Abstract
Objective
Genome-wide association studies (GWAS) have identified a large number of single nucleotide polymorphisms (SNPs) associated with a wide array of cancer sites. Several of these variants demonstrate associations with multiple cancers, suggesting pleiotropic effects and shared biological mechanisms across some cancers. We hypothesized that SNPs previously associated with other cancers may additionally be associated with colorectal cancer. In a large-scale study, we examined 171 SNPs previously associated with 18 different cancers for their associations with colorectal cancer.
Design
We examined 13,338 colorectal cancer cases and 40,967 controls from three consortia: Population Architecture using Genetics and Epidemiology (PAGE), Genetic Epidemiology of Colorectal Cancer (GECCO), and the Colon Cancer Family Registry (CCFR). Study-specific logistic regression results, adjusted for age, sex, principal components of genetic ancestry, and/or study specific factors (as relevant) were combined using fixed-effect meta-analyses to evaluate the association between each SNP and colorectal cancer risk. A Bonferroni-corrected p-value of 2.92×10−4 was used to determine statistical significance of the associations.
Results
Two correlated SNPs— rs10090154 and rs4242382—in Region 1 of chromosome 8q24, a prostate cancer susceptibility region, demonstrated statistically significant associations with colorectal cancer risk. The most significant association was observed with rs4242382 (meta-analysis OR=1.12; 95% CI: 1.07–1.18; P=1.74×10−5), which also demonstrated similar associations across racial/ethnic populations and anatomical sub-sites.
Conclusion
This is the first study to clearly demonstrate Region 1 of chromosome 8q24 as a susceptibility locus for colorectal cancer, thus adding colorectal cancer to the list of cancer sites linked to this particular multi-cancer risk region at 8q24.
Keywords: colorectal cancer, pleiotropy, genome-wide association study, single nucleotide polymorphism
INTRODUCTION
Since the first series of genome-wide association studies (GWAS) for cancer were published in 2007, several hundred common genetic variants have been associated with a wide array of cancer sites [1]. As GWAS continue to identify variants associated with cancer, patterns of pleiotropic associations have emerged that highlight key loci and shared pathways that affect multiple cancer sites. For instance, genetic variants at chromosome 8q24 have been associated with cancers of the prostate, colorectum, breast, bladder, and other sites [2, 3, 4, 5, 6, 7]. Similarly, genetic variants in and near the telomerase reverse transcriptase (TERT) gene, which encodes for telomerase activity, have been associated with glioma, lung, prostate, colorectal, and other cancers [5, 8, 9, 10, 11], emphasizing the importance of cellular aging in cancer development.
Pleiotropy occurs when a genetic locus is associated with multiple phenotypic traits. Accordingly, any genetic difference at a pleiotropic locus may have wide-ranging effects across different cell types. Evidence of pleiotropic associations can improve our understanding of disease etiology by identifying shared molecular components underlying disease risk and by validating the pathogenicity of variants at a locus [12]. To illustrate, a recent study of the genetic overlap between systematic lupus erythematosus and other autoimmune diseases found novel pleiotropic associations that support a role for T cell and innate immune response pathways, providing valuable evidence for dissecting the biological mechanisms that underlie their shared etiologies [13].
Previous analyses of shared genetic variants across cancers have focused primarily on hereditary disorders such as the Lynch and Li-Fraumeni syndromes. Although multiple cancer types are known to cluster within families [14], studies of shared genetic factors across various non-familial cancers have been limited. Given the numerous associations reported by GWAS of cancer, we now have an opportunity to assess pleiotropy across different cancers. These pleiotropic associations may have been missed in prior GWAS of colorectal cancer (CRC) due to smaller sample sizes and the stringent threshold of significance of testing hundreds of thousands to millions of SNPs in GWAS. For this study, we tested GWAS-identified risk variants of 18 other cancers for pleiotropic associations with CRC risk in a large-scale collaboration, including multiple racial/ethnic groups. Specifically, we conducted a meta-analysis study of 13,338 CRC cases and 40,967 controls from 16 studies of three consortia: Population Architecture using Genetics and Epidemiology (PAGE); Genetics and Epidemiology of Colorectal Cancer Consortium (GECCO); and the Colon Cancer Family Registry (CCFR).
METHODS
Study participants
Three consortia contributed data to this meta-analysis study: PAGE [15]; GECCO [11, 16]; and CCFR [17]. This collaboration comprised 13,338 CRC cases and 40,967 controls from 16 studies (Supplemental Table 1). Briefly, PAGE studies included: Atherosclerosis Risk in Communities (ARIC) [18], which is part of Causal Variants Across the Life Course (CALiCo); Epidemiologic Architecture for Genes Linked to Environment (EAGLE), which accesses the Vanderbilt University biorepository (BioVU) [19]; Multiethnic Cohort (MEC) [20]; and the Women’s Health Initiative (WHI). GECCO studies included: the french Association STudy Evaluating RISK for sporadic colorectal cancer (ASTERISK) [21]; Hawaii Colorectal Cancer Studies 2 & 3 (Colo2&3) [22]; Darmkrebs: Chancen der Verhütung durch Screening (DACHS) [22]; Diet, Activity, and Lifestyle Study (DALS) [23]; Health Professionals Follow-up Study (HPFS) [24]; Nurses’ Health Study (NHS); Ontario Familial Colorectal Cancer Registry (OFCCR) [25] [26]; Physicians’ Health Study (PHS) [27]; Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO) [28, 29]; Post-Menopausal Hormones Supplemental Study to the Colon Cancer Family Registry (PMH-CCFR) [30]; VITamins And Lifestyle (VITAL) study [31]; and the Women’s Health Initiative (WHI) [32, 33]. While WHI participates in both PAGE and GECCO, only WHI data as a part of GECCO was used. CCFR [17] included a population-based case-control subset.
Demographic, genetic and epidemiologic information was obtained by each study according to its enrollment, genotyping, and assessment protocols. Case and control definitions, as well as factors used in matching, differed by study (Supplemental Material, Supplemental Table 2). The majority of studies utilized incident CRC cases; controls had no diagnosis of CRC. Six GECCO studies (DACHS, DALS, HPFS, NHS, PLCO, and WHI) contained study-specific subsets that were genotyped and analyzed individually due to differences in sample collection, year of ascertainment, or controls used for each subset (Supplemental Material; Supplemental Table 2). This led to a total of 22 analytic subsets from the 16 studies. Supplemental Figure 1 shows the participating studies and overall study design. Institutional review board approval was obtained for all studies.
SNP selection and genotyping
A total of 171 SNPs previously associated with 18 cancers other than CRC were selected by PAGE researchers (Supplemental Table 3). These SNPs were identified to be associated with cancer, as of January 2010, from the NHGRI GWAS catalog (http://www.genome.gov/26525384) [1] as well as review of the cancer GWAS and fine-mapping literature [15]. References for each selected SNP are provided in Supplemental Table 3. The risk allele for each SNP was defined as the allele associated with an increased risk of cancer in prior publications. For SNPs associated with multiple cancer sites, the first reported GWAS was used in assigning the risk allele. These SNPs were genotyped using a custom panel in each PAGE study with the exception of ARIC. In ARIC, GECCO, and CCFR, genotype data were abstracted from previously generated GWAS data.
To control for potential bias due to population stratification (i.e. confounding due to racial/ethnic differences in allele frequencies and disease risk), 128 ancestry informative markers that capture the major continental genetic diversity [34] were genotyped in all PAGE studies with the exception of ARIC. Principal components were estimated from these markers by EIGENSTRAT [35] and included in regression models, providing objective quantitative estimates of genetic ancestry in comparison to self-reported race/ethnicity. In ARIC, CCFR, and GECCO, principal components of ancestry were derived from the GWAS dataset of each study using EIGENSTRAT [35].
In addition to directly genotyping, imputation for some of the 171 cancer risk variants was conducted in studies having GWAS data (ARIC study in PAGE and each study in GECCO) to estimate genotypes for untyped SNPs based on shared haplotypes and correlation with genotyped SNPs. Standard quality-assurance and quality-control measures were utilized to ensure genotyping quality. Further details are provided in the Supplementary Material. The majority of the 171 SNPs of interest were available across studies (97% SNPs were genotyped or imputed in all 22 analytic study sets; Supplemental Table 3).
Statistical analyses
For each study the association between each SNP and CRC was estimated using unconditional logistic regression. SNPs were coded additively with 0, 1, 2 referring to the number of risk alleles (or the allele dosage for imputed SNPs). Primary models were adjusted for age, sex, and the most relevant principal components of genetic ancestry to account for relevant population substructure for each study. A few studies were additionally adjusted for study center (CCFR, DALS, PLCO, and DACHS), study component (WHI), smoking (PHS), or batch effects (ASTERISK). To examine patterns of associations across race/ethnicity, each study with at least 80 CRC cases per race/ethnicity conducted analyses stratified by racial/ethnic population. Polytomous unconditional logistic regression was also performed in each study to examine associations across anatomical sub-site (colon versus rectum). This method allowed us to simultaneously examine the associations for colon and rectal cancer in a single regression model, providing an efficient approach and the ability to test for heterogeneity in effects by anatomical sub-site.
To examine whether the top associations found for the prostate cancer risk variants at Region 1 of chromosome 8q24 were independent from Region 3, an established colorectal risk region at 8q24, rs6983267 (a Region 3 CRC risk variant; meta-analysis OR=1.14; P=5×10−14) was included in the regression model with each Region 1 prostate cancer risk variant.
Log odds regression estimates were combined across studies using inverse-variance weighted, fixed-effect meta-analysis in METAL [36] for overall and stratified analyses. Heterogeneity P-values were estimated based on Cochran’s Q statistic. SNP associations demonstrating heterogeneity in associations across studies at P<0.05 were additionally examined using random-effects meta-analysis (Supplemental Table 4). A Bonferroni-corrected P=2.92×10−4 (nominal alpha/number of SNPs tested=0.05 /171) was used to determine the statistical significance of the overall association for each SNP with CRC.
RESULTS
The main characteristics of the 54,305 subjects (13,338 cases; 40,967 controls) are presented in Supplemental Table 1. The PAGE studies consisted of six different racial/ethnic populations, whereas the GECCO and CCFR consisted of European ancestry populations. In sum, the majority of the subjects were of European ancestry (80.6%) with the remainder comprising 7.0% African American, 4.5% Hispanic, 6.4% Asian, and 1.4% Pacific Islander or Native American ancestry. Most studies ascertained men and women (51.1% women overall), with the exception of WHI and NHS (women only) and HPFS and PHS (men only). Age varied across studies: ARIC ascertained younger adults (mean age of cases=55.8, controls=54.0) whereas the MEC ascertained older adults (mean age of cases=70.0, controls=68.4). Disease stage and anatomical sub-site also varied across studies: EAGLE-BioVU, a clinic-based collection of patients, had the largest proportions of advanced stage disease (59.2%) and rectal tumors (42%).
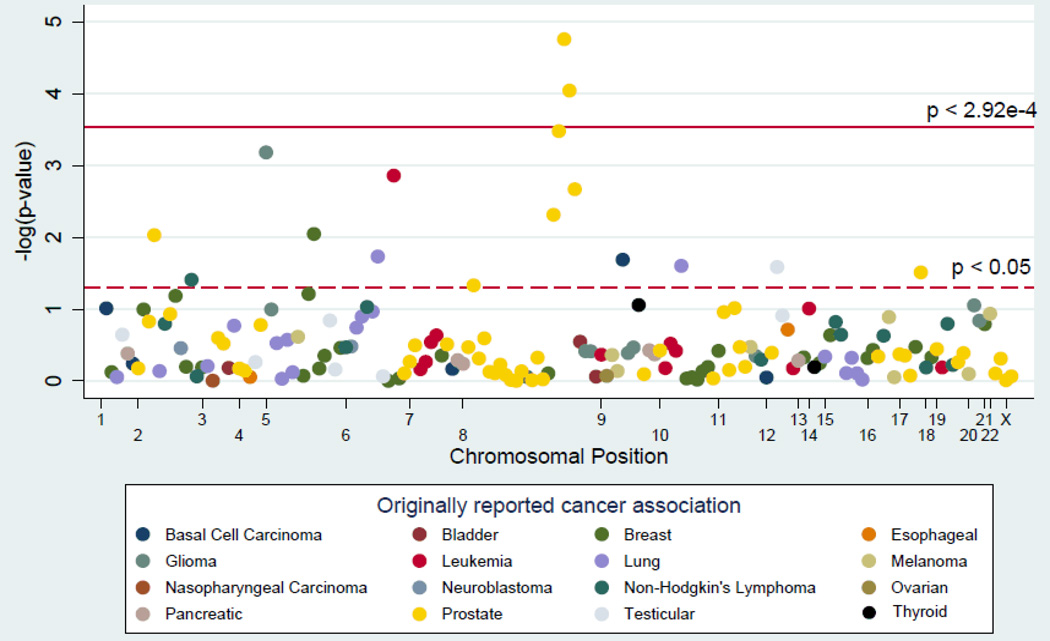
A total of 171 risk variants for 18 cancers other than CRC, representing 100 unique chromosomal regions, were tested in 13,338 cases and 40,967 controls from 16 studies across three consortia. Of the 171 risk variants, 16 variants were nominally associated with CRC at P<0.05 (Supplemental Table 3, Figure 1), which was more than the ∼9 associations expected by chance (171 SNPs x 0.05=8.55). These 16 risk variants consisted of 1 basal cell carcinoma SNP, 1 breast cancer SNP, 1 glioma SNP, 1 leukemia SNP, 2 lung cancer SNPs, 1 Non-Hodgkin’s Lymphoma SNP, 8 prostate cancer SNPs, and 1 testicular cancer SNP (Figure 1, Supplemental Table 3). Four of these 16 variants are correlated (8q24 Region 1 variants; r2>0.88 in HapMap CEU[37]) and may not represent independent results.
Figure 1. Manhattan plot of the meta-analysis association between risk variants of 18 other cancers and colorectal cancer.
The solid line is the Bonferroni-corrected significance threshold. Each association is colored according to the cancer for which the SNP was originally reported, and positioned on the x-axis according to its genomic position.
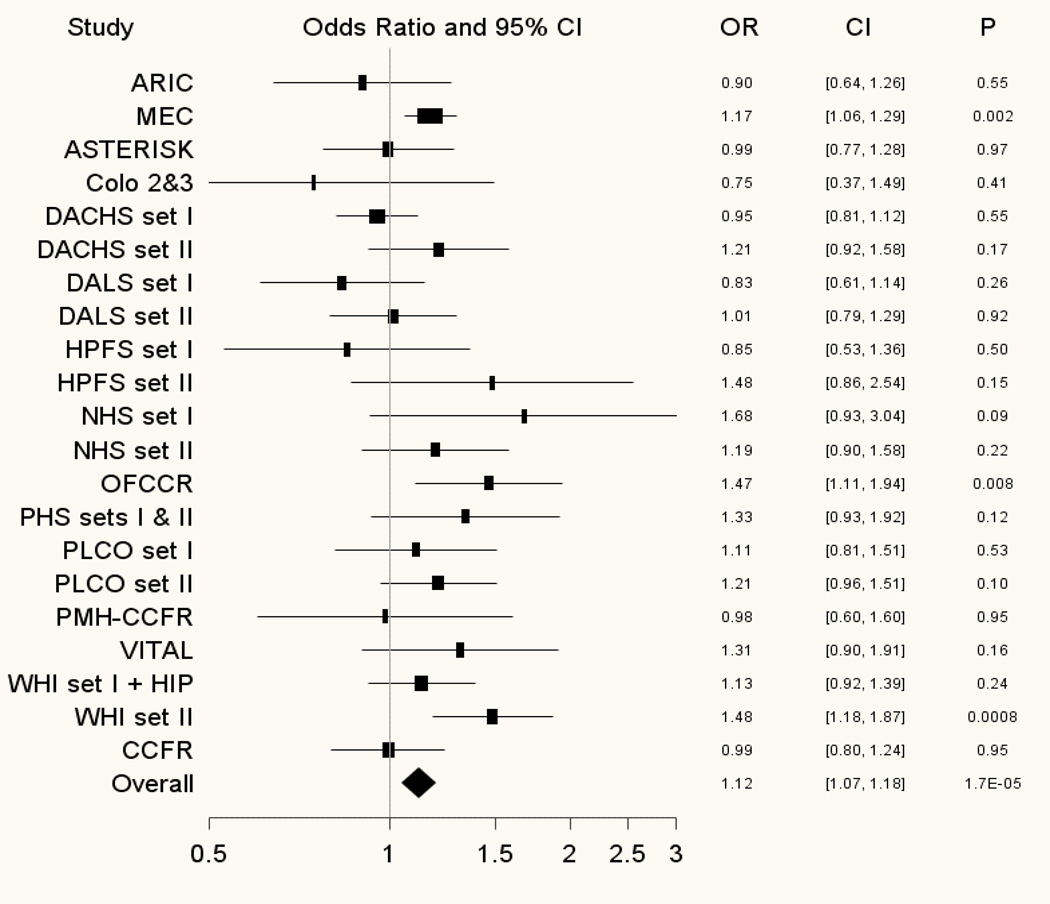
Two correlated prostate cancer risk variants (rs10090154 and rs4242382; r2=0.79 in CEU) in Region 1 of chromosome 8q24 (125.6–129.4 Mb [38]) demonstrated statistically significant associations with CRC, reaching a conservative Bonferroni-corrected criterion of significance (P< 2.92×10−4). For the most statistically significant association, rs4242382, we observed a 12%increased risk of CRC among CRC cases in comparison to controls (overall meta-analysis odds ratio (OR)=1.12, 95% Confidence Interval (CI)=1.07–1.18; P=1.74×10−5;Figure 2) and no evidence of heterogeneity across studies (Phet=0.07). Notably, the associations with rs10090154 and rs4242382 remained statistically significant when adjusting for rs6983267, a CRC risk variant in Region 3 of 8q24 (Region 3 adjusted meta-analysis ORrs10090154=1.11; P=5.0×10−5 and ORrs4242382=1.11; P=5.7×10−5). Two additional prostate cancer risk variants in Region 1 of 8q24 (rs7837688, rs1447295) and one in Region 3 (rs7000448) were also associated with CRC (P=3.32×10−4 – 4.85×10−3) though they did not reach our conservative threshold of statistical significance. These five prostate cancer SNPs demonstrated similar positive associations with CRC for the corresponding prostate cancer risk alleles. These SNPs are located upstream of MYC at chromosome 8q24, spanning ∼98 kb, and are in various amounts of linkage disequilibrium among HapMap Europeans. The Region 1 variants appear correlated with each other (r2>0.88) but not with the Region 3 variant (r2≤0.02; HapMap release 22 CEU).
Figure 2. Forest plot of the association between rs4242382 at Region 1 of chromosome 8q24 and colorectal cancer risk.
Study specific and meta-analysis associations are plotted, modeling the A risk allele for prostate cancer.
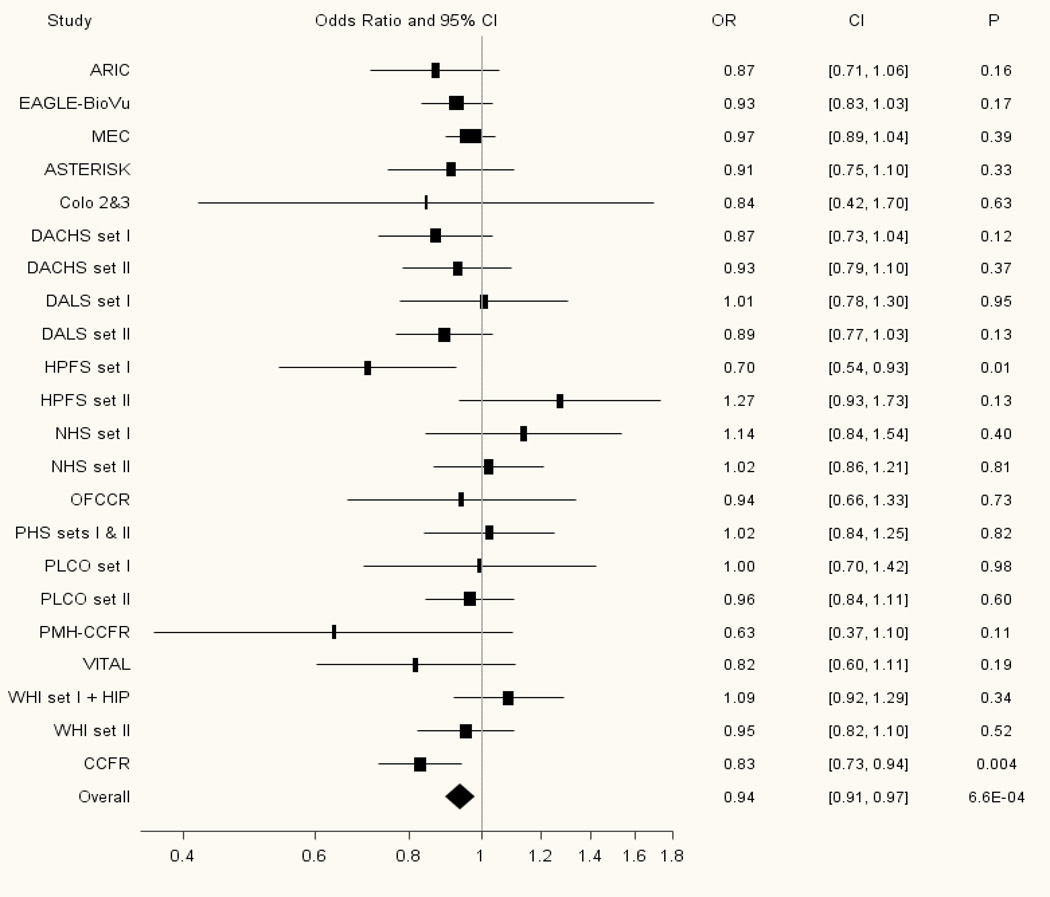
Outside of chromosome 8q24, we observed a marginally significant association with rs2736100, a glioma risk variant at the TERT locus at 5p15, and CRC (meta-analysis for the G allele OR=0.94; 95% CI: 0.91–0.97; P=6.57×10−4; Phet studies=0.31; Supplemental Table 3). This inverse association with CRC was in the opposite direction to that observed with the glioma G risk allele of this SNP (Figure 3). Another potentially interesting inverse association was observed with the A risk allele of rs981782, a breast cancer variant at the HCN1 locus at 5p12 (meta-analysis OR=0.96; 95% CI: 0.93–0.99; P=0.009; Phet studies=0.79; Supplemental Table 3).
Figure 3. Forest plot of the association between rs2736100 at the TERT locus at 5p15 and colorectal cancer risk.
Study specific and meta-analysis associations are plotted, modeling the G risk allele for glioma.
Next, we evaluated the 16 associations at P<0.05 for patterns of associations across race/ethnicity and anatomical sub-site (Supplemental Tables 5 and 6). We observed no evidence of heterogeneity in associations by race/ethnicity, with the exception of a potentially nominal association with rs7837688 (Phet=0.049). For the most statistically significant overall association, rs4242382, we observed consistent positive associations at P<0.05 for African American (OR=1.22; 95%CI: 1.03–1.45; P=0.024), Asian (OR=1.28; 95% CI: 1.09–1.51; P=3.06×10−3), and European ancestry populations (OR=1.10; 95% CI: 1.04–1.17; P=1.91×10−3). In addition, we observed generally similar directions of association in colon and rectal tumors (Supplemental Table 6). Nominal evidence of heterogeneity in associations by anatomical sub-site was observed for rs11155133 at chromosome 6q24 (Phet=0.03), where a stronger inverse association was observed for rectal cancer (meta-analysis OR=0.60; P=0.001) than colon cancer (meta-analysis OR=0.87; P=0.059).
DISCUSSION
In this large meta-analysis of 54,305 CRC cases and controls, we examined GWAS-identified risk variants of other cancers for their effects on CRC risk. To our knowledge, this is the first systematic analysis of pleiotropic associations of risk variants for other cancers with CRC. We identified two correlated SNPs—rs10090154 and rs4242382—at Region 1 of chromosome 8q24, a well-established prostate cancer susceptibility locus that demonstrated robust associations with CRC and reached a conservative criterion of statistical significance. We also observed a notable association at TERT, a key susceptibility locus for several cancers.
Chromosome 8q24 has been identified as an important risk locus for multiple cancers [2, 3, 4, 5, 6, 39, 40, 41, 42, 43], including CRC [44, 45, 46, 47, 48]. Distinct regions within this locus defined by their linkage disequilibrium structure have been associated with various cancers. SNPs within Region 3, initially identified as a 60kb region from 128.48 to 128.54 Mb at 8q24 [38], have been consistently associated with CRC in GWAS [44, 45, 46, 47, 48] and subsequent follow-up studies [11] [49, 50, 51, 52, 53]. Although associations between Region 3 of chromosome 8q24 and CRC risk are well-established, our findings appear to be the first demonstration of highly significant associations with Region 1. Prior candidate gene studies [49, 52, 54, 55, 56], all of smaller size, have not shown a statistically significant association between Region 1 and CRC, perhaps due to their limited statistical power as well as for prior GWAS of CRC [45, 46, 47, 48, 57, 58, 59, 60] and their stringent thresholds for genome-wide significance. Substantially large sample sizes are needed to have sufficient power to identify these small genetic associations, as seen here with the Region 1 variant rs4242382. While our study observed a modest increase in CRC risk (OR = 1.12) in 54,305 CRC cases and controls, the original finding for this SNP and prostate cancer observed a larger increase in risk (OR = 1.66) in 10,234 prostate cancer cases and controls [6]. By comparison, the largest pooled GWAS of CRC published to date included 27,809 CRC cases and controls [61]. Importantly, we were able to demonstrate that our most statistically significant associations at Region 1 of chromosome 8q24 were independent of the established Region 3 CRC risk variant, while maintaining a conservative threshold of statistical significance (P<5.7×10−5). Although not residing within a known gene, recent functional work indicates that these 8q24 regions contain long range tissue-specific enhancers that physically interact with the MYC oncogene [62], potentially influencing tumorigenesis. Furthermore, a recent study found that mice deficient in Myc-355, a putative regulatory element that contains the Region 3 rs6983267 variant, were resistant to induced intestinal tumors [63].
TERT, which encodes for telomerase reverse transcriptase, has been identified by GWAS as a susceptibility gene for several cancers [4, 5, 8, 10, 64, 65, 66, 67]. For example, the G allele of rs2736100, located in intron 2 of TERT, has been associated with an increased risk of lung adenocarcinoma and glioma and a decreased risk of testicular cancer in prior GWAS [5, 8, 9, 66]. These different directions of association across cancer sites may be due to context specific differences in regulation of nearby genes, just as transcription factors can serve as both oncogenes and tumor suppressors [68]. Our findings of an association between rs2736100 and CRC corroborates a recent study by Kinnersley et al. that reported a 7% increased risk of CRC with the T allele (P=2.49×10−5), using genotype data from six CRC cancer GWAS and an additional replication series [69]. As genotype data from the CCFR were used in both our study and this report [69], we further examined the association between rs2736100 and CRC without the CCFR: a similar nominally significant positive association was observed (meta-analysis OR for the T allele=1.05; 95% CI: 1.01–1.09; P=0.007). This provides further data for the involvement of TERT in CRC susceptibility. Additionally, an overall meta-analysis between our findings and those of Kinnersley et al. resulted in a more significant association between rs2736100 and CRC (meta-analysis OR for the T allele=1.06; 95% CI: 1.04–1.09; P=7.99×10−7).
The numerous risk loci identified by GWAS of cancer provide a valuable opportunity to assess similarities in the genetic susceptibility of different malignancies. Pleiotropic associations can underscore established etiologic links as well as uncover novel connections that provide new clues to shared molecular pathways [12]. Although cancer is a complex and heterogeneous disease with more than 200 different types, our findings identify shared genetic susceptibility variants between CRC and other cancers of the prostate, lung, breast, testis, and glioma. While the magnitudes of these associations are small, the cumulative effect of many such CRC risk variants may help explain the heritability of CRC [70]. Furthermore, these pleiotropic associations may indicate the biological importance of such shared genetic regions and suggest they should be prioritized for future functional and fine-mapping efforts. Specifically, our findings provide additional evidence for Region 1 of chromosome 8q24 and TERT as two such priority regions.
Our study is strengthened by the large number of subjects from well-designed CRC studies and the inclusion of multiple racial/ethnic populations. Limitations of this study include reduced study power for 6 SNPs that were not available across all studies. In addition, the smaller number of non-European ancestry participants limits our ability to fully explore generalizability across race/ethnicity. Finally, as more recent GWAS have identified several hundred new cancer risk loci, these variants remain to be evaluated for their pleiotropic effects with CRC.
In summary, our study indicates that several risk variants identified for other cancers also contribute to CRC risk. For the first time, these findings clearly demonstrate the importance of Region 1 at chromosome 8q24 in CRC susceptibility, and further bolster the evidence of this region as a multi-cancer risk locus. Further replication and future research into the biological mechanisms by which inherited differences in shared cancer risk loci influence CRC will expand our understanding of the key contributors to CRC development.
Supplementary Material
SUMMARY BOX.
What is known on this topic?
Several hundred common genetic variants have been associated with a wide array of cancer types.
Only a small proportion of the heritability of colorectal cancer can be explained by the currently identified risk loci from genome-wide association studies of colorectal cancer.
Identifying shared genetic associations between diseases (i.e. pleiotropy) is a useful approach to identify new risk loci and may elucidate common etiologies and help in risk prediction.
What this study adds?
This study clearly shows that two genetic variants in Region 1 of the 8q24 locus, a prostate cancer risk region, are also associated with colorectal cancer risk.
Furthermore, this study provides additional evidence that the TERT locus is associated with colorectal cancer.
How might it impact on clinical practice in the foreseeable future?
Colorectal risk variants may be used as part of a risk prediction model to define high-risk populations for targeted screening regimens and, possibly, inform clinical decision making.
ACKNOWLEDGEMENTS
The studies would like to thank all participants, staff, physicians, and investigators for making this project possible. We thank Dr. Bruno Buecher of ASTERISK; Ute Handte-Daub, Muhabbet Celik and Ursula Eilber of DACHS; Patrice Soule, Hardeep Ranu, Immaculata Devivo, David Hunter, Qin (Carolyn) Guo, Lixue Zhu, and Haiyan Zhang of HPFS, NHS and PHS; Christine Berg and Philip Prorok of PLCO; Tom Riley and staff of Information Management Services Inc.; Barbara O’Brien and staff of Westat Inc; Bill Kopp, Wen Shao and staff of SAIC-Frederick; WHI investigators ((see https://cleo.whi.org/researchers/SitePages/Write%20a%20Paper.aspx) and the GECCO Coordinating Center.
We would like to thank Awapuhi Lee and Kristine Winters for their technical assistance.
We would additionally like to thank the participants and staff of the Nurses’ Health Study and the Health Professionals Follow-up Study for their valuable contributions, as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY.
FUNDING
PAGE
The Population Architecture Using Genomics and Epidemiology (PAGE) program is funded by the National Human Genome Research Institute (NHGRI), supported by U01HG004803 (CALiCo), U01HG004798 (EAGLE), U01HG004802 (MEC), U01HG004790 (WHI), and U01HG004801 (Coordinating Center), and their respective NHGRI ARRA supplements. The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. The complete list of PAGE members can be found at http://www.pagestudy.org.
-
The data and materials included in this report result from collaboration between the following studies:
The "Epidemiologic Architecture for Genes Linked to Environment (EAGLE)" is funded through the NHGRI PAGE program (U01HG004798-01 and its NHGRI ARRA supplement). The dataset(s) used for the analyses described were obtained from Vanderbilt University Medical Center’s BioVU which is supported by institutional funding and by the Vanderbilt CTSA grant UL1 TR000445 from NCATS/NIH. The Vanderbilt University Center for Human Genetics Research, Computational Genomics Core provided computational and/or analytical support for this work.
The Multiethnic Cohort study (MEC) characterization of epidemiological architecture is funded through the NHGRI PAGE program (U01HG004802 and its NHGRI ARRA supplement). The MEC study is funded through the National Cancer Institute (R37CA54281, R01 CA63464, P01CA33619, U01CA136792, and U01CA98758).
Funding support for the “Epidemiology of putative genetic variants: The Women’s Health Initiative” study is provided through the NHGRI PAGE program (U01HG004790 and its NHGRI ARRA supplement). The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, and HHSN271201100004C. The authors thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. A full listing of WHI investigators can be found at: http://www.whiscience.org/publications/WHI_investigators_shortlist.pdf.
Funding support for the Genetic Epidemiology of Causal Variants Across the Life Course (CALiCo) program was provided through the NHGRI PAGE program (U01HG004803 and its NHGRI ARRA supplement). The following study contributed to this manuscript and is funded by the following agencies: The Atherosclerosis Risk in Communities (ARIC) Study is carried out as a collaborative study supported by supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research.
Assistance with phenotype harmonization, SNP selection and annotation, data cleaning, data management, integration and dissemination, and general study coordination was provided by the PAGE Coordinating Center (U01HG004801-01 and its NHGRI ARRA supplement). The National Institutes of Mental Health also contributes to the support for the Coordinating Center.
The PAGE consortium thanks the staff and participants of all PAGE studies for their important contributions.
GECCO and CCFR
NIH Funding
GECCO: U01 CA137088, R01 CA059045; DALS: R01 CA48998; Colo2&3: R01 CA60987; HPFS: U19 CA 055075, R01 137178, P50 CA 127003, UM1 CA167552; NHS: R01 137178, P50 CA 127003, and P01 CA 087969; OFCCR: U01 CA074783; PMH: R01 CA076366; PHS: CA42182; VITAL: K05 CA154337; WHI: HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, HHSN271201100004C and; PLCO: supported by Intramural Research Program of the DCEG and supported by contracts from the Division of Cancer Prevention, NIH. PLCO control samples were genotyped as part of the CGEMS prostate cancer scan, supported by the Intramural Research Program of the NCI, accessed through dbGaP accession number 000207v.1p1.c1 [43]. Control samples were also genotyped as part of the GWAS of Lung Cancer and Smoking [66] (Z01 CP 010200). Assistance with genotype cleaning, as well as with general study coordination, was provided by the Gene Environment Association Studies, GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438). The datasets used for the analyses described in this manuscript were obtained from dbGaP through accession number phs000093.
CCFR RFA # CA-95-011 and cooperative agreements with members of the CCFR. The genome-wide scans: U01 CA122839. Australasian Colorectal Cancer Family Registry: U01 CA097735; Familial Colorectal Neoplasia Collaborative Group: U01 CA074799; Mayo Clinic Cooperative Family Registry for Colon Cancer Studies: U01 CA074800; Ontario Registry for Studies of Familial Colorectal Cancer: U01 CA074783; Seattle Colorectal Cancer Family Registry: U01 CA074794; University of Hawaii Colorectal Cancer Family Registry: U01 CA074806
Contributions to this work by author Kocarnik were supported by grant R25CA94880 from NCI.
We would like to acknowledge the Colorectal Cancer Transdisciplinary (CORECT) Study, U19-CA148107 on behalf of the Genetic Associations and Mechanisms in Oncology (GAME-ON) Network
Non-NIH Funding
OFCCR: A GL2 grant from the Ontario Research Fund, the Canadian Institutes of Health Research, and the Cancer Risk Evaluation (CaRE) Program grant from the Canadian Cancer Society Research Institute. TJH and BWZ received Senior Investigator Awards from the Ontario Institute for Cancer Research, through generous support from the Ontario Ministry of Economic Development and Innovation.
DACHS: Deutsche Forschungsgemeinschaft (BR 1704/6-1, BR 1704/6-3, BR 1704/6-4 and CH 117/1-1), the German Federal Ministry of Education and Research (01KH0404 and 01ER0814).
ASTERISK: Funded by a Hospital Clinical Research Program (PHRC) and supported by the Regional Council of Pays de la Loire, the Groupement des Entreprises Françaises dans la Lutte contre le Cancer (GEFLUC), the Association Anne de Bretagne Génétique and the Ligue Régionale Contre le Cancer (LRCC).
Abbreviations
- GWAS
genome-wide association study
- PAGE
Population Architecture using Genetics and Epidemiology
- GECCO
Genetic Epidemiology of Colorectal Cancer
- CCFR
Colon Cancer Family Registry
- SNP
single nucleotide polymorphism
- TERT
telomerase reverse transcriptase
- CRC
colorectal cancer
- ARIC
Atherosclerosis Risk in Communities
- CALiCo
Causal Variants Across the Life Course
- EAGLE
Epidemiologic Architecture for Genes Linked to Environment
- BioVu
Vanderbilt University biorepository
- MEC
Multiethnic Cohort
- WHI
Women’s Health Initiative
- ASTERISK
Association STudy Evaluating RISK for sporadic colorectal cancer
- Colo 2&3
Hawaii Colorectal Cancer Studies 2 & 3
- DACHS
Darmkrebs: Chancen der Verhütung durch Screening (DACHS)
- DALS
Diet, Activity, and Lifestyle Study (DALS)
- HPFS
Health Professionals Follow-up Study
- NHS
Nurses’ Health Study
- OFCCR
Ontario Familial Colorectal Cancer Registry
- PHS
Physicians’ Health Study (PHS)
- PLCO
Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial
- PMH-CCFR
Post-Menopausal Hormones Supplemental Study to the Colon Cancer Family Registry
- VITAL
VITamins And Lifestyle part of GECCO
Footnotes
The Corresponding Authors have the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive license (or non exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd and its Licensees to permit this article (if accepted) to be published in Gut editions and any other BMJPGL products to exploit all subsidiary rights, as set out in our license.
CONTRIBUTIONS
Christian Caberto, Yi Lin, José Luis Ambite, Gowri Kumaraguruparan, Danielle Richardson, Robert J. Goodloe, Holli H. Dilks, Paxton Baker, Li Hsu, and Shuo Jiao were involved in data analysis. William S. Bush was involved in data analysis, data interpretation, and manuscript writing. Shelly-Anne Love was involved in data analysis and manuscript writing. Daniela Seminara and Chris S. Carlson were involved in data interpretation. Logan Dumitrescu, Noralane Lindor, Christy Avery, Andrew Chan, John Baron, Lucia A. Hindorff, Sungshim Lani Park, Fredrick R. Schumacher, David Duggan, Cara L. Carty, David V. Conti, Christopher A. Haiman, Carolyn Hutter, and Emily White were involved in data interpretation and manuscript writing. Thomas J. Hudson and Hermann Brenner were involved in data interpretation, monitoring data collected, provision of study subjects, and manuscript writing. Anne Butler, Lifang Hou, and Robert E. Schoen were involved in manuscript writing. Peter Kraft, Kristine Monroe, Sun Mann, Jing Ma, Edward Giovannucci, and Tabitha Harrison were involved in monitoring collected data. Charles Fuchs, Bette Caan, Mark Jenkins, Sébastien Küry, Brent Zanke, Mathieu Lemire, and Steven Gallinger were involved in monitoring data collected and provision of study subjects. Michael Hoffmeister was involved in monitoring data collected, provision of study subjects, and manuscript writing. Polly Newcomb, Richard Hayes, Stephen Chanock, Sonja Berndt, Richard Hayes, and Stephen Chanock were involved in the provision of study subjects. Stephane Bezieau was involved in provision of study subjects and manuscript writing. John Hopper, Robert Haile, Jenny Chang-Claude, Peter T. Campbell, Charles Kooperberg, Dana C. Crawford, Gerardo Heiss, John Potter, Martha L. Slattery, and Graham Casey were involved in the provision of study subjects, data interpretation, and manuscript writing. Loic Le Marchand and Ulrike Peters were involved in study design and conception, provision of study subjects, data interpretation, and manuscript writing. Iona Cheng, Jonathan M Kocarnik, and Lynne R Wilkens were involved in study design, data analysis, data interpretation, and manuscript writing.
COMPETING INTERESTS
AC: Partnership for Prevention: personal fees (board membership); Bayer Healthcare: personal fees (consultancy); Pfizer Inc.: personal fees (consultancy); Millennium Pharmaceuticals: personal fees (consultancy); Pozen Inc.: personal fees (consultancy).
REFERENCES
- 1.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crowther-Swanepoel D, Broderick P, Di Bernardo MC, Dobbins SE, Torres M, Mansouri M, et al. Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nature genetics. 2010;42:132–136. doi: 10.1038/ng.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rothman N, Garcia-Closas M, Chatterjee N, Malats N, Wu X, Figueroa JD, et al. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nature genetics. 2010;42:978–984. doi: 10.1038/ng.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shete S, Hosking FJ, Robertson LB, Dobbins SE, Sanson M, Malmer B, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nature genetics. 2009;41:899–904. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nature genetics. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 7.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nature genetics. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 8.Hu Z, Wu C, Shi Y, Guo H, Zhao X, Yin Z, et al. A genome-wide association study identifies two new lung cancer susceptibility loci at 13q12.12 and 22q12.2 in Han Chinese. Nature genetics. 2011;43:792–796. doi: 10.1038/ng.875. [DOI] [PubMed] [Google Scholar]
- 9.Turnbull C, Rapley EA, Seal S, Pernet D, Renwick A, Hughes D, et al. Variants near DMRT1, TERT and ATF7IP are associated with testicular germ cell cancer. Nature genetics. 2010;42:604–607. doi: 10.1038/ng.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haiman CA, Chen GK, Vachon CM, Canzian F, Dunning A, Millikan RC, et al. A common variant at the TERT-CLPTM1L locus is associated with estrogen receptor-negative breast cancer. Nature genetics. 2011;43:1210–1214. doi: 10.1038/ng.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters U, Hutter CM, Hsu L, Schumacher FR, Conti DV, Carlson CS, et al. Meta-analysis of new genome-wide association studies of colorectal cancer risk. Human genetics. 2012;131:217–234. doi: 10.1007/s00439-011-1055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sivakumaran S, Agakov F, Theodoratou E, Prendergast JG, Zgaga L, Manolio T, et al. Abundant pleiotropy in human complex diseases and traits. American journal of human genetics. 2011;89:607–618. doi: 10.1016/j.ajhg.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramos PS, Criswell LA, Moser KL, Comeau ME, Williams AH, Pajewski NM, et al. A comprehensive analysis of shared loci between systemic lupus erythematosus (SLE) and sixteen autoimmune diseases reveals limited genetic overlap. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amundadottir LT, Thorvaldsson S, Gudbjartsson DF, Sulem P, Kristjansson K, Arnason S, et al. Cancer as a complex phenotype: pattern of cancer distribution within and beyond the nuclear family. PLoS medicine. 2004;1 doi: 10.1371/journal.pmed.0010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matise TC, Ambite JL, Buyske S, Carlson CS, Cole SA, Crawford DC, et al. The Next PAGE in understanding complex traits: design for the analysis of Population Architecture Using Genetics and Epidemiology (PAGE) Study. American journal of epidemiology. 2011;174:849–859. doi: 10.1093/aje/kwr160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutter CM, Chang-Claude J, Slattery ML, Pflugeisen BM, Lin Y, Duggan D, et al. Characterization of gene-environment interactions for colorectal cancer susceptibility loci. Cancer research. 2012;72:2036–2044. doi: 10.1158/0008-5472.CAN-11-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newcomb PA, Baron J, Cotterchio M, Gallinger S, Grove J, Haile R, et al. Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2007;16:2331–2343. doi: 10.1158/1055-9965.EPI-07-0648. [DOI] [PubMed] [Google Scholar]
- 18.The Atherosclerosis Risk in Communities (ARIC) Study: design and objectives. The ARIC investigators. American journal of epidemiology. 1989;129:687–702. [PubMed] [Google Scholar]
- 19.Bush W, Boston J, Pendergrass SA, Dumitrescu L, Goodloe R, Brown-Gentry K, Wilson S, McClellan B, Jr, Torstenson E, Basford MA, Spencer KL, Ritchie MD, Crawford DC. Enabling high-throughput genotype-phenotype associations in the Epidemiologic Architecture for Genes Linked to Environment (EAGLE) project as part of the Population Architecture using Genomics and Epidemiology (PAGE) study. Pacific Symposium on Biocomputing. 2013;18:373–384. [PMC free article] [PubMed] [Google Scholar]
- 20.Kolonel LN, Henderson BE, Hankin JH, Nomura AM, Wilkens LR, Pike MC, et al. A multiethnic cohort in Hawaii and Los Angeles: baseline characteristics. American journal of epidemiology. 2000;151:346–357. doi: 10.1093/oxfordjournals.aje.a010213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kury S, Buecher B, Robiou-du-Pont S, Scoul C, Colman H, Lelievre B, et al. The thorough screening of the MUTYH gene in a large French cohort of sporadic colorectal cancers. Genetic testing. 2007;11:373–379. doi: 10.1089/gte.2007.0029. [DOI] [PubMed] [Google Scholar]
- 22.Le Marchand L, Hankin JH, Wilkens LR, Pierce LM, Franke A, Kolonel LN, et al. Combined effects of well-done red meat, smoking, and rapid N-acetyltransferase 2 and CYP1A2 phenotypes in increasing colorectal cancer risk. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2001;10:1259–1266. [PubMed] [Google Scholar]
- 23.Vossen CY, Hoffmeister M, Chang-Claude JC, Rosendaal FR, Brenner H. Clotting factor gene polymorphisms and colorectal cancer risk. J Clin Oncol. 2011;29:1722–1727. doi: 10.1200/JCO.2010.31.8873. [DOI] [PubMed] [Google Scholar]
- 24.Rimm EB, Stampfer MJ, Colditz GA, Chute CG, Litin LB, Willett WC. Validity of self-reported waist and hip circumferences in men and women. Epidemiology. 1990;1:466–473. doi: 10.1097/00001648-199011000-00009. [DOI] [PubMed] [Google Scholar]
- 25.Cotterchio M, Manno M, Klar N, McLaughlin J, Gallinger S. Colorectal screening is associated with reduced colorectal cancer risk: a case-control study within the population-based Ontario Familial Colorectal Cancer Registry. Cancer causes & control : CCC. 2005;16:865–875. doi: 10.1007/s10552-005-2370-3. [DOI] [PubMed] [Google Scholar]
- 26.Cotterchio M, McKeown-Eyssen G, Sutherland H, Buchan G, Aronson M, Easson AM, et al. Ontario familial colon cancer registry: methods and first-year response rates. Chronic diseases in Canada. 2000;21:81–86. [PubMed] [Google Scholar]
- 27.Belanger CF, Hennekens CH, Rosner B, Speizer FE. The nurses' health study. The American journal of nursing. 1978;78:1039–1040. [PubMed] [Google Scholar]
- 28.Prorok PC, Andriole GL, Bresalier RS, Buys SS, Chia D, Crawford ED, et al. Design of the Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial. Controlled clinical trials. 2000;21:273S–309S. doi: 10.1016/s0197-2456(00)00098-2. [DOI] [PubMed] [Google Scholar]
- 29.Gohagan JK, Prorok PC, Hayes RB, Kramer BS. The Prostate, Lung, Colorectal and Ovarian (PLCO) Cancer Screening Trial of the National Cancer Institute: history, organization, and status. Controlled clinical trials. 2000;21:251S–272S. doi: 10.1016/s0197-2456(00)00097-0. [DOI] [PubMed] [Google Scholar]
- 30.Newcomb PA, Zheng Y, Chia VM, Morimoto LM, Doria-Rose VP, Templeton A, et al. Estrogen plus progestin use, microsatellite instability, and the risk of colorectal cancer in women. Cancer research. 2007;67:7534–7539. doi: 10.1158/0008-5472.CAN-06-4275. [DOI] [PubMed] [Google Scholar]
- 31.White E, Patterson RE, Kristal AR, Thornquist M, King I, Shattuck AL, et al. VITamins And Lifestyle cohort study: study design and characteristics of supplement users. American journal of epidemiology. 2004;159:83–93. doi: 10.1093/aje/kwh010. [DOI] [PubMed] [Google Scholar]
- 32.Hays J, Hunt JR, Hubbell FA, Anderson GL, Limacher M, Allen C, et al. The Women's Health Initiative recruitment methods and results. Annals of epidemiology. 2003;13:S18–S77. doi: 10.1016/s1047-2797(03)00042-5. [DOI] [PubMed] [Google Scholar]
- 33.Design of the Women's Health Initiative clinical trial and observational study. The Women’s Health Initiative Study Group. Controlled clinical trials. 1998;19:61–109. doi: 10.1016/s0197-2456(97)00078-0. [DOI] [PubMed] [Google Scholar]
- 34.Kosoy R, Nassir R, Tian C, White PA, Butler LM, Silva G, et al. Ancestry informative marker sets for determining continental origin and admixture proportions in common populations in America. Hum Mutat. 2009;30:69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 36.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PI. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Waliszewska A, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nature genetics. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al Olama AA, Kote-Jarai Z, Giles GG, Guy M, Morrison J, Severi G, et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nature genetics. 2009;41:1058–1060. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 40.Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nature genetics. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 41.Gudmundsson J, Sulem P, Gudbjartsson DF, Blondal T, Gylfason A, Agnarsson BA, et al. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nature genetics. 2009;41:1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takata R, Akamatsu S, Kubo M, Takahashi A, Hosono N, Kawaguchi T, et al. Genome-wide association study identifies five new susceptibility loci for prostate cancer in the Japanese population. Nature genetics. 2010;42:751–754. doi: 10.1038/ng.635. [DOI] [PubMed] [Google Scholar]
- 43.Yeager M, Chatterjee N, Ciampa J, Jacobs KB, Gonzalez-Bosquet J, Hayes RB, et al. Identification of a new prostate cancer susceptibility locus on chromosome 8q24. Nature genetics. 2009;41:1055–1057. doi: 10.1038/ng.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cui R, Okada Y, Jang SG, Ku JL, Park JG, Kamatani Y, et al. Common variant in 6q26-q27 is associated with distal colon cancer in an Asian population. Gut. 2011;60:799–805. doi: 10.1136/gut.2010.215947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tenesa A, Farrington SM, Prendergast JG, Porteous ME, Walker M, Haq N, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nature genetics. 2008;40:631–637. doi: 10.1038/ng.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nature genetics. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 47.Tomlinson IP, Webb E, Carvajal-Carmona L, Broderick P, Howarth K, Pittman AM, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40:623–630. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 48.Zanke BW, Greenwood CM, Rangrej J, Kustra R, Tenesa A, Farrington SM, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nature genetics. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 49.Curtin K, Lin WY, George R, Katory M, Shorto J, Cannon-Albright LA, et al. Meta association of colorectal cancer confirms risk alleles at 8q24 and 18q21. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2009;18:616–621. doi: 10.1158/1055-9965.EPI-08-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsuo K, Suzuki T, Ito H, Hosono S, Kawase T, Watanabe M, et al. Association between an 8q24 locus and the risk of colorectal cancer in Japanese. BMC Cancer. 2009;9 doi: 10.1186/1471-2407-9-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haerian MS, Baum L, Haerian BS. Association of 8q24.21 loci with the risk of colorectal cancer: a systematic review and meta-analysis. J Gastroenterol Hepatol. 2011;26:1475–1484. doi: 10.1111/j.1440-1746.2011.06831.x. [DOI] [PubMed] [Google Scholar]
- 52.Cicek MS, Slager SL, Achenbach SJ, French AJ, Blair HE, Fink SR, et al. Functional and clinical significance of variants localized to 8q24 in colon cancer. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2009;18:2492–2500. doi: 10.1158/1055-9965.EPI-09-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haiman CA, Le Marchand L, Yamamato J, Stram DO, Sheng X, Kolonel LN, et al. A common genetic risk factor for colorectal and prostate cancer. Nature genetics. 2007;39:954–956. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ghoussaini M, Song H, Koessler T, Al Olama AA, Kote-Jarai Z, Driver KE, et al. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst. 2008;100:962–966. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kupfer SS, Torres JB, Hooker S, Anderson JR, Skol AD, Ellis NA, et al. Novel single nucleotide polymorphism associations with colorectal cancer on chromosome 8q24 in African and European Americans. Carcinogenesis. 2009;30:1353–1357. doi: 10.1093/carcin/bgp123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li M, Zhou Y, Chen P, Yang H, Yuan X, Tajima K, et al. Genetic variants on chromosome 8q24 and colorectal neoplasia risk: a case-control study in China and a meta-analysis of the published literature. PLoS One. 2011;6 doi: 10.1371/journal.pone.0018251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Broderick P, Wang Y, Vijayakrishnan J, Matakidou A, Spitz MR, Eisen T, et al. Deciphering the Impact of Common Genetic Variation on Lung Cancer Risk: A Genome-Wide Association Study. Cancer Research. 2009;69:6633–6641. doi: 10.1158/0008-5472.CAN-09-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heard A, Roder D, Luke C. Multiple primary cancers of separate organ sites: implications for research and cancer control (Australia) Cancer Causes Control. 2005;16:475–481. doi: 10.1007/s10552-004-8023-0. [DOI] [PubMed] [Google Scholar]
- 59.Houlston RS, Webb E, Broderick P, Pittman AM, Di Bernardo MC, Lubbe S, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40:1426–1435. doi: 10.1038/ng.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dunlop MG, Dobbins SE, Farrington SM, Jones AM, Palles C, Whiffin N, et al. Common variation near CDKN1A, POLD3 and SHROOM2 influences colorectal cancer risk. Nat Genet. 2012;44:770–776. doi: 10.1038/ng.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peters U, Jiao S, Schumacher FR, Hutter CM, Aragaki AK, Baron JA, et al. Identification of Genetic Susceptibility Loci for Colorectal Tumors in a Genome-Wide Meta-analysis. Gastroenterology. 2013;144:799–807. e24. doi: 10.1053/j.gastro.2012.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahmadiyeh N, Pomerantz MM, Grisanzio C, Herman P, Jia L, Almendro V, et al. 8q24 prostate, breast, and colon cancer risk loci show tissue-specific long-range interaction with MYC. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9742–9746. doi: 10.1073/pnas.0910668107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sur IK, Hallikas O, Vaharautio A, Yan J, Turunen M, Enge M, et al. Mice Lacking a Myc Enhancer That Includes Human SNP rs6983267 Are Resistant to Intestinal Tumors. Science. 2012 doi: 10.1126/science.1228606. [DOI] [PubMed] [Google Scholar]
- 64.Barrett JH, Iles MM, Harland M, Taylor JC, Aitken JF, Andresen PA, et al. Genome-wide association study identifies three new melanoma susceptibility loci. Nature genetics. 2011;43:1108–1113. doi: 10.1038/ng.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kote-Jarai Z, Olama AA, Giles GG, Severi G, Schleutker J, Weischer M, et al. Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nature genetics. 2011;43:785–791. doi: 10.1038/ng.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, et al. A genome-wide association study of lung cancer identifies a region of chromosome 5p15 associated with risk for adenocarcinoma. American journal of human genetics. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McKay JD, Hung RJ, Gaborieau V, Boffetta P, Chabrier A, Byrnes G, et al. Lung cancer susceptibility locus at 5p15.33. Nature genetics. 2008;40:1404–1406. doi: 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol. 2005;7:1074–1082. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]
- 69.Kinnersley B, Migliorini G, Broderick P, Whiffin N, Dobbins SE, Casey G, et al. The TERT variant rs2736100 is associated with colorectal cancer risk. British journal of cancer. 2012;107:1001–1008. doi: 10.1038/bjc.2012.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chatterjee N, Wheeler B, Sampson J, Hartge P, Chanock SJ, Park JH. Projecting the performance of risk prediction based on polygenic analyses of genome-wide association studies. Nature genetics. 2013;45:400–405. doi: 10.1038/ng.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.