Abstract
Alternative splicing (AS) enables programmed diversity of gene expression across tissues and development. We show here that binding in distal intronic regions (>500 nucleotides (nt) from any exon) by Rbfox splicing factors important in development is extensive and is an active mode of splicing regulation. Similarly to exon-proximal sites, distal sites contain evolutionarily conserved GCATG sequences and are associated with AS activation and repression upon modulation of Rbfox abundance in human and mouse experimental systems. As a proof of principle, we validated the activity of two specific Rbfox enhancers in KIF21A and ENAH distal introns and showed that a conserved long-range RNA-RNA base-pairing interaction (an RNA bridge) is necessary for Rbfox-mediated exon inclusion in the ENAH gene. Thus we demonstrate a previously unknown RNA-mediated mechanism for AS control by distally bound RNA-binding proteins.
The variety of alternative mRNA isoforms in higher eukaryotic transcriptomes indicates that a complex interplay among cis elements and trans factors exists to regulate splicing decisions. Splicing factors such as RNA-binding proteins (RBPs) often bind as complexes within precursor messenger RNA (pre-mRNA) sequences to promote or repress splice-site recognition1–3. Variation within trans factors or their binding sites leads to phenotypic diversity across mammalian evolution, and inherited or somatic genetic defects in these sites cause human diseases. The recent application of genome-scale immunoprecipitation and high-throughput sequencing in mammalian cells provides insights into the networks of interactions among RBPs and their RNA substrates4–14. It has long been known that splicing factors bind within constitutive and alternative exons and their proximal intronic regions to alter splicing5,15–18. Sequence information within 500 nt of alternative exons and their neighboring flanking exons has been extensively studied to derive a computational splicing regulatory code19. However, the genome-wide maps of RNA binding by proteins also show that a large fraction of binding sites are located much farther than 500 nt from potential target exons.
Published studies of distally located sequences that affect splicing allow only limited conclusions. The regulatory elements previously considered distal are often relatively close to the regulated exon or flanking exons and thus are not inconsistent with existing models of splicing regulation. Few distal intronic enhancers have been demonstrated biochemically20 or proposed on the basis of conservation21. For instance, the decoy 3′ splice acceptor site sequence in the mouse caspase-2 (Casp2) gene is located only ~200 nt downstream from the regulated exon22 and motifs that enhance splicing of rat Fn1 exon EIIIB are less than 500 nt from the downstream exon23,24. Another complicating aspect of these studies is that the splicing factors recognizing the distal sequences are not always known. For example, it is not known which RBPs bind a 526 nt segment of the intronic sequence downstream of an exon in the chicken PPP1R12A (also known as MYPT1) gene25. Similarly, it is unclear how a cis element in the first intron of the equine β-casein (CSN2) gene increases the inclusion of all weak exons in its pre-mRNA26. Finally, the mechanisms by which a splicing factor might act on a distant exon are largely obscure. For example, a distal Rbfox motif was found to affect exon N30 in human MYH10 (non-muscle myosin heavy chain B) gene, but how this occurred was unexplored20.
To examine the genome-wide relevance of distal regulatory sites in splicing, we examined the Rbfox family of RNA-binding proteins in vivo and in human cell lines. These proteins control tissue-specific AS of exons in brain, muscle, epithelial and mesenchymal cells, and embryonic stem cells5,27–32, and their binding sites are exceptionally highly conserved in sequence and position across vertebrate evolution16,33. RBFOX proteins interact with proteins mutated in spinal cerebellar ataxia types 1 and 2 (refs. 34,35), and individuals with mutations mapping to the RBFOX1 gene locus have a range of neurological deficits, such as mental retardation, epilepsy and autism-spectrum disorder (ASD)36–39. In muscle, post-transcriptional downregulation of RBFOX1 expression has a role in the pathology of facioscapulo-humeral muscular dystrophy (FSHD)40. Moreover, animal models with knockout or knockdown of Rbfox protein expression show extensive defects in both neuronal and muscle physiology28,29,31, further suggesting that this class of RNA-binding proteins plays key roles in normal development.
Here we used genome-wide cross-linking, immunoprecipitation and sequencing (CLIP-seq) assays in mammalian brain to show that more than half of Rbfox binding sites are located distally (>500 nt) from exons and that these distal sites are preserved through evolution. We used RNA-seq measurements of AS to show that distal Rbfox binding sites and distal conserved Rbfox motifs are preferentially associated with exons that are differentially spliced in experiments modeling Rbfox loss and gain. We experimentally demonstrated that these distal Rbfox binding sites directly control splicing in both endogenous genes and in minigene splicing reporters. Finally, we showed that long-range RNA-RNA secondary structures mediate distal splicing regulation by Rbfox. These results indicate that distal intronic regions are rich reservoirs of highly conserved RNA cis elements critical for splicing regulation.
RESULTS
Rbfox interacts in vivo with conserved and distal GCATGs
To generate genome-wide maps of Rbfox protein-RNA interactions in vivo, we used UV irradiation to crosslink protein–RNA complexes from adult mouse brain and immunoprecipitated them with antibodies specific for either Rbfox1 or Rbfox2 proteins (Supplementary Fig. 1a). Isolated RNA fragments representing Rbfox protein binding sites were sequenced and processed, resulting in 2,071,607 (Rbfox1) and 2,451,256 (Rbfox2) nonredundant alignments (Supplementary Table 1) to the mouse (mm9) genome. We used our CLIP-seq cluster-finding algorithm, CLIPper (available at https://github.com/YeoLab/clipper) to delineate clusters of reads representing regions in the transcriptome significantly (P < 0.01) associated with Rbfox binding. We identified 10,062 Rbfox1 clusters in 3,490 genes and 7,466 Rbfox2 clusters in 2,672 genes, with 1,901 genes containing both Rbfox1 and Rbfox2 clusters (Supplementary Fig. 1b).
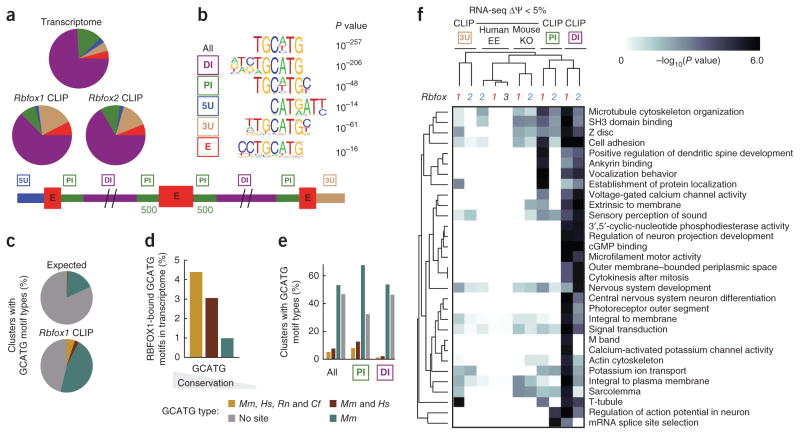
We found that the majority of Rbfox clusters (62%) were located within distal intronic regions, which we defined as intronic space >500 nt from any annotated exon (Fig. 1a). Rbfox clusters were also identified within 3′ UTRs and only a minority of clusters (8%) were located in proximal introns. The authenticity of distal clusters as bona fide Rbfox binding sites was supported by several observations. First, a de novo motif search with the HOMER algorithm, which demonstrated that both proximal and distal clusters showed statistically significant enrichment of the TGCATG motif (Rbfox1: P < 10−205 and P < 10−48 respectively, Rbfox2: P < 10−48 and P < 10−90 respectively), compared with the appropriate backgrounds selected from similar genic regions (Fig. 1b and see Supplementary Fig. 1c for other highly ranked motifs). An alternative method of statistical analysis (Z-score; ref. 5) confirmed a significant enrichment for TGCATG (P < 10−10) and hexamers that contained GCATG, for both proximal and distal clusters (Supplementary Fig. 1d). In addition, a GU-rich element previously observed in CLIP studies of RBFOX2 in human embryonic stem cells5 was present, likely representing other proteins that interact synergistically with Rbfox proteins. Second, we evaluated the evolutionary conservation of GCATG sequences within Rbfox CLIP-defined binding sites. Although only a small fraction (<15%) of bound sites contained a GCATG sequence that was evolutionarily conserved between mouse and human (Fig. 1c), GCATG sequences conserved across multiple genomes (mouse and human, rat or dog) were approximately 3.5 times more likely to be occupied in vivo by Rbfox than GCATG sequences present only in the mouse genome (Fig. 1d). Nevertheless, a statistically significant (P < 0.05) number of distal (and proximal) binding sites contained conserved GCATG motifs, as compared to clusters of similar sizes distributed randomly in distal introns (Fig. 1c,e). Third, as Rbfox1 and Rbfox2 proteins interact with the same sequence motif, we measured the correspondence in their binding sites as a measure of functional relevance. Notably, the level of overlap between distal Rbfox1 and Rbfox2 binding sites was similar to the proximal sites (Supplementary Fig. 1e). Furthermore, the overlap between both proximal and distal Rbfox1 and Rbfox2 binding sites increased as a function of degree of GCATG site conservation within clusters (Supplementary Fig. 1f).
Figure 1.
Characteristics of Rbfox binding in distal intronic regions. (a) Pie charts depict the fraction of Rbfox1 and Rbfox2 clusters, defined from CLIP-seq, within different genic regions, compared to the length distribution of each region across the transcriptome. A schematic of genic region definitions used in this paper is represented below. (b) De novo sequence motifs enriched above background that were similar to the canonical Rbfox motif are listed with their associated P value. (c) Pie charts depict the fraction of Rbfox1 clusters that contain (within 200 nt) the sequence GCATG, conserved in 1 (teal), 2 (burgundy) or 4 (goldenrod) species (species abbreviations: Mm, Mus musculus; Hs, Homo sapiens; Rn, Rattus norvegicus; Cf, Canis familiaris). (d) Bar plots show the fraction of GCATG motifs conserved in 1 (teal), 2 (burgundy) or 4 (orange) species occupied by Rbfox1 (overlapping within 200 nt). (e) Bar charts show the fraction of Rbfox1 CLIP-seq clusters that contain (within 200 nt) GCATG motifs conserved in 1, 2, or 4 species (as in c) is shown separately for all, proximal and distal regions. (f) A heat map shows a portion of gene ontology categories represented by distal binding (see Supplementary Fig. 1e). The intensity of gray corresponds to the −log10(P value) of a hypergeometric test for enrichment in gene ontology categories represented by genes bound in proximal intron (green boxed ‘PI’), distal intron (purple boxed ‘DI’) or 3′ UTR (brown boxed ‘3U’) or by genes exhibiting AS in the RNA-seq experiments in the Rbfox1 or Rbfox2 knockout (KO) or RBFOX ectopic expression (EE).
Last, we found that the ontologies of genes bound in distal intronic regions were similar to those associated with genes that contain exon-proximal binding sites, but also included some additional categories (Fig. 1f; see Supplementary Fig. 1g for the entire list of statistically significant gene ontology categories and Supplementary Table 2 for the list of genes within each category). Many of these genes bound by Rbfox in distal intronic regions, such as Shank1, previously implicated in autism41 (Supplementary Fig. 2a), are clearly important for neuronal function. Rbfox was observed to bind both proximal and distal regions downstream of the seizure-associated exon in Snap25 (Supplementary Fig. 2b and ref. 42) and the stress axis-related exon in the Kcnma1 gene, whose inclusion results in potassium channels that are more sensitive to Ca2+ (Supplementary Fig. 2c and ref. 43). We also identified distal Rbfox binding sites a kilobase away from the autoregulated exon encoding an RNA-recognition motif in each of the Rbfox1 and Rbfox2 genes, in addition to the previously known proximal sites44 (Supplementary Fig. 2d,e). Our genome-wide protein-RNA interaction maps of Rbfox proteins in mouse brains indicated that distal Rbfox sites have sequence conservation properties, gene targets and other features that support their functional roles in RNA regulation and disease etiology.
TGCATG is enriched and conserved distal to alternative exons
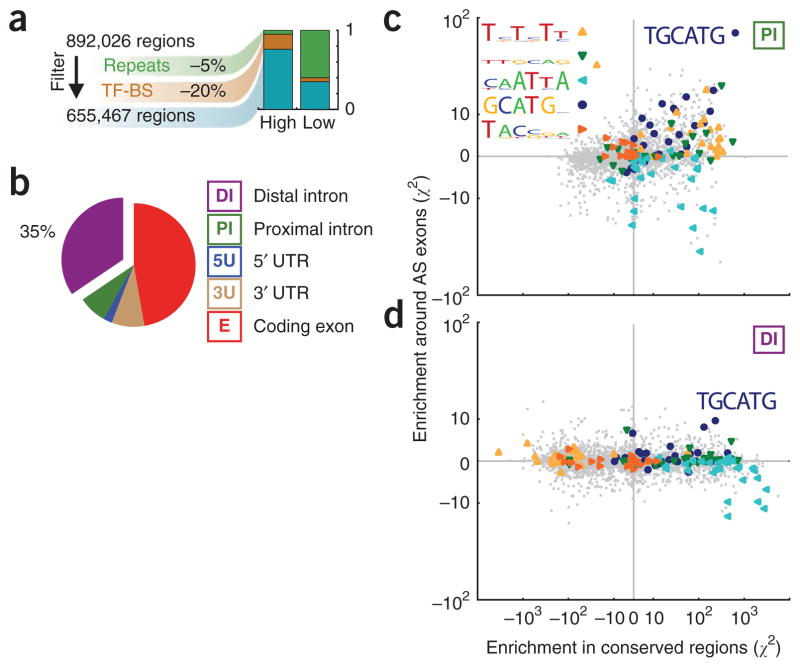
On the basis of our hypothesis that distal Rbfox splicing enhancers or repressors regulate mammalian exons important in development, we predicted that distal GCATG motifs, like proximal GCATG motifs5,18, are evolutionarily conserved and preferentially enriched in introns flanking AS exons. We tested whether the highly conserved distal regions have two properties expected for AS control regions: enrichment for known splicing regulatory motifs, and preferential association of these motifs with alternative exons more than constitutive exons. To achieve this, we modified a computational strategy (Online Methods; ref. 45) to score hexamers for their statistical enrichment in highly conserved intronic regions relative to weakly conserved intronic regions. We separately scored hexamer enrichment in conserved intronic regions flanking alternative cassette (single-exclusion) exons relative to regions flanking constitutively spliced exons, and then plotted these two scores against each other. This strategy infers function from evolutionary conservation, and relevance for AS regulation (as opposed to another function) from proximity to alternative exons. First, we computationally identified 655,467 highly conserved regions in the human transcriptome, of average length 51 bases, excluding repetitive DNA or RNA elements, microRNAs, snRNA, rRNAs and transcription factor binding sites identified by the ENCODE consortium46 (Fig. 2a). Although <1% of intronic space meets the criteria for high conservation, almost half (42%) of all highly conserved transcriptome regions are located within introns, with a great number of these falling in distal regions (35% of the total, or 224,813 regions; Fig. 2b and Supplementary Fig. 3a). We found that the cis-element composition of proximal conserved regions around AS exons (Fig. 2c) was not identical to that of distal regions (Fig. 2d and Supplementary Fig. 3b). For example, a CU-rich motif, which is a substrate for the polypyrimidine tract–binding protein (PTB) family of splicing factors, is enriched in conserved regions proximal to AS exons but depleted in distal conserved regions (Fig. 2c,d; yellow triangles), thus suggesting that these are under positive evolutionary selection in proximal regions, but negative selection in distal regions. Another motif (CAATTA) was found to be highly conserved in both proximal and distal regions, but preferentially depleted around AS exons, compared to constitutively spliced exons (Fig. 2c,d; light blue triangles). The Rbfox binding motif, TGCATG (Fig. 2c,d ; dark blue circles), was consistently the most enriched conserved cis element associated with AS exons in both proximal and distal regions (P < 0.01 by both criteria).
Figure 2.
The Rbfox binding motif TGCATG is the most enriched hexamer in conserved regions in distal intronic space around alternatively spliced exons. (a) A flowchart of the computational strategy used to identify highly conserved regions within the human transcriptome. (b) Pie chart showing the distribution of highly conserved regions among different genes. (c,d) Scatter plot of enrichment scores for 4,096 hexamers (gray points) in proximal (c) and distal (d) intronic regions. The y axis indicates the enrichment of each word within intronic regions proximal to cassette relative to constitutive exons. The x axis indicates the enrichment of each word within highly conserved regions, relative to weakly conserved regions. The five most enriched motifs in highly conserved regions proximal to AS exons compared to weakly conserved regions proximal to constitutive exons judged by an alternative de novo approach is inset in c (additional motif information is in Supplementary Fig. 3b). Words similar to these five motifs are highlighted with filled shapes, overlaid onto the scatter plot. TGCATG was significantly (P < 0.01) enriched in both proximal and distal conserved intronic regions flanking AS exons.
Distal Rbfox sites are associated with Rbfox-regulated exons
Having determined that in vivo distal intronic Rbfox binding sites contain conserved GCATG motifs, and that conserved distal intronic regions flanking annotated AS are enriched in Rbfox binding motifs in general, we next investigated the role of distal GCATG sites in Rbfox-dependent AS regulation. Rbfox-regulated exons were identified by strand-specific RNA-seq from homogenized whole mouse brain isolated from nestin-conditional Rbfox1−/ − and Rbfox2−/ − knockout (KO) animals and paired wild-type controls28,29, and human 293T cells ectopically expressing either RBFOX1, RBFOX2, RBFOX3 or empty-vector control plasmids. Both loss of Rbfox1 and Rbfox2, as well as ectopic expression of RBFOX in human 293T cells, had almost no effect on overall gene expression (Supplementary Fig. 4a–e and Supplementary Table 3).
To estimate the extent of Rbfox-dependent exon usage, we calculated percent-spliced-in (psi, Ψ) values for annotated AS exons. We identified 620 and 934 (379 in common) mouse exons that were alternatively spliced in brain (change in the absolute value of Ψ or |ΔΨ| ≥ 5%) upon loss of Rbfox1 and Rbfox2, respectively (see Supplementary Fig. 4f,g and Supplementary Table 4 for the list of regulated exons). The degree of differential splicing upon Rbfox loss correlated well with RT-PCR measurements in the publications describing these knockout mice (Supplementary Fig. 4h,i; refs. 28,29). In mouse brains, of the exons that were differentially spliced (|ΔΨ| ≥ 5%) in both experiments, only about half (210 of 379) changed in the same direction; in contrast, ectopic expression of each RBFOX in 293T cells resulted in AS of hundreds of cassette exons (Supplementary Fig. 4g), but the regulated changes in exon inclusion in these cell lines were more positively correlated (Supplementary Fig. 4j). RNA-splicing components such as Mbnl1, Mbnl2, Prpf18, Cwc22, Rsrc1, Raly, Thrap3, Rnps1 and Clk4 were themselves alternatively spliced upon Rbfox1 and Rbfox2 knockout, suggesting that some of the AS changes measured are indirect. Notably, categories of genes that undergo regulation by AS are more closely related to categories of genes bound in proximal and distal intronic regions by Rbfox1 and Rbfox2, as compared to ones bound in 3′ UTRs by Rbfox1 and Rbfox2 (Fig. 1f and Supplementary Fig. 1e), suggesting a separable biological function of Rbfox1 and Rbfox2 in 3′ UTR–mediated gene regulation.
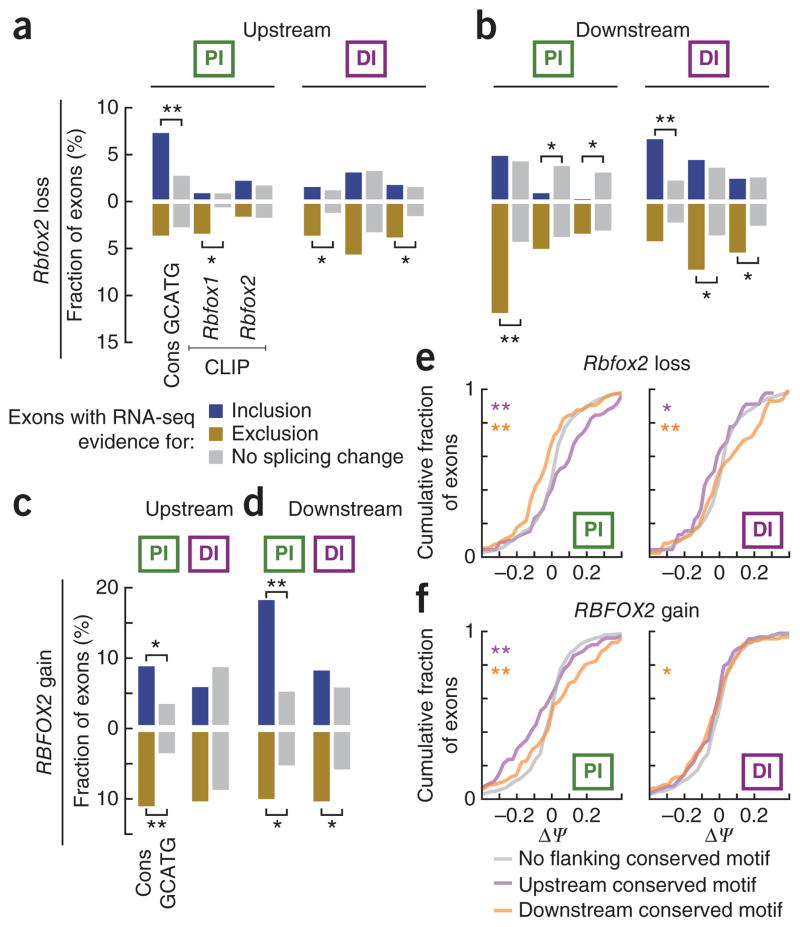
Cassette exons were divided into differentially included (ΔΨ > 5%), excluded (ΔΨ < −5%) and unaffected (|ΔΨ| < 2%) categories by Rbfox loss (in knockout mice compared to wild-type sibling pairs) or by RBFOX gain (in 293T cells ectopically expressing RBFOX compared to an empty-vector control) (Supplementary Fig. 4f–j). We determined the proportion of Rbfox-regulated and unaffected AS exons for which there was CLIP evidence for Rbfox binding or a GCATG motif, at different levels of evolutionary conservation, in the proximal or distal (‘PI’ and ‘DI’ columns of Fig. 3a–d), upstream (Fig. 3a,c) or downstream (Fig. 3b,d) intronic regions. Upon Rbfox2 loss in mouse brain, a statistically significantly higher fraction of differentially included AS exons (blue bars) than unaffected exons (gray bars) contain a conserved GCATG motif in the upstream proximal region (P < 1 × 10−3; Fig. 3a), whereas a higher fraction of excluded AS exons (golden bars) contain conserved GCATG motifs in the downstream proximal region (P < 4 × 10−6; Fig. 3b). Interestingly, exons included upon Rbfox2 loss are depleted of CLIP-defined binding sites in downstream proximal intronic regions (P < 4 × 10−3; Fig. 3b). As expected, inverse effects were observed when RBFOX2 was ectopically expressed in 293T cells (P < 3 × 10−4, P < 3 × 10−3 for downstream of included and upstream of excluded exons, respectively; Fig. 3c,d). RBFOX1 and RBFOX3 experiments had similar, but less dramatic, effects on splicing (Supplementary Fig. 5a,b). Therefore, Rbfox interaction within proximal intronic regions was associated with Rbfox regulation, confirming previous ‘position-dependent’ rules: that upstream Rbfox binding is associated with repression of exon recognition, whereas downstream binding correlates with exon inclusion5,18.
Figure 3.
Both proximal and distal Rbfox motifs regulate splicing. (a–d) Bar plots depict the fraction of cassette exons for which there is evidence (listed below the first column of each panel) for direct Rbfox regulation within proximal or distal intronic regions, up or downstream (listed above each panel). Exons were classified as differentially included (ΔΨ ≥ 5%; blue, on the positive y axis), excluded (ΔΨ ≤ −5%; goldenrod reflected on the negative y axis) or not changing (−2% < ΔΨ < 2%; gray, on the positive and y axis and also reflected on the negative y axis) according to Rbfox2 RNA-seq experiments in mouse (a,b) and human (c,d). *P < 0.05 and **P < 0.001, for a Fisher’s exact test comparing the relative proportion of changed versus unaffected exons which possess a particular feature in the indicated intronic region. A full accounting of analyses using other RNA-seq experiments and other features is in Supplementary Figure 5. (e,f) Cumulative distributions of ΔΨ values for mouse (e) and human (f) cassette exons which have conserved GCATG motifs (BL score > 0.2 in mouse BL score > 0.1 in human) in proximal (left) or distal (right) regions upstream (purple) or downstream (orange) or with no motifs at all in that region (gray).
We next examined the association of distal Rbfox interaction with splicing changes. In Rbfox2 loss, a statistically higher fraction of excluded exons than unaffected exons contained upstream, distal conserved motifs (P < 1 × 10−2; Fig. 3a) and CLIP-defined Rbfox2 binding sites (P < 2 × 10−2; Fig. 3a). Notably, unlike what we found in proximal regions, a higher proportion of differentially included exons than unaffected exons contained downstream, distal conserved motifs (P < 5 × 10−4; Fig. 3b). Also, a higher fraction of excluded than unaffected exons contained downstream, distal CLIP-defined Rbfox binding sites (P < 9 × 10−3 for Rbfox1 CLIP; P < 2 × 10−2 for Rbfox2 CLIP; Fig. 3b). In RBFOX2 ectopic expression in human cells, we found that a higher fraction (P < 4 × 10−2; Fig. 3d) of excluded than unaffected exons had downstream distal conserved motifs.
As further support for the hypothesis that distal Rbfox sites can elicit regulatory effects on AS, the cumulative distributions of ΔΨ values for mouse exons with either upstream or downstream, distal conserved GCATG motifs are statistically significantly different (P < 0.05 and P < 0.006 for up- and downstream motifs, respectively; two-sample Kolmogorov-Smirnov (KS) test) compared to the distribution of ΔΨ values for exons without intronic conserved GCATG motifs; Fig. 3e). In ectopic expression experiments, only the distribution of ΔΨ values for exons with downstream conserved motifs was significantly different from background (Fig. 3f). Although the directionality of AS changes mediated by distal sites is likely more complex than that for proximal sites, these two complementary approaches demonstrate that conserved GCATG motifs and in vivo distal Rbfox binding sites are active splicing regulatory elements associated with Rbfox-dependent AS changes.
Distal Rbfox sites regulate AS in vitro and in vivo
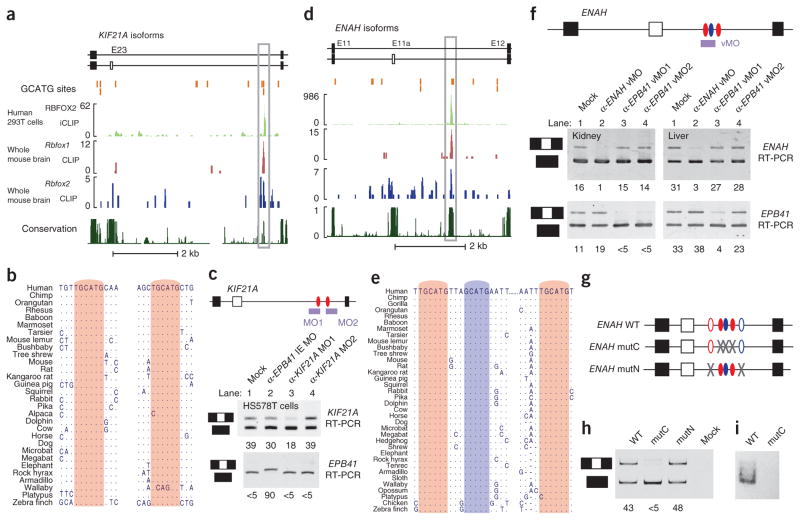
To demonstrate proof of principle that distal Rbfox motifs regulate AS, we investigated exons from two human genes that show RBFOX2-dependent AS21: KIF21A exon 23 (E23) and ENAH (also called MENA) exon 11a (E11a). Biochemical evidence demonstrates that distal intronic sites flanking these exons interact with Rbfox1 in mouse brains and Rbfox2 in both mouse brains and human 293T cells (Fig. 4a,d). Furthermore, binding sites are conserved across genomes (Fig. 4b,e). The distal TGCATG motifs in both exons are at least 500 base pairs away from any exon, allowing us to assess their functionality at long distances.
Figure 4.
Distal conserved regions containing Rbfox sites control splicing of upstream alternative exons. (a,d) Genomic regions showing KIF21A exon 23 (a) and ENAH exon 11a (d) and neighboring intronic and exonic regions. The location of GCATG sequences in both genes is marked by orange bars. Rbfox protein–RNA binding sites that overlap distal conserved GCATG motifs are outlined with a gray box, and match the highest density of CLIP-seq reads (graphed as continuous densities) for RBFOX2 in 293T cells (iCLIP; green track), Rbfox1 in mouse brain (CLIP-seq; red track) and Rbfox2 in mouse brain (CLIP-seq; blue track). PhastCons scores of evolutionary conservation are represented as continuous densities in dark green. (b,e) Phylogenetic conservation of TGCATG (red) and GCATG (blue) elements within the highlighted distal intronic regions in the KIF21A and ENAH gene (boxed in a and d). (c,f) Cartoon representations of binding sites for MOs or vMOs targeted to block distal RBFOX sites in KIF21A (c) and ENAH (f) are shown. Bottom panels show RT-PCR analysis of KIF21A and ENAH splicing in the presence or absence of morpholinos. Ψ is listed below each lane. Mock, transfection reagent only; mutC, mutated conserved sites; mutN, mutated nonconserved sites. (g) Three-exon minigenes consisting of E11-E11a-E12 are illustrated. Ovals represent conserved (filled) or nonconserved (unfilled) TGCATG (red) and GCATG (blue) sequences. (h) RT-PCR analyses of minigene-derived transcripts. Ψ is listed below each lane, as quantified by image densitometric analysis. (i) Pull-down assay measuring in vitro–translated RBFOX2 protein binding to biotinylated RNA containing consensus TGCATG motifs (lane 1) or mutated RBFOX motifs (lane 2).
KIF21A is a member of the kinesin superfamily that is overex-pressed in Down syndrome47 and mutated in congenital fibrosis of the extraocular muscles type 1 (ref. 48). Inclusion of a 21-nt exon (E23) in KIF21A is RBFOX2-dependent, and when we analyzed its downstream flanking intron, we found both proximal and distal conserved RBFOX motifs. The two distal sites are located 42 nt apart in a short region of homology ~3.3 kilobases (kb) downstream of the alternative exon and ~550 nt upstream of the next exon. To assess whether these distal conserved sites function in the context of endogenous transcripts to alter splicing, we tested the ability of anti-sense morpholino oligonucleotides (MOs) designed against these sites to alter E23 splicing in HS578T cells (Fig. 4c, top). Inclusion of E23 was reduced from ~39% in mock-treated cells to ~18% in cells treated with an MO against the first distal site, indicating that this TGCATG motif regulates E23 splicing from a distance. An MO complementary to the second distal site had no effect, either because this site does not regulate splicing in this cell line or because the site’s physical conformation inhibits MO efficacy. As a control, an MO directed against a heterologous event in the cytoskeletal gene EPB41 (ref. 49) altered splicing of its intended target transcript but did not affect E23 splicing (Fig. 4c, bottom). We concluded that one of the distal RBFOX motifs downstream of E23 is a strong distal splicing enhancer and is required for optimal splicing even in the presence of conserved proximal sites.
ENAH E11a shows notably reduced inclusion during epithelial- mesenchymal transition50, and is spliced in a breast cancer subtype– specific manner21. E11a splicing is regulated by RBFOX2 (refs. 5,21,51), and our genome-wide CLIP assays identified binding sites for Rbfox1 and Rbfox2 1.8 kb downstream of E11a in mouse brain, in human 293T cells (Fig. 4d) and in human embryonic stem cells5.
In contrast to KIF21A E23, the only conserved GCATG sequence motifs in the intron downstream of ENAH E11a are located distally, ~1.8 kb from the regulated exon. A group of three motifs is well conserved in at least 34 mammalian genomes and can also be found at orthologous positions in avian genomes (Fig. 4d). The absence of conserved GCATG sequences in the proximal flanking intron suggested that the conserved distal sites mediate RBFOX-dependent splicing enhancer activity. We investigated the function of these distal RBFOX sites in endogenous transcripts in an in vivo context using vivo-MOs (vMOs, MOs with chemical modifications that improve their efficiency in vivo; see Online Methods). E11a was partially included in ENAH mRNA isolated from kidney and liver of mice treated with a saline control (Fig. 4f, upper panel, mock). Injection of a single vMO complementary to two of the conserved RBFOX motifs (α-ENAH vMO) greatly reduced E11a inclusion in both tissues (Fig. 4f, upper panel). Control vMOs that target a heterologous event (α-EPB41 vMO1 and vMO2; ref. 49) showed splicing changes only in their intended targets. We conclude that RBFOX proteins regulate AS of E11a under physiological conditions from distal conserved binding sites.
To further validate the role of distal RBFOX sites and exclude off-target effects of the vMO, we transfected human breast cancer cell line HCC1954 with various three-exon minigene splicing reporters representing the E11-E11a-E12 region of the human ENAH gene (Fig. 4g). In strong support of our hypothesis that these distal sites are functional, inclusion of E11a was reduced to almost complete exon skipping by mutating the three conserved distal GCATG sequences (Fig. 4h, lanes 1 and 2). In contrast, mutation of two nonconserved GCATG sequences (open ovals) had little effect on splicing (Fig. 4h, lane 3). Co-precipitation of biotinylated RNA containing two of the distal RBFOX sites with in vitro translated RBFOX2 confirmed protein-RNA binding, which was lost when we mutated RBFOX sites (Fig. 4i). In summary, the above experiments functionally demonstrate that distal evolutionarily conserved GCAUG elements control splicing of exon E11a in the ENAH gene and E23 of the KIF21A gene and, more globally, there is strong statistical association between the presence of distal RBFOX sites and RBFOX-regulated exons. Next we addressed the mechanism of this molecular phenomenon.
A long-range RNA bridge mediates AS regulation by RBFOX
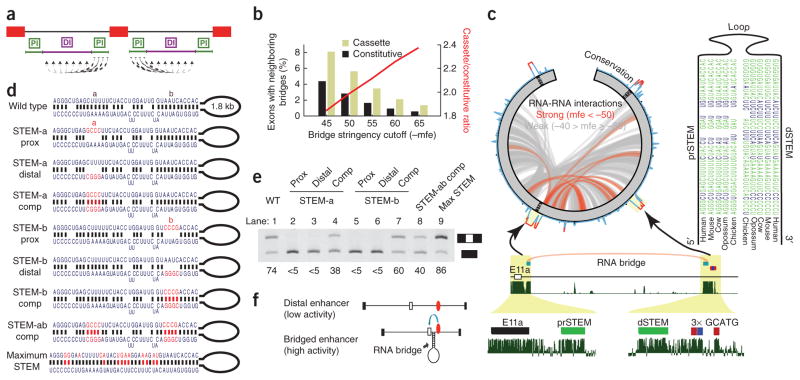
Distal Rbfox binding sites must be recruited to an alternative exon in order to productively enhance spliceosomal activity. We reasoned that RNA secondary structure might provide a ‘bridge’ that links distal cis elements with their exon targets. To investigate a role for such RNA bridges, we first scanned all cassette and constitutive exons for potential RNA-RNA interactions between exon-proximal and exon-distal intronic segments using RNAhybrid (Fig. 5a). By requiring candidate RNA bridges to be evolutionarily conserved, as expected for developmentally important structures, we pared the initial list of more than 2 million RNA bridges to approximately 24,000 conserved RNA bridges. These were significantly enriched near alternatively spliced exons, compared to constitutively spliced exons (P < 0.05 by χ2 test; Fig. 5b). Moreover, the relative enrichment of RNA bridges near alternative exons increased as a function of duplex stability, a variable not dependent on conservation levels, suggesting that conserved RNA bridges are more common and more thermodynamically stable around alternatively spliced exons than around constitutive exons (Fig. 5b, red line).
Figure 5.
An RNA bridge between ENAH E11a and a conserved distal RBFOX site is necessary for exon inclusion. (a) A schematic of the strategy used to find RNA bridges. Regions proximal to exons were tested for the ability to pair to all positions in the distal region in the same intron. (b) The fraction of cassette (light green) and constitutive (black) exons with neighboring predicted RNA bridges as the negative minimum free energy (mfe) threshold for defining an RNA bridge is made more stringent. The ratio between these fractions is depicted as a red line. (c) All predicted RNA-RNA interactions within E11-E11a-E12 (green) of the ENAH pre-mRNA are displayed in a circos plot (at left). Paired regions are classified as strong (mfe ≤ −50 kcal per mol; red) or weak (mfe > −50 kcal per mol; gray). PhastCons conservation scores are illustrated on the circumference of the circle. A stem-loop structure that is conserved across mammalian and even avian genomes is shown with base-paired nucleotides indicated in green. The location of this RNA bridge is shown in detail below. Vertebrate conservation from phastCons is represented as continuous density in dark green. (d) Wild-type and mutated (red letters) RNA duplex structures are shown. (e) RT-PCR analysis of ENAH structural mutants in transfected T47D cells. Labels above each numbered lane correspond to experiments using each of the structures in d. Ψ is listed below each lane. Prox, proximal; comp, compensatory mutations. (f) Model illustrating the function of the RNA bridge to position Rbfox sites (red ovals) close to an exon to regulate splicing.
We further enriched our search for RNA bridges that may play a role in distal Rbfox regulation by identifying structures that had a distal arm within 50 nt of a conserved GCATG site (BL score ≥ 0.3; see Online Methods). By this approach, we found 699 predicted RNA bridges around 125 exons (there are multiple potential bridges per exon). When we focused only on RNA bridges around exons that were altered upon ectopic expression of RBFOX (|ΔΨ| > 5% in any experiment), we identified 162 predicted RNA bridges around 19 exons. Among these, we found an RNA bridge connecting ENAH E11a to the distal site characterized above (Fig. 5c), but, notably, we did not predict an RNA bridge around the KIF21A E23 AS event that met the strict filtering criteria we applied. Some AS events, including those in the HnRNPR gene (not shown) and ENAH had predicted RNA bridges that met the above criteria and also had strong biochemical evidence for RBFOX2 binding from our iCLIP in 293T cells (Fig. 4a).
Downstream of ENAH E11a, our computational scan for RNA structures retrieved two overlapping RNA bridges predicted to connect a conserved region 30–120 nt immediately proximal to E11a to a similarly conserved region 10–100 nt upstream of the distal RBFOX cluster (Fig. 5c). The proximal and distal arms of the structure were separated by a putative loop of 1.1–1.7 kb (mammals) or 0.6 kb (chicken), and each arm consisted of two subdomains. This structure was conserved in most mammalian genomes and also in the chicken genome, with evidence for compensatory mutations that maintain structure but not primary sequence (Fig. 5c). We hypothesized that this stem-loop structure could bridge or recruit the distal RBFOX sites close to E11a, in effect recapitulating the classical example of Rbfox splicing regulation proximal to alternative exons.
To probe the role of this structure in regulating E11a splicing, we generated minigene constructs that contained either (i) disrupted RNA-RNA interactions in each subdomain of the structure, (ii) compensatory mutations that rescue RNA-RNA interactions but not primary sequence or (iii) a structure that had a perfectly complementary RNA bridge (Fig. 5d). We found a dramatic reduction in E11a inclusion upon mutation of any subdomain (referred to as STEM-a and STEM-b) of the predicted stem structure (Fig. 5e; lanes 2, 3, 5 and 6), even though the RBFOX sites remained intact. We combined the two STEM-a mutations to create construct STEM-a comp, which restored base pairing (Fig. 5e; lane 4) and rescued E11a inclusion to half of the normal level. Better rescue of E11a inclusion was observed in the STEM-b compensatory mutation (Fig. 5e; lane 7). Consistent with these results, we also found that a double-mutant construct (STEM-ab prox) completely abrogated inclusion (data not shown), whereas double-compensatory mutation (STEM-ab comp) recovered more than half of E11a inclusion (Fig. 5e; lane 8). Finally, a mutation that extended the original base pairing by creating a perfect 42-nt stem actually increased E11a splicing efficiency above normal levels (Fig. 5e; lane 9). On the basis of these results, we theorize that AS regulation can be mediated by long-range RNA-RNA interactions between paired sequences that form an ‘RNA bridge’ to recruit a distal RBFOX site close to its target exon (Fig. 5f).
DISCUSSION
Aberrant regulation of post-transcriptional RNA processing networks is increasingly recognized as a major cause of human genetic disease. Establishing the consequence of RBP interactions will be key to understanding the molecular basis of human diseases and the basic mechanisms that drive cellular processes. Our protein-RNA interaction maps reveal that Rbfox proteins bind not only proximally but also distally to exons and, furthermore, that these sites actively regulate alternative splicing. Minimally, this suggests there is a vast and untapped trove of information that could be used to predict the inclusion of exons according to cell state or environment. Aside from reaffirming the positional rules whereby proximal binding of Rbfox upstream of an exon suppresses and binding downstream of an exon enhances exon inclusion, we have identified hundreds of distal sites that are associated with Rbfox-regulated splicing. Furthermore, we provide evidence that long-range RNA-RNA interactions can function over kilobase distances in vivo to mediate activity of distal enhancers and that such RNA bridges may be common components of distal regulatory mechanisms in AS control.
RNA secondary structures have long been known to alter splicing patterns in yeast52–54, Drosophila55,56 and mammalian pre-mRNAs24,57. These have most often been observed to loop out exons or splice sites to induce their skipping56,58 (also reviewed in ref. 59). Early work in yeast also showed that intra-intronic base-pairing interactions could enhance the splicing of long introns52,54. In mammals, secondary structures have been shown to alter the activity of regulatory proteins by blocking or removing their binding sites60; here we show that secondary structures can also function as chaperones for RBP-mediated long-range splicing regulation. Our data indicate that RNA bridges can function by dramatically shortening the distance between a distally bound splicing regulator and its target exon; indeed, close examination of iCLIP-seq data in Figure 4d revealed evidence for RBFOX2 interactions at the proximal stem region of the bridge in ENAH intron 11a that lacks GCATG motifs and would not be expected to bind RBFOX2 directly. In summary, RNA-RNA interactions within introns can have major effects on splicing and provide a versatile mechanism for juxtaposing splicing controllers with synergistic or antagonistic relationships. Beyond that, it adds an additional layer of AS regulation through promotion or inhibition of RNA-bridge formation, for example, through RNA editing.
Our results suggest that long-distance regulation of splicing is more far-reaching than the small number of earlier reports might imply. Abundant regulatory information located deeper within introns also represents an underappreciated source of disease-causing mutations. As the number of sequenced human genomes increases, our catalogs of RNA binding sites and conserved regions will enable nucleotide-level, functional association of natural and disease variation with AS. We conclude that future studies of AS networks in normal development and in disease must consider both proximal and distal RNA binding sites and noncanonical molecular mechanisms that lead to distal enhancer activity in order to accurately predict RNA splicing. As we have demonstrated with vMOs, these cis-regulatory elements provide an important opportunity for targeted rationally designed therapeutic interventions, such as those that are proving effective and specific in the treatment of splicing-related diseases.
ONLINE METHODS
CLIP-seq library generation and analysis
CLIP-seq libraries were constructed as previously described7. Fresh brains from 8-week-old female C57Bl/6 mice were rapidly dissociated by forcing through a cell strainer with a pore size of 100 μm (BD Falcon) before ultraviolet cross-linking (400 mJ/cm2). Using antibodies against the proteins Rbfox1 (custom generated in the Black lab, UCLA) and Rbfox2 (Bethyl Laboratories) (10 μg antibody per ml tissue lysate), we generated CLIP-seq libraries that were sequenced on the Illumina GAIIx platform. Raw reads were subjected to custom trimming scripts that removed adaptor sequences and truncated reads at low-quality bases or 10-mer homopolymers. Reads were filtered through a catalog of consensus genomic elements by mapping with bowtie61 (parameters: -q -p 1 -e 100 -l 20). Reads that did not map to repetitive elements were mapped to the human (hg19) or mouse (mm9) reference genomes using GSNAP62. GSNAP was supplied with the location of exon junctions from Ensembl and UCSC genes and mapped with the following parameters: -t 2 -N 1 -n 10 -Q -B 5. CLIP-seq reads from replicate libraries were combined after quality checks and the analysis of clusters from each library revealed an enriched GCATG motif. CLIP-seq reads were collapsed to remove PCR artifacts using samtools rmdup63. A new cluster-identification algorithm, CLIPper (CLIP-seq peak enrichment; https://github.com/YeoLab/clipper), was developed to identify clusters representing binding sites for Rbfox1 and Rbfox2. For each pre-mRNA, we determined the minimum height of CLIP-seq reads required to satisfy a user-defined false discovery rate (FDR) of 0.05, based on our previous method5. Next, we interpolated the heights of reads across the length of the pre-mRNA using cubic splines. From the fitted curve, peaks, centers and widths that represented clusters were identified. The number of reads expected within each cluster was estimated by the Poisson distribution using the total number of reads that mapped within the entire length of the pre-mRNA64. In addition to this ‘pre-mRNA’ cutoff, for each cluster, we also recalculated these cutoffs for reads within 1 kb and included any putative clusters that were significant by either this local analysis or by the gene-wide analysis described above. Clusters were assigned to genic regions on the basis of the following order of priority: Exon > 3′ UTR > 5′ UTR > Proximal Intron > Distal Intron. Background regions to compute statistically significant motifs were selected four times for each cluster by deriving a random ‘cluster’ of equal length in a randomly selected location in the same type of genic region in any gene. The software packages pybedtools and bedtools65,66 were used to enumerate overlaps between clusters and motifs. To determine the statistical significance of the extent of overlap, regions were randomly located ten times, keeping the ratio of locations in the different genic regions the same. A Z score with corresponding P value was computed from the shuffled mean and s.d.
De novo motif analysis for Rbfox clusters
Parameters supplied for motif finding with HOMER’s findMotifs program were: -p 4 -rna -S 10 -len 5,6,7,8,9. Cluster sequences and background selected from the same genic region as real CLIP clusters were supplied as a fasta file.
Multispecies alignments
Alignments were obtained directly from MultiZ tracks (hg19 46-way and mm9 30-way alignments) available from the UCSC Genome Browser and avian genome alignments (Figs. 4 and 5), were manually adjusted for local accuracy where supplied alignments were unsatisfactory.
Branch-length (BL) scoring to measure conservation of GCATG motifs
Rbfox motifs were scored by walking through every position of the transcriptome and summing the edit distance from a TGCATG motif across all aligned orthologs. Edit distances were multiplied by the branch length (supplied by UCSC) to a given ortholog after we used a sigmoid function to severely penalize edit distances <0.8. Then we expressed this score as a fraction (0 to 1) of the maximum score possible if all orthologs contained an aligned perfect match to TGCATG. Last, we excluded positions where the target genome (either mouse or human) did not contain a GCATG pentamer.
De novo motif analysis for conserved regions
Parameters supplied for motif finding with HOMER’s findMotifsGenome program were: “-size given -S 100 -rna -float -bits -len 6 -nlen 0 -noweight -h -minlp 0”. Background locations supplied were weakly conserved regions flanking constitutive exons.
Identification and word frequency analysis of conserved regions
Contiguous regions within the human genome with phastCons score, S were divided into categories of low (0 ≤ S ≤ 0.3), moderate (0.3 < S ≤ 0.9) and high (0.9 < S ≤ 1.0) levels of evolutionary conservation. If a region is separated by only 1 nt with another region, both regions were combined. Conservatively, regions longer than 2 kb or shorter than 10 nt were eliminated from further consideration. Regions that overlapped RepeatMasker67 (http://www.repeatmasker.org/) annotated repeats or ribosomal RNA and microRNA genes were removed. We also removed any conserved regions overlapping with transcription-factor binding sites as defined by experiments conducted by the ENCODE Consortium (obtained from the UCSC genome browser here: http://hgdownload-test.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeRegTfbsClustered/wgEncodeRegTfbsClustered.bed.gz). We found that ~20% of our highly conserved regions overlapped with a transcription factor binding site. After defining conserved regions, they were assigned to functional genic categories (exon, 5′ untranslated region, 3′ untranslated region, proximal intron or distal intron). To compute the coverage of each genic region, we divided the number of nucleotides represented by each category of conservation with the total number of nucleotides in that region. Alternative and constitutive splicing annotations were defined using available transcripts from Ensembl compiled with previously published methods16. In total, we analyzed 23,982 annotated protein-coding genes, 18,551 cassette (or skipped) exons and 164,920 constitutive exons. A χ2 enrichment score for each motif was computed with the counts for each hexamer as follows: (1, x axis of Fig. 2) counts in highly conserved regions relative to weakly conserved regions and, (2, y axis of Fig. 2) counts in highly conserved regions associated with exons that were alternatively spliced, compared to hexamers in highly conserved regions associated with constitutively spliced exons. Background for each test was the number of other hexamers in these regions. Enrichment scores are multiplied by the direction of enrichment, which was determined as the sign of the difference between ratios of hexamer counts over background. The software package HOMER68 was used to identify degenerate motifs that were significantly enriched in highly conserved regions around cassette exons versus lowly conserved regions around constitutive exons, as inset into Figure 2c. To identify 6-mers that were similar to the HOMER-derived motif, we computed a Pearson correlation coefficient between the 6-mer and motif. A correlation coefficient of greater than 0.8 constituted a similar motif.
RNA-seq library generation and analysis
Total RNA from whole brain of Rbfox1 and Rbfox2 nestin-specific knockout one-month-old male and wild-type sib-pairs was extracted by Trizol as per manufacturer’s instructions. Total RNA from human 293T cells after ectopic expression of N-terminal Flag-tagged mouse Rbfox1 (NP_067452)69, human RBFOX2 (AAL67150)69 and mouse Rbfox3 (NM_ 001024931) in pcDNA3.1 (Life Technologies) 48 h after transfection using Lipofectamine 2000 (Life Technologies) was subjected to Trizol extraction. RBFOX1, RBFOX2 and RBFOX3 levels in 293T cells were measured by qRT-PCR and were evaluated to be six-, four- and fourfold higher than cells transfected with control vector pcDNA3.1 expressing only the Flag epitope, respectively. RNA-seq libraries were prepared using 8 μg of total RNA, and subjected to poly(A) selection. After strand-specific dUTP library preparation70, cDNA corresponding to 150–225 nt fragments was single-end sequenced with Illumina GAIIx to 101 nt. Raw reads were subjected to custom trimming scripts which removed adaptor sequences and truncated reads at low-quality bases or 10-mer homopolymers. Reads were filtered through a catalog of consensus genomic elements by mapping with bowtie61 (parameters: -q -p 1 -e 100 -l 20). Reads that did not map to repetitive elements were mapped to the human (hg19) or mouse (mm9) reference genomes using GSNAP62. We supplied GSNAP with the location of exon-junctions from Ensembl and UCSC genes and mapped with the following parameters: -t 2 -N 1 –n 10 –Q –B 5. Mapped reads were used to quantify gene-expression and splicing measurements as was done in ref. 7 with improvements that allowed us to use spliced reads mapped to the genome instead of an exon junction database. Our RNA-seq analysis verified reductions in mRNA levels of Rbfox1 (by 63%) and Rbfox2 (by 84%) in the Rbfox1 and Rbfox2 knockout mice, respectively. Also, as was previously observed by western blot28,29, we found reciprocal compensatory increases of 21% for Rbfox2 mRNA upon Rbfox1 loss and 10% for Rbfox1 upon Rbfox2 loss. Percent-spliced-in (Ψ) values were calculated as half the number of reads mapped to all inclusion isoforms over the number of reads mapped to all exclusion isoforms plus half the number of reads mapped to all inclusion. Only exons with evidence for alternative splicing from Ensembl annotations (human: GRCh37 v65, mouse: NCBIm37) were evaluated for differential splicing.
Gene ontology
Gene ontology analysis consisted of a hypergeometric test comparing the fraction of genes in each ontology category that appeared relevant (bound, differentially expressed or alternatively spliced) in a particular high-throughput experiment to the fraction of all expressed (RPKM > 0.5 in the species-appropriate RNA-seq experiment) genes in that category. Reported P values are Bonferroni-corrected for multiple-hypothesis testing and clustered along rows and columns using a Euclidean distance metric.
Distal association of Rbfox sites with regulated exons
We categorized cassette exons into classes undergoing three types of regulation. Exons were either included (ΔΨ > 5%), excluded (ΔΨ < −5%) or unaffected (|ΔΨ| < 2%) according to the RNA-seq experiments described above. Furthermore, exons were required to have ≥30 or ≥50 reads mapped across exon-junctions showing evidence for inclusion or exclusion in human and mouse experiments, respectively, and a flanking intron ≥ 1.5 kb in at least one direction. We tested several features we expected to be associated with exons that were regulated upon Rbfox depletion or ectopic expression. In mouse experiments, the features we examined were (i) Rbfox GCATG motifs, (ii) conserved Rbfox GCATG motifs with a BL score ≥ 0.1, (iii) conserved Rbfox GCATG motifs with a BL score ≥ 0.4, (iv) Rbfox1 CLIP clusters, and (v) Rbfox2 CLIP clusters. The features we tested in human were (i) Rbfox GCATG motifs, (ii) conserved Rbfox GCATG motifs with a BL score ≥ 0.1, and (iii) conserved Rbfox GCATG motifs with a BL score ≥ 0.2. We calculated significance by a Fisher’s exact test comparing the proportion of changing cassette exons with at least one of a given feature in a given region to the proportion of non-changing cassette exons with that feature in that region.
Splicing-reporter construction
The wild-type minigene contains a 7.3-kb fragment of the human ENAH gene encompassing the exon 11-11a-12 region, plus ~50 nt of upstream and downstream intron sequence upstream. Primers used to amplify the E11-E12 region were as follows:
Forward primer:
5′-tggaattctgcagatGTCTGGCATTGTGCAAATTAGA-3′;
Reverse primer:
5′-gccactgtgctggatCATTCAGGATCCATGTCAAAGA-3′.
The lower case nucleotides provided 15 nt overlaps with EcoRV-linearized pcDNA3.1 vector. Insert and vector were assembled together using the InFusion Advantage kit according to the manufacturer’s instructions (Clontech). In-Fusion technology was also used to introduce mutations at the deep intron RBFOX2 sites. First, the entire wild-type splicing reporter, except a small region containing the wild-type RBFOX sites, was PCR-amplified so as to generate a linearized construct opened at the intron enhancer region. Complementary 39-mer oligonucleotides containing the three mutated RBFOX sites, and 15 nt overlaps with the linearized splicing reporter, were annealed together and inserted into the vector by In-Fusion methods. Primers used to amplify the wild-type minigene:
Forward primer:
5′-TTAAAAATTTGACTGTTTCCACAATTG-TTTATTACA-3′;
Reverse primer:
5′-TCAGTCTAACAGTCAATCCATCACCACCACCACCAC-3′.
Oligonucleotides containing the mutated RBFOX sites (underlined):
Forward:
5′-TGACTGTTAGACTGAATTAAATTTTTAAAAATTTGACTG-3′;
Reverse:
5′-CAGTCAAATTTTT-AAAAATTTAATTCAGTCTAACAGTCA-3′.
The nonconserved RBFOX sites were modified using multisite mutagenesis to mutate both nonconserved sites in one reaction, using the following primers in which the mutated RBFOX motifs are represented in upper case:
Primer 1: 5′-catggtTGACTGtgcctgtgggaggctg-3′;
Primer 2: 5′-ggaataggtAGACTGagtgaatatatgaaataacatcc-3′.
Splicing analysis of endogenous transcripts and minigene reporters
Splicing reporter minigenes were transfected into human HCC1954 or T47D cells using FuGene HD Transfection Reagent (Roche). Total RNA was extracted from cells with Qiagen’s RNeasy Mini Kit, and then reverse-transcribed into cDNA using random primers and the Superscript III First Strand Synthesis System (Invitrogen). Subsequent PCR analysis was performed using AccuTaq polymerase (Sigma). Amplification of minigene E11a inclusion and exclusion products as a measure of splicing efficiency was done using primers in intron 10 (forward primer 5′-GCATTGTGCAAATTAGAGTCCTT-3′) and intron 12 (reverse primer 5′-CAGGATCCATGTCAAAGATATGC-3′). Amplification of mouse endogenous ENAH transcripts was performed using the following primers:
Forward primer: 5′-GCTGAGAAGGGATCAACAATAG-3′;
Reverse primer: 5′-GCTCTGCTTCAGCCTGTCATAG-3′
Splicing of human KIF21A was assayed using the following primers:
Forward primer: 5′-GAAATAACCAGTGCTACCCAAAAC-3′;
Reverse primer: 5′-GTTTAAAGGAGCATCCTCATCAGT-3′
Morpholino treatments
We obtained 25-nt antisense morpholinos sequences from Gene Tools, LLC (Philomath, OR). The vivo morpholino for mouse experiments contains a covalently linked octaguanidine dendrimer as a delivery moiety to facilitate entry into cells in vivo71. The enhancer blocking sequence for ENAH was as follows:
5′-TAATTCATGCTACCATGCAATCCAC-3′;
underlined sequences are complementary to two of the conserved RBFOX binding motifs. Mice received tail vein morpholino injections at 15 mg/kg on two consecutive days, then RNA was purified from selected tissues on the third day. Tissues were rinsed in 1× PBS and snap frozen in an ethanol and dry ice bath and stored at −80 °C. For KIF21A experiments, morpholinos (without a covalently linked octaguanidine dendrimer) blocking the distal RBFOX sites were delivered to HS578T cells in 12 well plates using 5 μl of morpholino and 6 μl of endoporter (Gene Tools, LLC). RNA was extracted 48 h after treatment. Morpholino sequences were as follows:
KIF21 distal1 5′-CATGCAACAGCTCTGTAAACTAATA-3′;
KIF21A distal2 5′-ACACATCAGCATGCAGCTCATTCAC-3′.
Uncropped images of gels are shown in Supplementary Figure 6.
Supplementary Material
Acknowledgments
The authors would like to thank A. Pasquinelli, N. Chi, K. Willert and L. Goldstein and members of the Yeo, Conboy and Goldstein labs for critical reading of the manuscript. M.T.L. is supported as a National Science Foundation GK12 Fellow. This work was supported by grants from the National Institute of Health to G.W.Y. (U54 HG007005, R01 HG004659, R01 GM084317 and R01 NS075449) and to J.G.C. (HL045182 and DK094699) and partially supported by grants to J.W.G. (CA112970 and CA126551). J.G.C. also acknowledges support from DK032094. This work was also supported by the Director, Office of Science, and Office of Biological & Environmental Research of the US Department of Energy under Contract No. DE-AC02-05CH1123. D.L.B. and L.T.G. were supported by US National Institutes of Health grant RO1 GM49662 to D.L.B. D.L.B. is an Investigator of the Howard Hughes Medical Institute. M.T.L. and G.W.Y. are grateful for a gift from P. Yang at Genentech that supported M.T.L. G.W.Y. is supported as an Alfred P. Sloan Research Fellow.
Footnotes
Accession codes. Raw .fastq files of RNA-seq and CLIP-seq data are available from the NCBI Sequence Read Archive under accession codes SRP029987 and SRP030031.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
M.T.L. and G.A.P. conducted the bioinformatics analyses. D.G., H.M., J.A., S.G., M.P., T.Y.L., T.J.S., S.H. and K.B.M. conducted biological experiments. L.T.G. and D.L.B. generated the Rbfox mutant mice and isolated brain RNA. J.W.G., J.G.C. and G.W.Y. designed the study. M.T.L., D.G., J.G.C. and G.W.Y. wrote the manuscript with input from all authors.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 2.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 3.Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Licatalosi DD, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeo GW, et al. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polymenidou M, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tollervey JR, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lagier-Tourenne C, et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15:1488–1497. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huelga SC, et al. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep. 2012;1:167–178. doi: 10.1016/j.celrep.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilbert ML, et al. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol Cell. 2012;48:195–206. doi: 10.1016/j.molcel.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zarnack K, et al. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013;152:453–466. doi: 10.1016/j.cell.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.König J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoell JI, et al. RNA targets of wild-type and mutant FET family proteins. Nat Struct Mol Biol. 2011;18:1428–1431. doi: 10.1038/nsmb.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ule J, et al. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 16.Yeo GW, Van Nostrand EL, Liang TY. Discovery and analysis of evolutionarily conserved intronic splicing regulatory elements. PLoS Genet. 2007;3:e85. doi: 10.1371/journal.pgen.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue Y, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang C, et al. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barash Y, et al. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 20.Guo N, Kawamoto S. An intronic downstream enhancer promotes 3′ splice site usage of a neural cell-specific exon. J Biol Chem. 2000;275:33641–33649. doi: 10.1074/jbc.M005597200. [DOI] [PubMed] [Google Scholar]
- 21.Lapuk A, et al. Exon-level microarray analyses identify alternative splicing programs in breast cancer. Mol Cancer Res. 2010;8:961–974. doi: 10.1158/1541-7786.MCR-09-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coté J, Dupuis S, Jiang Z, Wu JY. Caspase-2 pre-mRNA alternative splicing: Identification of an intronic element containing a decoy 3′ acceptor site. Proc Natl Acad Sci USA. 2001;98:938–943. doi: 10.1073/pnas.031564098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim LP, Sharp PA. Alternative splicing of the fibronectin EIIIB exon depends on specific TGCATG repeats. Mol Cell Biol. 1998;18:3900–3906. doi: 10.1128/mcb.18.7.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baraniak AP, Lasda EL, Wagner EJ, Garcia-Blanco MA. A stem structure in fibroblast growth factor receptor 2 transcripts mediates cell-type-specific splicing by approximating intronic control elements. Mol Cell Biol. 2003;23:9327–9337. doi: 10.1128/MCB.23.24.9327-9337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dirksen WP, Mohamed SA, Fisher SA. Splicing of a myosin phosphatase targeting subunit 1 alternative exon is regulated by intronic cis-elements and a novel bipartite exonic enhancer/silencer element. J Biol Chem. 2003;278:9722–9732. doi: 10.1074/jbc.M207969200. [DOI] [PubMed] [Google Scholar]
- 26.Lenasi T, Peterlin BM, Dovc P. Distal regulation of alternative splicing by splicing enhancer in equine beta-casein intron 1. RNA. 2006;12:498–507. doi: 10.1261/rna.7261206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim KK, Kim YC, Adelstein RS, Kawamoto S. Fox-3 and PSF interact to activate neural cell-specific alternative splicing. Nucleic Acids Res. 2011;39:3064–3078. doi: 10.1093/nar/gkq1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gehman LT, et al. The splicing regulator Rbfox2 is required for both cerebellar development and mature motor function. Genes Dev. 2012;26:445–460. doi: 10.1101/gad.182477.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gehman LT, et al. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat Genet. 2011;43:706–711. doi: 10.1038/ng.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeo GW, et al. Alternative splicing events identified in human embryonic stem cells and neural progenitors. PLoS Comput Biol. 2007;3:e196. doi: 10.1371/journal.pcbi.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallagher TL, et al. Rbfox-regulated alternative splicing is critical for zebrafish cardiac and skeletal muscle functions. Dev Biol. 2011;359:251–261. doi: 10.1016/j.ydbio.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venables JP, et al. RBFOX2 is an important regulator of mesenchymal tissue-specific splicing in both normal and cancer tissues. Mol Cell Biol. 2013;33:396–405. doi: 10.1128/MCB.01174-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minovitsky S, Gee SL, Schokrpur S, Dubchak I, Conboy JG. The splicing regulatory element, UGCAUG, is phylogenetically and spatially conserved in introns that flank tissue-specific alternative exons. Nucleic Acids Res. 2005;33:714–724. doi: 10.1093/nar/gki210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shibata H, Huynh DP, Pulst SM. A novel protein with RNA-binding motifs interacts with ataxin-2. Hum Mol Genet. 2000;9:1303–1313. doi: 10.1093/hmg/9.9.1303. [DOI] [PubMed] [Google Scholar]
- 35.Lim J, et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 36.Bhalla K, et al. The de novo chromosome 16 translocations of two patients with abnormal phenotypes (mental retardation and epilepsy) disrupt the A2BP1 gene. J Hum Genet. 2004;49:308–311. doi: 10.1007/s10038-004-0145-4. [DOI] [PubMed] [Google Scholar]
- 37.Martin CL, et al. Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:869–876. doi: 10.1002/ajmg.b.30530. [DOI] [PubMed] [Google Scholar]
- 38.Sebat J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis LK, et al. Rare inherited A2BP1 deletion in a proband with autism and developmental hemiparesis. Am J Med Genet A. 2012;158A:1654–1661. doi: 10.1002/ajmg.a.35396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pistoni M, et al. Rbfox1 downregulation and altered calpain 3 splicing by FRG1 in a mouse model of facioscapulohumeral muscular dystrophy (FSHD) PLoS Genet. 2013;9:e1003186. doi: 10.1371/journal.pgen.1003186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sato D, et al. SHANK1 deletions in males with autism spectrum disorder. Am J Hum Genet. 2012;90:879–887. doi: 10.1016/j.ajhg.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johansson JU, et al. An ancient duplication of exon 5 in the Snap25 gene is required for complex neuronal development/function. PLoS Genet. 2008;4:e1000278. doi: 10.1371/journal.pgen.1000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie J, McCobb DP. Control of alternative splicing of potassium channels by stress hormones. Science. 1998;280:443–446. doi: 10.1126/science.280.5362.443. [DOI] [PubMed] [Google Scholar]
- 44.Damianov A, Black DL. Autoregulation of Fox protein expression to produce dominant negative splicing factors. RNA. 2010;16:405–416. doi: 10.1261/rna.1838210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yeo GW, Nostrand EL, Liang TY. Discovery and analysis of evolutionarily conserved intronic splicing regulatory elements. PLoS Genet. 2007;3:e85. doi: 10.1371/journal.pgen.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maher B. ENCODE: the human encyclopaedia. Nature. 2012;489:46–48. doi: 10.1038/489046a. [DOI] [PubMed] [Google Scholar]
- 47.Salemi M, et al. KIF21A mRNA expression in patients with Down syndrome. Neurol Sci. 2013;34:569–571. doi: 10.1007/s10072-012-1183-x. [DOI] [PubMed] [Google Scholar]
- 48.Heidary G, Engle EC, Hunter DG. Congenital fibrosis of the extraocular muscles. Semin Ophthalmol. 2008;23:3–8. doi: 10.1080/08820530701745181. [DOI] [PubMed] [Google Scholar]
- 49.Parra MK, Gee S, Mohandas N, Conboy JG. Efficient in vivo manipulation of alternative pre-mRNA splicing events using antisense morpholinos in mice. J Biol Chem. 2011;286:6033–6039. doi: 10.1074/jbc.M110.158154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warzecha CC, et al. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J. 2010;29:3286–3300. doi: 10.1038/emboj.2010.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dittmar KA, et al. Genome-wide determination of a broad ESRP-regulated posttranscriptional network by high-throughput sequencing. Mol Cell Biol. 2012;32:1468–1482. doi: 10.1128/MCB.06536-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goguel V, Rosbash M. Splice site choice and splicing efficiency are positively influenced by pre-mRNA intramolecular base pairing in yeast. Cell. 1993;72:893–901. doi: 10.1016/0092-8674(93)90578-e. [DOI] [PubMed] [Google Scholar]
- 53.Plass M, Codony-Servat C, Ferreira PG, Vilardell J, Eyras E. RNA secondary structure mediates alternative 3′ss selection in Saccharomyces cerevisiae. RNA. 2012;18:1103–1115. doi: 10.1261/rna.030767.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rogic S, et al. Correlation between the secondary structure of pre-mRNA introns and the efficiency of splicing in Saccharomyces cerevisiae. BMC Genomics. 2008;9:355. doi: 10.1186/1471-2164-9-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raker VA, Mironov AA, Gelfand MS, Pervouchine DD. Modulation of alternative splicing by long-range RNA structures in Drosophila. Nucleic Acids Res. 2009;37:4533–4544. doi: 10.1093/nar/gkp407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kreahling JM, Graveley BR. The iStem, a long-range RNA secondary structure element required for efficient exon inclusion in the Drosophila Dscam pre-mRNA. Mol Cell Biol. 2005;25:10251–10260. doi: 10.1128/MCB.25.23.10251-10260.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pervouchine DD, et al. Evidence for widespread association of mammalian splicing and conserved long-range RNA structures. RNA. 2012;18:1–15. doi: 10.1261/rna.029249.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nasim FU, Hutchison S, Cordeau M, Chabot B. High-affinity hnRNP A1 binding sites and duplex-forming inverted repeats have similar effects on 5′ splice site selection in support of a common looping out and repression mechanism. RNA. 2002;8:1078–1089. doi: 10.1017/s1355838202024056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McManus CJ, Graveley BR. RNA structure and the mechanisms of alternative splicing. Curr Opin Genet Dev. 2011;21:373–379. doi: 10.1016/j.gde.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Warf MB, Diegel JV, von Hippel PH, Berglund JA. The protein factors MBNL1 and U2AF65 bind alternative RNA structures to regulate splicing. Proc Natl Acad Sci USA. 2009;106:9203–9208. doi: 10.1073/pnas.0900342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zisoulis DG, et al. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dale RK, Pedersen BS, Quinlan AR. Pybedtools: a flexible Python library for manipulating genomic datasets and annotations. Bioinformatics. 2011;27:3423–3424. doi: 10.1093/bioinformatics/btr539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smit A, Hubley R, Green P. RepeatMasker Open-3.0. 1996–2010 [Google Scholar]
- 68.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parkhomchuk D, et al. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009;37:e123. doi: 10.1093/nar/gkp596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morcos PA, Li Y, Jiang S. Vivo-Morpholinos: a non-peptide transporter delivers morpholinos into a wide array of mouse tissues. Biotechniques. 2008;45:613–623. doi: 10.2144/000113005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.