Significance
Human cytomegalovirus (HCMV)-infected cells and tumor cells produce similar alterations in glucose metabolism, including increasing glucose uptake and glycolysis and redirecting glucose carbon to support synthesis of biomolecules. We show that HCMV infection induces the glucose-responsive transcriptional factor carbohydrate-response element binding protein to reprogram glucose metabolism to support lipid and nucleotide synthesis. This study provides insight into viral mechanisms of pathogenesis.
Keywords: viral pathogenesis, glycolysis, lipid synthesis
Abstract
Carbohydrate-response element binding protein (ChREBP) plays a key role in regulating glucose metabolism and de novo lipogenesis in metabolic tissues and cancer cells. Here we report that ChREBP is also a critical regulator of the metabolic alterations induced during human cytomegalovirus (HCMV) infection. The expression of both ChREBP-α and ChREBP-β is robustly induced in HCMV-infected human fibroblasts; this induction is required for efficient HCMV infection. Depletion of ChREBP in HCMV-infected cells results in reduction of HCMV-induced glucose transporter 4 and glucose transporter 2 expression, leading to inhibition of glucose uptake, lactate production, nucleotide biosynthesis, and NADPH generation. We previously reported that HCMV infection induces lipogenesis through the activation of sterol regulatory element binding protein 1, which is mediated by the induction of PKR-like endoplasmic reticulum kinase. Data from the present study show that HCMV-induced lipogenesis is also controlled by the induction of ChREBP, in a second mechanism involved in the regulation of HCMV-induced de novo lipogenesis. These results suggest that ChREBP plays a key role in reprogramming glucose and lipid metabolism in HCMV infection.
Human cytomegalovirus (HCMV), a β-herpesvirus, is the leading cause of congenital viral infection in the developed countries, frequently leading to mental retardation, deafness, and developmental disability (1). Studies in recent years have shown that HCMV infection can induce dramatic changes in glucose and glutamine metabolism (2–4). HCMV-infected cells have a much higher rate of glycolysis (2), owing largely to the significant induction of glucose transporter (GLUT) 4 to increase glucose uptake (5). Unlike normal cells, the increased glucose flux in HCMV-infected cells is not used for energy production in the tricarboxylic acid (TCA) cycle; instead, a large amount of glucose-derived carbon supports lipid biosynthesis, which is essential for the viral infection (3). Our recent studies have shown that the increased lipid biosynthesis is mediated by the transcriptional factor sterol regulatory element binding protein 1 (SREBP1), which is activated by the induction of PKR-like endoplasmic reticulum kinase (PERK) in HCMV-infected cells (6, 7).
Glucose not only is a primary carbon source for anabolic and catabolic purposes in mammalian cells, but also serves as a signal in the regulation of metabolic gene expression. In this regard, the carbohydrate-response element binding protein (ChREBP) is a transcription factor that responds to glucose signaling (8, 9). ChREBP mRNA is most abundant in tissues in which lipogenesis is highly active (10). Human ChREBP-α, the full-length isoform, consists of 852 amino acids and contains several functional domains, including two nuclear export signals, a low-glucose inhibitory domain (LID), a nuclear localization signal, a glucose response conserved element, a proline-rich region, and a DNA-binding domain in sequence from the N terminus to the C terminus (9, 11). ChREBP-β is a truncated isoform (12). Its transcription starts from an alternative exon, exon 1b, and is controlled by an alternate upstream promoter that contains a carbohydrate-response element (ChoRE) sequence. Transcription from exon 1b results in loss of exon 1a found in ChREBP-α. Because no ATG is present in exon 1b, translation of ChREBP-β starts at the downstream ATG in exon 4, producing a short protein that is missing the first 177 amino acids, including two nuclear export signals, one LID, and one nuclear localization signal.
The lack of LID domain enhances the transcriptional activity of ChREBP-β in both low-glucose and high-glucose conditions (12, 13). In hepatocytes and adipocytes, ChREBP-α expression is induced by glucose (14, 15). In IL-3–dependent immortalized hematopoietic cells, ChREBP-α expression can be induced in response to mitogenic stimulation (16). Little is known about ChREBP-β, although reports suggest that it is expressed in a glucose- and ChREBP-dependent manner in which glucose-activated ChREBP-α initiates ChREBP-β transcription from the alternate promoter (12). ChREBP-α activity is regulated by phosphorylation-dependent mechanisms responsive to glucose levels (15, 17). Under low-glucose conditions, ChREBP-α is phosphorylated at Ser196 and Thr666 sites by protein kinase A and stays in the cytoplasm (17). A high-glucose condition leads to ChREBP-α dephosphorylation and translocation into the nucleus, where it forms a heterodimer with Max-like protein X (Mlx) and binds to ChoRE motifs in the promoters of glycolytic and lipogenic genes to activate transcription (17, 18).
The ChREBP/Mlx dimer and SREBP1 can synergistically up-regulate lipogenic gene transcription (14). In this regard, most mammalian lipogenic genes are regulated by both SREBP1 and ChREBP, and their full induction requires both factors. Compared with SREBP1, which has little effect on glycolytic gene expression (19), ChREBP has a much broader impact on metabolic gene expression. ChREBP regulates many genes encoding glycolytic enzymes, most genes involved in NADPH generation, and genes that play roles in regulating metabolic and energy homeostasis (10, 20). In summary, ChREBP/Mlx is the principal transcription factor in the up-regulation of genes (in a glucose-dependent manner) involved in glucose uptake, glycolysis, and lipogenesis in metabolic tissues, such as the liver and brown and white adipose tissue (10, 20). In tumor cells, the induction of ChREBP is required for efficient cell proliferation. Suppression of ChREBP reduces aerobic glycolysis, de novo lipogenesis, and nucleotide biosynthesis but stimulates mitochondrial respiration, suggesting that ChREBP plays a key role in redirecting glucose metabolism from oxidative phosphorylation pathways to anabolic pathways (16).
In this study, we report that ChREBP is also required for HCMV-induced glycolysis and lipogenesis. Expression of both ChREBP-α and ChREBP -β is significantly increased in response to HCMV infection. The induction of ChREBP by HCMV is critical for viral lytic infection. In HCMV-infected HFs, suppression of ChREBP results in inhibition of viral-induced lipogenic gene expression and lipid synthesis. The induction of ChREBP by HCMV also activates GLUT4 and GLUT2 expression, resulting in increased glucose uptake, lactate production, nucleotide biosynthesis, and NADPH generation. Our results demonstrate that ChREBP is a key regulator in altering glucose metabolism and lipogenesis in host cells in response to HCMV infection.
Results
HCMV Infection Induces ChREBP Expression.
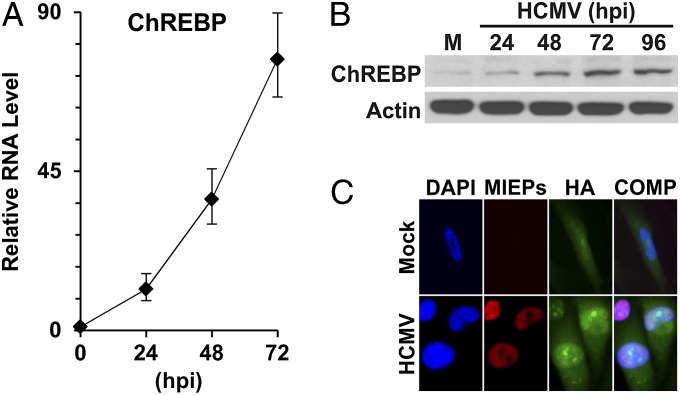
It was recently reported that genetically engineered mice overexpressing GLUT4 can induce lipogenesis through up-regulation of ChREBP in adipose tissues (12). Our early studies showed that GLUT4 expression is significantly induced during HCMV infection (5), suggesting that the expression of ChREBP may be activated. To evaluate this, we examined ChREBP mRNA and protein levels. Our quantitative PCR (qPCR) results showed that HCMV infection dramatically induced ChREBP expression over the time course of infection (Fig. 1A). Correspondingly, ChREBP-α protein levels increased steadily throughout the course of infection (Fig. 1B).
Fig. 1.
ChREBP expression in HCMV-infected HF cells. (A) ChREBP RNA levels during a HCMV infection time course. Total RNA was isolated at 0, 24, 48, and 72 hpi. Total ChREBP RNA levels were measured by qRT-PCR and normalized to actin RNA levels. (B) ChREBP-α protein levels in the time course of HCMV infection. ChREBP-α and actin protein levels were determined by Western blot analysis. M, mock infection. (C) Cellular localization of HA-tagged ChREBP-α in mock-infected and HCMV-infected cells. At 2 hpi, mock- and HCMV-infected primary HF cells were electroporated with a plasmid expressing an HA-tagged human ChREBP-α. At 1 d after electroporation, cells were serum-starved for overnight, fixed at 48 hpi and stained for nuclear DNA (DAPI, blue), HA (green), and the HCMV MIEPs (red). The DAPI, HA, and MIEP images are overlaid into a composite image (COMP).
To be functional, ChREBP protein must be present in the nucleus to allow activation of its target genes. Immunofluorescent microscopic studies using HA-tagged human full-length ChREBP have shown that HA-ChREBP is dispersed through the cell in mock infection (16). However, after HCMV infection, the level of nuclear-localized HA-ChREBP increases significantly (Fig. 1C), indicating that HCMV infection not only increases the amount of ChREBP, but also mediates its translocation to the nucleus.
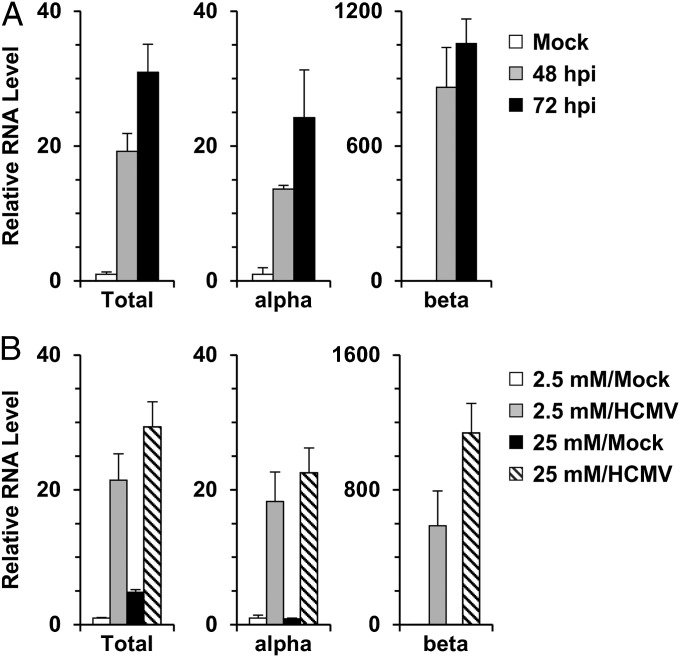
ChREBP-β expression is regulated by GLUT4 in adipose tissues (12). Overexpression of GLUT4 increases glucose flux, which posttranslationally modifies ChREBP-α to stimulate its nuclear translocation, thereby promoting its binding to ChoRE motifs in its upstream ChREBP-β promoter. Given that HCMV-infected cells have greatly increased GLUT4 expression (5), we asked whether HCMV infection could induce ChREBP-β expression. In agreement with Fig. 1A, a separate experiment showed that the RNA level of total ChREBP increased 19-fold at 48 hpi and 31-fold at 72 hpi (Fig. 2A). ChREBP-α RNA increased by approximately 14-fold at 48 hpi and 24-fold at 72 hpi. Surprisingly, ChREBP-β RNA increased by 860-fold at 48 hpi and by 1,050-fold at 72 hpi. The much greater fold increase of ChREBP-β RNA in response to HCMV infection is a reflection of the very low basal level of ChREBP-β expression in uninfected cells. This agrees with a recent report of a less abundant RNA level of ChREBP-β compared with ChREBP-α (12).
Fig. 2.
Expression of ChREBP isoforms in HCMV-infected cells. (A) RNA levels of ChREBP-α, ChREBP-β, and total ChREBP in HCMV-infected cells. Total RNA was isolated from mock- and HCMV-infected cells. RNA levels of ChREBP-α, ChREBP-β, and total ChREBP were quantified by qPCR using exon-specific primers. (B) RNA levels of total ChREBP, ChREBP-α, and ChREBP-β in HCMV-infected cells in response to high glucose level. After a 2-h incubation, mock- and HCMV-infected cells were cultured in either low-glucose (2.5 mM) or high-glucose (25 mM) medium. At 48 hpi, total RNA was isolated, and RNA levels of ChREBP-α, ChREBP-β, and total ChREBP were measured by qPCR. Total, total ChREBP; alpha, ChREBP-α; beta, ChREBP-β.
Although detectable levels of ChREBP-β protein have not been reported to date, the significant induction of ChREBP-β mRNA in HCMV-infected cells suggests that the protein may be detectable in infected cells. The anti-ChREBP antibody used in this study recognizes the common C terminus shared by both ChREBPs, and thus should be able to detect both α and β proteins. Along with ChREBP-α protein, a protein band of ∼75 kDa, (the predicted molecular weight of ChREBP-β) was detected in HCMV-infected cell lysates by the anti-ChREBP antibody (Fig. S1A, arrow). This band was diminished in HCMV-infected cells treated with a short hairpin (sh) RNA that targets both ChREBP RNAs, shChREBP1 (TRCN0000020078, Open Biosystems) (Fig. S1B, arrow).
Because ChREBP-β expression is induced by glucose-activated ChREBP-α (12), we asked whether the same mechanism is used in HCMV-infected cells. After HCMV infection, cells were cultured in either low-glucose (2.5 mM) or high-glucose (25 mM) medium, and ChREBP RNA levels were measured by qPCR at 48 hpi. Fig. 2B shows moderately increased ChREBP RNA levels in the high-glucose medium compared with the low-glucose medium, with the greatest effect a doubling of the ChREBP-β RNA level. The induction of ChREBP mRNAs was already very high in infected cells cultured in low-glucose medium, however, suggesting that ChREBP expression in HCMV-infected cells is induced largely by a glucose-independent mechanism and may require viral gene expression. Interestingly, ∼25% of the increase in total ChREBP mRNAs can be induced in cells infected with UV-irradiated HCMV (Fig. S2), suggesting that signaling due to attachment or tegument proteins may contribute to the induction.
Activation of ChREBP Expression Is Critical for HCMV Growth.
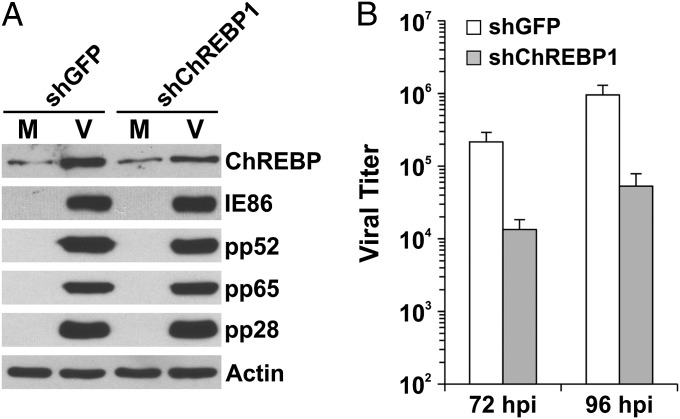
To assess the importance of ChREBP induction for HCMV growth, we introduced lentiviral vectors expressing shChREBPs to knock down ChREBP expression. Owing to a robust rise in ChREBP RNA levels after HCMV infection, the ChREBP protein level can be depleted only moderately in infected cells. Thus, the effects noted must be considered in the light of less-efficient depletion. Cells treated with shChREBP1 for 5 d before HCMV infection showed a 50–60% reduction in HCMV-induced ChREBP-α protein levels (Fig. 3A). The level of ChREBP depletion attained by shChREBP1 reduced the viral titer by nearly 20-fold at 72 hpi and 96 hpi (Fig. 3B), suggesting that ChREBP induction is critical for HCMV lytic infection. A similar viral growth inhibition result was obtained with another shRNA, shChREBP2 (Fig. S3A), which was used in a combination of two shRNAs targeting two coding regions of ChREBP different from shChREBP1 (21). Viral protein levels at 72 hpi were also tested in ChREBP-suppressed cells. The levels of an immediate-early protein (IE86), an early protein (pp52), and two late proteins (pp65 and pp28) were unaffected by ChREBP suppression (Fig. 3A). We previously reported similar findings in cells inhibited for SREBP activation (6, 7). The foregoing results suggest that the attenuation of HCMV growth is caused not by a disruption of viral protein synthesis, but rather by a disruption of lipid synthesis, which is required for virion formation.
Fig. 3.
Induction of ChREBP is critical for HCMV infection. (A) HCMV viral protein expression in ChREBP-depleted HF cells. At 72 hpi, whole-cell extracts were made from mock- and HCMV-infected HF cells pretreated with shGFP or shChREBP1, and levels of cellular and viral proteins were evaluated by Western blot analysis. M, mock infection; V, HCMV infection. (B) HCMV viral titers at 72 and 96 hpi in HF cells treated with shGFP or shChREBP1.
Suppression of ChREBP Leads to Inhibition of HCMV-Induced Lipogenesis, Glucose Uptake, and Glucose Metabolism.
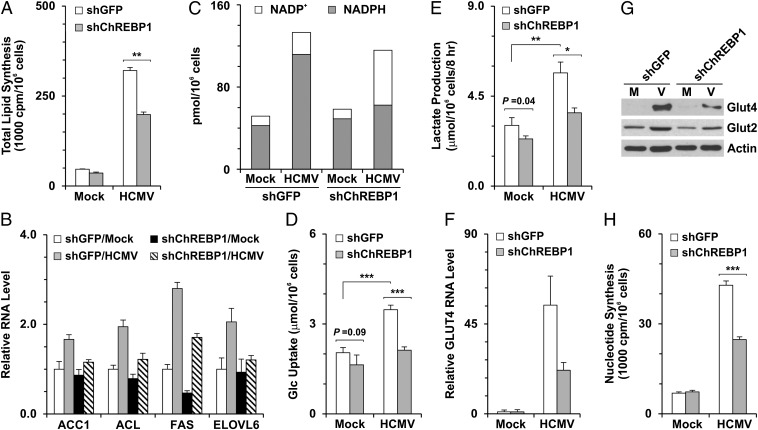
In ChREBP-deficient mice, expression levels of nearly every enzyme required for lipogenesis are reduced in the liver (10); thus, we predicted that lipogenesis induced during HCMV infection would be inhibited by suppression of ChREBP. The results of our total lipid synthesis assay support this prediction. As shown in Fig. 4A, HCMV infection significantly increased lipid synthesis in a control shRNA (shGFP)-treated cells. shChREBP1 treatment reduced lipid synthesis by approximately 20% in mock-infected cells and by 40% in HCMV-infected cells, whereas shChREBP2 treatment reduced it by approximately 7% in mock-infected cells and 34% in HCMV-infected cells (Fig. S3B). Previous laboratory studies have shown that lipogenic gene expression is induced in an SREBP1-dependent manner during HCMV infection (6, 7).
Fig. 4.
Metabolic effects of ChREBP depletion on HCMV-infected cells. HF cells were infected with lentiviral vector expressing shGFP or shChREBP1 for at least 4 d. The cells were then serum-starved for 1 d, followed by HCMV infection (MOI of 3). Metabolic gene expression levels and metabolic assays were performed at 48 hpi. (A) Total lipid synthesis in HF cells. Total lipid synthesis was assayed by measuring the incorporation of 14C-acetate into lipids. (B) RNA levels of key lipogenic genes—ACC1, ACL, ELOVL6, and FAS—quantified by qPCR. All RNA data were normalized to actin RNA levels. (C) NADP+/NADPH levels in mock- and HCMV-infected cells pretreated with shGFP or shChREBP1. (D) Glucose uptake. Glc, glucose. (E) Lactate production. Levels of glucose and lactate in the cultured medium were measured using the YSI 7100 Multiparameter Bioanalytical System. (F) Reduction of GLUT4 RNA levels in HCMV-infected cells from depletion of ChREBP. GLUT4 RNA levels were measured as described in B. (G) Protein levels of GLUT2 and GLUT4 in mock- and HCMV-infected cells pretreated with shGFP or shChREBP1. Whole-cell extracts were made at 48 hpi, and GLUT2, GLUT4, and actin protein levels were analyzed by Western blot analysis. (H) Reduction of nucleotide biosynthesis in infected cells from depletion of ChREBP. Total nucleotide synthesis was assayed by measuring the incorporation of d-[U-14C]-glucose into RNA at 48 hpi. M, mock infection; V, HCMV infection; *P < 0.01; **P < 0.005; ***P < 0.002.
To examine whether lipogenic gene expression is also regulated by ChREBP induction during HCMV infection, we measured mRNA levels of key lipogenic genes that are reportedly directly activated by ChREBP, including acetyl-CoA carboxylase 1 (ACC1), ATP-citrate lyase (ACL), elongase of very long chain fatty acids member 6 (ELOVL6), and fatty acid synthetase (FAS) (20, 22). The induction of ACC1, ACL, ELOVL6, and FAS by HCMV infection was inhibited by ChREBP suppression (Fig. 4B), which was confirmed by Western blot analysis of ACC1 and FAS (Fig. S4). These results are in concordance with the requirement for ChREBP to fully induce lipogenesis during HCMV infection.
The biosynthesis of fatty acid uses large amounts of NADPH, which is generated mainly in the pentose phosphate pathway (PPP) and the citrate-malate shuttle between the mitochondria and the cytosol catalyzed by malic enzyme in normal cells. Most genes involved in NADPH generation are targets of ChREBP/Mlx (20). To investigate the role of ChREBP induction in NADPH generation during HCMV infection, we measured NADP+ and NADPH levels in mock- and HCMV-infected cells pretreated with shGFP or shChREBP1. In mock-infected cells, NADPH levels were much higher than NADP+ levels, and shChREBP1 treatment had little effect on these levels. In shGFP-treated cells at 48 h after HCMV infection, the NADP+ and NADPH pool was increased compared with that in mock-infected cells, but the ratio of NADP+/NADPH (∼1:5) was similar in the two cell types. However, in HCMV-infected cells, ChREBP depletion reduced the NADPH level to almost that of mock-infected cells and significantly increased the NADP+ level, although the total NADP+ and NADPH pool remained high in infected cells (Fig. 4C).
These data suggest two conclusions. First, the induction of ChREBP in HCMV infection activates not only the expression of lipogenic genes, but also the pathways that produce NADPH to facilitate de novo lipid synthesis. Second, the increase in the total NADP+ and NADPH pool in infected cells is not mediated by ChREBP induction.
Along with lipogenic genes, ChREBP also up-regulates genes involved in glucose uptake and glycolysis. The loss of ChREBP reduces glucose uptake and glycolysis in the liver and in cancer cells (10, 16). To investigate whether the induction of ChREBP is responsible for the increased glucose uptake and glycolysis in HCMV infection, we measured the glucose and lactate levels in the cell culture medium. HCMV infection led to significant increases in glucose uptake and lactate excretion in cells treated with the control shGFP (Fig. 4 D and E and Fig. S3 C and D), as reported previously (3, 5). In infected cells, the shChREBP1 treatment reduced glucose uptake by 39% and lactate production by 35% (Fig. 4 D and E), whereas shChREBP2 treatment drceased both glucose uptake and lactate production by approximately 30% (Fig. S3 C and D). These findings indicate that the induction of ChREBP in HCMV-infected cells leads not only to increased glucose uptake, but also to an increased glycolytic rate.
Less significant reductions in glucose uptake and lactate production were observed in mock cells treated with shChREBP1 (Fig. 4 D and E), likely the result of a moderate decrease in GLUT1, the predominant GLUT in normal HF cells (5), in shChREBP1-treated mock cells (Fig. S5). We previously reported that HCMV infection eliminates GLUT1 and increases GLUT4 to replace GLUT1 in infected HF cells (5); thus, this slight reduction of GLUT1 by shChREBP1 would affect only normal cells, not infected cells.
As noted above, HCMV infection induces glucose uptake by greatly increasing expression of the GLUT4 gene, SLC2A4 (5), which is a direct target gene of ChREBP owing to a strong ChoRE motif in its promoter (20). Thus, it is possible that glucose uptake in HCMV-infected cells is up-regulated by ChREBP-mediated activation of GLUT genes. Examination of the mRNA level of GLUT4 showed that the increased GLUT4 expression in HCMV-infected cells was inhibited by ChREBP depletion (Fig. 4F and Fig. S3E), as reflected by the loss of GLUT4 protein (Fig. 4G). Several previous reports have suggested that ChREBP is capable of up-regulating the expression of GLUT2, as well as GLUT4, in various tissues (10, 20, 23). GLUT2 expression was modestly increased during HCMV infection (Fig. 4G). Depletion of ChREBP reduced GLUT2 protein levels in HCMV-infected cells. The results shown in Fig. 4 D, F, and G and Fig. S3 C–E indicate that ChREBP induction activates the expression of GLUT2 and GLUT4 to increase glucose uptake in HCMV-infected cells.
Suppression of ChREBP Reduces Nucleotide Biosynthesis in HCMV Infection.
The PPP, an alternative branch of glycolysis, is a primary anabolic pathway that uses glucose to generate ribose-5-phosphate for the synthesis of nucleotides, nucleic acids, and reducing equivalents in normal cells (24). Tumor cells deficient in ChREBP display reduced glucose flux through the PPP to RNA biosynthesis (16). To investigate the role of ChREBP in nucleotide biosynthesis during HCMV infection, we used the incorporation of d-[U-14C]-glucose into total RNA as a measure of nucleotide biosynthesis. By this measurement, nucleotide biosynthesis was increased approximately fivefold by HCMV infection in cells treated with the control shGFP (Fig. 4H and Fig. S3F), in agreement with a previous report (25) and metabolic flux data (3). In infected cells, depletion of ChREBP by shChREBP1 and shChREBP2 eliminated 40% and 32% of nucleotide biosynthesis, respectively (Fig. 4H and Fig. S3F), whereas there was little change in mock-infected cells (Fig. 4H and Fig. S3F).
Taken together, the data in Fig. 4 show that the induction of ChREBP in HCMV-infected cells initiates a transcriptional program that results in increased glucose uptake, increased glycolysis, and the induction of anabolic glucose metabolism (nucleotide biosynthesis and de novo lipogenesis). The data show that these effects of ChREBP are required for the formation of infectious progeny virions.
ChREBP Regulates Glucose Metabolism and Lipogenesis Independent of PERK/SREBP1 in HCMV-Infected Cells.
We previously reported that HCMV infection induces lipogenesis through PERK-mediated activation of SREBP1 (6, 7); however, in those studies, the depletion of SREBP1 or PERK did not completely inhibit lipid synthesis, suggesting the involvement of other lipogenic transcription factors. Previous reports also have suggested that SREBP1 activity alone does not fully activate lipogenesis; deletion of the SREBP1 gene was found to result in a <50% reduction in fatty acid synthesis (26). Furthermore, the expression of lipogenic genes in the liver is up-regulated by high-carbohydrate feeding even in mice lacking SREBP1c (27), again suggesting that other factors are involved. The expression of both ChREBP and PERK is significantly induced in HCMV infection; moreover, the expression of lipogenic enzymes and lipogenesis is inhibited in cells depleted of ChREBP or PERK. Thus, it is reasonable to ask whether there is any cross-talk between ChREBP-mediated and PERK-mediated lipogenesis in virally infected cells.
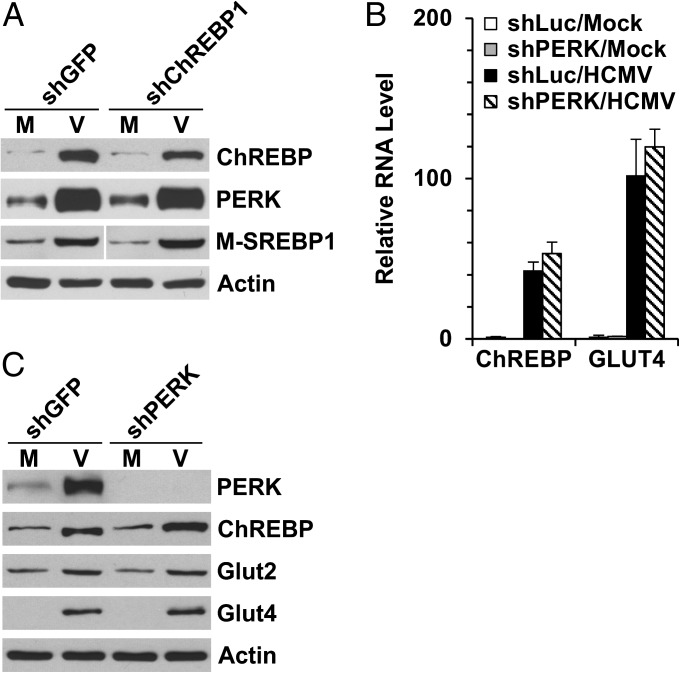
To examine whether ChREBP depletion disrupts SREBP1 activation, we analyzed the protein levels of PERK and the mature (i.e., transcriptionally active) form of SREBP1 (M-SREBP1) in ChREBP depleted cells. ChREBP depletion had little effect on either PERK induction or SREBP1 activation during HCMV infection (Fig. 5A). This finding agrees with a previous report showing that deletion of ChREBP gene in mice had no effect on the expression and activation of SREBP proteins (10).
Fig. 5.
Induction of ChREBP and PERK-mediated SREBP1 activation are independent pathways in HCMV-infected cells. (A) Depletion of ChREBP had little effect on the induction of PERK and the mature forms of SREBP1 during HCMV infection. Whole-cell extracts were made at 72 hpi, and protein levels were measured by Western blot analysis. The M-SREBP1 blot was spliced to match the order of other blots. (B and C) Depletion of PERK had little effect on the induction of ChREBP and GLUT4 during HCMV infection. (B) RNA levels of total ChREBP and GLUT4 in shLuc- and shPERK-treated cells. (C) Protein levels of ChREBP-α, GLUT2, and GLUT4 in PERK-depleted cells. M, mock infection; V, HCMV infection; M-SREBP1, mature forms of SREBP1.
We also analyzed ChREBP expression in PERK-depleted cells. The HCMV-induced increases in ChREBP mRNA and protein levels, as well as in mRNA and protein levels for ChREBP target genes (GLUT2 and GLUT4), were not significantly affected by PERK depletion (Fig. 5 B and C). These data suggest that the separate mechanisms coordinate the induction of PERK and ChREBP in HCMV-infected cells.
Discussion
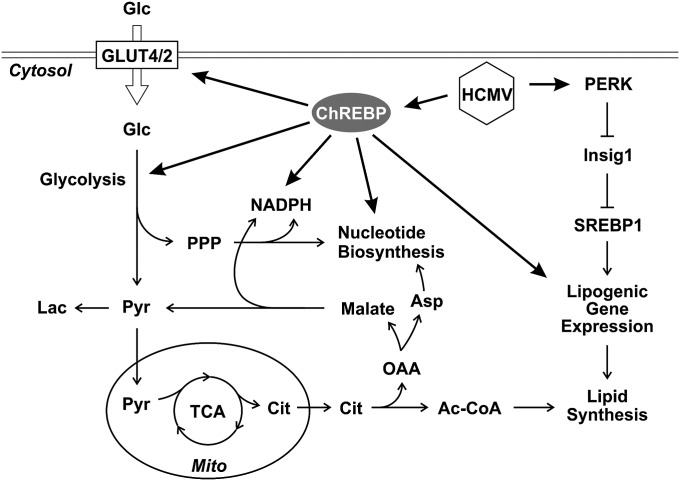
Although both ChREBP and SREBP proteins can up-regulate lipogenic gene expression, they regulate metabolism in different ways. ChREBP coordinately regulates genes required for the conversion of glucose to fatty acids and nucleotides. ChREBP deficiency reduces glucose uptake and glycolysis in HCMV-infected cells, resulting in decreased carbon flux for anabolic metabolism, such as lipid synthesis, NADPH generation, and nucleotide biosynthesis (Fig. 4 and Fig. S3). In contrast, SREBP proteins transactivate a highly specific subset of target genes. Thirty-three genes have been identified as SREBP target genes (19), including 29 involved in fatty acid synthesis, sterol metabolism, cholesterol synthesis and uptake and 4 with no known connection to lipid metabolism. Unlike ChREBP, SREBP proteins have little effect on glycolysis. In HCMV-infected HFs, suppression of ChREBP reduced the expression of GLUT4 and GLUT2, resulting in decreased glucose uptake and glycolysis (Fig. 4 D–G), whereas PERK depletion, which decreases SREBP activation, had no effect on GLUT4 and GLUT2 levels (Fig. 5 B and C). These results suggest that ChREBP and PERK-mediated induction of SREBPs are independent pathways, both of which must be induced in HCMV-infected cells to alter glucose metabolism and fully activate lipogenesis, as diagrammed in Fig. 6.
Fig. 6.
Diagram of glucose metabolism and lipogenesis in HCMV infection. Ac-CoA, acetyl-CoA; Asp, aspartate; Cit, citrate; Glc, glucose; Lac, lactate; Mito, mitochondria; OAA, oxaloacetate; Pyr, pyruvate.
Glucose uptake and phosphorylation are major controlling steps of glycolysis. Glucose uptake can limit the overall glycolytic rate when extracellular glucose concentration is low. Recent studies have shown that at low glucose uptake rates, glucose is used primarily for ATP production by oxidative phosphorylation in the mitochondria, and there is no significant lactate excretion. Conversely, high rates of glucose uptake, as in tumor cells, gradually activate aerobic glycolysis and reduce mitochondrial respiration (28), allowing the use of glucose carbon for biosynthesis.
HCMV infection causes dramatic alterations of intermediary metabolism, similar to those seen in tumor cells. In HCMV-infected cells, glucose is used biosynthetically instead of being broken down for ATP production in the TCA cycle. Our data indicate that the glucose responsive transactivator ChREBP is a key regulator that reprograms glucose metabolism in HCMV-infected cells. The induction of ChREBP in HCMV infection induces the expression of GLUT4 and GLUT2, as well as of glycolytic and lipogenic enzymes, resulting in increased glucose flux and a metabolic switch for glucose from energy production in the mitochondria to anabolic metabolism.
Materials and Methods
Cell Culture, Plasmids, Reagents, and Viruses.
Primary and life-extended human foreskin fibroblasts (29) were propagated and maintained as described previously (7). The plasmid expressing an HA-tagged human ChREBP-α was a gift from Xuemei Tong, Shanghai Jiaotong University, Shanghai, China (16). All HCMV experiments were performed in serum-starved HF cells and infection with the Towne strain of HCMV [multiplicity of infection (MOI) of 3] derived from a bacterial artificial chromosome clone modified to express GFP under control of the simian virus 40 early promoter. For immunofluorescent studies, the WT Towne strain of HCMV without a GFP-expressing cassette was used. Viral growth assays were performed as described previously (7).
Metabolic Assays.
For glucose and lactate assays, HF cells in six-well plates were cultured in serum-free DMEM containing 5.5 mM glucose and 4.0 mM glutamine, with no pyruvate. At 40 hpi, cells were refed with 1.5 mL of fresh medium. The culture medium was harvested at 48 hpi, and glucose and lactate levels in the culture medium were measured with the YSI 7100 Multiparameter Bioanalytical System. Nucleotide biosynthesis was measured by labeling cells in six-well plates with 5.0 μCi/mL d-[U-14C]-glucose in serum-free DMEM containing 200 μM glucose, with no pyruvate, for 2 h at 37 °C. Total RNA was isolated by TRIzol reagent (Life Technologies), and the incorporation of 14C into RNA was determined by scintillation counting. The data were normalized to cell number. Cellular NADP+/NADPH levels were measured using a NADP/NADPH quantitation kit (K347-100; Biovision), following the manufacturer’s protocol. The total lipid synthesis assay was performed by measuring the incorporation of 14C-acetate into lipids as described previously (6). P values were determined by the Student t test. A P value of <0.05 was considered statistically significant.
RNA Analysis.
Total RNA was isolated by TRIzol reagent according to the manufacturer's protocol. First-strand cDNA synthesis was performed with the SuperScript first-strand synthesis system (Life Technologies) using 2 µg of total RNA. For qPCR, input cDNA was analyzed in triplicate or quintuplicate. qPCR reactions were performed using the TaqMan Universal PCR Master Mix Kit or SYBR Green PCR Master Mix Kit and an ABI 7900 PCR system (Applied Biosystems). The following primer sets (Applied Biosystems) were used in the TaqMan PCR assays: ACC1 (Hs01046047_m1), ACL (Hs00153764_m1), β-actin (Hs99999903-m1), ChREBP (Hs00975714_m1), GLUT4 (Hs00168966_m1), and FAS (Hs00188012_m1). The following primers were used in the SYBR Green PCR assays: β-actin, ccagctcaccatggatgatg and GGAATCCTTCTGACCCATGC; ChREBP-α (12), AGTGCTTGAGCCTGGCCTAC and TTGTTCAGGCGGATCTTGTC; ChREBP-β (12), AGCGGATTCCAGGTGAGG and TTGTTCAGGCGGATCTTGTC; ChREBP-total, CACACCAGCGTTTTGACCAG and ACTCAAACAGAGGCCGGATG; and ELOVL6, GTCTCTGACCCTTGCAGTCTT and CCTGGTCACAAACTGACTGCT. Gene expression data were normalized to β-actin mRNA levels.
shRNA Experiments.
Lentiviral expression plasmids encoding shChREBP1 (clone ID: TRCN0000020078), shChREBP2 (clone IDs: V3LHS_334249 and V3LHS_313472) (21) and shPERK (clone ID: TRCN0000001401) were purchased from Open Biosystems. Lentiviral vectors expressing shRNA were made as described previously (30). For gene silencing experiments in primary HFs, subconfluent cells were infected with lentiviral vectors in the presence of 8 μg/mL polybrene for 2–3 h, followed by the addition of fresh medium containing 1.0 μg/mL puromycin, and then cultured for another 4 d before serum-starvation and HCMV infection. For experiments in life-extended human foreskin fibroblasts, cells were infected by lentivirus expressing shRNA, followed by a second lentiviral infection 2 d later. After incubation for another 3 d in fresh medium, cells were serum-starved for 1 d, then infected with HCMV (MOI of 3) in serum-free DMEM for the designed assays.
Western Blot Analysis.
Western blot analyses were performed as described previously (31). The following antibodies were used in this study: anti-ACC1, anti-actin, anti-FAS (6), anti-ChREBP (Abcam; ab157153), anti-ex2/3 [HCMV major immediate-early proteins (MIEPs)] (32), anti-GLUT2 and anti-GLUT4 (07-1402 and 07-1404; Millipore), anti-PERK, anti-SREBP1, and antibodies for viral proteins pp28, pp52, and pp65 (7).
Supplementary Material
Acknowledgments
We thank the members of the J.C.A. laboratory for helpful suggestions and guidance, and Kathryn Wellen for sharing equipment. This work was supported by National Institutes of Health Grants R01 CA157679 (to J.C.A.) and R21 AI105679 (to Y.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310779111/-/DCSupplemental.
References
- 1.Pass RF, Fowler KB, Boppana SB, Britt WJ, Stagno S. Congenital cytomegalovirus infection following first trimester maternal infection: Symptoms at birth and outcome. J Clin Virol. 2006;35(2):216–220. doi: 10.1016/j.jcv.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 2.Munger J, Bajad SU, Coller HA, Shenk T, Rabinowitz JD. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog. 2006;2(12):e132. doi: 10.1371/journal.ppat.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munger J, et al. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat Biotechnol. 2008;26(10):1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chambers JW, Maguire TG, Alwine JC. Glutamine metabolism is essential for human cytomegalovirus infection. J Virol. 2010;84(4):1867–1873. doi: 10.1128/JVI.02123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu Y, Maguire TG, Alwine JC. Human cytomegalovirus activates glucose transporter 4 expression to increase glucose uptake during infection. J Virol. 2011;85(4):1573–1580. doi: 10.1128/JVI.01967-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu Y, Maguire TG, Alwine JC. Human cytomegalovirus infection induces adipocyte-like lipogenesis through activation of sterol regulatory element binding protein 1. J Virol. 2012;86(6):2942–2949. doi: 10.1128/JVI.06467-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu Y, Pierciey FJ, Jr, Maguire TG, Alwine JC. PKR-like endoplasmic reticulum kinase is necessary for lipogenic activation during HCMV infection. PLoS Pathog. 2013;9(4):e1003266. doi: 10.1371/journal.ppat.1003266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Postic C, Dentin R, Denechaud PD, Girard J. ChREBP, a transcriptional regulator of glucose and lipid metabolism. Annu Rev Nutr. 2007;27:179–192. doi: 10.1146/annurev.nutr.27.061406.093618. [DOI] [PubMed] [Google Scholar]
- 9.Uyeda K, Repa JJ. Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab. 2006;4(2):107–110. doi: 10.1016/j.cmet.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 10.Iizuka K, Bruick RK, Liang G, Horton JD, Uyeda K. Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc Natl Acad Sci USA. 2004;101(19):7281–7286. doi: 10.1073/pnas.0401516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cairo S, Merla G, Urbinati F, Ballabio A, Reymond A. WBSCR14, a gene mapping to the Williams–Beuren syndrome deleted region, is a new member of the Mlx transcription factor network. Hum Mol Genet. 2001;10(6):617–627. doi: 10.1093/hmg/10.6.617. [DOI] [PubMed] [Google Scholar]
- 12.Herman MA, et al. A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature. 2012;484(7394):333–338. doi: 10.1038/nature10986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li MV, Chang B, Imamura M, Poungvarin N, Chan L. Glucose-dependent transcriptional regulation by an evolutionarily conserved glucose-sensing module. Diabetes. 2006;55(5):1179–1189. doi: 10.2337/db05-0822. [DOI] [PubMed] [Google Scholar]
- 14.Dentin R, et al. Hepatic glucokinase is required for the synergistic action of ChREBP and SREBP-1c on glycolytic and lipogenic gene expression. J Biol Chem. 2004;279(19):20314–20326. doi: 10.1074/jbc.M312475200. [DOI] [PubMed] [Google Scholar]
- 15.He Z, Jiang T, Wang Z, Levi M, Li J. Modulation of carbohydrate response element-binding protein gene expression in 3T3-L1 adipocytes and rat adipose tissue. Am J Physiol Endocrinol Metab. 2004;287(3):E424–E430. doi: 10.1152/ajpendo.00568.2003. [DOI] [PubMed] [Google Scholar]
- 16.Tong X, Zhao F, Mancuso A, Gruber JJ, Thompson CB. The glucose-responsive transcription factor ChREBP contributes to glucose-dependent anabolic synthesis and cell proliferation. Proc Natl Acad Sci USA. 2009;106(51):21660–21665. doi: 10.1073/pnas.0911316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawaguchi T, Takenoshita M, Kabashima T, Uyeda K. Glucose and cAMP regulate the L-type pyruvate kinase gene by phosphorylation/dephosphorylation of the carbohydrate response element binding protein. Proc Natl Acad Sci USA. 2001;98(24):13710–13715. doi: 10.1073/pnas.231370798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kabashima T, Kawaguchi T, Wadzinski BE, Uyeda K. Xylulose 5-phosphate mediates glucose-induced lipogenesis by xylulose 5-phosphate-activated protein phosphatase in rat liver. Proc Natl Acad Sci USA. 2003;100(9):5107–5112. doi: 10.1073/pnas.0730817100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horton JD, et al. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA. 2003;100(21):12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma L, Robinson LN, Towle HC. ChREBP*Mlx is the principal mediator of glucose-induced gene expression in the liver. J Biol Chem. 2006;281(39):28721–28730. doi: 10.1074/jbc.M601576200. [DOI] [PubMed] [Google Scholar]
- 21.Seo JY, Cresswell P. Viperin regulates cellular lipid metabolism during human cytomegalovirus infection. PLoS Pathog. 2013;9(8):e1003497. doi: 10.1371/journal.ppat.1003497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishii S, Iizuka K, Miller BC, Uyeda K. Carbohydrate response element binding protein directly promotes lipogenic enzyme gene transcription. Proc Natl Acad Sci USA. 2004;101(44):15597–15602. doi: 10.1073/pnas.0405238101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eissing L, et al. De novo lipogenesis in human fat and liver is linked to ChREBP-β and metabolic health. Nat Commun. 2013;4:1528. doi: 10.1038/ncomms2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wamelink MM, Struys EA, Jakobs C. The biochemistry, metabolism and inherited defects of the pentose phosphate pathway: A review. J Inherit Metab Dis. 2008;31(6):703–717. doi: 10.1007/s10545-008-1015-6. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka S, Furukawa T, Plotkin SA. Human cytomegalovirus stimulates host cell RNA synthesis. J Virol. 1975;15(2):297–304. doi: 10.1128/jvi.15.2.297-304.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimano H, et al. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J Clin Invest. 1997;100(8):2115–2124. doi: 10.1172/JCI119746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang G, et al. Diminished hepatic response to fasting/refeeding and liver X receptor agonists in mice with selective deficiency of sterol regulatory element-binding protein-1c. J Biol Chem. 2002;277(11):9520–9528. doi: 10.1074/jbc.M111421200. [DOI] [PubMed] [Google Scholar]
- 28.Vazquez A, Liu J, Zhou Y, Oltvai ZN. Catabolic efficiency of aerobic glycolysis: The Warburg effect revisited. BMC Syst Biol. 2010;4:58. doi: 10.1186/1752-0509-4-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bresnahan WA, Hultman GE, Shenk T. Replication of wild-type and mutant human cytomegalovirus in life-extended human diploid fibroblasts. J Virol. 2000;74(22):10816–10818. doi: 10.1128/jvi.74.22.10816-10818.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu Y, Alwine JC. Interaction between simian virus 40 large T antigen and insulin receptor substrate 1 is disrupted by the K1 mutation, resulting in the loss of large T antigen-mediated phosphorylation of Akt. J Virol. 2008;82(9):4521–4526. doi: 10.1128/JVI.02365-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu Y, Kudchodkar SB, Alwine JC. Effects of simian virus 40 large and small tumor antigens on mammalian target of rapamycin signaling: Small tumor antigen mediates hypophosphorylation of eIF4E-binding protein 1 late in infection. J Virol. 2005;79(11):6882–6889. doi: 10.1128/JVI.79.11.6882-6889.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harel NY, Alwine JC. Phosphorylation of the human cytomegalovirus 86-kilodalton immediate-early protein IE2. J Virol. 1998;72(7):5481–5492. doi: 10.1128/jvi.72.7.5481-5492.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.