Significance
The p53 family member deltaNp63 (ΔNp63) is required for transcriptional activation of the microprocessor complex subunit DGCR8 in epidermal cells, leading to terminal differentiation of tissues such as the epidermis. We show here that loss of ΔNp63 leads to the generation of cells with self-renewing but limited differentiation capacity. When DGCR8 is reexpressed in cells deficient for ΔNp63, these cells can terminally differentiate into all three germ layers. We dubbed these cells induced multipotent stem cells because of their remarkable plasticity and ability to differentiate into multiple cell lineages. Based on our results using human keratinocytes, we predict that epidermal cells can be extracted from patient skin biopsies and reprogrammed into multipotent stem cells by knockdown of ΔNp63 or DGCR8.
Abstract
The roles of microRNAs (miRNAs) and the miRNA processing machinery in the regulation of stem cell biology are not well understood. Here, we show that the p53 family member and p63 isoform, ΔNp63, is a transcriptional activator of a cofactor critical for miRNA processing (DGCR8). This regulation gives rise to a unique miRNA signature resulting in reprogramming cells to multipotency. Strikingly, ΔNp63−/− epidermal cells display profound defects in terminal differentiation and express a subset of markers and miRNAs present in embryonic stem cells and fibroblasts induced to pluripotency using Yamanaka factors. Moreover, ΔNp63−/− epidermal cells transduced with an inducible DGCR8 plasmid can differentiate into multiple cell fates in vitro and in vivo. We found that human primary keratinocytes depleted of ΔNp63 or DGCR8 can be reprogrammed in 6 d and express a unique miRNA and gene expression signature that is similar but not identical to human induced pluripotent stem cells. Our data reveal a role for ΔNp63 in the transcriptional regulation of DGCR8 to reprogram adult somatic cells into multipotent stem cells.
The factors required to reprogram adult somatic cells to induced pluripotent stem (iPS) cells is an area of intense research. The introduction of defined factors, such as octamer-binding transcription factor 4 (Oct4) sex determining region Y–box 2 (Sox2) kruppel-like factor 4 (Klf4), and the transcription factor c-myc, gives rise to the efficient reprogramming of fibroblasts to iPS cells (1). Cells deficient for p53 also show enhanced ability for reprogramming with the addition of Oct4 and Sox2 only (2–6). This enhanced reprogramming is thought to be due to loss of cell cycle checkpoints that lead to genomic instability of these iPS cells (7–9). In addition, overexpression of oncogenes or down-regulation of tumor suppressor genes, while leading to the generation of cells that are pluripotent, can also lead to the production of tumorigenic cells (4). Consequently, alternative methods for creating iPS cells or cells with stem-like properties from somatic cells are desirable. Here, we show that down-regulation of the p53 family member, ΔNp63, or down-regulation of its transcriptional target, DGCR8, in adult keratinocytes leads to the generation of induced multipotent stem (iMS) cells.
Both microRNAs (miRNAs) and the p53 family member, p63, have been implicated in processes that control stem cell proliferation and cell fate determination (10–14). As demonstrated using genetically engineered mice, p63 is critical for the development and maintenance of stratified epithelial tissues (11, 13). Previous studies using p63-deficient mice and human keratinocytes have shown that epidermal genes are suppressed by p63 and likely by ∆Np63, leading to the misexpression of mesodermal genes (15–17), indicating possible mechanisms for the failure of appropriate stratification of the epidermis in p63−/− mice. These previous studies did not make use of cellular or in vivo systems allowing dissection of the functions of the distinct isoforms of p63. Recent mouse models with deletions of specific p63 isoforms have allowed dissection of the functions of these isoforms (14, 18–21). There are two major groups of isoforms: those with a transactivation domain (TAp63) that structurally resemble p53 and those lacking this domain (ΔNp63); however, ΔNp63 also transcriptionally regulates unique target genes shown to be involved in limb and epidermal morphogenesis (18, 22). To further study the role of ΔNp63 in skin development, we generated ΔNp63 conditional KO mice (ΔNp63fl/fl) to specifically delete ΔNp63. We then generated ΔNp63 KO mice and found that in contrast to the skin of p63−/− mice, the ΔNp63−/− mice developed a disorganized epidermis that expressed some markers of terminal differentiation similar to the phenotype observed in another mouse model deficient for ΔNp63 (ΔNp63gfp/gfp) (19) or with in vivo siRNA knockdown of ΔNp63α (18). The ΔNp63gfp/gfp mice are born with a fragile epidermis that has accelerated differentiation in some areas of the epidermis and expression of keratin 8 (K8) and keratin 18 (K18) in other areas (19). The mice expressing an siRNA to knock down ΔNp63α exhibited skin that is hyperproliferative, and cells within the basal layer fail to exit the cell cycle (18). These observations are similar to the phenotypes observed in our allele of the ΔNp63−/− mice, which have areas of terminal differentiation in the epidermis, expression of K8 and K18, and hyperproliferative skin.
However, we found that epidermal cell lines derived from the epidermis of ΔNp63−/− mice morphologically resembled embryonic and induced pluripotent stem cells. Using a genome-wide analysis, we found that epidermal cell lines deficient for ΔNp63 express genes associated with pluripotency. We previously identified TAp63 as a transcriptional activator of Dicer (20) and hypothesized that ΔNp63 may similarly regulate enzymes required for miRNA biogenesis. Indeed, we found that ΔNp63 transcriptionally activates DGCR8 and in turn regulates a unique miRNA signature. Murine ΔNp63-deficient epidermal cell lines had the ability to self-renew and could be differentiated into multiple cell fates on reexpression of DGCR8. We recapitulated this stem cell–like morphology of ∆Np63−/− mouse epidermal cell lines in normal human epidermal keratinocytes (NHEKs) by deletion of ΔNp63 or DGCR8. Our data indicate that down-regulation of ΔNp63 or DGCR8 in mouse and human keratinocytes can be reprogrammed into multipotent stem cells.
Results and Discussion
Generation of a ΔNp63 Conditional KO Mouse.
To understand the roles of ΔNp63 in vivo, we generated a ΔNp63 conditional KO mouse (ΔNp63fn) using the cre-loxP system (SI Appendix, Fig. S1), allowing for tissue-specific deletion of the ΔNp63 isoforms and retention of the TAp63 isoforms. LoxP sites were inserted in to the p63 gene flanking exon 3′ (SI Appendix, Fig. S1 A and B). ΔNp63−/− mice were generated by intercrossing the ΔNp63 conditional KO mice (ΔNp63fn/fn) to FLPeR transgenic mice (23) to eliminate the intervening neo cassette (ΔNp63fl/fl) (SI Appendix, Fig. S1C). Resulting mice were intercrossed to female germ line–specific cre transgenic mice (Zp3-cre) (24) to generate ΔNp63+/− mice that were further intercrossed to generate ΔNp63−/− mice (SI Appendix, Fig. S1 B and C). We found that ΔNp63−/− mice are born at the proper Mendelian ratios but die within hours after birth similar to the p63−/− mice (13). Quantitative RT-PCR (qRT-PCR) performed on embryos at embryonic day (E)9.5 or on skin from embryos at E18.5 confirmed the absence of ΔNp63 mRNA (P < 0.0001; SI Appendix, Fig. S1D) and protein expression (SI Appendix, Fig. S1E) with maintenance of WT levels of TAp63 mRNA expression (SI Appendix, Fig. S1F).
Epidermis from ΔNp63−/− Displays Defects in Terminal Differentiation.
The phenotype of the ΔNp63−/− mice was reminiscent of the p63−/− mice (11, 13) (SI Appendix, Fig. S1 G–J). The ΔNp63−/− mice developed a fragile epidermis that easily detached from the dermis (SI Appendix, Fig. S1I). Microscopic analysis by H&E staining of E18.5 embryos (SI Appendix, Fig. S1 K–N) revealed the presence of rudimentary stratified epithelium with nests of basal epidermal cells that were apparent in patches over 20–30% of the embryo (SI Appendix, Fig. S1M). This phenotype is in contrast to the absence of a stratified epidermis in p63−/− embryos (SI Appendix, Fig. S1N). Interestingly, the ΔNp63+/− mice appeared to have excess folds of skin (SI Appendix, Fig. S1H). Analysis of H&E-stained cross sections of the epidermis of ΔNp63+/− mice revealed the presence of an expanded epidermal basal layer (SI Appendix, Fig. S1L).
Given that the ΔNp63+/− embryos had an expanded epidermis with basaloid cells above the basal epithelium and that the ΔNp63−/− embryos also developed a disorganized epidermis, we hypothesized that loss of one or both alleles of ΔNp63 leads to defects in epidermal differentiation. To test this hypothesis, we performed immunofluorescence (IF) for markers of epidermal differentiation assessing the expression of keratin 5 (K5) and keratin 14 (K14) in the basal layer, keratin 10 (K10) and keratin 1 (K1) in the spinous layer, and filaggrin (Fila) in the granular layer. All markers of epidermal differentiation were appropriately expressed in WT embryos (SI Appendix, Fig. S2 A, E, I, M, and Q). Interestingly, both ΔNp63+/− and ΔNp63−/− embryos expressed K5 and K14 in multiple epidermal layers (SI Appendix, Fig. S2 B, C, R, and S). The ΔNp63+/− embryos also expressed K10, K1, and Fila in multiple epidermal layers that were several cell layers thick (SI Appendix, Fig. S2 F, J, N, and R) in contrast to WT embryos, which expressed these markers in single or double cell layers (SI Appendix, Fig. S2 E, I, M, and Q). The ΔNp63−/− embryos also expressed K10 and Fila in a few scarce patches, but the expression was present only over 5–10% of the embryo (SI Appendix, Fig. S2 G, K, O, and S), suggesting a failure to terminally differentiate in the absence of ΔNp63. Given the apparent disorganization of the epidermis of ΔNp63+/− and ΔNp63−/− embryos, we asked whether cells in the epidermis expressed multiple differentiation markers simultaneously. We did this by performing double IF for K14 and K10. In WT embryos, K14 is expressed in the basal layer and K10 in the spinous layer of the epidermis (SI Appendix, Fig. S2Q). In both the ΔNp63+/− and ΔNp63−/− embryos, we detected overlapping expression of K14 and K10 in multiple layers of the epidermis (SI Appendix, Fig. S2 R and S), indicating that ΔNp63 is required for terminal epidermal differentiation. The epidermis of p63−/− embryos did not express any markers of terminal epidermal differentiation (SI Appendix, Fig. S2 D, H, L, P, T, X, and B′) as reported previously (13, 25).
Because of the appearance of basaloid cells in skin from ΔNp63+/− and ΔNp63−/− embryos, we asked whether skin from these mice expressed keratins of simple epithelia K8 and K18. Although we found a few positive cells in WT skin (SI Appendix, Fig. S2 U and Y), skin from ΔNp63+/− and ΔNp63−/− embryos had many areas of positive cells expressed in layers above the basal epithelium (SI Appendix, Fig. S2 V, W, Z, and A′).
ΔNp63-Deficient Epidermal Cells Are Hyperproliferative.
The presence of an expanded epidermis expressing markers associated with simple epithelia in ΔNp63 mutant mice suggested that cells within this tissue are hyperproliferative and may have the ability to self-renew. To ask whether there was a hyperproliferation of epidermal cells in the skin of ΔNp63+/− and ΔNp63−/− embryos, ΔNp63+/− and ΔNp63−/− embryos were labeled with bromodeoxyuridine (BrdU). By performing double immunofluorescence for BrdU and K5 (SI Appendix, Fig. S2 C′–E′) or BrdU and K10 (SI Appendix, Fig. S2 F′–H′), we found that the skin of ΔNp63+/− and ΔNp63−/− embryos have hyperproliferative and expanded basal and spinous layers as evidenced by the simultaneous expression of K5 and BrdU (SI Appendix, Fig. S2 D′, E′, and I′) and K10 and BrdU (SI Appendix, Fig. S2 G′, H′, and J′), respectively.
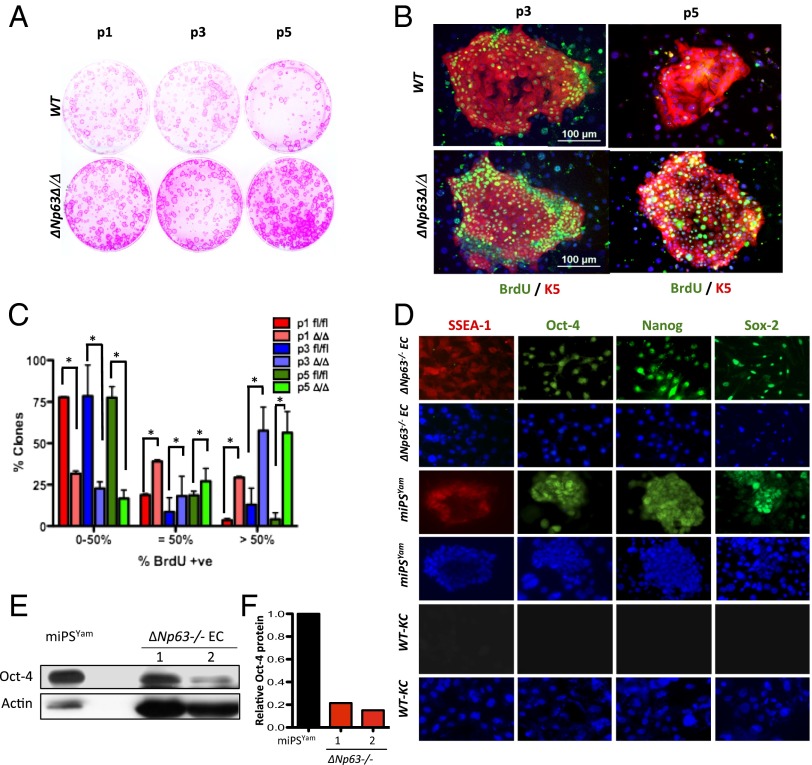
To ask whether epidermal cells deficient for ΔNp63 hyperproliferate, ΔNp63fl/fl (WT) and ΔNp63Δ/Δ (ΔNp63-deficient) epidermal cell lines were serially passaged under diluting conditions and stained with rhodamine B to score for the morphology of epidermal clones (Fig. 1A) (12, 14, 26). To quantify the proliferative capacity of these clones, we serially passaged WT and ΔNp63Δ/Δ cell lines (1,000 cells per passage) five times, followed by a pulse with BrdU after each passage. We then performed double IF using anti-BrdU and anti-K5 antibodies. We scored for BrdU positivity in each colony at each passage by counting the number of BrdU-positive cells within each of 50 colonies (SI Appendix, Table S1). Colonies of WT epidermal cell lines were initially proliferative (passage 1), but by passage 5, greater than 75% of these colonies incorporated very little BrdU (Fig. 1 B and C and SI Appendix, Table S1), indicating an inability to proliferate beyond passage 5. In contrast, colonies derived from ΔNp63-deficient epidermal cell lines incorporated high levels of BrdU even at passage 5 (Fig. 1 B and C and SI Appendix, Table S1), indicating the ability to hyperproliferate and self-renew.
Fig. 1.
ΔNp63-deficient epidermal cells are proliferative. (A) Epidermal colonies of the indicated genotypes stained with rhodamine B. Passages 1 (P1), 3 (P3), and 5 (P5) are shown. (B) Immunostaining for BrdU (green) and K5 (red) in P3 and P5 epidermal cells. (C) Quantification of BrdU incorporation in P1, P3, and P5 colonies after 8 d in culture. Asterisks indicate statistical significance (P < 0.001). (D) IF performed on the indicated cells using the indicated antibodies. (E) Western blot performed on lysates from miPSYam and ΔNp63−/− cells derived from two independent embryos (1 and 2) using the indicated antibodies. Actin was used as loading control. (F) Quantification of the Western blot in E.
ΔNp63-Deficient Epidermal Cell Lines Express High Levels of Some Factors Associated with Induced Pluripotency.
Based on the ability of ∆Np63-deficient epidermal cells to proliferate over several serial passages, we asked whether these cells express Oct4, Nanog, Sox2, and stage-specific embryonic antigen 1 (SSEA-1) by IF (Fig. 1D). Indeed, we found that ∆Np63−/− epidermal cell lines express Nanog, Sox2, and SSEA-1 at levels comparable to mouse induced pluripotent stem (iPS) cells generated by introduction of Yamanaka factors (miPSYam; Fig. 1D), whereas WT keratinocytes (WT-KCs) do not express these markers. We found that Oct4 expression in ∆Np63−/− epidermal cell lines is higher than WT-KCs but lower than miPSYam. Indeed, by Western blot analysis, we noted variability in the levels of Oct4 expression in the ∆Np63−/− epidermal cell lines (Fig. 1 E and F). These data indicate that ∆Np63−/− epidermal cell lines express factors associated with induced pluripotency and that different cell lines express these markers at different levels, which may be due to cell line to cell line variation or selection that occurs in cell culture. We therefore used the highest Oct4-expressing cell lines for our downstream analysis.
DGCR8 Is a Direct Transcriptional Target of ΔNp63.
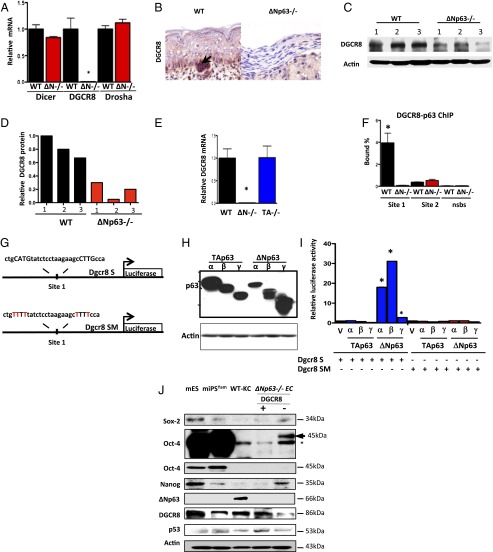
We showed previously that TAp63 transcriptionally regulates Dicer, a critical enzyme involved in miRNA biogenesis (20). We therefore asked whether ΔNp63 could be a transcriptional regulator of enzymes involved in miRNA biogenesis. To do this, we performed qRT-PCR for Dicer, DGCR8, and Drosha using total RNA derived from WT and ΔNp63−/− epidermal cell lines. Interestingly, DGCR8 mRNA expression was markedly decreased in the ΔNp63−/− and ΔNp63+/− epidermal cell lines (Fig. 2A and SI Appendix, Fig. S3 A and B). DGCR8 protein levels are also low in ΔNp63−/− epidermal cell lines and skin compared with their WT counterparts (Fig. 2 B–D). We found that, although the DGCR8 levels were reduced in ΔNp63−/− epidermal cell lines, there was variable expression of DGCR8 in different ∆Np63−/− epidermal cell lines (Fig. 2 C and D). We asked whether TAp63 may be similarly regulating DGCR8 and thus performed qRT-PCR for DGCR8 using total RNA isolated from TAp63−/− epidermal cell lines and compared it to the expression in ∆Np63−/− epidermal cell lines (Fig. 2E). We found that DGCR8 mRNA is expressed at WT levels in the absence of TAp63, indicating that ΔNp63 is a transcriptional regulator of DGCR8 and TAp63 is not.
Fig. 2.
DGCR8 is a transcriptional target of ΔNp63. (A) qRT-PCR for Dicer, DGCR8, and Drosha using total RNA from epidermal cell lines of the indicated genotypes. (B) IHC using an antibody for DGCR8 on skin samples from indicated mouse embryos. (C) Western blot analysis using lysates from WT and ΔNp63−/− epidermal cell lines derived from three independent embryos (1–3) using the indicated antibodies. Actin was used as loading control. (D) Quantification of the Western blot in C. All values were normalized to WT 1. (E) qRT-PCR for DGCR8 using total RNA from epidermal cell lines of the indicated genotypes. (F) Quantitative real-time PCR of DNA purified in ChIP assay using epidermal cell lines and p63-binding site (site 1), no binding of p63 to site 2, or nonspecific binding site (NSBS). (G) Schematic showing DGCR8 site 1 (Dgcr8 S) and DGCR8 mutant of site 1 (Dgcr8 SM) luciferase reporter genes. (H) Western blot analysis using lysates from p53−/−;p63−/− MEFs transfected with the indicated p63 isoforms. Actin was used as loading control. (I) Luciferase assay for DGCR8 in p53−/−;p63−/− MEFs transfected with the indicated plasmids. Each bar represents the average of the fold activation of three independent experiments. Values are normalized to p53−/−;p63−/− MEFs transfected with vector alone. The asterisks indicate statistical significance (P < 0.001). (J) Western blot analysis of mouse embryonic stem (mES) cells, mouse-induced pluripotent stem cells (miPSYam), WT-KCs, and ΔNp63−/− epidermal cell lines expressing DGCR8 (+) or not (−) using the indicated antibodies. Asterisk indicates nonspecific band. Upper Oct4 blot is a longer exposure of the one immediately below it. Actin was used as loading control.
To determine whether DGCR8 is a direct transcriptional target of ΔNp63, we performed ChIP analysis using p63 antibodies and primers specific for two putative p53/p63 binding sites that we identified within intron 1 of DGCR8 (SI Appendix, Table S2). We found that p63 robustly binds to one of these sites (site 1; Fig. 2 F and G), indicating that DGCR8 is a p63 target gene.
To determine whether p63 isoforms can transactivate a DGCR8-luciferase reporter gene, we cloned the site bound to p63 in the ChIP assay (site 1) into a vector containing the luciferase reporter gene (Fig. 2G). We cotransfected the DGCR8-luciferase reporter gene (Dgcr8 S-luc) (Fig. 2G) and each p63 isoform individually (TAp63α, β, γ, or ΔNp63α, β, γ) into p53−/−;p63−/− mouse embryonic fibroblasts (MEFs) (Fig. 2H). Only the ΔNp63 isoforms could transactivate the reporter gene with ΔNp63α and β exhibiting the highest transactivation activity (Fig. 2I). To ask whether the cloned p63 binding site is critical for transactivation of the DGCR8-luciferase reporter gene, we mutated the p63 consensus site from ctgCATGtat ctcctaaga agcCTTGcca to ctgTTTTtat ctcctaaga agcTTTTcca using site-directed mutagenesis, where the nucleotides in capital letters make up the core of the half site (Fig. 2G). None of the p63 isoforms transactivated the mutant DGCR8 reporter gene (Dgcr8 SM-luc; Fig. 2I), indicating that ΔNp63 transcriptionally activates DGCR8 by binding to site 1.
Oct4, Sox2, and Nanog Are Down-Regulated in ΔNp63−/− Epidermal Cell Lines Through Reexpression of DGCR8.
DGCR8 has been shown to be critical for repression of Oct4, Sox2, and Nanog expression (27), which are critical factors in the maintenance of stem cell pluripotency. To determine the expression levels of Oct4, Sox2, and Nanog in ΔNp63−/− epidermal cell lines expressing DGCR8, we performed immunoblotting using lysates from ΔNp63−/− epidermal cells (−DGCR8) and ΔNp63−/− epidermal cells expressing DGCR8 (+DGCR8) and compared them to WT-KCs, mES, and miPSYam cells (Fig. 2J). We found that ΔNp63−/− epidermal cell lines without DGCR8 expressed Sox2 and Nanog at levels higher than that of miPSYam cells and somewhat lower than levels expressed in mES cells (Fig. 2J). Reexpression of DGCR8 in ΔNp63−/− epidermal cell lines extinguished expression of Sox2 and Nanog (Fig. 2J). The pattern for Oct4 was similar in that it was expressed in ΔNp63−/− epidermal cell lines, albeit at lower levels than in mES and miPSYam cells, and expression was again extinguished in ΔNp63−/− epidermal cell lines with ectopic expression of DGCR8 (Fig. 2J). These data indicate that ΔNp63−/− epidermal cell lines express Sox2, Nanog, and Oct4 and that expression of these pluripotent markers is repressed after reexpression of DGCR8.
Because cells deficient for p53 have been shown to have an enhanced ability to be reprogrammed into iPS cells (2–6), we asked whether ΔNp63−/− epidermal cells express p53. We found that levels of p53 are comparable in ΔNp63−/− epidermal cells and in mES and miPS (Fig. 2J). We also assayed for expression of ΔNp63 in these cells and found that, although ΔNp63 is robustly expressed in WT-KCs, it is not expressed in mES or miPS cells (Fig. 2J), indicating that ΔNp63 is not expressed in stem cells.
Global Down-Regulation of miRNAs in ΔNp63−/− Epidermal Cell Lines.
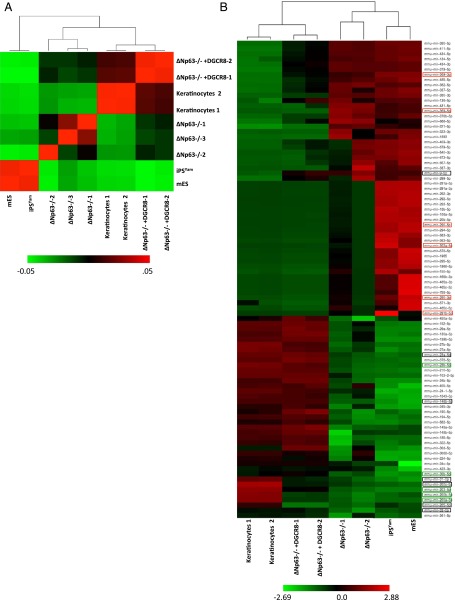
Because we found that ∆Np63 transcriptionally activates DGCR8, we determined the expression of miRNAs and mRNAs in the ∆Np63−/− epidermal cells using miRNA-Seq and RNA-Seq experiments. Importantly, there are critical microRNAs that have been shown to reprogram cells, such as miR-369 (28, 29). To determine the miRNA signature of the ∆Np63−/− epidermal cells, we performed miRNA-Seq experiments on RNA isolated from ∆Np63−/− epidermal cell lines and compared them to WT-KCs, iPSYam, and mouse ES cells (Fig. 3). Using Pearson’s correlation analysis, we found that the ∆Np63−/− epidermal cell lines clustered most closely with mouse iPSYam and ES cells (Fig. 3 A and B). We found a large number of microRNAs to be down-regulated in the ∆Np63−/− epidermal cell lines consistent with low levels of DGCR8 expression in these cells (Fig. 3B). We found a global down-regulation of miRNAs in the ∆Np63−/− epidermal cells including miR-200a, miR-200b, miR-200c, miR-141, miR-203, miR-205, miR-34a, and miR-34b (Fig. 3B). These miRNAs were also found to be down-regulated in iPSYam and ES cells. In addition, we found some miRNAs to be significantly up-regulated in ∆Np63−/− epidermal cell lines including miR-134, miR-9, and miR-369, which have been found to enhance reprogramming (28, 29) or are up-regulated in iPS cells reprogrammed with Yamanaka factors (30) (Fig. 3B).
Fig. 3.
miRNA signature of ΔNp63−/− epidermal cell lines. (A) Pearson’s correlation analysis from miRNA-Seq performed using the indicated samples. (B) Heat map showing supervised hierarchical clustering. Low miRNA expression is indicated in green and high expression in red. Boxes indicate miRNAs that were most significantly up- (red) or down- (green) regulated in the ∆Np63/iPS cell signature. The signature that was found to be most highly significant is boxed in black.
We next asked whether expression of DGCR8 could restore the miRNA signature of ∆Np63−/− epidermal cell lines to WT-KCs when cultured in keratinocyte media. Indeed, we found that ∆Np63−/− epidermal cell lines expressing DGCR8 had a very similar miRNA signature to WT-KCs (Fig. 3 A and B), indicating that critical miRNAs required for terminal differentiation of ∆Np63−/− epidermal cells into keratinocytes is controlled through DGCR8.
There were also a number of miRNAs that were differentially regulated in ∆Np63−/− epidermal cell lines that were distinct from the published ES cell and iPS cell miRNA profiles, indicating that ΔNp63−/− epidermal cells have a unique miRNA signature (Fig. 3B and SI Appendix, Fig. S4A). RNA-Seq using RNA isolated from WT-KCs, ∆Np63−/− epidermal cell lines, miPSYam, and mES cells revealed that the gene expression signature of the ∆Np63−/− epidermal cell lines partially resembled the miPSYam and mES cells (SI Appendix, Fig. S4B).
ΔNp63-Deficient Epidermal Cells Can Differentiate into Multiple Cell Fates in Vitro.
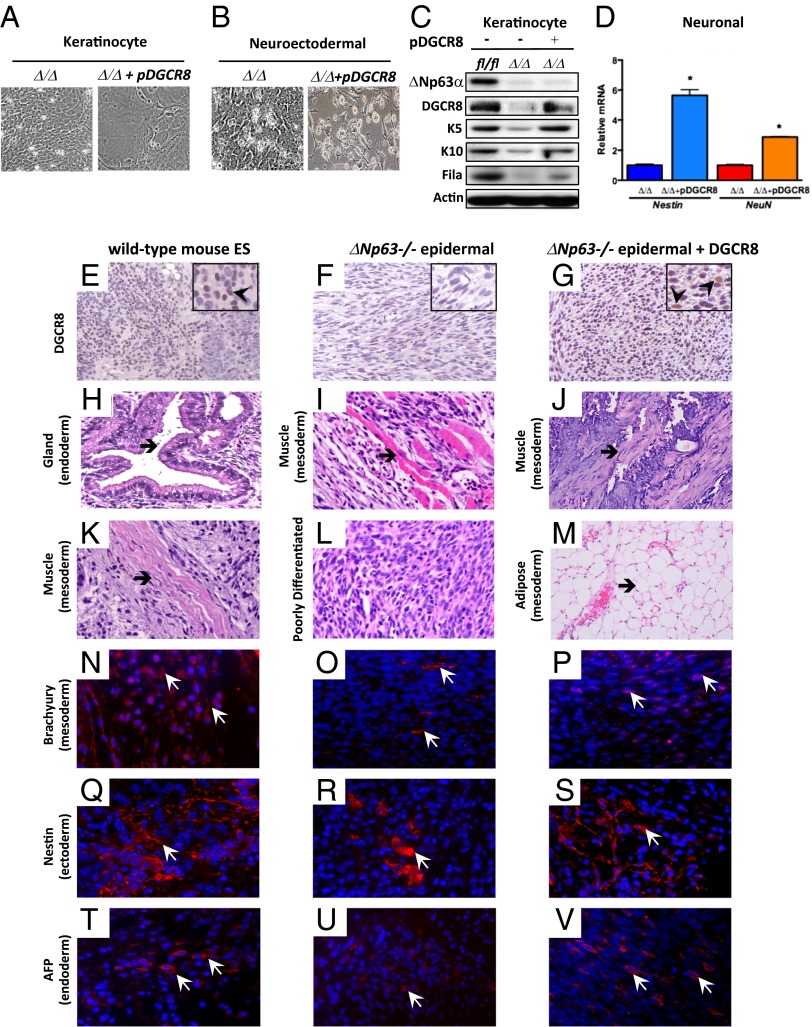
Because we have shown that activation of a miRNA signature involved in terminal differentiation in ΔNp63-deficient epidermal cells is dependent on expression of DGCR8, we cultured ∆Np63-deficient (ΔNp63Δ/Δ) epidermal cells without DGCR8 and with DGCR8 in keratinocyte media or neuroectodermal media (Fig. 4 A and B). Only ΔNp63-deficient epidermal cells expressing DGCR8 and cultured in keratinocyte or neuroectodermal media exhibited a terminally differentiated morphology (Fig. 4 A and B) and expressed markers of keratinocyte differentiation, similar to levels expressed in WT-KCs (Fig. 4C) or expressed markers of the neurons nestin and NeuN (Fig. 4D).
Fig. 4.
ΔNp63-deficient epidermal cell lines are multipotent. (A and B) ΔNp63-deficient epidermal cell lines (Δ/Δ) without or with DGCR8 (+pDGCR8) and cultured in keratinocyte media (A) or neuroectodermal media (B). (C) Western blot analysis of epidermal cell lines shown in A. (D) qRT-PCR for nestin and NeuN using total RNA from epidermal cell lines shown in B. (E–G) IHC analysis using teratomas of the indicated genotypes and an antibody for DGCR8 (200× magnification) and Insets (400× magnification). Arrowheads in Insets point to examples of positive cells. (H–M) H&E-stained cross sections of teratomas of the indicated genotypes. (N–V) IF of teratomas of the indicated genotypes. Arrowheads indicate examples of positive cells (red). Magnification, 200×.
ΔNp63-Deficient Epidermal Cells Expressing DGCR8 Can Differentiate into Multiple Cell Fates in Vivo.
To further determine whether ΔNp63−/− epidermal cell lines are pluripotent, we asked whether cells lacking ΔNp63 can form teratomas after injection into mice. We found that ΔNp63−/− epidermal cell lines alone can form poorly differentiated teratomas (Fig. 4 E and F). Further H&E analysis revealed that the teratomas generated with ∆Np63−/− epidermal cells have some structures like the muscle fibers shown in Fig. 4I; however, there were many areas consistent with poor terminal differentiation (Fig. 4L), suggesting that DGCR8 is required for ∆Np63−/− epidermal cells to fully terminally differentiate into multiple cell fates. To test this hypothesis, we transduced ΔNp63−/− epidermal cell lines with a tetracycline-inducible DGCR8 vector. These cells were injected into SCID mice, allowed to form teratomas for 4 wk, and fed doxycycline for 2 wk to induce DGCR8 expression. The ΔNp63−/− epidermal cell lines expressing DGCR8 formed well-differentiated teratomas (Fig. 4 G, J, and M) similar to what are observed using mouse ES cells (Fig. 4 E, H, and K). To better characterize these teratomas, we performed IF staining using markers for the mesoderm (brachyury), ectoderm (nestin), and endoderm (alpha-fetoprotein AFP). We found that teratomas generated using ΔNp63−/− epidermal cell lines contain a few cells that express the mesoderm marker, brachyury (Fig. 4O), the ectoderm marker, nestin (Fig. 4R), and the endoderm marker, AFP (Fig. 4U). On the other hand, teratomas generated using ∆Np63−/− epidermal cells expressing DGCR8 (Fig. 4G) form well-differentiated teratomas that contain many cells that express markers of the mesoderm (Fig. 4P), ectoderm (Fig. 4S), and endoderm (Fig. 4V), similar to the number of expressing cells seen in teratomas from WT mES cells (Fig. 4 N, Q, and T). These data indicate that ΔNp63−/− epidermal cells are multipotent stem cells that can be reprogrammed and terminally differentiated into multiple cell fates when DGCR8 is reexpressed.
ΔNp63-Deficient Epidermal Cells Contribute to Various Differentiated Tissues in Chimeric Mouse Embryos.
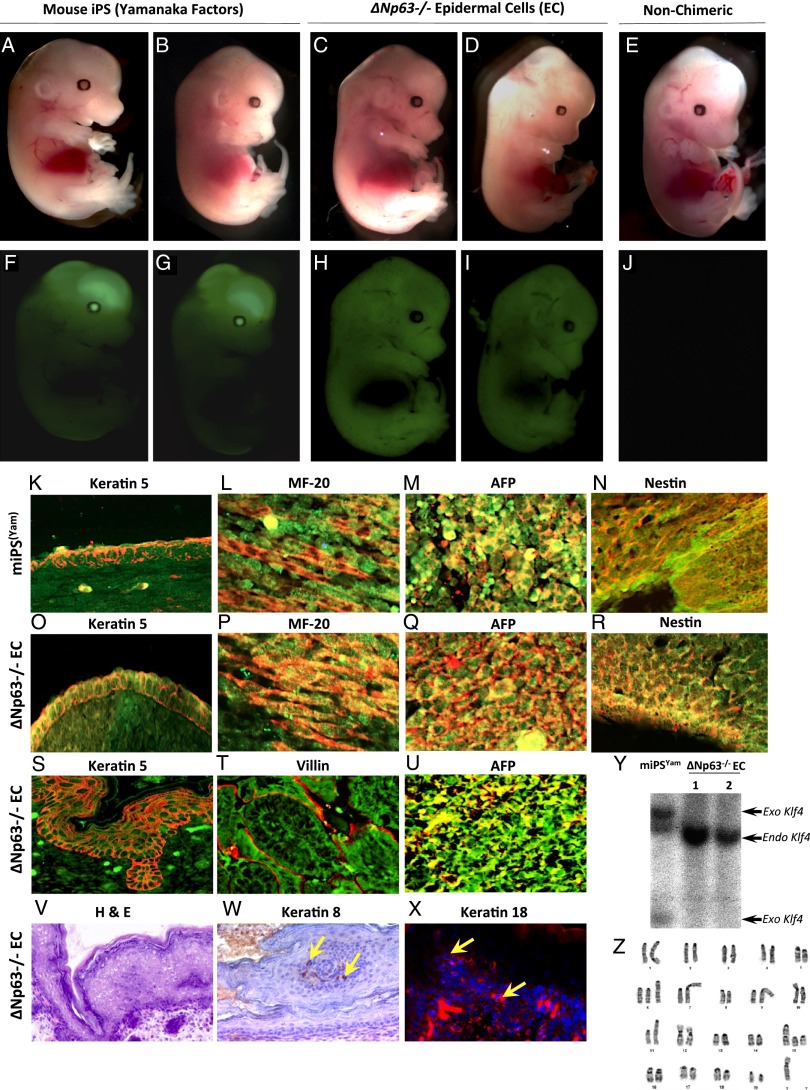
Because ∆Np63−/− epidermal cells can contribute to differentiated cells in teratomas, we next asked whether ΔNp63−/− epidermal cells could contribute to multiple differentiated tissues of the developing mouse. To do this, we performed blastocyst injections using iPSYam expressing GFP (iPS S3 cells) (31) and ΔNp63−/− epidermal cell lines expressing GFP (Fig. 5 A–J and SI Appendix, Tables S3 and S4). Indeed, we found that ΔNp63−/− epidermal cell lines expressing GFP form high contribution chimeric embryos at day E13.5 (Fig. 5 C, D, H, and I) comparable to what is seen with mouse iPS cells created using Yamanaka factors expressing GFP (Fig. 5 A, B, F, and G). We analyzed skin, muscle, liver, and brain from E13.5 chimeric embryos generated from iPSYam cells and ∆Np63−/− epidermal cell lines by co-IF for GFP expression (green) and the indicated markers of various differentiated tissues: K5 (red) for skin (Fig. 5 K and O), myosin II MF-20 (red) for muscle (Fig. 5 L and P), AFP (red) for liver (Fig. 5 M and Q), and nestin (red) for brain (Fig. 5 N and R). Both iPSYam cells and ∆Np63−/− epidermal cells contributed similarly to the tissues of these chimeric embryos (Fig. 5 K–R). We also analyzed E18.5 chimeric embryos generated from ∆Np63−/− epidermal cell lines marked by GFP (green) and found that these cells were also positive for keratin 5 (red) in the skin (Fig. 5S), villin (red) in the intestinal tract (Fig. 5T), and AFP (red) in the liver (Fig. 5U), again indicating that ∆Np63−/− epidermal cells contribute to multiple tissues. Within each embryo, we also detected areas where the tissues were GFP negative (SI Appendix, Fig. S5), and thus we found that the ∆Np63−/− epidermal cells contributed to 70–80% of the chimeric embryo. Although the ∆Np63−/− epidermal cell lines contributed to multiple tissues, we did detect abnormalities within the skin of E18.5 chimeric embryos. Specifically, these chimeras had a thickened epidermis (Fig. 5V) and inappropriate expression of K8 (Fig. 5W) and K18 (Fig. 5X) similar to ∆Np63+/− mice (SI Appendix, Figs. S1L and 2 V and Z). These abnormalities were further detected when we examined six litters of chimeric mice born from these cells at postnatal day 1 (P1) and found that they die shortly after birth due to skin defects (SI Appendix, Table S4) and are cannibalized by their mothers similar to the ΔNp63−/− mice. Given the remarkable contribution of ∆Np63−/− epidermal cells to many lineages in the embryos examined, we asked whether it was possible that the ∆Np63−/− epidermal cell lines had become contaminated with the mouse iPS cells made using Yamanaka factors (iPSYam). To address this, we performed Southern blot analysis for Klf4. We found that iPSYam cells expressed both endogenous and exogenous Klf4 DNA (Fig. 5Y), whereas ∆Np63−/− epidermal cell lines expressed endogenous Klf4 DNA only (Fig. 5Y), indicating that ∆Np63−/− epidermal cell lines are not contaminated with iPSYam cells. Some mouse and human iPSYam cells have been shown to have abnormal karyotype (2–9). Indeed, genomic instability and loss of cell cycle checkpoints have been shown to enhance reprogramming of iPS cells (2–9). To determine whether the ΔNp63−/− epidermal cell lines exhibit abnormalities, we performed karyotyping and found that these cells do exhibit chromosomal abnormalities, including duplications of chromosomes 6 and 15. These types of abnormalities are typical to cell culture models and are similar to those previously detected in other iPS cells (Fig. 5Z) (9).
Fig. 5.
Generation of chimeric mouse embryos from ΔNp63−/− epidermal cell lines. (A–E) Brightfield images of chimeric embryos at day 13.5 (E13.5) generated from mouse-induced pluripotent stem (miPS) cells expressing Yamanaka factors and GFP (Yamanaka factors) (A and B) or ΔNp63−/− epidermal cell lines expressing GFP (C and D). (E) Day E13.5 embryos from WT nonchimeric mice. (F–J) Fluorescent (GFP) images of chimeric embryos at E13.5 generated from Yamanaka miPS cells expressing GFP (Yamanaka factors) (F and G) or ΔNp63−/− epidermal cell lines expressing GFP (H and I). (J) Day E13.5 embryos from WT nonchimeric mice. (K–R) Double immunofluorescence for GFP (green) and the indicated tissue-specific markers for the basal cells of the epidermis (keratin5, red) (K and O), muscle (MF-20, red) (L and P), liver (AFP, red) (M and Q), and brain (nestin, red) (N and R) on day E13.5 embryos generated from Yamanaka miPS cells expressing GFP (Yamanaka factors) or ∆Np63−/− epidermal cell lines as indicated. (S–U) Double immunofluorescence of E18.5 chimeric embryos generated from ∆Np63−/− epidermal cell lines for GFP (green) and tissue-specific markers for the basal cells of the epidermis (keratin 5, red) (S), intestine (villin, red) (T), and liver (AFP, red) (U). DAPI (blue) or hematoxylin (purple) was used as counterstain. Magnification, 200×. (V–X) Cross sections from skin of E18.5 chimeric embryos generated from ∆Np63−/− epidermal cell lines and stained with H&E (V), keratin 8 (brown) (W), and keratin 18 (red) (X). Yellow arrows indicate examples of positive cells. (Y) Southern blot analysis using genomic DNA extracted from mouse iPS cells reprogrammed with Yamanaka factors (miPSYam) and ∆Np63−/− epidermal cell lines. A probe for Klf4 was used to indicate endogenous (endo) and exogenous (exo) Klf4 DNA. (Z) Representative image of karyotypic analysis of ∆Np63−/− epidermal cell lines.
Normal Human Epidermal Keratinocytes Depleted of ΔNp63 or DGCR8 Are Multipotent.
To determine whether knockdown of ΔNp63 or DGCR8 could induce pluripotency or multipotency in human epidermal keratinocytes, we knocked down ΔNp63 or DGCR8 with lentiviral shRNAs expressing GFP in NHEKs (SI Appendix, Fig. S6 A and B) and observed colonies with small tightly packed cells (cf. SI Appendix, Fig. S6 C–F). Six to 10 d subsequent to knock down of ΔNp63 or DGCR8 in NHEKs (SI Appendix, Fig. S6I), markers of pluripotency (Nanog, Tra-1–60, and Oct4) could be detected (Fig. 6 A–T). NHEKs transduced with Yamanaka factors (Oct4, Sox2, Klf4, and c-myc) expressed markers of pluripotency between days 7 and 10 (Fig. 6 K–O and SI Appendix, Fig. S6I). We next asked the efficiency of reprogramming of NHEKs with knock down of ΔNp63 or DGCR8 compared with NHEKs transduced with Oct4, Sox2, Klf4, and c-myc and found that NHEKs depleted of ΔNp63 or DGCR8 had a reprogramming efficiency of 0.07% and 0.1%, respectively, compared with 0.03% for NHEKs expressing Oct4, Sox2, Klf4, and c-myc (Fig. 6 A–J and SI Appendix, Table S5). These cells also efficiently formed embryoid bodies (SI Appendix, Fig. S6 G and H) comparable to those formed by human iPS cells expressing Yamanaka factors hiPSYam. Importantly, we also found that multiple NHEK clones stably transduced with sh∆Np63 or shDGCR8 retained silencing of ∆Np63 (SI Appendix, Fig. S6J) or DGCR8 (SI Appendix, Fig. S6K) and expression of Oct4, Sox2, and Nanog at equal or higher levels to human iPS (hiPS) cells (SI Appendix, Fig. S6 L–Q). NHEKs with knockdown of ΔNp63 (Fig. 6 A–E) or DGCR8 (Fig. 6 F–J) also expressed SSEA-4, Tra-1–60, Oct4, Sox2, and Nanog, similar to the expression in hiPSYam cells (Fig. 6 K–O), whereas NHEKs alone do not express these markers of pluripotency (Fig. 6 P–T). We next asked whether these cells could form teratomas in SCID mice (Fig. 6 U–T′). Indeed, we found that these cells form differentiated teratomas with structures representing the endoderm (Fig. 6 U and Y), mesoderm (Fig. 6 V, W, Z, and A′), and ectoderm (Fig. 6 X and B′). Additionally, we performed double IF using antibodies for GFP to mark human vs. mouse cells and antibodies for markers of the endoderm (AFP), mesoderm (MF-20), and ectoderm (K5). This analysis revealed that the human NHEK-sh∆Np63 and NHEK-shDGCR8, which express GFP, also had robust expression of AFP (Fig. 6 C′–H′), MF 20 (Fig. 6 I′–N′), and keratin 5 (Fig. 6 O′–T′). These data indicate that the NHEK-sh∆Np63 and NHEK-shDGCR8 cells contribute to differentiated cells within the teratomas. Taken together, these data indicate that human keratinocytes can be reprogrammed into multiple cell fates by knock down of ΔNp63 or DGCR8 in a rapid (6 d) and efficient manner.
Fig. 6.
NHEKs deficient for ∆Np63 or DGCR8 are pluripotent and form teratomas. (A–T) IF using normal human epidermal keratinocytes of the indicated genotypes and antibodies. (U–B′) Micrographs of H&E-stained cross sections. (C′–T′) Double IF performed on teratomas generated using an antibody against the indicated tissue-specific proteins (red) (C′, F′, I′, L′, O′, and R′) and GFP (green) (D′, G′, J′, M′, P′, and S′). Merged panels are shown for each (E′, H′, K′, N′, Q′, and T′). Magnification, 200×.
ΔNp63-Deficient Mouse and Human Epidermal Cells Have a Unique miRNA-mRNA Signature That Partially Overlaps with Human iPS Cells Generated Using Yamanaka Factors.
We performed miRNA-Seq and RNA-Seq using RNA isolated from NHEKs with knockdown of ΔNp63 or DGCR8 and compared them to mouse and human iPS cells generated using Yamanaka factors. Importantly, we found using Pearson’s correlation analysis and principal component analysis that mouse and human epidermal cells deficient of ∆Np63 or DGCR8 clustered most closely with mouse iPSYam, mouse ES, and human iPSYam cells (SI Appendix, Fig. S7 A and B). Moreover, we found that the seven most significantly differentially expressed miRNAs (miR-9, miR-146b, miR-30c, let-7e, miR-23a, miR141, miR-31, and miR-205) in mouse ∆Np63−/− epidermal cells were also differentially expressed in human cells (NHEKs) with knockdown of ∆Np63 or DGCR8 (SI Appendix, Fig. S7C). Both mouse and human cells deficient for ∆Np63 or DGCR8 expressed the same miRNA signature and clustered closely with mouse and human iPS cells (SI Appendix, Fig. S6 A–C). This miRNA signature indicates that these miRNAs are conserved between human and mouse cells reprogrammed through the ∆Np63/DGCR8 pathway.
Last, we performed RNA-Seq analysis and found that the gene expression signature of these human cells clustered most closely with ΔNp63−/− epidermal cells, miPSYam, and mES cells derived from the mouse using Pearson’s correlation analysis (SI Appendix, Fig. S7D), principal component analysis (SI Appendix, Fig. S7E), and supervised hierarchical clustering (SI Appendix, Fig. S8).
In summary, ΔNp63 is required for transcriptional activation of DGCR8 in epidermal cells leading to terminal differentiation of tissues like the epidermis. We show here that loss of ∆Np63 leads to cells that have self-renewing but limited terminal differentiation capacity. When DGCR8 is reexpressed in cells deficient for ∆Np63, these cells can terminally differentiate into all three germ layers. We dubbed these cells iMS cells because of their remarkable plasticity and ability to differentiate into multiple cell lineages. Based on our results using human keratinocytes, we predict that epidermal cells can be extracted from patient skin biopsies and reprogrammed into multipotent stem cells by knockdown of ΔNp63 or DGCR8. Indeed, inducible knockdown of ΔNp63 and/or DGCR8 may be preferable to the overexpression of potentially oncogenic factors such as c-myc or the down-regulation of the critical tumor suppressor gene, p53. In the future, understanding the mechanisms used by the individual p63 isoforms in maintaining progenitor and stem cells in various tissues is key to understanding its complex roles in stem cell maintenance, as well as cancer development and metastasis.
Methods
Teratoma Formation Assay.
∆Np63−/− epidermal cells were infected with the tet-inducible DGCR8 vector and the pTet-On Advanced vector (Clontech) as described previously (20). SCID mice were injected s.c. in the dorsal flank with WT mouse ES cells or ΔNp63−/− epidermal cells transduced with the tet-inducible DGCR8 vector as described previously (1). Mice were administered 2 mg/mL doxycycline in the drinking water to induce expression of DGCR8 3 wk postinjection after palpable tumors had formed. Another group of mice was administered water without doxycycline as controls for the same amount of time. Tumors were harvested 6 wk after injection and fixed in 10% (vol/vol) formalin. Paraffin-embedded cross sections were analyzed by hematoxylin. Teratomas from NHEKs transduced with shΔNp63 cells were performed by Applied StemCell. NHEK-shΔNp63 and NHEK-shDGCR8 cells were grown on feeders in ES cells media; 1 × 106 cells were injected into the testes or the kidney capsule of SCID mice.
Generation and Analysis of Chimeric Mice.
ΔNp63−/− cells (12–18 cells) expressing pLenti-GFP (Vector Development Laboratory, Baylor College of Medicine) or iPSYam (S3) cells (12–18 cells) expressing eGFP (31) were injected into albino B6 blastocysts and implanted in CD-1 pseudopregnant mice. Embryos at E18.5 days were analyzed for GFP expression using a Zeiss SteREO Lumar, V12 microscope, with fluorescent and bright field capability. Nonchimeric E18.5 embryos were used as negative controls. The embryos were fixed in 10% (vol/vol) formalin and embedded in paraffin, and IF was performed on cross sections using an anti-GFP antibody.
Supplementary Material
Acknowledgments
We thank A. Jain, M. Barton, B. Davis, J. Deng, R. Behringer, L. Fang, and K. Bissig for technical advice. We acknowledge the work of the Genetically Engineered Mouse Facility at The University of Texas M. D. Anderson Cancer Center (funded by National Cancer Institute Grant CA16672). This work was supported by the American Cancer Society Grant RSG-07-082-01-MGO, National Cancer Institute Grant R01CA134796, Cancer Prevention Research Institute of Texas Grant RP120124, the Hildegardo E. and Olga M. Flores Foundation, and a Career Development Award from the Genitourinary Cancer Specialized Programs of Research Excellence (National Cancer Institute Grant CA091846) (to E.R.F.). E.R.F. is a scholar of the Leukemia and Lymphoma Society of America. D.C. was funded by CPRIT Training Grant RP101502.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319743111/-/DCSupplemental.
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Hong H, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460(7259):1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawamura T, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460(7259):1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460(7259):1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marión RM, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460(7259):1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Utikal J, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460(7259):1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krizhanovsky V, Lowe SW. Stem cells: The promises and perils of p53. Nature. 2009;460(7259):1085–1086. doi: 10.1038/4601085a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, et al. The p53-PUMA axis suppresses iPSC generation. Nat Commun. 2013;4:2174. doi: 10.1038/ncomms3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayshar Y, et al. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7(4):521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 10.Koster MI, Kim S, Mills AA, DeMayo FJ, Roop DR. p63 is the molecular switch for initiation of an epithelial stratification program. Genes Dev. 2004;18(2):126–131. doi: 10.1101/gad.1165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mills AA, et al. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398(6729):708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 12.Senoo M, Pinto F, Crum CP, McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129(3):523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 13.Yang A, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398(6729):714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 14.Su X, et al. TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell. 2009;5(1):64–75. doi: 10.1016/j.stem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barbieri CE, Tang LJ, Brown KA, Pietenpol JA. Loss of p63 leads to increased cell migration and up-regulation of genes involved in invasion and metastasis. Cancer Res. 2006;66(15):7589–7597. doi: 10.1158/0008-5472.CAN-06-2020. [DOI] [PubMed] [Google Scholar]
- 16.De Rosa L, et al. p63 Suppresses non-epidermal lineage markers in a bone morphogenetic protein-dependent manner via repression of Smad7. J Biol Chem. 2009;284(44):30574–30582. doi: 10.1074/jbc.M109.049619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shalom-Feuerstein R, et al. ΔNp63 is an ectodermal gatekeeper of epidermal morphogenesis. Cell Death Differ. 2011;18(5):887–896. doi: 10.1038/cdd.2010.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koster MI, et al. p63 induces key target genes required for epidermal morphogenesis. Proc Natl Acad Sci USA. 2007;104(9):3255–3260. doi: 10.1073/pnas.0611376104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romano RA, et al. ΔNp63 knockout mice reveal its indispensable role as a master regulator of epithelial development and differentiation. Development. 2012;139(4):772–782. doi: 10.1242/dev.071191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su X, et al. TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature. 2010;467(7318):986–990. doi: 10.1038/nature09459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su X, et al. TAp63 is a master transcriptional regulator of lipid and glucose metabolism. Cell Metab. 2012;16(4):511–525. doi: 10.1016/j.cmet.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho MS, Chan IL, Flores ER. ΔNp63 transcriptionally regulates brachyury, a gene with diverse roles in limb development, tumorigenesis and metastasis. Cell Cycle. 2010;9(12):2434–2441. doi: 10.4161/cc.9.12.12051. [DOI] [PubMed] [Google Scholar]
- 23.Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28(3-4):106–110. [PubMed] [Google Scholar]
- 24.Lewandoski M, Wassarman KM, Martin GR. Zp3-cre, a transgenic mouse line for the activation or inactivation of loxP-flanked target genes specifically in the female germ line. Curr Biol. 1997;7(2):148–151. doi: 10.1016/s0960-9822(06)00059-5. [DOI] [PubMed] [Google Scholar]
- 25.Su X, et al. Rescue of key features of the p63-null epithelial phenotype by inactivation of Ink4a and Arf. EMBO J. 2009;28(13):1904–1915. doi: 10.1038/emboj.2009.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrandon Y, Green H. Three clonal types of keratinocyte with different capacities for multiplication. Proc Natl Acad Sci USA. 1987;84(8):2302–2306. doi: 10.1073/pnas.84.8.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39(3):380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heinrich EM, Dimmeler S. MicroRNAs and stem cells: Control of pluripotency, reprogramming, and lineage commitment. Circ Res. 2012;110(7):1014–1022. doi: 10.1161/CIRCRESAHA.111.243394. [DOI] [PubMed] [Google Scholar]
- 29.Miyoshi N, et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8(6):633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 30.Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27(5):459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng JM, et al. Generation of viable male and female mice from two fathers. Biol Reprod. 2011;84(3):613–618. doi: 10.1095/biolreprod.110.088831. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.