Significance
Vitamin C (also known as ascorbate), an essential nutrient for humans, plays an important role in protection against oxidative stress. The ascorbate-dependent oxidoreductase cytochrome b561 (Cyt b561) is a family of highly conserved, multipass transmembrane enzymes found only in eukaryotes. Cyt b561 plays a key role in ascorbate recycling and many other important physiological processes, such as iron absorption. The atomic structure and functional mechanism of Cyt b561 remain unknown. In this study, we report the high-resolution crystal structures of Cyt b561 in both ascorbate-free and ascorbate-bound states. Our structural and biochemical analyses identify a general functional mechanism for the Cyt b561 family.
Abstract
Vitamin C, also known as ascorbate, is required in numerous essential metabolic reactions in eukaryotes. The eukaryotic ascorbate-dependent oxidoreductase cytochrome b561 (Cyt b561), a family of highly conserved transmembrane enzymes, plays an important role in ascorbate recycling and iron absorption. Although Cyt b561 was identified four decades ago, its atomic structure and functional mechanism remain largely unknown. Here, we report the high-resolution crystal structures of cytochrome b561 from Arabidopsis thaliana in both substrate-free and substrate-bound states. Cyt b561 forms a homodimer, with each protomer consisting of six transmembrane helices and two heme groups. The negatively charged substrate ascorbate, or monodehydroascorbate, is enclosed in a positively charged pocket on either side of the membrane. Two highly conserved amino acids, Lys81 and His106, play an essential role in substrate recognition and catalysis. Our structural and biochemical analyses allow the proposition of a general electron transfer mechanism for members of the Cyt b561 family.
Vitamin C, an essential nutrient for humans (1), is important for the synthesis of collagen (2), carnitine (3), and the neurotransmitter norepinephrine (4). Vitamin C also plays an important role in protection against oxidative stress (5). Oxidation of ascorbate results in sequential production of monodehydroascorbate and dehydroascorbate through loss of one and two electrons, respectively (6). Ascorbate serves as an electron donor for various enzymes, such as prolyl and lysyl hydroxylase, dopamine β-hydroxylase, ascorbate peroxidase, and cytochrome b561 (Cyt b561). Cyt b561, initially identified in the chromaffin granules of bovine adrenal medullae about 40 y ago (7, 8), is a transmembrane ascorbate-dependent oxidoreductase (9–13) that plays a key role in ascorbate recycling and other physiological processes, such as iron absorption (14). To our knowledge, Cyt b561 is the only membrane-embedded oxidoreductase that relies on ascorbate as the electron donor.
Homologs of Cyt b561 are found only in eukaryotes (15). Mammalian chromaffin granule Cyt b561 (CGCyt b561), mammalian duodenal Cyt b561 (DCyt b561), and Zea mays Cyt b561 (ZmCyt b561) have been extensively investigated. CGCyt b561 resides in the chromaffin vesicle membrane and transfers electrons from cytoplasmic ascorbate to the intravesicular monodehydroascorbate radical for the regeneration of ascorbate (9, 13, 16), which is a substrate of dopamine β-hydroxylase for the synthesis of neurotransmitter norepinephrine. DCyt b561 is present in the duodenal mucosa cell membrane, where it relays electrons from cytoplasmic ascorbate to ferric-chelate in the intestinal lumen, yielding soluble ferrous ion for absorption via a Fe2+-transporter (17–20). Expression levels of DCyt b561 in the duodenal mucosa cell membrane are closely associated with iron metabolism disorders, such as chronic anemia and iron-deficiency anemia (19). Similar to CGCyt b561, ZmCyt b561 uses ascorbate and the monodehydroascorbate radical as the physiological electron donor and acceptor, respectively (11, 21, 22).
Despite rigorous investigation, there is no detailed structural information for any member of the Cyt b561 family. Consequently, the electron transfer path is yet to be identified and the molecular mechanisms of substrate recognition and catalysis remain largely mysterious. These aspects are crucial for understanding the functional mechanism of the Cyt b561 protein family. In this study, we answer these questions by elucidating the crystal structures of Cyt b561 in both ascorbate-free and -bound forms and by performing systematic, structure-guided biochemical analyses.
Results
Members of the Cyt b561 family share strong sequence homology and are functionally conserved, and structural information on any Cyt b561 enzyme is expected to reveal conserved structural features and functional insights for the entire family. To obtain Cyt b561 with a high expression level, we cloned and screened a number of orthologs from humans and plants. These proteins were also examined for solution behavior and crystallization. Based on preliminary analyses, we focused on the recombinant full-length WT Cyt b561-B protein (residues 1–230) from Arabidopsis thaliana, which gave rise to weak-diffracting crystals. Treatment with ferricyanide during protein purification, which presumably led to oxidation of the heme groups in Cyt b561, and hence more homogeneous protein, led to markedly improved diffraction for these crystals. The structure was determined by ferric-based, single-wavelength anomalous dispersion, and the atomic model was refined at a resolution of 1.7 Å (Table S1). The sulfur anomalous signals of internal methionine and cysteine residues were clearly detected (Fig. S1A).
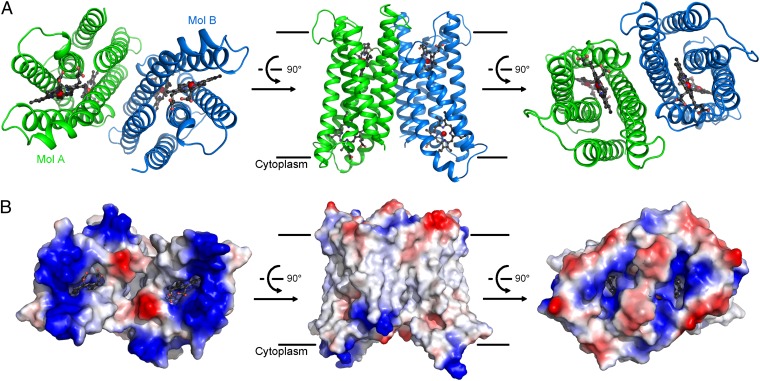
Each asymmetrical unit contains two molecules of Cyt b561, arranged as a pseudosymmetrical dimer through a twofold axis perpendicular to the plane of the lipid membrane (Fig. 1A). The two Cyt b561 protomers, named Mol A and Mol B, exhibit nearly identical structural features, with a pairwise rmsd of 0.49 Å over 210 aligned Cα atoms. Each Cyt b561 protomer comprises six transmembrane segments (TMs) (Fig. 1A), with both the N and C termini located in the cytoplasm based on prior knowledge (23, 24). Each protomer contains two heme groups, which are partially exposed to solvent (Fig. 1B). Notably, two cavities are present on each side of the membrane; each cavity is surrounded by positive charges, with a heme group located underneath. These analyses strongly suggest that these positively charged cavities may be the substrate-binding sites. The potential substrate-binding site on the cytoplasmic side of Mol A is occupied by a sulfate ion, which mediates crystal packing interactions (Fig. S1 B and C).
Fig. 1.
Overall structure of Cyt b561. (A) Overall structure of WT, full-length Cyt b561. The structure of Cyt b561 is shown in three successive views. There are two molecules of Cyt b561 in each asymmetrical unit, named Mol A (green) and Mol B (blue). (B) Surface features of the Cyt b561 homodimer by electrostatic potential. The three views shown correspond to those in A. Two cavities on either side are surrounded by positively charged amino acids. All structural figures were prepared using PyMOL Molecular Graphics System, Version 1.5 (Schrödinger, LLC).
The two Cyt b561 protomers associate with each other through a hydrophobic interface, resulting in a buried surface area of ∼1,410 Å2 (Fig. S2A). To examine the notion that Cyt b561 may exist as a homodimer, we sought to engineer a dimerization-dependent disulfide bond in Cyt b561. The Cα-Cα distance between Tyr115 in Mol A and Arg191 on Mol B is ∼5.8 Å (Fig. S2B, Left), which is ideally suited for the formation of a disulfide bond if these two residues are mutated to cysteine. The WT Cyt b561 protein appears mostly as a monomer on the denaturing SDS/PAGE; by contrast, a major proportion of the mutant Y115C/R191C protein formed a cross-linked homodimer even in the absence of the oxidation catalyst o-phenanthroline copper complex (25, 26) (Fig. S2B, Right, lanes 1 and 5). Incubation with the oxidation catalyst led to more complete formation of the cross-linked homodimer for Y115C/R191C but had little impact on WT Cyt b561 (Fig. S2B, Right, lanes 3 and 7). The presence of the reducing agent DTT reduced the proportion of cross-linked homodimer for Y115C/R191C (Fig. S2B, Right, lanes 6 and 8) but had no impact on WT Cyt b561 (Fig. S2B, Right, lanes 2 and 4). Further supporting this analysis, the disulfide-bonded Y115C/R191C was eluted from gel filtration with approximately the same elution volume as that for WT Cyt b561 (Fig. S2C). Taken together, our biochemical characterization demonstrates that the Cyt b561 protein exists as a homodimer.
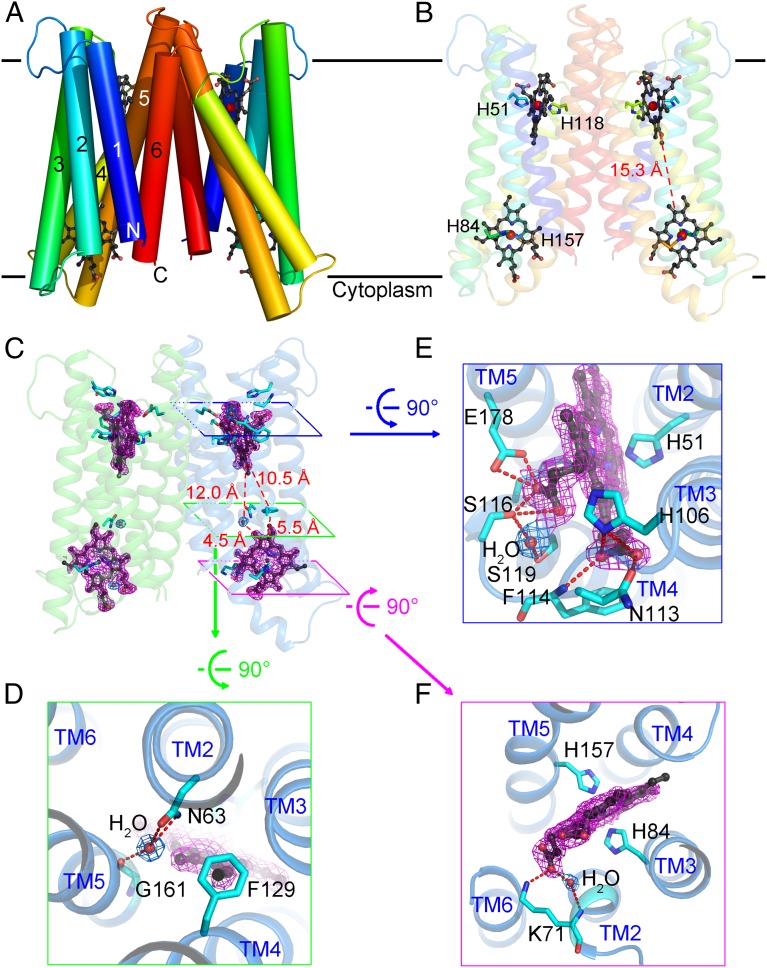
There are four heme groups in the Cyt b561 homodimer (Fig. 2A). Each heme is sandwiched by the same set of four transmembrane helices, TMs 2/3/4/5, and bound by two invariant His residues. The heme group on the cytoplasmic side is recognized by His84 from TM3 and His157 from TM5, whereas the heme on the noncytoplasmic side is coordinated by His51 from TM2 and His118 from TM4 (Fig. 2B and Fig. S3). The closest edge-to-edge distance between the two heme groups within the same Cyt b561 protomer is about 15.3 Å (Fig. 2B), out of the range (14 Å or less) for robust through-space electron transfer (27, 28). Intriguingly, a water molecule is located between the two heme centers, 4.5 Å from the cytoplasmic heme and 12 Å from the noncytoplasmic heme (Fig. 2C). This water molecule is hydrogen-bonded to the side chain of Asn63 from TM2 and the carbonyl oxygen atom of Gly161 from TM5 (Fig. 2D). In addition, the side chain of the conserved Phe129 is located between the two heme centers, 5.5 Å from the cytoplasmic heme and 10.5 Å from the noncytoplasmic heme (Fig. 2 C and D). All these distances are suitable for efficient electron transfer, suggesting potential roles for the water molecule and/or Phe129 in bridging a tunneling electron.
Fig. 2.
Heme-binding sites of Cyt b561. (A) Ribbon representation of the Cyt b561 homodimer. Cyt b561 is rainbow-colored, with its N terminus in blue and its C terminus in red. (B) Heme arrangement in Cyt b561. The heme groups are in a ball-and-stick configuration, with the iron atoms as red spheres. The four histidines that coordinate heme iron are shown. (C) Location of a water molecule and Phe129 between two heme groups in each protomer. The electron density maps, contoured at 1.0σ, are colored blue for waters and magenta for the heme groups. (D) Close-up view of the intervening water molecule and Phe129 between the two heme groups in each protomer of Cyt b561. H-bonds are indicated by red dashed lines. (E) Close-up view of the heme group on the noncytoplasmic side. (F) Close-up view of the heme group on the cytoplasmic side.
On the noncytoplasmic side, the two propanoate groups of the heme center mediate a number of hydrogen bonds (H-bonds) with surrounding amino acids (Fig. 2E). The A-propanoate interacts with the side chains of Ser116 and Glu178, whereas the D-propanoate is hydrogen-bonded to the amide nitrogen of Phe114 and the side chains of His106 and Asn113. On the cytoplasmic side, the A-propanoate of the heme center forms two H-bonds, one with the side chain of Lys71 in TM2 and the other through a water molecule to the amide nitrogen of Lys71, whereas the D-propanoate of the heme center is exposed to the solvent (Fig. 2F). The amino acids that use their side chains to form H-bonds with the heme groups are highly conserved among Cyt b561 family members (Fig. S3).
The crystal structure of Cyt b561 is related to that of the heme-containing protein cytochrome b6, a subunit of the cytochrome b6f complex (29, 30). In both cases, the central four TMs have similar conformations (Fig. S4). The four TMs of cytochrome b6 can be superimposed to those in Cyt b561, with an rmsd of 2.77 Å over 118 aligned Cα atoms. Despite this similarity, coordination of the heme group is different between these two proteins. In the case of Cyt b561, heme groups are bound by residues from all four TMs; in cytochrome b6, however, only residues from TM helices B and D participate in heme binding. In addition, Cyt b561 has unique defining features. Most importantly, the electron donor/acceptor pair for Cyt b561 is ascorbate/monodehydroascorbate or ascorbate/ferric-chelate, whereas the electron donor/acceptor pair for cytochrome b6f is hydroquinone/plastocyanin. The closest edge-to-edge distance between the two heme centers of Cyt b561 is considerably longer than that in cytochrome b6. Moreover, Cyt b561 contains two additional TMs and has a dimer arrangement different from that of cytochrome b6.
The primary sequences of A. thaliana Cytb561-B share 34% identity and 54% similarity with those of human CGCytb561 and 36% identity and 52% similarity with those of human DCytb561 (Fig. S3). This suggests that Cytb561-B may function similarly as human CGCytb561 or DCytb561 to participate in ascorbate recycling or ascorbate-dependent ferric-chelate reduction. To elucidate the mechanism of substrate recognition, we sought to determine the crystal structure of Cyt b561 bound to l-ascorbate by soaking Cyt b561 crystals in 1 M l-ascorbate for 10 min. The resulting crystals were used for X-ray data collection and structure determination at a resolution of 2.0 Å (Table S1).
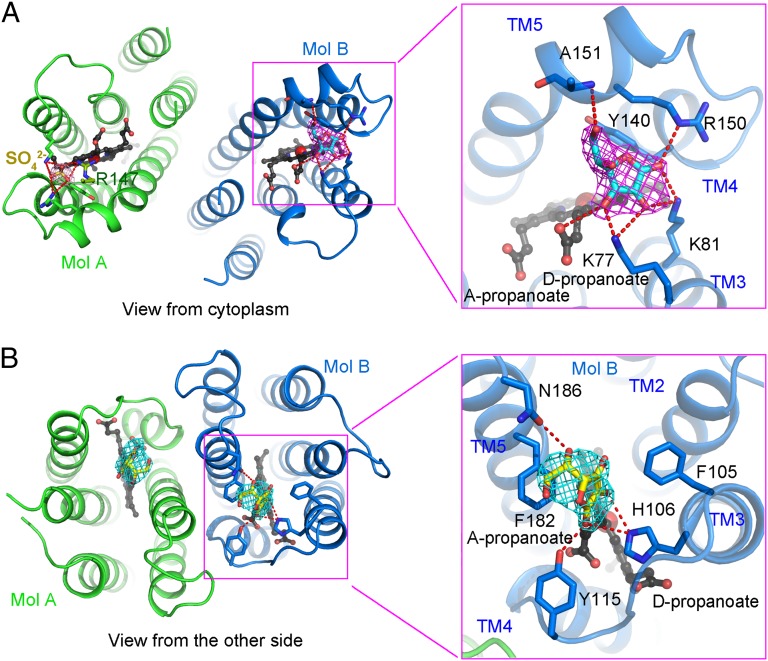
The overall structure of Cyt b561 is unaffected by ascorbate binding (Fig. S5A). After all protein atoms were modeled, the electron density allowed assignment for l-ascorbate in Mol B on the cytoplasmic side (Fig. 3A), but not on the noncytoplasmic side, under this condition. The same position in Mol A is occupied by a sulfate anion, which facilitated crystal packing. Ascorbate is caged in a positively charged pocket located in close proximity to the heme group on the cytoplasmic side (Fig. 3A and Fig. S5B). This binding mode is similar to that of the ascorbate–ascorbate peroxidase complex structure (31). The ketone group (position 1) (Fig. S5C) of ascorbate accepts two H-bonds from the side chains of Lys81 and Arg150, whereas the hydroxyl group at position 2 of ascorbate makes two H-bonds to the side chains of Lys77 and Lys81. The hydroxyl group at position 3 interacts with the side chain of Lys77 and the d-propanoate of the heme group. In addition, the 6-OH group of ascorbate mediates one H-bond with the main chain amide group of Ala151. The five-membered ring of ascorbate interacts with the heme group and Tyr140 from TM4 via van der Waals contacts. All residues that interact with ascorbate are highly conserved in Cyt b561 from plants to humans (Fig. S3).
Fig. 3.
Recognition of ascorbate by conserved amino acids. (A) Recognition of ascorbate on the cytoplasmic side. Ascorbate is bound on the cytoplasmic side of Mol B, coordinated by the heme group and residues from TM3, TM4, and TM5. The same position of Mol A is occupied by a sulfate ion and the side chain of Arg147 from a neighboring molecule, Mol A′. The Fobs − Fcalc electron density for ascorbate (magenta) is contoured at 2.0σ. (B) Recognition of monodehydroascorbate on the noncytoplasmic side. Monodehydroascorbate is bound on the noncytoplasmic side of the Cyt b561 dimer, coordinated by residues from TM3, Loop3, and TM5. The Fobs − Fcalc electron density for ascorbate (cyan) is contoured at 2.5σ.
To examine substrate binding on the noncytoplasmic side, we screened different substrate soaking times. Fortunately, upon soaking crystals in 1 M ascorbate for 40 min, we were able to determine the structure at a resolution of 2.2 Å (Table S1). Again, prolonged soaking had little impact on the overall structure (Fig. S5D) but produced two lobes of electron density in the positively charged pockets of the Cyt b561 dimer on the noncytoplasmic side (Fig. 3B and Fig. S5E). Because the monodehydroascorbate radical, the physiological substrate for human CGCyt b561 on the noncytoplasmic side (9, 10), is structurally indistinguishable from ascorbate at this resolution, we modeled two molecules of monodehydroascorbate, instead of ascorbate, to fit the density. Monodehydroascorbates are positioned above the heme groups on the noncytoplasmic side, each surrounded by two polar residues and three aromatic amino acids (Fig. 3B). The side chains of His106, Tyr115, and Asn186 mediate four potential H-bonds to monodehydroascorbate. In addition, monodehydroascorbate is stacked by the benzene rings of Phe105 and Phe182. Except for Asn186, all residues that interact with monodehydroascorbate are highly conserved among members of the Cyt b561 family. Notably, these substrate-binding residues observed in our crystal structures are different from the predicted substrate-binding motifs ALLVYR (residues 66–71) and SLHSW (residues 116–120) (14, 19, 32) (Fig. S3).
Cyt b561 is an oxidoreductase, transferring electrons from ascorbate to monodehydroascorbate or ferric-chelate. As previously reported (11, 33), ferricyanide-oxidized Cyt b561 shows a characteristic peak at 416 nm in its visible absorption spectra and, upon reduction by ascorbate, displays three unique peaks at 428, 531, and ∼561 nm (Figs. S6, Upper Left and S7A, Left). The oxidized Cyt b561 can be reduced by ascorbate but not by other electron donors, such as vitamin E, reduced glutathione, NADH, or NADPH (Fig. S6). Next, using a stopped-flow apparatus, we investigated the kinetics of ascorbate-mediated reduction of Cyt b561 by monitoring the absorbance change at 430 nm (Fig. S7A, Right). The result shows rapid reduction of Cyt b561 within 15 s.
The route of electron transfer might begin with ascorbate on either side of the membrane due to the two substrate-binding sites. To corroborate our structural finding, we generated 11 Cyt b561 variants, each containing one or multiple mutations targeting ascorbate-binding residues and purified these variants to homogeneity (Fig. S7B). Mutations on the cytoplasmic side only, K81A/R150A or Y140W, or mutations on the noncytoplasmic side only, F105W/H106E or Y115W, still allowed the oxidized Cyt b561 to be reduced by ascorbate (Fig. S7C). In sharp contrast, the Cyt b561 variant K81A/R150A/F105W/H106E, which carries ascorbate-binding mutations on both cytoplasmic and noncytoplasmic sides, completely lost its ability to be reduced by ascorbate (Fig. S7D). This analysis also suggests that two key mutations from either side, K81A/R150A on the cytoplasmic side or F105W/H106E on the noncytoplasmic side, are sufficient for abrogating electron transfer from the corresponding side. Notably, an important role of the amino acid that corresponds to Lys81 of Cyt b561 has previously been reported for ZmCyt b561 and bovine CGCyt b561, where mutation or chemical modification of the corresponding residue was shown to affect electron acceptance from ascorbate in a negative manner (11, 13, 22).
Next, we blocked the ascorbate-binding site on the noncytoplasmic side and examined the functional importance of the ascorbate-binding residues on the cytoplasmic side (Fig. S7E). Under the background of F105W/H106E, the variant K81A displayed a more severe loss of the ability to be reduced by ascorbate compared with the variant Y140W or R150A. Using a similar strategy, we examined the ascorbate-binding residues on the noncytoplasmic side (Fig. S7F). Under the background of K81A/R150A, the variant H106E, but not F105W or Y115W, nearly lost the ability to be reduced by ascorbate. Thus, consistent with our structural observations, Lys81 on the cytoplasmic side and His106 on the noncytoplasmic side play a particularly important functional role. This in vitro system allows efficient evaluation of the functional importance of select amino acids in Cyt b561; our observations strongly suggest that Cyt b561 indeed has two binding sites for ascorbate/monodehydroascorbate.
Discussion
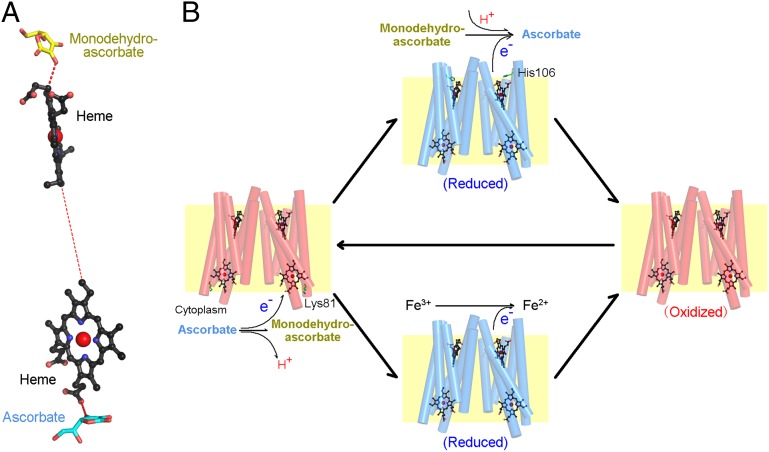
Our structural and biochemical analyses identify key elements of the electron transfer path for Cyt b561, which comprises two heme groups and two associated substrate molecules (Fig. 4A). The donor electron from ascorbate on the cytoplasmic side is transferred first to the heme group on the cytoplasmic side; then to the heme group on the noncytoplasmic side; and finally to the electron acceptor, monodehydroascorbate. To pinpoint the electron transfer route between the two heme groups, we performed in silico electron transfer modeling. Intriguingly, the results show that the energetically favored electron propagation occurs exclusively through bonding, via a number of covalent bonds and H-bonds (Fig. S8). Nonetheless, we cannot rule out a potential role by the en route water molecule and Phe129 in through-space electron tunneling.
Fig. 4.
Proposed model for electron transfer. (A) Proposed electron transfer path. This illustration shows the electron transfer path from ascorbate in the cytoplasm to monodehydroascorbate on the noncytoplasmic side. (B) Proposed mechanism of ascorbate recycling and ascorbate-dependent ferric reduction catalyzed by Cyt b561. The reactions occur on both sides of the membrane, with Cyt b561 catalyzing the transfer of electrons.
Cyt b561 family members are mainly involved in two physiological processes: ascorbate recycling and ferric-chelate reduction, as exemplified by CGCyt b561 and DCyt b561, respectively. Notably, CGCyt b561 also has ferrireductase activity (12). In addition, other members of the Cyt b561 family, such as ZmCyt b561 (11), Cyt b561-A (34), and Cyt b561-B from A. thaliana and lysosomal Cyt b561 (12), could be oxidized by ferric-chelates. On the other hand, reduced CGCyt b561 and ZmCyt b561 could react with the monodehydroascorbate radical (11, 13), consistent with our observation that Cyt b561 has two ascorbate/monodehydroascorbate-binding sites. Importantly, the substrate-binding and heme-coordinating residues are highly conserved, suggesting a similar functional mechanism for the entire family. These lines of evidence suggest that Cyt b561 family members may have electron-donating activities to both monodehydroascorbate and ferric-chelates (Fig. 4B).
In our model, one molecule of ascorbate binds to the conserved substrate-binding site on the cytoplasmic side (involving Lys81 in A. thaliana Cytb561-B) and donates an electron to Cyt b561, releasing a proton and generating one molecule of monodehydroascorbate. Next, on the other side of membrane, monodehydroascorbate or ferric-chelate may react with the reduced Cyt b561. Monodehydroascorbate likely binds to His106 of A. thaliana Cytb561-B or the corresponding residues in other homologs, accepting an electron, incorporating a proton, and producing one molecule of ascorbate. This model is consistent with the observations that the concentration of ascorbate is higher in the cytoplasm than on the other side (32), whereas the concentration of proton is the opposite (35). During the electron transfer process, Lys81 on the cytoplasmic side and His106 on the noncytoplasmic side might be involved not only in substrate recognition but in catalysis, perhaps through cycles of protonation and deprotonation. In addition, ferric-chelate might bind to the pocket on the noncytoplasmic side of Cyt b561, being reduced to a ferrous ion by accepting an electron from the heme center. The resulting oxidized Cyt b561 protein is now ready for another cycle of reduction and oxidation initiated by ascorbate in the cytoplasm.
Materials and Methods
All proteins in this study were expressed in Escherichia coli and purified by affinity chromatography and gel filtration. Crystals were obtained by the hanging drop, vapor diffusion method. Detailed methods describing protein preparation, crystallization, data collection, structure determination, and biochemical assays can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank J. He and S. Huang at the Shanghai Synchrotron Radiation Facility for onsite assistance. This work was supported by funds from the Ministry of Science and Technology (Grant 2009CB918801), National Natural Science Foundation (Grants 31021002 and 31130002), and Beijing Municipal Commissions of Education and Science and Technology.
Footnotes
The authors declare no conflict of interest.
Data deposition: The coordinates and structure factors for the ascorbate-free and ascorbate-bound structures have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4O6Y, 4O79, and 4O7G).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323931111/-/DCSupplemental.
References
- 1.Zetterström R. Nobel Prize 1937 to Albert von Szent-Györgyi: Identification of vitamin C as the anti-scorbutic factor. Acta Paediatr. 2009;98(5):915–919. doi: 10.1111/j.1651-2227.2009.01239.x. [DOI] [PubMed] [Google Scholar]
- 2.Peterkofsky B. Ascorbate requirement for hydroxylation and secretion of procollagen: Relationship to inhibition of collagen synthesis in scurvy. Am J Clin Nutr. 1991;54(6) Suppl:1135S–1140S. doi: 10.1093/ajcn/54.6.1135s. [DOI] [PubMed] [Google Scholar]
- 3.Rebouche CJ. Ascorbic acid and carnitine biosynthesis. Am J Clin Nutr. 1991;54(6) Suppl:1147S–1152S. doi: 10.1093/ajcn/54.6.1147s. [DOI] [PubMed] [Google Scholar]
- 4.Kaufman S. Dopamine-beta-hydroxylase. J Psychiatr Res. 1974;11:303–316. doi: 10.1016/0022-3956(74)90112-5. [DOI] [PubMed] [Google Scholar]
- 5.Padayatty SJ, et al. Vitamin C as an antioxidant: Evaluation of its role in disease prevention. J Am Coll Nutr. 2003;22(1):18–35. doi: 10.1080/07315724.2003.10719272. [DOI] [PubMed] [Google Scholar]
- 6.Lane DJ, Lawen A. Ascorbate and plasma membrane electron transport—Enzymes vs efflux. Free Radic Biol Med. 2009;47(5):485–495. doi: 10.1016/j.freeradbiomed.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Flatmark T, Terland O, Helle KB. Electron carriers of the bovine adrenal chromaffin granules. Biochim Biophys Acta. 1971;226(1):9–19. doi: 10.1016/0005-2728(71)90173-3. [DOI] [PubMed] [Google Scholar]
- 8.Terland O, Silsand T, Flatmark T. Cytochrome b-561 as the single heme protein of the bovine adrenal chromaffin granule membrane. Biochim Biophys Acta. 1974;359(2):253–256. doi: 10.1016/0005-2795(74)90222-0. [DOI] [PubMed] [Google Scholar]
- 9.Njus D, Knoth J, Cook C, Kelly PM. Electron transfer across the chromaffin granule membrane. J Biol Chem. 1983;258(1):27–30. [PubMed] [Google Scholar]
- 10.Kelley PM, Jalukar V, Njus D. Rate of electron transfer between cytochrome b561 and extravesicular ascorbic acid. J Biol Chem. 1990;265(32):19409–19413. [PubMed] [Google Scholar]
- 11.Nakanishi N, et al. Importance of the conserved lysine 83 residue of Zea mays cytochrome b(561) for ascorbate-specific transmembrane electron transfer as revealed by site-directed mutagenesis studies. Biochemistry. 2009;48(44):10665–10678. doi: 10.1021/bi9010682. [DOI] [PubMed] [Google Scholar]
- 12.Su D, Asard H. Three mammalian cytochromes b561 are ascorbate-dependent ferrireductases. FEBS J. 2006;273(16):3722–3734. doi: 10.1111/j.1742-4658.2006.05381.x. [DOI] [PubMed] [Google Scholar]
- 13.Takigami T, Takeuchi F, Nakagawa M, Hase T, Tsubaki M. Stopped-flow analyses on the reaction of ascorbate with cytochrome b561 purified from bovine chromaffin vesicle membranes. Biochemistry. 2003;42(27):8110–8118. doi: 10.1021/bi0267588. [DOI] [PubMed] [Google Scholar]
- 14.Tsubaki M, Takeuchi F, Nakanishi N. Cytochrome b561 protein family: Expanding roles and versatile transmembrane electron transfer abilities as predicted by a new classification system and protein sequence motif analyses. Biochim Biophys Acta. 2005;1753(2):174–190. doi: 10.1016/j.bbapap.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 15.Verelst W, Asard H. A phylogenetic study of cytochrome b561 proteins. Genome Biol. 2003;4(6):R38. doi: 10.1186/gb-2003-4-6-r38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Njus D, Kelley PM, Harnadek GJ. Bioenergetics of secretory vesicles. Biochim Biophys Acta. 1986;853(3-4):237–265. doi: 10.1016/0304-4173(87)90003-6. [DOI] [PubMed] [Google Scholar]
- 17.Tabuchi M, Yoshimori T, Yamaguchi K, Yoshida T, Kishi F. Human NRAMP2/DMT1, which mediates iron transport across endosomal membranes, is localized to late endosomes and lysosomes in HEp-2 cells. J Biol Chem. 2000;275(29):22220–22228. doi: 10.1074/jbc.M001478200. [DOI] [PubMed] [Google Scholar]
- 18.Fleming MD, et al. Microcytic anaemia mice have a mutation in Nramp2, a candidate iron transporter gene. Nat Genet. 1997;16(4):383–386. doi: 10.1038/ng0897-383. [DOI] [PubMed] [Google Scholar]
- 19.McKie AT, et al. An iron-regulated ferric reductase associated with the absorption of dietary iron. Science. 2001;291(5509):1755–1759. doi: 10.1126/science.1057206. [DOI] [PubMed] [Google Scholar]
- 20.McKie AT, et al. Molecular evidence for the role of a ferric reductase in iron transport. Biochem Soc Trans. 2002;30(4):722–724. doi: 10.1042/bst0300722. [DOI] [PubMed] [Google Scholar]
- 21.Rahman MM, et al. Roles of conserved Arg(72) and Tyr(71) in the ascorbate-specific transmembrane electron transfer catalyzed by Zea mays cytochrome b561. J Biosci Bioeng. 2013;115(5):497–506. doi: 10.1016/j.jbiosc.2012.11.013. [DOI] [PubMed] [Google Scholar]
- 22.Nakanishi N, et al. Inhibition of electron acceptance from ascorbate by the specific N-carbethoxylations of maize cytochrome b561: A common mechanism for the transmembrane electron transfer in cytochrome b561 protein family. J Biochem. 2009;146(6):857–866. doi: 10.1093/jb/mvp146. [DOI] [PubMed] [Google Scholar]
- 23.von Heijne G, Gavel Y. Topogenic signals in integral membrane proteins. Eur J Biochem. 1988;174(4):671–678. doi: 10.1111/j.1432-1033.1988.tb14150.x. [DOI] [PubMed] [Google Scholar]
- 24.Kent UM, Fleming PJ. Cytochrome b561 is fatty acylated and oriented in the chromaffin granule membrane with its carboxyl terminus cytoplasmically exposed. J Biol Chem. 1990;265(27):16422–16427. [PubMed] [Google Scholar]
- 25.Kobashi K. Catalytic oxidation of sulfhydryl groups by o-phenanthroline copper complex. Biochim Biophys Acta. 1968;158(2):239–245. doi: 10.1016/0304-4165(68)90136-0. [DOI] [PubMed] [Google Scholar]
- 26.Ma D, et al. Structure and mechanism of a glutamate-GABA antiporter. Nature. 2012;483(7391):632–636. doi: 10.1038/nature10917. [DOI] [PubMed] [Google Scholar]
- 27.Page CC, Moser CC, Dutton PL. Mechanism for electron transfer within and between proteins. Curr Opin Chem Biol. 2003;7(5):551–556. doi: 10.1016/j.cbpa.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 28.Page CC, Moser CC, Chen X, Dutton PL. Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature. 1999;402(6757):47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 29.Stroebel D, Choquet Y, Popot JL, Picot D. An atypical haem in the cytochrome b(6)f complex. Nature. 2003;426(6965):413–418. doi: 10.1038/nature02155. [DOI] [PubMed] [Google Scholar]
- 30.Kurisu G, Zhang H, Smith JL, Cramer WA. Structure of the cytochrome b6f complex of oxygenic photosynthesis: Tuning the cavity. Science. 2003;302(5647):1009–1014. doi: 10.1126/science.1090165. [DOI] [PubMed] [Google Scholar]
- 31.Sharp KH, Mewies M, Moody PC, Raven EL. Crystal structure of the ascorbate peroxidase-ascorbate complex. Nat Struct Biol. 2003;10(4):303–307. doi: 10.1038/nsb913. [DOI] [PubMed] [Google Scholar]
- 32.Asard H, Barbaro R, Trost P, Berczi A. Cytochromes b561: Ascorbate-mediated trans-membrane electron transport. Antioxid Redox Signal. 2012;19(9):1026–1035. doi: 10.1089/ars.2012.5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu W, et al. Purification and characterization of bovine adrenal cytochrome b561 expressed in insect and yeast cell systems. Protein Expr Purif. 2005;40(2):429–439. doi: 10.1016/j.pep.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 34.Bérczi A, Su D, Asard H. An Arabidopsis cytochrome b561 with trans-membrane ferrireductase capability. FEBS Lett. 2007;581(7):1505–1508. doi: 10.1016/j.febslet.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 35.Camacho M, Machado JD, Montesinos MS, Criado M, Borges R. Intragranular pH rapidly modulates exocytosis in adrenal chromaffin cells. J Neurochem. 2006;96(2):324–334. doi: 10.1111/j.1471-4159.2005.03526.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.