Abstract
Early immunological events during acute HIV infection are thought to fundamentally influence long-term disease outcome. Whereas the contribution of HIV-specific CD8 T cell responses to early viral control is well established, the role of HIV-specific CD4 T cell responses in the control of viral replication following acute infection is unknown. A growing body of evidence suggests that CD4 T cells - besides their helper function - have the capacity to directly recognize and kill virally infected cells. In a longitudinal study of a cohort of individuals acutely infected with HIV, we observed that subjects able to spontaneously control HIV replication in the absence of antiretroviral therapy showed a significant expansion of HIV-specific CD4 T cell responses—but not CD8 T cell responses–compared to subjects who progressed to a high viral set point (p=0.038). Strikingly, this expansion occurred prior to differences in viral load or CD4 T cell count and was characterized by robust cytolytic activity and expression of a distinct profile of perforin and granzymes at the earliest time point. Kaplan-Meier analysis revealed that the emergence of Granzyme A+ HIV-specific CD4 T cell responses at baseline was highly predictive of slower disease progression and clinical outcome (average days to CD4 T cell count <350/μl was 575 versus 306, p=0.001). These data demonstrate that HIV-specific CD4 T cell responses can be used during the earliest phase of HIV infection as an immunological predictor of subsequent viral set point and disease outcome. Moreover, these data suggest that expansion of Granzyme A+ HIV-specific cytolytic CD4 T cell responses early during acute HIV infection contributes substantially to the control of viral replication.
INTRODUCTION
Acute HIV infection results in the massive depletion of CD4 T cells throughout all compartments of the body. In particular, HIV-specific CD4 T cells are preferentially targeted, disrupting a central process for the successful coordination of the antiviral immune response (1). Vaccine design strategies eliciting these responses have consequently been met with skepticism due to the fear that the induction and activation of HIV-specific CD4 T cell responses may fuel, instead of prevent, viral replication. Effector CD4 T cell responses have therefore not traditionally been a primary focus of HIV research, and considerable attention has rather been focused on HIV-specific CD8 T cell responses. Indeed, studies in acute HIV infection have shown that there is a temporal association between the first emergence of HIV-specific CD8 T cell responses and a decrease in viral load to a set point (2, 3). Further work has shown that this early viral set point is a strong predictor of disease outcome (4, 5).
Nevertheless, an increasing number of reports have suggested that HIV-specific CD4 T cell responses may also play an important role in controlling viral replication in HIV infection (6–9). In particular, the results of the recent, modestly protective RV144 vaccine trial—which not only induced non-neutralizing antibodies, but also a robust HIV-specific CD4 T cell response (10, 11)—raised important questions regarding the contribution of HIV-specific CD4 T cells to the initial control of HIV viremia.
Besides governing the induction and maintenance of the CD8 T cell response, as well as B cell proliferation and antibody maturation, a growing body of evidence suggests that effector CD4 T cells can themselves display potent antiviral activity by directly killing infected targets (reviewed in (12)). In the context of infection by other viruses, including cytomegalovirus (13), influenza (14), and Friend virus (15), it has been demonstrated that cytolytic CD4 T cells are readily detectable ex vivo and can contribute to viral containment even in the absence of antigen-specific CD8 T cell or B cell responses. Interestingly, CD4 T cells from HIV infected patients have been shown to express large quantities of cytolytic effector molecules like perforin and granzymes, and HIV-specific CD4 T cell clones and cell lines can readily mediate target cell lysis and viral inhibition in vitro (16–18). Moreover, the phenotype of the CD4 T cell response observed in the RV144 trial showed cytolytic activity (11), suggesting that these cells may possibly play a part in the prevention of HIV acquisition.
In the present study, we assessed the dynamics and evolution of the HIV-specific CD4 T cell response in a cohort of highly acutely infected subjects in order to understand the role of HIV-specific cytolytic CD4 T cell responses during acute HIV infection and to determine their impact on subsequent disease outcome, early viral control, and establishment of the early viral set point.
RESULTS
HIV-specific CD4 T cell responses correlate with control of viral replication after acute HIV infection
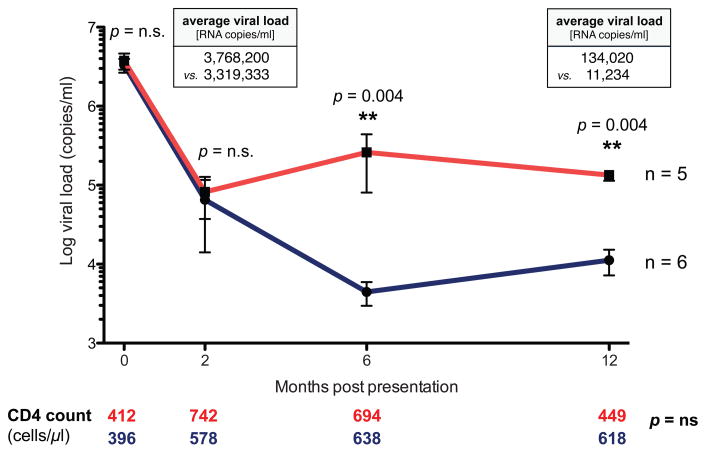
To assess the role of HIV-specific CD4 T cell responses during acute HIV infection and their subsequent impact on the early viral set point, we selected eleven individuals identified during peak viremia, prior to seroconversion, with exceedingly high viral loads averaging 3,523,364 HIV RNA copies/ml and a negative or indeterminate western blot test (≤3 bands; Table S1). The study group consisted of a very homogenous population in terms of race, gender, age, and risk factors. All individuals remained off therapy for at least one year, and were further divided into two groups based on their viral set points one year after acute HIV diagnosis. One group progressed to a low early viral set point (avg. 11,234 HIV RNA copies/ml), while the other group progressed to a significantly higher early viral set point (avg. 134,020 HIV RNA copies/ml; p=0.0043; Fig. 1). Interestingly, however, neither baseline peak viremia nor viral loads at the two month time point were significantly different between the two groups (avg. 3,768,200 vs. 3,319,333 HIV RNA copies/ml; and 82,232 copies/ml vs. 75,374 copies/ml, respectively). Yet, after six months post initial presentation viral loads diverged significantly (p=0.004) and remained lower in one group compared to the other. In addition, CD4 T cell counts for both groups did not differ throughout the study period, but began to decline in subjects who reached a high viral set point after 12 months of infection (449 vs. 618 cells/μl, p=n.s.). As it has been previously shown that certain HLA class I alleles are associated with lower viral loads, it is important to note that none of the subject groups were enriched for protective HLA class I alleles (19) (Table S2).
Figure 1. Dynamics of viral load during acute HIV infection.
11 acutely infected individuals were split into two groups based on their early viral set points. 5 individuals progressed to a high viral set point of 134,020 HIV RNA copies/ml one year after presentation (red); 6 subjects progressed to a significantly lower viral set point (11,234 copies/ml, p = 0.004, Mann-Whitney test) one year after presentation (blue). Average CD4 T cell counts for subjects progressing to high or low viral set points are denoted below the graph in red or blue, respectively, at each analysis time point.
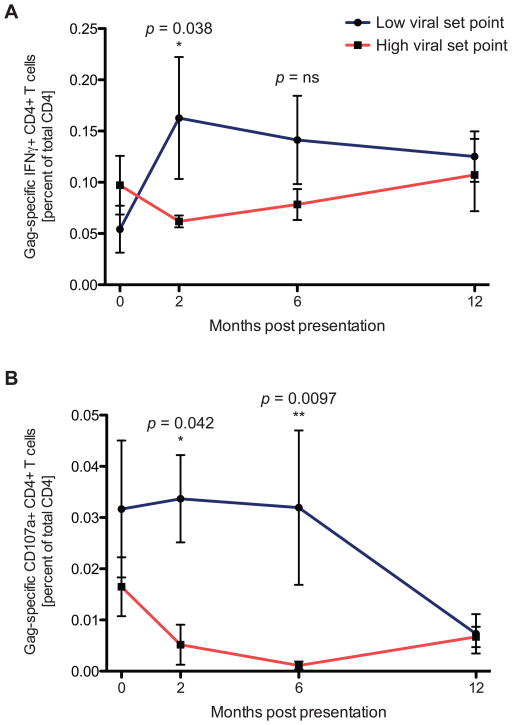
To evaluate the impact of HIV-specific CD4 T cell responses on the early viral set point, we longitudinally assessed whether the early emergence of HIV-specific CD4 T cell responses could positively impact the viral load trajectories of these two untreated, acutely infected subject groups or whether—as previously hypothesized—they would have a negative impact due to the generation of new target cells for viral infection. In the high viral set point group, we observed an initial decrease in the Gag-specific interferon-γ (IFNγ)+ CD4 T cell response that was maintained at a lower level over the course of the study period. In contrast, in subjects who progressed to a low early viral set point, we observed an early expansion of the HIV-specific CD4 T cell response two months post infection (p=0.038; Fig. 2). Strikingly, this increase was observable before viral loads between the groups diverged, suggesting that the expansion of the Gag-specific CD4 T cell responses plays a positive role in the early control of viral replication. In contrast, no significant difference in the HIV-specific CD8 T cell response between the groups was observed (Fig. S1). Comparable behavior of the HIV-specific CD4 T cell responses to other viral proteins was also detected, but these proteins were observed to be less consistently targeted by CD4 T cells, similar to findings in previous reports (6, 20). Importantly, the expression of the CCR5 co-receptor on the HIV-specific CD4 T cells was not significantly different between the two subject groups, suggesting that an elevated activation status of the HIV-specific CD4 T cells did not lead to their preferential depletion in patients progressing to high one-year viral set points (Fig. S2), as previously speculated (21). Moreover, no difference in proviral HIV DNA levels could be detected within sorted HIV-specific CD4 T cells, with no virus detectable in cells from either subject group (limit of detection: <1 HIV DNA copies/ml). Rather, the simultaneous expansion of virus-specific IFNγ+ CD4 T cell responses and initial decline of viremia preceded the point where differences in viral load became evident between the two groups. This temporal association suggests a possible contribution of HIV-specific CD4 T cell responses to the subsequent control of HIV replication to a lower viral set point.
Figure 2. HIV-specific CD4 T cell responses during acute HIV infection.
HIV-specific CD4 T cell responses expand in acute HIV infection in subjects progressing to lower viral set points. (A) The Gag-specific IFNγ+ CD4 T response was evaluated longitudinally in individuals who progressed to a lower viral set point (blue, n=6) and in those who progressed to a high viral set point (red, n=5). A significant difference in the IFNγ response levels was noted at 2 months post presentation (p=0.038, Mann-Whitney test) (B) Longitudinal monitoring of the Gag-specific CD4 T cell degranulation (CD107a) response was performed following acute HIV infection in subjects progressing to low (blue, n=6) and high (red, n=5) viral set points. Significant differences in CD107a expressing CD4 T cells between patient groups are observable at both 2(0.034 versus 0.005; P = 0.042, Mann-Whitney test) and 4 months after presentation (0.032 versus 0.001; P = 0.0097, Mann-Whitney test).
Early HIV-specific CD4 T cell responses exhibit cytolytic activity in subjects who control viral replication
Recent studies of other viral infections like Epstein-Barr virus (22) have indicated that CD4 T cells may also contribute to viral control through potentially independent cytolytic effector functionality, raising the possibility that a similar mechanism may be taking place during acute HIV infection. Indeed, cytolytic CD4 T cells have not only been described in HIV infection (17), but have also been shown to have the ability to kill virally infected cells (16, 18, 23, 24). Therefore, to assess the direct cytolytic activity of HIV-specific CD4 T cell responses during acute infection, we first longitudinally evaluated CD107a expression after HIV peptide stimulation in each patient group as a surrogate measure of degranulatory activity (25). As with HIV-specific IFNγ-secretion, in individuals who progressed to a high viral set point, we observed an early decline in the HIV-specific CD107a response (Fig. 2B). This was in stark contrast to individuals who spontaneously controlled viral replication and progressed to a low viral set point. In these individuals, there was a significant early expansion of CD107a responses to a level significantly higher both at 2 months and 6 months post infection (p=0.042 and p=0.0097, respectively; Fig. 2). This difference in CD107a expression became evident prior to the divergence of viral load or CD4 T cell count, again suggesting a causal relationship between the specific enrichment of HIV-specific CD107a+CD4 T cells and viral load in individuals who progress to a low viral set point. As before, no significant difference was observed for the HIV-specific CD8 T cell response at any time point (Fig. S1). Differences in CD4 T cell CD107a expression were also not due to a general enrichment of degranulatory CD4 T cell responses in one subject group over the other, as no significant differences were observed between patient groups at any time point in the ability of bulk CD4 T cells to degranulate after polyclonal stimulation with PMA/ionomycin (Fig. S3A).
CD4 T cells from HIV infected individuals mediate direct cytolytic activity
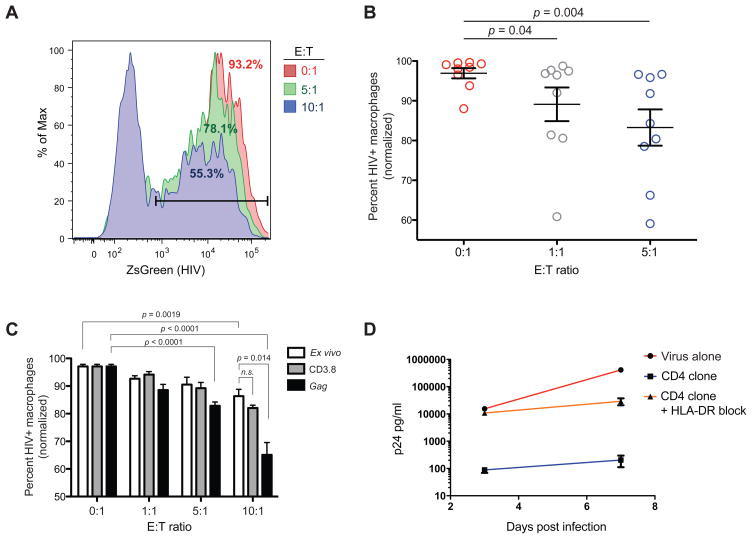
To establish that HIV-specific CD4 T cells from acutely HIV-infected patients can exert a cytolytic effect that directly contributes to the control of HIV replication, we developed a single-cycle, flow based viral inhibition assay optimized for the detection of antiviral CD4 T cell function. This assay makes use of autologous monocyte-derived macrophages as optimal target cells (18) and expanded CD4 T cells as effectors. Macrophages were infected with a single-cycle, ZsGreen expressing, pseudotyped HIV to facilitate a high level of synchronized target cell infection and to prevent any reverse infection of the effector cell population. Expanded CD4 T cells from the chronic phase of the infection from nine subjects within our cohort were purified and then co-incubated with autologous, infected macrophages at different effector to target (E:T) ratios. Interestingly, we observed a significant, dose-dependent suppressive effect of up to 40% reduction in HIV-infected macrophages mediated by CD4 T cells (E:T 5:1 range: 5–40%; p=0.004; Fig. 3A and 3B). To determine whether short term HIV-specific expansion could augment the CD4 T cell antiviral capacity, we expanded purified CD4 T cells from one individual for 7 days by culture with Gag peptide pools. Indeed, this short term antigen specific expansion resulted in a significant increase in the suppressive capacity of the CD4 T cells from 14% to 35% compared to direct ex vivo viral suppression (E:T 10:1; p = 0.014; Fig. 3C). To further confirm our findings, we generated HIV-specific CD4 T cell clones from the same individual specific for an epitope within p24/Gag (TAPPEESFRFGEETTTPSQK). In a 7 day viral inhibition assay using HLA-DR matched H9 CD4 T cell line targets, these HIV-specific CD4 T cell clones were able to inhibit viral replication up to 1000-fold. Moreover, this effect was almost completely abrogated when HLA-DR was blocked using a neutralizing antibody (Fig. 3D). Our data therefore suggest that the HIV-specific CD4 T cells in our cohort may indeed have direct cytolytic activity against HIV infected cells and directly contribute to the control of viral replication.
Figure 3. CD4 T cells from HIV-infected subjects mediate viral suppression.
Functional assays were performed to assess the ability of CD4 T cells from HIV-infected subjects to directly exhibit an antiviral effect. (A) Representative example of the results of one single-cycle macrophage inhibition assay; CD4 T cell effector cells were tested for their ability to reduce the number of macrophages infected with a ZsGreen reporter HIV at various effector to target (E:T) ratios. Percentages represent the number of HIV+ macrophages remaining at the end of the assay period. (B) CD4 T cells from a subset of the cohort of subjects analyzed longitudinally (n=9) were assessed for suppressive capacity in the single-cycle inhibition assay. Results are expressed as percentage of HIV+ autologous macrophages remaining and have been normalized to the respective maximum for each set of conditions. (C) CD4 T cells from a chronically HIV-infected patient were used as effectors in the single-cycle macrophage inhibition assay ex vivo or following non-specific (CD3.8) or Gag-specific expansion. (D) CD4 T cell clones were generated from the same chronically infected patient and assessed for their ability to inhibit viral replication in a standard 7 day viral inhibition assay using infected HLA-DR matched H9 cells as targets.
Individuals who progress to low viral set point exhibit a unique HIV-specific cytolytic CD4 phenotype at baseline
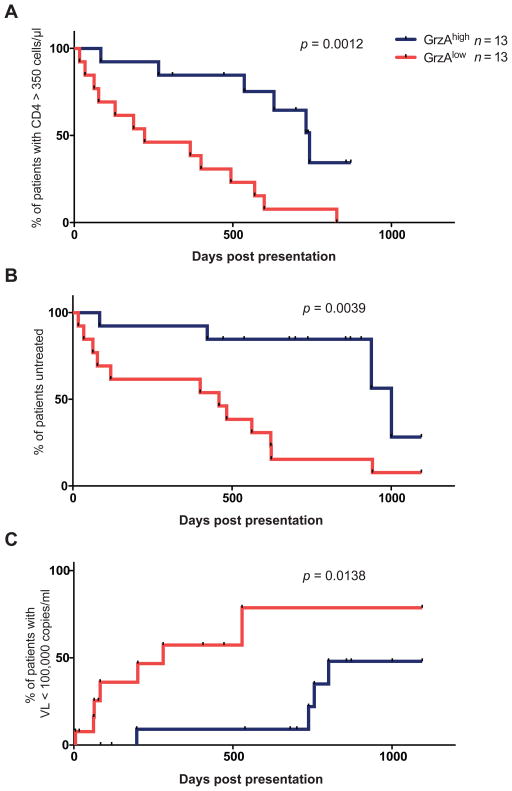
To further investigate the differences in the HIV-specific cytolytic CD4 T cells in both groups, we examined the cytolytic phenotype of the HIV-specific CD4 T cell response at each time point. Using intracellular cytokine staining, we assessed the presence of cytolytic effector molecules granzyme A, granzyme B, granzyme K, and perforin in HIV-specific IFNγ-secreting CD4 T cell responses (Fig 4A). Using a boolean gating strategy, we observed that the HIV-specific IFNγ+ CD4 T cell response at baseline was unexpectedly cytolytic—nearly 75% of the IFNγ responding cells contained at least one type of granzyme or perforin. Interestingly, however, the baseline HIV-specific IFNγ response in subjects who progressed to a low viral set point was substantially different than in those who progressed to a high set point (Fig 4B), at a time when viral loads and CD4 T cell counts were not significantly different. The baseline response in the low set point group not only contained an increased number of responses expressing all four cytolytic molecules but was also strikingly dominated by granzyme A (GrzA). Expression of GrzA—but not any other measured effector molecule—was significantly associated with lower viral set point (p<0.0001, Fig. 4C). These differences were only observed at baseline and the cytolytic profile of the responses at later time points did not differ substantially between the two subject groups. Additional analysis revealed that these differences were HIV-specific and were not due to a general elevation of GrzA in IFNγ-responsive CD4 T cells in individuals who progressed to a low viral set point. Indeed, no difference was observed in the baseline cytolytic phenotype of IFNγ positive cells after PMA/ionomycin stimulation in both groups (Fig. S3). Further examination of HIV-specific GrzA+ CD4 T cell responses revealed an almost exclusive co-secretion of IFNγ; GrzA+ CD4 T cells expressed other markers like IL-2, TNFα or CD40L to a lesser extent (Fig. S4A). Nonetheless, a second analysis performed using antigen-specific CD40L upregulation to define HIV-specific CD4 T cells provided further verification that the presence of HIV-specific GrzA+ CD4 T cells at baseline is significantly enriched in those individuals subsequently controlling viremia (p=0.038; Fig. S4B). To confirm a role for GrzA expressing HIV-specific CD4 T cells in the initial control of HIV viremia, we next examined the cytolytic phenotype of the HIV-specific IFNγ+CD4 T cell responses at baseline in an expanded cohort of 26 acutely infected individuals without prior knowledge of their clinical course. This cohort consisted of the original subjects studied longitudinally as well as an additional group of individuals who met the same criteria for acute infection (Table S1). We observed a striking association between the proportion of the Gag-specific IFNγ+ CD4 T cells expressing GrzA (relative to the other cytolytic effector molecules) and ultimate clinical outcome (Fig. 5). Kaplan-Meier analysis revealed that patients who exhibited GrzAhigh Gag-specific CD4 T cell responses at the time of initial presentation with acute HIV infection maintained CD4 T cell counts above the treatment initiation threshold of 350 cells/μl (US Department of Health and Human Services (26)) significantly longer than subjects with GrzAlow responses (avg. 575 days vs. 306 days; Log-rank p=0.0012, Wilcoxon p=0.0019; Fig. 5A). Similarly, analysis of the time after initial presentation until initiation of antiretroviral therapy demonstrated that subjects with GrzAhigh Gag-specific CD4 T cell responses at baseline remained off therapy significantly longer than individuals with GrzAlow responses (avg. 716 days vs. 423 days; Log-rank p = 0.0039, Wilcoxon p=0.0026; Fig. 5B). Moreover, subjects with elevated HIV-specific GrzA+ CD4 T cell responses at baseline demonstrated significantly longer control of viral load to levels below 100,000 HIV RNA copies/ml (BII recommendation for treatment initiation (26)) compared to subjects with lowered responses (649 vs. 258 days before reaching this threshold; Log-rank p=0.0138, Wilcoxon p=0.007; Fig. 5C). Our data therefore demonstrate that the expansion of specific GrzA-expressing, HIV-specific IFNγ+ CD4 T cell responses during the earliest phase of HIV infection is highly predictive of subsequent clinical outcome and may potentially represent a prognostic tool to provide an early estimate for long-term disease progression risk prior to the establishment of viral set point.
Figure 4. Baseline HIV-specific CD4 T cell responses and viral control.
Baseline HIV-specific CD4 T cell responses enriched in Granzyme A are associated with viral control.(A) Representative example of flow cytometric analysis of the expression of the cytolytic effector molecules granzymes A, B, K, and perforin in HIV-specific IFNγ+ CD4 T cells. (B) Co-expression analysis of the expression of granzymes and perforin in Gag-responding CD4 T cells was performed to determine if differences in the cytolytic profile of these cells could be detected in the cohort of subjects analyzed longitudinally (n=11). Pie slices are colored according to the number of cytolytic molecules expressed in the Gag-specific CD4 T cell response (0: pattern; 1: yellow; 2: green; 3: blue, 4: red); the orange colored arc represents the fraction of the total HIV-specific IFNγ+ CD4 T cells expressing granzyme A. (C) Phenotypic analysis was performed to determine the ratio of Granzyme A to other cytolytic effector molecules within HIV-specific IFNγ-secreting CD4 T cells expressing at least one cytolytic enzyme in both patient groups (low set point, 43.2%, versus high set point, 13.4%; P = 0.019, Mann-Whitney test).
Figure 5. Cytolytic phenotype of HIV-specific CD4 T cells predicts clinical outcome.
The cytolytic phenotype of the HIV-specific CD4 IFNγ response was measured at baseline in an expanded cohort of 26 patients comprised of the original patients evaluated longitudinally and additional patients meeting the same criteria for acute HIV infection. Subjects were stratified into two groups based on the presence of GrzAhigh CD4 T cell responses (blue lines, n = 13) or GrzAlow CD4 T cell responses (red lines, n = 13). Kaplan-Meier analysis was then performed to determine if differences between the two groups were present in (A) the time until CD4 counts declined to 350 cells/μl, (B) the time subjects remained off antiretroviral treatment, or (C) the length of time individuals were able to control viremia to levels below 100,000 HIV RNA copies/ml. P values denoted on the graphs represent Log-rank test results.
DISCUSSION
The contribution of CD4 T cells to the HIV-specific immune response has remained unclear. Although CD4 T cells and the helper and effector functions they employ are recognized to be necessary for optimal antiviral responses, the fact that HIV-specific CD4 T cells are preferentially depleted has cast doubt on their ability to effectively contribute to HIV control. Whereas HIV-specific CD4 T cell activity has previously been associated with viral control in studies of chronically infected patients, the kinetics and character of these responses have not been fully assessed, especially during primary HIV infection when initial immune control results in establishment of the viral set point. Here, we therefore investigated the HIV-specific CD4 T cell response longitudinally following acute HIV infection in two patient groups with very similar baseline characteristics but progressing to divergent viral set points, with a specific focus on the cytolytic CD4 T cell response. We observed that the HIV-specific CD4 T cell response, and not the HIV-specific CD8 T cell response, was enhanced in individuals progressing to a lower viral set point following acute HIV infection. In these subjects, greater CD4 T cell cytolytic activity was observed, reflected both as an expansion of CD107a+ CD4 T cell responses as well as an enrichment of granzyme A expressing cytolytic CD4 T cells at baseline. These findings raise the possibility that the involvement of HIV-specific CD4 T cell responses in controlling viral replication may be particularly important in the context of a less effective HIV-specific CD8 T cell response. Our cohort was not enriched for protective HLA class I alleles that have been previously associated with HIV control, suggesting that inefficient CD8 T cell activity can potentially be compensated for by a strong HIV-specific CD4 T cell response. Conversely, in elite HIV controllers who have protective HLA class I alleles such as HLA-B57 and -B27, it is possible that the role of HIV-specific cytolytic CD4 T cell responses is comparatively attenuated and that viral control may be mediated dominantly through CD8 T cell responses.
Our results are in agreement with previous, albeit smaller, studies suggesting that individuals who control HIV replication have more robust HIV-specific CD4 T cell responses following acute infection (27, 28). Enhanced levels of IFNγ production by HIV-specific CD4 T cells and particularly high avidity CD4 T cells may provide important helper signals to aid in the antiviral response mediated by CD8 T cells or B cells (29). However, the ability of HIV-specific CD4 T cells to degranulate and release cytolytic effector molecules may also have special significance for HIV pathogenesis, especially during the first phases of infection. During acute infection, the virus irreversibly establishes viral reservoirs in long-lived cells such as memory CD4 T cells or macrophages. Due to their residence in the tissues, macrophages in particular may represent critical early targets for viral infection and dissemination (30, 31). By virtue of their expression of high levels of MHC class II molecules, they may also be important targets for lysis by cytolytic CD4 T cells. Our inhibition assays using short-term expanded CD4 T cells suggest that macrophages can consistently be targeted by suppressive CD4 T cell responses. Moreover, our data demonstrate dose-dependent cytolytic CD4 T cell activity against HIV infected macrophages that was significantly enhanced after HIV-specific expansion, in line with previous reports (18, 24). HIV-specific cytolytic CD4 T cells may therefore be uniquely suited to lysing macrophages (as well as other MHC class II expressing cells) and, with CD8 T cells, provide a second route of cytolytic pressure on the virus. Further evidence for the importance of these cells is provided by the SIV model. Not only does the presence of strong virus-specific CD4 T cell responses correlate with protection after vaccination, but cytolytic SIV-specific CD4 T cells that are able to degranulate and express perforin are also associated with enhanced viral control in the context of attenuated SIV infection (32–34). Additionally, a recent study by Ortiz et al has demonstrated a lack of post peak viral decline in SIV infected macaques when CD4 T cells were depleted prior to SIV infection. Interestingly, no association with humoral or CD8 T cell mediated factors was observed, suggesting that CD4 T cells may indeed play an important direct antiviral role during acute SIV infection (35). Although further studies are required to evaluate the mechanisms used by HIV-specific cytolytic CD4 T cells, our results nonetheless suggest that cytotoxicity may be an important CD4 T cell function in the context of the early control of HIV infection. However, it is important to note that the contribution of HIV-specific CD4 T cell responses in the control of HIV infection is most likely multifactorial and includes not only direct cytolytic activity, but also help for B cells and CD8 T cells. Our data suggest a critical role for HIV-specific cytolytic CD4 T cells early during acute infection that is maintained into chronic HIV infection but ultimately lost, suggesting that alternate factors may play a role in the continued control of viral replication in later stages. In particular, broadly neutralizing antibody responses that have been shown to emerge late in chronic HIV infection might additionally contribute to the containment of viral replication (36). Thus, the enhanced early HIV-specific CD4 T cell response we observe may also play an important role in priming later immune function for chronic control of viremia. Further studies will be necessary to examine the interplay of the various short and long-term antiviral effects mediated by virus specific CD4 T cells during acute HIV infection.
The relevance of enriched GrzA expression in cytolytic CD4 T cells to the control of HIV replication is unclear. Although the expression of granzyme B and perforin in CD8 T cells has been linked to efficient cytotoxic functionality and to HIV control (37, 38), granzyme A is nonetheless the protease most prevalent in the granules of cytotoxic cells (39). Although its specific functions remain incompletely characterized, GrzA has been shown to be important for recovery from poxvirus infections in the murine model, for example (40). The enzyme is known to act at least in part by proteolytically destroying elements of the SET complex, a group of proteins which plays an important role in stress-response and DNA-damage repair pathways (39). Interestingly, it has recently been shown that the SET complex facilitates chromosomal integration of the HIV-1 genome by preventing abortive autointegration events (41). It is therefore possible that GrzA produced by HIV-specific cytolytic CD4 T cells may be uniquely able to inhibit this complex, preventing HIV-integration in newly infected cells and reducing the early establishment of viral reservoirs. Indeed, the strongest cytolytic activity observed in the subjects in our cohort was directed towards Gag peptides, which can be presented as early as two hours after infection in infected cells, prior to the occurrence of HIV integration (42). GrzA additionally has been shown to mediate extracellular and pro-inflammatory activity in certain contexts. Additional research will be required to investigate the functional effects of granzyme A produced by cytolytic CD4 T cells and determine how this protease may facilitate viral control.
Our findings showing robust IFNγ+ and cytolytic CD4 T cell responses in individuals who spontaneously control viral replication to a lower viral set point after acute infection have important implications regarding the role of CD4 T cells in the early control of HIV. The fact that the expansion of the HIV-specific CD4 T cell response—but not CD8 T cell response—was evident before viral load differences and correlated with lower subsequent viral set point suggests the possibility that HIV-specific CD4 T cell activity, and especially cytolytic CD4 T cell activity, may indeed have a contributing role in ultimately determining viral set point. Here, in particular, the granzyme A status of HIV-specific CD4 T cell response at baseline appears to predict subsequent clinical outcome, suggesting that GrzA may have a unique role in shaping the early antiviral response against HIV. Thus, our findings not only provide an additional example of the importance of cytolytic CD4 T cells in the human antiviral response, but will also help guide future HIV vaccine design strategies as the field builds on the results of the RV144 trial.
MATERIALS AND METHODS
Subjects
All primary subjects were identified during acute HIV infection (Table S1) and were recruited as subjects following written informed consent. Subjects were enrolled at either the Jessen-Jessen-Stein clinic in Berlin, Germany, the Fenway Community Health Center in Boston, MA, or the Massachusetts General Hospital in Boston, MA. For all subjects, acute infection was defined clinically as a negative or indeterminate (less than 3 bands) HIV western blot test and positive HIV RNA.
Stimulation
Cryopreserved PBMCs were thawed and allowed to rest overnight at 37° C, 5% CO2 at a concentration of 2×106/ml in R10 media (RPMI-1640 supplemented with 10% heat-inactivated fetal calf serum, 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10 mM HEPES). The following day, PBMCs were washed and resuspended at a concentration of 5–10 million cells/ml in R10 medium containing anti-CD28/49d costimulatory antibodies (1 μg/ml, clones L293 and L25, BD Biosciences). For detection of CD107a, a pre-titered amount of PE-Cy5 conjugated antibody to CD107a (clone H4A3, BD Biosciences) was added to the medium. 1–2 million cells were stimulated for six hours with 18-mer overlapping peptide pools comprising HIV clade B Gag, Pol, Nef, Gp120, or Gp41 at a concentration of 2 μg/ml. As a positive control, cells were stimulated with a combination of PMA and ionomycin. An unstimulated (medium only) sample served as a negative control. 30 minutes into the stimulation, the transport inhibitors brefeldin A (Sigma) and monensin (BD Biosciences) were added as previously described (43) to facilitate detection of T cell responses. For detection of CD40L, cells were stimulated in H10 medium (RPMI-1640 supplemented with 10% heat-inactivated human AB serum [Gemcell], 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 10 mM HEPES) without CD107a-PECy5 and without costimulatory antibodies (to minimize CD40L background).
Flow cytometric staining
Longitudinal analysis. Following stimulation, cells were washed with phosphate buffered saline (PBS) and stained with a UV-excitable, amine-reactive viability dye (LIVE/DEAD Blue, Invitrogen). Cells were subsequently washed with staining buffer (PBS containing 2% fetal calf serum and 0.09% sodium azide) and stained with CD4-BD Horizon v450 (clone RPA-T4, BD Biosciences), CD8-eFluor 650NC (clone RPA-T8, eBioscience), and CCR5-APC-Cy7 (clone 2D7/CCR5, BD Biosciences). After surface staining, cells were fixed and permeabilized (FIX/PERM, Invitrogen) and stained intracellularly using the following antibodies: CD3-Qdot 605 (clone UCHT1, Invitrogen), IFNγ-PE-Cy7 (clone B27, Biolegend), Granzyme A-Alexa 647 (clone CB9, Biolegend), Granzyme B-Alexa 700 (clone GB11, BD Biosciences), Granzyme K-FITC (clone GM6C3, Santa Cruz Biotechnology), and Perforin-PE (clone B-D48, Santa Cruz Biotechnology). CD40L analysis. Cells were stained as described above, with the following modifications: CD107a-PE-Cy5 and CCR5-APC-Cy7 were excluded; CD4 was stained with CD4-Qdot 705 (clone S3.5); CD40L and CD69 were stained for intracellularly using CD154-Brilliant Violet 421 (clone 24–31, Biolegend) and CD69-APC-Cy7 (clone FN50, Biolegend), respectively. Polyfunctionality analysis. Cells were stained as described above, with a modified antibody panel. Surface staining was performed with CD4-BD Horizon v450 (clone RPA-T4, BD Biosciences) and CD8-eFluor 650NC (clone RPA-T8, eBioscience). Cells were stained intracellularly using CD3-Qdot 605 (clone UCHT1, Invitrogen), IFNγ-PE-Cy7 (clone B27, Biolegend), Granzyme A-Alexa 647 (clone CB9, Biolegend), IL-2 FITC (clone 5344.111, BD Biosciences), TNFα Alexa Fluor 700 (clone MAb11, BD Biosciences), and CD40L PE (clone TRAP1, BD Biosciences).
Flow cytometric analysis
Flow cytometric data was collected using a special order 5-laser LSR Fortessa and FACSDiva software (BD Biosciences). Compensation was performed using single-stained antibody capture beads (CompBeads, BD Biosciences) and amine-dye reactive beads (ArC, Invitrogen). Cytometer settings were standardized and tracked between runs using multi-fluorescent calibration beads (Rainbow Fluorescent Particles, Spherotech). Data were analyzed using FlowJo version 9.2 (TreeStar). Initial gating was performed using a forward scatter (area) vs side scatter (area) lymphocyte gate, followed by a forward scatter (area) vs. forward scatter (height) doublet exclusion gate. CD4 cells consisted of live (viability stain negative), CD3+ T cells, CD4+, CD8- T cells (Fig. S5). For analysis of CD40L expression, CD40L positive CD4 T cells were defined as CD69+CD40L+ CD4 T cells. All response data shown have been background subtracted based on the unstimulated control for each sample set. Coexpression analysis of Granzyme A, B, K, and Perforin was performed using a Boolean gating strategy and the PESTLE and SPICE software suite (NIH/Mario Roederer (44)).
Detection of cellular HIV-1 DNA
Cryopreserved PBMC were thawed and stimulated as described above. Following stimulation, cells were labeled with LIVE/DEAD Violet (Invitrogen) and surface stained with CD3-Alexa 700 (clone UCHT1, BD Biosciences), CD4-FITC (clone RPA-T4, Biolegend), CD8-APC-Cy7 (clone SK1, Biolegend), and CD45RO-APC (clone UCHL1, Biolegend). Following fixation and permeabilization (FIX/PERM, Invitrogen), cells were stained intracellularly with IFNγ-PECy7 (clone B27, Biolegend). HIV-specific IFNγ+ CD4 T cells and CD45RO+ memory CD4 T cell subsets were sorted using a FACS Aria IIu using FACSDiva software (BD Biosciences). Sorted cells were then treated with 25 μL of a 1:100 dilution of proteinase K (Roche, Indianapolis, IN) in 10mM Tris buffer. Quantitative PCR was carried out using 5 μL of each of cell lysate per reaction as template, as previously described (45). Thermal cycling was carried out as follows: 95°C holding stage for 5 minutes, and 50 cycles of 95°C for 15 seconds followed by 60°C for 1 minute using the Taq DNA polymerase kit (Invitrogen, Carlsbad, CA). The sequence of the forward primer for HIV is GGTGCGAGAGCGTCAGTATTAAG. The reverse primer sequence is AGCTCCCTGCTTGCCCATA. The probe sequence is AAAATTCGGTTAAGGCCAGGGGGAAAGAA. For cell number quantification, albumin was measured as previously described (45). The qPCR instrument used was the StepOne Plus (Applied Biosystems); analysis was performed using StepOne software (Applied Biosystems).
Single-cycle viral inhibition assay
Generation of macrophages and CD4 effector cells. CD14+ monocytes were purified from cryopreserved PBMC samples by positive magnetic selection (EasySep, Stemcell technologies). Monocytes were differentiated into macrophages by culture for seven days in H10 supplemented with 50 ng/ml recombinant human M-CSF (R&D systems) in ultra-low adhesion flasks (Corning). CD4 T cells were expanded from the CD14-depleted PBMC fraction by seven day culture in H10 supplemented with 100 U/ml IL-2 (NIH AIDS Reagent Program), 5 μM nevirapine (NIH AIDS Reagent Program), and 0.5 μg/ml bispecific CD3.8 antibody (Dr. Johnson Wong, Massachusetts General Hospital), which activates CD4 T cells and depletes CD8 T cells (46). For the inhibition assay, macrophages were transferred into R10 and plated at 15,000 cells per well in an ultra-low adhesion 96-well plate (Corning). Effector cells were enriched for high purity CD4 T cells by negative magnetic enrichment prior to use (EasySep, StemCell technologies). Viruses and infection. Production of HIV-1 vectors and SIVmac virus-like particles (VLPs) has been described previously (47). Briefly, VSV-G-pseudotyped HIV-1 vectors were produced by co-transfecting 293T cells (ATCC) with pAGM, psPAX2, and pMD2.G using Lipofectamine 2000 (Invitrogen). VSV-G-pseudotyped SIVmac251 VLPs were produced by co-transfecting 293T cells with pSIV3+ and pMD2.G using Lipofectamine 2000. SIVmac VLPs were harvested 48 h post transfection, clarified by centrifugation at 200 x g, filtered through a 0.45 micron syringe filter (Whatman), and added to macrophages 3 h prior to overnight challenge with HIV-1 vectors. After extensive washing of the targets, expanded CD4 effectors were added at 1:1, 5:1 and 10:1 effector:target ratios and incubated for 36 hours at 37°C. At least three replicates were performed for each condition. To assess infectivity, cell mixtures were stained with APC conjugated anti-CD11b antibody (clone ICRF44, Biolegend) in the presence of 2 mM EDTA for 15 minutes and fixed. Samples were acquired on an LSR Fortessa (BD Biosciences) and data were analyzed using FlowJo (Treestar). Macrophages were identified by scatter properties and high CD11b expression; percent infection was determined by analyzing the percentage of ZsGreen positive events in the macrophage gate. ZsGreen gates were established based on uninfected controls for each individual. Inhibition data for each subject were normalized to the maximum respective positive control (HIV-infected, no effector) condition.
Traditional viral inhibition assay
CD4 T cell cloning. CD4 T cell clones were generated from cryopreserved PBMC samples by stimulation with the HIV Gag peptide TAPPEESFRFGEETTTPSQK in the presence of IL-2followed by limiting dilution cloning. Inhibition assay. Viral inhibition assays using the CD4 T cell clones were performed as previously described (8) using the CD4 T cell line H9 as the target population, which express the matching restricting HLA-DR molecule DRB1*0401 (48). Briefly, H9 target cells were infected with NL4-3 at an MOI of 0.01. Effector cells were added at a 1:1 effector:target ratio and incubated for 7 days. Supernatants were sampled on days 3 and 7 and assayed for p24 level using the Alliance p24 ELISA kit (Perkin-Elmer). To block the MHC class II HLA-DR presentation pathway, purified HLA-DR blocking antibody (low-endotoxin, azide-free, clone L243, Biolegend) was added to the culture medium at a concentration of 100 μg/ml.
Statistical analysis
Statistical analysis was performed using Prism (GraphPad). Parameters and responses were compared between subject groups using a Mann-Whitney analysis. Kaplan-Meier survival outcomes were compared by the Log-rank (Mantel-Cox) and Gehan-Breslow-Wilcoxon tests.
Supplementary Material
HIV-specific CD8 T cell responses during acute HIV infection are not associated with better viral control.
CCR5 expression on Gag-specific CD4 T cells does not differ between subjects progressing to high or low viral set points
Cytolytic CD4 T cell responses following PMA/ionomycin stimulation do not differ between set point groups.
Alternate markers of HIV-specificity may be used to define Granzyme A+ CD4 T cells
Flow cytometry gating strategy
Subject characteristics.
Subject HLA class I and class II genotypes
Acknowledgments
Funding: This study was funded by a supplement to 5P01AI074415-03. H.S. is funded by 1R01AI091450-01 and 1R01AI094602-01. A.B and T.P. are also funded by the Charles H. Hood Foundation.
Footnotes
Author contributions: D.Z.S. and H.S. designed the experiments. D.Z.S., M.F., and S.C. performed the ex vivo cytometric studies. D.Z.S. and J.M.B. performed the analysis of cell associated HIV DNA. D.Z.S., M.F., T.P., A.L.B, K.S.D., and A.P.T. participated in the viral inhibition experiments. H.J., K.L, J.R., and E.S.R. participated in patient care and cohort management. D.Z.S., S.R., M.L., I.D., and H.S. participated in study analysis. D.Z.S., B.D.W., and H.S. wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
References
- 1.Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, Grossman Z, Dybul M, Oxenius A, Price DA, Connors M, Koup RA. HIV preferentially infects HIV-specific CD4+ T cells. Nature. 2002 May 2;417:95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- 2.Borrow P, Lewicki H, Hahn BH, Shaw GM, Oldstone MB. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. Journal of Virology. 1994 Sep;68:6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koup RA, Safrit JT, Cao Y, Andrews CA, McLeod G, Borkowsky W, Farthing C, Ho DD. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. Journal of Virology. 1994 Jul;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mellors JW, Rinaldo CR, Jr, Gupta P, White RM, Todd JA, Kingsley LA. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996 May 24;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 5.Lyles RH, Munoz A, Yamashita TE, Bazmi H, Detels R, Rinaldo CR, Margolick JB, Phair JP, Mellors JW. Natural history of human immunodeficiency virus type 1 viremia after seroconversion and proximal to AIDS in a large cohort of homosexual men. Multicenter AIDS Cohort Study. The Journal of infectious diseases. 2000 Mar;181:872–880. doi: 10.1086/315339. [DOI] [PubMed] [Google Scholar]
- 6.Pitcher CJ, Quittner C, Peterson DM, Connors M, Koup RA, Maino VC, Picker LJ. HIV-1-specific CD4+ T cells are detectable in most individuals with active HIV-1 infection, but decline with prolonged viral suppression. Nat Med. 1999 May;5:518–525. doi: 10.1038/8400. [DOI] [PubMed] [Google Scholar]
- 7.Rosenberg ES, Billingsley JM, Caliendo AM, Boswell SL, Sax PE, Kalams SA, Walker BD. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science. 1997 Nov 21;278:1447–1450. doi: 10.1126/science.278.5342.1447. [DOI] [PubMed] [Google Scholar]
- 8.Chevalier MF, Julg B, Pyo A, Flanders M, Ranasinghe S, Soghoian DZ, Kwon DS, Rychert J, Lian J, Mueller M, Cutler S, McAndrew E, Jessen H, Pereyra F, Rosenberg ES, Altfeld M, Walker BD, Streeck H. HIV-1-specific IL-21+ CD4+ T cell responses contribute to durable viral control through the modulation of HIV-specific CD8+ T cell function. J Virol. 2010 Nov 3; doi: 10.1128/JVI.02030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harari A, Petitpierre S, Vallelian F, Pantaleo G. Skewed representation of functionally distinct populations of virus-specific CD4 T cells in HIV-1-infected subjects with progressive disease: changes after antiretroviral therapy. Blood. 2004 Feb 1;103:966–972. doi: 10.1182/blood-2003-04-1203. [DOI] [PubMed] [Google Scholar]
- 10.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009 Dec 3;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 11.de Souza M, Ratto-Kim S, Cheuenarom W, Schuetz A, Chantakulkij S, Nuntapinit B, Valencia-Micolta A, Thelian D, Nitayaphan S, Pittisuttihum P, Paris R, Kaewkungwal J, Michael N, Rerks-Ngarm S, Mathieson B, Marovich M, Currier J, Kim J, M-T, et al. The ALVAC-HIV prime, AIDSVAX boost vaccine regimen (RV144) induces T cell responses that are predominantly CD4+ and to HIV Envelope. 2011 Manuscript submitted to J. Exp. Med. [Google Scholar]
- 12.Soghoian DZ, Streeck H. Cytolytic CD4(+) T cells in viral immunity. Expert Rev Vaccines. 2010 Dec;9:1453–1463. doi: 10.1586/erv.10.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casazza JP, Betts MR, Price DA, Precopio ML, Ruff LE, Brenchley JM, Hill BJ, Roederer M, Douek DC, Koup RA. Acquisition of direct antiviral effector functions by CMV-specific CD4+ T lymphocytes with cellular maturation. J Exp Med. 2006 Dec 25;203:2865–2877. doi: 10.1084/jem.20052246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown DM, Dilzer AM, Meents DL, Swain SL. CD4 T cell-mediated protection from lethal influenza: perforin and antibody-mediated mechanisms give a one-two punch. J Immunol. 2006 Sep 1;177:2888–2898. doi: 10.4049/jimmunol.177.5.2888. [DOI] [PubMed] [Google Scholar]
- 15.Pike R, Filby A, Ploquin MJ, Eksmond U, Marques R, Antunes I, Hasenkrug K, Kassiotis G. Race between retroviral spread and CD4+ T-cell response determines the outcome of acute Friend virus infection. Journal of Virology. 2009 Nov;83:11211–11222. doi: 10.1128/JVI.01225-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Norris PJ, Moffett HF, Yang OO, Kaufmann DE, Clark MJ, Addo MM, Rosenberg ES. Beyond Help: Direct Effector Functions of Human Immunodeficiency Virus Type 1-Specific CD4+ T Cells. Journal of Virology. 2004 Aug 1;78:8844. doi: 10.1128/JVI.78.16.8844-8851.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Appay V, Zaunders JJ, Papagno L, Sutton J, Jaramillo A, Waters A, Easterbrook P, Grey P, Smith D, McMichael AJ, Cooper DA, Rowland-Jones SL, Kelleher AD. Characterization of CD4(+) CTLs ex vivo. J Immunol. 2002 Jun 1;168:5954–5958. doi: 10.4049/jimmunol.168.11.5954. [DOI] [PubMed] [Google Scholar]
- 18.Sacha JB, Giraldo-Vela JP, Buechler MB, Martins MA, Maness NJ, Chung C, Wallace LT, León EJ, Friedrich TC, Wilson NA, Hiraoka A, Watkins DI. Gag- and Nef-specific CD4+ T cells recognize and inhibit SIV replication in infected macrophages early after infection. Proceedings of the National Academy of Sciences. 2009 May 16;106:9791–9796. doi: 10.1073/pnas.0813106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pereyra F, Addo MM, Kaufmann DE, Liu Y, Miura T, Rathod A, Baker B, Trocha A, Rosenberg R, Mackey E, Ueda P, Lu Z, Cohen D, Wrin T, Petropoulos CJ, Rosenberg ES, Walker BD. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. The Journal of infectious diseases. 2008 Feb 15;197:563–571. doi: 10.1086/526786. [DOI] [PubMed] [Google Scholar]
- 20.Kaufmann D, Bailey P, Sidney J, Wagner B, Norris P, Johnston M, Cosimi L, Addo M, Lichterfeld M, Altfeld M, Frahm N, Brander C, Sette A, Walker B, Rosenberg E. Comprehensive Analysis of Human Immunodeficiency Virus Type 1-Specific CD4 Responses Reveals Marked Immunodominance of gag and nef and the Presence of Broadly Recognized Peptides. The Journal of Virology. 2004 May 1;78:4463. doi: 10.1128/JVI.78.9.4463-4477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Appay V, Sauce D. Immune activation and inflammation in HIV-1 infection: causes and consequences. J Pathol. 2008 Jan;214:231–241. doi: 10.1002/path.2276. [DOI] [PubMed] [Google Scholar]
- 22.Adhikary D, Behrends U, Moosmann A, Witter K, Bornkamm GW, Mautner J. Control of Epstein-Barr virus infection in vitro by T helper cells specific for virion glycoproteins. J Exp Med. 2006 Apr 17;203:995–1006. doi: 10.1084/jem.20051287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norris PJ, Sumaroka M, Brander C, Moffett HF, Boswell SL, Nguyen T, Sykulev Y, Walker BD, Rosenberg ES. Multiple Effector Functions Mediated by Human Immunodeficiency Virus-Specific CD4+ T-Cell Clones. Journal of Virology. 2001 Oct 15;75:9771–9779. doi: 10.1128/JVI.75.20.9771-9779.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng N, Fujiwara M, Ueno T, Oka S, Takiguchi M. Strong ability of Nef-specific CD4+ cytotoxic T cells to suppress human immunodeficiency virus type 1 (HIV-1) replication in HIV-1-infected CD4+ T cells and macrophages. J Virol. 2009 Aug;83:7668–7677. doi: 10.1128/JVI.00513-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Betts MR, Brenchley JM, Price DA, De Rosa SC, Douek DC, Roederer M, Koup RA. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. Journal of immunological methods. 2003 Oct 1;281:65–78. doi: 10.1016/s0022-1759(03)00265-5. [DOI] [PubMed] [Google Scholar]
- 26.Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. Department of Health and Human Services; 2011. pp. 1–166. [Google Scholar]
- 27.Gloster SE, Newton P, Cornforth D, Lifson JD, Williams I, Shaw GM, Borrow P. Association of strong virus-specific CD4 T cell responses with efficient natural control of primary HIV-1 infection. AIDS. 2004 Mar 26;18:749–755. doi: 10.1097/00002030-200403260-00005. [DOI] [PubMed] [Google Scholar]
- 28.Oxenius A, Fidler S, Brady M, Dawson SJ, Ruth K, Easterbrook PJ, Weber JN, Phillips RE, Price DA. Variable fate of virus-specific CD4(+) T cells during primary HIV-1 infection. European journal of immunology. 2001 Dec;31:3782–3788. doi: 10.1002/1521-4141(200112)31:12<3782::aid-immu3782>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 29.Vingert B, Perez-Patrigeon S, Jeannin P, Lambotte O, Boufassa F, Lemaitre F, Kwok WW, Theodorou I, Delfraissy JF, Theze J, Chakrabarti LA. HIV controller CD4+ T cells respond to minimal amounts of Gag antigen due to high TCR avidity. PLoS pathogens. 2010 Feb;6:e1000780. doi: 10.1371/journal.ppat.1000780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pope M, Haase AT. Transmission, acute HIV-1 infection and the quest for strategies to prevent infection. Nature medicine. 2003 Jul;9:847–852. doi: 10.1038/nm0703-847. [DOI] [PubMed] [Google Scholar]
- 31.Shen R, Richter HE, Clements RH, Novak L, Huff K, Bimczok D, Sankaran-Walters S, Dandekar S, Clapham PR, Smythies LE, Smith PD. Macrophages in vaginal but not intestinal mucosa are monocyte-like and permissive to human immunodeficiency virus type 1 infection. Journal of Virology. 2009 Apr;83:3258–3267. doi: 10.1128/JVI.01796-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gauduin MC, Yu Y, Barabasz A, Carville A, Piatak M, Lifson JD, Desrosiers RC, Johnson RP. Induction of a virus-specific effector-memory CD4+ T cell response by attenuated SIV infection. The Journal of experimental medicine. 2006 Nov 27;203:2661–2672. doi: 10.1084/jem.20060134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Letvin NL, Mascola JR, Sun Y, Gorgone DA, Buzby AP, Xu L, Yang ZY, Chakrabarti B, Rao SS, Schmitz JE, Montefiori DC, Barker BR, Bookstein FL, Nabel GJ. Preserved CD4+ central memory T cells and survival in vaccinated SIV-challenged monkeys. Science. 2006 Jun 9;312:1530–1533. doi: 10.1126/science.1124226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reynolds MR, Weiler AM, Weisgrau KL, Piaskowski SM, Furlott JR, Weinfurter JT, Kaizu M, Soma T, Leon EJ, MacNair C, Leaman DP, Zwick MB, Gostick E, Musani SK, Price DA, Friedrich TC, Rakasz EG, Wilson NA, McDermott AB, Boyle R, Allison DB, Burton DR, Koff WC, Watkins DI. Macaques vaccinated with live-attenuated SIV control replication of heterologous virus. The Journal of experimental medicine. 2008 Oct 27;205:2537–2550. doi: 10.1084/jem.20081524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ortiz AM, Klatt NR, Li B, Yi Y, Tabb B, Hao XP, Sternberg L, Lawson B, Carnathan PM, Cramer EM, Engram JC, Little DM, Ryzhova E, Gonzalez-Scarano F, Paiardini M, Ansari AA, Ratcliffe S, Else JG, Brenchley JM, Collman RG, Estes JD, Derdeyn CA, Silvestri G. Depletion of CD4+ T cells abrogates post-peak decline of viremia in SIV-infected rhesus macaques. The Journal of clinical investigation. 2011 Nov 1;121:4433–4445. doi: 10.1172/JCI46023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF. The immune response during acute HIV-1 infection: clues for vaccine development. Nature reviews Immunology. 2010 Jan;10:11–23. doi: 10.1038/nri2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harari A, Enders FB, Cellerai C, Bart PA, Pantaleo G. Distinct profiles of cytotoxic granules in memory CD8 T cells correlate with function, differentiation stage, and antigen exposure. J Virol. 2009 Apr;83:2862–2871. doi: 10.1128/JVI.02528-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hersperger AR, Pereyra F, Nason M, Demers K, Sheth P, Shin LY, Kovacs CM, Rodriguez B, Sieg SF, Teixeira-Johnson L, Gudonis D, Goepfert PA, Lederman MM, Frank I, Makedonas G, Kaul R, Walker BD, Betts MR. Perforin expression directly ex vivo by HIV-specific CD8 T-cells is a correlate of HIV elite control. PLoS pathogens. 2010 May;6:e1000917. doi: 10.1371/journal.ppat.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lieberman J. Granzyme A activates another way to die. Immunological reviews. 2010 May;235:93–104. doi: 10.1111/j.0105-2896.2010.00902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mullbacher A, Ebnet K, Blanden RV, Hla RT, Stehle T, Museteanu C, Simon MM. Granzyme A is critical for recovery of mice from infection with the natural cytopathic viral pathogen, ectromelia. Proceedings of the National Academy of Sciences of the United States of America. 1996 Jun 11;93:5783–5787. doi: 10.1073/pnas.93.12.5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan N, Cherepanov P, Daigle JE, Engelman A, Lieberman J. The SET complex acts as a barrier to autointegration of HIV-1. PLoS pathogens. 2009 Mar;5:e1000327. doi: 10.1371/journal.ppat.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sacha JB, Chung C, Rakasz EG, Spencer SP, Jonas AK, Bean AT, Lee W, Burwitz BJ, Stephany JJ, Loffredo JT, Allison DB, Adnan S, Hoji A, Wilson NA, Friedrich TC, Lifson JD, Yang OO, Watkins DI. Gag-specific CD8+ T lymphocytes recognize infected cells before AIDS-virus integration and viral protein expression. Journal of immunology. 2007 Mar 1;178:2746–2754. doi: 10.4049/jimmunol.178.5.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Streeck H, Brumme ZL, Anastario M, Cohen KW, Jolin JS, Meier A, Brumme CJ, Rosenberg ES, Alter G, Allen TM, Walker BD, Altfeld M. Antigen load and viral sequence diversification determine the functional profile of HIV-1-specific CD8+ T cells. PLoS Med. 2008 May 6;5:e100. doi: 10.1371/journal.pmed.0050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roederer M, Nozzi JL, Nason MC. SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A. 2011 Feb;79:167–174. doi: 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brenchley JM, Ruff LE, Casazza JP, Koup RA, Price DA, Douek DC. Preferential infection shortens the life span of human immunodeficiency virus-specific CD4+ T cells in vivo. Journal of Virology. 2006 Jul;80:6801–6809. doi: 10.1128/JVI.00070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong JT, Eylath AA, Ghobrial I, Colvin RB. The mechanism of anti-CD3 monoclonal antibodies. Mediation of cytolysis by inter-T cell bridging. Transplantation. 1990 Oct;50:683–689. doi: 10.1097/00007890-199010000-00030. [DOI] [PubMed] [Google Scholar]
- 47.Pertel T, Hausmann S, Morger D, Zuger S, Guerra J, Lascano J, Reinhard C, Santoni FA, Uchil PD, Chatel L, Bisiaux A, Albert ML, Strambio-De-Castillia C, Mothes W, Pizzato M, Grutter MG, Luban J. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011 Apr 21;472:361–365. doi: 10.1038/nature09976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arthur LO, Bess JW, Jr, Urban RG, Strominger JL, Morton WR, Mann DL, Henderson LE, Benveniste RE. Macaques immunized with HLA-DR are protected from challenge with simian immunodeficiency virus. Journal of Virology. 1995 May;69:3117–3124. doi: 10.1128/jvi.69.5.3117-3124.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HIV-specific CD8 T cell responses during acute HIV infection are not associated with better viral control.
CCR5 expression on Gag-specific CD4 T cells does not differ between subjects progressing to high or low viral set points
Cytolytic CD4 T cell responses following PMA/ionomycin stimulation do not differ between set point groups.
Alternate markers of HIV-specificity may be used to define Granzyme A+ CD4 T cells
Flow cytometry gating strategy
Subject characteristics.
Subject HLA class I and class II genotypes