Abstract
Historically, animal viruses have been classified on the basis of the presence or absence of an envelope – an external lipid bilayer membrane typically carrying one or more viral glycoproteins. However, growing evidence indicates that some ‘non-enveloped’ viruses circulate in the blood of infected individuals enveloped in host-derived membranes that provide protection from neutralizing antibodies. In this opinion piece, we discuss this novel strategy for virus survival and consider how it contributes to the pathogenesis of acute viral hepatitis. The acquisition of an envelope by ‘non-enveloped’ viruses profoundly influences their interaction with the host at both the cellular and system level, and challenges how we think about vaccine protection against these infections.
Keywords: hepatitis A virus, hepatitis E virus, membrane hijacking, ESCRT, late domain
To have and have not: viruses and envelopes
Virologists, like all scientists, are drawn to classifying the things they study. As of 2012, the International Committee on the Taxonomy of Viruses (ICTV) had agreed on the existence of 2618 species of viruses, spread across 420 genera, 96 families and 7 orders [1]. The classification of viruses increasingly rests upon evolutionary relationships inferred from phylogenetic analyses of virus sequences, but for many decades viruses were classified based on a small number of key physical attributes: size, shape, type of nucleic acid, and the presence or absence of an envelope as reflected in susceptibility to ether or bile-salt inactivation. Old habits die slowly and even today most virologists maintain a dichotomous view of viruses, placing them into one of two bins: those with, and those without an envelope, i.e., an outer lipid bilayer derived from cellular membranes in which host membrane proteins have been largely replaced by viral glycoproteins (peplomers). The envelope is essential for those viruses that possess one, because the peplomers embedded within it are typically key determinants of cell tropism and entry. On the other hand, the absence of an envelope can confer specific advantages, including greater environmental stability. Such attributes have substantial impact on how viruses are transmitted, and how they are seen by the host immune system.
Life is never simple, however, and there are now examples of viruses that exist outside the cell in two equally infectious guises: one with, and one without an envelope. How and why do these viruses do it? And what does this mean for our ability to prevent these infections with a vaccine? We focus here on two such viruses, hepatitis A virus (HAV) and hepatitis E virus (HEV), both considered by virologists for many years to be ‘non-enveloped’ viruses, but recently recognized to have such a complicated modus operandi. HAV, a member of the non-enveloped family Picornaviridae, has evolved a unique strategy by which it hijacks cellular membranes to exit cells fully cloaked in a lipid membrane [2]. HEV, which may have evolved from an ancestral enveloped virus [3], appears to have acquired the ability to shed its envelope and the liabilities it poses for transmission through the environment [4]. Their unusual lifestyle allows these viruses to evade neutralizing antibodies and facilitates their spread within infected hosts while maximizing opportunities for inter-host transmission.
HAV and HEV, viruses with dual personalities
HAV and HEV are phylogenetically unrelated, small RNA viruses, each with a single- stranded, positive-sense genome (Figure 1). Both infect the liver and cause acute inflammatory hepatitis [5, 6]. Remarkably, they circulate in the blood during acute infection in a membrane- enveloped form, but are shed in feces as non-enveloped viruses [2, 4]. Both types of particles, enveloped and non-enveloped, appear to be equally infectious, but in both cases the enveloped viruses are highly resistant to neutralizing antibodies, while non-enveloped particles are not [2, 4]. This remarkable convergence of phenotypes likely relates in part to the unique setting of the liver within which these viruses replicate, and for which the biliary tract provides an efficient conduit to the outside environment (Figure 2A).
Figure 1.
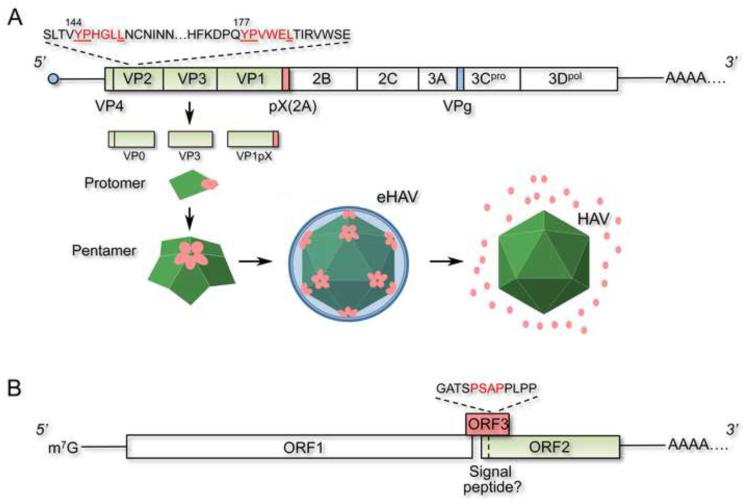
Organization of the 3’ poly-adenylated, positive-strand RNA genomes of HAV and HEV. (A) The HAV genome contains one large ORF (box) that encodes a giant polyprotein that is processed into 9 or more mature proteins. The three major capsid proteins, VP0, VP3 and VP1 (shaded in green) are processed from the polyprotein by the viral protease, 3Cpro, and form protomers that then assemble into pentamers in a process dependent upon pX, an 8 kd carboxy- terminal extension of VP1 (pink). Pentamers subsequently assemble into an icosahedral capsid that packages the viral genome and is released from cells wrapped in cellular membranes (eHAV). pX is stable within the eHAV particle, but is degraded following dissolution of the membrane. Tandem YPX3L motifs (red font) in VP2 likely interact with the ESCRT-associated protein Alix, and together with pX direct the membrane envelopment of the capsid. (B) The HEV genome contains 3 ORFs, the largest and most 5’ of which (ORF1) encodes nonstructural proteins that direct its replication. The most 3’, ORF2, encodes the capsid protein that appears to have an amino-terminal signal sequence. ORF3 is required for the release of virus from cells enveloped in membranes, and contains a PSAP late domain mediating interactions with the ESCRT-associated protein, TSG101. The HEV genome terminates at the 5’ end with a m7G cap, while the HAV genome is covalently linked to a small viral protein (VPg) and is translated under the control of an internal ribosome entry site in its 5’ untranslated RNA segment.
Figure 2.
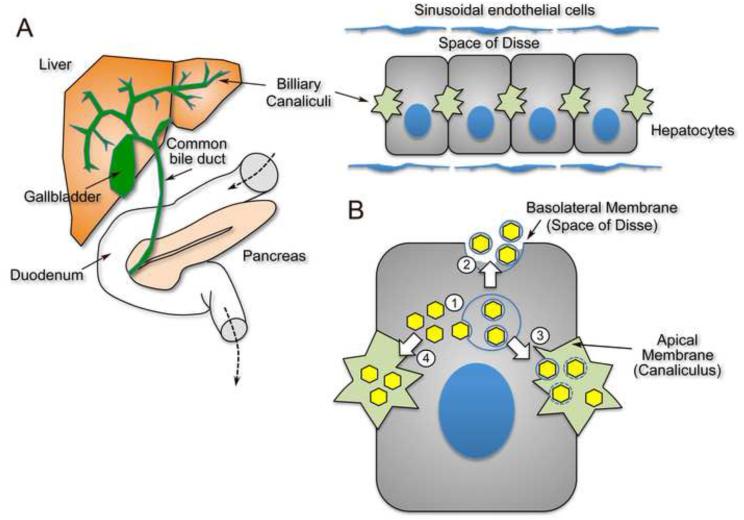
Hepatic origin of fecally-shed HAV and HEV. (A) Replication of HAV and HEV occurs primarily within cords of polarized hepatocytes (right) that form the architecture of the liver. Virus is released across the canalicular (apical) membrane into the biliary canaliculus and to a lesser extent across the basolateral membrane into the space of Disse and the hepatic sinusoids that are bathed by blood. It is likely that most virus shed in feces is replicated within hepatocytes and passes to the duodenum via the hepatobiliary system (left). (B) Cellular egress and trafficking of HAV from hepatoctyes. (1) Enveloped virions (eHAV, yellow hexagons) are likely formed by budding into MVBs mediated by interactions of the capsid with Alix and the ESCRT system, and (2) are then released across the basolateral membrane into the space of Disse resulting in systemic circulation of eHAV. Virus shed in feces lacks an envelope, and could exit hepatocytes across the canalicular membrane either (3) by a similar mechanism with loss of the envelope due to the detergent action of bile salts, or (4) by an independent mechanism resulting in vectorial apical transport of non-enveloped capsids. Yet a third possibility (not shown) is that eHAV released from the basolateral membrane may re-enter the hepatocyte and undergo trans-cytosis with subsequent release across the canalicular membrane as non- enveloped virions.
HAV is an ancient human pathogen and a common cause of enterically-transmitted acute viral hepatitis [5]. Its genome organization and phylogeny classify it squarely as the sole member of a distinct genus, the Hepatoviruses, within the family Picornaviridae [7]. As with all picornaviruses, the genome is released from the HAV capsid during the process of viral entry, with an internal ribosome entry site in the 5’ untranslated RNA segment then initiating the translation of a giant polyprotein that is subsequently processed into 9 mature proteins as well as several functional processing intermediates [8] (Figure 1A). The major capsid proteins, VP0, VP3 and VP1pX, assemble in the cytoplasm as pentamers, with 12 pentamers subsequently forming a viral capsid. ‘pX’ is an 8 kDa carboxy-terminal extension of VP1 that is cleaved from it by an unknown cellular protease late in maturation of the virion [9-11]. HAV replication is slow and generally noncytolytic in cell culture. Infectious virus is released into supernatant fluids [12], but how this occurs in the absence of cell lysis was unexplained for many years.
Recent work reveals that the virus released into supernatant fluids comprises two populations of infectious particles with distinctly different morphology, buoyant density, and resistance to neutralizing antibodies [2]. Under the electron microscope, virus banding at ~1.22 g/cm3 in isopycnic iodixanol gradients has the size and shape of HAV particles first identified by Feinstone et al. in 1973 in human feces [13]. However, a greater proportion of the virus bands at a much lighter density, ~1.08 g/cm3, and is comprised of similar appearing capsids enveloped in a single, amorphous lipid bilayer [2]. Most of these vesicle-like structures contain 1 or 2 capsids, but a few contain as many as 4, suggesting the membrane is acquired after capsid assembly. Surprisingly, viruses in the light and dense fractions appear to have equivalent infectivity (infectious focus-forming units/genome copy). Consistent with being fully enveloped in membranes, the light particles are resistant to neutralizing antibodies [2]. Their infectivity is virtually eliminated by extraction with chloroform, as the membrane-associated virus partitions into the interface and is lost from the aqueous phase. Given that HAV has been classified for many years among the picornaviruses, a large and diverse family of non-enveloped viruses, it was surprising to find that most virions released into the supernatant fluids of infected hepatoma cell cultures were enveloped in this fashion. Even more surprising, only the enveloped form of the virus (eHAV) was found in the blood of patients with acute hepatitis A [2]. In contrast, virus shed in feces lacks an envelope, similar to any standard picornavirus.
Less is known about HEV. It causes acute enterically-transmitted hepatitis, similar to HAV, and has been associated with massive water-borne outbreaks of disease in developing countries [6, 14]. In recent years, however, there has been growing awareness of its ability to cause both persistent infections and liver disease in immunocompromised persons, especially solid organ transplant recipients [15]. The HEV genome is organized very differently from HAV, with 3 separate open reading frames (ORFs), the most 3’ of which (designated ORF2) encodes a single capsid protein [16] (Figure 1B). HEV is currently classified together with a closely related avian virus in a free-standing virus family (no assigned order), the Hepeviridae. However, its closest phylogenetic relationship is with the rubiviruses (Rubella virus) [3], a genus in the Togaviridae - so named for the envelope that cloaks viruses in this family. Despite this relatedness, HEV particles shed in the feces of infected individuals lack a lipid envelope. Thus, since it was first visualized by electron microscopy 30 years ago [17], HEV has been considered a non-enveloped virus.
However, recent work has shown that HEV circulates in the blood during acute infection (or is released from infected cell cultures) entirely enveloped in host membranes and, as with HAV, highly resistant to neutralizing antibody [4, 18]. Both its association with membranes and the release of HEV from infected cell cultures is dependent upon the small, centrally placed ORF3 (Figure 1B), deletion of which ablates release without affecting replication of the RNA [19]. Remarkably, detergent-treated membrane-associated HEV (by analogy with eHAV, ‘eHEV’) infects cultured cells with an efficiency that is nearly identical to untreated virions [4]. Immune sera or monoclonal antibodies against ORF2 neutralize the infectivity of non-enveloped HEV derived from feces, but have no effect on eHEV found in serum or released from infected cell cultures [18, 20].
Thus, both HAV and HEV engage in ‘membrane hijacking’: a process by which an apparently ‘non-enveloped’ virus hijacks cellular membranes as it is released from the infected cell into the extracellular milieu (in the case of eHAV and eHEV, the hepatic sinusoid) (Figure 2A), facilitating its escape from neutralizing antibodies directed at the capsid. At the same time, virus shed from the host into the external environment (in feces, with both HAV and HEV) lacks an envelope, providing for a high level of stability and promoting transmission to other individuals. The lifestyles adopted by these two hepatotropic viruses thus resemble a game of peek-a-boo, where first you see the virus, then you don’t. It is remarkable that these two, phylogenetically distinct, enterically-transmitted hepatitis viruses have evolved such similar yet unusual lifestyles. How do they do it, and what are the implications for viral evolution and disease prevention?
Membrane hijacking and the ESCRT system
Many enveloped viruses rely on the host endosomal sorting complex required for transport (ESCRT) for the budding of their envelope from cell membranes [21]. The ESCRT system plays central roles in the biogenesis of multivesicular bodies (MVBs, see Glossary), cytokinesis, and autophagy [22]. It consists of four multi-protein complexes (ESCRT-0, -I, -II, III) and several associated proteins, among them tumor suppressor gene 101 (TSG101) and programmed cell death-6 interacting protein (PDCD6IP, better known as Alix). At the simplest level, these proteins work in a coordinated fashion to sort and load protein cargos into vesicles within MVBs for transport to the lysosome or the plasma membrane [22]. The protein cargos are typically ubiquitylated and also contain a ‘late domain’ with a defined sequence motif, P(S/T)XP, YPX1-3L, or PPXY, which specifies interactions with an ESCRT-associated protein such as TSG101 or Alix. ESCRT complexes are assembled and generate a local membrane invagination (a bud) to which the cargo is moved. Vacuolar protein sorting 4 homolog A and B (VPS4A and VPS4B), related AAA type ATPases, are recruited in the final step of the budding process and facilitate removal and re-cycling of ESCRT components [22]. Structural proteins of many enveloped viruses contain late domains that function in the budding of their envelopes [21, 23]. Two well-known examples are the Gag protein of human immunodeficiency virus (HIV) and the VP40 protein of Ebola virus [24], both of which express proteins containing late domains that recruit TSG101 to facilitate their egress from cells enveloped in membranes.
HAV and HEV have evolved similar strategies to direct their release from cells as enveloped viruses. A search for potential late domains in the HAV polyprotein identified two tandem YPX3L motifs in the VP2 capsid protein. Tyr to Ala substitutions within either motif abrogated capsid assembly [2], making it difficult to determine whether the capsid interacts with Alix, an ESCRT-I/III associated protein that binds this late domain motif. However, anti-Alix antibodies precipitated encapsidated viral RNA, and silencing of Alix expression blocked release of the enveloped virus. Depletion of VPS4B also led to a significant reduction in eHAV release [2]. In contrast, depletion of beclin-1, a key factor in autophagosome formation, had no effect on eHAV release, excluding autophagosome-mediated exit as proposed for non-cytolytic release of poliovirus [25]. These data are consistent with a role for the ESCRT system, Alix, and the candidate VP2 late domains in viral assembly and release. However, other signals are likely to be involved in membrane hijacking by HAV, as the eHAV capsid contains unprocessed VP1pX (Figure 1A), while it is fully processed to VP1 in non-enveloped virions [2]. This suggests that the pX extension of VP1 contributes to the envelopment of HAV by membranes, but exactly how is uncertain. eHEV also contains a viral protein not present in the non-enveloped virus, as the ORF3 protein is found in enveloped virions released from infected cells but not non-enveloped virions shed in feces [26]. The ORF3 protein possesses a PSAP motif that directs interactions with TSG101, and disrupting this motif or depleting TSG101 results in a significant reduction in viral release [26, 27]. Corroborating the notion that HEV, like HAV, usurps the ESCRT system for budding, overexpression of a dominant-negative VPS4 mutant inhibits HEV release.
pX is not expressed on the surface of enveloped eHAV particles, and becomes accessible to proteinases only after removal of the membranes by detergent [2]. Whether ORF3 is expressed on the eHEV particle is not clear. Some published data suggest that antibodies to ORF3 protein precipitate and neutralize eHEV infectivity, while other data from the same group suggest this is not the case [4, 18]. At present there is no evidence that either of these membrane-wrapped viruses have surface-exposed virally-encoded glycoprotein peplomers, a feature that distinguishes them from conventional enveloped viruses. However, there may be more to this story than we know so far, as the HEV ORF2 protein (the major capsid protein) possesses an N-terminal signal sequence (Figure 1B) and may undergo glycosylation [28]. The available data suggest the ORF2 protein is not accessible on the surface of the eHEV particle, however, and how this might function in the context of assembly and release of the HEV particle remains uncertain.
The presence of a signal sequence and potential glycosylation of the capsid protein distinguish HEV from HAV, as does the phylogenetic relatedness of HEV to the togaviruses. Taken together, these observations suggest the interesting possibility that HEV may be an ‘enveloped’ virus that has evolved the capacity to survive extracellularly and mediate its cellular entry in the absence of its envelope. HAV, in contrast, is a ‘non-enveloped’ virus that cloaks itself in an envelope by hijacking cellular membranes, thereby providing it with a survival advantage within the host. There is little understanding of how the enveloped HAV and HEV particles, both of which are as infectious as their non-enveloped counterparts, enter cells [2, 4]. In the case of HAV, this occurs via a late endosome pathway and may have some similarities with exosome entry [2].
Membrane hijacking and disease pathogenesis
Virologists have known for almost 75 years that some viruses (conventional ‘enveloped’ viruses, such as the togaviruses) are readily inactivated by bile salts, while others (‘non- enveloped’ viruses such as poliovirus) are not [29]. The detergent action of bile salts may account for the lack of membranes surrounding fecally shed HAV and HEV particles, as both are likely to be produced primarily within hepatocytes and secreted into the gastrointestinal tract via the biliary system (Figure 2A). This is a very attractive hypothesis, but it remains to be demonstrated. There was no loss of the membranes enveloping eHAV upon being placed directly into 98% fresh porcine bile [2]. However, bile salt concentrations and detergent action may be higher immediately adjacent to the canalicular membrane of the hepatocyte across which bile salts [and virus] are transported [30] (Figure 2B). An alternative possibility is that these viruses have evolved distinct bi-polar mechanisms of egress from the highly polarized hepatocyte, exiting across the canalicular (apical) membrane into the bile as non-enveloped particles, but being released from basolateral membranes into the hepatic sinusoid enveloped in membranes (Figure 2B). While this seems unnecessarily complicated and contrary to Ockham’s razor, the vesicular transport and secretion pathways of the hepatocyte are highly complex and very specialized [31].
How do these viruses benefit from membrane hijacking? The most obvious answer is the resistance to neutralizing antibodies displayed by the membrane-enveloped forms of both viruses that circulate in the blood [2, 4]. Indeed, the coexistence of infectious virus and neutralizing antibodies in serum during acute hepatitis A had puzzled investigators for many years [32]. Although the kinetic of the neutralizing antibody response to HEV infection is not well characterized, antibodies to the capsids of HAV and HEV do not develop until several weeks after infection is established in the liver [33-35]. This delay in the host B cell response could result from the relevant antigens being masked from recognition by the immune system, allowing time for the virus to be shed in feces and transmitted to the next host. An additional advantage that may result from capsid envelopment is wider cellular tropism. Both viruses have been associated with extra-hepatic disease, and some reports suggest neurological involvement in HEV infection [36]. However, there is no direct evidence for replication of either virus outside the liver. A wider capacity to enter cells might also work against the virus, as recent studies indicate that plasmacytoid dendritic cells (pDCs), which play a key role in host recognition of viral invasion, sense eHAV but not non-enveloped virus (unpublished work). Further work is needed to define how envelopment alters host cell range, and the impact of that on the pathogenesis of both HAV and HEV infections.
Although probably not related to its ability to cloak itself in membranes, HAV efficiently evades early host innate immune responses [33, 37, 38]. This could also contribute to delays in the adaptive immune response. There are no direct comparative studies, but HEV appears to engender a more robust interferon response [35].
Implications of membrane hijacking for disease prevention
Efficacious vaccines have been developed for the prevention of both hepatitis A and hepatitis E, although commercial development of the latter has lagged [39-41]. In both cases, the vaccine immunogen is capsid antigen. There is strong evidence that vaccine-induced antibodies play a primary role in vaccine prevention of hepatitis A [42, 43], and it is likely that this is so with the HEV vaccine also. But, how can this be, if the virus circulating in the blood in both cases is completely enveloped in membranes and therefore resistant to neutralizing antibody? For HAV, the answer appears to lie in the fact that the enveloped virus can be neutralized after its entry into the cell, most likely within a late endosomal compartment [2]. This was demonstrated experimentally by a substantial reduction in viral growth when cells were treated with antibody as late as 6 hrs after removal of an eHAV inoculum [2]. Monoclonal IgA and IgG antibodies to the capsid effectively neutralized eHAV infectivity under these conditions, although they demonstrated no ability to reduce the number of infectious virions when mixed with eHAV prior to infection. eHAV enters cells via an acidification-dependent endosomal pathway [2], and is likely to lose its membrane in a late endosome/lysosome compartment. This compartment may be the site of ‘intracellular’ neutralization. This could also help explain why passively transferred antibodies are capable of preventing HAV-related disease even when administered long after exposure, when virus replication is robust within the liver [33, 42]. Membrane-enveloped HEV may be subject to a similar fate. Many questions remain, however, including whether specific cellular immunoglobulin receptors are required for such neutralization (Box 1). Whatever the mechanism of protection, the striking discordance between high-level clinical protection by vaccines and the inability of the dominant circulating virus to be neutralized in quantal neutralization assays calls into question common paradigms for how antibodies work.
Concluding remarks
Striking similarities exist between HAV, a ‘non-enveloped’ virus that has learned how to steal an envelope from host cell membranes, and HEV, most likely an ‘enveloped’ virus that has adapted to an extracellular existence without one. The absence of an envelope in fecally-shed HAV and HEV allows for enhanced stability, and contributes to virus spread and the potential for epidemic transmission. On the other hand, the membranes wrapping the HAV and HEV capsids circulating extracellularly within the body protect against neutralizing antibodies and likely contribute to the prolonged course and slow antibody responses to these otherwise acute, mostly self-limited infections. At a cellular level, membrane hijacking provides a mechanism for non-enveloped viruses to exit cells without lysis. At a system level, it provides for immune escape. These viral survival strategies may not be unique to these hepatitis viruses, as parallels can be found among poxviruses [44]. The take-home message is that the presence or absence of an envelope is not a fixed attribute of a particular virus family, but rather one that is subject to the selective forces that shape their evolution.
BOX 1. Outstanding questions.
Where does eHAV acquire its membrane: at the plasma membrane or in MVBs?
Are enveloped forms of HAV and HEV secreted differentially from basolateral vs. apical membranes of hepatocytes, or does the virus lose its envelope during passage through the biliary tract?
How do the enveloping membranes affect the cell tropism of these viruses?
What proteins reside on the surface of the enveloped forms of HAV and HEV, and do these forms of the virus interact with specific cellular receptors to facilitate entry?
Does ‘intracellular’ neutralization of eHAV suggest novel strategies for immunization against other viruses that undergo acidification-dependent endosomal entry?
Highlights.
Hepatitis A virus (HAV) and hepatitis E virus (HEV) are phylogenetically unrelated but possess similar dual lifestyles.
Both circulate in vivo cloaked in membranes and hidden from antibodies, but are shed in feces as stable, non-enveloped virions.
Enveloped and non-enveloped forms of HAV and HEV have similar infectivity in cell culture.
Whether members of a viral family possess or lack an envelope is dependent upon evolutionary forces.
Acknowledgments
This work was supported by NIH grant RO1-AI103083.
Glossary
- Exosomes
small 50-120 nm single-membrane vesicles released from numerous cell types via fusion of MVBs to the plasma membrane that carry a variety of biologically active protein and RNA cargos and that are increasingly recognized to play important roles in intercellular communication.
- Multivesicular bodies (MVBs)
specialized late endosomes that contain multiple internal vesicles formed by inward budding of the endosomal membrane in a process mediated by ESCRT complexes. MVBs may traffic to and fuse with lysosomes, or move to the plasma membrane where fusion of their outer membrane results in release of their internal vesicles as exosomes.
- Ockham’s razor
a philosophical and scientific tenet attributed to a 14th century friar that states in effect that the simplest explanation for a particular phenomenon is to be preferred over more complicated explanations with unnecessary assumptions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.King AMQ, et al. Ninth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press; 2011. Virus taxonomy. [Google Scholar]
- 2.Feng Z, et al. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature. 2013;496:367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koonin EV, Dolja VV. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi M, et al. Hepatitis E Virus (HEV) strains in serum samples can replicate efficiently in cultured cells despite the coexistence of HEV antibodies: characterization of HEV virions in blood circulation. Journal of Clinical Microbiology. 2010;48:1112–1125. doi: 10.1128/JCM.02002-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemon SM. Type A viral hepatitis: new developments in an old disease. N Engl J Med. 1985;313:1059–1067. doi: 10.1056/NEJM198510243131706. [DOI] [PubMed] [Google Scholar]
- 6.Purcell RH, Emerson SU. Hepatitis E: an emerging awareness of an old disease. J Hepatol. 2008;48:494–503. doi: 10.1016/j.jhep.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 7.Cohen JI, et al. Complete nucleotide sequence of an attenuated hepatitis A virus: comparison with wild-type virus. Proc Nat'l Acad Sci USA. 1987;84:2497–2501. doi: 10.1073/pnas.84.8.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin A, Lemon SM. The molecular biology of hepatitis A virus. In: Ou J, editor. In Hepatitis Viruses. Kluwer Academic Publishers; 2002. pp. 23–50. [Google Scholar]
- 9.Anderson DA, Ross BC. Morphogenesis of hepatitis A virus: Isolation and characterization of subviral particles. J Virol. 1990;64:5284–5289. doi: 10.1128/jvi.64.11.5284-5289.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graff J, et al. Hepatitis A virus capsid protein VP1 has a heterogeneous C terminus. Journal of Virology. 1999;73:6015–6023. doi: 10.1128/jvi.73.7.6015-6023.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin A, et al. Maturation of the hepatitis A virus capsid protein VP1 is not dependent on processing by the 3Cpro proteinase. J Virol. 1999;73:6220–6227. doi: 10.1128/jvi.73.8.6220-6227.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Binn LN, et al. Primary isolation and serial passage of hepatitis A virus strains in primate cell cultures. J Clin Microbiol. 1984;20:28–33. doi: 10.1128/jcm.20.1.28-33.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feinstone SM, et al. Hepatitis A: detection by immune electron microscopy of a viruslike antigen associated with acute illness. Science. 1973;182:1026–1028. doi: 10.1126/science.182.4116.1026. [DOI] [PubMed] [Google Scholar]
- 14.Emerson SU, Purcell RH. Hepatitis E virus. Rev Med Virol. 2003;13:145–154. doi: 10.1002/rmv.384. [DOI] [PubMed] [Google Scholar]
- 15.Kamar N, et al. Factors associated with chronic hepatitis in patients with hepatitis E virus infection who have received solid organ transplants. Gastroenterology. 2011;140:1481–1489. doi: 10.1053/j.gastro.2011.02.050. [DOI] [PubMed] [Google Scholar]
- 16.Tam AW, et al. Hepatitis E virus (HEV): Molecular cloning and sequencing of the full-length viral genome. Virology. 1991;185:120–131. doi: 10.1016/0042-6822(91)90760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sreenivasan MA, et al. Non-A, non-B epidemic hepatitis: visualization of virus-like particles in the stool by immune electron microscopy. J Gen Virol. 1984;65:1005–1007. doi: 10.1099/0022-1317-65-5-1005. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi M, et al. Monoclonal antibodies raised against the ORF3 protein of hepatitis E virus (HEV) can capture HEV particles in culture supernatant and serum but not those in feces. Arch Virol. 2008;153:1703–1713. doi: 10.1007/s00705-008-0179-6. [DOI] [PubMed] [Google Scholar]
- 19.Yamada K, et al. ORF3 protein of hepatitis E virus is essential for virion release from infected cells. J Gen Virol. 2009;90:1880–1891. doi: 10.1099/vir.0.010561-0. [DOI] [PubMed] [Google Scholar]
- 20.Takahashi M, et al. Production of monoclonal antibodies against hepatitis E virus capsid protein and evaluation of their neutralizing activity in a cell culture system. Arch Virol. 2008;153:657–666. doi: 10.1007/s00705-008-0045-6. [DOI] [PubMed] [Google Scholar]
- 21.Chen BJ, Lamb RA. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology. 2008;372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hurley JH. The ESCRT complexes. Crit Rev Biochem Mol Biol. 2010;45:463–487. doi: 10.3109/10409238.2010.502516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren X, Hurley JH. Proline-rich regions and motifs in trafficking: from ESCRT interaction to viral exploitation. Traffic. 2011;12:1282–1290. doi: 10.1111/j.1600-0854.2011.01208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin-Serrano J, et al. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 25.Taylor MP, et al. Role of microtubules in extracellular release of poliovirus. J Virol. 2009;83:6599–6609. doi: 10.1128/JVI.01819-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagashima S, et al. Tumour susceptibility gene 101 and the vacuolar protein sorting pathway are required for the release of hepatitis E virions. J Gen Virol. 2011;92:2838–2848. doi: 10.1099/vir.0.035378-0. [DOI] [PubMed] [Google Scholar]
- 27.Nagashima S, et al. A PSAP motif in the ORF3 protein of hepatitis E virus is necessary for virion release from infected cells. J Gen Virol. 2011;92:269–278. doi: 10.1099/vir.0.025791-0. [DOI] [PubMed] [Google Scholar]
- 28.Zafrullah M, et al. Mutational analysis of glycosylation, membrane translocation, and cell surface expression of the hepatitis E virus ORF2 protein. Journal of Virology. 1999;73:4074–4082. doi: 10.1128/jvi.73.5.4074-4082.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith W. The action of bile salts on viruses. J Path Bact. 1939;48:557–571. [Google Scholar]
- 30.Nicolaou M, et al. Canalicular ABC transporters and liver disease. J Pathol. 2012;226:300–315. doi: 10.1002/path.3019. [DOI] [PubMed] [Google Scholar]
- 31.Weisz OA, Rodriguez-Boulan E. Apical trafficking in epithelial cells: signals, clusters and motors. J Cell Sci. 2009;122:4253–4266. doi: 10.1242/jcs.032615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asher LV, et al. Pathogenesis of hepatitis A in orally inoculated owl monkeys (Aotus trivirgatus) J Med Virol. 1995;47:260–268. doi: 10.1002/jmv.1890470312. [DOI] [PubMed] [Google Scholar]
- 33.Lanford RE, et al. Acute hepatitis A virus infection is associated with a limited type I interferon response and persistence of intrahepatic viral RNA. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1101939108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemon SM, Binn LN. Serum neutralizing antibody response to hepatitis A virus. J Infect Dis. 1983;148:1033–1039. doi: 10.1093/infdis/148.6.1033. [DOI] [PubMed] [Google Scholar]
- 35.Yu C, et al. Pathogenesis of hepatitis E virus and hepatitis C virus in chimpanzees: similarities and differences. J Virol. 2010;84:11264–11278. doi: 10.1128/JVI.01205-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheung MC, et al. Review of the neurological manifestations of hepatitis E infection. Ann Hepatol. 2012;11:618–622. [PubMed] [Google Scholar]
- 37.Yang Y, et al. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc Natl Acad Sci U S A. 2007;104:7253–7258. doi: 10.1073/pnas.0611506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qu L, et al. Disruption of TLR3 signaling due to cleavage of TRIF by the hepatitis A virus protease-polymerase processing intermediate, 3CD. PLoS Pathog. 2011;7(9):e1002169. doi: 10.1371/journal.ppat.1002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Werzberger A, et al. A controlled trial of a formalin-inactivated hepatitis A vaccine in healthy children. N Engl J Med. 1992;327:453–457. doi: 10.1056/NEJM199208133270702. [DOI] [PubMed] [Google Scholar]
- 40.Shrestha MP, et al. Safety and efficacy of a recombinant hepatitis E vaccine. N Engl J Med. 2007;356:895–903. doi: 10.1056/NEJMoa061847. [DOI] [PubMed] [Google Scholar]
- 41.Zhu FC, et al. Efficacy and safety of a recombinant hepatitis E vaccine in healthy adults: a large-scale, randomised, double-blind placebo-controlled, phase 3 trial. Lancet. 2010;376:895–902. doi: 10.1016/S0140-6736(10)61030-6. [DOI] [PubMed] [Google Scholar]
- 42.Victor JC, et al. Hepatitis A vaccine versus immune globulin for postexposure prophylaxis. The New England journal of medicine. 2007;357:1685–1694. doi: 10.1056/NEJMoa070546. [DOI] [PubMed] [Google Scholar]
- 43.Lemon SM, et al. Immunoprecipitation and virus neutralization assays demonstrate qualitative differences between protective antibody responses to inactivated hepatitis A vaccine and passive immunization with immune globulin. J Infect Dis. 1997;176:9–19. doi: 10.1086/514044. [DOI] [PubMed] [Google Scholar]
- 44.Benhnia MR, et al. Vaccinia virus extracellular enveloped virion neutralization in vitro and protection in vivo depend on complement. J Virol. 2009;83:1201–1215. doi: 10.1128/JVI.01797-08. [DOI] [PMC free article] [PubMed] [Google Scholar]