Significance
Both posttranscriptional RNA editing and tetrapyrrole metabolism are important processes in land plants and animals. A direct link between these two distinct programs had hitherto not been established. This study reveals an unexpected function for protoporphyrinogen IX oxidase 1 from model plant Arabidopsis thaliana in regulating plastid RNA editing through interacting with and modulating the stability of multiple organellar RNA editing factors. In addition to furthering our knowledge of the composition of the plant organellar editing apparatus, this research provides insight into both the conserved and divergent roles of enzymes in the tetrapyrrole metabolism during evolution.
Keywords: metabolism, organelle, editosome
Abstract
RNA editing is a posttranscriptional process that covalently alters the sequence of RNA molecules and plays important biological roles in both animals and land plants. In flowering plants, RNA editing converts specific cytidine residues to uridine in both plastid and mitochondrial transcripts. Previous studies identified pentatricopeptide repeat (PPR) motif-containing proteins as site-specific recognition factors for cytidine targets in RNA sequences. However, the regulatory mechanism underlying RNA editing was largely unknown. Here, we report that protoporphyrinogen IX oxidase 1 (PPO1), an enzyme that catalyzes protoporphyrinogen IX into protoporphyrin IX in the tetrapyrrole biosynthetic pathway, plays an unexpected role in editing multiple sites of plastid RNA transcripts, most of which encode subunits of the NADH dehydrogenase-like complex (NDH), in the reference plant Arabidopsis thaliana. We identified multiple organellar RNA editing factors (MORFs), including MORF2, MORF8, and MORF9, that interact with PPO1. We found that two conserved motifs within the 22-aa region at the N terminus of PPO1 are essential for its interaction with MORFs, its RNA editing function, and subsequently, its effect on NDH activity. However, transgenic plants lacking key domains for the tetrapyrrole biosynthetic activity of PPO1 exhibit normal RNA editing. Furthermore, MORF2 and MORF9 interact with three PPRs or related proteins required for editing of ndhB and ndhD sites. These results reveal that the tetrapyrrole biosynthetic enzyme PPO1 is required for plastid RNA editing, acting as a regulator that promotes the stability of MORF proteins through physical interaction.
RNA editing, the process of covalently altering the sequence of an RNA molecule, generates protein diversity in eukaryotes (1, 2). Generally, in land plants, RNA editing highly specifically converts cytidine to uridine nucleotides in transcripts of both plastid and mitochondrial genes (3); 34 cytidine residues in plastids and more than 500 residues in mitochondria have been reported to be editing target sites in Arabidopsis thaliana (4, 5). A series of studies identified members of the PLS subfamily of pentatricopeptide repeat (PPR) motif-containing proteins as the site-specific recognition factors for cytidine targets (6). These specific PPR trans-acting proteins recognize cis elements within a region of ∼30 nt within the sites to be edited (1, 6–11). Although the DYW domains of some PPR factors contain several conserved residues with a cytidine deaminase motif (12), the enzyme that executes the editing reaction is elusive. Two recent reports documented that members of multiple organellar RNA editing factors (MORFs)/RNA editing factor interacting proteins (RIPs) widely affect RNA editing sites in both mitochondria and plastids (13, 14). Organelle RNA recognition motif protein 1, which contains two truncated RIP domains, is also essential for plastid RNA editing (15). These studies reveal additional components of the plant organellar editing apparatus. However, the mechanism by which these proteins edit RNA and the complete composition of the editing machinery remain largely unknown.
Tetrapyrrole biosynthesis is a universal metabolic process, occurring in all kingdoms of life. In plants, tetrapyrroles play critical roles in various biological processes, including photosynthesis and respiration. They serve as cofactors for essential proteins involved in a wide variety of crucial cellular functions (16, 17). Tetrapyrrolic metabolites, such as Mg protoporphyrin and heme, have also been suggested to act as signaling molecules that coordinate organellar functions within the cell (18, 19). The tetrapyrrole biosynthesis pathway in plants consists of two major branches, the Mg and Fe branch, and is thought to take place almost exclusively within the plastid (16, 17). Over 20 enzymes directly involved in tetrapyrrole biosynthesis have been identified, some of which have been functionally characterized by biochemical and genetic approaches (reviews in refs. 16 and 20). It was hitherto unclear whether these enzymes have biological functions that extend beyond their well-established roles as catalysts in the tetrapyrrole pathway.
In this study, we show that disruption of protoporphyrinogen IX oxidase 1 (PPO1), which encodes the last enzyme in the common pathway to chlorophyll and heme biosynthesis (21), causes RNA editing defects in 18 of 34 known plastid RNA targets, especially those encoded by NADH dehydrogenase-like complex (ndh) genes. PPO1 interacts with plastid-localized MORF proteins, which in turn, additionally interact with two PPR proteins, CHLORORESPIRATORY REDUCTION 28 (CRR28) and ORGANELLE TRANSCRIPT PROCESSING 82 (OTP82), and a DYW domain-containing protein, DYW1. We found that the interaction with MORFs is critical for the RNA editing function of PPO1. Our study not only elucidates a role for PPO1 that does not involve tetrapyrrole biosynthesis but also, identifies a distinct regulator of plastid RNA editing in higher plants.
Results
Disruption of PPO1 Severely Impairs Seedling Growth, Chlorophyll Synthesis, and NDH Complex Accumulation.
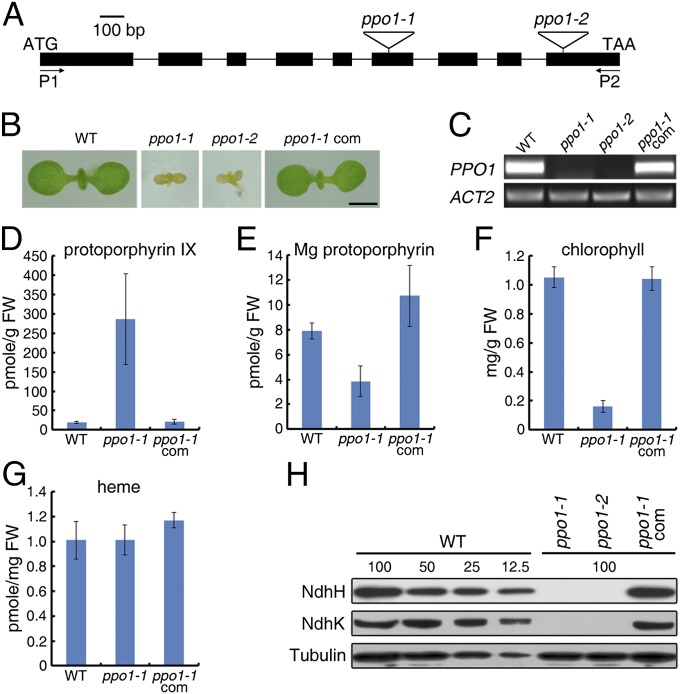
The PPO enzymes are encoded by PPO1 (At4g01690) and PPO2 (At5g14220) in the A. thaliana genome. PPO1 is induced by light and ubiquitously expressed in plant tissues, whereas PPO2 transcripts are barely detectable (Fig. S1 A and B). To determine the biological function of PPO1 in Arabidopsis, we obtained and analyzed two independent T-DNA mutant alleles (Fig. 1A). Homozygosity for either the ppo1-1 or ppo1-2 null mutation resulted in etiolated cotyledons and eventually, seedling lethality (Fig. 1 B and C).
Fig. 1.
Seedling phenotype, tetrapyrrole contents, and NDH accumulation of ppo1 mutants. (A) Diagram of PPO1 and the insertion sites of the T-DNA mutants. Black boxes represent exons, and lines between the boxes indicate introns. Triangles denote T-DNA insertions. P1 and P2 are the positions of primers used for PCR genotyping and RT-PCR assays. (B) Seedling lethality phenotype of the ppo1 mutants. Seedlings were grown in Murashige and Skoog (MS) medium for 7 d. ppo1-1 com, ppo1-1 complemented with ProPPO1:PPO1. (Scale bar: 1 mm.) (C) RT-PCR confirmation of the ppo1 mutants. Amplification of ACT2 served as an equal loading control. (D–G) HPLC analysis of (D) protoporphyrin IX, (E) Mg protoporphyrin, (F) chlorophyll, and (G) heme contents. Plants were grown in MS medium for 10 d before harvesting. FW, fresh weight. Data are mean ± SD of biological triplicates. (H) Immunoblot analysis of NdhH and NdhK subunits. Ten micrograms total protein extract from 7-d-old seedlings were separated by SDS/PAGE. The lanes were loaded with a series of dilutions as indicated. Immunoblotting against the tubulin antibody served as a loading control.
To test how the tetrapyrrole biosynthetic pathway was affected by the ppo1 mutation, several key intermediates in this pathway were determined by HPLCy. Protoporphyrin IX was drastically increased in ppo1-1 compared with the WT, consistent with the loss of PPO1 function in the mutant (Fig. 1D). Importantly, the steady state levels of Mg protoporphyrin and chlorophyll were decreased in ppo1-1 (Fig. 1 E and F). However, the heme level was not influenced by the PPO1 mutation (Fig. 1G). These results indicate that PPO1 dominantly affects the Mg branch of the tetrapyrrole biosynthesis pathway and likely provides the substrate for Mg chelation. Transgenic expression of the PPO1 coding sequence under the control of its own promoter fully compensated for the defects of the ppo1-1 mutant (Fig. 1).
We next determined the protein levels of thylakoid complexes, including photosystem I (PSI), PSII, cytochrome b6f, ATP synthase, light harvesting complex of PSI, and light harvesting complex of PSII. The content of some representative subunits of these complexes was drastically decreased in the ppo1 alleles (Fig. S2A), indicating that loss of PPO1 widely affects stability of the thylakoid proteins. Most strikingly, NdhH and NdhK, two subunits of the chloroplast NDH, were absent in ppo1 (Fig. 1H). The absence of the NDH complex is unlikely caused by changes in transcriptional control, because the transcript level of ndhH, ndhK, ndhB, and ndhD was not affected by the PPO1 mutations (Fig. S2B). Moreover, it is also unlikely the result of secondary effects of seedling lethality, because the NDH complex is present, even in etioplasts.
Loss of PPO1 Affects Multiple Editing Sites in Plastids.
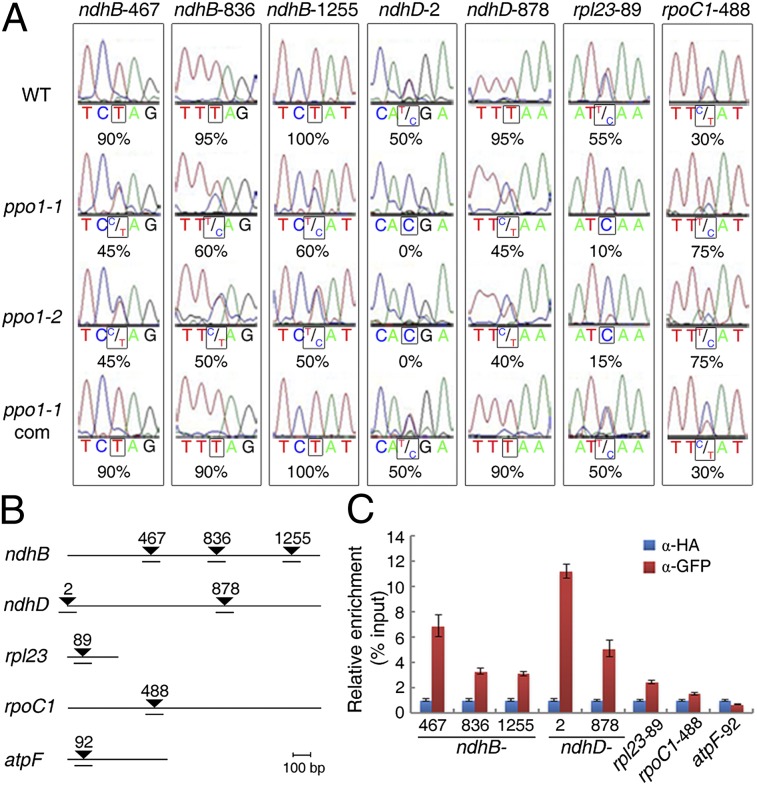
Previous studies revealed that 16 of 34 known editing sites in plastids occur in transcripts encoding multiple subunits of the NDH complex (8). We tested whether the RNA editing status in these sites was altered in ppo1 mutants. Interestingly, except for the ndhB-746 site, the editing efficiencies of all sites in ndhB, ndhD, ndhF, and ndhG transcripts were reduced to different extents in ppo1 compared with the WT (Fig. 2A and Fig. S3A). Strikingly, disruption of PPO1 led to a complete loss of editing of the ndhD-2 site, where ACG was partly edited into the translation start codon AUG in the WT (Fig. 2A). In addition, we screened 18 other known plastid editing sites (4) and found that the editing efficiency of rpl23-89 exhibited a significant reduction, whereas the editing efficiency of petL-5 showed a moderate reduction in ppo1. However, editing activity at the rpoC1-488 site was markedly increased (Fig. 2A and Fig. S3A). No editing variation was observed for the remaining plastid transcripts analyzed (Fig. S3B). Therefore, PPO1 widely affects plastid RNA editing.
Fig. 2.
Loss of PPO1 affects multiple editing sites in plastids. (A) Sequencing analysis showing the editing efficiency (percent) of sites severely affected in the ppo1 mutants. (B) Diagram of genes analyzed in the RNA immunoprecipitation assay. Triangles indicate RNA editing sites, and numbers shown above are their positions from the ATG start codon. Regions analyzed by PCR are underlined. (C) RNA immunoprecipitation followed by a quantitative PCR assay using the GFP antibody and 35S:PPO1-GFP plants, showing relative enrichment against the input of various RNA targets. Precipitation using the HA antibody served as a control. Data are mean ± SD of triplicates.
Our hypothesis that PPO1 is recruited to the editing target transcripts was supported by an RNA immunoprecipitation analysis. There was a pronounced enrichment of sites in ndhB-467, ndhB-836, ndhB-1255, ndhD-2, ndhD-878, rpl23-89, and rpoC1-488 (but not of the atpF-92 control) in RNA samples isolated from 35S:PPO1-GFP transgenic plants and pulled down with the GFP antibody (but not in samples pulled down with the HA antibody control) (Fig. 2 B and C), indicating that PPO1 is specifically associated with these target sequences in the transcripts. Consistently, PPO1 localized to the chloroplasts of 35S:PPO1-GFP transgenic plants (Fig. S1C). An immunoblot assay of these plants using GFP antibody also showed that PPO1 is targeted to both the stroma and the thylakoid membrane (Fig. S1D). In agreement with these findings, a PPO1 construct lacking the coding sequence of the chloroplast transit peptide failed to rescue the ppo1 mutant phenotype (Fig. S1E), indicating that targeting of PPO1 to chloroplasts is required for its function.
PPO1 Interacts with Plastid-Localized MORF Proteins.
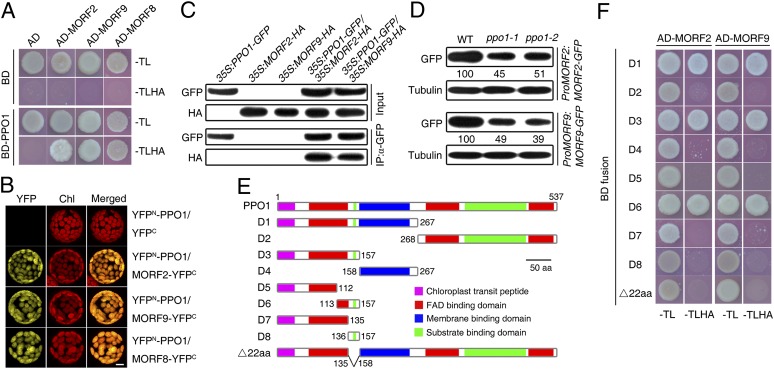
To establish how PPO1 affects the RNA editing of plastid-encoded genes, we used PPO1 as bait in a yeast two-hybrid screen and identified a putative interaction partner, MORF2 (Fig. 3A). MORF2 was reported to be essential for RNA editing in plastids (13). Notably, PPO1 also interacted with two MORF2 homologs, MORF9 and RIP1/MORF8 (13, 14), in a yeast system (Fig. 3A). Our failure to identify coding sequences for MORF9 and MORF8 in our initial yeast two-hybrid screen might have been caused by their low abundance in the cDNA library. Direct interaction between PPO1 and MORF2, MORF9, or MORF8 was verified using a bimolecular fluorescence complementation assay. Coexpression of the N-terminal YFP fusion of PPO1 (YFPN-PPO1) and the C-terminal YFP fusion of MORF2, MORF9, or MORF8 (i.e., MORF2-YFPC, MORF9-YFPC, or MORF8-YFPC) reconstituted a functional YFP in chloroplasts (Fig. 3B). Next, we generated transgenic plants coexpressing 35S:PPO1-GFP and 35S:MORF2-HA or 35S:MORF9-HA and performed coimmunoprecipitation analysis using extracts from these plants. The GFP antibodies were able to precipitate the MORF2-HA and MORF9-HA fusion proteins in vivo (Fig. 3C). Moreover, glycerol gradient fractionation followed by immunoblot analysis showed that PPO1 partly comigrated with MORF2 or MORF9, and PPO1 mutation seems to influence the complex size of MORF2 (Fig. S4). Interestingly, the protein levels of MORF2 and MORF9 were reduced in the ppo1 mutant compared with the WT plants transformed with MORF2-GFP or MORF9-GFP under the control of their corresponding native promoter (Fig. 3D), indicating that loss of PPO1 affects the stability of MORF2 and MORF9.
Fig. 3.
PPO1 directly interacts with MORF2/8/9 proteins. (A) Yeast two-hybrid assay. AD, GAL4 activation domain; BD, GAL4 DNA binding domain. -TL and -TLHA indicate SD/-Trp-Leu and SD/-Trp-Leu-His-Ade dropout plates, respectively. The ability to grow on -TLHA plates indicates an interaction between two proteins. (B) Bimolecular fluorescence complementation assay showing that YFPN-PPO1 interacts with MORF2-YFPC (or MORF9-YFPC or MORF8-YFPC) to produce YFP fluorescence in the chloroplasts. Chl, chlorophyll autofluorescence. (Scale bar: 5 μm.) (C) Coimmunoprecipitation assay showing that the GFP antibody could precipitate MORF2-HA or MORF9-HA in samples isolated from the indicated transgenic plants. (D) Immunoblot analysis of MORF proteins in the ppo1 mutants. Homozygous transgenic plants harboring ProMORF2:MORF2-GFP or ProMORF9:MORF9-GFP in the WT or ppo1-1 or ppo1-2 mutant backgrounds were immunoblotted with the GFP antibody. Immunoblotting against the tubulin antibody served as a loading control. Relative amounts of GFP fusion proteins normalized to tubulin are shown below. (E) Diagram of the domain structures of PPO1 and various PPO1 deletions (D1–D8) and a truncation (Δ22aa). The domains were predicated based on an alignment with tobacco PPO2 (27). (F) Yeast two-hybrid assay between an AD fusion of MORF2 or MORF9 and different versions of PPO1 tagged with the BD domain.
The PLS subfamily of PPR proteins recognizes distinct targets for RNA editing (6–11). A number of PPR factors, including CRR4, CRR28, OTP80, and OTP82, and a DYW domain-containing protein, DYW1, are known to be involved in the editing of ndhB and ndhD transcripts (22–26), which we show in this study to contain editing sites severely affected in ppo1. To examine whether PPO1 promotes RNA editing by associating with these PPR proteins and DYW1, we carried out a yeast two-hybrid assay. Surprisingly, there was no interaction between PPO1 and these proteins (Fig. S5A). However, MORF2 and MORF9 interacted with CRR28, OTP82, and DYW1 (Fig. S5). Moreover, we showed that the C-terminal region containing the MORF/RIP domain of MORF2 and MORF9 interacted with these PPR proteins and DYW1 (Fig. S6). Similarly, the N terminus of RIP1, including the MORF/RIP domain, interacted with the PPR protein REQUIRED FOR AccD RNA EDITING 1 (14). Therefore, the MORF/RIP domain is most likely responsible for the interaction with PPR or related proteins. However, these MORFs did not interact with CRR4 or OTP80 (Fig. S5A). In line with this finding, organelle RNA recognition motif protein 1 and several mitochondria-localized MORF proteins interact selectively with some PPRs (13, 15).
The crystal structure of mitochondrial PPO2 from Nicotiana tabacum (NtPPO2) has been solved (27). Arabidopsis PPO1 shows 23.5% amino acid identity with NtPPO2, and the 3D structure of reconstituted PPO1 is highly similar to that of NtPPO2 (Fig. S7). To determine which domain of PPO1 is responsible for interacting with MORF proteins, we constructed a series of PPO1 truncations (named D1-8) and examined their interactions with MORF2 and MORF9 in yeast cells (Fig. 3E). Importantly, we found that the N-terminal portion (amino acid residues 113–157; D6) of PPO1 was sufficient for the interaction with MORF2 and MORF9 and that the deletion of amino acid residues 136–157 (Δ22aa) completely abolished this interaction (Fig. 3F), indicating that this 22-aa region of PPO1 is critical for the interaction with MORF proteins. However, the 22-aa fragment alone (D8) is not sufficient for the interaction with MORFs (Fig. 3F). Furthermore, MORF2 and MORF9 interacted with PPO1 through their N-terminal fragments in the absence of a known domain (Fig. S6).
RNA Editing Function of PPO1 Depends on Its Interaction with MORFs.
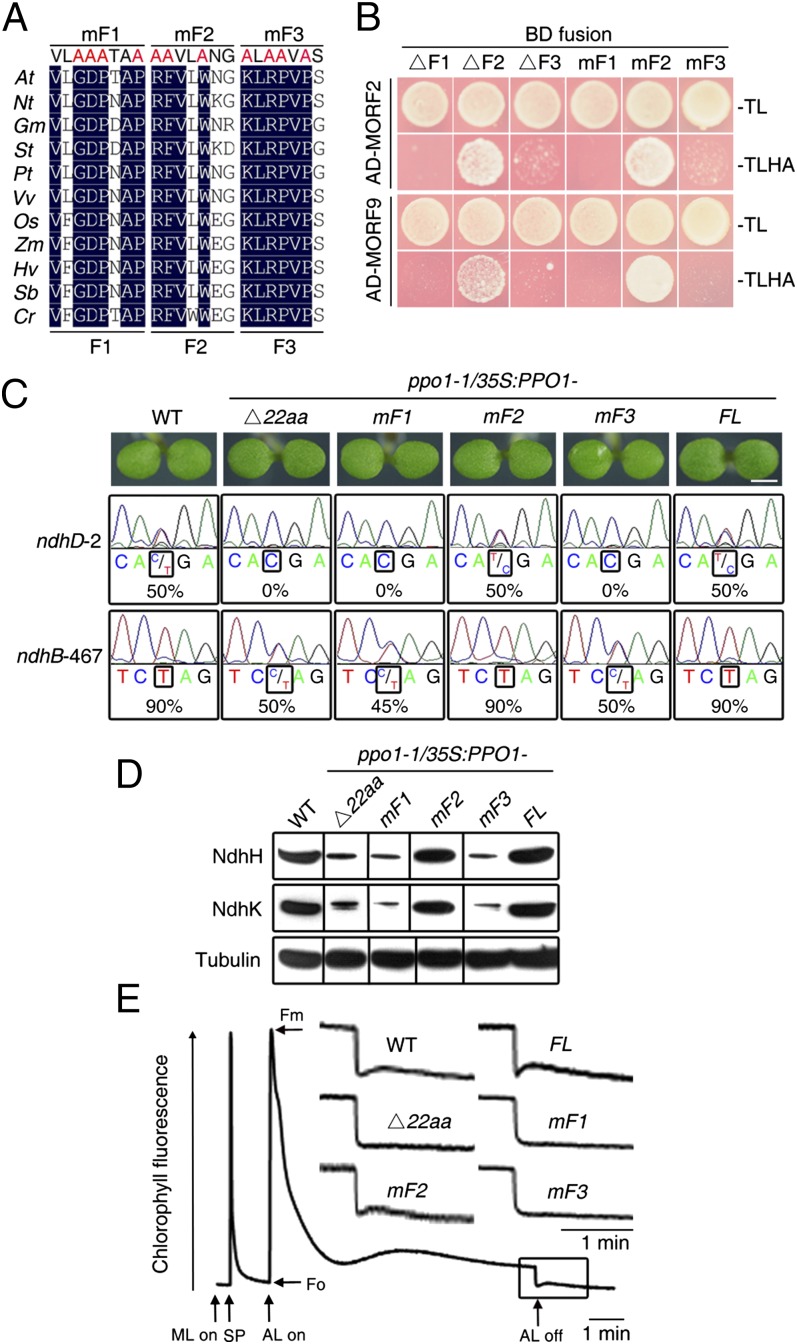
We further dissected the 22-aa interacting region of PPO1 into three motifs, F1 (amino acid residues 136–143), F2 (amino acid residues 144–150), and F3 (amino acid residues 151–157), and explored the functions of these regions individually (Fig. 4A). Interestingly, deleted or mutated (containing alanine substitutions) versions of the F1 or F3 regions failed to interact with MORF2 and MORF9, whereas those versions of F2 did not (Fig. 4B), suggesting that F1 and F3 are the interaction motifs of PPO1. In support of this conclusion, structural remodeling of PPO1 revealed that the F2 motif is a β-strand inside the protein, whereas the F1 motif belongs to a loop preceding the F2 strand; additionally, the F3 motif consists of a loop and a β-strand antiparallel to F2 and is exposed on the protein surface (Fig. S7), thus providing the point of contact with MORF proteins.
Fig. 4.
The interaction with MORFs is essential for the RNA editing function of PPO1. (A) Amino acid alignment of the 22-aa interacting motif corresponding to amino acid residues 136–157 in Arabidopsis PPO1. This region was further divided into three parts: F1, F2, and F3. In these regions, some of the conserved amino acid residues among species were mutated to alanine (red) and designated as mF1, mF2, and mF3, respectively. At, A. thaliana; Cr, Chlamydomonas reinhardtii; Gm, Glycine max; Hv, Hordeum vulgare; Nt, N. tabacum; Os, Oryza sativa; Pt, Populus trichocarpa; Sb, Sorghum bicolor; St, Solanum tuberosum; Vv, Vitis vinifera; Zm, Zea mays. (B) A yeast two-hybrid assay between AD-fused MORFs and deletions or mutations of PPO1 tagged with BD. ΔF1, ΔF2, and ΔF3 represent deletions in the F1, F2, and F3 regions of PPO1, respectively. (C) Seedling phenotype and editing efficiency (percent) of the WT and various transgenic plants harboring PPO1-based constructs in a ppo1-1 background. Seedlings were grown on MS medium for 5 d. FL, full-length coding sequence of WT PPO1. (Scale bar: 1 mm.) (D) Immunoblot analysis of NdhH and NdhK subunits in WT and various transgenic plants. Immunoblotting against the tubulin antibody served as a loading control. Samples were run side-by-side with those in Fig. S8C. Dividing lines indicate noncontiguous lanes in the gel. (E) Chlorophyll fluorescence showing NDH activity in the WT and various transgenic plants. The curve shows a characteristic transient rise in chlorophyll fluorescence ascribed to NDH activity after the offset of actinic light (AL) in WT plants. Insets are magnified traces of the boxed areas. Fo, minimum fluorescence yield; Fm, maximum fluorescence yield; ML, measuring light; SP, saturating pulse.
We generated a series of vectors containing various mutant PPO1 sequences and tested their ability to complement ppo1-1. Constructs harboring mutations in F1 (mF1) or F3 (mF3) or a deletion of F1 (ΔF1), F3 (ΔF3), or the entire 22-aa region (Δ22aa) completely rescued the seedling lethality phenotype of ppo1-1 (Fig. 4C and Fig. S8A). Intriguingly, the RNA editing efficiency of the ndhD-2 and ndhB-467 sites was not recovered in these transgenic plants (Fig. 4C and Fig. S8A). Consistently, the NDH complex was only weakly detectable, and NDH activity, as monitored by chlorophyll fluorescence, was impaired in transgenic plants harboring mutated or truncated forms of PPO1 compared with transgenic plants harboring the full-length control (Fig. 4 D and E and Fig. S8 B and C). The chlorophyll fluorescence of these transgenic plants was identical to that of ndh KO mutants (28). It should be noted that loss of the NDH complex in Arabidopsis does not result in a visible plant phenotype. However, deletion (ΔF2) or mutation (mF2) of the F2 motif completely rescued the seedling lethality, RNA editing, and NDH activity of ppo1 (Fig. 4 C–E and Fig. S8). Therefore, we conclude that the F1 and F3 motifs of PPO1 are critical for the interaction with MORFs and that this interaction between these proteins is required for the stability of the NDH complex, likely because of the editing of mRNAs, including ndhB and ndhD, by means of PPO1.
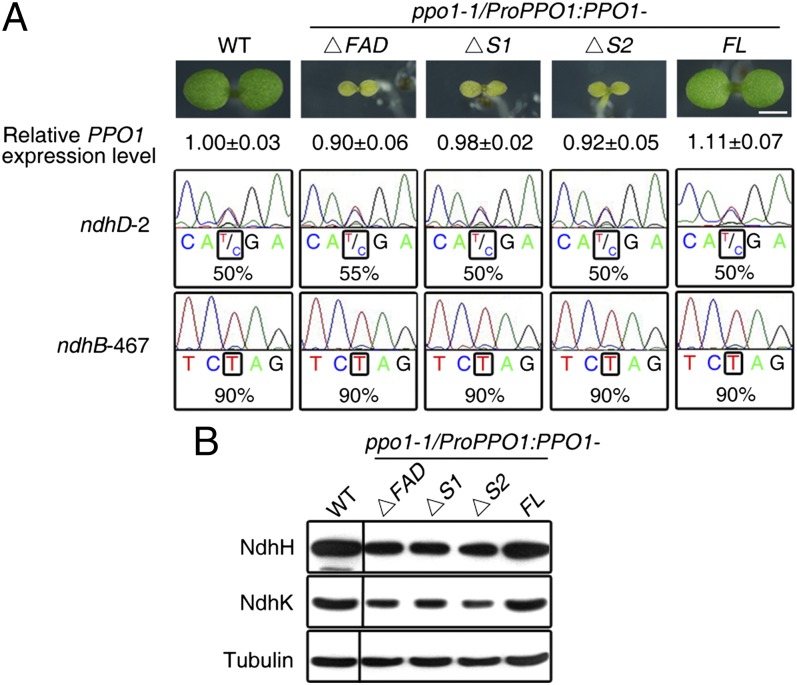
Loss of FAD or Substrate Binding of PPO1 Does Not Affect RNA Editing.
Because of their importance in catalyzing protoporphyrinogen IX oxidation, the FAD, membrane, and substrate binding domains of PPO1 and PPO2 are largely conserved in species from plants to animals (Fig. S9). To assess how the FAD and substrate binding activities affect the function of PPO1, we introduced versions of PPO1 with truncations in the FAD binding domain (amino acids 63–69, ΔFAD) or two substrate binding sites (amino acids 389–395, ΔS1; amino acids 403–409, ΔS2) into ppo1-1. None of the transgenes could rescue the lethal phenotype of the ppo1-1 homozygote, although the transcript level of mutant PPO1 was similar to that of endogenous PPO1 in the WT (Fig. 5A), confirming that the catalytic activity of PPO1 requires efficient FAD and protoporphyrinogen IX binding and indicating that PPO2 expression does not compensate for the loss of PPO1 function. Surprisingly, the extent of RNA editing of ndhD-2 and ndhB-467 in the ppo1-1 plants harboring these truncated PPO1 genes was largely restored to levels observed in ppo1-1 transformed with full-length WT PPO1 (Fig. 5A). In agreement with this finding, the levels of the analyzed NDH subunits, NdhH and NdhK, were also largely restored (Fig. 5B). These data together suggest that the FAD and substrate binding domains of PPO1 are critical for tetrapyrrole biosynthesis but not RNA editing.
Fig. 5.
The FAD and substrate binding domains of PPO1 are not required for RNA editing. (A) Seedling phenotype and editing efficiency in various PPO1 transgenic plants and the WT. Seedlings were grown in MS medium for 5 d. ΔFAD, deletion from amino acid residues 63–69 in the FAD binding domain of PPO1; ΔS1 and ΔS2, deletion from amino acid residues 389–395 and 403–409, respectively, in the substrate binding domain of PPO1. Relative PPO1 expression level was quantified by real-time RT-PCR. FL, full-length WT PPO1. (Scale bar: 1 mm.) (B) Immunoblot analysis of NdhH and NdhK subunits in the WT and various transgenic plants. Immunoblotting against tubulin served as a loading control. Dividing lines indicate noncontiguous lanes in the gel.
Discussion
Several previous studies reported that PPO1 mediates the plant defense response, resistance to peroxidizing herbicide, and drought tolerance (29–31). These roles of PPO1 are directly linked to the photosensitizing and oxidizing properties of porphyrins, which are synthesized in the tetrapyrrole metabolic pathway. This study, however, reveals an unexpected function for PPO1 in regulating RNA editing in plastids. We show that more than one-half of the plastid editing sites, including 15 sites in transcripts that encode multiple NDH subunits, display editing defects in the ppo1 mutants, supporting the broad effect of PPO1 on plastid RNA editing. Consistently, the NDH complex was abolished by mutation of PPO1. However, the deficiency of other tested thylakoid protein complexes likely results from the disturbed tetrapyrrole function, because ppo1 largely impairs chlorophyll biosynthesis.
Three pieces of evidence support the notion that PPO1 is truly involved in RNA editing. First, there is no pronounced difference in ndhB and ndhD transcript abundance or pattern between the ppo1 mutants and the WT (Fig. S2B). This observation is significant, because altered RNA stability or RNA processing can indirectly influence editing efficiency (32, 33). Second, 18 of 34 known editing sites in chloroplast transcripts are affected by the ppo1 mutations, and transcripts with multiple editing sites (e.g., ndhB and ndhD) exhibit different editing efficiencies (Fig. 2 and Fig. S3), indicating that RNA editing defects in ppo1 are site-specific. Variations in the extent of editing might represent a mechanism for controlling the amount of active NDH or changing the properties or functions of the NDH complex (34). Third, PPO1 associates with transcripts in the vicinity of specific sites to be edited (Fig. 2 B and C).
We collected in vitro and in vivo evidence that PPO1 directly interacts with MORF proteins (including MORF2, MORF8, or MORF9) through two short peptide regions comprising amino acid residues 136–143 and 151–157 in the N terminus of PPO1 (Fig. 3). The interaction between PPO1 and MORFs is essential for its RNA editing function, because constructs with mutations or deletions of these two regions fail to rescue the editing defects at ndhB and ndhD and the loss of NDH stability in ppo1 (Fig. 4). MORF2 and MORF9 also have a high affinity for PPR proteins (including CRR28 and OTP82) and DYW1, which lacks PPR motifs (Fig. S5). However, REQUIRED FOR AccD RNA EDITING 1 interacts with RIP1 through its PPR domain (14). Hence, the regions of PPR or related proteins required for interacting with MORF/RIP factors are diverse and likely depend on the editing factors and the editing sites that they control. Accordingly, MORF2 and MORF9 prefer to interact with DYW1 but not CRR4, although both proteins are essential for editing of the ndhD-2 site (26).
The current consensus RNA editing model predicts that PPR proteins specifically recognize cis elements near the target cytidine residue and provide a platform for molecular attachment of other editing factors, such as MORF proteins (1, 7, 10, 11, 35). MORFs act as bridges that physically link PPR or related proteins to PPO1 through specific regions (Fig. S6). Because MORF proteins are required for the complete editing of almost all sites in plastid transcripts and many sites in mitochondrial transcripts (13), they likely serve as universal factors in the editing apparatus. However, 18 editing sites in plastid transcripts require the expression of PPO1, indicating that there is a selective recruitment of PPO1 to target transcripts. The questions of how this specific recruitment is organized and why PPO among the tetrapyrrole enzymes is recruited for the control of RNA editing deserve additional investigation. Nevertheless, PPO1 represents a component of the plastid RNA editosome and adds a regulatory layer to the editing process, in which an enzyme involved in tetrapyrrole biosynthesis interacts with MORFs and affects their stability. Therefore, PPR or related proteins, MORFs, PPO1, and possibly some other factors function together to mediate RNA editing in plastids. A similar regulatory mechanism possibly also applies to mitochondrial RNA editing.
Interestingly, our extensive structure function analysis reveals that the FAD and substrate binding domains are essential for the catalytic activity of PPO1 in the tetrapyrrole biosynthetic pathway, whereas the MORF-interacting motifs are required for RNA editing. Accordingly, the FAD and substrate binding domains are more conserved than the overall sequences of PPO1 among species, whereas the stretch of amino acid residues responsible for interacting with MORFs diverges during the evolution of PPO in plants and animals (Fig. S9). The functions of both types of domains are mutually dispensable, indicating that PPO1 regulates RNA editing independently of the enzymatic oxidization of protoporphyrinogen IX. The localization of PPO1 in both the thylakoid membrane and the stroma may enable it to have dual functions in chloroplasts. Although PPO1 is critical for tetrapyrrole biosynthesis in both plants and animals, plants have chloroplasts that require RNA editing of particular transcripts. Thus, PPO1 might enable plants to contend with the sharp increase in editing sites that emerged in flowering plants during the course of evolution. Consistent with this speculation, genes encoding MORFs are only present in flowering plant lineages (13).
In addition, the sequence of the MORF-interacting domain of PPO1 shares low similarity (∼13.6%) with the corresponding region of the mitochondrially localized PPO2 (Fig. S9). Therefore, this study indicates that RNA editing is a plant-specific function of the PPO1 isoform that differs from that of PPO2, which is most likely derived from the progenitors of mitochondria and their tetrapyrrole pathway.
Materials and Methods
Plant Materials and Growth Conditions.
The ppo1-1 (Salk_143057) and ppo1-2 (Salk_017634) mutants were obtained from the Arabidopsis Biological Resource Center and are of the Columbia (Col) ecotype. The mutants were screened by PCR genotyping, and their T-DNA insertion sites were verified by sequencing. After sterilization, seeds were sown on MS medium containing 1% sucrose and 0.8% agar, and plants were grown at 22 °C under 16 h light/8 h darkness.
Extraction and Analysis of Tetrapyrroles.
Protoporphyrin IX, Mg porphyrins, and heme were extracted and analyzed by HPLC as described previously (36). During porphyrin extraction, the substrate protoporphyrinogen IX is instantaneously oxidized to protoporphyrin IX. Porphyrins and Mg porphyrins were identified and quantified by fluorescence detection using authentic standards (Frontier Scientific). Heme absorbance was monitored at 398 nm using a millimolar extinction coefficient of 144 (37).
Analysis of RNA Editing.
Plant total RNA was isolated using an RNAprep Pure Plant Kit (Tiangen), and the first-strand cDNA was synthesized using reverse transcriptase (Invitrogen). The RT-PCR products were obtained with specific primers surrounding the editing sites and then used as templates for sequencing. The levels of RNA editing were estimated by the relative heights of the peaks of the nucleotide in the sequence analyzed. The primer sequences are listed in Table S1.
Bimolecular Fluorescence Complementation Assay.
Plasmids containing N- and C-terminal fusions of YFP were cotransformed into Arabidopsis protoplasts as previously described (38). The protoplasts were then incubated under light for 12–16 h, and the YFP fluorescence was determined using a confocal microscope (Olympus).
Yeast Two-Hybrid Assay.
The yeast two-hybrid assay was performed according to the Yeast Protocols Handbook (Clontech). Briefly, the respective combinations of GAL4 DNA binding domain and GAL4 activation domain fusions were cotransformed into the yeast strain Y2H Gold (Clontech). The transformants were grown on SD/-Trp-Leu or SD/-Trp-Leu-His-Ade dropout plates.
Plasmid Construction.
A detailed description of the plasmid construction method is in SI Materials and Methods.
Chlorophyll Fluorescence Analysis.
The transient increase in chlorophyll fluorescence after turning off actinic light was monitored as previously described (39) using a MINI-PAM Portable Chlorophyll Fluorometer (Waltz).
Coimmunoprecipitation and Immunoblot Assays.
The procedures used for coimmunoprecipitation and immunoblot assays were described previously (38). Proteins were separated on a 10% (vol/vol) SDS/PAGE gel and detected by immunoblotting with anti-GFP (Abcam) and anti-HA (Invitrogen) antibodies. The procedures for isolation of chloroplasts, thylakoid membranes, and stroma proteins, immunoblot analysis, and glycerol density gradient centrifugation were as previously described (40).
Supplementary Material
Acknowledgments
We thank Arabidopsis Biological Resource Center for providing the T-DNA mutants. This work was supported by National Natural Science Foundation of China Grant 31170221 (to R.L.) and a Chinese Academy of Sciences Grant (to R.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1316183111/-/DCSupplemental.
References
- 1.Shikanai T. RNA editing in plant organelles: Machinery, physiological function and evolution. Cell Mol Life Sci. 2006;63(6):698–708. doi: 10.1007/s00018-005-5449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang W, Fei Y, Page M. Biological significance of RNA editing in cells. Mol Biotechnol. 2012;52(1):91–100. doi: 10.1007/s12033-012-9498-7. [DOI] [PubMed] [Google Scholar]
- 3.Castandet B, Araya A. RNA editing in plant organelles. Why make it easy? Biochemistry (Mosc) 2011;76(8):924–931. doi: 10.1134/S0006297911080086. [DOI] [PubMed] [Google Scholar]
- 4.Chateigner-Boutin AL, Small I. A rapid high-throughput method for the detection and quantification of RNA editing based on high-resolution melting of amplicons. Nucleic Acids Res. 2007;35(17):e114. doi: 10.1093/nar/gkm640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bentolila S, Elliott LE, Hanson MR. Genetic architecture of mitochondrial editing in Arabidopsis thaliana. Genetics. 2008;178(3):1693–1708. doi: 10.1534/genetics.107.073585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lurin C, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16(8):2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmitz-Linneweber C, Small I. Pentatricopeptide repeat proteins: A socket set for organelle gene expression. Trends Plant Sci. 2008;13(12):663–670. doi: 10.1016/j.tplants.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Fujii S, Small I. The evolution of RNA editing and pentatricopeptide repeat genes. New Phytol. 2011;191(1):37–47. doi: 10.1111/j.1469-8137.2011.03746.x. [DOI] [PubMed] [Google Scholar]
- 9.Zehrmann A, Verbitskiy D, Härtel B, Brennicke A, Takenaka M. PPR proteins network as site-specific RNA editing factors in plant organelles. RNA Biol. 2011;8(1):67–70. doi: 10.4161/rna.8.1.14298. [DOI] [PubMed] [Google Scholar]
- 10.Yagi Y, et al. Pentatricopeptide repeat proteins involved in plant organellar RNA editing. RNA Biol. 2013;10(9):1419–1425. doi: 10.4161/rna.24908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shikanai T, Fujii S. Function of PPR proteins in plastid gene expression. RNA Biol. 2013;10(9):1446–1456. doi: 10.4161/rna.25207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salone V, et al. A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett. 2007;581(22):4132–4138. doi: 10.1016/j.febslet.2007.07.075. [DOI] [PubMed] [Google Scholar]
- 13.Takenaka M, et al. Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proc Natl Acad Sci USA. 2012;109(13):5104–5109. doi: 10.1073/pnas.1202452109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bentolila S, et al. RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing. Proc Natl Acad Sci USA. 2012;109(22):E1453–E1461. doi: 10.1073/pnas.1121465109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun T, et al. An RNA recognition motif-containing protein is required for plastid RNA editing in Arabidopsis and maize. Proc Natl Acad Sci USA. 2013;110(12):E1169–E1178. doi: 10.1073/pnas.1220162110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka R, Tanaka A. Tetrapyrrole biosynthesis in higher plants. Annu Rev Plant Biol. 2007;58:321–346. doi: 10.1146/annurev.arplant.57.032905.105448. [DOI] [PubMed] [Google Scholar]
- 17.Mochizuki N, et al. The cell biology of tetrapyrroles: A life and death struggle. Trends Plant Sci. 2010;15(9):488–498. doi: 10.1016/j.tplants.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 18.Strand A, Asami T, Alonso J, Ecker JR, Chory J. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature. 2003;421(6918):79–83. doi: 10.1038/nature01204. [DOI] [PubMed] [Google Scholar]
- 19.Pogson BJ, Woo NS, Förster B, Small ID. Plastid signalling to the nucleus and beyond. Trends Plant Sci. 2008;13(11):602–609. doi: 10.1016/j.tplants.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 20.Warren MJ, Smith AG. Tetrapyrroles Birth, Life and Death. New York: Landes Bioscience, Austin, Texas; Springer Science+Business Media; 2009. [Google Scholar]
- 21.Poulson R, Polglase WJ. The enzymic conversion of protoporphyrinogen IX to protoporphyrin IX. Protoporphyrinogen oxidase activity in mitochondrial extracts of Saccharomyces cerevisiae. J Biol Chem. 1975;250(4):1269–1274. [PubMed] [Google Scholar]
- 22.Kotera E, Tasaka M, Shikanai T. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature. 2005;433(7023):326–330. doi: 10.1038/nature03229. [DOI] [PubMed] [Google Scholar]
- 23.Okuda K, et al. Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. Plant Cell. 2009;21(1):146–156. doi: 10.1105/tpc.108.064667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammani K, et al. A study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites. Plant Cell. 2009;21(11):3686–3699. doi: 10.1105/tpc.109.071472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohtani S, et al. Targeted gene disruption identifies three PPR-DYW proteins involved in RNA editing for five editing sites of the moss mitochondrial transcripts. Plant Cell Physiol. 2010;51(11):1942–1949. doi: 10.1093/pcp/pcq142. [DOI] [PubMed] [Google Scholar]
- 26.Boussardon C, et al. Two interacting proteins are necessary for the editing of the NdhD-1 site in Arabidopsis plastids. Plant Cell. 2012;24(9):3684–3694. doi: 10.1105/tpc.112.099507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koch M, et al. Crystal structure of protoporphyrinogen IX oxidase: A key enzyme in haem and chlorophyll biosynthesis. EMBO J. 2004;23(8):1720–1728. doi: 10.1038/sj.emboj.7600189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rumeau D, et al. New subunits NDH-M, -N, and -O, encoded by nuclear genes, are essential for plastid Ndh complex functioning in higher plants. Plant Cell. 2005;17(1):219–232. doi: 10.1105/tpc.104.028282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molina A, et al. Inhibition of protoporphyrinogen oxidase expression in Arabidopsis causes a lesion-mimic phenotype that induces systemic acquired resistance. Plant J. 1999;17(6):667–678. doi: 10.1046/j.1365-313x.1999.00420.x. [DOI] [PubMed] [Google Scholar]
- 30.Lermontova I, Grimm B. Overexpression of plastidic protoporphyrinogen IX oxidase leads to resistance to the diphenyl-ether herbicide acifluorfen. Plant Physiol. 2000;122(1):75–84. doi: 10.1104/pp.122.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phung T-H, et al. Porphyrin biosynthesis control under water stress: Sustained porphyrin status correlates with drought tolerance in transgenic rice. Plant Physiol. 2011;157(4):1746–1764. doi: 10.1104/pp.111.188276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu B, Hanson MR. A single nuclear gene specifies the abundance and extent of RNA editing of a plant mitochondrial transcript. Nucleic Acids Res. 1992;20(21):5699–5703. doi: 10.1093/nar/20.21.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chateigner-Boutin AL, Hanson MR. Cross-competition in transgenic chloroplasts expressing single editing sites reveals shared cis elements. Mol Cell Biol. 2002;22(24):8448–8456. doi: 10.1128/MCB.22.24.8448-8456.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chateigner-Boutin AL, Small I. Plant RNA editing. RNA Biol. 2010;7(2):213–219. doi: 10.4161/rna.7.2.11343. [DOI] [PubMed] [Google Scholar]
- 35.Okuda K, Nakamura T, Sugita M, Shimizu T, Shikanai T. A pentatricopeptide repeat protein is a site recognition factor in chloroplast RNA editing. J Biol Chem. 2006;281(49):37661–37667. doi: 10.1074/jbc.M608184200. [DOI] [PubMed] [Google Scholar]
- 36.Czarnecki O, Peter E, Grimm B. Methods for analysis of photosynthetic pigments and steady-state levels of intermediates of tetrapyrrole biosynthesis. Methods Mol Biol. 2011;775:357–385. doi: 10.1007/978-1-61779-237-3_20. [DOI] [PubMed] [Google Scholar]
- 37.Castelfranco PA, Jones OT. Protoheme turnover and chlorophyll synthesis in greening barley tissue. Plant Physiol. 1975;55(3):485–490. doi: 10.1104/pp.55.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jing Y, et al. Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell. 2013;25(1):242–256. doi: 10.1105/tpc.112.105742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shikanai T, et al. Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc Natl Acad Sci USA. 1998;95(16):9705–9709. doi: 10.1073/pnas.95.16.9705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong LL, et al. Chloroplast small heat shock protein HSP21 interacts with plastid nucleoid protein pTAC5 and is essential for chloroplast development in Arabidopsis under heat stress. Plant Cell. 2013;25(8):2925–2943. doi: 10.1105/tpc.113.111229. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.