SUMMARY
The spatiotemporal organization and dynamics of chromatin play critical roles in regulating genome function. However, visualizing specific, endogenous genomic loci remains challenging in living cells. Here, we demonstrate such an imaging technique by repurposing the bacterial CRISPR/Cas system. Using an EGFP-tagged endonuclease-deficient Cas9 protein and a structurally optimized small guide (sg) RNA, we show robust imaging of repetitive elements in telomeres and coding genes in living cells. Furthermore, an array of sgRNAs tiling along the target locus enables the visualization of non-repetitive genomic sequences. Using this method, we have studied telomere dynamics during elongation or disruption, the subnuclear localization of the MUC4 loci, the cohesion of replicated MUC4 loci on sister chromatids, and their dynamic behaviors during mitosis. This CRISPR imaging tool has potential to significantly improve the capacity to study the conformation and dynamics of native chromosomes in living human cells.
INTRODUCTION
The functional output of human genome is determined by its spatial organization and dynamic interactions with protein and RNA regulators. For example, the subnuclear positioning of genomic elements can modulate gene expression, heterochromatin formation, and cell replication (Misteli, 2007; Misteli, 2013). To elucidate the mechanisms that relate genome function to its spatiotemporal organization, a method to image specific DNA sequences in living cells would be indispensable. So far, such studies have mostly relied on fluorescently tagged DNA-binding proteins. However, because of their fixed target sequence and limited choices of native DNA-binding proteins, this approach has been restricted to imaging artificial repetitive sequences inserted into the genome (Robinett et al., 1996) or specialized genomic elements such as the telomeres (Wang et al., 2008), centromeres (Hellwig et al., 2008) and, in bacteria, H-NS binding loci (Wang et al., 2011). Imaging arbitrary, endogenous genes and genomic loci remains challenging. Although fluorescence in situ hybridization (FISH) (Langer-Safer et al., 1982; Lichter et al., 1990) brings in target sequence flexibility through base paring of the nucleic acid probes, it is incompatible with live imaging due to sample fixation and DNA denaturation. Thus, we sought to develop a genome imaging technique that combines the flexibility of nucleic acid probes and the live imaging capability of DNA-binding proteins.
The type II CRISPR (clustered regularly interspaced short palindromic repeats) system derived from Streptococcus pyogenes (Barrangou et al., 2007; Deltcheva et al., 2011; Wiedenheft et al., 2012) provides a promising platform to accomplish this goal. CRISPR uses a Cas9 protein to recognize DNA sequences, with target specificity solely determined by a small guide (sg) RNA and a protospacer adjacent motif (PAM) (Jinek et al., 2012). Upon binding to target DNA, the Cas9-sgRNA complex generates a DNA double-stranded break. Harnessing this RNA-guided nuclease activity, recent work has demonstrated that CRISPR can be repurposed to edit the genomes of a broad range of organisms (Cong et al., 2013; Mali et al., 2013; Wang et al., 2013). Furthermore, a repurposed, nuclease-deactivated Cas9 (dCas9) protein has been used to regulate endogenous gene expression by controlling the RNA polymerase activity or by modulating promoter accessibility when fused with transcription factors (Gilbert et al., 2013; Qi et al., 2013). Going beyond gene editing and regulation, we sought to use the CRISPR system as a universal and flexible platform for the dynamic imaging of specific genomic elements in living mammalian cells.
Here we report a CRISPR-based technique for sequence-specific visualization of genomic elements in living human cells. Our imaging system consists of an EGFP-tagged, endonuclease-deactivated dCas9 protein and a structurally optimized sgRNA that improves its interaction with the dCas9 protein. We show that this optimized CRISPR system enables robust imaging of repetitive elements in both telomeres and protein-coding genes such as the Mucin genes in human cells. Furthermore, we use multiple sgRNAs to tile along the target locus to visualize non-repetitive genomic sequences in the human genome. This CRISPR imaging method allows easy and reliable tracking of the telomere dynamics during telomere elongation or disruption, and enables us to observe chromatin organization and dynamics throughout the cell cycle. The CRISPR technology offers a complementary approach to FISH or the use of DNA-binding proteins for imaging, providing a general platform for the study of native chromatin organization and dynamics in living human cells.
RESULTS
An optimized CRISPR system enables visualization of telomeres and enhances gene regulation
To engineer the CRISPR system for imaging endogenous genomic sequences, we fused a dCas9 protein lacking the endonucleolytic activity to an enhanced green fluorescent protein (EGFP). Co-expression of dCas9-EGFP and sequence-specific sgRNAs should allow the enrichment of fluorescent signal at the targeted genomic loci for imaging (Figure 1A). To better target the dCas9-EGFP protein into the nucleus, we tested different dCas9 and EGFP fusions carrying two copies of nuclear localization signal (NLS) sequences (Figure S1A). A fully nuclear-localized version (# 4) was selected (see Extended Experimental Procedures for the dCas9-EGFP sequence). We then created clonal RPE, HeLa and UMUC3 cell lines that stably expressed dCas9-EGFP from an inducible Tet-On 3G system using lentiviral vectors (Figure 1B). To reduce the background fluorescence that arises from unbound dCas9-EGFP, we performed subsequent imaging experiments at the basal level of dCas9-EGFP expression without doxycycline induction (Figure S1B).
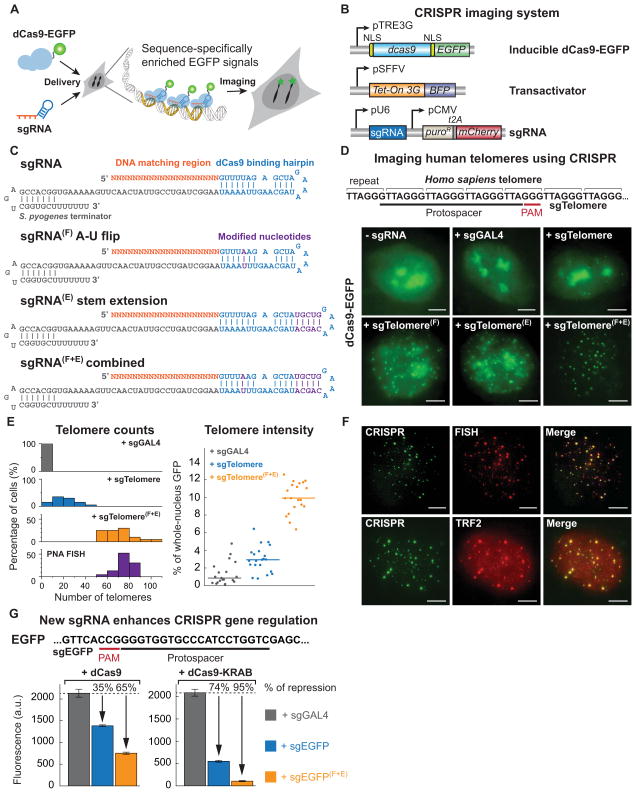
Figure 1. An optimized CRISPR/Cas system for visualizing genomic sequences in living mammalian cells.
(A) Overview of CRISPR imaging. Sequence-specific enrichment of fluorescence signals by sgRNA-directed dCas9-EGFP allows the imaging of genomic elements in living cells. (B) The three components of the CRISPR imaging: a doxycycline-inducible dCas9-EGFP fusion protein, a Tet-on 3G transactivator, and target-specific sgRNAs expressed from a murine U6 promoter. (C) Optimized sgRNA designs. sgRNA(F), A-U pair flip; sgRNA(E), a 5-bp extension of the hairpin; sgRNA(F+E), combination of both modifications. Target basepairing region (orange), dCas9 binding hairpin (blue), the S. pyogenes-derived terminator (grey), and nucleotide modifications (purple) are shown. (D) CRISPR imaging of human telomeres in RPE cells using different sgRNA designs. The sgRNA target sequence (black line) and the PAM (red line) are shown. sgGAL4 is used as the negative control. (E) Histograms of telomere counts and telomere intensity (measured as % of whole-nucleus GFP) in single cell using sgTelomere and sgTelomere(F+E). The telomere number detected by PNA FISH is also shown. n = 20. (F) Colabeling of telomeres using dCas9-EGFP (green) & PNA FISH (top, red), or dCas9-EGFP & antibody to TRF2 (bottom, red). (G) Optimized sgRNA design improves gene regulation efficiency using dCas9 alone (left) or dCas9-KRAB fusion protein (right). The target sequence (black line) and the PAM (red line) are shown. The data are displayed as mean ± standard deviation for 3 independent experiments. All scale bars: 5 μm. See also Figure S1.
We started by imaging human telomeres, specialized chromatin structures composed of 5 to 15 kilo base pair (kb) tracts of TTAGGG repeats and associated proteins (Moyzis et al., 1988). Such repeats allow the recruitment of multiple dCas9-EGFP proteins to the same locus using a single sgRNA sequence. Following a previously reported sgRNA design (Jinek et al., 2012; Qi et al., 2013), we created an sgRNA (sgTelomere) containing a 22 nt telomere targeting sequence (Figure 1C and 1D; see Extended Experimental Procedures for sgRNA sequences). We infected stable dCas9-EGFP RPE cells with a lentivirus that expressed sgTelomere from a murine polymerase III U6 promoter (Figure 1B). An sgRNA that had no cognate target in the human genome (sgGAL4) was used as the negative control. At 48 h post-infection, about 80% sgTelomere-expressing cells showed fluorescent puncta resembling telomeres in addition to bright regions resembling nucleoli. In contrast, sgGAL4-expressing cells only contained nucleolar signal (Figure 1D). Nevertheless, the observed number of telomere puncta, typically 10 to 40 per cell, was substantially lower than the expected telomere number in human cells (146 for our RPE cells, see later karyotyping results), indicating that the system was sub-optimal.
Previous work has indicated that the sgRNA expression level limits CRISPR/Cas9 function in human cells (Jinek et al., 2013). Indeed, the observed nucleolus-like signal possibly came from dCas9 proteins that were not bound to sgRNA. Therefore, we modified the sgRNA design to increase its stability and to enhance its assembly with the dCas9 protein (Figures 1C and S1C). To avoid premature termination of U6 Pol-III transcription, we removed a putative Pol-III terminator (4 consecutive U’s) in the sgRNA stem-loop by an A-U basepair flip (Figures 1C, sgRNA(F)) (Nielsen et al., 2013). To improve sgRNA-dCas9 assembly, we extended the dCas9-binding hairpin structure (Figures 1C, sgRNA(E)). Both sgRNA designs produced increased puncta numbers as well as decreased background and nucleolar signals. Further enhanced imaging efficiency was achieved by combining the A-U flip and hairpin extension (Figures 1C and 1D, sgRNA(F+E)), which increased the observable puncta number by 2-fold and the signal-to-background intensity ratio by 5-fold (Figure 1E).
To verify the specificity and efficiency of telomere imaging by CRISPR, we performed two-color imaging with telomere-specific FISH using Cy5-tagged peptide nucleic acid (PNA) probes, or with immunofluorescence for endogenous TRF2, a protein in the shelterin complex that binds to the telomeric DNA repeats (Griffith et al., 1999) (Figure 1F). For brighter CRISPR puncta (top 1/3) we observed nearly perfect (95%) colocalization with either PNA or TRF2 puncta. However, we had to use a modified PNA FISH protocol to preserve dCas9-EGFP signal (see Extended Experimental Procedures), which resulted in reduced efficiency for PNA to detect shorter telomeres. The relatively high background of TRF2 immunofluorescence also hindered short telomere detection. Therefore, we measured the total telomere number identified by CRISPR or standard PNA FISH to compare the labeling efficiencies. The two numbers perfectly matched (Figure 1E), indicating a similar efficiency for CRISPR and PNA FISH. In addition, the finding that no puncta were detected in the negative sgGAL4 control suggests a minimal off-target effect for CRISPR imaging.
Interestingly, this optimized sgRNA design also greatly enhanced gene regulation by CRISPR interference (CRISPRi) (Qi et al., 2013). We observed that the new sgRNA(F+E) design significantly improved transcriptional repression of a genomic EGFP reporter in HEK293 cells that stably expressed dCas9 or dCas9-KRAB (Figure 1G). We also tested additional sgRNA designs which consisted of a polymerase-III SINE poly-adenylation signal sequence at the 3′ end, alternative A-U flips, or alternative ways of hairpin extension (Figure S1B). No further improvement of transcriptional repression was observed comparing to the three designs described above (Figure S1D). Thus, the optimized sgRNA(F+E) design improves efficiencies for both imaging and gene regulation.
CRISPR imaging allows visualization of repetitive sequences in endogenous protein-coding genes
To expand the applications of CRISPR imaging, we tested whether we could use CRISPR to image protein-coding genes. Specifically, we chose the MUC4 gene on chromosome 3 that encodes a glycoprotein important for protecting mucus in diverse epithelial tissues and during tumor formation (Hollingsworth and Swanson, 2004). The MUC4 gene contains a region with variable number tandem repeats (100 to 400 repeats of a 48-bp sequence) in the second exon (Figure 2A) (Nollet et al., 1998). To image the MUC4 exon, we designed three sgRNAs (See Extended Experimental Procedures for sgRNA sequences) targeting this repetitive region (sgMUC4-E1, E2, E3). We observed that the labeling efficiency depended on the target site. The best one, sgMUC4-E3, showed 2 or more puncta in all cells even when the original sgRNA design was used (Figures 2B and 2C). We confirmed the specificity of the CRISPR signal by colabeling with oligo-DNA FISH of the same repetitive region in fixed cells (Figure 2D, see Extended Experimental Procedures for DNA-FISH probe sequence).
Figure 2. CRISPR imaging of endogenous genes in different human cell lines.
(A) Schematic of the human MUC4 gene showing two repeated regions in exon 2 (blue) and intron 3 (yellow). The target sequence (black line) and the PAM (red line) are shown. (B) CRISPR labeling of MUC4 loci (arrows) in RPE cells by targeting the exon 2 repeats or the intron 3 repeats with different sgRNAs. The arrow pairs in the bottom right image indicate replicated MUC4 loci. (C) Histograms of MUC4 loci counts by CRISPR labeling (n = 20). (D) Colocalization of dCas9-EGFP labeling (green) and Oligo DNA FISH labeling (red) for MUC4. (E) CRISPR imaging of MUC4 and telomeres in HeLa cells. sgGAL4 is used as the negative control. (F) CRISPR labeling of the MUC1 loci (arrows) in RPE cells. Schematic of the human MUC1 gene shows the repeat region in exon 3 and intron 3. The target sequence (black line) and the PAM (red line) are shown. All scale bars: 5 μm. See also Figures S2 and S3.
The MUC4 gene also contains ~90 repeats of a 15 bp sequence in the third intron. For this tract, we designed two sgRNAs with different lengths of complementarity (23 bp and 13 bp). In this case, the sgRNA(F+E) design was critical for visualizing the MUC4 intron (Figure 2B and 2C). Surprisingly, the 13 nt sgMUC4-I2(F+E) showed higher labeling efficiency, suggesting that its DNA-binding affinity might not be lower than that of the longer sgMUC4-I1(F+E), as both sgRNAs have a footprint of two 15-bp repeats with the 3 nt PAM included. This result possibly suggests a shorter sgRNA basepairing length requirement for imaging compared to that for efficient gene editing or gene regulation (Jinek et al., 2012; Qi et al., 2013).
We noticed that increasing the dosage of sgRNA lentivirus could reduce the nucleolar signal from dCas9-EGFP. For example, for both MUC4 and telomere labeling, infecting cells with 1:3-diluted lentivirus effectively reduced the nucleolar signal (Figure S2A). Furthermore, the optimized sgRNA(F+E) design not only allowed efficient labeling of target sites using a lower viral dosage (Figure S2B), but also decreased the nucleolar signal (Figure S2C). These observations further confirmed that sgRNA is a limiting factor for CRISPR imaging: low expression or suboptimal design potentially contributes to off-target clustering of dCas9-EGFP in nucleoli.
We saw 3 labeled MUC4 loci in the majority of cells by both exon and intron labeling. Indeed, whole-cell karyotyping revealed aneuploidy of our RPE cell line (Figure S3A), and chromosome 3 trisomy was further confirmed by FISH staining of two different regions on chromosome 3 (Figure S3B). Concomitantly, in ~ 15% of cells we also observed 6 CRISPR puncta with either MUC4 exon or intron labeling, suggesting that these cells had replicated these genomic loci (Figure 2B & C). These results demonstrated that CRISPR imaging is capable of detecting gene copy numbers in living cells.
To demonstrate the generality of CRISPR imaging in different cell types, we imaged telomeres and MUC4 in HeLa cells using sgTelomere(F+E) and sgMUC4-E3. In both cases, we observed effective labeling of the target genomic loci (Figure 2E). We similarly observed three copies of MUC4 loci in our HeLa cell line. FISH experiments confirmed that these cells were also trisomic for chromosome 3 (Figure S3B). To test the ability to image repetitive elements in other genes, we designed sgRNAs to visualize the MUC1 gene on chromosome 1 (Figure 2F) (Gendler et al., 1990). The MUC1 gene contains a polymorphic region with a variable number of 60-bp repeats in the third exon and intron, and the sgRNAs were designed to target within each repeat (sgMUC1-E1, E2, E3). We similarly observed multiple distinct MUC1 loci in RPE cells, and again observed that the labeling efficiency varied by target sequences.
To test whether CRISPR imaging affects gene expression, we performed qPCR to quantify MUC4 transcription in RPE cells labeled with sgMUC4-E1, sgMUC4-E3(F+E), sgMUC4-I2(F+E), or both sgMUC4-E3(F+E) and sgMUC4-I2(F+E) (Figure S3C). Only in the presence of both sgMUC4-E3(F+E) and sgMUC4-I2(F+E), a weak (~45%) repression was observed. This phenomenon is consistent with previous observations that targeting downstream sequences of the transcription start site is less effective for CRISPRi gene silencing (Qi et al., 2013). In contrast, labeling MUC1 using sgMUC1-E1(F+E) or sgMUC1-E3(F+E) both repressed MUC1 transcription by ~80% (Figure S3D), likely due to the fact that the target sites were close (~1 kb) to the transcription start site. Thus, although CRISPR imaging may perturb gene expression, this perturbation could be minimized by targeting the far downstream region or upstream region of the promoter (but not the enhancers).
CRISPR allows imaging of arbitrary non-repetitive genomic sequences
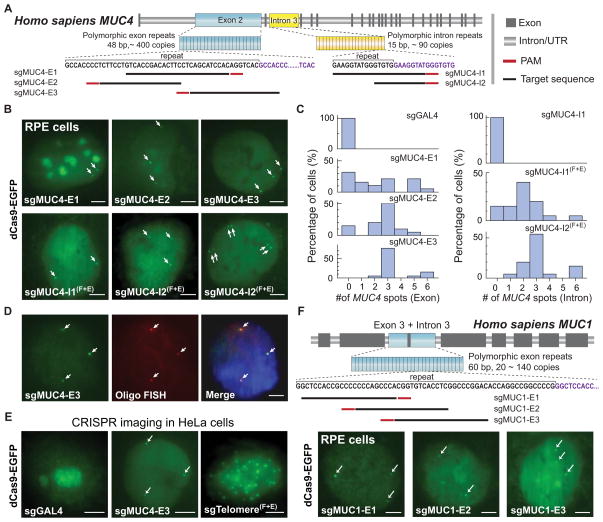
Most sequences in the human genome are non-repetitive. Unlike other DNA binding proteins, the target specificity of dCas9 is determined by sgRNA, which allows easy labeling of non-repetitive sequences by targeting multiple adjacent sites with a single dCas9 protein. To demonstrate this capability of CRISPR to image non-repetitive genomic loci, we designed 73 sgRNAs targeting both DNA strands spanning a 2 kb non-repetitive region in the first intron of MUC4 gene (Figure 3A, see Extended Experimental Procedures for target sequences). We produced lentiviral cocktails, each containing 5 to 6 sgRNAs, and infected different numbers of sgRNAs (16, 26, 36 or 73) into RPE cells. The total virus dosage was twice as much as used for repetitive sequence imaging. We observed effective labeling of the MUC4 loci using 36 sgRNAs. Increasing the number of sgRNAs to 73 did not improve the labeling efficiency, but reducing the sgRNA number to 16 resulted in no detectable puncta (Figure 3A and 3B). Our results suggest that 26 ~ 36 sgRNAs are sufficient to detect a non-repetitive genomic locus using CRISPR. Further reduction of the required number of sgRNAs could be implemented by decreasing the background level of dCas9-EGFP or improving the imaging sensitivity (Gaj et al., 2013).
Figure 3. CRISPR imaging of non-repetitive genomic sequences and multiple gene loci.
(A) CRISPR labeling of the non-repetitive region of MUC4 intron 1 using multiple optimized sgRNAs. With 26, 36 or 73 sgRNAs, 1 to 3 spots (arrows) can be detected. (B) Histograms of MUC4 loci counts by CRISPR imaging of the non-repetitive MUC4 sequence. (C) Colabeling of the MUC4 exon 2 and intron 3. The physical proximity (~1 kb) of the two target regions does not increase the puncta number as shown in the histograms. (D) Colabeling of MUC1 and MUC4 genes. Labeling two distal genes (MUC1 on chromosome 1 and MUC4 on chromosome 3 respectively) increases the puncta count as shown in the histograms (n = 20). All scale bars: 5 μm. See also Movie S3
We also used multiple sgRNAs to co-label either the same MUC4 gene or both MUC1 and MUC4 genes. Labeling the MUC4 gene using two sgRNAs, sgMUC4-E3 and sgMUC4-I2(F+E), did not result in more puncta compared to using sgMUC4-E3 alone (Figure 3C), which can be explained by the close proximity (~ 1 kb) of these two loci. In contrast, using both sgMUC4-E3 and sgMUC1-E1 led to 6 ~ 9 puncta observed in 45% of cells (Figure 3D). Unlike the MUC4-only images where the 6 puncta formed pairs, the MUC1+MUC4 images showed mostly unpaired spots. These results demonstrate the potential of using CRISPR for the simultaneous and multiplexing labeling of many genomic elements.
CRISPR imaging monitors telomere length
The ability of using CRISPR to label telomeres prompted us to test whether it allowed direct detection of the telomere length in living cells. Indeed, in two-color images of telomeres in RPE cells (Figure 1F), the intensity of individual telomere puncta detected using CRISPR and PNA FISH showed good linear correlation (Figure S4A). Linear correlation was also found between CRISPR labeling and TRF2 immunolabeling of the telomeres (Figure S4B).
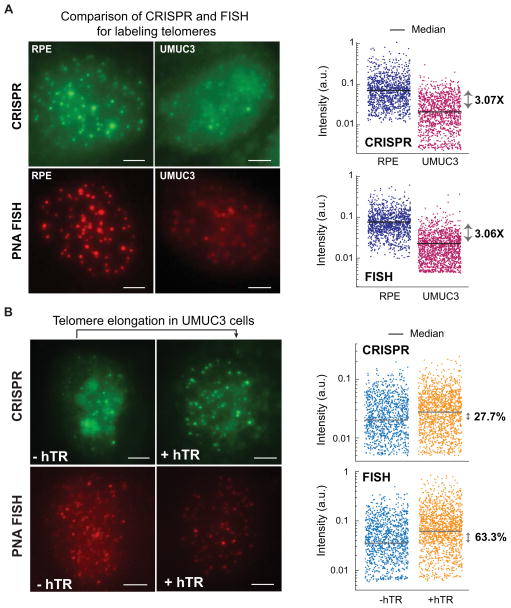
Although two-color imaging directly assesses whether CRISPR intensity accurately measures the length of individual telomeres, these experiments had the caveats of low efficiency in detecting short telomeres by TRF2 immunofluorescence and our modified PNA FISH protocol. Therefore, we also analyzed the median telomere puncta intensity labeled by CRISPR imaging or by PNA FISH, a common method for quantifying telomere length (Figure 4A) (Hultdin et al., 1998). We compared telomere images of RPE cells and those of the UMUC3 human bladder cancer cell line. The median CRISPR puncta intensity in RPE cells was 3.1 times as high as that in UMUC3 cells, which exactly matched the intensity ratio measured by PNA FISH (Figure 4A). In both RPE and UMUC3 cells, we detected similar numbers of telomeres using CRISPR imaging or PNA FISH (Figures S4C). Moreover, the telomere length in UMUC3 cells can be conditionally elongated (from 1.6~5 kb to 3~10 kb) by transfection with a human telomerase RNA (hTR) gene (Xu and Blackburn, 2007). In this study, six days after hTR lentiviral infection, we detected a 63% increase of median PNA FISH intensity, while the median CRISPR intensity also increased by 28% (Figure 4B). The correlation of both measured intensity and telomere counts suggests that CRISPR imaging is a method comparable to PNA FISH for detecting telomere length, with the added feature of labeling in living cells.
Figure 4. CRISPR imaging detects telomere length.
(A) Comparison of telomere length in RPE and UMUC3 cells using CRISPR imaging (upper panels) or PNA FISH (lower panels). The log-scale scatter plot displays the intensity of each identified telomere in RPE (navy) and UMUC3 (purple) cells. (B) hTR-induced telomere elongation in UMUC3 cells visualized by CRISPR (upper panels) or PNA FISH (lower panels). The log-scale scatter plot displays the intensity of each identified telomere without (orange) and with (blue) hTR expression. At least 20 cells were analyzed for each case. All images are maximum z projections. All scale bars: 5 μm. See also Figure S4.
CRISPR imaging monitors telomere dynamics
CRISPR imaging offers a unique platform to track native genetic elements in living cells without introducing artificially inserted sequences. We performed high-frequency (0.2 s per frame) time-lapse microscopy to track the movement of telomeres in living RPE cells. Single particle tracking revealed the confined diffusion of telomeres, which is occasionally overlaid with a slow directed motion (Figure 5A and Movie S1). To test if CRISPR labeling could affect telomere dynamics, we compared telomere movement labeled by CRISPR or TRF1, one of the major telomeric binding proteins (Wang et al., 2008). We saw very similar mean-squared displacement (MSD) curves using two methods, demonstrating that CRISPR does not disrupt telomere dynamics (Figure 5B). The microscopy diffusion coefficient of individual telomeres displayed a negative correlation with the fluorescence intensity (Figure 5C), which is consistent with the previous study showing that longer telomeres have slower movement (Wang et al., 2008), This result was further supported by our tracking of telomeres in UMUC3 cells, wherein the elongation of telomeres by hTR overexpression induced a slow-down of telomere movement (Figure 5D).
Figure 5. Tracking of telomere dynamics in live cells by CRISPR imaging.
(A) CRISPR imaging of telomeres in RPE cells (scale bar: 5 μm) and trajectories of three telomeres with different movement modes (scale bars: 200 nm). The trajectory lengths are 600 frames for 1 & 3 and 260 frames for 2. See Movie S1. (B) Comparison of telomere dynamics using CRISPR (blue) and EGFP-TRF1 (red) labeling in RPE cells. The data are displayed as mean ± standard error. (C) Scatter plot of the CRISPR foci intensity and their microscopic diffusion coefficients. (D) The average MSD curves of telomeres in UMUC3 cells without (blue) and with (orange) hTR. The data are displayed as mean ± standard error. (E) Averaged MSD curves of CRISPR-labeled telomeres in RPE cells measured with scrambled shRNA (blue), TIN2 shRNA (green), or co-expression of TIN2 shRNA and the long (L, red) or short (S, purple) isoform of TIN2. At least 15 cells are analyzed in each case. The data are displayed as mean ± standard error. See also Figure S5 and Movie S1.
As telomere damage has previously been shown to enhance telomere movement, likely to facilitate DNA repair (Dimitrova et al., 2011; Wang et al., 2008), we examined the movement of telomeres after shRNA knockdown of TIN2, which disrupts the telomere shelterin complex (Kim et al., 1999). We confirmed the resulting DNA damage localized to telomeres by immunostaining of 53BP1, a protein recruited to sites of DNA damage (d’Adda di Fagagna et al., 2003) (Figure S5A). We note that dCas9 binding did not apparently affect telomere integrity because we observed an almost negligible increase in telomerically localized 53BP1 compared to TIN2 knockdown (Figures S5A and S5B). As shown by the MSD curves detected by CRISPR (Figure 5E), we saw an expected increase in microscopic diffusion speed after TIN2 knockdown but not with a scrambled shRNA as the negative control. Simultaneous overexpression of exogenous TIN2 alleviated the DNA damage and restored near-wild type telomere movement. These results demonstrate the power of CRISPR to directly visualize the movement of endogenous genomic elements.
CRISPR imaging reveals the organization and dynamics of MUC4 loci
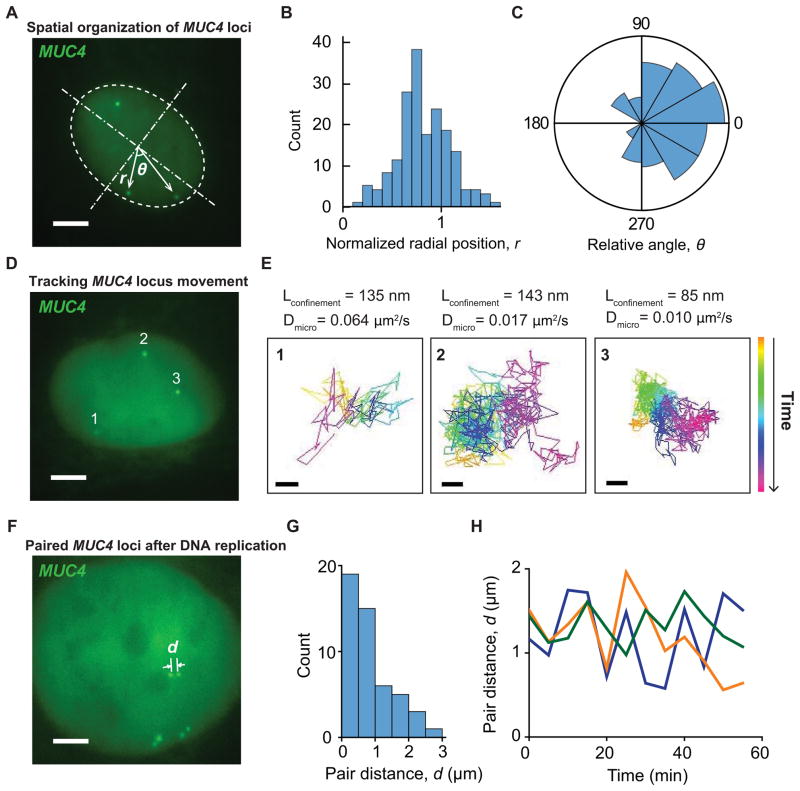
In addition to special genetic elements such as the telomere, CRISPR imaging also allows us to examine the spatiotemporal dynamics of protein-encoding DNA sequences in live cells. By tagging MUC4 exon 2 and intron 3 simultaneously using two sgRNAs, we measured the position of MUC4 loci by approximating the shape of the nucleus as an oval (Figure 6A). The distribution of normalized MUC4 radial position peaked near the nuclear envelope (Figure 6B), indicating that MUC4 loci preferentially locate at the nuclear periphery. Furthermore, by calculating the angle between any two MUC4 loci (Figure 6C), we found that MUC4 loci exhibited polarized spatial organization, with the three allelic loci clustering in the same half of the nucleus. These live cell observations support the notion of non-random spatial organization of genes and chromosomes (Cremer and Cremer, 2010).
Figure 6. Spatial organization and dynamics of the MUC4 gene in RPE cells.
(A) Scheme for analyzing the nuclear localization of the MUC4 gene using both sgMUC4-E3 and sgMUC4-I2(F+E). The nucleus is modeled as an oval and then normalized to a round circle to measure MUC4 positions. (B) The histogram of the normalized MUC4 radial position, r. The nuclear envelope is at the unity position. (C) The histogram of the relative angle of MUC4 loci with respect to the center of the nucleus, θ. 50 cells are analyzed for (B) and (C). (D) Single particle tracking of MUC4 loci movement. See Movies S2 and S3. (E) Trajectories of the three loci in (D), which show different confinement sizes, Lconfinement, and microscopic diffusion coefficients, Dmicro (scale bars: 200 nm). The trajectory lengths are 900 frames for 2 & 3 and 115 frames for 1. (F) Paired MUC4 loci after DNA replication. See Movie S4. (G) Histogram of the distance between two MUC4 loci in a pair. 20 cells are analyzed. (H) Long term 3D tracking to measure the pair distances of three MUC4 pairs within a cell. (A), (D), and (F) are 20-frame averages of live recording images. Scale bars: 5 μm. See also Figure S6 and Movies S2 and S4.
Next, we monitored the movement of MUC4 loci (Figure 6D and Movie S2). Similar to telomeres, trajectories of these loci displayed confined movement at short (< 5 s) time scales, with additional macroscopic diffusion or directional transport observed over longer time scales (Figure S6A). The short-time-scale confinement sizes and the microscopic diffusion coefficients were highly heterogeneous (Figures 6E, S6B and S6C). The median values of both parameters were comparable to those measured using LacO arrays on bacterial artificial chromosomes in CHO cells (Levi et al., 2005). Using 36 sgRNAs simultaneously, we also performed live imaging of non-repetitive sequences of the MUC4 gene. Despite slightly lower signal, we observed similar movement behaviors, comparable to the movement of repetitive sequences of MUC4 (Movie S3).
Finally, we characterized replicated MUC4 loci during late S phase and G2 phase, which appeared as closely located pairs of dCas9-GFP puncta (Figure 6F and Movie S3). The distance between such paired MUC4 loci on sister chromatids often reached over 1 μm (Figure 6G), and was similar for all three pairs within the same cell (Figure 6F). Although each individual MUC4 punctum in the pair underwent fast diffusive movement (Movie S4), the pair distance remained relatively constant over several hours (Figure 6H). These results suggest a stable but dispersed distribution along the genomic DNA of factors such as cohesin, which physically holds the two sister chromatids together (Nasmyth and Haering, 2009).
CRISPR imaging reports chromosome dynamics during mitosis
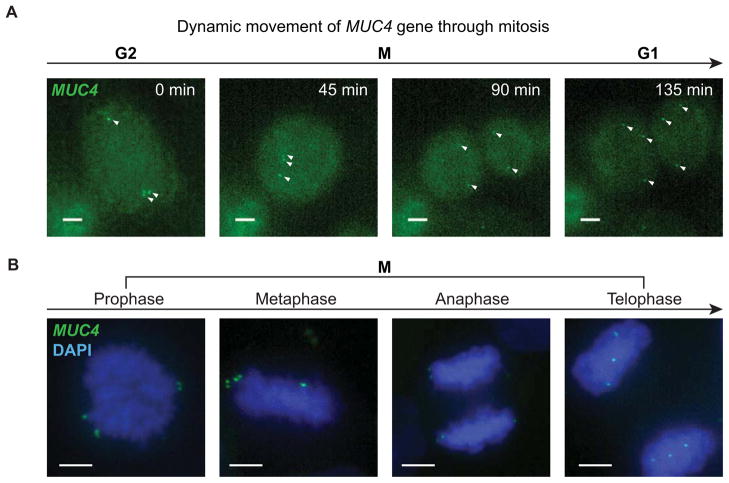
By labeling specific genomic loci with dCas9-EGFP, we could also investigate chromosome reorganization during cell division. For this purpose, we tagged MUC4 exon 2 and intron 3 simultaneously in HeLa cells. Using time lapse imaging, we recorded cell division from G2 through cytokinesis (Figure 7A and Movie S5). To examine the detailed relationship between the MUC4 loci and the chromosomes, we stained fixed HeLa cells with DAPI and performed two-color imaging to capture cells at different stages of mitosis (Figure 7B). During prophase and metaphase, MUC4 puncta localized to the end of the chromosome arm, precisely reflecting the telomere-proximal position of the MUC4 gene (at 195.5 Mb on the ~ 200 Mb chromosome 3). The separation of the two MUC4 puncta on sister chromatids was discernible but small, indicating that chromosome arm cohesion was maintained during metaphase (Onn et al., 2008). Separation of the pairs of MUC4 puncta initiated at anaphase. As the result of the symmetric separation of sister chromosomes, in telophase, the position of MUC4 loci at the two poles of the spindle nearly mirrored each other. This mirror-image relationship was maintained when the two daughter cells formed in both fixed and live cell experiments (Figure 7A). This phenomenon could lead to further studies of the mirror symmetry of initial chromosome packaging in the two daughter cells (Gerlich et al., 2003).
Figure 7. Dynamics of the MUC4 loci through mitosis.
(A) Snapshots from a MUC4 image sequence in which a HeLa cell undergoes mitosis, showing z maximum projections of 4 μm depth. The arrows indicate the MUC4 loci, which are not completely captured during mitosis because the cell thickness exceeds the z range. See Movie S5. (B) CRISPR labeled HeLa cells fixed and stained with DAPI (blue) to image the relationship between MUC4 loci (green) and the chromosomes. Cells at different stages of mitosis are displayed, showing z maximum projections of 18 μm. Scale bars: 5 μm. See also Movie S5.
DISCUSSION
CRISPR provides a robust and flexible platform for dynamic visualization of arbitrary genomic sequences
Systematic characterization of the relationship between genome spatiotemporal organization and its functional output depends on the ability to visualize genomic elements in living cells. Here we report a new imaging technique based on an optimized CRISPR/Cas system to fluorescently label specific genomic loci. We have shown that, simply by using site-specific sgRNAs, an EGFP-tagged dCas9 allows highly flexible and effective detection of both repetitive and nonrepetitive elements in the human genome. This genomic labeling by CRISPR is non-destructive, allowing the observation of native chromatin dynamics. Our data suggests that CRISPR imaging enables a new approach to study chromatin conformation and dynamics in both short time frames and long term processes such as mitosis.
While FISH requires denaturation of the DNA and is thus incompatible with live imaging, CRISPR imaging allows direct recording of real-time dynamic events. With flexible DNA sequence recognition, CRISPR imaging does not rely on targeted insertion of artificial sequences such as LacO or TetO arrays, which are often challenging to implement and maintain. Compared to transcription activator-like effector (TALE) based systems that have recently been applied to image repetitive sequences in telomere and satellite DNA (Miyanari et al., 2013), the Watson-Crick base pairing mechanism for CRISPR targeting makes our method powerful enough to detect non-repetitive sequences. The target sequence flexibility of CRISPR may enable genome-wide imaging studies in living cells. Although previous studies have reported off-target binding and editing of the CRISPR system in the human genome (Hsu et al., 2013), our method could filter such sporadic off-target events through a local enrichment of the fluorescence signal. This conclusion is supported by our observation of no puncta in the sgGAL4 negative control. As an even stronger piece of evidence, none of the MUC4 or MUC1 images contained a puncta number higher than the actual gene copy number despite the wide variety of sgRNAs used.
The improved CRISPR system enhances the efficiency of imaging, gene regulation, and likely genome editing
Our CRISPR imaging technique has also provided opportunities to understand and improve the CRISPR system itself. In our experiments, we revealed that unbound dCas9 is enriched in the nucleolus, presumably by nonspecific interaction with other RNAs or genomic loci. Such nucleolar signal was reduced by using ours optimized sgRNA design for more efficient expression and better assembly with the dCas9 protein. This observation provides strong evidence that a major limitation of CRISPR efficacy in mammalian cells is sgRNA stability and folding. Furthermore, the re-designed sgRNA also improved the CRISPRi effects by up to 5 fold. It is possible that this new sgRNA, by eliminating non-specific binding in the nucleolus, can also improve the efficacy for gene editing, with less off-target effects.
Cas9 variants from different bacterial species pair only with their cognate guide RNAs, and each guide RNA can be designed to recognize a distinct target sequence (Esvelt et al., 2013). Fusion of dCas9 variants with different fluorescent proteins therefore should allow labeling of multiple genomic sequences within a genome, enabling multicolor imaging for multiplexed detection of genetic events. Future engineering of the CRISPR system may also enable the detection of RNAs in addition to genomic DNAs.
CRISPR imaging allows direct visualization of genetic element dynamics
As demonstrated by our analysis of MUC4 loci position distribution (Figure 6A), a straightforward application of CRISPR imaging is to monitor the position of specific genomic loci in the nucleus, which is an important mechanism for gene regulation (Misteli, 2007). While this measurement has been traditionally done by FISH or LacO labeling (Heun et al., 2001), CRISPR imaging enables continuous tracking of endogenous loci over a long time period. Colocalization analysis with other nuclear landmarks such as transcription factories, nuclear pore complex, nuclear lamina, and heterochromatin markers may provide further insights into how the spatial organization of genes regulates its expression.
The uncovering of the aneuploidy of our RPE and HeLa cell lines by CRISPR imaging (Figure 2) illustrates its capability to monitor the gene copy number in living cells. This ability could provide a way to visualize gene deletions or duplications, events commonly occurring in cancers. Transposition and chromosome translocation events (Roukos et al., 2013) could conceivably also be recorded.
The human genome contains large numbers of repetitive elements such as telomeres, centromeres, and satellite DNAs. Our study has shown the application of CRISPR imaging to follow the dynamics of telomeres during telomere elongation (Figure 4). Previously, such live telomere experiments have relied on introduction of fluorescently tagged telomere binding proteins such as TRF1 (Wang et al., 2008), which could potentially perturb binding or localization of other proteins in the same complex. The same is true for imaging centromeres and other genomic loci bound by multi-protein complexes. CRISPR imaging allows direct detection of these loci without overexpression of individual DNA-binding components, avoiding perturbation of the stoichiometry.
CRISPR imaging provides a powerful tool to study chromatin architecture and nuclear organization
How chromatin ultrastructure regulate gene expression is an unsolved question in cell biology. Given our capability to simultaneously label multiple positions within the same MUC4 gene locus (Figure 3C), we should be able to characterize the local compaction state of the labeled chromatin segment. Such experiments have previously been limited to LacO labeled bacterial artificial chromosomes (Sinclair et al., 2010). In contrast, CRISPR imaging will shed light on the packaging of endogenous genomic loci, and may eventually enable mapping of the whole genome with an sgRNA library. Super-resolution microscopy will further unveil sequence-specific chromatin ultrastructure. Of note, our imaging of the highly heterochromatic telomeres also suggests that dCas9 can access heterochromatin regions of the genome. Therefore, CRISPR imaging offers a powerful tool to understand the control of heterochromatin formation (Grewal and Jia, 2007). For example, at the whole-chromosome scale, CRISPR imaging can be instrumental in the study of X chromosome inactivation (Augui et al., 2011; Meyer, 2010).
Our simultaneous imaging of MUC1 and MUC4 gene loci (Figure 3D) illustrates the capability of CRISPR imaging to monitor the spatial relationship between different genomic elements. In many cases, long range DNA interactions are involved in regulating gene expression (Fraser and Bickmore, 2007). CRISPR imaging provides an opportunity to visualize such interactions between a target gene and distant regulatory elements, allowing investigation of the underlying driving forces. Such studies will be further enhanced by the development of multicolor imaging capability. For this application, CRISPR imaging is fully complementary to chromatin conformation capture (3C) and its derived methods 5C, hi-C, etc. (van Steensel and Dekker, 2010). Although CRISPR imaging has lower sequence throughput and sequence resolution, it readily measures individual cell-to-cell variations and adds superior spatiotemporal resolution.
Using CRISPR imaging, we have imaged the MUC4 loci at different times through the cell cycle. We were able to distinguish replicated MUC4 loci, the pairing of sister chromatid MUC4 loci, and the dynamics of MUC4 loci during mitosis. These observations may allow the measurement of replication timing, sister chromatid cohesion, as well as chromosome condensation and decondensation during mitosis. Labeling different genomic loci with CRISPR could map specific DNA sequences for genomic organization during cell division. Moreover, studies of homologous pairing and recombination in meiosis should also be amenable using this approach.
A unified CRISPR system for genome engineering including editing, regulation and imaging
CRISPR has recently been developed for genome editing and gene expression in a broad range of organisms. In addition to modifying the genome sequence and modulating gene expression, here we add a new application of CRISPR: its use to directly image the spatial organization and temporal interactions of chromatin. The use of the same type II CRISPR system might greatly simplify the rules and efforts for different tasks in genome engineering and imaging, as the same set of sgRNAs can be modularly combined with different versions of Cas9 – a nuclease Cas9 (genome engineering), a transcription factor-fused dCas9 (gene regulation), or a fluorescent protein-tagged dCas9 (live chromatin imaging). Furthermore, with the characterization of orthogonal Cas9 proteins, it is possible to create a unified CRISPR platform for using different Cas9s and cognate sgRNAs to perform these various tasks of genomic manipulation and observation in the same cell. We believe that such molecular tools will be invaluable to understand, interrogate and engineer genomes, and are suitable for numerous applications for biomedical research and clinical therapies.
EXPERIMENTAL PROCEDURE
Plasmid construction
The DNA sequence encoding the dCas9 gene with inactivating D10A and H840A mutations was fused with EGFP and two copies of SV40 nuclear localization sequence (NLS). Using standard ligation-independent cloning, we cloned these fusion proteins into a lentiviral vector containing an inducible promoter PTRE3G (Tet-on 3G inducible expression system, Clontech). sgRNAs were cloned into a lentiviral U6-based expression vector derived from pSico, which coexpresses mCherry and a puromycin resistance cassette from a CMV promoter (Larson et al., 2013). For sgRNA design and cloning, see the Extended Experimental Procedures for details.
Cell culture
Human embryonic kidney (HEK) cell line HEK293T, human renal cancer cell line UMUC3 and HeLa cells were maintained in Dulbecco’s modified Eagle medium (DMEM) with high glucose (UCSF Cell Culture Facility) in 10% Tet system approved FBS (Clontech). Human retinal pigment epithelium (RPE) cells were maintained in DMEM with GlutaMAX1 (Life Technologies) in 10% Tet system approved FBS. All cells were maintained at 37°C and 5% CO2 in a humidified incubator.
Lentiviral production and stable expression of dCas9 and sgRNA
For viral production, HEK293T cells were seeded into T75 flask one day prior to transfection. 1 μg of pMD2.G plasmid, 8 μg of pCMV-dR8.91 and 9 μg of the lentiviral vector (Tet-on 3G, dCas9-EGFP, GFP-TRF1, sgRNA or TIN-2 shRNA) were co-transfected into HEK293T cells using FuGENE (Promega) following the manufacture’s recommended protocol. Virus was harvested 48 hours post-transfection. For viral transduction, cells were incubated with culture medium-diluted viral supernatant supplemented with 5μg/ml polybrene for 12 hours. RPE, UMUC3 and HeLa cell lines stably expressing dCas9-EGFP were generated by co-infecting cells with a lentiviral cocktail containing viruses encoding both dCas9-EGFP and the Tet-on 3G transactivator protein (Clontech). Clonal cell lines expressing dCas9-EGFP were generated by picking a single cell colony. The clones with low basal level expression of dCas9-EGFP were selected for CRISPR imaging. See the Extended Experimental Procedures for details.
Gene regulation assay
1 μg of each sgRNA plasmid was transfected into 50,000 HEK293 cells stably expressing both the SV40-GFP reporter, and dCas9-BFP-KRAB in a 24 well plate. 72 hours or 6 days following transfection, cells were trypsinized and analyzed by flow cytometry using an LSR-II (BD Biosciences) and/or replated for the 6 day time point. mCherry bright cells were gated and EGFP levels were measured in this population.
Immunostaining
Cells were fixed in 4% paraformaldehyde, permeabilized with 0.5% NP-40 in phosphate buffered saline (PBS) for 10 minutes, washed with PBS for 5 minutes, blocked in 0.2% cold water fish gelatin and 0.5% bovine serum albumin (BSA) for 20 minutes, incubated with the primary antibody in blocking buffer at 4 degree overnight, washed three times and then incubated with Alexa647-conjugated secondary antibody at room temperature for 1 hour, washed again and stained with DAPI. Primary and secondary antibodies used in this study were anti-TRFP2 (E-20, sc-32106, Santa Cruz Biotechnology) and anti-53BP1 (Novex, NB100-304).
Supplementary Material
Highlights.
An optimized CRISPR enables live imaging and better gene regulation in human cells
CRISPR imaging visualizes either repetitive or non-repetitive genomic sequences
CRISPR imaging reports telomere length change and telomere movements
CRISPR imaging monitors the dynamics of gene loci throughout the cell cycle
Acknowledgments
The authors thank Cell Line Genetics for karyotype analysis. B.C., W.Z. and B.H. acknowledge support from the California Institute for Quantitative Biomedical Research (QB3) and the UCSF Program for Breakthroughs in Biomedical Research. L.S.Q. acknowledges support from the UCSF Center for Systems and Synthetic Biology, NIH Office of The Director (OD), and National Institute of Dental & Craniofacial Research (NIDCR). J.S. acknowledges support from a Boehringer Ingelheim Fonds Ph.D. fellowship. This work was in part supported by NIH P50 (grant GM081879, L.S.Q.), NIH Director’s Early Independence Award (grant OD017887, L.S.Q.), NIH R01 (grant DA036858, L.S.Q. and J.S.W.), NIH P50 (grant GM102706, J.S.W), NIH U01 (grant CA168370, J.S.W), NIH R01 (grant CA096840, E.H.B. and B.A.C.), the Leukemia and Lymphoma Society (L.A.G.), the Helen Hay Whitney Foundation (G.W.L.), NIH Pathway to Independence Award (GM105913, G.W.L.), and the Howard Hughes Medical Institute (L.A.G., G.L., J.P. and J.S.W).
Footnotes
Supplemental Information includes Extended Experimental Procedures, six Supplementary Figures and five Supplementary Movies can be found with this article online at XXX.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES AND NOTES
- Augui S, Nora EP, Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat Rev Genet. 2011;12:429–442. doi: 10.1038/nrg2987. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2:a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d’Adda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova N, Chen YC, Spector DL, de Lange T. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature. 2008;456:524–528. doi: 10.1038/nature07433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendler SJ, Lancaster CA, Taylor-Papadimitriou J, Duhig T, Peat N, Burchell J, Pemberton L, Lalani EN, Wilson D. Molecular cloning and expression of human tumor-associated polymorphic epithelial mucin. J Biol Chem. 1990;265:15286–15293. [PubMed] [Google Scholar]
- Gerlich D, Beaudouin J, Kalbfuss B, Daigle N, Eils R, Ellenberg J. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell. 2003;112:751–764. doi: 10.1016/s0092-8674(03)00189-2. [DOI] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- Hellwig D, Munch S, Orthaus S, Hoischen C, Hemmerich P, Diekmann S. Live-cell imaging reveals sustained centromere binding of CENP-T via CENP-A and CENP-B. J Biophotonics. 2008;1:245–254. doi: 10.1002/jbio.200810014. [DOI] [PubMed] [Google Scholar]
- Heun P, Laroche T, Shimada K, Furrer P, Gasser SM. Chromosome dynamics in the yeast interphase nucleus. Science. 2001;294:2181–2186. doi: 10.1126/science.1065366. [DOI] [PubMed] [Google Scholar]
- Hollingsworth MA, Swanson BJ. Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer. 2004;4:45–60. doi: 10.1038/nrc1251. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hultdin M, Gronlund E, Norrback K, Eriksson-Lindstrom E, Just T, Roos G. Telomere analysis by fluorescence in situ hybridization and flow cytometry. Nucleic Acids Res. 1998;26:3651–3656. doi: 10.1093/nar/26.16.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Kaminker P, Campisi J. TIN2, a new regulator of telomere length in human cells. Nat Genet. 1999;23:405–412. doi: 10.1038/70508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc. 2013;8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi V, Ruan Q, Plutz M, Belmont AS, Gratton E. Chromatin dynamics in interphase cells revealed by tracking in a two-photon excitation microscope. Biophys J. 2005;89:4275–4285. doi: 10.1529/biophysj.105.066670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichter P, Tang CJ, Call K, Hermanson G, Evans GA, Housman D, Ward DC. High-resolution mapping of human choromsome 11 by in situ hybridization with cosmid clones. Science. 1990;247:64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- Langer-Safer PR, Levine M, Ward DC. Immunological method for mapping genes on Drosophila polytene chromosomes. Proc Nat Acad Sci USA. 1982;79:4381–4385. doi: 10.1073/pnas.79.14.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BJ. Targeting X chromosomes for repression. Curr Opin Genet Dev. 2010;20:179–189. doi: 10.1016/j.gde.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Misteli T. The cell biology of genomes: bringing the double helix to life. Cell. 2013;152:1209–1212. doi: 10.1016/j.cell.2013.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanari Y, Ziegler-Birling C, Torres-Padilla ME. Live visualization of chromatin dynamics with fluorescent TALEs. Nat Struct Mol Biol. 2013;20:1321–1324. doi: 10.1038/nsmb.2680. [DOI] [PubMed] [Google Scholar]
- Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K, Haering CH. Cohesin: its roles and mechanisms. Annu Rev Genet. 2009;43:525–558. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- Nielsen S, Yuzenkova Y, Zenkin N. Mechanism of eukaryotic RNA polymerase III transcription termination. Science. 2013;340:1577–1580. doi: 10.1126/science.1237934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollet S, Moniaux N, Maury J, Petitprez D, Degand P, Laine A, Porchet N, Aubert JP. Human mucin gene MUC4: organization of its 5′-region and polymorphism of its central tandem repeat array. Biochem J. 1998;332(Pt 3):739–748. doi: 10.1042/bj3320739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roukos V, Voss TC, Schmidt CK, Lee S, Wangsa D, Misteli T. Spatial dynamics of chromosome translocations in living cells. Science. 2013;341:660–664. doi: 10.1126/science.1237150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair P, Bian Q, Plutz M, Heard E, Belmont AS. Dynamic plasticity of large-scale chromatin structure revealed by self-assembly of engineered chromosome regions. J Cell Biol. 2010;190:761–776. doi: 10.1083/jcb.200912167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovei I, Cavallo A, Schermelleh L, Jaunin F, Scasselati C, Cmarko D, Cremer C, Fakan S, Cremer T. Spatial preservation of nuclear chromatin architecture during three-dimensional fluorescence in situ hybridization (3D-FISH) Exp Cell Res. 2002;276:10–23. doi: 10.1006/excr.2002.5513. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Dekker J. Genomics tools for unraveling chromosome architecture. Nat Biotechnol. 2010;28:1089–1095. doi: 10.1038/nbt.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Li GW, Chen C, Xie XS, Zhuang X. Chromosome organization by a nucleoid-associated protein in live bacteria. Science. 2011;333:1445–1449. doi: 10.1126/science.1204697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Kam Z, Carlton PM, Xu L, Sedat JW, Blackburn EH. Rapid telomere motions in live human cells analyzed by highly time-resolved microscopy. Epigenetics Chromatin. 2008;1:4. doi: 10.1186/1756-8935-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- Xu L, Blackburn EH. Human cancer cells harbor T-stumps, a distinct class of extremely short telomeres. Mol Cell. 2007;28:315–327. doi: 10.1016/j.molcel.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.