Abstract
IL-17+ CD4+ T (Th17) cells contribute to the pathogenesis of several human inflammatory diseases. Here we demonstrate that TNF-inhibitor (TNFi) drugs induce the anti-inflammatory cytokine IL-10 in CD4+ T cells including IL-17+ CD4+ T cells. TNFi-mediated induction of IL-10 in IL-17+ CD4+ T cells is Treg/Foxp3 independent, requires IL-10 and is overcome by IL-1β. TNFi-exposed IL-17+ CD4+ T cells are molecularly and functionally distinct, with a unique gene signature characterised by expression of IL10 and IKZF3 (encoding Aiolos). We show that Aiolos binds conserved regions in the IL10 locus in IL-17+ CD4+ T cells. Furthermore, IKZF3 and IL10 expression levels correlate in primary CD4+ T cells and Aiolos overexpression is sufficient to drive IL10 in these cells. Our data demonstrate that TNF-α blockade induces IL-10 in CD4+ T cells including Th17 cells and suggest a role for the transcription factor Aiolos in the regulation of IL-10 in CD4+ T cells.
INTRODUCTION
IL-17 producing CD4+ T cells (often referred to as Th17 cells) are considered critical contributors to the pathogenesis of several human inflammatory diseases1. IL-17+ CD4+ T cells have potent pro-inflammatory effects, are enriched at sites of inflammation and correlate with markers of disease activity in inflammatory diseases1-3. Results from recent clinical trials using IL-17 blocking drugs further underscore the pathogenic role of Th17 cells in human inflammatory disease4.
The polarizing conditions for Th17 cell differentiation in vitro are increasingly well-defined, however accumulating evidence indicates that once differentiated, CD4+ effector T cell lineages display a considerable degree of plasticity and diversity5, 6. Human CD4+ T cells can co-express IL-17 and IFN-γ, particularly at sites of inflammation3, 7. Foxp3+ CD4+ regulatory T cells (Tregs) can gain IL-17 expression and cells co-expressing RORγt and Foxp3 can be detected in vivo8. Furthermore, mouse models revealed that Th17 cells can gain expression of the anti-inflammatory cytokine IL-10 and become non-pathogenic when stimulated in the presence of IL-6 and TGF-β, whilst stimulation in the presence of IL-23 maintains their pathogenicity9, 10. The small intestine was shown to play a key role in the induction of immunosuppressive IL-17+IL-10+ CD4+ T cells11. A recent study in humans demonstrated that depending on the pathogen encountered (Candida albicans vs. Staphylococcus aureus), Th17 cells can co-express significant amounts of IFN-γ or IL-10, respectively12. The ability to co-express IL-10 indicates that Th17 cells have an intrinsic ability to self-regulate. Understanding how IL-10 induction in pro-inflammatory Th17 cells is regulated is therefore of considerable immunological and clinical interest.
TNF-inhibitor (TNFi) drugs have been used to treat millions of patients worldwide, thereby transforming the treatment of immune-mediated inflammatory diseases. Here we use treatment with TNFi drugs as a model system to investigate whether the resulting changes in the pro-inflammatory environment affect the function and phenotype of human IL-17+ CD4+ T cells. We demonstrate that a counterintuitive rise in the percentage of IL-17+ CD4+ T cells following TNFi therapy may be explained by the acquisition of anti-inflammatory properties by these typically pathogenic cells. We show that TNFi-exposed IL-17+ CD4+ T cells express high levels of IL-10, which is Treg/Foxp3 independent, and relies on an IL-10 positive feedback loop, and is overcome by the addition of the pro-inflammatory cytokine IL-1β. Interestingly, TNFi-exposed IL-17+ CD4+ T cells are functionally and molecularly distinct, with increased expression of IKZF3 encoding the transcription factor Aiolos, which binds conserved regions in the IL10 locus in IL-17+ CD4+ T cells. Our data provide evidence to suggest that the transcription factor Aiolos may be a regulator of IL-10 expression in human CD4+ T cells.
RESULTS
TNFi drugs increase IL-17+ and IL-10+ CD4+ T cells
We have previously shown that patients with rheumatoid arthritis (RA) have an increased percentage of IL-17+IFN-γ-CD4+ T cells in their peripheral blood compared to healthy controls3. When patients with RA were separated based on their treatment regimen, i.e. disease-modifying anti-rheumatic drug (DMARD) therapy, or TNF-inhibitor (TNFi) therapy, a significantly higher percentage of peripheral IL-17+ CD4+ T cells was observed in patients receiving TNFi therapy (median [IQR] 1.4% [0.8-2.4]) relative to those receiving DMARD (0.6% [0.4-1.1]) or healthy controls (0.4% [0.3-0.7]) (Figure 1a; gating strategy shown in Supplementary Fig. 1). The increase in the percentage of IL-17+ CD4+ T cells was not related to differences in clinical parameters of disease (disease activity score (DAS) 28, erythrocyte sedimentation rate (ESR) or C-reactive protein (CRP)) or patient characteristics (rheumatoid factor positivity, age, gender) between the two treatment groups (Supplementary Fig. 2). Interestingly, we also observed a concurrent increase in the percentage of CD4+ T cells expressing the anti-inflammatory cytokine IL-10 in the peripheral blood of TNFi-treated patients (Figure 1b).
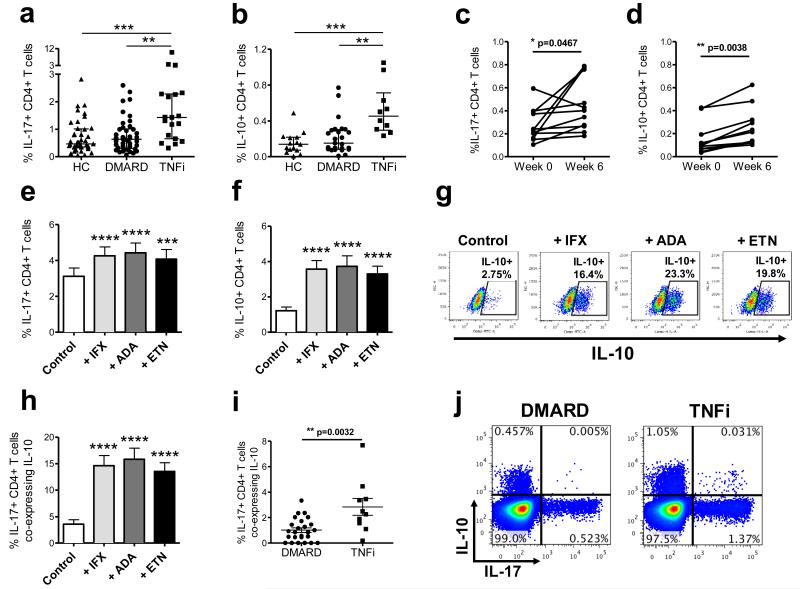
Figure 1. TNFi drugs increase the percentages of IL-17+ and IL-10+ CD4+ T cells in vivo and in vitro.
Percentages (median with interquartile range) of IL-17+ (a) and IL-10+ (b) within the total peripheral CD4+ T cell population were analysed in healthy controls (HC, n=15-41), and patients with RA on DMARD (n=25-42) vs. TNFi therapy (n=10-19). Data analysed by ANOVA (Kruskal-Wallis test) followed by Dunn’s Multiple Comparison Test. ** p<0.01, *** p<0.001. Percentages of IL-17+ (c) and IL-10+ (d) cells within peripheral blood CD4+ T cells of patients with SpA (n=11) before (week 0) and 6 weeks after (week 6) the start of TNFi therapy (analysed by Wilcoxon matched-pairs signed rank test or paired t-test). (e-h) CD4+ T cells and CD14+ monocytes from healthy controls were co-cultured with anti-CD3 mAb (Control) with or without 1 μg/ml of infliximab (IFX), adalimumab (ADA) or etanercept (ETN), followed by PMA/ionomycin restimulation on day 3. Percentages of IL-17+ (e) and IL-10+ (f) cells within total CD4+ T cells are shown (mean ± SEM, n=15). Representative dot plots (g) and cumulative data (h) of the same cultures showing the percentage of IL-17+ CD4+ T cells co-expressing IL-10. Data analysed by repeated measures ANOVA followed by Bonferroni’s Multiple Comparison test. *** p<0.001, **** p<0.0001 relative to control. Cumulative data (i) and representative dot plots (j) showing the percentage of IL-17+ CD4+ T cells co-expressing IL-10 in the peripheral blood of patients with RA on DMARD (n=25) or TNFi therapy (n=10). Data shown as mean ± SEM, analysed by Mann Whitney test.
In order to determine whether the increase in IL-17+ and IL-10+ CD4+ T cells was a drug-induced effect, we measured IL-17+ and IL-10+ CD4+ T cell frequencies longitudinally in cryopreserved PBMC from a cohort of patients with peripheral spondyloarthritis pre/post TNFi therapy13. Significantly higher levels of IL-17+ and IL-10+ CD4+ T cells were detected in samples collected 6 weeks after the start of TNFi therapy compared to samples taken at the start of therapy (Figure 1c and d). These data indicate that TNFi therapy leads to an increase in the percentages of both IL-17+ and IL-10+ CD4+ T cells in the peripheral blood in vivo.
To further examine the increase in IL-17+ and IL-10+ CD4+ T cells, in vitro co-cultures of CD4+ T cells and autologous CD14+ monocytes from healthy donors in the presence of anti-CD3 mAb were set up, a system previously shown by our group to induce IL-17 responses in human memory CD4+ T cells14, 15. Cells were cultured in the absence or presence of 1 μg/ml of infliximab (IFX), adalimumab (ADA) or etanercept (ETN), TNFi drugs routinely used in clinical practice. After three days, cells were pulsed with PMA/ionomycin in the presence of GolgiStop and stained intracellularly for the presence of cytokines. In vitro addition of each of the three TNFi drugs led to a significant increase in the percentages of both IL-17+ and IL-10+ CD4+ T cells relative to control-treated cells (Figure 1e and f).
Interestingly, when added in vitro, all three TNFi drugs (IFX, ADA, ETN) also stimulated a significant proportion of IL-17+ CD4+ T cells to co-express IL-10 (on average 15%) (Figure 1g and h), suggesting that in the presence of TNFi drugs Th17 cells acquire anti-inflammatory properties. Addition of non-specific IgG1 antibodies did not increase the percentage of IL-17+ CD4+ T cells co-expressing IL-10, indicating that this was a drug-specific effect (Supplementary Fig. 3). Cross-sectional analysis of patients with RA treated with TNFi therapy also revealed a significant increase in IL-17+ CD4+ T cells co-expressing IL-10 when compared to patients receiving DMARD therapy (Figure 1i and j). These data indicate that the counterintuitive increase in IL-17+ CD4+ T cells upon treatment with TNFi drugs may be counterbalanced by the acquisition of an anti-inflammatory potential by these cells.
IL-10 promotes and IL-1β counteracts TNFi-induced IL-10
Having demonstrated that TNFi drugs can stimulate the expression of IL-10 in otherwise inflammatory IL-17+ CD4+ T cells, we next sought to identify the cellular mechanism that promotes this response. It has been suggested previously that the presence of TNF-α can inhibit CD4+CD25+ Treg function and that TNFi drugs enhance Treg-mediated suppression16, 17. To determine whether the presence of Tregs was critical for the induction of IL-10 expression in IL-17+ CD4+ T cells, we separated PBMC into total CD4+ T cells and Treg-depleted responder cells (CD4+CD25− T cells). Irrespective of the presence or absence of CD4+CD25+ Tregs, the proportion of IL-17+ T cells co-expressing IL-10 was increased in the presence of adalimumab (Figure 2a, Supplementary Fig. 4a). IL-10 co-expression by IL-17+ CD4+ T cells occurred within the memory CD4+ T cell population, suggesting that it did not involve inducible Treg generation from naive CD4+ T cells (Supplementary Fig. 4a, b). Furthermore, we found that the majority (>80%) of IL-17+ T cells were Foxp3-, whilst only a very small percentage (on average <5%) of the total Foxp3+ T cells expressed IL-17 or IL-10 in either TNFi-exposed or control conditions (Figure 2b and Supplementary Fig. 4c, d). Finally, no increase in the percentage of Foxp3+ cells within the CD4+ T cell population as a whole or within the IL-17+IL-10+ CD4+ T cells was observed upon treatment with TNFi drugs (Figure 2c, d). Together, these data indicate that the TNFi-mediated induction of IL-10 co-expressing IL-17+ CD4+ T cells occurs independently of Tregs and Foxp3.
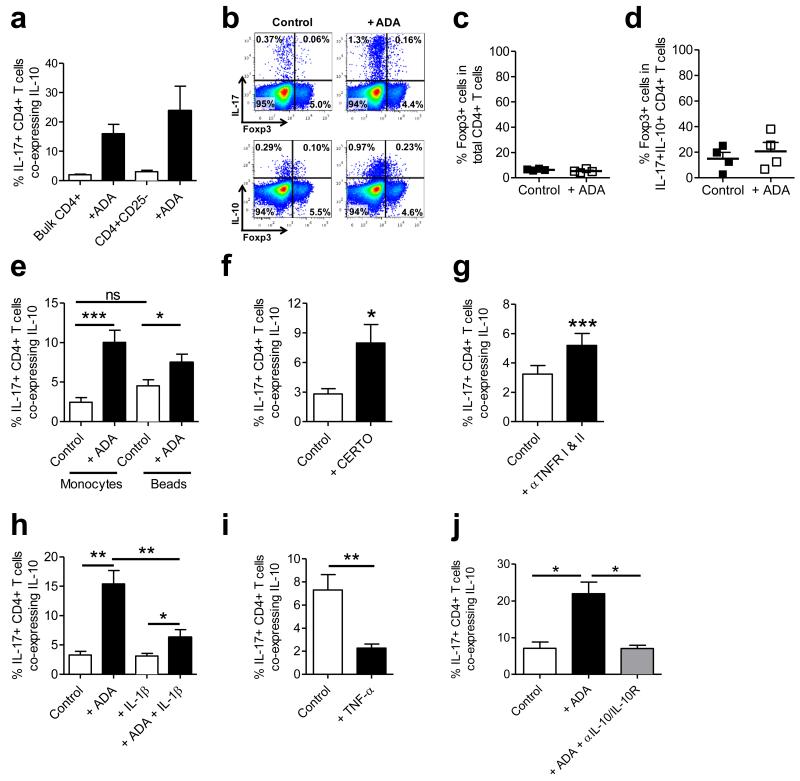
Figure 2. Dissecting the cellular mechanisms underlying the TNFi-mediated induction of IL-10 co-expression by IL-17+ CD4+ T cells.
(a) PBMC were separated into bulk CD4+ T cells or Treg-depleted CD4+CD25-T cells (n=3) and co-cultured with monocytes with anti-CD3 mAb +/− ADA for three days. Cells were stimulated with PMA/ionomycin/GolgiStop and stained for IL-17 and IL-10. (b-d) Bulk CD4+ T cells were cultured with monocytes and anti-CD3 mAb in the absence or presence of ADA for three days followed by stimulation and staining for IL-17, IL-10 and Foxp3. Representative dot plots (b) and the percentage of Foxp3+ cells within total CD4+ T cells (c) or within IL-10+ IL-17+ CD4+ T cells are shown (d) (median ± IQR, n=4). (e) CD4+ T cells were cultured for three days without/with ADA, with either anti-CD3 and CD14+ monocytes or anti-CD3/CD28 microbeads (n=12). The percentage of IL-17+ CD4+ T cells co-expressing IL-10 is shown. (f) CD4+ T cells and monocytes were co-cultured with anti-CD3 mAb with/without 1 μg/ml certolizumab (CERTO, n=7). (g) Co-cultures were set up in the absence/presence of blocking Abs against TNFRI and II (n=8). (h) Co-cultures were set up with/without ADA in the absence or presence of hrIL-1β (n=9). (i, j) Memory enriched CD4+ T cells were co-cultured with CD14+ monocytes and anti-CD3 in the absence or presence of TNF-α (n=8) (i) or in the absence or presence of adalimumab +/− neutralising Abs against IL-10 and IL-10R (n=6) (j). All data are shown as mean ± SEM. Data were analysed by repeated measures ANOVA followed by Bonferroni’s or Dunn’s Multiple Comparison test, or by paired t-test or Wilcoxon matched-pairs signed-rank test. * p<0.05; ** p<0.01; *** p<0.001.
To examine the importance of monocytes in the induction of IL-10 co-expression by IL-17+ CD4+ T cells, co-cultures were set up with either monocytes and anti-CD3 mAb, or with anti-CD3/CD28 Ab-coated beads in the presence or absence of adalimumab. As shown before, the addition of TNFi drugs in the presence of monocytes led to a strong increase in the percentage of IL-17+ CD4+ T cells co-expressing IL-10, however there was also a significant increase in IL-17+ CD4+ T cells co-expressing IL-10 in the presence of anti-CD3/CD28 beads (Figure 2e), albeit to a lesser degree, suggesting that this process is not entirely monocyte dependent. It should however be noted that in the presence of beads the total percentage of IL-17+ CD4+ T cells was considerably reduced (Supplementary Fig. 4e and14) highlighting the importance of monocytes for the induction of IL-17 responses in human CD4+ T cells. Reduced induction of IL-10 co-expression by IL-17+ CD4+ T cells was observed when CD14-depleted accessory cells were added instead of CD14+ monocytes (Supplementary Fig. 4f) or when cell-contact between monocytes and T cells was prevented (Supplementary Fig. 4g). Together, these data suggest that although the presence of monocytes is not an absolute requirement for IL-10 expression in IL-17+ CD4+ T cells, monocytes do potentiate this effect.
The three TNF inhibitors used thus far (infliximab, adalimumab and etanercept) all contain Fc domains, raising the possibility that binding or signalling through the FcγR might be involved in driving the responses observed. The induction of IL-10 expression in IL-17+ CD4+ T cells was however found to be FcγR-independent, as shown by the addition of certolizumab, a PEGylated Fab’ fragment of humanized anti-TNF-α mAb (Figure 2f) and by the addition of FcγR blocking reagent (Supplementary Fig. 4h). Instead, the increase in IL-10 co-expression in IL-17+ CD4+ T cells appeared to be due to a loss of TNF-α signalling through its receptor, as addition of blocking Abs to TNFRI and II also significantly increased the percentage of IL-17+ CD4+ T cells co-expressing IL-10 (Figure 2g). Interestingly, the addition of hrIL-1β, a potent pro-inflammatory cytokine, led to a significant reduction in TNFi-induced IL-10 co-expression in IL-17+ CD4+ T cells (Figure 2h). Furthermore, addition of hrTNF-α significantly reduced the percentage of IL-17+ CD4+ T cells co-expressing IL-10 in memory CD4+ T cells (Figure 2i). Mechanistically, the co-expression of IL-10 by IL-17+ CD4+ T cells was found to rely on an IL-10 positive feedback loop as addition of blocking antibodies to IL-10 and IL-10R decreased the induction of IL-17+ CD4+ T cells co-expressing IL-10 (Figure 2j). Together, these data indicate that the induction of IL-10 co-expression in human IL-17+ CD4+ T cells is IL-10 dependent, and can be overcome by inflammatory cytokines.
TNFi-exposed IL-17+ CD4+ T cells are functionally distinct
Next we examined the functional consequences of TNFi exposure on IL-17+ CD4+ T cells. Control and TNFi-exposed IL-17+ CD4+ T cells were sorted using a cytokine secretion assay (Supplementary Fig. 5) and cultured overnight. Supernatants from TNFi-exposed IL-17+ CD4+ T cells contained significantly increased levels of IL-10, whilst maintaining high levels of IL-17 and IFN-γ (Figure 3a). These data suggest that TNFi-exposed IL-17+ CD4+ T cells acquire regulatory potential rather than lose their inflammatory capacity.
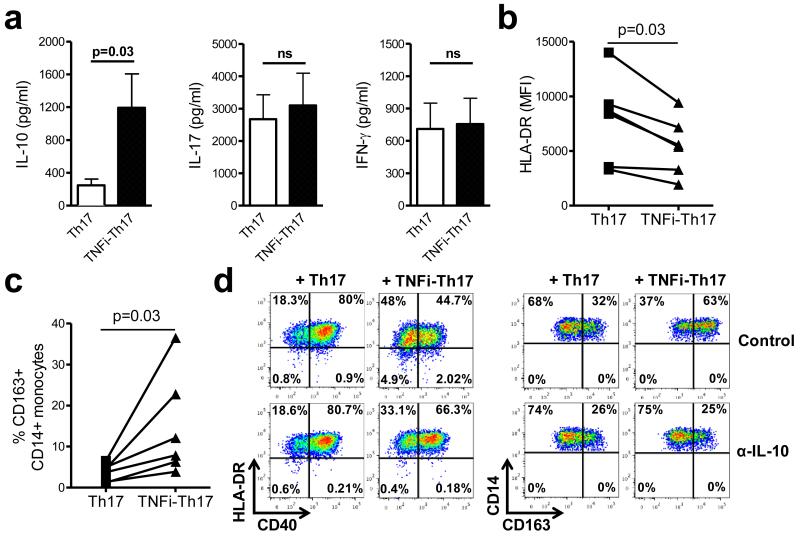
Figure 3. TNFi-exposed Th17 cells are functionally distinct.
CD4+ T cells and monocytes were co-cultured with anti-CD3 mAb in the absence or presence of adalimumab for three days. (a) Sorted IL-17+CD4+ T cells (100,000 cells/well) from both conditions were cultured overnight and cell culture supernatants collected to test for the presence of the indicated cytokines (mean ± SEM, n=6). (b, c) Sorted control-treated (Th17) or TNFi-exposed (TNFi-Th17) IL-17+ CD4+ T cells were added to autologous or allogeneic CD14+ monocytes at a 1:2 ratio and monocyte phenotype was assessed after 20 hours by flow cytometry. Graphs show the expression of HLA-DR (MFI) (b) and the percentage of CD163+CD14+ monocytes (c) (n=6). (d) Sorted Th17 cells were added to monocytes in the absence or presence of neutralizing anti-IL-10 mAb (10 μg/ml) and monocyte phenotype was measured; one of two experiments shown. Data in (a-c) were analysed by Wilcoxon matched-pairs signed-rank test.
To determine whether the increased IL-10 production by TNFi-exposed IL-17+ CD4+ T cells had functional consequences, control or TNFi-exposed IL-17+ CD4+ T cells were sorted and added to freshly isolated CD14+ monocytes. Monocyte phenotype was examined at 20 hrs. Whilst control IL-17+ CD4+ T cells induced an upregulation of HLA-DR and CD40 expression on monocytes, TNFi-exposed IL-17+ CD4+ T cells were significantly reduced in this capacity (Figure 3b, d). Instead, in the presence of TNFi-exposed IL-17+ CD4+ T cells, an increase was observed in the percentage of CD14+ monocytes expressing the haptoglobin/haemoglobin scavenger receptor CD163, a molecule known to be regulated by IL-1018 (Figure 3c, d). The effect of TNFi-exposed IL-17+ CD4+ T cells on monocyte phenotype was reversed when neutralising anti-IL-10 mAbs were added (Figure 3d). Together, these data indicate that TNFi-exposed IL-17+ CD4+ T cells are functionally distinct and produce biologically active IL-10.
TNFi-exposed IL-17+ CD4+ T cells are molecularly distinct
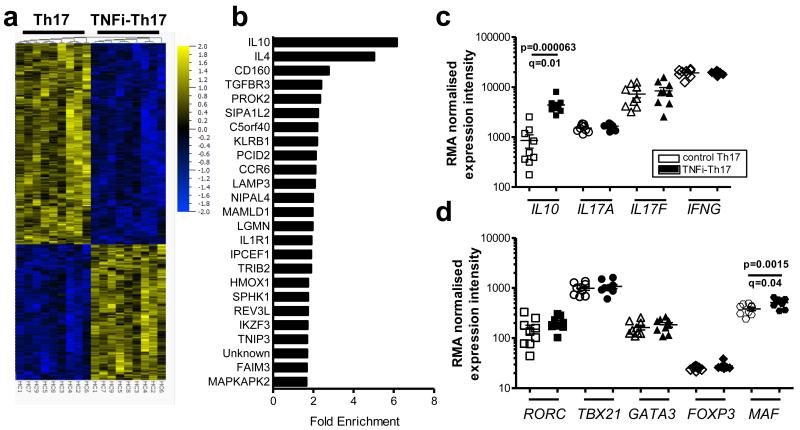
Next, we performed gene expression profiling of control and TNFi-exposed IL-17+ CD4+ T cells. Analysis of nine paired samples revealed 42 differentially expressed genes when analysed at a 1% false-discovery rate (FDR) and 813 genes at a 5% FDR (Figure 4a), indicating that TNFi-induced Th17 cells are molecularly distinct, with a unique gene expression profile. The fold enrichment of the top 25 upregulated genes is shown in Figure 4b. Amongst these genes, we found significantly increased levels of IL10 (p=0.000063 (paired t-test), q=0.01 (adjusted p-values using the Benjamini-Hochberg procedure) (Figure 4c), confirming our flow cytometry and cytokine secretion data. No significant differences were detected in the expression of IL17A, IL17F and IFNG (Figure 4c) or the transcription factors RORC, TBX21, GATA3 and FOXP3 (Figure 4d). A very small but significant increase in MAF expression was detected in TNFi-exposed IL-17+ CD4+ T cells (Figure 4d), which could contribute to the increase in IL-10 expression19.
Figure 4. TNFi-exposed Th17 cells are molecularly distinct.
CD4+ T cells and monocytes were co-cultured with anti-CD3 mAb in the absence (Th17) or presence of adalimumab (TNFi-Th17). IL-17+ T cells were re-sorted on day 3 for gene expression profiling. (a) Heat map of differentially expressed genes (5% FDR) in control-treated vs. TNFi-exposed IL-17+ CD4+ T cells. (b) The top 25 upregulated genes are shown with their average fold enrichment. (c, d) RMA normalised expression levels of indicated genes (mean ± SEM, n=9 independent healthy donors; q-values indicate p-values corrected for multiple testing (Benjamini Hochberg method)).
Correlation between IKZF3 and IL10 expression
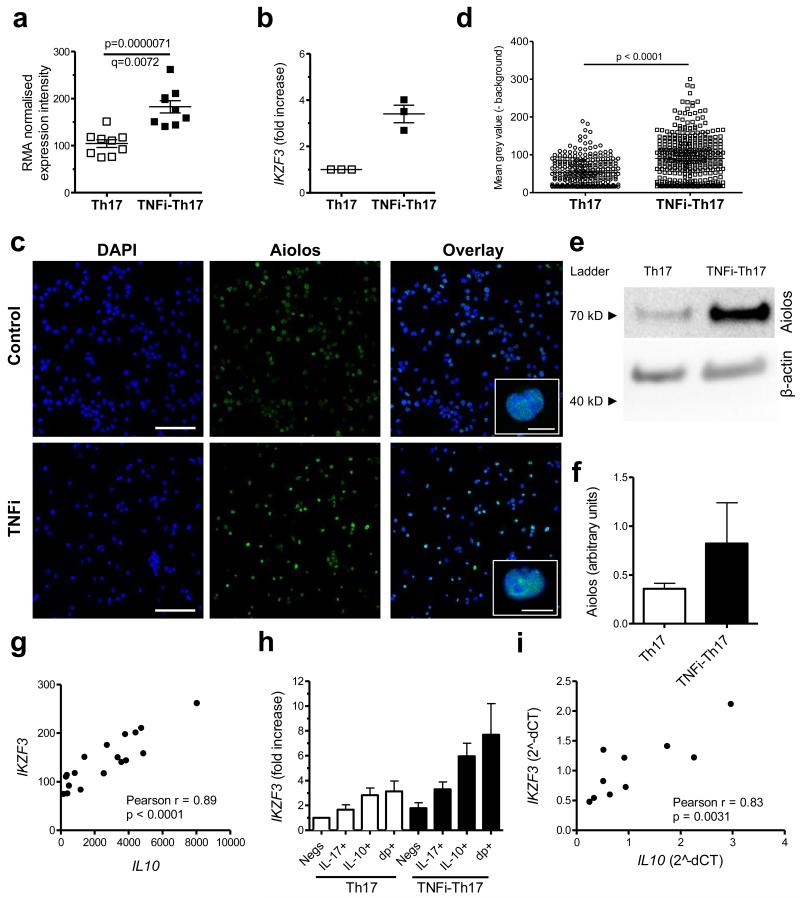
One of the genes that was most significantly upregulated at 1% FDR in TNFi-exposed IL-17+ CD4+ T cells was IKZF3, encoding the transcription factor Aiolos (p=0.0000071, q=0.0072) (Figure 5a). IKZF3 is a member of the IKAROS family of zinc finger domain containing transcription factors, however other members, IKZF1 (Ikaros), IKZF2 (Helios), IKZF4 (Eos) and IKZF5 (Pegasus) were not significantly increased (Supplementary Fig. 6). The increased IKZF3 expression in TNFi-exposed IL-17+ CD4+ T cells was confirmed by real-time PCR in independent samples (Figure 5b). We also analysed protein expression of Aiolos using a monoclonal anti-Aiolos Ab that recognizes the unique N-terminal part of Aiolos that is not conserved among other IKAROS family members, as confirmed by peptide blocking assay (Supplementary Fig. 7). We found enhanced Aiolos expression in TNFi-exposed IL-17+ CD4+ T cells in 3 out of 3 donors by confocal imaging (Figure 5c, d) and in 3 out of 4 donors by Western blot analysis (Figure 5e, f and Supplementary Fig. 8). Furthermore, confocal imaging indicated that Aiolos expression was restricted to the nucleus (Figure 5c).
Figure 5. TNFi-exposed IL-17+ CD4+ T cells express increased levels of the transcription factor IKZF3 associated with IL10 expression.
CD4+ T cells and monocytes were co-cultured with anti-CD3 mAb in the absence or presence of adalimumab, and IL-17+ CD4+ T cells were sorted on day 3. (a) The RMA normalised expression intensity for IKZF3 is shown for n=9 paired samples (mean with SEM). (b) IKZF3 expression (mean with SEM) was determined in 3 independent samples by real-time PCR, normalised to GAPDH. (c) Confocal microscopy of sorted control and TNFi-exposed IL-17+ CD4+ T cells stained for Aiolos (green). Nuclei were stained with DAPI (blue). Inserts (right) show single cells, demonstrating co-localization of Aiolos with DAPI in both conditions. Scale bars: 40 μm; 10 μm in inserts. (d) Quantification (mean grey value with 95% CI, corrected for background) of Aiolos staining per nucleus from confocal microscopy images. The experiment was repeated in three independent donors with similar results. (e, f) Western blot analysis of lysates from sorted control and TNFi-exposed IL-17+ CD4+ T cells probed with mAbs against Aiolos or β-actin. A single blot (e) and cumulative data (mean with SEM) quantified by densitometry (n=4. normalised to actin by densitometry) (f) are shown. Full gels shown in Supplementary Fig. 8. (g) Correlation between IKZF3 and IL10 expression in array samples (see Figure 4 for description), analysed by Pearson test. (h) IKZF3 expression (normalised to PPIA, mean with SEM, n=3) was measured by real-time PCR in sorted control or TNFi-exposed IL-17-IL-10- (Negs), IL-17+IL-10- (IL-17+), IL-17-IL-10+ (IL-10+) and IL-17+IL-10+ (dp+) CD4+ T cells. (i) Correlation between IKZF3 and IL10 expression in primary CD4+ T cells isolated from healthy donors as measured by real-time PCR (normalised to SDHA) and analysed by Pearson test.
Expression of IKZF3 was found to correlate very strongly with IL10 expression in the array samples (Pearson test, r=0.89, p<0.0001) (Figure 5g). To characterise which subpopulation of cells in the TNFi-exposed IL-17+ CD4+ T cell population expressed IKZF3, we cultured CD4+ T cells and monocytes with anti-CD3 mAb in the absence or presence of adalimumab for three days and sorted CD4+ T cells based on IL-17 and/or IL-10 expression (Supplementary Fig. 9). IKZF3 was expressed at the highest levels in the IL-10 expressing subsets (in both IL-10+IL-17− and IL-10+IL-17+ populations), and this was further enhanced following TNFi treatment (Figure 5h). To further investigate the relationship between IL-10 and Aiolos, we measured the mRNA expression of their genes in freshly isolated primary CD4+ T cells from healthy donors, and found a strong correlation (Figure 5i). Together these data suggested to us that the transcription factor Aiolos, encoded by IKZF3, may be involved in regulation of IL-10 expression in human CD4+ T cells.
We wished to investigate whether Aiolos may also regulate IL-10 in IFN-γ+ CD4+ T cells (Th1 cells), because we found that these cells also showed an increase in IL-10+ cells following TNFi exposure (Supplementary Fig. 10a, b). In a similar approach as we had employed for IL-17+ CD4+ T cells, we cultured CD4+ T cells and monocytes with anti-CD3 mAb in the absence or presence of adalimumab. After 3 days we sorted the IFN-γ+ IL-17− CD4+ T cells and measured the expression of IKZF3 by qPCR and Aiolos by Western blot. Although IKZF3/Aiolos expression was increased in some individuals, overall we could not detect a consistent increase in TNFi-exposed Th1 cells (Supplementary Fig. 8 and Supplementary Fig. 10c, d). These data indicate to us that there is a strong relationship between Aiolos and IL-10 expression in IL-17+ CD4+ T cells, whilst this relationship is less consistent in IFN-γ+ CD4+ T cells.
Aiolos drives IL10 in primary CD4+ effector T cells
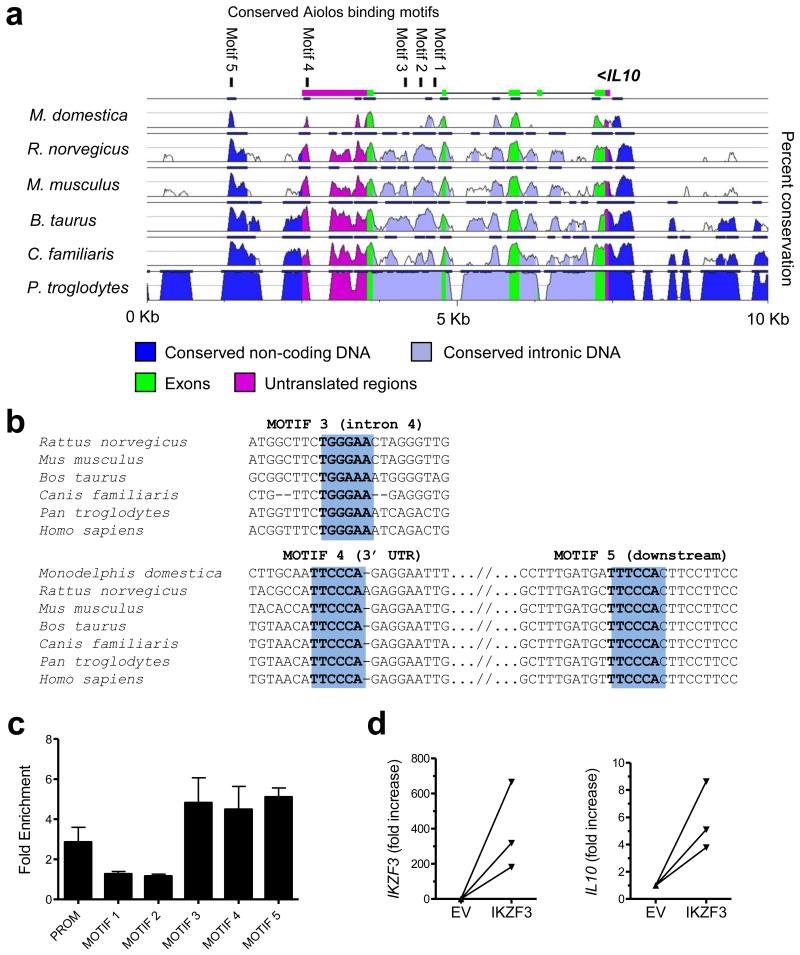
The strong association between IL10 and IKZF3 in IL-17+ CD4+ T cells suggested to us that Aiolos may play a role in the regulation of IL-10 expression in TNFi-exposed IL-17+ CD4+ T cells. To investigate this further, we first carried out an in depth bioinformatical analysis of the IL10 genomic region (Figure 6a, b). We found considerable evolutionary conservation in the IL10 locus and could detect 5 conserved putative Aiolos binding sites (TGGGAA). Interestingly, all of these putative Aiolos binding motifs are located either in the last intron or in the downstream region. To find out whether Aiolos interacts with these sites in TNFi-exposed IL-17+ CD4+ T-cells, we employed chromatin IP (ChIP). Chromatin from TNFi-exposed IL-17+ CD4+ T cells was fragmented to 150-200 bp fragments, enabling us to distinguish between individual binding sites. We detected strong Aiolos binding to motifs in the last intron (motif 3), in the 3′ UTR (motif 4) and the downstream non-coding region (motif 5) (Figure 6c). Of particular interest are motifs 4 and 5, which are located in narrow, but evolutionarily ultraconserved regions, placed amidst diverged genomic territories (Figures 6a, b). Conserved motifs 1 and 2, which are located in the 5′ end of the last intron, did not show significant Aiolos enrichment in our assay. The previously identified Ikaros binding site in the IL10 promoter20 demonstrated only a weak interaction with Aiolos. Overall these data provide evidence for Aiolos binding to evolutionarily conserved regions in the last intron and downstream of IL10.
Figure 6. Aiolos binds to the IL10 genomic region in TNFi-exposed IL-17+ CD4+ T cells and drives IL10 expression in primary CD4+ effector T cells.
(a) Analysis of sequence conservation in genomic IL10. Pair-wise sequence alignment to the human genome is depicted (50% lower cut-off), with evolutionarily conserved regions identified where identity above 70% over 100 bp was scored. These conserved genomic regions were searched for conserved consensus Aiolos DNA binding motifs, shown at the top of the graph and numbered from 1-5. (b) Multiple sequence alignment of putative Aiolos motifs 3, 4 and 5. The consensus binding site is shown in bold and highlighted in blue. (c) ChIP-PCR of sorted TNFi-exposed IL-17+ CD4+ T cells shows Aiolos enrichment at motifs 3, 4 and 5. Analysed DNA motifs are demonstrated on the horizontal axes and numbered according to Figure 6a. PROM indicates the previously found Ikaros binding sites in the promoter of mouse IL10. The values were normalized to two negative control regions (titin exons) to find enrichment of Aiolos binding at a region of interest over background signal (fold enrichment is shown ± SEM). (d) Memory CD4+ T cells were co-transfected with a CMV-eGFP reporter vector and either CMV-IKZF3 or CMV-empty vector (EV). At 48hrs eGFP+ cells were sorted and IKZF3 and IL10 expression determined by real-time PCR (data normalised to SDHA, mean ± SEM, n=3).
Finally, to determine whether Aiolos is sufficient to drive IL10 expression, we co-transfected primary memory CD4+ T cells from healthy controls with constructs encoding eGFP and IKZF3, and sorted the eGFP+ cells after 2 days. IL10 expression was increased in eGFP+ cells that were transfected with the IKZF3 construct as compared to the empty vector (EV) control (Figure 6d). Together, these data suggest that Aiolos is a transcriptional regulator of IL-10 expression in human IL-17+ CD4+ T cells, and potentially in other T helper cells.
DISCUSSION
Aiolos was identified in 1997 as the second member of the IKAROS family of zinc-finger transcription factors to which Ikaros, Helios, Eos and Pegasus belong21. Like other IKAROS gene family members, Aiolos has multiple isoforms and can control gene expression by interacting with other family members and different transcriptional regulators, including chromatin remodeling complex components22. Aiolos deficiency leads to alterations in B cells and increased levels of serum immunoglobulins and autoantibodies, B cell hyperproliferation and lymphomas23. Although T cell development in Aiolos-null animals appeared relatively unaffected, an increase in TCR-mediated proliferative responses in thymocytes and peripheral T cells, and an increased percentage of IFN-γ or IL-2 producing peripheral T cells has been reported23. Recently, Quintana et al. demonstrated that Aiolos silences the Il2 locus in mice, and that Aiolos-deficient mice show reduced Th17 and increased Th1 cell differentiation in vitro and in vivo24. The authors suggested that Aiolos, through its repressive effects on Il2 transcription, promotes Th17 differentiation, consistent with previous observations that IL-2 inhibits Th17 differentiation in mice25. Whether IL-2 exerts similar inhibitory effects on human Th17 cell development is not firmly established; in fact, addition of IL-2 seems a prerequisite in human in vitro Th17 differentiation/expansion protocols26, 27. Moreover, we found a small increase rather than a decrease in IL-2 in conditions where IKZF3 was increased (Supplementary Fig. 11). An important difference between the study by Quintana et al. and our study is that we observed the effects of Aiolos in effector CD4+ T cells, whilst Quintana et al. studied the role of Aiolos in naive CD4+ T cell differentiation. Taken together, these findings suggest that Aiolos may affect CD4+ T helper cells differently depending on their differentiation stage.
Our findings indicate that Aiolos may act as a direct transcriptional regulator of IL10 in CD4+ T cells. Using sorted TNFi-exposed IL-17+ CD4+ T cells, we show that Aiolos binds in the 3′ region of the IL10 locus. There is strong Aiolos binding to evolutionarily conserved regions with consensus motifs located 2192 bp downstream of the STOP codon, 959 bp from the start of the 3′ UTR and 584 bp from the beginning of intron 4. Interestingly, analysis of large-scale chromatin modification and transcription factor-DNA interaction screens, made available by the ENCODE consortium28, 29, demonstrate that our identified binding regions are possibly active in other cell types. For example, both intronic and downstream regions are covered by H3K27 acetylation signature, signifying an active, open state of chromatin30, 31. The downstream region is also located in a DNAse hypersensitivity cluster and occupied by p300, further suggesting transcription factor recruitment at this site32. Indeed, the ENCODE ChIP data on cultured cell-lines illustrate binding of SP1 to intron 4 and the downstream conserved region of IL10, and binding of MEF2A to these conserved genomic regions as well as to the 3′ UTR of IL10. These two factors have previously been linked with IL10 transcriptional regulation33, 34. Based on ENCODE data IRF4 binding could be detected in IL10 intron 4 and STAT3 binding in the downstream region. Both of these factors have also been associated with IL10 regulation in different cell types and conditions35-37. ENCODE results demonstrate NFκB binding in intron 4, the UTR and the downstream conserved region of IL10. Finally, Ikaros can be bound at these conserved regions in lymphoblastoid cell lines28, 29. Future efforts are needed to understand how many of the transcription factor-DNA binding events that were identified in cultured cell lines (ENCODE) occur in vivo in CD4+ T cells and how the different factors interact to fine-tune IL-10 expression and CD4+ T cell behaviour. Overall, the emerging evidence suggests that IL10 transcription relies on transcription factor recruitment to downstream enhancers. In addition to finding Aiolos binding regions in the IL10 locus we show here the functional consequence of this transcription factor recruitment, since IKZF3 overexpression was sufficient to drive IL10 expression in CD4+ T cells. IKAROS family members have been characterized mostly as transcriptional repressors38-40. Our results suggest a transcriptional activator function for Aiolos in CD4+ T cells.
It is well established that IL-10 can be expressed by multiple CD4+ T cell subsets, supporting the concept that IL-10 functions as a negative regulator following immune cell activation6. IL-10 co-expression in Th1 cells can be mediated by various pathways including STAT4, ERK, c-MAF and Notch6, 41. We observed that TNFi-exposed IFN-γ+ CD4+ T cells also showed an increase in the percentage of IL-10+ cells, however this was not associated with a consistent upregulation of IKZF3/Aiolos. It is possible that IL-10 regulation is differentially regulated in different Th subsets. We do not exclude the possibility however that Aiolos is involved in IL-10 regulation in cell types other than IL-17+ CD4+ T cells, as our data indicate that following TNFi exposure IKZF3 is increased in both IL-10+IL-17- and IL-10+IL-17+ cells. We also found that IKZF3 and IL10 expression are strongly correlated in primary CD4+ T cells ex vivo, and that IKZF3 overexpression results in increased IL10 expression in bulk memory CD4+ T cells. Together, our results suggest that increased expression of Aiolos in CD4+ T cells allows these cells to gain regulatory potential in the form of IL-10 production. In this regard, it is of interest to note that in the study by Quintana et al. the upregulated Aiolos expression was found in Th17 cells that were differentiated in the presence of IL-6 and TGF-β1, a differentiation protocol which leads to the generation of non-pathogenic Th17 cells9, 24, 42. Indeed, Lee et al. recently demonstrated that IKZF3 expression was part of the transcriptional signature of non-pathogenic Th17 cells43. An immunoregulatory role of Aiolos is furthermore supported by reports that Aiolos is expressed in inducible Tregs24.
IL-17 and IL-17+ CD4+ T cells are considered to be important contributors to synovial inflammation and bone erosion in experimental and clinical arthritis3, 44-48. Although IL-17+ CD4+ T cells constitute a relatively small population in human blood, these cells are enriched in the inflamed joint, and can have potent pro-inflammatory activity3, 44, 49. The acquisition of anti-inflammatory function through IL-10 expression by a substantial proportion of these cells may therefore aid in reducing inflammation. Although the IL-17+IL-10+ CD4+ T cell population is relatively small in size, studies in animal models have shown their potential in regulating inflammation, thus demonstrating their in vivo relevance10, 11. Given that TNFi treatment promotes IL-10 expression not only in IL-17+ CD4+ T cells but also in IFN-γ+ T cells, and possibly other effector cells, the concerted increase in IL-10 production is likely to result in a less inflammatory environment.
Our findings that IL-10+ CD4+ cells are increased following TNFi therapy in vitro and in vivo is in line with earlier work on mouse transgenic T cells and human tetanus-specific T cell clones which showed that chronic TNF exposure decreased IL-10 production whilst TNF blockade increased IL-1050, 51. Recently, several research groups have investigated the effect of TNFi treatment specifically on peripheral IL-17+ CD4+ T cells in vivo, with studies reporting increased52-54 as well as decreased55 percentages. The increase in peripheral IL-17+ CD4+ T cells was suggested to be due to impaired Th17 homing to the joint53. Our findings that TNFi drugs increase the percentage of IL-10+ cells within the IL-17+ CD4+ T population provides an additional explanation as to why an increase in IL-17+ CD4+ T cells does not necessarily lead to an increase in pathogenic potential and may therefore co-exist with successful TNFi therapy. Further large-scale studies are required to identify whether Aiolos is upregulated upon treatment in vivo and whether this can be linked to response, although a recent genome-wide association study of genetic predictors of TNFi treatment efficacy in RA did not identify IKZF3 as a significant locus56. It is interesting to note that the co-expression of IL-10 by IL-17+ CD4+ T cells in our in vitro studies could be blocked by the addition of the pro-inflammatory cytokine IL-1β. This finding corroborates a recent report that IL-1β can regulate IL-10 expression in human IL-17+ CD4+ T cells12, and suggests that depending on the inflammatory environment IL-17+ CD4+ T cells may be more or less pathogenic.
In conclusion, our data provide further evidence for the intrinsic ability of CD4+ effector T cells to have regulatory activity through the expression of the anti-inflammatory cytokine IL-10. We demonstrate that TNF-α blockade results in increased IL-10 responses in human CD4+ T cells and propose a role for Aiolos as a transcriptional regulator of this process.
METHODS
Patient and healthy control samples
Blood samples from patients with RA were obtained from the Rheumatology outpatient clinic at Guy’s Hospital. Disease activity score (DAS28), erythrocyte sedimentation rate (ESR), C-reactive protein (CRP), rheumatoid factor (RF) seropositivity, age, sex, and disease duration were documented on the day of sample collection (see Supplementary Fig. 2). Healthy control samples were obtained from local healthy volunteers. Analysis of blood samples from patients with peripheral spondyloarthritis (SpA) was performed on cryopreserved PBMC13. All clinical investigation was conducted according to Declaration of Helsinki principles. The work in this study was approved by the Bromley Research Ethics Committee and the local Ethics Committee of the AMC. Written informed consent was received from participants prior to inclusion in the study.
Peripheral blood mononuclear cell and cell subset isolation
Peripheral blood mononuclear cells (PBMC) were isolated by density gradient centrifugation (Lymphocyte separation media, PAA, Pasching, Austria). Cell subsets were purified by magnetic cell separation according to the manufacturer’s instructions (Miltenyi Biotec, Bergisch-Gladbach, Germany) and purity was confirmed by flow cytometry. Monocytes (average purity >95%) were isolated by positive selection using anti-CD14 microbeads. CD4+ T cells were isolated via negative depletion resulting in an average purity of >99%. In some experiments, CD4+ T cells were separated into CD45RO- and CD45RO+ cells using CD45RO beads (average purities 97% and 89%, respectively). CD4+CD25-T cells (average purity 85% CD25-) were obtained by CD4+CD25+ T cell depletion using CD25 MicroBeads.
Cell culture
CD4+ T cells (0.5×106) and CD14+ monocytes (0.5×106) were co-cultured in 48-well plates in culture medium (RPMI-1640 (Gibco, Camarillo, CA, USA), supplemented with 1% penicillin/streptomycin, 1% L-glutamine (Gibco) and 10% heat-inactivated fetal calf serum (Sigma, UK), in the presence of 100 ng/ml anti-CD3 mAb (OKT3, Janssen-Cilag Ltd, High Wycombe, UK). The TNFi drugs infliximab (Janssen Biologics), adalimumab (Abbott), etanercept (Pfizer) and certolizumab (UCB Pharma) were purchased from Guy’s Hospital Pharmacy, prepared according to manufacturers’ instructions, and frozen in aliquots at −20°C or kept at 4°C (certolizumab). All TNFi drugs were added in vitro at 1 μg/ml based on previous work15; human IgG1 Fc (R&D Systems, UK) and human IgG1 (Abcam, UK) were added at the same concentration as TNFi drugs. In some experiments, anti-CD3/CD28 beads (Invitrogen, Paisley, UK) were added to T cells at a ratio of 0.2:1, or PBMC depleted of CD14+ monocytes and CD4+ T cells were added as accessory cells at a 1:1 ratio. Transwell experiments were performed using a 0.4 μm semi-permeable membrane insert (Corning). Where indicated, FcγR blocking reagent (10 μl/ml, Miltenyi Biotec), blocking TNFRI/II Ab (5 μg/ml, R&D Systems), neutralizing anti-IL-10 mAb (clone 23738, 10 μg/ml, R&D Systems), blocking IL-10R mAb (clone 37607, 10μg/ml, R&D Systems), hrTNF-α (10ng/ml, R&D Systems) or hrIL-1β (10 ng/ml, R&D Systems) were added at the start of the co-culture.
Functional characterization of IL-17-secreting cells
CD4+ T cells and monocytes were co-cultured at a 2:1 ratio with anti-CD3 mAb +/− adalimumab (1 μg/ml). After 3 days, cells were stimulated with PMA (50 ng/ml, Sigma-Aldrich, St. Louis, MO, US) and ionomycin (750 ng/ml, Sigma-Aldrich) for 1.5 hours, followed by an IL-17 and/or IL-10 cytokine secretion assay, or where indicated an IFN-γ secretion assay (Miltenyi Biotec), according to the manufacturer’s instructions. CD14− cells were sorted using a FACSAria II (BD, Franklin Lakes NJ, USA) based on expression of IL-17-PE and/or IL-10-APC. Re-isolated total IL-17+ cells (20,000-200,000 cells) were stored in TRIzol (Invitrogen) for gene expression profiling, or RNA was isolated using the RNeasy mini plus extraction kit (Qiagen, Germantown, MD, USA) for real-time PCR, or cells were cultured overnight in culture medium (100,000 cells/well) for cytokine secretion. In some experiments, IL-17+ cells (50,000 cells) were added to autologous or allogeneic CD14+ monocytes (100,000 cells) in the absence or presence of neutralizing anti-IL-10 mAb (10 μg/ml) and monocyte phenotype was assessed after 40 hours by flow cytometry.
Flow cytometry
For ex vivo analysis, 1-5×106 PBMC were cultured in 24-well plates for 3 hrs in the presence of PMA, ionomycin and GolgiStop (according to the manufacturer’s instructions; BD, Oxford, UK). Cells were stained for cell surface markers using CD3-PE-Cy7 (1:100) and CD14-APC-Cy7 (1:100) (BioLegend, Cambridge, UK), fixed in 2% paraformaldehyde, permeabilized with 0.5% Saponin and stained for CD4-PacificBlue (1:100) in combination with IL-17-PE (1:20), IFN-γ-PerCP-Cy5.5 (1:200), IL-10-AF488 (1:20), and TNF-α-APC (1:100) (all BioLegend). Co-cultures were re-stimulated at day three with PMA/ionomycin for 6 hrs, with GolgiStop present during the last 3 hrs. Cells were stained for cell surface markers using CD2-Pacific Blue (1:1,000) (BioLegend), CD14-APC-Cy7, fixed in 2% paraformaldehyde, and permeabilized with 0.5% Saponin and stained for IL-17-PE, IFN-γ-PerCP-Cy5.5, IL-10-AF488, and TNF-α-APC. Foxp3 was measured using Foxp3-AF647 (1:20) (BioLegend) according to manufacturer’s instructions. Monocyte phenotype was determined by staining with antibodies to CD14-APC-Cy7 (BioLegend), HLA-DR-PerCP-Cy5.5 (1:50) (BD), CD40-PE (1:50) (AbD Serotec, Kidlington, UK), and CD163-FITC (1:50) (SantaCruz, Santa Cruz, CA, USA). Cells were acquired using a FACSCantoII (BD) and analyzed using FlowJo software (TreeStar, Inc).
Detection of soluble cytokines
Monocyte/T cell co-culture supernatants were collected at day 3 after stimulation with PMA/ionomycin and stored at −80°C. Cytokine levels were determined by ELISA using R&D Systems DuoSets for IL-17A, IL-10 and IFN-γ according to manufacturer’s instructions. Cytokine levels in supernatants from sorted IL-17-secreting T cells were determined using a Human Cytokine 25-Plex Panel (Invitrogen, Carlsbad, CA, USA) according to manufacturer’s instructions.
Gene expression profiling and analysis
Sorted IL-17+ cell samples were lysed in 500 μl of TRIzol (Invitrogen). Chloroform (0.2 ml) was added to 1 ml TRIzol cell homogenate and whirl mixed for 15 sec. Homogenates were incubated 2-3 min at RT and centrifuged 15 min at 10,000g (4°C). The water phase containing the RNA was further purified using RNeasy MinElute Cleanup Kit (Qiagen). RNA integrity was confirmed on an Agilent 2100 Bioanalyzer using total RNA nano chips (Agilent Technologies, Santa Clara, CA, USA). An amount of 100 ng of total RNA was used to prepare targets by 3′ IVT Express kit (Affymetrix, Santa Clara, CA, USA) following manufacturer’s instructions. Hybridization cocktails were hybridized onto Human Genome U133 Plus 2.0 Gene Chips® (Affymetrix) at 45°C for 17 h (60 rpm) in a Hybridization Oven 640 (Affymetrix). GeneChips® were washed and stained in a GeneChip® fluidics station 450 using the fluidics protocol “EukGE-WS2v5_450” (Affymetrix). Chips were scanned in a GeneChip® scanner 3000 (Affymetrix). Microarray data were normalized and gene expression measures derived using the RMA algorithm and the Bioconductor package “Affy” (http://www.bioconductor.org). Custom CDF (chip definition file) from brainarray.mbni.med.umich.edu was used. Qlucore Omics Explorer 2.2 (Qlucore AB, Sweden) was used for the statistical analysis of the normalized data.
Real time PCR
RNA was extracted with the RNeasy mini or RNeasy micro plus kit (Qiagen) and cDNA transcribed using a Reverse Transcription kit (High Capacity cDNA RT Kit, Applied Biosystems, Paisley, UK) according to manufacturers’ instructions. Real-time PCR reactions were performed in multiple replicates and run on an ABI Prism 7900HT sequence detection system (Applied Biosystems) using a SYBR Green PCR mastermix (Applied Biosystems). All primers are listed in Supplementary Table 1. Results were calculated using both the dCt method and the standard curve method and were normalized to two housekeeping genes.
Confocal microscopy
Control and TNFi-exposed IL-17+ CD4+ T cells were sorted and allowed to adhere onto poly-L-lysine coated cover slips for 45 minutes at 37 °C. Cells were fixed with 4% PFA and permeabilised in the presence of triton-X-100 (Sigma). Staining with rabbit anti-human Aiolos mAb (clone EPR9342(B), Epitomics, 1:10,000 dilution) (or an appropriate isotype control) was followed by incubation with goat anti-rabbit AF488 (Invitrogen) and DAPI (Invitrogen). Coverslips were mounted onto slides and slides imaged using an inverted Leica TCS SP5 DMI6000 confocal laser-scanning microscope (63x1.4NA oil immersion objective). To minimize fluorophore overlap DAPI stain (excited at 405 nm) and Aiolos (488 nm) were acquired sequentially. For quantification, Z projections (sum of slices) from eight fields of view were acquired for each condition and DAPI staining used to define a region of interest around the nucleus. Ten background measurements were collected per field and subtracted from the mean grey intensity of each cell measured.
Western blotting
Cells were washed with cold PBS followed by direct lysis in Laemmli buffer (Sigma-Aldrich). Lysates were boiled for 5 min at 95°C and resolved by electrophoresis using 4–12% Bis-Tris NuPAGE gels (Life Technologies Ltd, Paisley, UK). Proteins were transferred onto PVDF membranes (Merck Millipore, Billerica, MA, USA) which were cut horizontally near the 50 kD marker and probed with either anti-IKZF3 (1:5,000) (EPR9342(B), Epitomics) or anti-beta-actin (1:5,000) (13E5, Cell Signaling Technology, Danvers, MA), followed by horseradish peroxidase-conjugated polyclonal swine anti-rabbit immunoglobulins (1:3,333) (Dako, Glostrup, Denmark). Blots were developed with SuperSignal West Pico Substrate (Thermo Scientific, Rockford, IL, USA). Signals were acquired with the ChemiDoc XRS+ System using Image Lab software (Version 3.0, both Bio-Rad, Hemel Hempstead, UK) and quantified using Image J (Version 1.48b, NIH, Bethesda, MA).
Chromatin immunoprecipitation
TNFi-exposed IL-17+ CD4+ T cells (4×106) were sorted, washed in PBS, fixed at room temperature for 15 min in 1% formaldehyde and incubated 10 min in 0.125M Glycine. Cells were washed 2x in PBS, resuspended in 42 μl lysis buffer (100mM NaCl, 50 mM Tris-HCl pH 8.1, 5mM EDTA, 1% SDS) and incubated on ice for 15 min. 88 μl of dilution buffer (167 mM NaCl, 16.7 mM Tris-HCl pH 8.1, 1.2 mM EDTA, 1.1% Triton X-100, 0.01% SDS) was added to lysis buffer and the sample was transferred to a Covaris microtube. Chromatin was sheared with Covaris E220 to achieve average fragment sizes between 150-200 bp. After chromatin shearing the sample was diluted further by dilution buffer (total volume 420 μl) and centrifuged at maximum speed to remove insoluble cell debris. Millipore Magna ChIP Protein A-G beads were blocked with blocking buffer (0.2 mg/ml glycogen, 0.2 mg/ml BSA, 0.2 mg/ml yeast RNA in dilution buffer) overnight at 4 °C. Sheared chromatin was precleared with blocked beads for 1 h at 4 °C and centrifuged at 13,000 g for 5 min, after which 10% of the sample was removed for input control. 400 μl of the precleared chromatin was transferred to a new tube to bind 3 μg of antibody on a rotating platform (6 h at 4 °C). After 15 min centrifugation at 13,000g the antibody-chromatin solution was transferred to blocked magnetic beads and incubated overnight at 4 °C on a rotating platform. Next, the bead-DNA-antibody complex was washed twice in ChIP dilution buffer, once in low salt buffer (standard ChIP buffer compositions), twice in high salt buffer, once in LiCl buffer and twice in TE. Reverse crosslinking was carried out overnight at 65 °C and DNA purification was performed with IPure magnetic beads (Diagenode). qPCR for Aiolos enhancers and negative control regions was performed in duplicate with primers listed in Supplementary Table 1. The efficiency of ChIP was estimated for each chromatin region as a percent from the total input chromatin sample. The values were normalized to two negative control regions (titin exons) to find enrichment of Aiolos binding at a region of interest over background signal (fold enrichment is shown ± SEM).
Transfection
Primary memory CD4+ T cells from healthy donors were stimulated with 10 ng/ml PMA and 150 ng/ml ionomycin for 18-20 hrs. Cells were co-transfected with 5 μg cmv-eGFP and 5 μg p-CMV-SPORT-IKZF3 (IRATp970D0655D, Source BioScience LifeSciences) or 5 μg p-CMV-SPORT (made by excising IKZF3 from the p-CMV-SPORT-IKZF3 vector using Not1 and Sal1) using the Amaxa™ Human T Cell Nucleofector™ kit (Lonza), according to the manufacturer’s instructions. Cells were then cultured for 48 hrs followed by sorting of the GFP+ cells for qPCR. Viability post-transfection was ~60%, and the transfection efficiency in viable cells was 11-30%.
Statistical and bioinformatical analysis
Statistical testing was performed with GraphPad 5.03 (GraphPad, San Diego, CA, USA). Datasets were tested for normality using the D’Agostino & Pearson omnibus normality test, followed by statistical significance testing using the appropriate statistical tests as indicated in the legends. Datasets with n-values less than 8 were always tested non-parametrically. For analysis of the microarray data, for each gene, a paired t-test was performed to compare the expression levels between the conditions (Adalimumab and control), using Qlucore Omics Explorer 2.2 (Qlucore AB, Sweden). Adjusted p-values (or q-values) were computed using the Benjamini-Hochberg procedure. P-values <0.05 were considered to be statistically significant. Evolutionarily conserved regions (70% similarity over 100 bp) were identified with ECR Browser57 aligned by Mulan and searched for conserved Aiolos motifs by multiTF58, 59.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the IMI JU funded project BTCure 115142-2, Arthritis Research UK (GJW, grant 19307), the Guy’s and St Thomas’ Charity Prize PhD Programme in Biomedical and Translational Science (CAR and JS) and the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. Work in FG lab (UR, FG) was funded by MRC G0900867 and ERC 261299 to FG. The authors would like to acknowledge Ms Jacqueline Paramarta (AMC), Dr Bina Menon (GSTT) and Ms Cristina Blanco-Gil (GSTT) for help in patient recruitment, Dr Jie Li (KCL) for testing antibodies, Dr Megha Rajasekhar (KCL) for assistance in growing plasmids and cell sorting, and Dr Susan John (KCL) and Dr Susan Chan (IGBMC) for helpful discussions.
Footnotes
Conflict of interest
JGG and KSF are employees of Novo Nordisk A/S. BWK received financial support or fees from UCB Pharma, Roche, Abbott, Pfizer, Bristol-Myers Squibb and Chugai. All other authors declare no competing financial interests.
Accession codes
The microarray data are deposited in Gene Expression Omnibus (GEO) with accession number GSE51540.
REFERENCES
- 1.Miossec P, Korn T, Kuchroo VK. Interleukin-17 and type 17 helper T cells. N Engl J Med. 2009;361:888–898. doi: 10.1056/NEJMra0707449. [DOI] [PubMed] [Google Scholar]
- 2.Pene J, et al. Chronically inflamed human tissues are infiltrated by highly differentiated Th17 lymphocytes. J Immunol. 2008;180:7423–7430. doi: 10.4049/jimmunol.180.11.7423. [DOI] [PubMed] [Google Scholar]
- 3.Gullick NJ, et al. Linking Power Doppler ultrasound to the presence of Th17 cells in the rheumatoid arthritis joint. PLoS ONE. 2010;5:e12516. doi: 10.1371/journal.pone.0012516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel DD, Lee DM, Kolbinger F, Antoni C. Effect of IL-17A blockade with secukinumab in autoimmune diseases. Ann Rheum Dis. 2013;72(Suppl 2):ii116–123. doi: 10.1136/annrheumdis-2012-202371. [DOI] [PubMed] [Google Scholar]
- 5.O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saraiva M, O’Garra A. The regulation of IL-10 production by immune cells. Nat Rev Immunol. 2010;10:170–181. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- 7.Annunziato F, et al. Phenotypic and functional features of human Th17 cells. J Exp Med. 2007;204:1849–1861. doi: 10.1084/jem.20070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Afzali B, Mitchell P, Lechler RI, John S, Lombardi G. Translational Mini-Review Series on Th17 Cells: Induction of interleukin-17 production by regulatory T cells. Clin Exp Immunol. 2010;159:120–130. doi: 10.1111/j.1365-2249.2009.04038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghoreschi K, et al. Generation of pathogenic TH17 cells in the absence of TGF-β signalling. Nature. 2010;467:967–971. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGeachy MJ, et al. TGF-[beta] and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain TH-17 cell-mediated pathology. Nat Immunol. 2007;8:1390–1397. doi: 10.1038/ni1539. [DOI] [PubMed] [Google Scholar]
- 11.Esplugues E, et al. Control of TH17 cells occurs in the small intestine. Nature. 2011;475:514–518. doi: 10.1038/nature10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zielinski CE, et al. Pathogen-induced human TH17 cells produce IFN-γ or IL-10 and are regulated by IL-1β. Nature. 2012;484:514–518. doi: 10.1038/nature10957. [DOI] [PubMed] [Google Scholar]
- 13.Paramarta JE, et al. Efficacy and safety of adalimumab for the treatment of peripheral arthritis in spondyloarthritis patients without ankylosing spondylitis or psoriatic arthritis. Ann Rheum Dis. 2013;72:1793–1799. doi: 10.1136/annrheumdis-2012-202245. [DOI] [PubMed] [Google Scholar]
- 14.Evans HG, Suddason T, Jackson I, Taams LS, Lord GM. Optimal induction of T helper 17 cells in humans requires T cell receptor ligation in the context of Toll-like receptor-activated monocytes. Proc Natl Acad Sci U S A. 2007;104:17034–17039. doi: 10.1073/pnas.0708426104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans HG, et al. In vivo activated monocytes from the site of inflammation in humans specifically promote Th17 responses. Proc Natl Acad Sci U S A. 2009;106:6232–6237. doi: 10.1073/pnas.0808144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ehrenstein MR, et al. Compromised function of regulatory T cells in rheumatoid arthritis and reversal by anti-TNF-α therapy. J Exp Med. 2004;200:277–285. doi: 10.1084/jem.20040165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valencia X, et al. TNF down-modulates the function of human CD4+CD25hi T regulatory cells. Blood. 2006;108:253–261. doi: 10.1182/blood-2005-11-4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buechler C, et al. Regulation of scavenger receptor CD163 expression in human monocytes and macrophages by pro- and antiinflammatory stimuli. J Leukoc Biol. 2000;67:97–103. [PubMed] [Google Scholar]
- 19.Xu J, et al. c-Maf regulates IL-10 expression during Th17 polarization. J Immunol. 2009;182:6226–6236. doi: 10.4049/jimmunol.0900123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Umetsu SE, Winandy S. Ikaros is a regulator of IL10 expression in CD4+ T cells. J Immunol. 2009;183:5518–5525. doi: 10.4049/jimmunol.0901284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morgan B, et al. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J. 1997;16:2004–2013. doi: 10.1093/emboj/16.8.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitt C, et al. Aiolos and Ikaros: regulators of lymphocyte development, homeostasis and lymphoproliferation. Apoptosis. 2002;7:277–284. doi: 10.1023/a:1015372322419. [DOI] [PubMed] [Google Scholar]
- 23.Wang J-H, et al. Aiolos regulates B cell activation and maturation to effector state. Immunity. 1998;9:543–553. doi: 10.1016/s1074-7613(00)80637-8. [DOI] [PubMed] [Google Scholar]
- 24.Quintana FJ, et al. Aiolos promotes Th17 differentiation by directly silencing Il2 expression. Nat Immunol. 2012;13:770–777. doi: 10.1038/ni.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laurence A, et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26:1–11. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 26.Acosta-Rodriguez EV, Napoletani G, Lanzavecchia A, Sallusto F. Interleukins 1beta and 6 but not transforming growth factor-beta are essential for the differentiation of interleukin 17-producing human T helper cells. Nat Immunol. 2007;8:942–949. doi: 10.1038/ni1496. [DOI] [PubMed] [Google Scholar]
- 27.Manel N, Unutmaz D, Littman DR. The differentiation of human Th17 cells requires transforming growth factor-β and induction of the nuclear receptor RORγt. Nat Immunol. 2008;9:641–649. doi: 10.1038/ni.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Consortium TEP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Consortium TEP. A User’s Guide to the Encyclopedia of DNA Elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ong C-T, Corces VG. Enhancers: emerging roles in cell fate specification. EMBO Rep. 2012;13:423–430. doi: 10.1038/embor.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liopeta K, et al. cAMP regulates IL-10 production by normal human T lymphocytes at multiple levels: A potential role for MEF2. Mol Immunol. 2009;46:345–354. doi: 10.1016/j.molimm.2008.10.025. [DOI] [PubMed] [Google Scholar]
- 34.Tone M, Powell MJ, Tone Y, Thompson SAJ, Waldmann H. IL-10 Gene Expression Is Controlled by the Transcription Factors Sp1 and Sp3. J Immunol. 2000;165:286–291. doi: 10.4049/jimmunol.165.1.286. [DOI] [PubMed] [Google Scholar]
- 35.Cretney E, et al. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat Immunol. 2011;12:304–311. doi: 10.1038/ni.2006. [DOI] [PubMed] [Google Scholar]
- 36.Samarasinghe R, et al. Induction of an anti-inflammatory cytokine, IL-10, in dendritic cells after toll-like receptor signaling. J Interferon Cytokine Res. 2006;26:893–900. doi: 10.1089/jir.2006.26.893. [DOI] [PubMed] [Google Scholar]
- 37.Benkhart EM, Siedlar M, Wedel A, Werner T, Ziegler-Heitbrock HWL. Role of Stat3 in Lipopolysaccharide-Induced IL-10 Gene Expression. J Immunol. 2000;165:1612–1617. doi: 10.4049/jimmunol.165.3.1612. [DOI] [PubMed] [Google Scholar]
- 38.Brown KE, et al. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 39.Naito T, Gomez-del Arco P, Williams CJ, Georgopoulos K. Antagonistic interactions between Ikaros and the chromatin remodeler Mi-2beta determine silencer activity and CD4 gene expression. Immunity. 2007;27:723–734. doi: 10.1016/j.immuni.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 40.Koipally J, Renold A, Kim J, Georgopoulos K. Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. EMBO J. 1999;18:3090–3100. doi: 10.1093/emboj/18.11.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cope A, Le Friec G, Cardone J, Kemper C. The Th1 life cycle: molecular control of IFN-γ to IL-10 switching. Trends Immunol. 2011;32:278–286. doi: 10.1016/j.it.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 42.McGeachy MJ, et al. TGF-beta and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain Th-cell-mediated pathology. Nat Immunol. 2007;8:1390–1397. doi: 10.1038/ni1539. [DOI] [PubMed] [Google Scholar]
- 43.Lee Y, et al. Induction and molecular signature of pathogenic Th17 cells. Nat Immunol. 2012;13:991–999. doi: 10.1038/ni.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leipe J, et al. Role of Th17 cells in human autoimmune arthritis. Arthritis Rheum. 2010;62:2876–2885. doi: 10.1002/art.27622. [DOI] [PubMed] [Google Scholar]
- 45.Kirkham BW, et al. Synovial membrane cytokine expression is predictive of joint damage progression in rheumatoid arthritis: A two-year prospective study (the DAMAGE study cohort) Arthritis & Rheumatism. 2006;54:1122–1131. doi: 10.1002/art.21749. [DOI] [PubMed] [Google Scholar]
- 46.Sato K, et al. Th17 functions as an osteoclastogenic helper T cell subset that links T cell activation and bone destruction. J. Exp. Med. 2006;203:2673–2682. doi: 10.1084/jem.20061775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koenders MI, et al. Blocking of Interleukin-17 during Reactivation of Experimental Arthritis Prevents Joint Inflammation and Bone Erosion by Decreasing RANKL and Interleukin-1. Am J Pathol. 2005;167:141–149. doi: 10.1016/S0002-9440(10)62961-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lubberts E, et al. Treatment with a neutralizing anti-murine interleukin-17 antibody after the onset of collagen-induced arthritis reduces joint inflammation, cartilage destruction, and bone erosion. Arthritis & Rheumatism. 2004;50:650–659. doi: 10.1002/art.20001. [DOI] [PubMed] [Google Scholar]
- 49.van Hamburg JP, et al. Th17 cells, but not Th1 cells, from patients with early rheumatoid arthritis are potent inducers of matrix metalloproteinases and proinflammatory cytokines upon synovial fibroblast interaction, including autocrine interleukin-17A production. Arthritis & Rheumatism. 2011;63:73–83. doi: 10.1002/art.30093. [DOI] [PubMed] [Google Scholar]
- 50.Cope AP, et al. Chronic exposure to tumor necrosis factor (TNF) in vitro impairs the activation of T cells through the T cell receptor/CD3 complex; reversal in vivo by anti-TNF antibodies in patients with rheumatoid arthritis. J Clin Invest. 1994;94:749–760. doi: 10.1172/JCI117394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cope AP, et al. Chronic tumor necrosis factor alters T cell responses by attenuating T cell receptor signaling. J Exp Med. 1997;185:1573–1584. doi: 10.1084/jem.185.9.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen D-Y, et al. Increasing levels of circulating Th17 cells and interleukin-17 in rheumatoid arthritis patients with an inadequate response to anti-TNF-alpha therapy. Arthritis Res Ther. 2011;13:R126. doi: 10.1186/ar3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aerts NE, et al. Increased IL-17 production by peripheral T helper cells after tumour necrosis factor blockade in rheumatoid arthritis is accompanied by inhibition of migration-associated chemokine receptor expression. Rheumatol. 2010;49:2264–2272. doi: 10.1093/rheumatology/keq224. [DOI] [PubMed] [Google Scholar]
- 54.Alzabin S, et al. Incomplete response of inflammatory arthritis to TNFα blockade is associated with the Th17 pathway. Ann Rheum Dis. 2012;71:1741–1748. doi: 10.1136/annrheumdis-2011-201024. [DOI] [PubMed] [Google Scholar]
- 55.Aravena O, et al. Anti-TNF therapy in patients with rheumatoid arthritis decreases Th1 and Th17 cell populations and expands IFN-γ-producing NK cell and regulatory T cell subsets. Immunobiology. 2011;216:1256–1263. doi: 10.1016/j.imbio.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 56.Plant D, et al. Genome-wide association study of genetic predictors of anti-tumor necrosis factor treatment efficacy in rheumatoid arthritis identifies associations with polymorphisms at seven loci. Arthritis Rheum. 2011;63:645–653. doi: 10.1002/art.30130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loots GG, Ovcharenko I. Mulan: multiple-sequence alignment to predict functional elements in genomic sequences. Methods Mol Biol. 2007;395:237–254. [PMC free article] [PubMed] [Google Scholar]
- 58.Ovcharenko I, et al. Mulan: Multiple-sequence local alignment and visualization for studying function and evolution. Genome Research. 2005;15:184–194. doi: 10.1101/gr.3007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucl Acids Res. 2004;32:W280–W286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.