Abstract
Flexible regions of proteins play an important role in catalysis, ligand binding, and macromolecular interactions. Because of its enhanced sensitivity to motional narrowing, two-dimensional coupling constant J-correlated 1H NMR may be used to observe these regions selectively. Dynamic filtering is an intrinsic feature of this experiment because cross-peak amplitude decays rapidly as linewidths approach the coupling constant. We demonstrate here the flexibility of the NH2-terminal arm of phage lambda repressor, which is thought to wrap around the double helix in the repressor-operator complex. The assignment of arm resonances is made possible by the construction of mutant repressor genes containing successive NH2-terminal deletions.
Full text
PDF
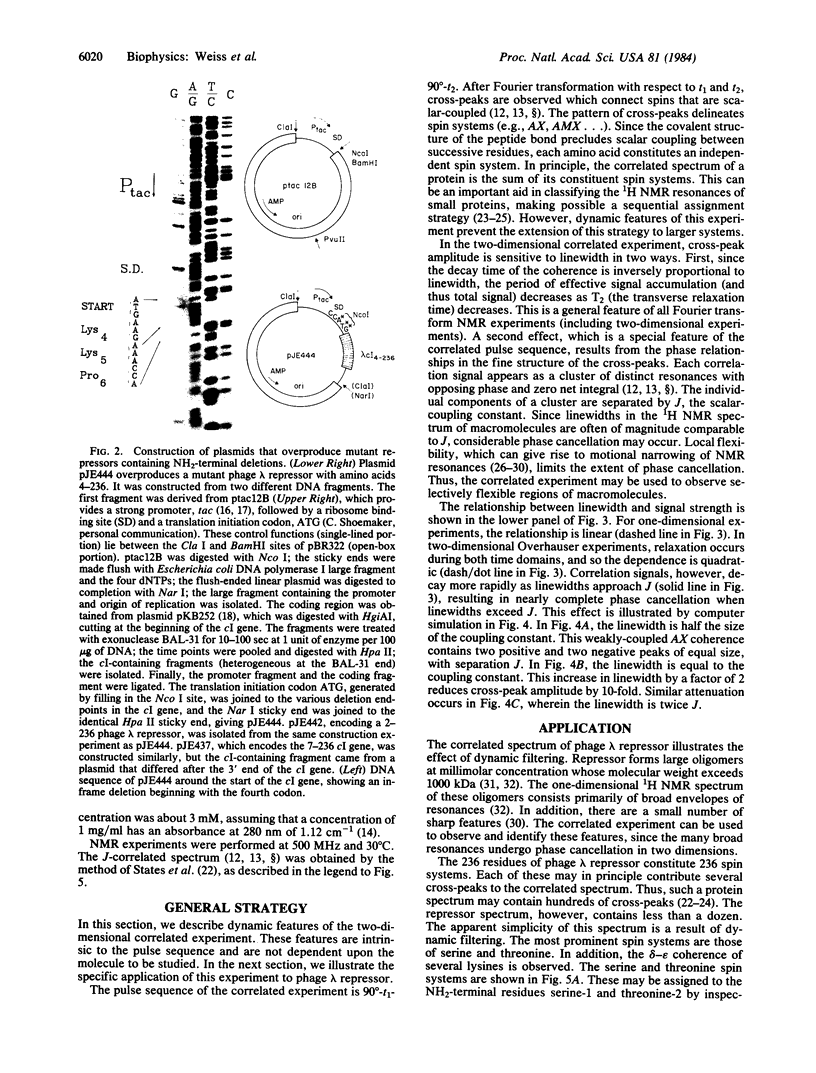
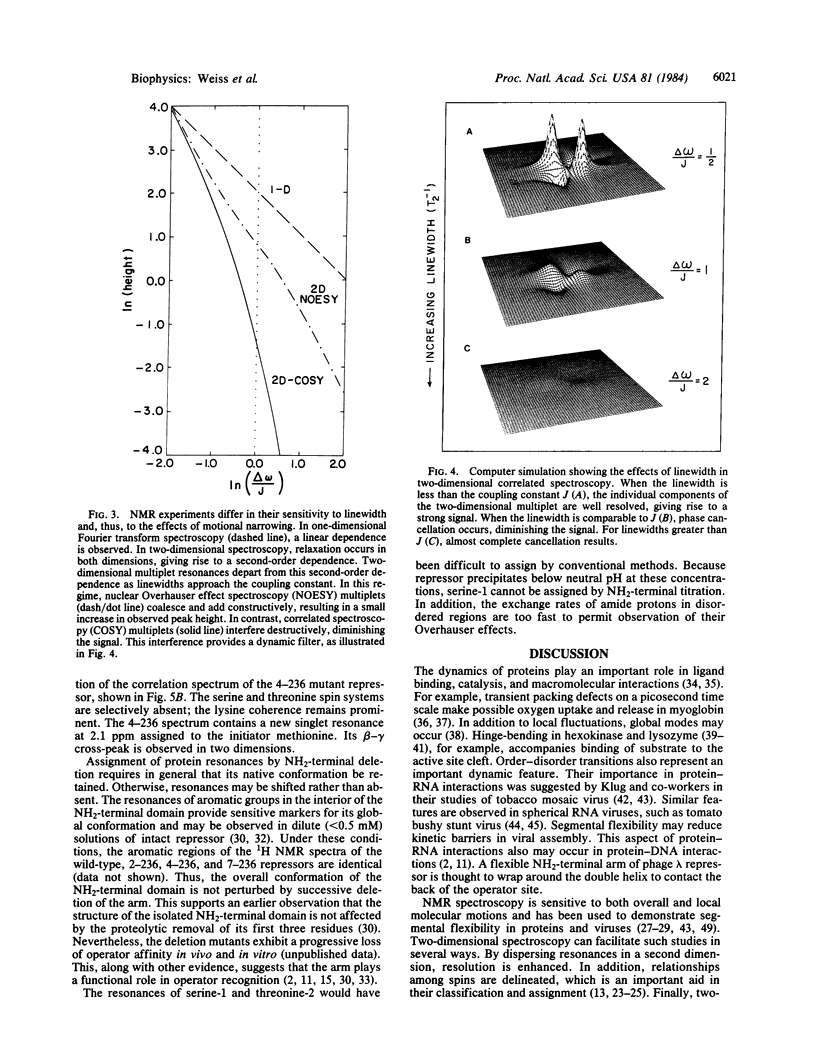
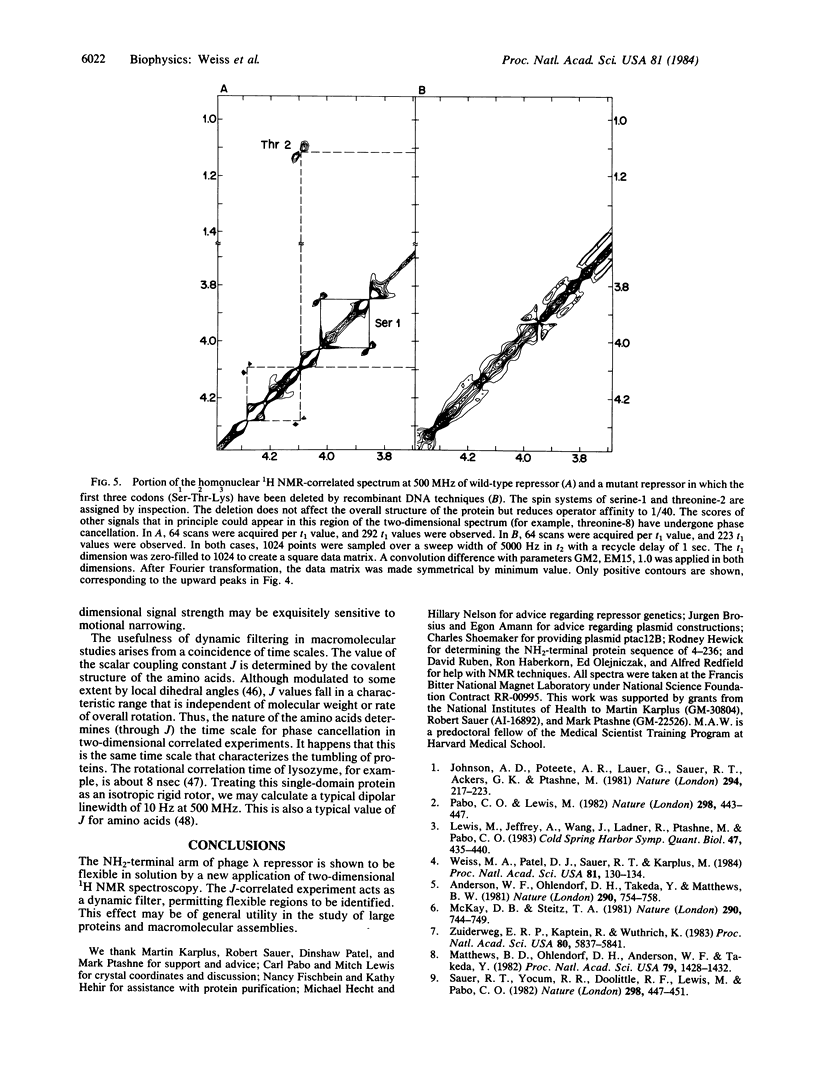

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amann E., Brosius J., Ptashne M. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene. 1983 Nov;25(2-3):167–178. doi: 10.1016/0378-1119(83)90222-6. [DOI] [PubMed] [Google Scholar]
- Anderson C. M., Zucker F. H., Steitz T. A. Space-filling models of kinase clefts and conformation changes. Science. 1979 Apr 27;204(4391):375–380. doi: 10.1126/science.220706. [DOI] [PubMed] [Google Scholar]
- Anderson W. F., Ohlendorf D. H., Takeda Y., Matthews B. W. Structure of the cro repressor from bacteriophage lambda and its interaction with DNA. Nature. 1981 Apr 30;290(5809):754–758. doi: 10.1038/290754a0. [DOI] [PubMed] [Google Scholar]
- Backman K., Ptashne M., Gilbert W. Construction of plasmids carrying the cI gene of bacteriophage lambda. Proc Natl Acad Sci U S A. 1976 Nov;73(11):4174–4178. doi: 10.1073/pnas.73.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G., Dobberstein B. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol. 1975 Dec;67(3):835–851. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boublik M., Bradbury E. M., Crane-Robinson C., Rattle H. W. Proton magnetic resonance ttudies of the interactions of histones F1 and F2B with DNA. Nat New Biol. 1971 Feb 3;229(5):149–150. doi: 10.1038/newbio229149a0. [DOI] [PubMed] [Google Scholar]
- Brooks B., Karplus M. Harmonic dynamics of proteins: normal modes and fluctuations in bovine pancreatic trypsin inhibitor. Proc Natl Acad Sci U S A. 1983 Nov;80(21):6571–6575. doi: 10.1073/pnas.80.21.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell I. D., Dobson C. M., Williams R. J., Wright P. E. Pulse methods for the simplification of protein NMR spectra. FEBS Lett. 1975 Sep 1;57(1):96–99. doi: 10.1016/0014-5793(75)80160-8. [DOI] [PubMed] [Google Scholar]
- Case D. A., Karplus M. Dynamics of ligand binding to heme proteins. J Mol Biol. 1979 Aug 15;132(3):343–368. doi: 10.1016/0022-2836(79)90265-1. [DOI] [PubMed] [Google Scholar]
- Harrison S. C. Protein interfaces and intersubunit bonding. The case of tomato bushy stunt virus. Biophys J. 1980 Oct;32(1):139–153. doi: 10.1016/S0006-3495(80)84930-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht M. H., Nelson H. C., Sauer R. T. Mutations in lambda repressor's amino-terminal domain: implications for protein stability and DNA binding. Proc Natl Acad Sci U S A. 1983 May;80(9):2676–2680. doi: 10.1073/pnas.80.9.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewick R. M., Hunkapiller M. W., Hood L. E., Dreyer W. J. A gas-liquid solid phase peptide and protein sequenator. J Biol Chem. 1981 Aug 10;256(15):7990–7997. [PubMed] [Google Scholar]
- Jardetzky O., Akasaka K., Vogel D., Morris S., Holmes K. C. Unusual segmental flexibility in a region of tobacco mosaic virus coat protein. Nature. 1978 Jun 15;273(5663):564–566. doi: 10.1038/273564a0. [DOI] [PubMed] [Google Scholar]
- Johnson A. D., Pabo C. O., Sauer R. T. Bacteriophage lambda repressor and cro protein: interactions with operator DNA. Methods Enzymol. 1980;65(1):839–856. doi: 10.1016/s0076-6879(80)65078-2. [DOI] [PubMed] [Google Scholar]
- Johnson A. D., Poteete A. R., Lauer G., Sauer R. T., Ackers G. K., Ptashne M. lambda Repressor and cro--components of an efficient molecular switch. Nature. 1981 Nov 19;294(5838):217–223. doi: 10.1038/294217a0. [DOI] [PubMed] [Google Scholar]
- Karplus M., McCammon J. A. The internal dynamics of globular proteins. CRC Crit Rev Biochem. 1981;9(4):293–349. doi: 10.3109/10409238109105437. [DOI] [PubMed] [Google Scholar]
- Klug A. The assembly of tobacco mosaic virus: structure and specificity. Harvey Lect. 1980;74:141–172. [PubMed] [Google Scholar]
- Kurachi K., Sieker L. C., Jensen L. H. Structures of triclinic mono- and di-N-acetylglucosamine: lysozyme complexes--a crystallographic study. J Mol Biol. 1976 Feb 15;101(1):11–24. doi: 10.1016/0022-2836(76)90063-2. [DOI] [PubMed] [Google Scholar]
- Lewis M., Jeffrey A., Wang J., Ladner R., Ptashne M., Pabo C. O. Structure of the operator-binding domain of bacteriophage lambda repressor: implications for DNA recognition and gene regulation. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):435–440. doi: 10.1101/sqb.1983.047.01.051. [DOI] [PubMed] [Google Scholar]
- Lingappa V. R., Katz F. N., Lodish H. F., Blobel G. A signal sequence for the insertion of a transmembrane glycoprotein. Similarities to the signals of secretory proteins in primary structure and function. J Biol Chem. 1978 Dec 25;253(24):8667–8670. [PubMed] [Google Scholar]
- Matthews B. W., Ohlendorf D. H., Anderson W. F., Takeda Y. Structure of the DNA-binding region of lac repressor inferred from its homology with cro repressor. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1428–1432. doi: 10.1073/pnas.79.5.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McCammon J. A., Gelin B. R., Karplus M. Dynamics of folded proteins. Nature. 1977 Jun 16;267(5612):585–590. doi: 10.1038/267585a0. [DOI] [PubMed] [Google Scholar]
- McKay D. B., Steitz T. A. Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature. 1981 Apr 30;290(5809):744–749. doi: 10.1038/290744a0. [DOI] [PubMed] [Google Scholar]
- Nagayama K. Two-dimensional NMR spectroscopy: an application to the study of flexibility of protein molecules. Adv Biophys. 1981;14:139–204. [PubMed] [Google Scholar]
- Nelson H. C., Hecht M. H., Sauer R. T. Mutations defining the operator-binding sites of bacteriophage lambda repressor. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):441–449. doi: 10.1101/sqb.1983.047.01.052. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Krovatin W., Jeffrey A., Sauer R. T. The N-terminal arms of lambda repressor wrap around the operator DNA. Nature. 1982 Jul 29;298(5873):441–443. doi: 10.1038/298441a0. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Lewis M. The operator-binding domain of lambda repressor: structure and DNA recognition. Nature. 1982 Jul 29;298(5873):443–447. doi: 10.1038/298443a0. [DOI] [PubMed] [Google Scholar]
- Perutz M. F. Stereochemistry of cooperative effects in haemoglobin. Nature. 1970 Nov 21;228(5273):726–739. doi: 10.1038/228726a0. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Anderegg R. Primary structure of the lambda repressor. Biochemistry. 1978 Mar 21;17(6):1092–1100. doi: 10.1021/bi00599a024. [DOI] [PubMed] [Google Scholar]
- Sauer R. T., Yocum R. R., Doolittle R. F., Lewis M., Pabo C. O. Homology among DNA-binding proteins suggests use of a conserved super-secondary structure. Nature. 1982 Jul 29;298(5873):447–451. doi: 10.1038/298447a0. [DOI] [PubMed] [Google Scholar]
- States D. J., Dobson C. M., Karplus M. A new two-disulphide intermediate in the refolding of reduced bovine pancreatic trypsin inhibitor. J Mol Biol. 1984 Apr 5;174(2):411–418. doi: 10.1016/0022-2836(84)90345-0. [DOI] [PubMed] [Google Scholar]
- Wagner G., Wüthrich K. Sequential resonance assignments in protein 1H nuclear magnetic resonance spectra. Basic pancreatic trypsin inhibitor. J Mol Biol. 1982 Mar 5;155(3):347–366. doi: 10.1016/0022-2836(82)90009-2. [DOI] [PubMed] [Google Scholar]
- Weber I. T., McKay D. B., Steitz T. A. Two helix DNA binding motif of CAP found in lac repressor and gal repressor. Nucleic Acids Res. 1982 Aug 25;10(16):5085–5102. doi: 10.1093/nar/10.16.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M. A., Karplus M., Patel D. J., Sauer R. T. Solution NMR studies of intact lambda repressor. J Biomol Struct Dyn. 1983 Oct;1(1):151–157. doi: 10.1080/07391102.1983.10507431. [DOI] [PubMed] [Google Scholar]
- Weiss M. A., Patel D. J., Sauer R. T., Karplus M. Two-dimensional 1H NMR study of the lambda operator site OL1: a sequential assignment strategy and its application. Proc Natl Acad Sci U S A. 1984 Jan;81(1):130–134. doi: 10.1073/pnas.81.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüthrich K., Wider G., Wagner G., Braun W. Sequential resonance assignments as a basis for determination of spatial protein structures by high resolution proton nuclear magnetic resonance. J Mol Biol. 1982 Mar 5;155(3):311–319. doi: 10.1016/0022-2836(82)90007-9. [DOI] [PubMed] [Google Scholar]
- Zuiderweg E. R., Kaptein R., Wüthrich K. Secondary structure of the lac repressor DNA-binding domain by two-dimensional 1H nuclear magnetic resonance in solution. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5837–5841. doi: 10.1073/pnas.80.19.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer H. A., Comstock L. J., Vasser M. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc Natl Acad Sci U S A. 1983 Jan;80(1):21–25. doi: 10.1073/pnas.80.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]