SUMMARY
Autophagy is reported to be an important innate immune defence against the intracellular bacterial pathogen Group A Streptococcus (GAS). However, the GAS strains examined to-date belong to serotypes infrequently associated with human disease. We find that the globally disseminated serotype M1T1 clone of GAS can evade autophagy and replicate efficiently in the cytosol of infected cells. Cytosolic M1T1 GAS (strain 5448), but not M6 GAS (strain JRS4), avoids ubiquitylation and recognition by the host autophagy marker LC3 and ubiquitin-LC3 adaptor proteins NDP52, p62 and NBR1. Expression of SpeB, a streptococcal cysteine protease, is critical for this process, as an isogenic M1T1 ΔspeB mutant is targeted to autophagy and attenuated for intracellular replication. SpeB degrades p62, NDP52 and NBR1 in vitro and within the host cell cytosol. These results uncover a proteolytic mechanism utilized by GAS to escape the host autophagy pathway which may underpin the success of the M1T1 clone.
INTRODUCTION
Autophagy is a highly conserved cellular process that targets cytosolic components, including protein aggregates, damaged organelles and intracellular bacteria for lysosomal degradation, thus playing important roles in homeostasis and innate immunity (Deretic, 2010). Autophagy is an important cytosolic innate immune defence against bacterial infections (Huang and Brumell, 2009), and successful intracellular bacterial pathogens avoid autophagy by replicating in membrane-bound vacuoles or by camouflaging their surface with host or bacterial-derived proteins (Dortet et al., 2011; Ogawa et al., 2005; Yoshikawa et al., 2009). Intracellular bacteria can be targeted to autophagy by a number of adaptor proteins that recognise polyubiquitylated bacteria in the cytosol or damaged bacteria-containing vacuoles (Kirkin et al., 2009; Thurston et al., 2009; Thurston et al., 2012). These adaptor proteins, which include p62 (SQSTM1), NDP52 (CALCOCO2), NBR1, and optineurin, direct cargo to nascent LC3-positive phagophores and ultimately to degradation by the lysosomal pathway (Chong et al., 2012; Thurston et al., 2009; Wild et al., 2011; Zheng et al., 2009).
Group A Streptococcus (GAS) is an obligate human pathogen and the fourth most common bacterial cause of human mortality (Carapetis et al., 2005). The GAS disease burden ranges from superficial infections (pharyngitis, impetigo), to life-threatening invasive conditions (toxic shock, necrotizing fasciitis), to post-infectious immune disorders (rheumatic fever, glomerulonephritis) (Cole et al., 2011). A number of GAS strains are efficiently internalized into epithelial cells where they can be targeted to autophagy and cleared; however, these strains belong to serotypes M6 (Joubert et al., 2009; Nakagawa et al., 2004; Sakurai et al., 2010), M49 (Joubert et al., 2009) and M89 (Thurston et al., 2009), which are not representative of the prevalent serotypes associated with contemporary human disease epidemiology (Cole et al., 2011; Steer et al., 2009). Here, we show that the globally disseminated serotype M1T1 clone of group A Streptococcus can replicate efficiently in the cytosol of infected cells through a process that involves proteolysis of the host proteins that target intracellular bacteria to autophagy.
RESULTS
M1T1 strain 5448 replicates within epithelial cells and avoids autophagy
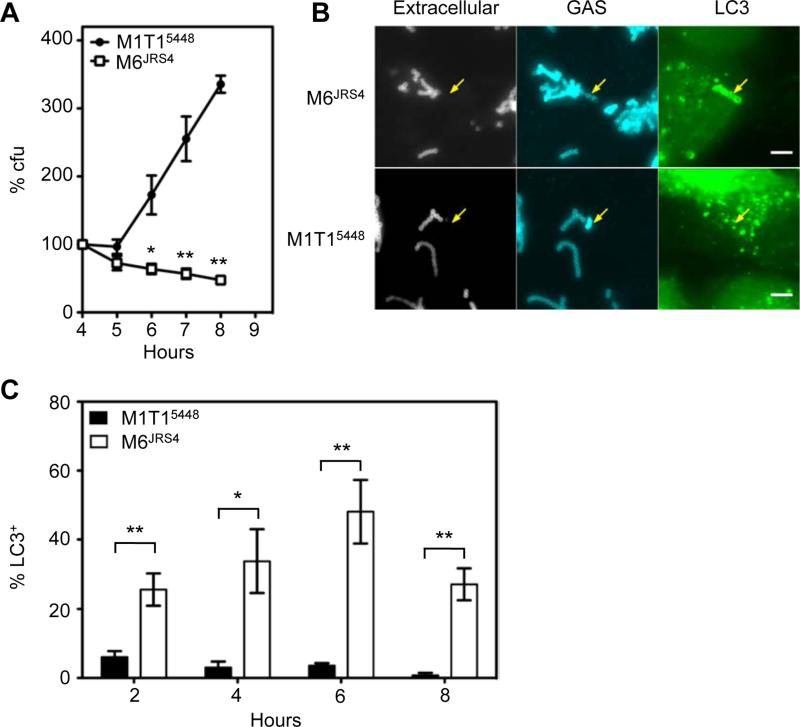
While GAS has served as a model organism to unravel the complex molecular events that lead to anti-bacterial autophagy, the strains examined belong to serotypes infrequently associated with human disease. We therefore compared the intracellular survival of one such laboratory-adapted M6 strain (strain JRS4, hereafter M6JRS4) (Nakagawa et al., 2004), with a recent clinical isolate of the globally-disseminated serotype M1T1 clone (strain 5448, hereafter M1T15448) that has been the single leading cause of both pharyngitis and severe invasive GAS infections during the last three decades. Intracellular viability of GAS following entry into human HEp-2 epithelial cells was monitored over time by measuring colony-forming units (cfu) (Figure 1A). Consistent with prior studies, the viability of the M6JRS4 strain decreased over time, as only 47% of cfu present at 4 h post infection remained at 8 h post infection. In contrast, recoverable cfu of the M1T15448 strain tripled from 4 to 8 h post infection, revealing a capacity of this clinically important strain to not only survive, but replicate, within epithelial cells.
Figure 1. M1T15448 replicates within epithelial cells and avoids autophagy.
(A) Ability of M1T15448 and M6JRS4 GAS to survive following internalization into HEp-2 epithelial cells. Data is represented as mean ± SEM of three independent experiments. *, p < 0.05; **, p < 0.01; one-tailed paired t-test.
(B) Association of intracellular M6JRS4 and M1T15448 with GFP-LC3 at 4 h post-infection. Arrows indicate intracellular M6JRS4 associated with GFP-LC3 and intracellular M1T15448 devoid of GFP-LC3. Bar = 5 μm.
(C) Percentage of intracellular GAS contained within LC3-positive compartments. Values represent the mean ± SEM for three independent experiments. *, p < 0.05; **, p < 0.01; one-tailed unpaired t-test.
To determine whether M1T15448 intracellular replication reflected resistance to autophagy, we performed immunofluorescence microscopy to quantitate intracellular M1T15448 or M6JRS4 GAS that co-localized with the autophagy marker GFP-LC3 (Figures 1B and 1C). M6JRS4 GAS were efficiently targeted to autophagy, with 48.1 ± 9.2% of intracellular M6JRS4 found in LC3-positive vacuoles at 6 h post infection. In contrast, only low numbers of M1T15448 GAS were transiently associated with GFP-LC3 at 2 h post infection and by 6 h post infection the vast majority (96.4 ± 0.7%) were negative for GFP-LC3, demonstrating that the virulent human M1T1 isolate avoids targeting to autophagy to replicate within human epithelial cells.
M1T15448 GAS replicate in the cytosol of epithelial cells
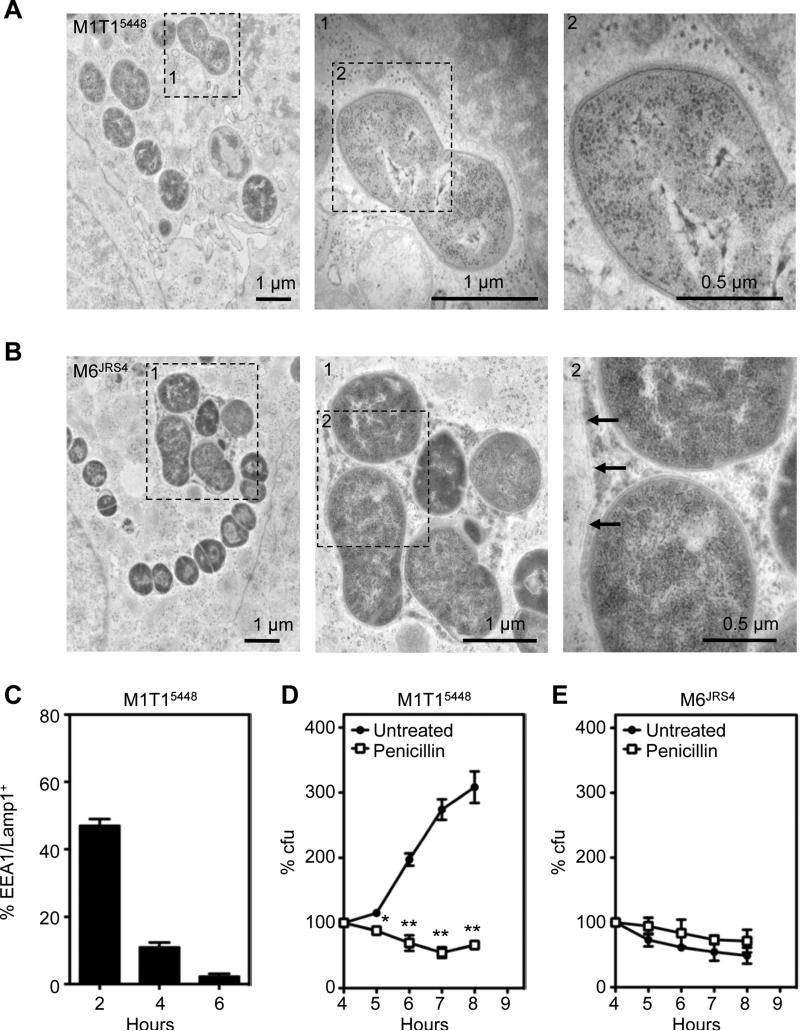
Autophagy primarily targets bacteria in the cytosol or in damaged membrane compartments, and bacterial pathogens such as Salmonella Typhimurium avoid autophagy by replicating within modified vacuoles (Birmingham et al., 2006). We therefore explored whether M1T15448 avoids autophagy by replicating within an intact vacuole. To directly visualize intracellular M1T15448 and M6JRS4 GAS, we performed transmission electron microscopy on GAS-infected HEp-2 cells at 6 h post-infection (Figures 2A and 2B). The M1T15448 strain was abundantly present in the cytosol of infected cells, whereas the M6JRS4 strain was contained within a membrane-bound compartment. To confirm that M1T15448 was not associated with endosomal membranes, we performed immunofluorescence microscopy to quantitate the association of M1T15448 with markers of early (EEA1) and late (Lamp1) endosomes (Figures 2C and S1). While transiently associated with endosomes at 2 h post-infection, only 2.3 ± 0.7% of intracellular M1T15448 were EEA1 or Lamp1-associated at 6 h post infection, suggesting that M1T15448 escapes the endosomal pathway to replicate. We additionally examined the susceptibility of intracellular M1T15448 to penicillin. Penicillin is an antibiotic that rapidly enters the cytosol but not components of the endocytic pathway (Renard et al., 1987), a property that has been used to select for Listeria monocytogenes mutants that fail to escape the endocytic pathway (Camilli et al., 1989). Penicillin treatment abolished the intracellular replication of WT M1T15448 with viability diminishing over the course of the experiment (Figure 2D). In contrast, penicillin treatment had minimal affect on the intracellular viability of M6JRS4 (Figure 2E). Taken together, these results suggest that the intracellular replication of M1T15448 likely correlates to their successful entry into the cytosol of infected cells.
Figure 2. M1T15448 replicate efficiently in the cytosol of epithelial cells.
(A and B) Transmission electron micrographs of HEp-2 cells infected with M1T15448 (A) and M6JRS4 (B) GAS at 6 h post-infection. The M6JRS4 GAS is entrapped within a membrane compartment with arrowheads indicating the membrane (zoomed micrograph on right); In contrast, the M1T15448 GAS is entirely exposed to the cytosol.
(C) Percentage of intracellular M1T15448 contained within EEA1/Lamp1-positive compartments. Values represent the mean ± SEM for three technical replicates.
(D and E) Susceptibility of intracellular M1T15448 (D) and M6JRS4 (E) GAS to penicillin. Values represent the mean ± SEM of three independent experiments. *, p < 0.05; **, p < 0.01; one-tailed paired t-test. See also Figure S1.
Intracellular M1T15448 GAS avoids ubiquitylation and the ubiquitin-LC3 adaptor proteins NDP52, p62 and NBR1
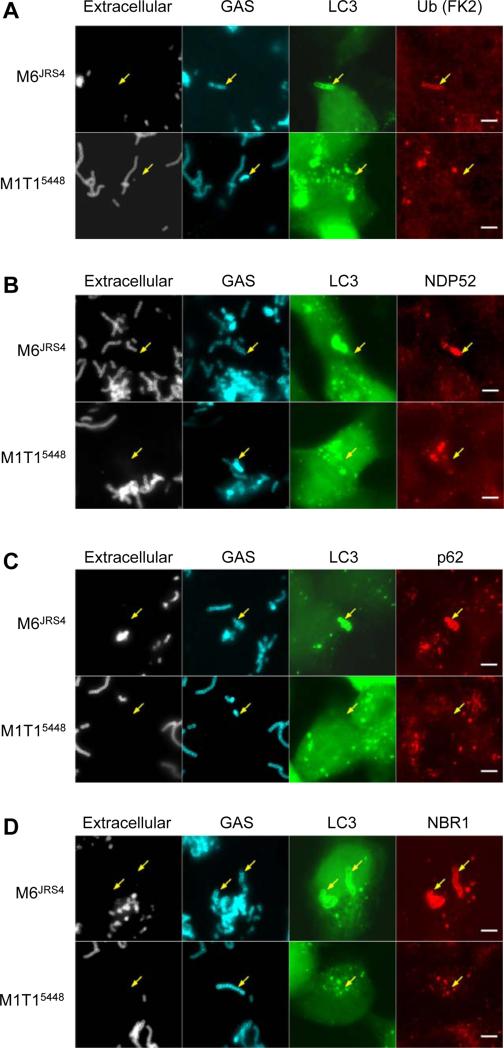
With few exceptions, ubiquitylation is a critical step in selective autophagy (Kirkin et al., 2009). LC3 is targeted to ubiquitylated bacteria via ubiquitin-LC3 adaptor proteins, which include p62, NDP52, NBR1 and optineurin (Chong et al., 2012; Deretic, 2010; Mostowy et al., 2011; Thurston et al., 2009; Wild et al., 2011). To determine whether M1T15448 escapes autophagy by avoiding these pathways, we performed immunofluorescence microscopy on M1T15448 and M6JRS4 infected cells to determine their respective co-localization with ubiquitylated proteins (Figure 3A), NDP52 (Figure 3B), p62 (Figure 3C) and NBR1 (Figure 3D). Paralleling previous observations (Thurston et al., 2009), M6JRS4 GAS were found associated with NDP52, with the proportion of GFP-LC3-positive M6JRS4 GAS associated with NDP52 gradually increasing from 13.7 ± 3.0% at 2 h post infection to 33.3 ± 4.3% at 8 h post infection. However, the majority of GFP-LC3-positive M6JRS4 GAS were found associated with p62 (82.5 ± 3.7% to 96.3 ± 3.7%) and NBR1 (92.2 ± 4.0% to 98.4 ± 1.6%) at all time points examined (Table S1 available online). In contrast, intracellular M1T15448 GAS bacteria were not found in association with ubiquitylated proteins, NDP52, p62 nor NBR1 (Figure 3), suggesting that intracellular M1T15448 actively evades the autophagy pathways in epithelial cells.
Figure 3. Intracellular M1T15448 avoids ubiquitylation and the ubiquitin-LC3 adaptor proteins NDP52, p62 and NBR1.
(A to D) Association of M1T15448 and M6JRS4 GAS with ubiquitylated proteins (A), NDP52 (B), p62 (C) and NBR1 (D). Quantitative data at 2, 4, 6 and 8 h post-infection is provided in Table S1. Arrows indicate intracellular M6JRS4 associated with GFP-LC3 and adaptors and intracellular M1T15448 devoid of GFP-LC3 and adaptors at 4 h post-infection. Bar = 5 μm. See also Table S1.
SpeB cysteine protease is required for efficient intracellular replication of M1T15448 GAS
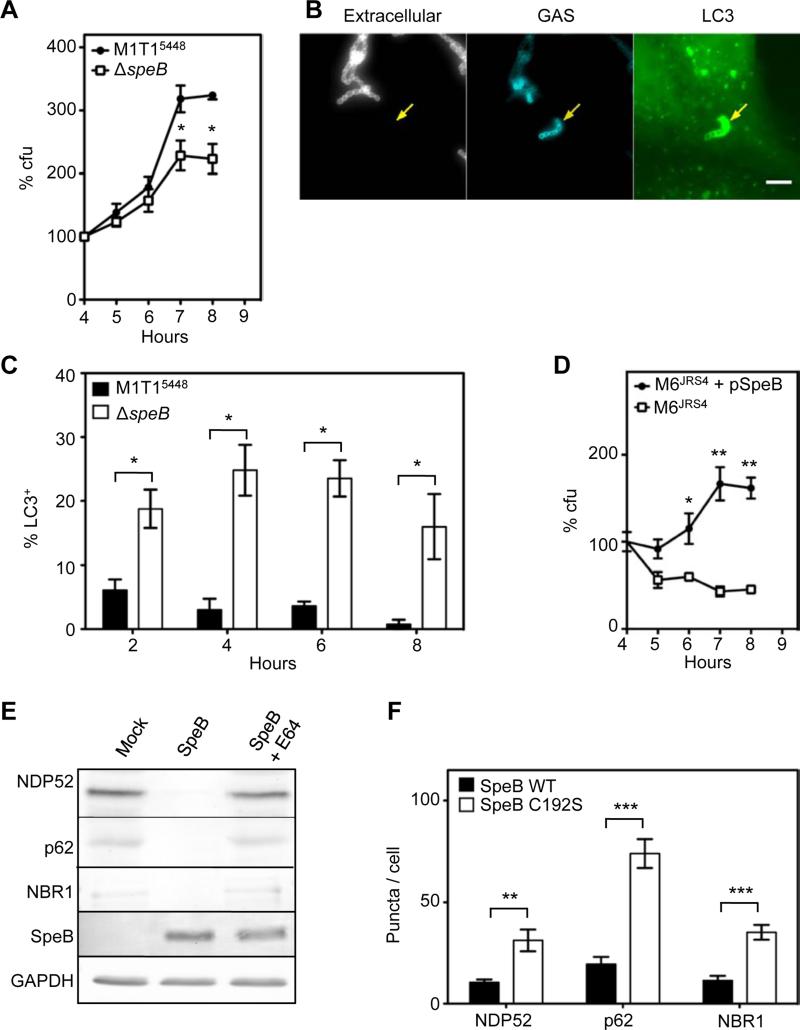
The M6JRS4 strain examined by Nakagawa et al. (Nakagawa et al., 2004) was previously shown to be defective in expression of the extracellular cysteine protease SpeB (Lyon et al., 2001) (Figure S2A), and is avirulent in a murine model of GAS infection (Figure S2B). Given that SpeB is a secreted and surface-associated protease (Hytonen et al., 2001) whose expression varies amongst GAS strains, we hypothesized that this virulence determinant may play a role in avoidance of the ubiquitylation system. Thus, we examined the ability of WT M1T15448 and an isogenic ΔspeB mutant to replicate within epithelial cells. Comparison of the genome sequences of M1T15448 and M1T15448 ΔspeB revealed that these strains differ by only 2 SNPs (Figure S3). Compared to the WT parent strain, the M1T15448 ΔspeB mutant was attenuated for intracellular replication (Figure 4A), but not for replication in THY broth in vitro (Figure S3A). Furthermore, immunofluorescence microscopy revealed that, in comparison to wild type M1T15448, targeting of the M1T15448 ΔspeB mutant to autophagy was much more efficient (Figures 4B and 4C), with 25.3 +/- 3.9% intracellular M1T15448 ΔspeB associated with GFP-LC3 at 6 hours post-infection.
Figure 4. SpeB cysteine protease degrades ubiquitin-LC3 adaptor proteins and is required for efficient intracellular replication of GAS.
(A) Ability of M1T15448 ΔspeB to replicate within HEp-2 epithelial cells. Data is represented as mean ± SEM of three independent experiments. *, p < 0.05; **, p < 0.01; one-tailed paired t-test.
(B) Association of M1T15448 ΔspeB with GFP-LC3 at 4 h post-infection. Arrows indicate intracellular M1T15448 ΔspeB mutant associated with GFP-LC3. Bar = 5 μm.
(C) Percentage of intracellular GAS contained within LC3-positive compartments. Values represent the mean ± SEM of three independent experiments. *, p < 0.05; one-tailed paired t-test. The M1T15448 data was taken from Figure 1C and is included for comparison.
(D) Intracellular replication of the SpeB-expressing M6JRS4 + pSpeB strain compared with the SpeB-negative M6JRS4 strain. Data is represented as mean ± SEM of three technical replicates. *, p < 0.05; **, p < 0.01; one-tailed unpaired t-test.
(E) Western immunoblot showing that purified SpeB cysteine protease degrades NDP52, p62 and NBR1 in HEp-2 cell lysates.
(F) Ectopically expressed SpeB degrades cytosolic NDP52, p62 and NBR1. Bars represent the number of puncta ± SEM in at least 15 transfected cells and is representative of 2 experimental replicates. **, p < 0.01; ***, p < 0.001; one-tailed paired t-test. Representative images used for the quantitation are shown in Figure S2H to J. See also Figure S2.
To further investigate the role of SpeB in resistance to autophagy, we engineered a M6JRS4 strain that expresses SpeB. In contrast to M6JRS4, M6JRS4 + pSpeB replicated efficiently within HEp-2 cells (Figures 4D), and was only rarely associated with GFP-LC3 (Figure S2C). Similar results were also obtained with the SpeB-expressing strains M6MGAS10394 (Banks et al., 2004) and M12HKU16 (Tse et al., 2012), which replicated efficiently, versus the naturally occurring SpeB-negative M4NS244 strain (McKay et al., 2004), which failed to replicate (Figure S2D).
SpeB degrades ubiquitin-LC3 adaptor proteins NDP52, p62 and NBR1
To determine whether SpeB conferred resistance to autophagy by its broad-spectrum cysteine protease activity, we purified SpeB from M1T15448 (Figure S2E), and examined its ability to degrade components of the host ubiquitylation system. Purified SpeB efficiently degraded NDP52, p62 and NBR1 (Figure 4E), as well as ubiquitylated proteins (Figure S2F) from HEp-2 epithelial cell extracts. Similar results were obtained with bacterial culture supernatants from wild-type M1T15448, but not the isogenic ΔspeB strain (Figure S2G). To confirm that these effects were specific to SpeB, we performed parallel experiments in the presence of the cysteine protease inhibitor E64 (Cole et al., 2007) (Figures 4E, S2F and S2G). In a manner comparable to the mock treated samples, the E64-treated purified SpeB was deficient in the proteolytic activities described above.
We propose the ability of SpeB to act as a defence against host ubiquitylation components relies on its proteolytic activity within the environment of the host cytosol. To confirm that SpeB was enzymatically active in the host cytosol, we transfected HEp-2 cells with plasmids encoding either codon-optimised SpeB or a catalytically-inactive C192S derivative and compared their affect on NDP52, p62 and NBR1 (Figures 4F and S2H to S2J). The plasmid encoding wild-type SpeB significantly reduced the number of NDP52, p62 and NBR1 puncta within transfected cells when compared to the catalytically inactive C192S mutant, demonstrating that wild-type SpeB is enzymatically active within the host cell cytosol. Taken together, these results demonstrate that GAS SpeB protease is necessary and sufficient to degrade ubiquitylation components within the host cytosol that normally serve to direct the bacterium to autophagy.
DISCUSSION
Autophagy primarily targets bacteria in the cytosol or in damaged membrane vacuoles. Some intracellular bacterial pathogens, such as Salmonella Typhimurium (Birmingham et al., 2006) and Staphylococcus aureus (Schnaith et al., 2007), avoid autophagy by replicating within modified endocytic or autophagic compartments. Other bacterial pathogens, such as Shigella flexneri and Listeria monocytogenes, escape the endocytic pathway and replicate in the cytosol of infected cells, avoiding ubiquitylation by camouflaging their surface with bacterial or host-derived proteins (Dortet et al., 2011; Ogawa et al., 2005; Yoshikawa et al., 2009). Here we provide several lines of evidence that GAS employ a proteolytic mechanism to evade autophagy and replicate in the cytosol of infected cells: (1) genetic deletion of the gene encoding SpeB significantly reduced the intracellular replication of M1T15448, (2) genetic deletion of the gene encoding SpeB from the M1T15448 strain significantly increased the frequency of the recruitment of LC3 to the surface of the intracellular bacteria, (3) expression of SpeB by M6JRS4 promoted intracellular replication and abolished recruitment of LC3 to the bacterial surface, (4) purified SpeB degrades the ubiquitin-LC3 adaptor proteins p62, NDP52 and NBR1, (5) ectopic expression of codon-optimized SpeB in HEp-2 cells reduced the amounts of p62, NDP52 and NBR1, (6) wild-type SpeB-expressing strains replicate efficiently in epithelial cells while naturally-occurring SpeB-defective strains fail to replicate. Therefore, we propose that production of a bacterial protease that degrades the host proteins responsible for targeting bacteria to autophagy, in addition to proteolytic degradation of ubiquitylated bacterial surface proteins, constitutes a previously unrecognized mechanism employed by a bacterial pathogen to evade autophagy.
GAS is a highly successful pathogen of its human host, causing a wide array of superficial and invasive diseases. The transition from superficial to invasive disease involves a genetic switch that abolishes SpeB expression while concomitantly increasing expression of numerous virulence factors required for growth in deeper tissues (Cole et al., 2011). As such, there is a strong correlation between SpeB expression and superficial disease (Cole et al., 2011; Ikebe et al., 2010). SpeB is a broad-spectrum cysteine protease required for virulence (Cole et al., 2006) that has been shown to degrade a number of immunologically important human proteins, including immunoglobulins, chemokines, proinflammatory cytokines and cathelicidin, , and to proteolytically modify GAS surface proteins such as M protein, which is important for host colonisation (Johansson et al., 2008; Nelson et al., 2011). Additionally, SpeB has been shown to bind to a variety of host proteins including laminin (Hytonen et al., 2001), and integrins (Stockbauer et al., 1999), which may have undescribed influences on host cell differentiation and signaling. Together, these properties suggest that SpeB expression is required by GAS for multiple aspects of epithelial colonisation that includes resistance to autophagy and other innate defences.
In addition to SpeB expression, there is also a strong correlation of certain GAS serotypes with superficial disease in developed countries; M1T1 and M12 strains are the most prevalent in published epidemiological studies (Shulman et al., 2009). In contrast, M6 (Joubert et al., 2009; Nakagawa et al., 2004; Sakurai et al., 2010), M49 (Joubert et al., 2009) and M89 (Thurston et al., 2009) that have been used in prior GAS autophagy studies are much less frequently associated with human disease. It is likely that M1T1 and other prevalent serotypes possess other genetic traits that enhance their success at colonising epithelial surfaces (Maamary et al., 2012). In agreement with this is our observation that the M1T15448 ΔspeB strain was not as attenuated as M6JRS4, suggesting that M1T15448 has other intrinsic mechanisms of resistance to ubiquitylation and autophagy. Such mechanisms may include additional surface-bound proteins that mask the bacterial surface or subvert autophagy or, alternatively, the absence of bacterial targets for ubiquitylation. In addition, our data also illustrate the importance of investigating virulent strains of bacterial pathogens rather than relying exclusively on lab-adapted strains such as M6JRS4, as lab-adapted strains may not provide a true representation of the phenotype of a bacterial pathogen in a particular disease model.
In conclusion, we report here a previously unrecognized strategy employed by an intracellular bacterial pathogen to manipulate the host autophagy pathway. By production of a protease that degrades host proteins that target bacteria to autophagy, a strain of the globally disseminated M1T1 GAS serotype can evade autophagy and replicate efficiently in the cytosol of infected epithelial cells. We propose that SpeB is required by GAS strains to establish successful colonization of epithelial tissues, by allowing such strains to escape an important component of host innate defence.
EXPERIMENTAL PROCEDURES
Bacterial Growth
GAS strains M6JRS4 (Nakagawa et al., 2004), M1T15448 (Kansal et al., 2003), M1T15448ΔspeB (Kansal et al., 2003), M6MGAS10394 (Banks et al., 2004), M12HKU16 (Tse et al., 2012) and M4NS244 (McKay et al., 2004) were grown in Todd-Hewitt medium with 0.2% yeast extract (THY) at 37°C. Strain M6JRS4 + pSpeB was constructed by introducing plasmid pSpeB (Korotkova et al., 2012) into M6JRS4 by electroporation.
Cell culture and transfections
Cell lines were cultured at 37°C in 5% CO2. HEp-2 cells were originally obtained from ATCC and grown in DMEM (Dulbecco's modified Eagle medium, Gibco) containing 10% foetal bovine serum (FBS, Gibco). Plasmid DNA was introduced into HEp-2 cells using Attractene reagent (Qiagen).
GAS intracellular growth assay
For GAS infection, early stationary phase bacteria (OD600 = 1.0-1.2) were harvested and washed once with Dulbecco's phosphate buffered saline (DPBS; pH 7.4). The bacteria were diluted into 1% FBS/DMEM and added to confluent cell culture monolayers at a multiplicity of infection = 0.8 (8 bacteria for every 10 cells). After 2 h, cells were washed once with 10% FBS/DMEM, and then incubated in 10% FBS/DMEM with 100 μg/ml gentamicin to kill extracellular bacteria. Where indicated, 100 U/ml penicillin G was also included. After an appropriate incubation time, infected cells were washed twice with PBS, treated with 200 μl trypsin, and lysed by the addition of 0.025% Triton X-100. Cell lysates were serially diluted and plated on THY agar for bacterial enumeration.
Construction of mRFP-LC3
A plasmid encoding LC3 fused to the C-terminus of mRFP was constructed by inverse-PCR using primers delEGFP-S (5′-CGAGCTGTACAAGTCCGGACTCAGATCTC-3′) and delEGFPA (5′-GCATAGATCTCTTGCTCACCATGGTGGCGAC-3′) and a plasmid encoding mRFPGFP-LC3 (Kimura et al., 2007) as template DNA. The resulting PCR amplicon was digested with BglII and ligated with T4 DNA ligase to create pmRFP-LC3.
Antibodies
Primary antibodies used were rabbit anti-group A polysaccharide (PAB13831, Abnova), rabbit anti-GAPDH (2275-PC-100, R&D Systems), rabbit anti-SpeB (PBI222, Toxin Technology), mouse anti-EEA1 (610457, BD Transduction Laboratories), mouse anti-Lamp1 (CD107a; 555798, BD Pharmingen), mouse anti-p62 (TA502239, OriGene), mouse anti-NDP52 (TA501971, OriGene), mouse anti-NBR1 (B01P, Abnova), mouse anti-ubiquitylated proteins (FK2, Millipore), mouse anti-ubiquitin (P4D1, Cell Signaling Technology) and rabbit anti-myc (Novus Biologicals). Secondary antibodies used for immunofluorescence microscopy were AlexaFluor-conjugated goat anti-mouse or goat anti-rabbit antibodies (Invitrogen). Secondary antibodies used for western immunoblots were IRDye680LT-conjugated goat anti-rabbit or IRDye800CW-conjugated goat anti-mouse (Li-Cor).
Immunofluorescence microscopy
HEp-2 cells cultured on glass coverslips were infected with GAS strains as described above. At the indicated times post-infection, coverslips were washed four times with 500 μl DPBS and fixed in 4% paraformaldehyde (PFA). The fixed cells were blocked in DPBS containing 2% bovine serum albumin and 0.02% sodium azide. Extracellular bacteria were stained with anti-group A polysaccharide antiserum. Coverslips were again fixed in 4% PFA, permeabilised with 0.1% Triton X-100 in PBS for 10 min and blocked in DPBS containing 2% bovine serum albumin, 0.1% Triton X-100 and 0.02% sodium azide (PBS-BT). Coverslips were stained with primary antibodies and AlexaFluor-conjugated secondary antibodies (Invitrogen) in PBS-BT, mounted onto glass slides and imaged on a Personal Deltavision inverted microscope (Applied Precision). Maximum projection of acquired images was performed using the ImageJ Fiji software package (http://fiji.sc/wiki/index.php/Fiji).
SpeB activity assay
HEp-2 cell lysates were prepared by lysing confluent monolayers in T75 flasks (~8 × 106 cells) in 150 μl DPBS containing 1% Triton X-100, 0.1% SDS and EDTA-free complete protease inhibitor (Roche). Lysates were passed ten times through a 27-guage needle and insoluble material removed by centrifuging at 14,000 × g for 15 min. The resulting supernatant was dialyzed extensively against DPBS before use.
SpeB was purified from stationary phase M1T15448 culture supernatants using a modification of the protocol described by Berge and Bjorck (Berge and Bjorck, 1995). Briefly, SpeB was precipitated from a 500 ml 24 h culture supernatant of M1T15448 with 80% w/v ammonium sulphate. The resuspended precipitate was dialysed against 5 mM MES pH=6.0 and purified by affinity chromatography on SP-sephadex column eluted with a 5-250 mM MES, pH=6.0 stepwise gradient and dialysed extensively against PBS.
Susceptibility of ubiquitin components to SpeB was measured using a modification of the method described by Collin and Olsen (Collin and Olsen, 2000). Filtered 24 h culture supernatants were incubated for 30 min at 40°C with an equal volume of activation buffer (20 mM DTT in DPBS). Purified SpeB (644 nM final concentration following addition of HEp-2 cell lysate) was activated in activation buffer for 30 min at 40°C. After activation, an equal volume of HEp-2 cell lysate was added, and 10 μl was removed for a loading control. Reactions were incubated for 120 min at 37°C, and stopped by the addition of 1 × SDS-PAGE loading buffer and 100 mM DTT followed by heating at 95°C for 7 min. Samples were separated by SDS-PAGE, transferred to Immobilon-FL PVDF membranes and probed with specific antibodies. Blots were visualized using IRdye680LT and IRdye800CW-conjugated secondary antibodies (Li-Cor) and scanned on an Odyssey infra-red scanner (Li-Cor).
Ectopic SpeB expression
Human codon-optimised open reading frames encoding the mature form of SpeB and a catalytically-inactive SpeB-C192S derivative were purchased from Genscript as N-myc-tagged expression constructs in pcDNA3.1(+). HEp2 cells grown on glass cover slips were transfected with myc-SpeB or myc-SpeB-C192S. At 6 h post-transfection, cells were fixed in 4% PFA and permeabilised with 0.1% TritonX-100. SpeB was detected with anti-myc antisera; NDP52, p62 and NBR1 were detected with mouse monoclonal antibodies. Nuclei were stained with DAPI. Confocal z-stacks were acquired using inverted confocal microscope Zeiss LSM 710 (Plan-Apochromat 63×/1.4 Oil DIC objective) operated by ZEN2009 acquisition software. Intracellular structures positive for respective antigen were quantified using image segmentation and particle analysis in ImageJ 1.46a (Wayne Rasband, NIH, USA).
Transmission Electron Microscopy
HEp-2 cells cultured on glass coverslips were infected with GAS strains as described above at a multiplicity of infection = 8. At 6 h post-infection, the cells were fixed in glutaraldehyde, postfixed in osmium tetroxide, dehydrated in ethanol series, flat-embedded in EPON resin and polymerized at 60°C. Thin sections were cut on Leica EM UC6 microtome and contrasted with uranyl acetate and lead citrate. Images were acquired on Jeol EM 1011 electron microscope operating at 100kV.
Statistics
For statistical analysis, the mean ± SEM for three independent experiments is shown in figures unless otherwise stated, and p values were calculated using a one-tailed Student's t-test. A p value of less than 0.05 was determined to be statistically significant.
Supplementary Material
HIGHLIGHTS.
M1T1 Group A Streptococcus (GAS) evade autophagy and replicate intracellularly
M1T1 GAS evade ubiquitylation and host ubiquitin-LC3 adaptors NBR1, p62 and NDP52
M1T1 ΔspeB is targeted to autophagy and has reduced intracellular replication
GAS cysteine protease SpeB degrades ubiquitin-LC3 adaptors in vitro and in vivo
ACKNOWLEDGMENTS
Microscopy was carried out at the Australian Cancer Research Foundation (ACRF)/ Institute for Molecular Bioscience Dynamic Imaging Facility for Cancer Biology. Electron microscopy was carried out at The Centre for Microscopy and Microanalysis (the Queensland Node of the Australian Microscopy and Microanalysis Research Facility). We thank Ericka Anderson for additional experimental assistance. This work was supported by funding from the National Health and Medical Research Council (NHMRC) of Australia (1041294, 631386, 635250, 565526, 1041929, 606788) and the National Institutes of Health (NIH) of the United States of America.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes three figures and one table and can be found with this article online.
REFERENCES
- Banks DJ, Porcella SF, Barbian KD, Beres SB, Philips LE, Voyich JM, DeLeo FR, Martin JM, Somerville GA, Musser JM. Progress toward characterization of the group A Streptococcus metagenome: complete genome sequence of a macrolide-resistant serotype M6 strain. J. Infect. Dis. 2004;190:727–738. doi: 10.1086/422697. [DOI] [PubMed] [Google Scholar]
- Berge A, Bjorck L. Streptococcal cysteine proteinase releases biologically active fragments of streptococcal surface proteins. J. Biol. Chem. 1995;270:9862–9867. doi: 10.1074/jbc.270.17.9862. [DOI] [PubMed] [Google Scholar]
- Birmingham CL, Smith AC, Bakowski MA, Yoshimori T, Brumell JH. Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J. Biol. Chem. 2006;281:11374–11383. doi: 10.1074/jbc.M509157200. [DOI] [PubMed] [Google Scholar]
- Camilli A, Paynton CR, Portnoy DA. Intracellular methicillin selection of Listeria monocytogenes mutants unable to replicate in a macrophage cell line. Proc. Natl. Acad. Sci. USA. 1989;86:5522–5526. doi: 10.1073/pnas.86.14.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect. Dis. 2005;5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- Chong A, Wehrly TD, Child R, Hansen B, Hwang S, Virgin HW, Celli J. Cytosolic clearance of replication-deficient mutants reveals Francisella tularensis interactions with the autophagic pathway. Autophagy. 2012;8:1342–1356. doi: 10.4161/auto.20808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JN, Aquilina JA, Hains PG, Henningham A, Sriprakash KS, Caparon MG, Nizet V, Kotb M, Cordwell SJ, Djordjevic SP, et al. Role of group A Streptococcus HtrA in the maturation of SpeB protease. Proteomics. 2007;7:4488–4498. doi: 10.1002/pmic.200700626. [DOI] [PubMed] [Google Scholar]
- Cole JN, Barnett TC, Nizet V, Walker MJ. Molecular insight into invasive group A streptococcal disease. Nat. Rev. Microbiol. 2011;9:724–736. doi: 10.1038/nrmicro2648. [DOI] [PubMed] [Google Scholar]
- Cole JN, McArthur JD, McKay FC, Sanderson-Smith ML, Cork AJ, Ranson M, Rohde M, Itzek A, Sun H, Ginsburg D, et al. Trigger for group A streptococcal M1T1 invasive disease. FASEB J. 2006;20:1745–1747. doi: 10.1096/fj.06-5804fje. [DOI] [PubMed] [Google Scholar]
- Collin M, Olsen A. Generation of a mature streptococcal cysteine proteinase is dependent on cell wall-anchored M1 protein. Mol. Microbiol. 2000;36:1306–1318. doi: 10.1046/j.1365-2958.2000.01942.x. [DOI] [PubMed] [Google Scholar]
- Deretic V. Autophagy in infection. Curr. Opin. Cell Biol. 2010;22:252–262. doi: 10.1016/j.ceb.2009.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortet L, Mostowy S, Louaka AS, Gouin E, Nahori MA, Wiemer EA, Dussurget O, Cossart P. Recruitment of the major vault protein by InlK: a Listeria monocytogenes strategy to avoid autophagy. PLoS Pathog. 2011;7:e1002168. doi: 10.1371/journal.ppat.1002168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Brumell JH. Autophagy in immunity against intracellular bacteria. Curr. Top. Microbiol. Immunol. 2009;335:189–215. doi: 10.1007/978-3-642-00302-8_9. [DOI] [PubMed] [Google Scholar]
- Hytonen J, Haataja S, Gerlach D, Podbielski A, Finne J. The SpeB virulence factor of Streptococcus pyogenes, a multifunctional secreted and cell surface molecule with strepadhesin, laminin-binding and cysteine protease activity. Mol. Microbiol. 2001;39:512–519. doi: 10.1046/j.1365-2958.2001.02269.x. [DOI] [PubMed] [Google Scholar]
- Ikebe T, Ato M, Matsumura T, Hasegawa H, Sata T, Kobayashi K, Watanabe H. Highly frequent mutations in negative regulators of multiple virulence genes in group A streptococcal toxic shock syndrome isolates. PLoS Pathog. 2010;6:e1000832. doi: 10.1371/journal.ppat.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson L, Thulin P, Sendi P, Hertzen E, Linder A, Akesson P, Low DE, Agerberth B, Norrby-Teglund A. Cathelicidin LL-37 in severe Streptococcus pyogenes soft tissue infections in humans. Infect. Immun. 2008;76:3399–3404. doi: 10.1128/IAI.01392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubert PE, Meiffren G, Gregoire IP, Pontini G, Richetta C, Flacher M, Azocar O, Vidalain PO, Vidal M, Lotteau V, et al. Autophagy induction by the pathogen receptor CD46. Cell Host Microbe. 2009;6:354–366. doi: 10.1016/j.chom.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Kansal RG, Nizet V, Jeng A, Chuang WJ, Kotb M. Selective modulation of superantigen-induced responses by streptococcal cysteine protease. J. Infect. Dis. 2003;187:398–407. doi: 10.1086/368022. [DOI] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol. Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- Korotkova N, Hoff JS, Becker DM, Quinn JK, Icenogle LM, Moseley SL. SpyA is a membrane-bound ADP-ribosyltransferase of Streptococcus pyogenes which modifies a streptococcal peptide, SpyB. Mol. Microbiol. 2012;83:936–952. doi: 10.1111/j.1365-2958.2012.07979.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon WR, Madden JC, Levin JC, Stein JL, Caparon MG. Mutation of luxS affects growth and virulence factor expression in Streptococcus pyogenes. Mol. Microbiol. 2001;42:145–157. doi: 10.1046/j.1365-2958.2001.02616.x. [DOI] [PubMed] [Google Scholar]
- Maamary PG, Ben Zakour NL, Cole JN, Hollands A, Aziz RK, Barnett TC, Cork AJ, Henningham A, Sanderson-Smith M, McArthur JD, et al. Tracing the evolutionary history of the pandemic group A streptococcal M1T1 clone. FASEB J. 2012;26:4675–4684. doi: 10.1096/fj.12-212142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay FC, McArthur JD, Sanderson-Smith ML, Gardam S, Currie BJ, Sriprakash KS, Fagan PK, Towers RJ, Batzloff MR, Chhatwal GS, et al. Plasminogen binding by group A streptococcal isolates from a region of hyperendemicity for streptococcal skin infection and a high incidence of invasive infection. Infect. Immun. 2004;72:364–370. doi: 10.1128/IAI.72.1.364-370.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S, Sancho-Shimizu V, Hamon MA, Simeone R, Brosch R, Johansen T, Cossart P. p62 and NDP52 proteins target intracytosolic Shigella and Listeria to different autophagy pathways. J. Biol. Chem. 2011;286:26987–26995. doi: 10.1074/jbc.M111.223610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, et al. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- Nelson DC, Garbe J, Collin M. Cysteine proteinase SpeB from Streptococcus pyogenes - a potent modifier of immunologically important host and bacterial proteins. Biol. Chem. 2011;392:1077–1088. doi: 10.1515/BC.2011.208. [DOI] [PubMed] [Google Scholar]
- Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- Renard C, Vanderhaeghe HJ, Claes PJ, Zenebergh A, Tulkens PM. Influence of conversion of penicillin G into a basic derivative on its accumulation and subcellular localization in cultured macrophages. Antimicrob. Agents Chemother. 1987;31:410–416. doi: 10.1128/aac.31.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai A, Maruyama F, Funao J, Nozawa T, Aikawa C, Okahashi N, Shintani S, Hamada S, Ooshima T, Nakagawa I. Specific behavior of intracellular Streptococcus pyogenes that has undergone autophagic degradation is associated with bacterial streptolysin O and host small G proteins Rab5 and Rab7. J. Biol. Chem. 2010;285:22666–22675. doi: 10.1074/jbc.M109.100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaith A, Kashkar H, Leggio SA, Addicks K, Kronke M, Krut O. Staphylococcus aureus subvert autophagy for induction of caspase-independent host cell death. J. Biol. Chem. 2007;282:2695–2706. doi: 10.1074/jbc.M609784200. [DOI] [PubMed] [Google Scholar]
- Shulman ST, Tanz RR, Dale JB, Beall B, Kabat W, Kabat K, Cederlund E, Patel D, Rippe J, Li Z, et al. Seven-year surveillance of north american pediatric group A streptococcal pharyngitis isolates. Clin. Infect. Dis. 2009;49:78–84. doi: 10.1086/599344. [DOI] [PubMed] [Google Scholar]
- Steer AC, Law I, Matatolu L, Beall BW, Carapetis JR. Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. Lancet Infect. Dis. 2009;9:611–616. doi: 10.1016/S1473-3099(09)70178-1. [DOI] [PubMed] [Google Scholar]
- Stockbauer KE, Magoun L, Liu M, Burns EH, Jr., Gubba S, Renish S, Pan X, Bodary SC, Baker E, Coburn J, et al. A natural variant of the cysteine protease virulence factor of group A Streptococcus with an arginine-glycine-aspartic acid (RGD) motif preferentially binds human integrins alphavbeta3 and alphaIIbbeta3. Proc. Natl. Acad. Sci. USA. 1999;96:242–247. doi: 10.1073/pnas.96.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston TL, Ryzhakov G, Bloor S, von Muhlinen N, Randow F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 2009;10:1215–1221. doi: 10.1038/ni.1800. [DOI] [PubMed] [Google Scholar]
- Thurston TL, Wandel MP, von Muhlinen N, Foeglein A, Randow F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature. 2012;482:414–418. doi: 10.1038/nature10744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse H, Bao JY, Davies MR, Maamary P, Tsoi HW, Tong AH, Ho TC, Lin CH, Gillen CM, Barnett TC, et al. Molecular characterization of the 2011 Hong Kong scarlet fever outbreak. J. Infect. Dis. 2012;206:341–351. doi: 10.1093/infdis/jis362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild P, Farhan H, McEwan DG, Wagner S, Rogov VV, Brady NR, Richter B, Korac J, Waidmann O, Choudhary C, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science. 2011;333:228–233. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa Y, Ogawa M, Hain T, Yoshida M, Fukumatsu M, Kim M, Mimuro H, Nakagawa I, Yanagawa T, Ishii T, et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat. Cell Biol. 2009;11:1233–1240. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- Zheng YT, Shahnazari S, Brech A, Lamark T, Johansen T, Brumell JH. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J. Immunol. 2009;183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.