: 10.1371/journal.pone.0016637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rubin GM, Lewis EB. A brief history of Drosophila's contributions to genome research. Science. 2000;24:2216–2218. doi: 10.1126/science.287.5461.2216. [DOI] [PubMed] [Google Scholar]
- 60.Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 61.Driever W, Nüsslein-Volhard C. The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell. 1988;54:95–104. doi: 10.1016/0092-8674(88)90183-3. [DOI] [PubMed] [Google Scholar]
- 62.Frohnhöfer HG, Nüsslein-Volhard C. Maternal genes required for the anterior localization of bicoid activity in the embryo of Drosophila. Genes Dev. 1987;1:880–890. [Google Scholar]
- 63.Lucchetta EM, Lee JH, Fu LA, Patel NH, et al. Dynamics of Drosophila embryonic patterning network perturbed in space and time using microfluidics. Nature. 2005;434:1134–1138. doi: 10.1038/nature03509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kenis PJA, Ismagilov RF, Whitesides GM. Microfabrication inside capillaries using multiphase laminar flow patterning. Science. 1999;285:83–85. doi: 10.1126/science.285.5424.83. [DOI] [PubMed] [Google Scholar]
- 65.Takayama S, Ostuni E, LeDuc P, Naruse K, et al. Subcellular positioning of small molecules. Nature. 2001;411:1016. doi: 10.1038/35082637. [DOI] [PubMed] [Google Scholar]
- 66.Lucchetta EM, Munson MS, Ismagilov RF. Characterization of the local temperature in space and time around developing Drosophila embryo in a microfluidic device. Lab Chip. 2006;6:185–190. doi: 10.1039/b516119c. [DOI] [PubMed] [Google Scholar]
- 67.Dagani GT, Monzo K, Fakhoury JR, Chen CC, et al. Microfluidic self-assembly of live Drosophila embryos for versatile high-throughput analysis of embryonic morphogenesis. Biomed Microdevices. 2007;9:681–694. doi: 10.1007/s10544-007-9077-z. [DOI] [PubMed] [Google Scholar]
- 68.Bernstein RW, Zhang X, Zapp S, Fish M, et al. Characterization of fluidic microassembly for immobilization and positioning of Drosophila embryos in 2-D arrays. Sens Actuators A. 2004;114:191–196. [Google Scholar]
- 69.Chun K, Hashiguchi G, Toshiyoshi H, Fujita H. Fabrication of array of hollow microcapillaries used for injection of genetic materials into animal/plant cells. Jpn J Appl Phys. 1999;38:L279–L281. [Google Scholar]
- 70.Zhang X, Scott MP, Quate CF, Solgaard O. Microoptical characterization of piezoelectric vibratory microinjections in Drosophila embryos for genome-wide RNAi screen. J Microelectromech Syst. 2006;15:1–11. [Google Scholar]
- 71.Chung K, Kim Y, Kanodia JS, Gong E, et al. A microfluidic array for large-scale ordering and orientation of embryos. Nat Methods. 2011;8:171–176. doi: 10.1038/nmeth.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dahm R, Geisler R. Learning from small fry: The zebrafish as a genetic model organism for aquaculture fish species. Mar Biotechnol. 2006;8:329–345. doi: 10.1007/s10126-006-5139-0. [DOI] [PubMed] [Google Scholar]
- 73.Kinkel MD, Prince VE. On the diabetic menu: Zebrafish as a model for pancreas development and function. Bioessays. 2009;31:139–152. doi: 10.1002/bies.200800123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bretaud S, Allen C, Ingham PW, Bandmann O. p53-dependent neuronal cell death in a DJ-1-deficient zebrafishmodel of Parkinson's disease. J Neurochem. 2007;100:1626–1636. doi: 10.1111/j.1471-4159.2006.04291.x. [DOI] [PubMed] [Google Scholar]
- 75.Ramesh T, Lyon AN, Pineda RH, Wang C, et al. A genetic model of amyotrophic lateral sclerosis in zebrafish displays phenotypic hallmarks of motoneuron disease. Dis Model Mech. 2010;3:652–662. doi: 10.1242/dmm.005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lieschke GJ, Currie PD. Animal models of human disease: Zebrafish swim into view. Nat Rev Genet. 2007;8:353–367. doi: 10.1038/nrg2091. [DOI] [PubMed] [Google Scholar]
- 77.Penberthy WT, Shafizadeh E, Lin S. The zebrafish as a model for human disease. Front Biosci. 2002;7:1439–1453. doi: 10.2741/penber. [DOI] [PubMed] [Google Scholar]
- 78.Tamplin OJ, White RM, Jing L, Kaufman CK, et al. Small molecule screening in zebrafish: Swimming in potential drug therapies. WIREs Dev Biol. 2012;1:459–468. doi: 10.1002/wdev.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Funfak A, Brösing A, Brand M, Köhler JM. Micro fluid segment technique for screening and development studies on Danio rerio embryos. Lab Chip. 2007;7:1132–1138. doi: 10.1039/b701116d. [DOI] [PubMed] [Google Scholar]
- 80.Yang F, Chen Z, Pan J, Li X, et al. An integrated microfluidic array system for evaluating toxicity and teratogenicity of drugs on embryonic zebrafish developmental dynamics. Biomicrofluidics. 2011;5:024115. doi: 10.1063/1.3605509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu D, Wang L, Zhong R, Li B, et al. Parallel microfluidic networks for studying cellular response to chemical modulation. J Biotechnol. 2007;131:286–292. doi: 10.1016/j.jbiotec.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 82.Jeon NL, Dertinger SKW, Chiu DT, Choi IS, et al. Generation of solution and surface gradients using microfluidic systems. Langmuir. 2000;16:8311–8316. [Google Scholar]
- 83.Choudhury D, Noort D v, Iliescu C, Poon BZKL, et al. Fish and Chips: a microfluidic perfusion platform for monitoring zebrafish development. Lab Chip. 2012;12:892–900. doi: 10.1039/c1lc20351g. [DOI] [PubMed] [Google Scholar]
- 84.Shen Y-c, Li D, Al-Shoaibi A, Bersano-Begey T, et al. Zebrafish in education: A student team in a University of Michigan biomedical engineering design course constructs a microfluidic bioreactor for studies of zebrafish development. Zebrafish. 2009;6:201–213. doi: 10.1089/zeb.2008.0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wielhouwer EM, Ali S, Al-Afandi A, Blom MT, et al. Zebrafish embryo development in a microfluidic flow-through system. Lab Chip. 2011;11:1815–1824. doi: 10.1039/c0lc00443j. [DOI] [PubMed] [Google Scholar]
- 86.Lammer E, Kamp HG, Hisgen V, Koch M, et al. Development of a flow-through system for the fish embryo toxicity test (FET) with the zebrafish (Danio rerio) Tox in Vit. 2009;23:1436–1442. doi: 10.1016/j.tiv.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 87.Akagi J, Khoshmanesh K, Evans B, Hall CJ, et al. Miniaturized embryo array for automated trapping, immobilization and microperfusion of zebrafish embryos. PLoS ONE. 2012;7:e36630. doi: 10.1371/journal.pone.0036630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huang KS, Lin YC, Su KC, Chen HY. An electroporation microchip system for the transfection of zebrafish embryos using quantum dots and GFP genes for evaluation. Biomed Microdevices. 2007;9:761–768. doi: 10.1007/s10544-007-9087-x. [DOI] [PubMed] [Google Scholar]
- 89.Son SU, Garrell RL. Transport of live yeast and zebrafish embryo on a droplet (“digital”) microfluidic platform. Lab Chip. 2009;9:2398–2401. doi: 10.1039/b906257b. [DOI] [PubMed] [Google Scholar]
- 90.Noori A, Selvaganapathy PR, Wilson J. Microinjection in a microfluidic format using flexible and compliant channels and electroosmotic dosage control. Lab Chip. 2009;9:3202–3211. doi: 10.1039/b909961a. [DOI] [PubMed] [Google Scholar]
- 91.Ghazali IE, Saqrane S, Carvalho AP, Ouahid Y, et al. Compensatory growth induced in zebrafish larvae after pre-exposure to a Microcystis aeruginosa natural bloom extract containing microcystins. Int J Mol Sci. 2009;10:133–146. doi: 10.3390/ijms10010133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pardo-Martin C, Chang TY, Koo BK, Gilleland CL, et al. High-throughput in vivo vertebrate screening. Nat Methods. 2010;7:634–636. doi: 10.1038/nmeth.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chang TY, Pardo-Martin C, Allalou A, Wählby C, et al. Fully automated cellular-resolution vertebrate screening platform with parallel animal processing. Lab Chip. 2012;12 doi: 10.1039/c1lc20849g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huisken J, Swoger J, Bene FD, Wittbrodt J, et al. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305:1007–1009. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- 95.Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EHK. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science. 2008;322:1065–1069. doi: 10.1126/science.1162493. [DOI] [PubMed] [Google Scholar]
- 96.Keller PJ, Schmidt AD, Santella A, Khairy K, et al. Fast, high-contrast imaging of animal development with scanned light sheet-based structured-illumination microscopy. Nat Methods. 2010;7:637–642. doi: 10.1038/nmeth.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.O’Brien GS, Rieger S, Martin SM, Cavanaugh AM, et al. Two-photon axotomy and time-lapse confocal imaging in live zebrafish embryos. J Vis Exp. 2009;24:e1129. doi: 10.3791/1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li J, Mack JA, Souren M, Yaksi E, et al. Early development of functional spatial maps in the zebrafish olfactory bulb. J Neurocsci. 2005;25:5784–5795. doi: 10.1523/JNEUROSCI.0922-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mohler WA. Visual reality: Using computer reconstruction and animation to magnify the microscopist’s perception. Mol Biol Cell. 1999;10:3061–3065. doi: 10.1091/mbc.10.10.3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhao T, Xie J, Amat F, Clack N, et al. Automated reconstruction of neuronal morphology based on local geometrical and global structural models. Neuroinform. 2011;9:247–261. doi: 10.1007/s12021-011-9120-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Long F, Peng H, Liu X, Kim SK, et al. A 3D digital atlas of C. elegans and its application to single-cell analyses. Nat Methods. 2009;6:667–672. doi: 10.1038/nmeth.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bao Z, Murray JI, Boyle T, Ooi SL, et al. Automated cell lineage tracing in Caenorhabditis elegans. Proceed Nat Acad Sci. 2006;103:2707–2712. doi: 10.1073/pnas.0511111103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Murray JI, Bao Z, Boyle TJ, Boeck ME, et al. Automated analysis of embryonic gene expression with cellular resolution in C. elegans. Nat Methods. 2008;5:703–709. doi: 10.1038/nmeth.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nahabedian JF, Qadota H, Stirman JN, Lu H, et al. Bending amplitude – A new quantitative assay of C. elegans locomotion: Identification of phenotypes for mutants in genes encoding muscle focal adhesion components. Methods. 2012;56:95–102. doi: 10.1016/j.ymeth.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Swierczek NA, Giles AC, Rankin CH, Kerr RA. High-throughput behavioral analysis in C. elegans. Nat Methods. 2011;8:592–598. doi: 10.1038/nmeth.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mo W, Chen F, Nechiporuk A, Nicolson T. Quantification of vestibular-induced eye movements in zebrafish larvae. BMC Neurosci. 2010;11:110. doi: 10.1186/1471-2202-11-110. [DOI] [PMC free article] [PubMed] [Google Scholar]


