Significance
Deficiencies in genome maintenance genes (so-called mutator genes) result in increased mutagenesis that impacts cell evolvability. How the mutational processes drive the evolution of genome structure is not well understood. Here, we used high-throughput sequencing to characterize the mutation events (from punctual mutations to large chromosomal rearrangements) that occurred in mutation accumulation lines derived from a broad set of yeast mutator strains. Establishing genome-wide mutation profiles revealed that each mutator exhibits a unique and sometimes complex mutational signature. Our results also show how the dynamics of mutation accumulation can generate different genomes.
Keywords: genetic instability, mutation profile, MUTome, genome drift, mutation accumulation lines
Abstract
The acquisition of mutations is relevant to every aspect of genetics, including cancer and evolution of species on Darwinian selection. Genome variations arise from rare stochastic imperfections of cellular metabolism and deficiencies in maintenance genes. Here, we established the genome-wide spectrum of mutations that accumulate in a WT and in nine Saccharomyces cerevisiae mutator strains deficient for distinct genome maintenance processes: pol32Δ and rad27Δ (replication), msh2Δ (mismatch repair), tsa1Δ (oxidative stress), mre11Δ (recombination), mec1Δ tel1Δ (DNA damage/S-phase checkpoints), pif1Δ (maintenance of mitochondrial genome and telomere length), cac1Δ cac3Δ (nucleosome deposition), and clb5Δ (cell cycle progression). This study reveals the diversity, complexity, and ultimate unique nature of each mutational spectrum, composed of punctual mutations, chromosomal structural variations, and/or aneuploidies. The mutations produced in clb5Δ/CCNB1, mec1Δ/ATR, tel1Δ/ATM, and rad27Δ/FEN1 strains extensively reshape the genome, following a trajectory dependent on previous events. It comprises the transmission of unstable genomes that lead to colony mosaicisms. This comprehensive analytical approach of mutator defects provides a model to understand how genome variations might accumulate during clonal evolution of somatic cell populations, including tumor cells.
Transmission of the genetic material through mitotic clonal cell expansion or sexual reproduction is extremely accurate, with an estimated single nucleotide substitution rate in yeast of less than 10−9 per cell division, with rarely observed chromosomal structural variations (1, 2). However, mutants arising from cellular metabolism and environmental insults, as well as genetic or epigenetic defects, are biologically and evolutionarily important. Certain mutants may confer fitness defects that are removed by natural selection or generate an immediate, if not long term, advantage for adaptive evolution.
The cornerstone for genetic control of mutagenesis is the existence of numerous genes and pathways that prevent mutagenesis to occur during replication, repair, recombination, and other processes that ensure proper chromosome segregation, avoiding chromosome number variations or cells from becoming polyploidic. These genes are called mutators when their defects increase the mutation rates compared with the WT strain.
Classical methods to measure mutagenic events in yeast use genetic reporter assays. The canavanine resistance (Canr) forward assay allows detection of all kinds of mutations, inactivating this arginine permease gene (3). Other assays, using specific strain constructs, detect a narrower spectrum of mutations: the lys2-Bgl and hom3-10 reversion assays detect mutational events that revert a four-base insertion in the LYS2 gene and a +1 T insertion in a stretch of six Ts in the HOM3 gene, respectively (4, 5). Differently, a subset of gross chromosomal rearrangements (GCRs) can be detected by measuring the rate of the simultaneous loss of the CAN1 and URA3 markers located in the terminal nonessential part of chromosome V (6). Likewise, chromosome segregation fidelity (aneuploidy) can also be assayed by measuring the rate of loss of a yeast artificial chromosome (7). All these assays are convenient but have limitations. First, they do not measure phenotypically silent mutations. Second, they do not take into account the rate of spontaneous DNA damages and the variable rate of repair efficiencies across the genome. Chromosome plasticity is affected by sequence composition, responsible for mutagenic hotspots and a larger genomic context that can modulate the frequency and the pattern of mutations, according to the structural elements of the genome organization, such as repeated sequences prone to ectopic homologous recombination and direct or indirect effects of gene transcription and chromatin structure. Third, to approach an exhaustive mutational profile requires the use of several specific mutation assays and different strains.
To overcome the above functional and technical limitations, we investigated the mutational landscape of haploid Saccharomyces cerevisiae mutator strains at a genome-wide level based on examination of parallel mutation accumulation (MA) lines through bottleneck passages by using high-throughput sequencing coupled with a comprehensive bioinformatics pipeline, Sanger sequencing, pulsed-field gel electrophoresis (PFGE), and comparative genome hybridization arrays (aCGH) (Fig. 1 and Fig. S1). We chose to examine several mutagenic processes, each represented by the inactivation of a key gene function (Table 1). Compared with WT, we analyzed the following mutant strains: pol32Δ and rad27Δ (replication) (8, 9), msh2Δ (mismatch repair, MMR) (10, 11), tsa1Δ (oxidative stress) (12), mre11Δ (recombination) (13), mec1Δ tel1Δ (DNA damage/S-phase checkpoints) (14–17), pif1Δ (maintenance of the mitochondrial genome and telomere length) (18, 19), cac1Δ cac3Δ (nucleosome deposition) (20), and clb5Δ (cell cycle progression) (21, 22). All of these genes have well-conserved orthologs in mammals that are associated with genetic diseases and/or cancers (Table 1). Using the Canr and/or GCR assays, these mutants exhibited a relatively weak or strong mutator phenotype (Table 1).
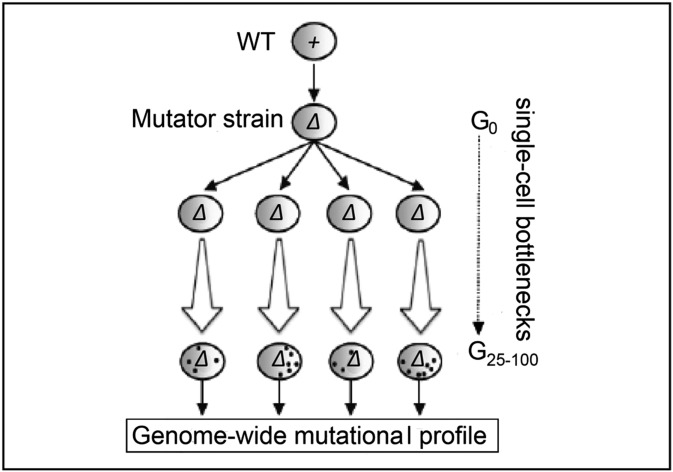
Fig. 1.
Experimental strategy. The haploid BY4741 (MATa) WT strain was transformed to delete the listed mutator gene(s) (Table 1). Next, four parallel long-term mutation accumulation (MA) lines were derived from each parental strain, by performing up to 100 single cell bottleneck passages (G100), on growth on a nonselective rich medium [yeast extract/peptone/dextrose (YPD) plate] at 30 °C. The spots present in the G25-100 lines symbolically represent the acquired mutations. Next-generation sequencing (NGS) of the WT and final passaged strains was performed (Fig. S1).
Table 1.
Activity of the genes analyzed
| Deleted yeast gene | Human homolog | Biological function | Associated mammalian syndome | Yeast mutator genetic assays | |
| CAN1r [rate (×10−7)*] | GCR assay [rate (×10−9)*,†] | ||||
| WT | — | — | — | 2.80 (1) | 0.35 (1) |
| CAC1 and CAC3 | CAF1 subunits | Chromatin-assembly factor | CAF-1 is overexpressed in breast tumors | 1.16 (0.4) | 690 (1971) |
| PIF1 | hPIF1 | Helicase with telomerase inhibitor activity; involved in maintenance of mitochondrial genome | Unknown | 20.41 (7) | 353 (1,009) |
| POL32 | POLD3 | Subunit of polymerase δ involved in DNA replication | Unknown | 2.81 (1) | 0.65 (2) |
| TSA1 | PRDX1 | Thioredoxin peroxidase involved in oxidative stress response | Lymphoma, carcinoma, and sarcoma | 19.25 (7) | 6 (17) |
| MSH2 | hMSH2 | Protein playing a key role in the mismatch repair process | Hereditary nonpolyposis colorectal cancer | 59.60 (21) | 4 (11) |
| RAD27 | FEN1 | Flap endonuclease involved in DNA replication and repair | Lung adenoma and adenocarcinoma | 95.87 (34) | 320 (914) |
| MEC1 and TEL1‡ | ATR and ATM | DNA damage/S-phase checkpoints | Ataxia telangiectasia | 482.49 (172) | 4,500 (12,857) |
| CLB5 | CCNB1 | B-type cyclin involved in DNA replication during S-phase | Unknown | 2.28 (0.8) | 0.22 (6) |
| MRE11 | hMRE11 | Protein involved in DSB repair, NHEJ, and telomere metabolism | Ataxia telangiectasia-like disorder (ATLD) | 20.81 (7) | 220 (629) |
Mammalian orthologs and diseases associated with their deficiencies are indicated.
Average mutation rate of the WT and parental mutator strains (G0) in the CANr (present results) and GCR (6) reporter assays. The relative fold increase over the WT strain is reported in parentheses.
The sml1Δ mutation is necessary to suppress the lethality of the mec1Δ mutation (47). In the text, the mec1Δ tel1Δ sml1Δ genotype is abbreviated to mec1Δ tel1Δ.
Our genome-wide study of yeast mutation events shows that each mutator strain is characterized by a unique and sometimes complex mutational spectrum composed of punctual and large chromosomal structural variations. Our results provide insights into the dynamic mutational processes occurring during the clonal evolution of somatic cell populations.
Results and Discussion
Sequencing of Yeast MA Lines.
To obtain a significant number of mutations per strain and characterize their mode of accumulation, we generated MA lines over 25–100 bottleneck passages (G25–100), on growth on a nonselective rich medium, as illustrated in Fig. 1. For each strain, we performed bottleneck passages by picking at least four individual colonies from the original WT or freshly transformed mutant strain (SI Materials and Methods). These G0 colonies were independently restreaked on plates to isolate colony-forming units (G1 passage) (each CFU corresponds to ∼25 generations), and subsequent passages from single cells to CFU were performed up to 100 times (G100). The specific parental mutator phenotype (Canr assay) did not change between the G0 and final passages (Table 1 and Table S1). Thus, the mutagenic trajectory could be functionally attributed to deficiencies in the starting mutator gene, with a rare contribution of secondary arising mutator phenotypes (see below). The MA lines were sequenced with high coverages (∼100-fold) and analyzed with a robust set of bioinformatic tools (Fig. S1) allowing the identification of all types of mutational events: base substitutions (SNP), small insertions/deletions (InDels, from −11 to +3 bp), intermediate-sized and large structural variants (SVs, from 50 bp to 5 kb and >5 kb, respectively), and aneuploidies. As repeated regions of the genome are a source of read multialignments and their polymorphisms a source of false-positive calls, variants found in these regions were mostly filtered out, except SNPs that could be efficiently detected in repeated regions (SI Materials and Methods). To further validate and resolve any ambiguous situations, we conducted extensive complementary molecular (Figs. S1 and S2) and phenotypic analyses (Table S2). Importantly, the accuracy of our NGS pipelines was high because the validation rate of mutation calling reached 95% (125/131 calls) for the SNPs and 98% (215/220 calls) for small InDels, including the most difficult detection of small InDels in homopolymers and microsatellites. Calls for structural variants including large deletions/amplifications and aneuploidies were all confirmed by PFGE and aCGH analyses.
Mutational Landscapes.
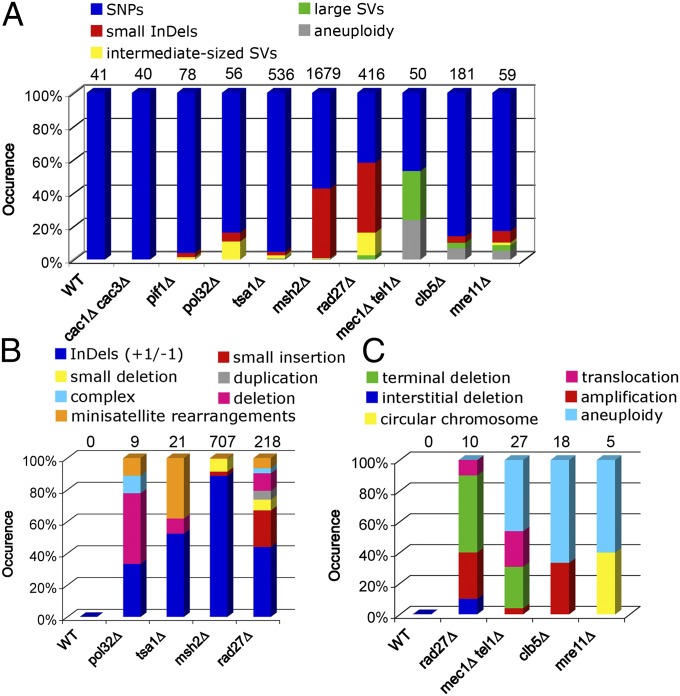
Altogether, 3,319 mutations were identified (individually annotated for the WT and mutant strains in Datasets S1–S10). Around 60% (1,878/2,988) of SNPs and small InDels were found in yeast ORFs; 17% (315/1,878) of these small mutations were in essential genes that represent 21% (1,079/5,064) of the characterized yeast ORFs. Therefore, only mutations resulting in the inactivation of essential genes could not be recovered. The number of mutations per parallel line was roughly similar (Table 2), but the total number of mutations produced per strain genotype varied by tens of folds and defined specific mutational landscapes (Table 2 and Fig. 2A). At G100, the four WT lines accumulated a total of 41 single nucleotide substitutions (SNPs) but no other types of mutations (0.36 × 10−9 substitutions per site per generation), in close agreement to previous estimates (1, 2). Our first class of mutational landscapes is similar to WT cells, as observed in the cac1Δ cac3Δ, pif1Δ, and pol32Δ strains that accumulate few base substitutions (Table 2 and Fig. 2A). Mechanistically, we can envisage that this background level of mutagenesis results from stochastic replication polymerase errors and mutagenic repair of oxidative damages, likely to occur in any proliferative strain.
Table 2.
Mutational behaviors
| Mutation type | No. of mutations | WT | cac1Δ cac3Δ | pif1Δ | pol32Δ | tsa1Δ | msh2Δ | rad27Δ | mec1Δ tel1Δ | clb5Δ | mre11Δ |
| Parallel lines | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 8 | 4 | 4 | |
| Bottlenecks | 100 | 75 | 100 | 100 | 100 | 100 | 100 | 25 | 100 | 75 | |
| SNPs | Transition | ||||||||||
| A:T→G:C | 6 | 9 | 7 | 12 | 60 | 229 | 26 | 4 | 21 | 6 | |
| G:C→A:T | 13 | 9 | 13 | 9 | 194 | 461 | 36 | 4 | 34 | 14 | |
| Transversion | |||||||||||
| A:T→T:A | 6 | 5 | 7 | 6 | 58 | 41 | 28 | 0 | 17 | 7 | |
| G:C→T:A | 8 | 7 | 36 | 10 | 68 | 191 | 42 | 6 | 32 | 12 | |
| A:T→C:G | 4 | 1 | 2 | 2 | 65 | 22 | 17 | 5 | 18 | 6 | |
| G:C→C:G | 4 | 9 | 10 | 8 | 66 | 21 | 26 | 5 | 34 | 3 | |
| Total number of mutations | 41 [8–14] | 40 [8–14] | 75 [12–26] | 47 [9–15] | 512* [50-227] | 965* [153–473] | 175* [33–54] | 24 [1–5] | 156* [32–53] | 49 [9–14] | |
| Mutation rate | 0.36 | 0.47 | 0.65 | 0.41 | 4.47 | 8.42 | 1.53 | 0.42 | 1.36 | 0.57 | |
| Relative increase | 1 | 1 | 2 | 1 | 12 | 23 | 4 | 1 | 4 | 2 | |
| Small InDels (−11 to +3 bp) | Number of mutations | 0 | 0 | 2 [0–1] | 3 [0–2] | 11 [2–4] | 703 [133–215] | 174 [25–54] | 0 | 7 [1–2] | 4 [1–2] |
| Relative increase | 1 | 1 | >2 | >3 | >11 | >703 | >174 | 1 | >7 | >5 | |
| Intermediate-sized SVs (50 bp to 5 kb) | Number of mutations | 0 | 0 | 1 | 6 [1–3] | 10 [2–3] | 4 [1–4] | 57 [12–18] | 0 | 0 | 1 |
| Relative increase | 1 | 1 | >1 | >6 | >10 | >4 | >57 | 1 | 1 | >1 | |
| Large SVs (>5 kb) | Number of mutations | 0 | 0 | 0 | 0 | 2 [0–1] | 6 | 10 [1–3] | 14 [1–3] | 6 [0–3] | 2 [0–1] |
| Relative increase | 1 | 1 | 1 | 1 | >2 | >6 | >10 | >28 | >6 | >3 | |
| Aneuploid chromosomes | Number of mutations | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 12 [1–2] | 12 [1–6] | 3 [0–1] |
| Relative increase | 1 | 1 | 1 | 1 | >1 | >1 | 1 | >24 | >12 | >4 | |
| Total number of mutations | 41 | 40 | 78 | 56 | 536 | 1,679 | 416 | 50 | 181 | 59 | |
| Relative increase | 1 | 1 | 2 | 1 | 13 | 41 | 10 | 2 | 4 | 2 | |
The number of parallel lines for each strain and the number of bottlenecks per strain are indicated (Fig. 1). For each mutation type and strain, the total number of mutations accumulated is indicated. The number of mutations observed in the two most variant lines is provided within brackets. Base substitution mutation rates were calculated as follows: total number of mutations identified/number of parallel lines/number of bottlenecks/25 (estimated number of generation per midsize colonies)/1.15 × 107 (total number of nucleotides in the attainable S. cerevisiae genome). For example, in the WT strain, 41/4/100/25/1.15 × 107 = 0.36 × 10−9. The relative increase over the WT strain is also provided. Aneuploidies correspond to the number of additional chromosome copies that accumulated in the total set of lines. The list of mutations per line per mutant is reported in Datasets S1–S10.
Statistical analysis: based on the Wilcoxon Mann–Whitney test the number of SNPs accumulated in the clb5Δ, msh2Δ rad27Δ, and tsa1Δ mutant lines is significantly higher compared with WT lines (P = 0.028).
Fig. 2.
Mutational profiles. (A) Overview of the spectrum of mutations accumulated in each of the sequenced haploid MA lines. The total number of mutations per strain is shown at the top of the panels. (B) Type of InDels. (C) Type of large SV mutations. In B and C, only the mutator strains that accumulated more than eight events are depicted.
Another type of mutational landscape is defined by a massive accumulation of base substitutions and/or small InDels as observed in the tsa1Δ, msh2Δ, and rad27Δ mutants. These mutants were top ranked as single gene mutators in the Canr assay (12) (Table 1). Consistently, in the present genome-wide analysis, they accumulate the highest number of mutations (Table 2), corresponding to a 41-, 13-, and 10-fold increase in mutation rates, respectively. Tsa1-deficient lines only enhanced base substitutions (12-fold increase; Fig. 2A). The excess of G:C→A:T transitions (Table 2 and Fig. S3A) can be explained by the defects in repairing DNA oxidative damage (23). Two other mutants, clb5Δ and pif1Δ, exhibited a slight four- and twofold increase of base substitutions with an excess of G:C→C:G (22% vs. 10% for WT) and G:C→T:A transversions (48% vs. 20% for WT), respectively (Table 2 and Fig. S3A). Interestingly, because clb5Δ lines became aneuploid (Table 2, see below), the accumulation of base substitutions in clb5Δ cells can be attributed to the associated mechanism of aneuploid-induced genomic instability (7). How it differentially impacts transitions and transversions remains to be elucidated. The approximate fivefold excess of G:C→T:A transversions in pif1Δ might reflect a defect in the repair of 8-oxoguanines, possibly related to the incapacity of pif1Δ cells to maintain mitochondrial DNA (18, 24) (Table S2).
The msh2Δ strain, compromised for both Msh3- and Msh6-dependent mismatch repair pathways, exhibits a dual mutational signature: it produces a large number of base substitutions (965) and small InDels (703), reaching a mutation rate of 8.4 × 10−9 per bp per generation (11) (Table 2). Transitions are observed in large excess (72% compared with 46% in WT) (Table 2 and Fig. S3A), with no apparent sequence-specific context. In contrast, InDels mostly occur in specific motifs: a vast majority within homopolymer (HP) tracts (663/703) and others within di-/trinucleotide tracts (microsatellites) (40/703). Almost all small InDels we found consisted of single nucleotide variations, with a sixfold excess of −1 to +1 bp (535 vs. 93, respectively; Fig. 2B) (11). Across the genome, the frequency of +1/−1 mutations in both A:T or G:C tracts increases by several orders of magnitude from 2 to 10 nt HPs and then plateaus (Fig. S3B). The rad27Δ lines accumulate less base substitutions and small InDels than msh2Δ but in a similar proportion (175 vs.174; Table 2). Also in contrast to msh2Δ, rad27Δ InDels are enriched in microsatellites (73) vs. HP tracts (89). These results reflect the overall propensity of rad27Δ cells to destabilize all types of tandem repeats (Fig. 2B) (25, 26). The overwhelming strength of these replication- and repair-related mutators and the abundance of vulnerable HP tracts (223,528 HP tracts 5–14 nt long across the reference S. cerevisiae genome) lead to heavily drifted clones and the acquisition of numerous phenotypes (Table S2). Interestingly, the presence of a poly(A) tract of 5 nt long in the coding region of the MSH3 gene has given rise to a −1 nt frameshift in the G100 msh2Δ A line (Dataset S6). msh3Δ is a weak mutator according to the Canr and hom3-10 assays (27) and has no synergistic effect with msh2Δ as they belong to the same MMR pathway. Consequently, the inactivation of MSH3 had no aggravating consequence on the final G100 mutational rate measured with the Canr assay (Table S1). In contrast, in the same msh2Δ A line, the arising of a −2 frameshift mutation in the 10 nt poly(A) tract located in the PIF1 ORF (Dataset S6) created an additional secondary mutator phenotype that likely led to the deficiency of the G100 cell growth on YPGlycerol media, due to the role of Pif1 in the maintenance of the mitochondrial genome (Table S2).
rad27Δ Cells Exhibit a Complex Mutation Signature.
Another important class of genome alterations consists of intermediate (50 bp to 5 kbp) and large scale (>5 kbp) SVs, including both deletion and insertion events. rad27Δ lines accumulated 57 intermediate SVs (Table 2 and Fig. 2 A and B). Remarkably, and as previously discovered with the Canr assay (9), seven events contained a unique mutational signature, namely small tandem duplications ranging from 13 to 4,068 bp, flanked by short 5- to 10-bp direct repeats but also complex intermediate size rearrangements (Fig. 2B and Dataset S7). The other events contained 15 deletions between distant repeats (a hallmark of a Single-Strand Annealing mechanism) (28) and 14 expansions or contractions of the endogenous minisatellite loci (Table S3). In addition, the rad27Δ lines yielded five chromosomal terminal deletions healed by de novo telomere addition, a large deletion, and three large tandem duplications, ranging from 15 to 150 kbp that can be explained by the mechanism of rereplication-induced gene amplification (RRIGA) (29) and a reciprocal translocation (Figs. 2C and 3D). Unexpectedly tsa1Δ also alters the size of the minisatellite loci (eight cases) (Fig. 2B and Table S3). No gene known to play a role in minisatellite stability was mutated (25, 26, 30, 31). Thus, Tsa1 now appears to play a role in two apparently unrelated mutagenic pathways: the elimination of H2O2 and specific minisatellite (mostly A:T rich) instability.
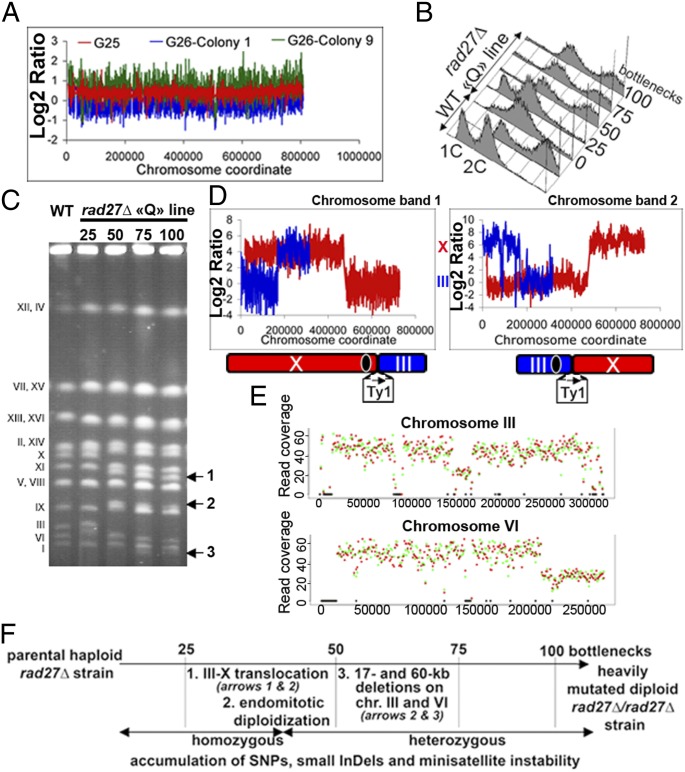
Fig. 3.
Dynamics of mutation accumulation. (A) mec1Δ tel1Δ O line. Ratio of aCGH signal on chromosome II from cells at G25 (red; ratio of 1.4) and two independent colonies at G26 (green; 1.8 ratio and blue;1.0 ratio). (B–F) G100 rad27Δ Q line. (B) FACS analysis to determine the evolution of cell ploidy. The haploid (1C) rad27Δ Q line becomes diploid (2C) between G26 to G50 by endomitotic diploidization. (C) Evolution of yeast chromosome sizes visualized by PFGE through the bottleneck passages. Arrows indicate the chromosome bands of altered length. (D) Band CGH microarray analysis of the altered chromosome bands 1 and 2 excised from PFGE lane G100 in C. A genomic microarray reveals a reciprocal translocation between chromosomes III and X (ratio of hybridization of the probes located on chromosomes III (blue) and X (red). The structure of the reciprocal translocation mediated by Ty1 retrotransposon elements is described below (for more details, see Dataset S7). (E) NGS read coverage of the G100 line that became diploid. The twofold decrease of coverage at the end of chromosome VI and in the center of chromosome III reveals a heterozygous 60-kb terminal deletion (chromosome band 3) and a 17-kb interstitial deletion, respectively. Red and green points indicate the read coverage calculated in a 1-kb window size on forward and reverse strands, respectively. Black bars indicate multialigned regions. (F) Diagram indicating the chronological order in which chromosomal rearrangements accumulated in the haploid G100 rad27Δ Q line.
pif1Δ Mutational Landscape.
To our surprise, besides the loss of the mitochondrial genome (observed at G0) and the slight increase of base substitutions described above, the deletion of this gene had no detectable additional effect (Table 2). This result is surprising because the elimination of the Pif1 helicase causes a 1,000-fold increase in the rate of GCRs according to the chromosome V assay (32) (Table 1). Because this experiment was performed in a different strain background, we introduced the GCR assay in our pif1Δ haploid strain (derivative of BY4741; SI Materials and Methods). Clearly, in both strain backgrounds, the GCR rate is highly increased in pif1Δ cells compared with WT and reaches similar rates: 4.82 × 10−7 vs. 3.53 × 10−7 in the BY4741 and RDKY backgrounds, respectively. Alternatively, the rarity of GCR events in our pif1Δ MA lines (and in other strains) can result from the massive counterselection of such rearrangements in haploid cells, but this seems unlikely because GCR events involving different chromosomes were detected in other haploid BY4741 MA lines with similar chromosome V GCR rate (rad27Δ and mre11Δ). Nevertheless, to compare haploid and diploid cells, we derived 16 homozygous diploid pif1Δ/pif1Δ MA lines for 25 bottleneck passages (equivalent to the four haploid lines derived for 100 passages) and analyzed potential variations in chromosome size by PFGE. As illustrated in Fig. S2, we detected only a single rearrangement (a chromosome II-XII translocation characterized in detail in Dataset S3). This rare event reasonably excludes the possibility that the lack of Pif1 yields a large number of structural variations. Thus, the GCR effect of pif1Δ might be specific to the chromosome V construct and eventually to other chromosomal regions but not necessarily general. It reveals a regional chromosomal context that, for example, can be prone to stimulate initiation events. This situation could also be envisaged for the cac1Δ cac3Δ mutant strain, which exhibits an enhanced GCR rate (2,000-fold) (33) but appeared WT-like in our genome-wide analyses (Table 2 and Fig. 2A).
Mutants Enhancing Aneuploidies and Chromosome Rearrangements.
Aneuploidies also greatly contribute to genome variation and are associated with severe functional abnormalities, partly due to large-scale imbalances of gene expression (34, 35). Chromosome mis-segregation during mitosis can result from defects in chromosome attachment to the mitotic spindle (36) or from problems in DNA replication or repair. In particular, it depends on the DNA damage/S phase checkpoint controlled by the Mec1/ATR and Tel1/ATM kinases and the activity of the Clb5 B-type cyclin that functionally controls and coordinates the progression of the cell cycle (22, 37). Clearly, the mec1Δ tel1Δ cell lines do not produce either an abnormal level of base substitutions or small to intermediate-sized InDels (Table 2). However, and as anticipated (17), the haploid mec1Δ tel1Δ MA lines yield large SVs and chromosome aneuploidies (average of three per line at G25; Table 2 and Fig. 2 A and C), confirmed by detailed chromosome PFGE analyses (Fig. S2). Similarly, the clb5Δ mutant produced amplifications and aneuploidies. Remarkably, the clb5Δ, rad27Δ, and mre11Δ mutants reveal chromosome rearrangement junctions mostly localized within ∼0.3 kb LTR or 5.9 kb Ty elements (Fig. S4A), likely arising from intrachromosomal or interchromosomal exchanges in dispersed Ty retrotransposons. In contrast, only two Ty elements were found in the vicinity of the 18 translocation breakpoints detected in the mec1Δ tel1Δ lines. In this case, the translocation junctions predominantly occurred in interstitial nonrepeated regions of the yeast genome or in subtelomeric and telomeric chromosomal regions (Fig. S4B). The junctions shared 1–3 nt of microhomology, suggesting the participation of the nonhomologous end joining (NHEJ) mechanism (17). The structure of these translocations indicates that they likely result from chromosome fusion events (16). Although many large SVs were found in rad27Δ, clb5Δ, and mec1Δ tel1Δ MA lines, only two occurred in the four mre11Δ MA lines (G75). These events were unusual as they correspond to a large duplication (32 and 65 kb), with one copy carried on a circular chromosome (Fig. 2C and Fig. S5). They probably result from homologous recombination events between Ty elements that copied the intervening region containing the centromere and one or several origins of replication, allowing stable episomal maintenance.
With respect to aneuploidies, most clb5Δ and mec1Δ tel1Δ MA lines (examined by sequencing, PFGE, and/or aCGH) contain additional chromosome copies (Table 2 and Fig. 2C). We observed di- and trisomies involving 10 of the 16 yeast chromosomes, with a biased 46% overrepresentation of chromosomes III and XI (Table S4) that can result from preferential defects in proper mitotic segregation at anaphase or a better tolerance to the outcomes of gene overexpression.
Genetic mosaicisms reflect genome heterogeneity from cell to cell, a common feature observed in tumor samples that complicates NGS analyses (38). We found clonal heterogeneities in both clb5Δ and mec1Δ tel1Δ MA lines. For example, in the G25 mec1Δ tel1Δ O line, we saw that the average chromosome index was 1.0 (0.87–1.08 per chromosome), except for chromosomes II, XI, and XII, which varied between 1.44 and 1.76 (Table S5). To determine whether this reflects stable cell-to-cell heterogeneities, we examined G24 and G26 by aCGH. The chromosome II copy number evolved from 1.01 at G24 to 1.44 at G25 and fluctuated between 0.99 and 1.82 in the 10 sibling G26 colonies (Fig. 3A). This genome heterogeneity reveals an inherited and unstable genomic situation (17, 39). Extending the degree of mosaicisms, in two of the G26 colonies, chromosome IX became overrepresented (1.44 ratio), indicative of an early subclonal event, now captured in a single bottleneck analysis (Table S5). Mechanistically, the maintenance of full chromosomes in these mutants does not evoke a chromosome break-fusion process but rather reveals a failure in the mitotic checkpoint, perhaps amplified by the aneuploidy-induced process of chromosome instability (7).
Dynamics of Mutation Accumulation.
Finally, we mined the temporal signature of the various mutations to determine their order of occurrence. An astonishing case is observed by examining the historical evolution of the G100 haploid rad27Δ Q line, which is uniquely landmarked by the occurrence of an endomitotic diploidization event between G26 and G50 (Fig. 3B and Dataset S7). Meanwhile, between G26 and G50, a balanced translocation (Fig. 3 C and D) preceded the genome endoduplication event, and later other large deletions occurred (Fig. 3E). The chronological order of such complex mutational drifts is summarized in Fig. 3F. Remarkably, at passage G100, each rad27Δ MA line had accumulated >100 heterozygous and homozygous distinct mutations and various phenotypes (Table 2 and Table S2). Similarly, in the msh2Δ lines, we examined the appearance over time of four 3–5 nt deletions (HP tracts) present at G100. All arose from temporally distinct single nt deletion events. Convincingly, the snapshot detection of mutagenic events helps to resolve the cumbersome complexity of mutagenic events once they have accumulated.
In summary, our yeast MUTome data uncover the uniqueness of each mutator gene signature at the genome-wide level. Globally, mutational profiles differ by both the type and the frequency of acquired mutation events; a high or low mutational frequency is related to the number of target sites. For example, the msh2Δ and rad27Δ mutants share overlapping mutation spectra characterized by HP tracts and microsatellite instabilities, although with a distinct ratio. Additional hallmarks of Rad27/FEN1-deficient cells are the unique small and large duplication events occurring between small repeats (9), minisatellite instability, and chromosome terminal deletions. Similarly, the mec1Δ tel1Δ and clb5Δ mutants exhibit a marked increase in aneuploidy frequency, but their mutation spectra can be distinguished by the frequency of terminal deletions, translocations (mec1Δ tel1Δ), and amplifications (clb5Δ). It enlights the diversity of potential genome variations and emphasizes the tremendous impact of several single gene activities to prevent the massive drift of parental genetic information. Most likely, the accumulation of mutations will decrease the cell fitness, but it has the advantage to increase the space of genetic information on which Darwinian selection acts.
Pursuing this approach in model organisms and in human cells should contribute to the development of our understanding of genome evolution, help to identify the current chromosomal regions at higher risks of variation, and contribute to elucidate the mechanisms and dynamics of clonal genome evolution in cancer cell populations (40–43). The uniqueness of each mutational profile can serve as a resource to functionally analyze the cancer somatic mutation data in guiding the identification of human mutator candidates and ultimately in deconvoluting the history of tumor genome evolution.
Materials and Methods
Yeast Mutation accumulation lines.
Four parallel mutation accumulation lines were derived from each mutant strain (BY4741 background) and passed through 75–100 single-cell bottlenecks. See SI Materials and Methods for details.
Mutation Detection.
Accumulated mutations were identified by using bioinformatics pipelines (Fig. S1), CGHa and PFGE (Fig. S2). See SI Materials and Methods for details.
Supplementary Material
Acknowledgments
We thank A. Piazza, R. Laureau, A. Holmes, and O. Delattre for fruitful discussions and critical reading of the manuscript. We also thank T. Rio-Frio from the Institut Curie Next Generation Sequencing (IC-NGS) platform. We are indebted to S. Lair, A. Lermine, and E. Barillot for technical help in bioinformatics. We thank E. Louis, S. Thoraval, and T. Scarcez from the Life Technology Company for help in applying SOLiD technology. The NGS raw data are available on demand. This work was funded by l’Agence Nationale de la Recherche (ANR) (MUTome Grant ANR-09-BLAN-0278-01) and the Ligue Nationale Contre le Cancer (Equipe Labelisée EL2010). C.J. received a partial doctoral grant from the Fondation ARC pour la Recherche sur le Cancer and the Cellule de l’Enseignement de l‘Institut Curie. The IC-NGS platform received financial support from the ANR (investissements d'avenir ANR-10-EQPX-03 and ANR10-INBS-09-08), the Canceropôle, the Région Ile de France, and France Génomique.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314423111/-/DCSupplemental.
References
- 1.Lynch M, et al. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc Natl Acad Sci USA. 2008;105(27):9272–9277. doi: 10.1073/pnas.0803466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishant KT, et al. The baker’s yeast diploid genome is remarkably stable in vegetative growth and meiosis. PLoS Genet. 2010;6(9):e1001109. doi: 10.1371/journal.pgen.1001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williamson MS, Game JC, Fogel S. Meiotic gene conversion mutants in Saccharomyces cerevisiae. I. Isolation and characterization of pms1-1 and pms1-2. Genetics. 1985;110(4):609–646. doi: 10.1093/genetics/110.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Steele DF, Jinks-Robertson S. An examination of adaptive reversion in Saccharomyces cerevisiae. Genetics. 1992;132(1):9–21. doi: 10.1093/genetics/132.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prolla TA, Christie DM, Liskay RM. Dual requirement in yeast DNA mismatch repair for MLH1 and PMS1, two homologs of the bacterial mutL gene. Mol Cell Biol. 1994;14(1):407–415. doi: 10.1128/mcb.14.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C, Kolodner RD. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet. 1999;23(1):81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- 7.Sheltzer JM, et al. Aneuploidy drives genomic instability in yeast. Science. 2011;333(6045):1026–1030. doi: 10.1126/science.1206412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burgers PM. Eukaryotic DNA polymerases in DNA replication and DNA repair. Chromosoma. 1998;107(4):218–227. doi: 10.1007/s004120050300. [DOI] [PubMed] [Google Scholar]
- 9.Tishkoff DX, Filosi N, Gaida GM, Kolodner RD. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997;88(2):253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 10.Tran HT, Keen JD, Kricker M, Resnick MA, Gordenin DA. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol Cell Biol. 1997;17(5):2859–2865. doi: 10.1128/mcb.17.5.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zanders S, et al. Detection of heterozygous mutations in the genome of mismatch repair defective diploid yeast using a Bayesian approach. Genetics. 2010;186(2):493–503. doi: 10.1534/genetics.110.120105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang ME, Rio AG, Nicolas A, Kolodner RD. A genomewide screen in Saccharomyces cerevisiae for genes that suppress the accumulation of mutations. Proc Natl Acad Sci USA. 2003;100(20):11529–11534. doi: 10.1073/pnas.2035018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haber JE. The many interfaces of Mre11. Cell. 1998;95(5):583–586. doi: 10.1016/s0092-8674(00)81626-8. [DOI] [PubMed] [Google Scholar]
- 14.Ritchie KB, Mallory JC, Petes TD. Interactions of TLC1 (which encodes the RNA subunit of telomerase), TEL1, and MEC1 in regulating telomere length in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1999;19(9):6065–6075. doi: 10.1128/mcb.19.9.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myung K, Datta A, Kolodner RD. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell. 2001;104(3):397–408. doi: 10.1016/s0092-8674(01)00227-6. [DOI] [PubMed] [Google Scholar]
- 16.Pennaneach V, Kolodner RD. Recombination and the Tel1 and Mec1 checkpoints differentially effect genome rearrangements driven by telomere dysfunction in yeast. Nat Genet. 2004;36(6):612–617. doi: 10.1038/ng1359. [DOI] [PubMed] [Google Scholar]
- 17.McCulley JL, Petes TD. Chromosome rearrangements and aneuploidy in yeast strains lacking both Tel1p and Mec1p reflect deficiencies in two different mechanisms. Proc Natl Acad Sci USA. 2010;107(25):11465–11470. doi: 10.1073/pnas.1006281107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foury F, Kolodynski J. pif mutation blocks recombination between mitochondrial rho+ and rho- genomes having tandemly arrayed repeat units in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1983;80(17):5345–5349. doi: 10.1073/pnas.80.17.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schulz VP, Zakian VA. The saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76(1):145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 20.Loyola A, Almouzni G. Histone chaperones, a supporting role in the limelight. Biochim Biophys Acta. 2004;1677(1-3):3–11. doi: 10.1016/j.bbaexp.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 21.Donaldson AD, et al. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol Cell. 1998;2(2):173–182. doi: 10.1016/s1097-2765(00)80127-6. [DOI] [PubMed] [Google Scholar]
- 22.Schwob E, Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 1993;7(7A):1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- 23.Wang D, Kreutzer DA, Essigmann JM. Mutagenicity and repair of oxidative DNA damage: Insights from studies using defined lesions. Mutat Res. 1998;400(1-2):99–115. doi: 10.1016/s0027-5107(98)00066-9. [DOI] [PubMed] [Google Scholar]
- 24.Cheng X, Dunaway S, Ivessa AS. The role of Pif1p, a DNA helicase in Saccharomyces cerevisiae, in maintaining mitochondrial DNA. Mitochondrion. 2007;7(3):211–222. doi: 10.1016/j.mito.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 25.Kokoska RJ, et al. Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase delta (pol3-t) Mol Cell Biol. 1998;18(5):2779–2788. doi: 10.1128/mcb.18.5.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopes J, Ribeyre C, Nicolas A. Complex minisatellite rearrangements generated in the total or partial absence of Rad27/hFEN1 activity occur in a single generation and are Rad51 and Rad52 dependent. Mol Cell Biol. 2006;26(17):6675–6689. doi: 10.1128/MCB.00649-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marsischky GT, Filosi N, Kane MF, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10(4):407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 28.Ivanov EL, Sugawara N, Fishman-Lobell J, Haber JE. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142(3):693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finn KJ, Li JJ. Single-stranded annealing induced by re-initiation of replication origins provides a novel and efficient mechanism for generating copy number expansion via non-allelic homologous recombination. PLoS Genet. 2013;9(1):e1003192. doi: 10.1371/journal.pgen.1003192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ribeyre C, et al. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5(5):e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelly MK, Brosnan L, Jauert PA, Dunham MJ, Kirkpatrick DT. Multiple pathways regulate minisatellite stability during stationary phase in yeast. G3. 2012;2(10):1185–1195. doi: 10.1534/g3.112.003673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myung K, Chen C, Kolodner RD. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature. 2001;411(6841):1073–1076. doi: 10.1038/35082608. [DOI] [PubMed] [Google Scholar]
- 33.Myung K, Pennaneach V, Kats ES, Kolodner RD. Saccharomyces cerevisiae chromatin-assembly factors that act during DNA replication function in the maintenance of genome stability. Proc Natl Acad Sci USA. 2003;100(11):6640–6645. doi: 10.1073/pnas.1232239100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torres EM, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317(5840):916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 35.Pavelka N, et al. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature. 2010;468(7321):321–325. doi: 10.1038/nature09529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8(5):379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 37.Harrison JC, Haber JE. Surviving the breakup: The DNA damage checkpoint. Annu Rev Genet. 2006;40:209–235. doi: 10.1146/annurev.genet.40.051206.105231. [DOI] [PubMed] [Google Scholar]
- 38.Jacobs KB, et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet. 2012;44(6):651–658. doi: 10.1038/ng.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vernon M, Lobachev K, Petes TD. High rates of “unselected” aneuploidy and chromosome rearrangements in tel1 mec1 haploid yeast strains. Genetics. 2008;179(1):237–247. doi: 10.1534/genetics.107.086603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loeb LA. Human cancers express mutator phenotypes: Origin, consequences and targeting. Nat Rev Cancer. 2011;11(6):450–457. doi: 10.1038/nrc3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campbell PJ, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467(7319):1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463(7278):191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nik-Zainal S, et al. Breast Cancer Working Group of the International Cancer Genome Consortium Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149(5):979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Motegi A, Kuntz K, Majeed A, Smith S, Myung K. Regulation of gross chromosomal rearrangements by ubiquitin and SUMO ligases in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26(4):1424–1433. doi: 10.1128/MCB.26.4.1424-1433.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Myung K, Datta A, Chen C, Kolodner RD. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat Genet. 2001;27(1):113–116. doi: 10.1038/83673. [DOI] [PubMed] [Google Scholar]
- 46.Putnam CD, et al. Bioinformatic identification of genes suppressing genome instability. Proc Natl Acad Sci USA. 2012;109(47):E3251–E3259. doi: 10.1073/pnas.1216733109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2(3):329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.